Abstract
Background: The pathogenesis of hidradenitis suppurativa (HS) remains unclear. In order to develop effective treatment strategies, a deeper understanding of pathophysiology is needed. This is impaired by multiple small studies with inconsistent methodologies and the impact of co-occurring pro-inflammatory conditions such as smoking and obesity.
Methods: This systematic review aimed to collate all published reports of cytokine studies in tissue, blood, serum and exudate. It was registered with PROSPERO (Registration number CRD42018104664) performed in line with the PRISMA checklist.
Results: 19 studies were identified comprising 564 individual HS patients and 198 control patients examining 81 discrete cytokines. Methodology was highly varied and the quality of studies was generally low. There was a large degree of variance between the measured levels of cytokines. 78.2% of cytokines demonstrated heterogeneity by the chi-squared test for homogeneity and hence meta-analysis was not deemed appropriate. However, a strong and significant IL-17 signalling component was identified.
Conclusions: Cytokines consistently elevated in lesional, peri-lesional and unaffected tissue are identified and discussed. Areas for further investigation include the role of dendritic cells in HS; the contribution of obesity, smoking, diabetes and the microbiome to cytokine profiles in HS; and examining the natural history of this disease through longitudinal measurements of cytokines over time.
Keywords: Hidradenitis Suppurativa, Cytokines, Inflammation, Pathogenesis, IL-17, TNF-alpha
Introduction
Hidradenitis Suppurativa (HS) is a chronic inflammatory disease, the exact pathophysiology of which remains poorly defined 1. Dysregulation of the T h17: Treg axis 2, IL-36 signalling pathways 3 and keratinocyte-mediated inflammatory cytokines 4 have been demonstrated in lesional skin, blood, serum, and exudate 5– 8 although contradictory results exist 4, 9. Given the variable and incomplete response of patients to treatment, including monoclonal antibodies 1, some authors have proposed clinical 10, 11, and immunological 5 subtypes of HS in an effort to better predict treatment outcome and response. Thus far, no current schema accurately predicts treatment efficacy.
In order to develop and implement effective treatment strategies in HS, a deeper understanding of the underlying inflammatory pathophysiology is needed. However, due to the heterogeneity of sampling methods, laboratory processing methods and data analysis, comparison across studies is problematic and potentially biased or inaccruate 12. Heterogeneity of tissue sampling and laboratory techniques alone may explain the inconsistent and conflicting results regarding specific cytokines, 4, 9 however, no systematic analysis of cytokine studies has been undertaken to compare results, methodology, and analytical techniques.
An additional complicating factor is that clinical comorbidities, which are strongly associated with disease activity in HS, such as obesity 13, diabetes 14, inflammatory bowel disease 15, and smoking 16, also produce pro-inflammatory cytokines, which affect multiple organ systems including the skin 15, 17– 19. Hence, it remains unclear whether the presence or absence of these conditions confound the findings of cytokine studies in HS, and whether clinical stratification of patients is necessary to identify significant pathogenic pathways, which may be amenable to pharmacological intervention. Critical evaluation and analysis of existing studies may also enable meta-analysis, which may identify cytokines, which, in smaller studies, do not have sufficient power to meet statistical significance when compared to controls.
Objectives
The objectives of this systematic review are:
-
1)
To collate and describe all published reports of human cytokine studies in HS including those in skin, blood, serum and exudate.
-
2)
To critically evaluate the sampling, laboratory and analysis techniques used in each study to assess whether comparisons can be made across individual studies.
-
3)
To analyze the heterogeneity of published studies enable meta-analysis
Methods
This systematic review was registered with PROSPERO 20 (Registration number CRD42018104664) and was conducted in line with the PRISMA checklist 21
Data sources
Information sources for this review included PubMed (1946-July 1 2018), Scopus (2004- July 1 2018) and Web of Science (1990-July 1 2018) as shown in Figure 1. Search strategy is presented in Table 1
Figure 1. PRISMA Flowchart.
Table 1. Search Strategy.
| Resources: | |
|---|---|
| 1) Pubmed (1946-July 1 2018),
2) Scopus (2004- July 1 2018) 3) Web of Science (1990-July 1 2018) 4) Published Abstracts 5) Contact with Authors for abstracts without full text for clarification of data and methodology |
|
| Pubmed Search Strategy: | |
| acne inversa OR apocrine acne OR apocrinitis OR Fox-den disease OR hidradenitis axillaris OR HS OR
pyodermia sinifica fistulans OR Velpeau’s disease OR Verneuil’s disease OR Hidradenitidis Suppurative AND Cytokine OR chemokine OR inflammatory mediator |
|
Study eligibility criteria
Eligibility criteria for this review included cohort studies, case-control studies and other observational studies with no restrictions of patient age, sex, ethnicity or language of publication. Eligible studies included:
-
1)
Studies reporting the results of cytokine investigations (in cutaneous tissue, serum, blood or exudate) in human subjects clinically diagnosed with hidradenitis suppurativa.
Studies deemed not eligible included those which:
-
1)
Provide no new data but a review or summary of previously published data
-
2)
Provide no comparison with controls or non-lesional tissue
Appraisal and synthesis methods
Data collection was performed independently by 2 authors (JWF & JEH), with any disagreements regarding inclusion of citations being referred to a third author (JGK) for mediation. Information was collected using a standardized data collection form (available as Extended data 22) with the principal outcomes of interest being the cytokine of interest, measured level of cytokine in lesional HS skin or serum. Comparison data against either peri-lesional, unaffected or control skin or serum was also collated. If data from individual patients was not available then the aggregate data including average change and statistical analyses of the significance of change was collected.
For each individual cytokine, where more than one study reported results, heterogeneity was assessed using the chi-squared tests for homogeneity. Homogeneity was defined as a chi squared value >0.05. All statistical analysis was undertaken using R (version 3.5.1)
Potential sources of bias in the identified studies are acknowledged including the small size of patient cohorts, the variability in sampling, laboratory techniques and the inclusion of patients being treated with a wide-variety of medications including immunosuppressants. Bias was also assessed using the NIH quality assessment tool for observational studies 23.
Results
A total of 367 non-duplicated citations were identified in the literature review ( Figure 1). 343 of these articles were removed upon review of titles and abstracts against the pre-defined eligibility criteria. Full text review of the remaining 24 articles excluded 5 review articles providing no new data. The remaining 19 studies 2– 9, 24– 33 included the results of 564 individual HS patients and 198 control patients, which were included in this systematic review.
Demographics
The summarized demographic data of the patients and controls comprising this review are included in Table 2. The 564 reported cases comprised of 231 males (40.9% reported cases) and 333 females (59.0%). 24 cases were unreported (4.1%). The average age was 38.5 years (n=560, 18 cases unreported). 141 individuals were current smokers (82.4% reported cases), 8 ex-smokers (4.7% reported cases), 22 non-smokers (12.8% reported cases) and 407 unreported. Obesity (BMI>30) was reported in 85 individuals (42.5% reported cases), with 115 (57.5%) individuals non-obese (BMI<30) and unreported in 378 cases. 8 cases reported diabetes mellitus out of 24 reports (33% of reported cases). 12/38 cases reported a positive family history of HS (31.6% reported cases). Hurley Stage was reported as stage 1 in 68 individuals (17.4% reported), stage 2 in 199 individuals (51% reported cases) and stage 3 in 123 individuals (31.6% reported cases) with 188 cases going unreported. The average mHSS (modified hidradenitis suppurativa score) was 78.1 (n=247 cases). Biopsies were largely taken from the axillae (n=32, 43.8%) and groin (n=35, 48.0%), with a minority of samples being taken from the genital and perianal region (n=6, 8.2%). At the time of sampling patients were on treatment including Clindamycin+ Rifampicin (n=18); adalimumab (n=26); Metformin (n=2); levothyroxine (n=1); MABp1 (n=10); tetracyclines (n=12) Infliximab (n=2); other antibiotics (n=4). Treatment was not specified in 74 cases, with no treatment in 86 individuals and treatment withheld in 85 patients.
Table 2. Demographic data of included studies.
| Number
of HS Patients |
Male | Female | Mean Age (Years) | Comorbidities | Biopsy Sites | Hurley
Staging |
mHSS Score (Mean) | Therapy | Study
Reference |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Smoking | Obesity
(BMI>30) |
Diabetes | Family
History |
Axillae | Groin | Genital | ||||||||
| 17 | 1 | 45 | Ex | Y | NR | NR | Serum Measurements | 2 | NR | Thyroxine | 2 | |||
| 1 | 39 | Y | N | NR | NR | 2 | NR | N | ||||||
| 1 | 24 | N | N | NR | NR | 2 | NR | N | ||||||
| 1 | 41 | Y | Y | NR | NR | 2 | NR | N | ||||||
| 1 | 23 | Ex | Y | NR | NR | 1 | NR | N | ||||||
| 1 | 35 | Y | Y | NR | NR | 2 | NR | N | ||||||
| 1 | 30 | Y | N | NR | NR | 2 | NR | Metformin | ||||||
| 1 | 41 | Y | Y | NR | NR | 3 | NR | Clindamycin, Rifampicin | ||||||
| 1 | 35 | Y | Y | NR | NR | 3 | NR | Metformin | ||||||
| 1 | 47 | Y | N | NR | NR | 3 | NR | N | ||||||
| 1 | 19 | N | N | NR | NR | 1 | NR | N | ||||||
| 1 | 34 | Y | N | NR | NR | 2 | NR | Adalimumab | ||||||
| 1 | 47 | N | N | NR | NR | 3 | NR | Adalimumab, Doxycycline | ||||||
| 1 | 32 | Y | N | NR | NR | 2 | NR | Adalimumab | ||||||
| 1 | 38 | Y | N | NR | NR | 3 | NR | Adalimumab, Doxycycline | ||||||
| 1 | 24 | Y | Y | NR | NR | 2 | NR | Adalimumab | ||||||
| 1 | 26 | E | Y | NR | NR | 2 | NR | Adalimumab | ||||||
| 18 | 11 | 7 | (Range 19–62) | NR | NR | NR | NR | NR | NR | NR | 24 | |||
| 15 | 6 | 9 | 38.7 | NR | NR | NR | NR | N=9 | N=4 | N=2 | Stage 1=0
Stage 2=10 Stage 3=5 |
N | 3 | |
| 18 | 1 | 1 | 38 | N | Y | NR | N | NR | NR | NR | 3 | 54 | N | 4 |
| 1 | 42 | Y | N | NR | N | NR | NR | NR | 3 | 56 | N | |||
| 1 | 30 | N | Y | NR | Y | NR | NR | NR | 3 | 57 | Tetracycline | |||
| 1 | 43 | Y | N | NR | N | NR | NR | NR | 1 | 11 | Tetracycline | |||
| 1 | 32 | N | Y | NR | Y | NR | NR | NR | 1 | 14 | Tetracycline | |||
| 1 | 14 | N | N | NR | N | NR | NR | NR | 3 | 65 | Rifampicin, Clindamycin | |||
| 1 | 47 | Y | N | NR | N | NR | NR | NR | 3 | 44 | Tetracycline | |||
| 1 | 43 | Y | Y | NR | N | NR | NR | NR | 3 | 22 | N | |||
| 1 | 21 | Y | N | NR | N | NR | NR | NR | 1 | 13 | Tetracycline | |||
| 1 | 47 | N | N | NR | N | NR | NR | NR | 1 | 11 | Tetracycline | |||
| 1 | 27 | Y | N | NR | N | NR | NR | NR | 2 | 7 | Tetracycline | |||
| 1 | 22 | N | N | NR | Y | NR | NR | NR | 3 | 68 | N | |||
| 1 | 50 | Y | N | NR | Y | NR | NR | NR | 2 | 46 | N | |||
| 1 | 23 | N | N | NR | Y | NR | NR | NR | 2 | 22 | N | |||
| 1 | 19 | Y | Y | NR | N | NR | NR | NR | 2 | 26 | N | |||
| 1 | 44 | Y | N | NR | Y | NR | NR | NR | 2 | 14 | N | |||
| 1 | 22 | Y | N | NR | N | NR | NR | NR | 3 | 23 | N | |||
| 1 | 20 | N | N | NR | Y | NR | NR | NR | 2 | 21 | Tetracycline | |||
| 1 | 48 | Y | N | NR | N | 1 | 3 | NR | Rifampicin, Clindamycin | |||||
| 1 | 25 | Y | N | NR | N | 1 | 2 | NR | Amoxicillin+ Clav Acid | |||||
| 1 | 20 | N | N | NR | N | 1 | 2 | NR | N | |||||
| 1 | 31 | N | Y | NR | N | 1 | 3 | NR | Adalimumab | |||||
| 1 | 40 | NA | NA | NR | NA | 3 | NR | N | ||||||
| 1 | 46 | Y | N | NR | N | 1 | 3 | NR | Tetracycline | |||||
| 1 | 26 | Y | N | NR | N | 1 | 2 | NR | Azithromycin | |||||
| 1 | 36 | Y | N | NR | N | 1 | 2 | NR | Amoxicillin+ Clav Acid | |||||
| 1 | 29 | N | N | NR | y | 1 | 2 | NR | Amoxicillin+ Clav Acid | |||||
| 24 | 8 | 16 | 36.5 (Range 21–51) | NR | NR | NR | NR | NR | NR | NR | Mean=2.29
(SD=0.62) |
NR | Untreated | 7 |
| 74 | 36 | 38 | 37.4 (SD=12.0) | NR | N=32
(43.2%) |
NR | NR | Serum Measurements | Stage 1= 11
Stage 2=47 Stage 3=16 |
All on treatment
(Not further elaborated) |
8 | |||
| 8 | 4 | 4 | 41.61 (SD=13.81) | N=5
Y=2 Ex=1 |
NR | N=4 | NR | Exudate Measurements | Stage 1=0
Stage 2=3 Stage 3=5 |
68.88 (SD=41.45) | NR | 6 | ||
| 19
19 |
11 | 8 | 45.6 (SD=10.7) | N=14
(74%) |
N=13
(68.4%) |
NR | NR | Serum Measurements | Stage 1=0
Stage 2=9 Stage 3=10 |
82.79 (SD 41.0) | NR | 25 | ||
| 34.5 (SD 43.5) | Adalimumab | |||||||||||||
| 120 | 43 | 77 | 37.3 (SD=5.9) | NR | NR | NR | NR | Serum Measurements | Stage 1=39
Stage2=52.4 Stage 3=44 |
28.1 (SD=20.2)
52.4 (SD=24.9) 129.3 (SD=79.2) |
NR | 5 | ||
| 44 | 13 | 31 | 39.1 (SD=11.4) | Y=34
Ex=4 |
N=16 | NR | NR | NR | NR | NR | Stage 1=5
Stage 2=27 Stage 3=12 |
NR | N=15 Rifampicin,
Clindamycin N=1 Minocycline N=2 Adalimumab n=2 Infliximanb n=24 untreated |
31 |
| 22 | 10 | 12 | 38.2 (Range 19-60) | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | 30 |
| 3 | 1 | 54 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | 9 | |
| 1 | 36 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | |||
| 1 | 59 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | |||
| 10 | 5 | 5 | 42 (Range 21–49) | NR | NR | NR | NR | 1 | 1 | N | Stage 2
(100%) |
NR | Treatment Withheld | 32 |
| 20 | 8 | 12 | 37.5 (Range 21–51) | N=18 | N=10 | NR | NR | NR | NR | NR | NR | NR | Treatment Withheld
(8 weeks prior) |
29 |
| 25 | 9 | 16 | 36 (Range 18–51) | NR | NR | NR | NR | NR | NR | NR | Mean =2.16
(SD=0.55) |
NR | Treatment Withheld
(3 weeks prior) |
28 |
| 47 | 19 | 28 | 42.3 (Range 22–54) | NR | Serum Measurements | 48.3 (Range 8–144) | NR | 27 | ||||||
| 11 | 9 | 2 | 39.6 (Range 18–61) | NR | NR | NR | NR | NR | NR | NR | “Mod-Severe
Disease” |
NR | NR | |
| 20 | 6 | 14 | 40 (SD=15) | 19 | 27.6 (4.1) | NR | NR | 7 | 12 | 1 | Stage 1=4
Stage 2=11 Stage 3=5 |
Treatment withheld
3 weeks prior |
26 | |
| 10 | 1 | 9 | 38 (SD=15) | 10 | 28.9 (SD
4.5) |
NR | NR | 3 | 7 | 0 | Stage1=2
Stage2=7 Stage3=1 |
Treatment Withheld
3 weeks prior |
||
| 10 | 7 | 3 | 46.6 (SD=15.1) | 10 | 29.4 (4.7) | 3 | 2 | Serum | Stage 3=10 | 195.6 (SD=97.9) | MABp1 | 33 | ||
| 10 | 6 | 4 | 49.3 (SD=9.8) | 8 | 27.9 (7.1) | 1 | 2 | Stage 2=2
Stage 3=8 |
124.9 (SD=73.7) | No Treatment | ||||
| TOTAL:
564 |
231 | 333 | 38.5 | 141 | 85 (0f 200) | 8 (of 24) | 12 | 32 | 35 | 6 | Stage 1= 68
Stage 2=199 Stage 3=123 |
Average =78.1
(n=247) |
Clindamycin+
Rifampicin=18; Adalimumab=26; Metformin=2; Treatment withheld= 85; Thyroxine=1; MABp1=10; Tetracycylines=12; No Treatment=86; Not Specified=74; Infliximab=2; Antibiotics=4; Not Reported=258 |
|
BMI= Body Mass Index mHSS= modified Hidradenitis Suppurativa Score (Sartorius Score) NR= Not Reported SD= Standard Deviation Y= Yes N=No Ex= Ex Smoker
Only 5/19 (26.3%) studies analysed both lesional tissue and serum levels of cytokines, enabling direct comparison between these two compartments. 8/19 (42.1%) studies provided age and sex matched controls, 5/15 (33.3%) studies stratified by disease severity and no studies stratified by lesion site or comorbidities. 8/19 (42.1%) studies stratified or accounted for treatment or reported discontinuing treatment up to 3 weeks prior to sample collection ( Table 3).
Table 3. Critical evaluation of methodology of studies included in this review.
| Cytokines Measured | Number
of HS Patients |
Number of
Controls |
Samples
Analyzed |
Age/Sex
Matched Controls |
Timing of
Samples |
Stratified
by severity |
Stratified
by lesion site |
Stratified
by Co- morbidities |
Stratified by
Treatment |
Sample
Storage Time |
Sample Types | Study
Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-17 IL-22 IFNg IL-2 IL-10 GM-CSF | 17 | 9 | L, PL, U,
C, S |
Y | NR | NR | N | N | Y | NR | Skin, Serum | 2 |
| S100A7 Lysozyme LL37 hBD3 α-MSH
MIF TNF-α IL-8 MHC1 |
18 | 12 | L | N | NR | NR | N | N | N | NR | Skin | 24 |
| IL-36α IL-36β IL-36g | 15 | 15 | L, PL | NR | NR | NR | N | N | N | NR | Skin | 3 |
| IL-17 IL-22 IFNg CCl20 CCL27
S100A7 S100A8 IL-1B CCL5 IP10 IL-8 IL-6 TNF-α |
18 | 18 | L, PL, S | Y | NR | Y | N | N | N | NR | Skin, Serum | 4 |
| LL37 IL-17 TNF-α IL-23 IL-1b IL-10
IL-32 |
24 | 9 | L | Y | NR | NR | NR | N | Y (untreated) | NR | Skin | 7 |
| IL-6 IL-23 TNF-α R1 IL-1β IL-8 IL-10
IL-12p70 IL17A TNFR2 CRP ESR |
74 | 22 | Serum only | N | NR | Y | NR | N | N | NR | Serum | 8 |
| IFNg, IL-12p70,IL-1β IL-1α IL-17A
IL-6 TNF-α TNF-β IL-16 IL-12/23p40 IL-10 IL-4 IL-13 IL-2 IL-15 IL-7 IL-5 GM-CSF VEGF |
8 | 8 | Wound
Exudate |
Y | NR | N | N | N | N | NR | Wound Exudate | 6 |
| IL-1B IL-6 IL-8 IL-10 IL-17A IL-23
TNFR1 TNFR2 |
19 | 19 | Serum only | N | Y
(Fasting) |
N | N | N | Y (Adalimumab) | NR | Serum only | 25 |
| TNF-α, IL-1B, IL-6 IL-10 IL-17 IL-22
IL-1RA |
120 | 24 | Serum and
Pus |
Y | N | Y | N | N | Y (Etanercept) | NR | Serum
Pus |
5 |
| IL-17 IL-1B IL-10 TNF-α | 44 | 5 | L, PL, U | N | N | N | N | N | N | NR | Skin | 31 |
| IL-17 Caspase1 NLRP3 S100A8
S100A9 |
22 | Yes (NR) | L, PL, U, C | NR | NR | N | N | N | N | NR | Skin | 30 |
| TNF-α IL-1β IL-6 IFNg IL-17A IL-22 | 3 | (Unknown) | S | Y | NR | N | N | N | N | NR | Serum | 9 |
| IL1-2p70 IL-23p19 IL-17 | 10 | 8 | L, C | N | NR | N | N | N | Y (ceased 3/25 prior) | NR | Skin | 32 |
| IL-32 IL-32α IL-32β IL-32d IL-32g
IFNg IL-17 IL-13 |
20 | 10 | L, C, S | N | NR | Y | N | N | Y (ceased 8/52 prior) | NR | Skin, Serum | 29 |
| IL-36α IL-36β IL-36g IL-36RA | 25 | 7 | L, C, S | N | NR | N | N | N | Y (ceased 3/25 prior) | NR | Skin Serum | 28 |
| TNF-α IFNg IL-1β IL-6 IL-10 IL-19,
IL-17A IL-22 IL-36b IL-12/23p40 IL-22 E Selectin P Selectin CXCL6 CXCL11 CX3CL1 CCL2 CCL18 CXCL9 sVEGFR1 MMP2 Cystatin C LCN2 |
10 | 16 | L | Y | NR | N | N | N | N | NR | Skin Serum | 27 |
| IL-1β IL-2 IL-4 IL-5 IL-6 IL-8 IL-10 IL-
12p70 TNF-α IFNg |
20 | 6 | L, PL, C | N | NR | Y | N | N | N | NR | Skin | 26 |
| IL-1α, IL-8 | 10 | 10 | S | N | NR | N | N | N | Y | NR | Serum | 33 |
Table 2: Critical Evaluation of Methodology of Studies Included in This Review Key:L= Lesional, PL= Perilesional, U= Uninvolved, C= Control S=Serum, Y=Yes, N=No, NR= Not Reported,
Cytokine analysis
A total of 81 discrete cytokines were analysed over the 19 studies (presented in Table 4). 6 studies provided a total of 78 outcomes from tissue of lesional or peri-lesional biopsies, 4 studies provided a total of 30 results from serum analysis and 1 study provided 15 results from exudate analysis. The remaining 8 studies did not provide quantification of cytokine levels but did provide analysis of the change and significance between lesion and control samples. The degree of change between lesional and control samples varied widely from 1.5 times the control level (IL-1RA p=0.0112) to 149 times the control level (IL-17 p<0.05). 33 cytokines were evaluated in more than one study. Only IL-1β, IL-6, IL-8, IL-17A and TNF-α had data from 5 or more separate studies.
Table 4. Reported cytokine results of studies included in this systematic review.
| Target
Cytokine |
Mean Level
in Patient Serum (pg/mL) |
Mean Level in
Control Serum (pg/mL) |
Mean Level in
Lesional Tissue (pg/mL) |
Mean Level
in Perilesional Tissue (pg/mL) |
Mean
Uninvolved Tissue Levels (pg/mL) |
Mean Control
Tissue Levels (pg/mL) |
Fold
Increase |
Comparison and
Significance |
Comparison and
Significance |
Study Reference |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-1α | 1126 | 2549 | Le:Ce | P= 0.53 | 6 | ||||||||
| 0.2 | 0.1 | NR | L:C | NS | 26 | ||||||||
| 772.0 | 697.2 | HSs:Cs | NS | 33 | |||||||||
| IL-1RA | 44.0 | 29.6 | 1.5 | L:C | P= 0.0112 | 26 | |||||||
| IL-1β | 0.9 | 0.4 | HSs:Cs | P=0.801 | 8 | ||||||||
| 862.5 | 1503 | Le:Ce | P= 0.69 | 6 | |||||||||
| L:C | NS | Lpa:C | NS | 25 | |||||||||
| SERUM ONLY | HSs:Cs | P= 0.044 | 5 | ||||||||||
| 100 | 10 | 3 | 1 | 115 fold | L:C | P= 0.001 | PL:C | 0.05 | 31 | ||||
| L:U | P= 0.01 | U:C | NS | ||||||||||
| R=0.7 # | L:C | NS | 7 | ||||||||||
| 1.6 | 0.0 | 54.4 | L:C | P= 0.0028 | 26 | ||||||||
| IL-4 | 6.56 | 9.77 | Le:Ce | P= 0.54 | 6 | ||||||||
| 0.0 | 0.1 | L:C | NS | 7 | |||||||||
| IL-5 | 0.2 | 0.2 | L:C | NS | 7 | ||||||||
| 30.15 | 9.314 | Le:Ce | P= 0.17 | 6 | |||||||||
| IL-6 | L:C
*
L:C ** L:C *** |
NS
NS NS |
4 | ||||||||||
| 6.2 | 0.6 | HSs:Cs | P= 0.001 | 8 | |||||||||
| 2377 | 5451 | Le:Ce | NS | 6 | |||||||||
| L:C | P= 0.05 | Lpa:C | 0.05 | 25 | |||||||||
| SERUM ONLY | HSs:
Cs +++ |
P= 0.002 | 5 | ||||||||||
| 124.4 | 101.9 | L:C | NS | 7 | |||||||||
| sIL-6R | 16.3 | 4.4 | 3.7 | L:C | P= 0.0028 | 7 | |||||||
| IL-8 | NR | NR | i69.6 / s67.6 | 64.9 | Li:C | P<0.01 | Ls:C | P<0.001 | 24 | ||||
| L:C
*
L:C ** L:C *** |
NS
NS NS |
4 | |||||||||||
| 27.9 | 36.3 | HSs:Cs | NS | 8 | |||||||||
| L:C | P= 0.05 | Lpa:C | NS | 25 | |||||||||
| 1401 | 12.0 | L:C | NS | 7 | |||||||||
| 1000 | 3000 | L:C | P= 0.049 | 33 | |||||||||
| IL-10 | L:C | P<0.05 | 4 | ||||||||||
| 3.4 | 3.3 | HSs:Cs | NS | 8 | |||||||||
| 19.85 | 34.74 | Le:Ce | NS | 6 | |||||||||
| L:C | P= 0.05 | Lpa:C | 0.05 | 25 | |||||||||
| SERUM ONLY | HSs:Cs + | P= 0.0001 | 5 | ||||||||||
| SERUM ONLY | HSs:Cs ++ | P= 0.0001 | 5 | ||||||||||
| 3.8 | 1.1 | 0.4 | 3-4 | L:C | P= 0.01 | PL:C | NS | 31 | |||||
| L:U | P= 0.01 | U:C | NR | ||||||||||
| 3 | 2 | HSs:Cs | NS | 27 | |||||||||
| 19.2 | 1.3 | 14.8 | L:C | P= 0.0028 | 7 | ||||||||
| IL-11 | 78.6 | 7.2 | 11.0 | L:C | P= 0.0056 | 7 | |||||||
| IL-12p40 | 488.3 | 97.86 | Le:Ce | P= 0.07 | 6 | ||||||||
| 75 | 75 | HSs:Cs | NS | 27 | |||||||||
| 0.5 | 0.4 | L:C | NS | 7 | |||||||||
| IL-12p70 | 3.4 | 0.6 | HSs:Cs | P= 0.427 | 8 | ||||||||
| 9.412 | 15.02 | Le:Ce | P= 0.609 | 6 | |||||||||
| 0.0 | 0.0 | L:C | NS | 7 | |||||||||
| IL-13 | 70.98 | 55.61 | Le:Ce | P= 0.56 | 6 | ||||||||
| 0.0 | 0.1 | L:C | NS | 7 | |||||||||
| IL-15 | 24.5 | 5.61 | Le:Ce | P= 0.18 | 6 | ||||||||
| 1.9 | 2.9 | L:C | NS | 7 | |||||||||
| IL-16 | 15277 | 15586 | Le:Ce | P= 0.97 | 6 | ||||||||
| 22.3 | 4.2 | 5.3 | L:C | P= 0.0028 | 7 | ||||||||
| IL-17 | S:C | P<0.005 | 4 | ||||||||||
| SERUM
ONLY |
SERUM ONLY | SERUM
ONLY |
HSs:Cs + | 0.014 | 5 | ||||||||
| SERUM
ONLY |
SERUM ONLY | SERUM
ONLY |
HSs:Cs ++ | 0.005 | 5 | ||||||||
| 150 | 45 | 1 | 1 | 149 fold | L:C | P= 0.05 | PL:C | 0.05 | 31 | ||||
| L:PL | NS | U:C | 0.05 | ||||||||||
| No Quantification | L:C | ↑(NS) | L:PL | No Diff | 30 | ||||||||
| R=0.66 # | NS | 27 | |||||||||||
| IL-17A | L:C | P<0.005 | 4 | ||||||||||
| 5.6 | 0.3 | HSs:Cs | NS | 8 | |||||||||
| 1006 | 32.7 | Le:Ce | NS | 6 | |||||||||
| L:C | P= 0.05 | Lpa:C | NS | 25 | |||||||||
| 4 | 5 | HSs:Cs | NS | 27 | |||||||||
| 8.1 | NR | 1.1 | 7.3 | L:C | P= 0.0056 | 26 | |||||||
| IL-22 | L:C | NS | 4 | ||||||||||
| 8.8 | 0.0 | HSs:Cs | NS | 8 | |||||||||
| IL-23 | L:C | NS | Lpa:C | 0.05 | 25 | ||||||||
| R=0.68 # | NS | 7 | |||||||||||
| IL-32 | 50ng/mL | 1ng/mL | Only Normalised Values Provided | 4 (skin)
50 (serum) |
L:C | P= 0.01 | HSs:
Cs |
p<0.05 | 29 | ||||
| IL-32α | 3 fold | L:C | P= 0.01 | 29 | |||||||||
| IL-32β | 2 fold | L:C | P= 0.05 | 29 | |||||||||
| IL-32g | Not
elevated |
L:C | P= 0.001 | 29 | |||||||||
| IL-32d | 3 fold | L:C | NS | 29 | |||||||||
| IL-36α | 0.4 | 0.02 | 0.02 | L:C | P=0.0174 | PL:C | NS | 3 | |||||
| 250 | 0 | 1 | 45.07 fold | L:C | P= 0.01 | 28 | |||||||
| IL-36b | 4.33 | 3.00 | 0.51 | L:C | P= 0.0001 | PL:C | 0.0035 | 3 | |||||
| 15 | 4 | 1 | 1.45 fold | L:C | P= 0.25 | 28 | |||||||
| IL-36g | 3.64 | 0.83 | 0.49 | L:C | P= 0.0161 | PL:L | 0.0302 | 3 | |||||
| 100 | 20 | 1 | 1.96 fold | L:C | P= 0.07 | 28 | |||||||
| IL-36RA | 0.46 | 0.28 | 0.06 | L:C | P= 0.0001 | PL:C | 0.0003 | 3 | |||||
| 50 | 100 | No Quantificaiton | No
Increase |
L:C | P= 0.10 | 28 | |||||||
| IL-37 | 3.24 | 14.7 | 1.81 | PL:L | P= 0.0002 | PL:C | 0.0001 | 3 | |||||
| IL-38 | 0.09 | 0.19 | 0.06 | L:C | P= 0.0230 | PL:C | 0.0069 | 3 | |||||
| TNF-α | i69.4 | 66.6 s | NR | 65.8 | NR | Li:C | NS | Ls:C | NS | 24 | |||
| L:C
*
L:C ** L:C *** |
NS
NS NS |
4 | |||||||||||
| 83.26 | 65.74 | Le:Ce | P= 0.7 | 6 | |||||||||
| SERUM ONLY | SERUM
ONLY |
HSs:Cs + | P=0.021 | 5 | |||||||||
| 2.2 | 1.3 | 0.6 | 0.7 | L:C | P=0.01 | PL:C | 0.01 | 31 | |||||
| L:PL | NS | U:C | NS | ||||||||||
| 0.3 | 0.2 | 1.6 | L:C | P=0.0336 | 26 | ||||||||
| TNF-β | 9.24 | 1.65 | Le:Ce | P=0.03 | 6 | ||||||||
| 0.4 | 0.4 | NR | L:C | NS | 26 | ||||||||
| sTNFR1 | 879.8 | 325.9 | HSs:Cs | P <0.001 | 8 | ||||||||
| L:C | NS | Lpa:C | 0.05 | 25 | |||||||||
| 78.0 | 40.2 | 1.9 | L:C | P= 0.0112 | 26 | ||||||||
| sTNFR2 | 927.9 | 527.4 | HSs:Cs | P= 0.053 | 8 | ||||||||
| L:C | P= 0.05 | Lpa:C | 0.05 | 25 | |||||||||
| 47.0 | 8.1 | 5.8 | L:C | P= 0.0028 | 26 | ||||||||
| hBD1 | 0.019
0.021 0.018 |
0.058
0.077 0.095 |
0.3
0.3 0.2 |
L:C
*
L:C ** L:C *** |
P= 0.240
P= 0.132 P= 0.026 |
4 | |||||||
| hBD2 | 0.013
0.019 0.058 |
0.011
0.018 0.067 |
1.1
1.1 0.9 |
L:C
*
>L:C ** L:C *** |
P= 0.937
P= 0.699 P= 0.937 |
4 | |||||||
| hBD3 | 76.9 i | 75.7 s | 72.5 | NR | Li:C | P<0.05 | Ls:C | NS | 24 | ||||
| 0.33
0.33 0.379 |
0.117
0.125 0.203 |
2.8
2.6 1.9 |
L:C
*
L:C ** L:C *** |
P= 0.485
P= 0.394 P= 0.485 |
4 | ||||||||
| S100A7 | i84.8 | 77.8 s | 71.5 | NR | Li:C | P<0.001 | Ls:C | P<0.05 | 24 | ||||
| 1.516
1.625 2.297 |
0.177
0.354 0.707 |
8.6
4.6 3.2 |
L:C
*
L:C ** L:C *** |
P= 0.009
P= 0.180 P= 0.132 |
4 | ||||||||
| S100A8 | 24.251
25.992 24.251 |
4.925
11.314 10.556 |
4.9
2.3 2.3 |
L:C
*
L:C ** L:C *** |
P= 0.240
P= 0.537 P= 0.393 |
4 | |||||||
| NR | L:C | ↑ (NS) | L:PL | ↑ (NS) | 30 | ||||||||
| S100A9 | 0.003
0.005 0.003 |
0.002
0.004 0.006 |
1.7
1.1 0.6 |
L:C
*
L:C ** L:C *** |
NS
NS NS |
4 | |||||||
| L:C | ↑ (NS) | L:PL | ↑ (NS) | 30 | |||||||||
| LL37 | 84.1 i /80.9 s | 75.8 | Li:C | P<0.05 | Ls:C | NS | 24 | ||||||
| Lyzozyme | 55.2 i / 52.7 s | 59.6 | Li:C | NS | Ls:C | P<0.05 | 24 | ||||||
| MIF | 77.8 i/ 77.8 s | 70.7 | Li:C | NS | Ls:C | P<0.01 | 24 | ||||||
| αMSH | NR | i74.6 i / 73.1 s | NR | 70.9 | Li:C | P<0.01 | Ls:C | P<0.01 | 24 | ||||
| MHC1 | 75.5 i/74.7 s | 74.4 | Li:C | NS | Ls:C | NS | 24 | ||||||
| RNase7 | 0.435
0.330 0.574 |
0.063
0.077 0.109 |
7.0
4.3 5.3 |
L:C
*
L:C ** L:C *** |
P= 0.145
P= 0.589 P= 0.179 |
4 | |||||||
| IP10 |
89.9 |
12.6 |
L:C
*
L:C ** L:C *** |
P<0.05
P<0.005 P<0.05 |
4 | ||||||||
| CCL3 | 0.4 | 0.2 | 2.0 | L:C | P= 0.0196 | 26 | |||||||
| CCL5 | -
46.1 - |
-
6.2 - |
L:C
*
L:C ** L:C *** |
P<0.05
P<0.05 NS |
4 | ||||||||
| 7.6 | 1.4 | 5.4 | L:C | P= 0.0112 | 26 | ||||||||
| CCL20 | L:C | P<0.005 | 4 | ||||||||||
| CCL27 | L:C | P<0.05 | 4 | ||||||||||
| CRP | 13.4 | 1.2 | HSs:Cs | p<0.001 | 8 | ||||||||
| L:C | P= 0.05 | Lpa:C | 0.05 | 25 | |||||||||
| ESR | 29.5 | 10.2 | HSs:Cs | <0.001 | 8 | ||||||||
| L:C | P= 0.05 | Lpa:C | 0.05 | 25 | |||||||||
| IFNg | R=0.7 | L:C | NS | 7 | |||||||||
| <5%
Normal |
HSs:Cs | ↑ (NS) | 9 | ||||||||||
| 1418 | 102.5 | Le:Ce | P= 0.027 | 6 | |||||||||
| HSs:Cs | P<0.05 | L:C | P<0.05 | 4 | |||||||||
| GMCSF | 78.45 | 82.13 | Le:Ce | P= 0.96 | 6 | ||||||||
| 0.4 | 0.0 | NR | L:C | NS | 26 | ||||||||
| VEGF | 632.1 | 1544 | Le:Ce | P= 0.23 | 6 | ||||||||
| sVEGFR1 | 60 | 60 | HSs:Cs | NS | 27 | ||||||||
| Caspase 1 | No Quanti | No Quanti | L:C | ↑ (NS) | L:PL | ↑ (NS) | 30 | ||||||
| NLRP3 | No Quanti | No Quanti | L:C | ↑ (NS) | L:PL | NS | 30 | ||||||
| CAMP | 4 | L:C | NS | 7 | |||||||||
| Uteroglobulin | 20 | 20 | HSs:Cs | NS | 27 | ||||||||
| Cystatin C | 0.85 | 0.8 | HSs:Cs | 27 | |||||||||
| LCN2 | 90 | 40 | 0.5 | 0.02 | HSs:Cs | <0.001 | L:C | <0.001 | 27 | ||||
| BD2 | 0.9 | 1 | HSs:Cs | NS | 27 | ||||||||
| MMP2 | 200 | 210 | HSs:Cs | <0.05 | 27 | ||||||||
| BLC | 8.1 | 0.58 | 10.5 | L:C | P= 0.0056 | 26 | |||||||
| ICAM-1 | 98.7 | 31.9 | 3.1 | L:C | P= 0.0028 | 26 | |||||||
| Eotaxin | 0.1 | 0.1 | NR | L:C | NS | 26 | |||||||
| Eotaxin2 | 3.9 | 2.5 | NR | L:C | NS | 26 | |||||||
| CXCL6 | 160 | 140 | NS | 27 | |||||||||
| CXCL9 | 219.8 | 13.8 | 16 | L:C | P= 0.0028 | 26 | |||||||
| CXCL11 | 0.4 | 0.4 | NS | 27 | |||||||||
| CX3CL1 | 0.9 | 1 | NS | 27 | |||||||||
| I-309 | 0.4 | 0.3 | NR | L:C | NS | 26 | |||||||
| MCP1 | 47.5 | 37.1 | NR | L:C | NS | 26 | |||||||
| M-CSF | 0.4 | 0.2 | NR | L:C | NS | 26 | |||||||
| MIP1b | 16.1 | 5.8 | NR | L:C | NS | 26 | |||||||
| MIP1d | 0.1 | 0.1 | NR | L:C | NS | 26 | |||||||
| PDGF | 0.5 | 0.2 | NR | L:C | NS | 26 | |||||||
| TIMP1 | 260.1 | 166.2 | NR | L:C | NS | 26 | |||||||
| TIMP2 | 989.2 | 997.3 | NR | L:C | NS | 26 | |||||||
Key: L= Lesional ; PL= Perilesional; C= Control; NS= Not Significant ; HSs= HS Serum; Cs= Control Serum; HSe= HS Exudate; Ce= Control Exudate; I = Inflamed lesional skin, S= Scarred lesional skin, #= Vs CAMP, *= NT (Non-Treated) Samples ,** = Stimulation by Pam2CSK4 Lipopeptide,*** Stimulation by Muramyl Dipeptide (MDP), + Heat Killed Candida Albicans; ++ Heat Killed Staph Aureus, +++ Lipopolysaccharide;
Cytokines and inflammatory proteins which were elevated in more than one study in lesional tissue included IL-1β, IL-6R, IL-10, IL-17A, IL-36α, IL-36β, IL-36 γ, IL-36RA, TNF-α, sTNFR2, hBD1, hBD2, hBD3, s100A7, LL37/Cathelicidin, CCL3, CCL5, CCL27 and BLC. Cytokines and inflammatory proteins elevated in peri-lesional tissue included IL-1β, IL-17, IL-36β, IL-36RA, IL-37, IL-38 and TNF-α. IL-37 was the only cytokine identified which showed significant differences between lesional and peri-lesional tissue, with a 1.81 times elevation in lesional compared to peri-lesional tissue (p=0.0002) 3. IL-17 was elevated in unaffected HS tissue compared to control patient tissue (p<0.05) in one study 31. In HS tissue, S100A9, hBD1 and hBD2 were reduced but this data did not meet statistical significance. Two studies measuring IL-1β levels showed no statistically significant difference between lesional and control skin 7, 25. No significant elevation of IL-6 was seen in lesional tissue compared to control with the exception of 1 study 25. IL-8 levels only just made significance in two studies 5, 7, with one study showing significant elevation of IL-8 in lesional compared to control tissue 24. Two additional studies showed no significant difference 4, 8. TNF-α levels were significantly elevated compared to control tissue in two studies 7, 31 but not significantly in 2 additional studies 4, 24. sTNFR1 was significantly elevated in one study 26 whilst showing a non-significant difference in a second study 25. CCL5 was significant in 2 studies in lesional tissue compared with controls 4, 26. One methodology using muramyl dipeptide (MDP) did not reach statistical significance compared to stimulation with Pam2CSK4 Lipopeptide, and non-treated (NT) cells. IFN- γ was elevated in lesional tissue with no significance in one study 28 and significance in another 4.
Elevated cytokines and inflammatory proteins in HS serum included IL-1β, IL-6, IL-8, IL-10, IL-12p70, IL-17, TNF-α, sTNFR1, CRP, ESR, LC2, and MMP2. TNF-β, and IFN-γ were elevated in wound exudate from active HS lesions. IFN-γ was noted to be decreased in HS patient serum compared to healthy control serum, despite the elevation in wound exudate. Conflicting results were seen in serum findings in IL-10, IL-17 and IFN-γ. One study demonstrated elevated serum IL-10 levels compared to control 5 whereas two other studies 8, 27 showed no significant difference. Whilst two studies 4, 5 illustrated elevated IL-17 Serum levels in HS patients, one study 7 showed no significant difference between patients and controls. IFN-γ showed no statistically significant decrease in the serum of HS patients compared to control in one study 9 but a significant difference in a larger, higher powered study 4.
Because adalimumab improves HS through TNF antagonism 1, 2, this cytokine must be classified as pathogenic. TNF mediates inflammation in a classic “sepsis” cascade in tissues—in this pathway LPS from gram negative bacteria activates TNF release from cells, and then TNF stimulates production of IL-1b, IL-6, and IL-8, leading to neutrophil attraction into sites of infection 2, 4. Increases in IL-1β and IL-8 measured in HS, as well as neutrophil accumulation, could result from this pathway. Alternatively, in psoriasis, TNF is a major cytokine that acts on the IL-23/Type 17 T-cell pathway at two points. First TNF induces IL-23 synthesis in myeloid (CD11c+) dendritic cells in the skin 34. Second, TNF (as well as other cytokines that also activate NF-kB) act synergistically with IL-17A or IL-17F to increase synthesis of many other cytokines, chemokines, and inflammatory molecules in keratinocytes and other cell types. There are several clues that an IL-23/Type17 T-cell pathway may be active in HS which include detection of T h17 T-cells in skin infiltrates, increased production of IL-17A, and increased production of LL-37/cathlecidin, S100A7, S100A8, S100A9, LCN2, IL-8, beta-defensins and IL-36; which are all molecules induced by IL-17 in keratinocytes, as also the presence of psoriasis-like epidermal hyperplasia in some reports. The increased production of CCL20 4, would be predicted to increase tissue infiltration of both T h17 T-cells and CD11c+ DCs, which have both been observed in HS, and increased production of TGF-β could increase differentiation of T h17 T-cells from precursors and/or influence scarring in skin lesions. If IL-17 is driving inflammation in HS, one would expect to see increased production of additional chemokines that regulate neutrophil chemoattraction (CXCL1, CXCL2, CXCL3). Epidermal hyperplasia is not presently explained in HS, but this could be related potentially to increased expression of IL-19, IL-20 or IL-22, which are associated with the IL-23/Type 17 T-cell axis. If IL-22 is produced in HS lesions, this would implicate T h22 T-cells as a T-cell type also associated with the IL-23/Type 17 T-cell axis. There is an uncertain role for other T-cell subsets in HS. Increased production of CXCL9 and IP-10 (CXCL10) are often linked to production of IFN-γ from T h1 T-cells in inflammatory sites, but IL-26 or IL-29, which are also cytokines produced by T h17 T-cells are alternative activators of STAT1 and CXCL9 production. IL-32 production in HS may also be linked to a T-cell subset that produces this cytokine. Low production of T h2 associated cytokines (IL-4, IL-5, or IL-13) has been measured in HS, suggesting an unlikely role of this T-cell subset. Likewise, the presence and function of T regulatory cells (Tregs) in HS lesions needs further study. IL-10 which is elevated in HS could be produced by either Tregs or the cDC1 (BDCA3+) DC subset, but levels may be inadequate to control tissue inflammation. At present, dendritic cell subsets are also incompletely characterized in HS. Potential sources of IL-12 or IL-23 are CD11c+ DCs, which includes the tissue resident BDCA-1+ (cDC2) subset and less mature inflammatory DCs, which are abundant cells in inflammatory lesions of psoriasis or atopic dermatitis but have not been investigated in HS. Cytokine contributions by other cell types such as innate lymphoid cells, macrophages, mast cells, and other leukocytes also remains to be determined.
Cytokine analysis methods
The methodologies of cytokine analysis varied widely ( Table 5). 92 results were produced using electrochemical luminescence (ECL) procedures from three separate systems and manufacturers. 62 results were produced using ELISA. 18 results 4 were performed with either ELISA or ECL but not further specified. 15 results were produced using polymerase chain reaction (PCR) with three separate systems from three manufacturers. Four discrete cytokines (IL-10, IL-17, TNF-α and IFN-γ) were analysed using all three techniques (ECL, ELISA and PCR), whilst 15 discrete cytokines (IL-6, IL-8, IL12p40, IL-17A, IL-22, IL-23, S100A7, S100A8, S100A9, RNAse7, IP-10, CCL5, CCL20, CCL27) were analysed using ELISA and ECL only. We note IL-17 levels may well be below the lower limit of quantification with ELC and ELISA based approaches, with only the Singulex platform having the ability to quantify levels of IL-17 present in blood and serum of normal subjects.
Table 5. Cytokine analysis methodology of studies included in this review.
| Cytokine | Method | Details | Study |
|---|---|---|---|
| IL-1α | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| IL-1ra | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| IL-1β | ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 | |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 5 | |
| PCR | IL10, IL17A, IL1Β, IL18 and NLRP3 was performed with predesigned Taqman gene expression assays (Applied Biosystems) on a Roche Light Cycler
(Roche, Pleasanton, CA, U.S.A.) |
31 | |
| PCR | (Hs01555410_m1), ABI-Prism 7300 Sequence Detector System (Applied Biosystems | 7 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| IL-4 | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-5 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| IL-6 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 | |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA). The Milliplex MAP multiplex assay | 25 | |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 5 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| sIL-6R | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 |
| IL-8 | ELISA | pABG AHC0881 1:50 rabbit antihuman | 24 |
| ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 | |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 | |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 33 | |
| IL-10 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 | |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 | |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 5 | |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 5 | |
| PCR | IL10, IL17A, IL1Β, IL18 and NLRP3 was performed with predesigned Taqman gene expression assays (Applied Biosystems) on a Roche Light Cycler
(Roche, Pleasanton, CA, U.S.A.) |
31 | |
| ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-11 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 |
| IL-12p40 | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-12p70 | ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-13 | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-15 | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-16 | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 7 | |
| IL-17 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 5 | |
| PCR | IL10, IL17A, IL1Β, IL18 and NLRP3 was performed with predesigned Taqman gene expression assays (Applied Biosystems) on a Roche Light Cycler
(Roche, Pleasanton, CA, U.S.A.) |
31 | |
| PCR | IL-17 (clone AF-317-NA; R&D Systems, Wiesbaden, Germany), | 30 | |
| PCR | IL-17 (Hs00174383_m1), ABI-Prism 7300 Sequence Detector System | 27 | |
| IL-17A | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). eBioscience, Paris, France | 4 |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 | |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 | |
| ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| IL-22 | ELISA | ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). eBioscience, Paris, France | 4 |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 | |
| IL-23 | ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 |
| PCR | (Hs00992441_m1) ABI-Prism 7300 Sequence Detector System (Applied Biosystems | 7 | |
| IL-32 | PCR | IL-32 (Hs00992441_m1), ABI-Prism 7300 Sequence Detector System | 29 |
| IL-32α | PCR | IL-32a (Hs04353657_gH), ABI-Prism 7300 Sequence Detector System | 29 |
| IL-32β | PCR | IL-32b (Hs04353658_gH), ABI-Prism 7300 Sequence Detector System | 29 |
| IL-32g | PCR | IL-32c (Hs04353656_g1), ABI-Prism 7300 Sequence Detector System | 29 |
| IL-32d | PCR | IL-32d (Hs04353659_gH), ABI-Prism 7300 Sequence Detector System | 29 |
| IL-36α | ELISA | Rabbit polyclonal anti-IL-36a (C-terminal; ab180909), from Abcam, Cambridge, U.K. at 1 : 500 dilution. | 3 |
| ELISA | IL-36a AF1078, RnD | 28 | |
| IL-36β | ELISA | Rabbit polyclonal anti- IL-36b (C-terminal; ab180890) from Abcam, Cambridge, U.K. at 1 : 500 dilution. | 3 |
| ELISA | AF1099, RnD | 28 | |
| IL-36g | ELISA | Mouse monoclonal anti-IL-36c ab156783; (Abcam, Cambridge, U.K.) at 1 : 500 dilution. | 3 |
| ELISA | AF2320, RnD | 28 | |
| IL-36RA | ELISA | Rabbit polyclonal from Abcam, Cambridge, U.K. at 1 : 500 dilution. | 3 |
| ELISA | AF1275, RnD | 28 | |
| IL-37 | ELISA | Rabbit polyclonal Abcam, Cambridge, U.K. at 1 : 500 dilution. | 3 |
| IL-38 | ELISA | Rabbit polyclonal Abcam, Cambridge, U.K. at 1 : 500 dilution. | 3 |
| TNF-α | ELISA | TNF-alpha: 559071 mABG 1:10 mouse antihuman | 24 |
| ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 | |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ELISA | Cytokines were measured in duplicate by ELISA (R&D Minneap- olis, USA). | 5 | |
| PCR | Taqman gene expression assays (Applied Biosystems) on a Roche Light Cycler | 31 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| TNF-β | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| sTNFR1 | ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| sTNFR2 | ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 8 |
| ECL | xMAP technology (Luminex Corporation, Austin, TX, USA) | 25 | |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| hBD1 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| hBD2 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| hBD3 | ELISA | ELISA 1 : 400; rabbit antihuman | 24 |
| ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 | |
| S100A7 | ELISA | Psoriasin HL15-4 mAbG 1:20,000 mouse antihuman | 24 |
| ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 | |
| S100A8 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| ELISA | S100A8 and S100A9 (monospecific affinity-purified rabbit antisera to S100A8 and to S100A9 | 30 | |
| S100A9 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| ELISA | S100A8 and S100A9 (monospecific affinity-purified rabbit antisera to S100A8 and to S100A9 | 30 | |
| LL37 | ELISA | Cathelicidin ab64892 pAbG 1:1000 rabbit antihuman | 24 |
| Lyzozyme | ELISA | Lysozyme A0099 pAbG 1:100 rabbit antihuman | 24 |
| MIF | ELISA | MIF MAB289 mABG 1:100 mouse antihuman | 24 |
| αMSH | ELISA | alpha MSH M09393 mABG 1:500 rabbit antihuman | 24 |
| MHC1 | ELISA | MHC1 W6/32 mABG 1:50 mouse antihuman | 24 |
| RNase7 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| IP10 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| CCL3 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| CCL5 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 | |
| CCL20 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| CCL27 | ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 |
| CRP | ECL | xMAP luminex Luminex Corporation, Austin, TX, USA | 8 |
| ECL | xMAP luminex Luminex Corporation, Austin, TX, USA | 25 | |
| ESR | ECL | xMAP luminex Luminex Corporation, Austin, TX, USA | 8 |
| ECL | xMAP luminex Luminex Corporation, Austin, TX, USA | 25 | |
| IFNg | PCR | (Hs00174143_m1), ABI-Prism 7300 Sequence Detector System (Applied Biosystems) | 7 |
| ELISA | ELISA kits from Sanquin (Amsterdam, The Nether- lands) | 9 | |
| ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 | |
| ELISA/
ECL |
ELISA (Quantikine; R&D Systems) or Luminex assay (Millipore, Billerica, MA). | 4 | |
| GMCSF | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| ELISA | Quantibody Human Inflammation array 3 (RayBiotech Inc., Norcross, GA, U.S.A.). | 26 | |
| VEGF | ECL | Meso Scale Discovery electrochemiluminescent assay (MSD, Meso Scale Diagnostics, Rockville, MD MSD V-Plex cytokine panel 1 and the V-plex
proinflammatory panel 1 |
6 |
| sVEGFR1 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| Caspase 1 | ELISA | Kelly et al. Caspase-1 fluorochrome inhibitor of caspases (FLICA) (ImmunoChemistry Technologies, Bloomington, MN, U.S.A. | 30 |
| NLRP3 | PCR | Kelly IL10, IL17A, IL1
Β, IL18 and NLRP3 was performed with predesigned Taqman gene expression assays (Applied Biosystems) on a Roche Light Cycler
(Pleasanton, CA, U.S.A.) |
30 |
| CAMP | PCR | (Hs00189038_m1) ABI-Prism 7300 Sequence Detector System (Applied Biosystems) | 7 |
| Uteroglob | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| Cystatin C | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| LCN2 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| BD2 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| MMP2 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| BLC | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| ICAM-1 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| Eotaxin | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| Eotaxin2 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| CXCL6 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| CXCL9 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| CXCL11 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| CX3CL1 | ELISA | Quantikine enzyme-linked immunosorbent assay (ELISA) systems from Bio-Techne | 27 |
| I-309 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| MCP1 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| M-CSF | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| MIP1b | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| MIP1d | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| PDGF-BB | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| TIMP1 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
| TIMP2 | ECL | (CBA Human Inflammation kit and CBA Human TH1⁄TH2 Cytokine kit; BD Biosciences, Franklin Lakes, NJ, U.S.A.) Analyssi: FACSCalibur (BD Biosciences) | 26 |
Table 4: Antibodies Used for Identification of Cytokines in Studies Included in this Systematic Review. ECL: Electrochemicoluminescence
Assessment of bias
Assessment of bias is presented in Table 6. Two of the 14 questions regarding participation rate and loss to follow up were considered not applicable. All included studies identified clear objectives and a clearly defined study population. No clear inclusion or exclusion criteria were specified for 17 of the 19 studies. Power estimation was made for one study 33, and recording of all exposures (disease activity, comorbidities etc) were made prior to assessment of the outcomes (cytokine levels). The timeframe of analysis was sufficient to identify an association, but only 10 of the 19 studies (52.6%) documented different levels of exposures (disease severity, metabolic comorbidities, family history etc). There were no serial measures of cytokine levels in the majority of studies. Only three studies 5, 25, 33, examining cytokine levels after monoclonal antibody administration has measurements at two distinct time points. Outcomes of interest (cytokine levels) were measured consistently within studies, however there was great variance in the methods of measurement and analysis between studies ( Table 5). No studies took into account known confounding variables into analysis of their results by stratification or regression analyses.
Table 6. Risk of bias across studies included in this review.
| Study
Reference |
1. Was
the research question or objective in this paper clearly stated? |
2. Was
the study population clearly specified and defined? |
3. Was the
participation rate of eligible persons at least 50%? |
. Were all
the subjects selected or recruited from the same or similar populations (including the same time period)? Were inclusion and exclusion criteria for being in the study prespecified and applied uniformly to all participants? |
5. Was a
sample size justification, power description, or variance and effect estimates provided? |
6. For the
analyses in this paper, were the exposure(s) of interest measured prior to the outcome(s) being measured? |
7. Was the
timeframe sufficient so that one could reasonably expect to see an association between exposure and outcome if it existed? |
8. For
exposures that can vary in amount or level, did the study examine different levels of the exposure as related to the outcome (e.g., categories of exposure, or exposure measured as continuous variable)? |
9. Were the
exposure measures (independent variables) clearly defined, valid, reliable, and implemented consistently across all study participants? |
10. Was the
exposure(s) assessed more than once over time? |
11. Were the
outcome measures (dependent variables) clearly defined, valid, reliable, and implemented consistently across all study participants? |
12 Were the
outcome assessors blinded to the exposure status of participants? |
13. Was
loss to follow- up after baseline 20% or less? |
14. Were key
potential confounding variables measured and adjusted statistically for their impact on the relationship between exposure(s) and outcome(s)? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Moran
et al. 2 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Emelianov
et al. 24 |
Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Hessam
et al. 3 |
Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Hotz et al. 4 | Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Thomi
et al. 7 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Jimenez-
Gallo et al. 8 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Banerjee
et al. 6 |
Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Jimenez-
Gallo et al. 25 |
Y | Y | N/A | Y | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Kanni et al. 5 | Y | Y | N/A | N | N | Y | Y | Y | Y | Y | Y | NR | N/A | N |
| Kelly et al. 31 | Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Lima et al. 30 | Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Ten Oever
et al. 9 |
Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Schlapbach
et al. 32 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Thomi
et al. 29 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Thomi
et al. 28 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Wolk et al. 27 | Y | Y | N/A | N | N | Y | Y | N | Y | N | Y | NR | N/A | N |
| Van der Zee
et al. 26 |
Y | Y | N/A | N | N | Y | Y | Y | Y | N | Y | NR | N/A | N |
| Kanni
et al. 33 |
Y | Y | N/A | Y | Y | Y | Y | Y | Y | N | Y | NR | N/A | N |
Key: Y = Yes; N= No, NR= Not Reported N/A = Not Applicable
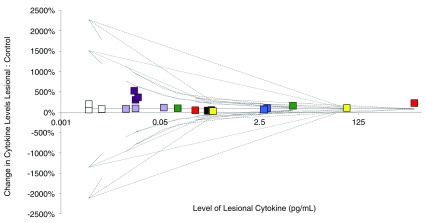
Assessment of heterogeneity
36 of the 81 identified cytokines or inflammatory proteins were assessed by more than 1 study. 23 of those cytokines had raw data available. No studies had sufficient measures of spread in order to calculate I 2measure of heterogeneity and so chi-squared statistic was used as an alternate marker of heterogeneity ( Table 7) along with a funnel plot ( Figure 3). In total, 18 individual cytokines (78.2%) were found to demonstrate heterogeneity. Only eight cytokines (Serum IL-10, Lesional IL-1α, IL-12p70, hBD1, hBD2, hBD3, S100A9 and GMCSF) illustrated homogeneity. Due to this high level of heterogeneity and concerns regarding the methodological quality of included studies, meta-analysis was not deemed appropriate to perform.
Table 7. Table of heterogeneity of cytokine studies by chi-squared tests for homogeneity.
| Cytokine | Chi Squared | P |
|---|---|---|
| IL1a Lesional | 0.3525 | p=0.552705 |
| IL1b Lesional | 153.5947 | p<0.00001 |
| IL4 Lesional | 4.3992 | P=0.035955 |
| IL5 Lesional | 15.1692 | P=0.000098 |
| IL6 Lesional | 461.9724 | P<0.00001 |
| IL8 Lesion | 846.6251 | P<0.0001 |
| IL8 Serum | 94.4212 | P<0.0001 |
| IL10 Lesion | 90.3211 | P<0.0001 |
| IL10 Serum | 0.1595 | P=0.689624 |
| IL12p40 Lesional | 4.9618 | P=0.025913 |
| IL12p70 Lesional | 2.2116 | P=0.136973 |
| IL13 Lesional | 5.4163 | P=0.019949 |
| IL15 Lesional | 39.2837 | P<0.00001 |
| IL16 Lesional | 126.1959 | P<0.00001 |
| IL17A Lesional | 22.6668 | P<0.00001 |
| IL17A Serum | 19.1621 | P=0.000012 |
| TNFa Lesional | 6.9761 | P=0.030561 |
| TNFb Lesional | 7.4004 | P=0.006521 |
| hBD1 Lesional | 2.3317 | P=0.311656 |
| hBD2 Lesional | 0.6488 | P=0.722954 |
| hBD3 Lesional | 1.0314 | P=0.597084 |
| S100A7 Lesional | 621.2537 | P<0.00001 |
| S100A8 Lesional | 19.6371 | P=0.000054 |
| S100A9 Lesional | 1.27 | P=0.529927 |
| RNAse 7 | 6.7263 | P=0.034626 |
| GMCSF Lesional | 1.9405 | P=0.163611 |
Discussion
The overall quality of reporting in the identified studies was low with little consistency between methodologies and cytokines examined. There was also great variability in the ages, genders, comorbidities, associated conditions and treatments of the patients included in these studies. This was again reflected in the high number of cytokines with statistical heterogeneity ( Table 7). The studies presenting conflicting data are often those studies with lower numbers of patients as well as lack of matched controls and/or lack of stratification by treatment. Meta-analysis using individual patient data would be required in order to account for these factors and re-assess the relationship between lesional and control cytokine levels.
In assessing the relationship between lesional and peri-lesional tissue, it has been demonstrated by many authors that different cytokines are present in peri-lesional tissue as opposed to lesional tissue. The definition of peri-lesional tissue is fairly consistent in the studies examined being 2cm from an active HS nodule on unaffected skin. However, no studies reported ultrasound examination of the peri-lesional skin to ensure that subclinical extension of the adjacent nodule (either in the dermis or the subcutaneous tissue) was being inadvertently sampled. This is an important differentiation to make in terms of identifying the subclinical pathogenic processes that precipitate this disease.
The raw data collated illustrates a number of paradoxically elevated levels of control cytokines (IL-15, IL-16) ( Table 4). Many of these control readings lie near the lower detection limit of specific assays in individual papers, and thus the possibility of erroneously elevated control readings cannot be excluded. The wide interquartile ranges of studies which did report individual patient data 7, suggest that analyzing aggregate data is not optimal and is prone to misrepresentation of the relationship between clinical disease, comorbidities and cytokine levels. Furthermore, high levels of heterogeneity within the measurements of individual cytokines suggest that examination of and correction for other variables or confounders is required.
Methodological quality
Regarding methods of cytokine analysis, a number of authors have identified variability in cytokine levels measured with different forms of multiplex assays as well as traditional ELISA methods 35– 39. Different methods of cytokine analysis are known to be prone to variability, with some cytokines more sensitive than others. For example, IFN- γ and IL-1β were overestimated compared with ELISA methods 37, whilst IL-6 levels were underestimated 37. IL-6 levels when compared across four different multiplex assays showed significant variation in detectable range, accuracy and responsiveness 36. The correlation of TNF-α between ELISA and Multiplex assays was also poor (r=0.31) 36. Issues also exist with minimum detectable levels of cytokines with specific bead-based arrays 36 As an example, minimal detectable dose readings reported for IL-12p70 using some multiplex arrays 39 are higher than the levels reported in lesional HS samples 6. Therefore, whilst the general trends in the level of consistently elevated or suppressed cytokines in HS are reliable, the quantification of individual cytokines as well as the relationship between comorbidities and cytokine levels requires further research with consistent, reliable and accurate methodologies in order to further dissect the inflammatory cascade in this disease.
Keratinocyte mediated inflammatory pathways
The majority of elevated cytokines and inflammatory proteins identified in lesional skin of HS (TNF-α, IL-1β, IL-6, IL-8, IL-11, IL-23, IL-17A, IL-33, IL-36, LL-37, S100A7, S100A8, S100A9, GM-CSF, TGF-β, hBD2, hBD3, CCL3, CXCL9, CXCL11, PDGF, CCL5, CCL-20, MIF, GM-CSF and LCN2) are those known to be produced by keratinocytes, as well as perpetuating a self-amplification pathway 34 ( Figure 2). Additionally T-cells produce IL-17A, IL-17F, IL-26, IL-29, and IFN-γ; dendritic cells produce IL-12, IL-23 and possibly IL-39; neutrophils produce S100A8 and S100A9 (calgranulin); and innate lymphoid cells also contribute IFN-γ, IL-17A and IL-17F. This inflammatory model has been well documented and explored in both psoriasis and atopic dermatitis 34, 40. The psoriasiform epidermal hyperplasia seen in HS (mediated by IL-17 and maintained by IL-23-mediated T h17 stimulation) 34 reflects this common inflammatory pathway.
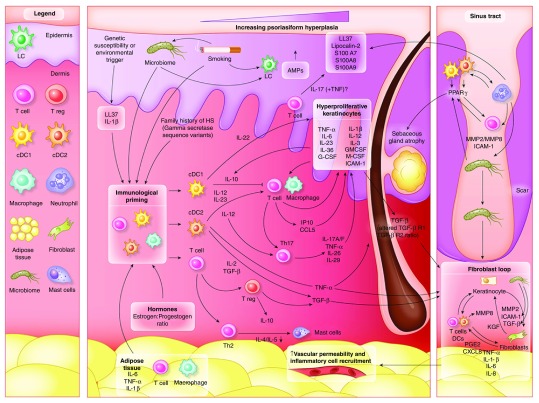
Figure 2. Inflammatory pathways in hidradenitis suppurativa, a schematic representation of the results identified in this systematic review.
Immunological ‘priming’ occurs due to the contribution of adipose tissue, genetic susceptibility, smoking-related inflammatory mediators and obesity related pro-inflammatory signals and the composition of the microbiome. Increased activity of cDC1, cDC2 and T cells lead to both keratinocyte hyperplasia via the actions of IL-12 and IL-23, as well as a Th17 predominant immune response. Alterations of antimicrobial peptides (AMP’s) also occur throughout the epidermis. The dermal inflammation interacting with the hyperplastic epidermis result leads to a self-perpetuating inflammatory feed forward mechanism mediated by IL-36, Il-1B and TNF-a. The development of scarring and sinus tracts is associated with MMP2, ICAM-1 and TGF-Beta, with possible augmentation of ICAM-1 and TGF-B signaling via specific components of the microbiome. TNF-a, PGE2 and CXCL2 then lead to additional feed forward mechanisms perpetuating the inflammatory cycle.
Figure 3. Funnel plot of selected cytokine in lesional and control samples of hidradenitis suppurativa.
IL-1a = Red, IL-10 = Blue, IL-12p70 = Green, hBD1 = Purple, hBD2 = light purple, hBD3 = Black, S100A9 = White, GMCSF = Yellow.
The other elevated non-keratinocyte produced cytokines in HS (IL-4, IL-5, IL-10, IL-16, IL-17A, IL-22, IL-32, IL-36, hBD1), are produced by a combination of dendritic cells, monocytes, neutrophils and CD4+ T cells. IL-4 and IL-5 as key cytokines in the T h2 axis are consistent with the findings of Mast cells in HS 41, as well as the pruritus, which is frequently reported by patients. IL-10 in HS is produced by Treg cells 2 (although dendritic cells may also be a source), and whilst quantitatively the IL-10 signal appears paradoxically elevated, it can be explained by the up-regulation of T cells including Treg cells, which although significantly elevated from baseline, are not elevated enough in comparison to T H17/IL-17/IL-22 signal to counteract this strong pro-inflammatory cascade 2. Further exploration of these cytokines may reveal the initial trigger(s) of the inflammatory cascade in HS, or correlations with known pro-inflammatory comorbidities.
Insights into pathogenesis of HS
In light of investigations in psoriasis and atopic dermatitis, the role of dendritic cells in HS needs to be clarified, as dendritic cell influx has been reported in histological studies 41, 42, and they may contribute to the high IL-10 and IL-15 levels reported. IL-32 is a second cytokine produced by dendritic cells, but has only been reported in one study 29. Further research into the functional role of IL-32 in the activity of dendritic cells in HS would be of value. The role of IL-20, IL-22, IL-24 and IL-26 needs further clarification. IL-19, TSLP and CCL17 (TARC) have not yet been examined in HS and this is required in order to further explore the role of dendritic cell, monocyte and T cell activation and migration in this disease.
It is well established that smoking, obesity and diabetes are strongly associated with HS 13– 19, 42, 43. The immunological effects of smoking include increase in number and responsiveness of dendritic cells, altered function of Treg cells and activation of Th17 pathways 44, whilst obesity and diabetes can result in production of IL-1β, IL-6 and TNF-α through activated macrophages in adipose tissue 45, 46. These potential mechanistic pathways (which may prime or contribute towards inflammation in HS) require validation in functional studies. However, if they are a significant contributor to inflammation, the presence or absence of these comorbidities need to be considered in future cytokine studies as confounding variables in order to identify significant biochemical markers independent of these other pro-inflammatory states that reflect the pathogenesis of HS.
The role of the microbiome 42, 43 in stimulating chronic inflammation has parallels in diabetes 47 and colonic inflammation 48 and the presence of Porphyromonas and Peptoniphilus species has been associated with a subpopulation of patients with HS 42. Porphyromonas has been associated with systemic inflammation and atherosclerosis through aberrant toll-like-receptor 4 signalling 48 and is not part of the natural cutaneous flora 43. Altered cutaneous and gastrointestinal microbiome can also act via microbiome metabolites (including lipopolysaccharides, short chain fatty acids and bile salts) 49 through stimulation of myeloid dendritic cells via G Protein Coupled Receptors (including GPR41, GPR43 and GPR109A) 49, 50. The microbiome may be implicated as a trigger factor for the initial inflammatory cascade in HS in a proportion of patients. Similarly, the presence of genetic polymorphisms as reported in HS 51 have the potential to up-regulate inflammatory activity through shedding of IL-6R, IL-15R, TNF-α 52 as well as up-regulating the response of dendritic cells to LPS stimulation via ADAM17 (which has been demonstrated to be elevated in a published gene expression study of HS) 53. These pathways may be involved prior to the activation of keratinocyte-mediated inflammation, and hence, may reveal novel targets for new interventions to control the disease prior to the onset of destructive inflammation.
Limitations, interpretation and generalisability
The limitations to this study include the high degree of methodological variability ( Table 5) and high impact of bias ( Table 6) within the included studies. The lack of individual patient data has also prevented any further analysis into the contribution of comorbidities such as smoking and obesity to variable levels of cytokines in lesional tissue and/or serum. This, along with the high level of heterogeneity in many cytokines ( Table 7), has resulted in analyses of the collated data being limited to descriptive analyses only and limited the generalisability of results.
Conclusions
Through this review we have catalogued the various cytokines that have been reported as elevated in lesional, peri-lesional tissue, serum or exudate of HS patients. We have also identified those cytokines with inconsistent results and identified methodological factors that may explain variability in findings. We have identified a number of missing links in disease pathogenesis with respect to cytokine actions and pathways that must be addressed in future work. Areas for further investigation include the role of dendritic cells in HS, the contribution of obesity, smoking, diabetes and the microbiome to cytokine profiles in HS, and examining the natural history of the disease through longitudinal measurements of cytokines over time.
Data availability
All data underlying the results are available as part of the article and no additional source data are required.
Extended data
OSF: Extend data. Data Collection Sheet Cytokine. Review HS. https://doi.org/10.17605/OSF.IO/N2E7A 22
License: CC0 1.0 Universal
Reporting guidelines
OSF: PRISMA checklist for ‘A systematic review and critical evaluation of inflammatory cytokine associations in hidradenitis suppurativa’. https://doi.org/10.17605/OSF.IO/N2E7A 22
License: CC0 1.0 Universal
Funding Statement
Supported in part by a grant from the National Center for Advancing Translational Sciences (NCATS) [UL1 TR001866], National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved
References
- 1. Hoffman LK, Ghias MH, Lowes MA: Pathophysiology of hidradenitis suppurativa. Semin Cutan Med Surg. 2017;36(2):47–54. 10.12788/j.sder.2017.017 [DOI] [PubMed] [Google Scholar]
- 2. Moran B, Sweeney CM, Hughes R, et al. : Hidradenitis Suppurativa is characterized by Dysregulation of the Th17:Treg Cell Axis, which is Corrected by Anti-TNF Therapy. J Invest Dermatol. 2017;137(11):2389–2395. 10.1016/j.jid.2017.05.033 [DOI] [PubMed] [Google Scholar]
- 3. Hessam S, Sand M, Gambichler T, et al. : Interleukin-36 in hidradenitis suppurativa: evidence for a distinctive proinflammatory role and a key factor in the development of an inflammatory loop. Br J Dermatol. 2018;178(3):761–767. 10.1111/bjd.16019 [DOI] [PubMed] [Google Scholar]
- 4. Hotz C, Boniotto M, Guguin A, et al. : Intrinsic Defect in Keratinocyte Function Leads to Inflammation in Hidradenitis Suppurativa. J Invest Dermatol. 2016;136(9):1768–1780. 10.1016/j.jid.2016.04.036 [DOI] [PubMed] [Google Scholar]
- 5. Kanni T, Tzanetakou V, Savva A, et al. : Compartmentalized Cytokine Responses in Hidradenitis Suppurativa. PLoS One. 2015;10(6):e0130522. 10.1371/journal.pone.0130522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banerjee A, McNish S, Shanmugam VK: Interferon-gamma (IFN-γ) is Elevated in Wound Exudate from Hidradenitis Suppurativa. Immunol Invest. 2017;46(2):149–158. 10.1080/08820139.2016.1230867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thomi R, Schlapbach C, Yawalkar N, et al. : Elevated levels of the antimicrobial peptide LL-37 in hidradenitis suppurativa are associated with a Th1/Th17 immune response. Exp Dermatol. 2018;27(2):172–177. 10.1111/exd.13482 [DOI] [PubMed] [Google Scholar]
- 8. Jiménez-Gallo D, de la Varga-Martínez R, Ossorio-García L, et al. : The Clinical Significance of Increased Serum Proinflammatory Cytokines, C-Reactive Protein, and Erythrocyte Sedimentation Rate in Patients with Hidradenitis Suppurativa. Mediators Inflamm. 2017;2017:2450401. 10.1155/2017/2450401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ten Oever J, van de Veerdonk FL, Joosten LA, et al. : Cytokine Production Assays Reveal Discriminatory Immune Defects in Adults with Recurrent Infections and Noninfectious Inflammation. Clin Vacc Immunol. 2014:21(8):1061–1069. 10.1128/CVI.00152-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canoui-Poitrine F, Le Thuaut A, Revuz JE, et al. : Identification of three hidradenitis suppurativa phenotypes: latent class analysis of a cross-sectional study. J Invest Dermatol. 2013;133(6):1506–11. 10.1038/jid.2012.472 [DOI] [PubMed] [Google Scholar]
- 11. van der Zee HH, Jemec GB: New Insights into the diagnosis of hidradenitis suppurativa: Clinical presentations and phenotypes. J Am Acad Dermatol. 2015;73(5 Suppl 1):S23–26. 10.1016/j.jaad.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 12. Zhou X, Fragala MS, McElhaney JE, et al. : Conceptual and Methodological Issues relevant to Cytokine and inflammatory marker measurements in clinical research. Curr Opin Clin Nutr Metab Care. 2010;13(5) :541–547. 10.1097/MCO.0b013e32833cf3bc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ergun T: Hidradenitis Suppurativa and the Metabolic Syndrome. Clin Dermatol. 2018;36(1):41–47. 10.1016/j.clindermatol.2017.09.007 [DOI] [PubMed] [Google Scholar]
- 14. Garg A, Birabaharan M, Strunk A: Prevalence of type 2 diabetes mellitus among patients with hidradenitis suppurativa in the United States. J Am Acad Dermatol. 2018;79(1):71–76. 10.1016/j.jaad.2018.01.014 [DOI] [PubMed] [Google Scholar]
- 15. Lukach AJ, Saul MI, Ferris LK, et al. : Risk Factors for Hidradenitis Suppurativa in Patients with Inflammatory Bowel Disease. Dig Dis Sci. 2018;63(3):755–760. 10.1007/s10620-018-4919-5 [DOI] [PubMed] [Google Scholar]
- 16. Garg A, Papagermanos V, MIdura M, et al. : Incidence of Hidradenitis Suppurativa among tobacco smokers: a population-based retrospective analysis in the U.S.A. Br J Dermatol. 2018;178(3):709–714. 10.1111/bjd.15939 [DOI] [PubMed] [Google Scholar]
- 17. Sgambato JA, Jones BA, Caraway JW, et al. : Inflammatory profile analysis reveals differences in cytokine expression between smokers, moist snuff users, and dual users compared to non-tobacco consumers. Cytoine. 2018;107:43–51. 10.1016/j.cyto.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 18. Spoto B, Di Betta E, Mattace-Raso F, et al. : Pro- and anti-inflammatory cytokine gene expression in subcutaneous and visceral fat in severe obesity. Nutr Metab Cardiovasc Dis. 2014;24(10):1137–43. 10.1016/j.numecd.2014.04.017 [DOI] [PubMed] [Google Scholar]
- 19. Rehman K Akash MS: Mechanisms of inflammatory responses and development of insulin resistance: how are they interlinked? J Biomed Sci. 2016;23(1):87. 10.1186/s12929-016-0303-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frew J, Hawkes J Krueger J: Cytokine Studies in Hidradenitis Suppurativa: A Systematic Review of Observational Studies.PROSPERO 2018:CRD42018104664, Accessed 30 July 2018. Reference Source [Google Scholar]
- 21. Moher D, Liberati A, Tetzlaff J, et al. : Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frew J: A Systematic Review and Critical Evaluation of Inflammatory Cytokine Associations in Hidradenitis Suppurativa.2018. 10.17605/OSF.IO/N2E7A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. NIH NIoH: Quality Assessment Tool for Observational Cohort and Cross Sectional Studies.2018; Accessed 30 July 2018. Reference Source [Google Scholar]
- 24. Emelianov VU, Bechara FG, Gläser R, et al. : Immunohistological pointers to a possible role for excessive cathelicidin (LL-37) expression by apocrine sweat glands in the pathogenesis of hidradenitis suppurativa/acne inversa. Br J Dermatol. 2012;166(5):1023–1034. 10.1111/j.1365-2133.2011.10765.x [DOI] [PubMed] [Google Scholar]
- 25. Jiménez-Gallo D, de la Varga-Martínez R, Ossorio-García L, et al. : Effects of adalimumab on T-helper-17 lymphocyte- and neutrophil-related inflammatory serum markers in patients with moderate-to-severe hidradenitis suppurativa. Cytokine. 2018;103:20–24. 10.1016/j.cyto.2017.12.020 [DOI] [PubMed] [Google Scholar]
- 26. van der Zee HH, Laman JD, de Ruiter L, et al. : Adalimumab (antitumour necrosis factor-α) treatment of hidradenitis suppurativa ameliorates skin inflammation: an in situ and ex vivo study. Br J Dermatol. 2012;166(2):298–305. 10.1111/j.1365-2133.2011.10698.x [DOI] [PubMed] [Google Scholar]
- 27. Wolk K, Wenzel J, Tsaousi A, et al. : Lipocalin-2 is expressed by activated granulocytes and keratinocytes in affected skin and reflects disease activity in acne inversa/hidradenitis suppurativa. Br J Dermatol. 2017;177(5):1385–1393. 10.1111/bjd.15424 [DOI] [PubMed] [Google Scholar]
- 28. Thomi R, Kakeda M, Yawalkar N, et al. : Increased expression of the interleukin-36 cytokines in lesions of hidradenitis suppurativa. J Eur Acad Dermatol Venereol. 2017;31(12):2091–2096. 10.1111/jdv.14389 [DOI] [PubMed] [Google Scholar]
- 29. Thomi R, Yerly D, Yawalkar N, et al. : Interleukin-32 is highly expressed in lesions of hidradenitis suppurativa. Br J Dermatol. 2017;177(5):1358–1366. 10.1111/bjd.15458 [DOI] [PubMed] [Google Scholar]
- 30. Lima AL, Karl I, Giner T, et al. : Keratinocytes and neutrophils are important sources of proinflammatory molecules in hidradenitis suppurativa. Br J Dermatol. 2016;174(3):514–521. 10.1111/bjd.14214 [DOI] [PubMed] [Google Scholar]
- 31. Kelly G, Hughes R, McGarry T, et al. : Dysregulated cytokine expression in lesional and nonlesional skin in hidradenitis suppurativa. Br J Dermatol. 2015;173(6):1431–1439. 10.1111/bjd.14075 [DOI] [PubMed] [Google Scholar]
- 32. Schlapbach C, Hänn T, Yawalkar N, et al. : Expression of the IL-23/Th17 pathway in lesions of hidradenitis suppurativa. J Am Acad Dermatol. 2011;65(4):790–8. 10.1016/j.jaad.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 33. Kanni T, Argyropoulou M, Spyridopoulos T, et al. : MABp1 Targeting IL-1α for Moderate to Severe Hidradenitis Suppurativa Not Eligible for Adalimumab: A Randomized Study. J Invest Dermatol. 2018;138(4):795–801. 10.1016/j.jid.2017.10.030 [DOI] [PubMed] [Google Scholar]
- 34. Hawkes JE, Chan TC, Krueger JG: Psoriasis pathogenesis and the development of novel targeted immune therapies. J Allergy Clin Immunol. 2017;140(3):645–653. 10.1016/j.jaci.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bastarache JA, Koyama T, Wickersham NE, et al. : Validation of a multiplex electrochemiluminescent immunoassay platform in human and mouse samples. J Immunol Methods. 2014;408:13–23. 10.1016/j.jim.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson DK, Huffman KM, Kraus WE, et al. : Critical appraisal of four IL-6 immunoassays. PLoS One. 2012;7(2):e30659. 10.1371/journal.pone.0030659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Koning L, Liptak C, Shkreta A, et al. : A multiplex immunoassay gives different results than singleplex immunoassays which may bias epidemiologic associations. Clin Biochem. 2012;45(10–11):848–851. 10.1016/j.clinbiochem.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 38. Tighe PJ, Ryder RR, Todd I, et al. : ELISA in the multiplex Era: Potentials and Pitfalls. Proteomics Clin Appl. 2015;9(3–4):406–22. 10.1002/prca.201400130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. dupont NC, Wang K, Wadhwa PD, et al. : Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66(2):175–191. 10.1016/j.jri.2005.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brunner PM, Guttman-Yassky E, Leung DY: The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J Allergy Clin Immunol. 2017;139(4S):S65–S76. 10.1016/j.jaci.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Zee HH, de Ruiter L, Boer J, et al. : Alterations in leucocyte subsets and histomorphology in normal-appearing perilesional skin and early and chronic hidradenitis suppurativa lesions. Br J Dermatol. 2012;166(1):98–106. 10.1111/j.1365-2133.2011.10643.x [DOI] [PubMed] [Google Scholar]
- 42. Ring HC, Thorsen J, Saunte DM, et al. : The Follicular Skin Microbiome in Patients With Hidradenitis Suppurativa and Healthy Controls. JAMA Dermatol. 2017;153(9):897–905. 10.1001/jamadermatol.2017.0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guen-Revillet H, Jais JP, Ungeheuer MN, et al. : The Microbiological Landscape of Anaerobic Infections in Hidradenitis Suppurativa: A Prospective Metagenomic Study. Clin Infec Dis. 2017;65(2):282–291. 10.1093/cid/cix285 [DOI] [PubMed] [Google Scholar]
- 44. Qiu F, Liang CL, Liu H, et al. : Impacts of cigarette smoking on immune responsiveness: Up and down or upside down? Oncotarget. 2017;8(1):268–284. 10.18632/oncotarget.13613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kothari V, Galdo JA, Mathews ST: Hypoglycemic agents and potential anti-inflammatory activity. J Inflamm Res. 2016;9:27–38. 10.2147/JIR.S86917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Richards JL, Yap YA, McLeod KH, et al. : Dietary metabolites and the gut microbiota: an alternative approach to control inflammatory and autoimmune diseases. Clin Transl Immunology. 2016;5(5):e82. 10.1038/cti.2016.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Basson A, Trotter A, Rodruiguez-Palacios A, et al. : Mucosal Interactions between Genetics, Diet, and Microbiome in Inflammatory Bowel Disease. Front Immunol. 2016;7:290. 10.3389/fimmu.2016.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kramer CD, Genco CA: Microbiota, Immune Subversion, and Chronic Inflammation. Front Immunol. 2017;8:255. 10.3389/fimmu.2017.00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim CH: Immune regulation by microbiome metabolites. Immunology. 2018;154(2):220–229. 10.1111/imm.12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tan JK, McKenzie C, Marino E, et al. : Metabolite-Sensing G Protein-Coupled Receptors-Facilitators of Diet-Related Immune Regulation. Annu Rev Immunol. 2017;35:371–402. 10.1146/annurev-immunol-051116-052235 [DOI] [PubMed] [Google Scholar]
- 51. Frew JW, Vekic DA, Woods J, et al. : A systematic review and critical evaluation of reported pathogenic sequence variants in hidradenitis suppurativa. Br J Dermatol. 2017;177(4):987–998. 10.1111/bjd.15441 [DOI] [PubMed] [Google Scholar]
- 52. Gooz M: ADAM-17: the enzyme that does it all. Crit Rev Biochem Mol Biol. 2010;45(2):146–9. 10.3109/10409231003628015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blok JL, Li K, Brodmerkel C, et al. : Gene expression profiling of skin and blood in hidradenitis suppurativa. Br J Dermatol. 2016;174(6):1392–4. 10.1111/bjd.14371 [DOI] [PubMed] [Google Scholar]