Abstract
A number of differentially expressed proteins (DEPs) were identified in comparative two-dimensional gel electrophoresis analysis of dry and 24-h water-imbibed seeds of maize F1 hybrid DHM 117 (BML 6 × BML 7) and its parental inbreds. Of the DEPs, 53.4% (86/161) in dry seeds and 58% (127/219) in water-imbibed seeds exhibited a nonadditive pattern in the F1 hybrid as compared to parental inbreds. A total of 30 DEPs were categorized into different biological processes, most of which were related to metabolism and energy (34%), followed by storage proteins (27%), stress response (23%), transcription and translation (7%), cell cycle (3%), and hormone biosynthesis (3%). The transcript accumulation pattern of 8 selected genes corresponding to DEPs was examined using qRTPCR. Interestingly, LEA protein Rab28 showed higher accumulation in dry seeds at both protein and transcript levels, whereas indole3-acetaldehyde oxidase showed lower accumulation in water-imbibed seeds of the F1 hybrid than the female parent at the protein level. Thus, the DEPs particularly involved in metabolic and energy processes, as well as hormone biosynthesis in the F 1 hybrid, might be responsible for heterotic seed germination in the F1 hybrid. The DEPs identified in this study provide a scope for improving the seed germination trait of agricultural crops.
Keywords: Heterosis, two-dimensional gel electrophoresis, differentially expressed proteins, F 1 hybrid, seed germination
1. Introduction
Mendel’s law of inheritance has been extensively exploited by plant breeders for bringing genes controlling agriculturally important traits from two different parents, whose outcome can be seen in the form of hybrid vigor (heterosis). This phenomenon has been exploited for improvement of phenotypic and agronomic traits such as growth vigor, increased plant height, size, biomass, speed of development, fertility, resistance to biotic/abiotic stress, flowering time, and high yield (Paschold et al., 2012; Guo et al., 2013; Fu et al., 2015) . The heterotic trait of F1 heterozygous hybrids can be quantified not only in adult plants; it has also been observed in the early embryo, primary root, plumule, and coleoptile development (Hoecker et al., 2008; Marcon et al., 2010; Jin et al., 2014) . Despite its agronomic importance, the molecular principles underlying heterosis are still poorly understood due to the molecular complexity and environmental inuflence on the expression of genes (Zhang et al., 2012; Fu et al., 2014) .
Several genetic and molecular studies have shed light on the molecular mechanisms underlying the process of heterosis in crops like maize, rice, wheat, Arabidopsis, etc. (Baranwal et al., 2012) . Heterosis is likely caused by genetic interactions between alleles of parental genomes that change the regulatory network of related genes instead of new genes (Wang et al., 2015) . At the gene expression level, the variation of transcripts and protein abundance in the F1 hybrid is regulated by interactions between parental genomes (Xing et al., 2016) . There are reports showing that changes at transcript levels do not necessarily correlate with protein levels as many posttranslational modifications are crucial for the regulation of protein function, and these might be important molecular determinants for heterosis (Song et al., 2007; Dahal et al., 2012) . Hence, there is a need to thoroughly investigate genome-wide protein changes between hybrids and their parents to determine their functional relations to heterosis (Guo et al., 2014) .
Seed germination involves a relay of events, which commences with water inhibition by quiescent dry seeds followed by the protuberance of the radicle towards the end. This phenomenon is regulated by a balance of two antagonistic phytohormones, gibberellic acid (GA) and abscisic acid (ABA), as positive and negative regulators (Shuai et al., 2017) , respectively. Water inhibition by the dry seeds induces gene expression in response to GA and ABA hormones to overcome the effect of dormancy, hence assisting seed germination (Schoonheim et al., 2007; Nonogaki, 2014) . The plant seed is the reservoir of carbohydrates, oils, and proteins, which maintains its viability during dormancy and provides primary substances for the growth of the embryo during germination (Han et al., 2013) . Moreover, higher oxygen uptake during inhibition can lead to oxidative stress generating reactive oxygen species (ROS) in rehydrated seeds; therefore, ROS scavenging enzymes are needed to reduce the cell damage and promote seed germination (Schopfer et al., 2001; Gomes and Garcia, 2013) . Seed germination is a tightly regulated process and an important step toward seedling establishment where we need to find the proteins responsible for initiating seed germination by overcoming dormancy, thereby helping the embryo to resume its growth while keeping ROS levels under check.
Maize is one of the most important cereal crops cultivated globally for food, feed, energy, and forage throughout the world (Klopfenstein et al., 2013) . It contributes 9% of the total food grains production in India. Maize serves as a superior model system for exploring the molecular mechanism of heterosis because it exhibits high levels of phenotypic, genomic, transcriptional, and translational variation (Fu et al., 2011) . In maize, F1 hybrid seeds exhibit strong heterosis in terms of faster seed germination trait and vigorous seedling growth as compared to their parental inbred lines. The rapid seedling establishment effects the crop yield and seed quality, which is governed by the speed of seed germination and root length during seedling growth (Jin et al., 2014) . Hence, faster and synchronous germination followed by seedling emergence should be the prime consideration to improve the crop production worldwide.
In the recent past, several proteomic approaches have been successfully used to identify the heterosis-related differentially expressed proteins (DEPs) in different tissues and/or stages, such as the embryos (Marcon et al., 2010; Guo et al., 2013) , plumules (Jin et al., 2014) , internodes (Chen et al., 2018), roots (Hoecker et al., 2008; Rockenbach et al., 2018) , and leaves (Guo et al., 2014) of maize F1 hybrids and parents. For example, Fu et al. (2011) reported DEPs showing nonadditive expression patterns as a reason for the major manifestation of heterosis during seed germination in five sets of maize hybrids along with their parental lines. Guo et al. (2013) observed significant differences for nonadditively expressed proteins between dry and 24-h water-imbibed embryos, suggesting that the expression pattern of the proteins depends on the developmental stage and plant organ analyzed. In view of the few proteomic reports available about heterotic seed germination, more studies are needed for better understanding of molecular processes of heterosis, particularly during early stages of plant development. Such studies might help in the development of protein markers for early prediction of heterosis.
Several maize hybrids have been developed and cultivated commercially in India; however, no attempts have been made to investigate heterosis at the translational level in them. Thus, this study aimed at identifying differentially expressed proteins using a two-dimensional (2-D) gel electrophoresis-based approach during heterotic seed germination in a high-yielding commercially grown F1 maize hybrid by comparing it with its parental inbreds. We investigated the divergence of differentially expressed proteins, i.e. additive and nonadditive proteins during seed germination heterosis, and compared our results with the previous literature. Furthermore, the transcript levels of genes corresponding to DEPs were analyzed using qRT-PCR. Thus, the study has added more information on DEPs that are associated with rapid seed germination in F1 hybrids and broadened the knowledge of molecular processes of heterotic seed germination.
2. Materials and methods
2.1. Plant material and seed treatment
Seeds of a commercially grown maize F1 hybrid (DHM 117) and its female parent (BML 6) and male parent (BML 7) were generated by hand pollination and harvested at physiological maturity between July and September 2015 on the experimental farm of Prof. Jayashankar Telangana State Agricultural University, Rajendranagar, Hyderabad (17°19′N, 78°28′E; altitude 536 m) under a natural photoperiod and temperature regime. The soil of experimental farm was clay loam in texture with pH 7.7 and average temperatures were 28.4 to 34.8 °C (daytime). The purity of the maize F1 hybrid and its parental inbreds was ascertained using four gene-specific simple sequence repeats (SSRs) before using the samples for proteomic studies (Supplementary Table 1). The SSR primers that detected polymorphism in the parental inbreds were used in the study, which confirmed the purity of the F 1 hybrid and its parents (Supplementary Figure 1). The weight of 100 seeds and the germination rates of both parents and the F1 hybrid were determined. The seeds were imbibed in distilled water for 24 h in the dark at 26 ± 2 °C and then were placed with the embryo side down on moist double-layered filter paper in a petri dish with 24 h of dark incubation for germination. Seeds were examined at regular intervals for determining the onset of germination and were considered as germinated based on the emergence of the visible radicle. The germination percentage was recorded at 30, 36, 48, and 60 h after water inhibition. The data on 100-seed weight represent an average of five replicates. The germination experiments were repeated three times with each experiment consisting of five replicates and 50 seeds were used in each replicate. The data on seed weight and germination percentage were analyzed statistically by oneway analysis of variance (ANOVA) followed by Newman– Keuls multiple comparison tests at the 5% probability level using SigmaPlot for Windows (Systat Software, Inc., Bangalore, India). For proteomics analysis, dry and 24-h water-imbibed seeds were used for analyzing the changes in the protein pattern of the F1 hybrid in comparison to the parental inbreds.
2.2. Total soluble protein extraction
Protein extraction from whole dry and 24-h waterimbibed seeds was carried out according to Faurobert et al. (2007) . A sample of 25 seeds (dry and 24-h waterimbibed) was brieyfl ground in a mortar in the presence of liquid nitrogen. The homogenate (300 mg) was suspended in 800 µL of ice-cold extraction buffer [50 mM Tris-HCl (pH 8.5), 5 mM ethylenediaminetetraacetic acid (EDTA), 100 mM potassium chloride (KCl), 1% (w/v) dithiothreitol (DTT), 30% (w/v) sucrose] and vortexed for 20 min. Eight hundred microliters of ice-cold Tris-buffered phenol (pH 8.0) was added and the samples were vortexed for 10 min at room temperature. To separate insoluble material from aqueous and organic phases, the samples were centrifuged (10 min, 5500 × g at 4 °C) and the phenolic phase was collected into a fresh Eppendorf tube. This phenol phase was back-extracted with 800 µL of extraction buffer and vortexed for 3 min followed by centrifugation (10 min, 5500 × g at 4 °C). The resultant phenolic phase containing the soluble proteins was precipitated with five volumes of 100 mM ammonium acetate in methanol at –20 °C overnight. The precipitate was centrifuged (10 min, 5500 × g at 4 °C), and the pellet was washed three times with icecold precipitation solution (100 mM ammonium acetate) followed by washing with ice-cold acetone. Finally, the pellets were air-dried and resuspended in 200 µL of rehydration buffer containing 7 M urea, 2 M thiourea, 4% (w/v) 3-(3-cholamidopropyl) dimethylammonio1-propanesulfonate (CHAPS), 0.8% immobilized pH gradient (IPG) buffer (pH 3-10, GE Healthcare, USA), and 1.0% (w/v) DTT. The completely dissolved protein samples were centrifuged at 5500 × g for 10 min at 4 °C for removing the insoluble fractions. The supernatant was stored at –20 °C for further use. Proteins were quantified by Bradford protein assay (Sigma, USA) using bovine serum albumin (BSA) as a standard.
2.3. Isoelectric focusing and 2-D gel electrophoresis of proteins and staining
For first-dimensional isoelectric focusing (IEF), 800 µg of protein in 320 µL of rehydration buffer was applied on 18cm IPG strips with linear gradients of pH 3–10 (Immobiline Dry Strips, GE Healthcare) in a rehydration tray (GE Healthcare). The strips were passively rehydrated at 20 °C overnight. An Ettan IPG Phor 3 IEF unit (GE Healthcare) at 20 °C was used for IEF under the following conditions: 50 V for 1 h, 250 V for 1 h, 500 V for 3 h, 10,000 V gradient for 2.5 h, 10,000 V for 6 h to reach a total of 60,000 Vh, and 500 V for 20 h. Aeftr IEF, the strips were incubated twice at room temperature in equilibration buffer [6 M urea, 30% (v/v) glycerol, 2% (w/v) sodium dodecyl sulfate (SDS), 50 mM Tris-HCl (pH 8.8)] with 1% (w/v) DTT followed by 2.5% (w/v) 2-iodoacetamide (IAA), respectively, each for 20 min. Equilibrated strips were placed on 12% vertical SDS-polyacrylamide gel electrophoresis (PAGE) for second-dimensional electrophoresis and sealed with 0.5% agarose gel in the running buffer with a trace amount of bromophenol blue. SDS-PAGE was performed in an Ettan DALTsix chamber (GE Healthcare) with running buffer (0.3% Tris, 1.44% glycine, and 0.1% SDS) at 10 mA per gel for 1 h and then at 38 mA per gel for 6–8 h until the bromophenol blue dye reached the bottom. Aeftr electrophoresis, gels were stained with modified 0.1% colloidal Coomassie Brilliant Blue G-250 overnight on an orbital shaker and then destained with distilled water. Gels were scanned with a calibrated densitometric scanner (GE Healthcare).
2.4. Analysis of protein expression pattern
In the scanned gels, spots were detected automatically with ImageMaster 2D Platinum Software Version 7.2 (GE Healthcare) with the parameters smooth, minimum area, and saliency set to 5, 1, and 5, respectively, followed by manual spot editing, such as artificial spot deletion, spot splitting, and merging that was done manually. The experiments were carried out in triplicate. For each genotype, only those protein spots that could be detected consistently in all three replicated gels were considered for analysis. All the gels were matched to the reference gel in an automated mode combined with manual pair correction. DEPs between the hybrid and its parental lines were determined by fold change greater than 1.5 and the Student t-test (P ≤ 0.05) for relative spot volumes. The DEPs that deviated from the midparent value (MPV; the average value of the parental inbred lines) were considered as nonadditively expressed proteins, whereas others were considered as additively expressed proteins.
2.5. In-gel protein digestion and mass spectrometry
Thirty DEP spots (15 from dry and 15 from 24-h waterimbibed seeds) that showed high intensity and clear resolution were excised manually from the 2-D gels for analysis and transferred to 1.5-mL microcentrifuge tubes. Each excised protein spot was destained with 200 µL of 50% acetonitrile (ACN) in 50 mM ammonium bicarbonate (NH4HCO3) buffer (pH 7.8–8.0) with repeated vortexing until completely destained. Thereafter, the opaque white gel slices were reduced with 10 mM DTT in 50 mM ammonium bicarbonate buffer at 56 °C for 60 min and subsequently alkylated with 55 mM IAA in 50 mM ammonium bicarbonate buffer for 45 min in the dark at room temperature (26 ± 2 °C). Then gel slices were washed with 25 mM ammonium bicarbonate and ACN and dried in a vacuum centrifuge at ambient temperature. The gel slices were rehydrated with 15 µL of 25 ng/µL sequencing grade trypsin (Promega, USA) in 25 mM ammonium bicarbonate buffer for 10 min at 4 °C followed by incubation at 37 °C overnight for enzymatic digestion (Wu et al., 2011) . Aeftr incubation, the resulting tryptic fragments were eluted by diffusion into 100% ACN and 0.1% triufloroacetic acid (TFA) (1:1, v/v). One microliter of digested protein sample was mixed with 1 µL of α-cyano-4-hydroxycinnamic acid (CHCA) matrix in 50% ACN and 1% TFA (1:1, v/v) and then 2 µL of sample was spotted onto a matrix-assisted laser desorption/ionization (MALDI) plate and air-dried at room temperature for mass spectrometry (MS). Subsequently, the MALDI plate was placed in a MALDI-tandem time of flight (TOF/ TOF) mass spectrometer (Bruker Autoflex III Smartbeam; Bruker Daltonics, Germany) and MALDI TOF-MS/MS analysis was performed according to Shevchenko et al. (1996) with minor modifications (Kumar et al., 2015) . The peptide mass data were acquired in positive ion reflector mode with accelerating voltage of 20 kV by Flex Control 3.0 (Build 100) software using batched processing and automatic switching between MS and MS/MS modes. All monoisotopic peak masses were collected within the mass range of m/z 800–4000 Da with a signal-to-noise ratio greater than 10 and a local noise window width of m/z 250 excluding contaminants such as keratins and trypsin autolytic products. For MS/MS sequencing analysis, the MS spectra of the eight most abundant precursors were selected with collision-induced dissociation (CID) at 1 kV positive mode and ±4 precursor mass windows.
2.6. Protein identification through peptide mass fingerprinting
For protein identification, peptide mass data were analyzed with the public Mascot search engine (http:// www.matrixscience.com) employing BioTools 3.1 software (Bruker Daltonics) against the National Center for Biotechnology Information nonredundant (NCBInr) database and Swiss-Prot databases. The search was performed on the following parameters: taxonomic, Viridiplantae (Green plants); monoisotopic peptide mass (MH+); one missed cleavage per peptide; enzyme, trypsin; precursor-ion mass tolerance on an average 200 ppm; MS/MS fragment-ion mass tolerance, 2 Da; variable modifications like carbamidomethylation (C) for cysteine and oxidation for methionine (M) were allowed. Mascot search results of protein equal to or higher than the minimum significant score (P ≤ 0.05) were considered as a positive identification. The nomenclature of identified proteins was done according to the corresponding annotations of NCBI and Swiss-Prot.
2.7. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Maize seeds of dry and 24-h water-imbibed samples from parents and the F1 hybrid were ground into fine powder in a mortar and pestle with liquid nitrogen. Total RNA was extracted by a modified sodium dodecyl sulfate/TRIzol method (Wang et al., 2012) . The quality of extracted RNA was checked on 1.2% agarose gel prepared in TBE (Tris-borate-EDTA) buffer and quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, USA). For each sample, 2 µg of RNA was used to synthesize the first-strand cDNA using a Prime-Script 1st Strand cDNA Synthesis Kit (Takara Bio Inc., Japan) according to the manufacturer’s instructions. Quality of cDNA was confirmed for all samples by performing semiquantitative PCR amplification of the internal control, GAPDH (accession no. NM_001111943.1). The expression of GAPDH in semi-qPCR did not vary in the samples analyzed. First-strand cDNA samples were diluted 2.5 times and 1 µL of the diluted reaction mixture was taken as a qRT-PCR template in 10 µL of total reaction volume containing 0.4 µM gene-specific primers and 5 µL of SYBR Premix Ex Taq II (TliRNase H Plus) with ROX (Takara Bio Inc.) and the samples were appraised in three technical replicates. PCR analysis was carried out in a Realplex amplifier (Eppendorf, Germany) with the following cycle parameters: 95 °C for 5 min; 40 cycles of 95 °C for 20 s, 57 °C for 20 s, and 72 °C for 20 s; followed by a melting curve to ensure that each amplicon was a single product. The relative fold change in RNA expression was estimated using the ΔΔCT method (Livak and Schmittgen, 2001) and GAPDH (accession no. NM_001111943.1) with primer sequences 5’-GCCATCACTGCCACACAGAA-3’ and 5’-GGAACACGGAAGGACATACCA- 3’ used as internal controls to normalize the real-time amplification data.
3. Results
3.1. Heterosis during germination
The pattern of heterosis was examined in a maize F 1 hybrid (DHM 117) in comparison to its parental inbreds (BML 6 and BML 7) during early stages of seed germination. Germination was initiated in F1 hybrid seeds after 30 h with 38% germination achieved at 36 h compared to 17.3%–19.3% germination observed in parental inbreds (Figure 1). The rapid germination was associated with vigorous growth in the F1 hybrid throughout the germination period until all the seeds were germinated. Germination percentage of 100% and 96% was achieved in the F1 hybrid and female parent, respectively, at 60 h, which was significantly higher than that of the male parent. Thus, heterosis was manifested during seed germination in the F1 hybrid (DHM 117) as evidenced by early radicle emergence along with the vigorous growth of the seedlings compared to the parental inbreds. In the present study, we have examined the dry weight of all genotypes and found that the dry 100-seed weight of the F1 hybrid (18.5 ± 1.0 g) was significantly lower than that of the male parent (22.0 ± 0.9 g) but did not differ from that of the female parent (18.2 ± 0.9 g). Thus, rapid seed germination and superior seedling growth observed in the F1 hybrid did not correlate with seed weight in this study.
Figure 1.
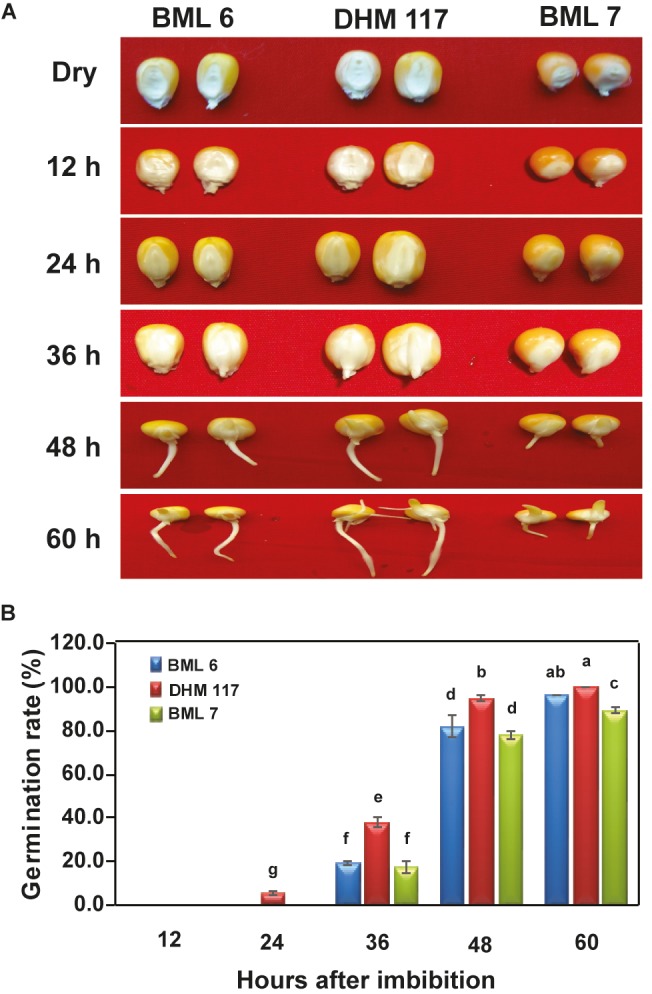
Germination pattern (A) and germination rate (B) of seeds of maize F1 hybrid DHM 117 and its parental inbreds (BML 6 and BML 7) at different time points of culture. The seeds were imbibed in distilled water for 24 h and then placed on moist filter paper at 26 ± 2 °C in the dark. Significance of differences was analyzed by one-way ANOVA and Newman–Keuls multiple comparison tests. Means followed by the same letter are not significantly different (P < 0.05).
3.2. Analysis of DEPs during heterosis
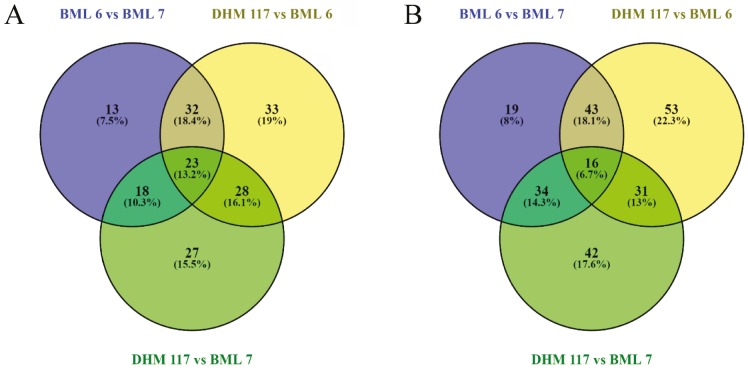
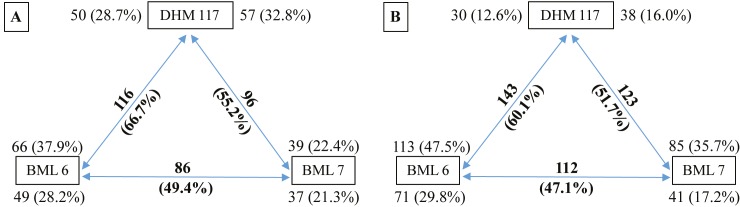
In order to characterize the dynamic proteome changes associated with heterosis in maize during early seed germination, a comparative proteome analysis between an F1 hybrid (DHM 117) and its parental inbreds (BML 6 and BML 7) was performed using 2-D gel electrophoresis with total soluble protein extracts of dry and 24-h waterimbibed seeds. A total of 856 and 1082 protein spots were reproducibly detected in three biological replicates of dry and 24-h water-imbibed seed proteome maps of the three genotypes, respectively (Figure 2). To elucidate the DEPs, pairwise comparisons between the parents or between the parents and F1 hybrid were performed, among which 174 and 238 protein spots were found to be significantly differentially expressed (fold change >1.5, P ≤ 0.05) in dry and 24-h water-imbibed seeds, respectively (Figure 3). In the above analysis, 49.4% and 47.1% of proteins between parental inbreds BML 6 and BML 7 were observed as DEPs, fewer than the parents and F1 hybrid in both dry and water-imbibed seeds. Comparison between DHM 117 and BML 6 revealed 116 (66.7%) and 143 (60.1%) DEPs in dry and 24-h water-imbibed seeds, respectively. In the case of DHM 117 and BML 7, 96 (55.2%) and 123 (51.7%) protein spots were differentially expressed in dry and 24-h water-imbibed seeds, respectively (Figure 4).
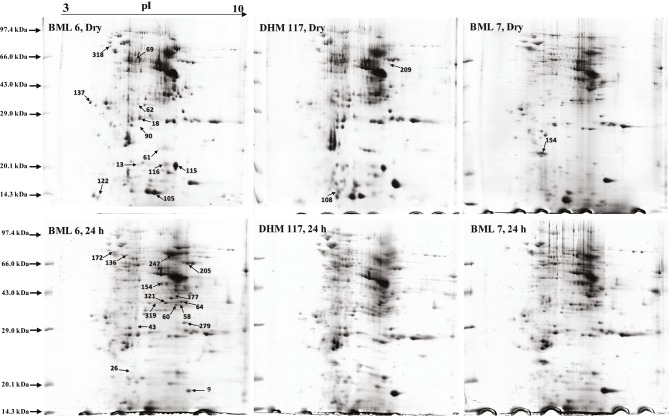
Figure 2.
Two-dimensional gels of total soluble proteins extracted from dry and 24-h water-imbibed seeds of F1 hybrid (DHM 117) and its parents (BML 6 and BML 7). Isoelectric focusing of the protein extracts in the first dimension was performed on linear IPG strips of pH 3–10. In the second dimension, proteins were separated according to their molecular masses on 12% SDS-PAGE gels. Proteins were stained with colloidal Coomassie Brilliant Blue G-250. Proteins spots that accumulated differentially in the hybrid compared to parental inbreds are numbered on the 2-D gels.
Figure 3.
Venn diagram of significantly differentially expressed proteins (FC > 1.5 and P < 0.05) between F1 hybrid DHM 117 and its parental inbreds (BML 6 and BML 7) in dry (A) and 24-h water-imbibed seeds (B).
Figure 4.
Differentially expressed proteins in each comparison between parental inbreds and F1 hybrid of dry (A) and 24-h waterimbibed seeds (B). Numbers on arrows (bold) indicate the total number and contribution of DEPs between pairwise comparison among the total 174 and 238 proteins of dry and 24-h water-imbibed seeds, respectively. For each pairwise comparison, the numbers at the end of the arrow indicate the upregulated proteins in each pair of genotypes.
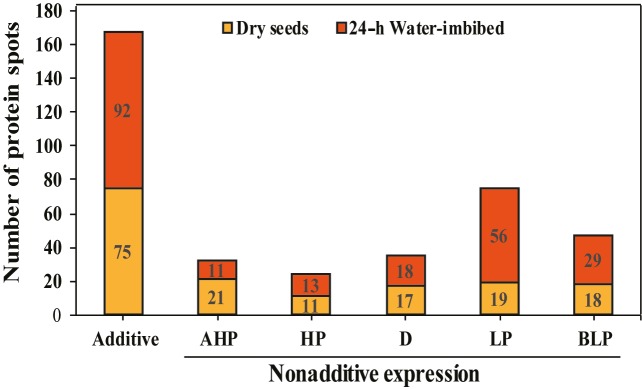
The DEP spots between the F 1 hybrid and its parents were divided into additive and nonadditive categories on the basis of pairwise comparison between the F1 hybrid and the MPV. Of the total, 46.6% (75/161) of proteins in dry seeds and 42% (92/219) in 24-h water-imbibed seeds were expressed additively, while 53.4% (86/161) and 58% (127/219) of DEPs were expressed nonadditively in dry and 24-h water-imbibed seeds, respectively (Figure 5). The nonadditively expressed proteins were categorized into five classes according to the system suggested by Stupar and Springer (2006) . Among the nonadditive protein spots, above high parent (AHP) or high parent (HP) dominance expression was higher in dry seeds compared to 24-h water-imbibed seeds (32/161 and 24/219, respectively). Interestingly, 85 protein spots out of 219 were found to be below or equal to the low parent (LP) dominance in 24-h water-imbibed seeds, which was double that of dry seeds (37/161) (Figure 5).
Figure 5.
Classification of differentially expressed proteins in dry and 24-h waterimbibed seeds of the F1 hybrid and its parental inbreds. AHP, Above high parent expression; HP, high parent expression; D, different from additive, not belonging to any other class; LP, low parent expression; BLP, below low parent expression.
3.3. Functional analysis of DEPs during heterosis
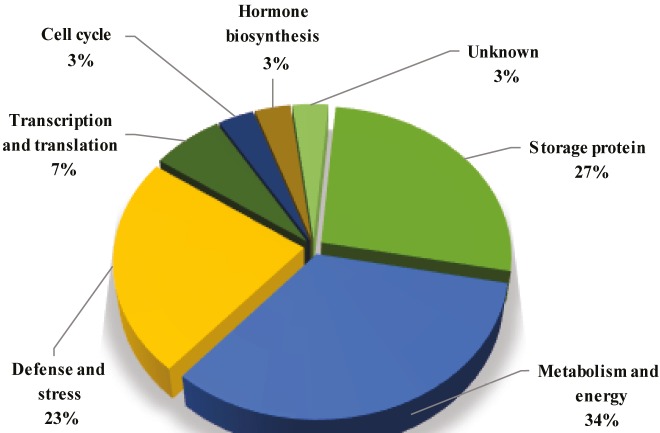
In total, 30 DEP spots (15 from each treatment) that showed clear difference with respect to intensity and high resolution between parental inbreds and F1 hybrid were eluted from the representative 2-D gels and identified with MALDI TOF-MS/MS spectrometer (Tables 1 and 2). Among these DEPs, two spots corresponding to storage proteins were identified twice, including globulin-1 S allele (spot nos. 205 and 247) and globulin-1 S allele-like (spot nos. 90 and 43) that were represented by the same GenBank accession, which might be due to the isoform of spots or different posttranslational modifications of the same gene product. Some proteins such as zein-beta (spot nos. 9 and 115) and oil body-associated protein (spot nos. 62 and 279) were identified twice with different GenBank accessions that might also represent closely related members of gene families. For further analysis, all 30 DEPs were classified into seven classes according to functional annotation including storage protein, metabolism and energy, defense and stress, transcription and translation, cell cycle, hormone biosynthesis, and unknown with the help of UniProtKB (http://www.uniprot.org/uniprot/). As shown in Figure 6, metabolism and energy process (34%) were the largest categories of DEPs, followed by storage protein (27%), defense and stress (23%), transcription and translation (7%), cell cycle (3%), hormone biosynthesis (3%), and unknown (3%). Thus, metabolism and detoxifying proteins might be influencing heterotic seed germination in the hybrid. Further analysis revealed that 11 DEPs overlapped with those of previous reports on heterosis (Supplementary Table 2). The trend in expression pattern of three proteins spots correlated with previous reports, whereas it varied for other protein spots in similar types of work.
Table 1.
Mass spectrometric identification and annotation of proteins isolated from dry seeds of maize that displayed differential accumulation in the hybrid DHM 117 compared to its parental inbred lines (BML 6 and BML 7) after separation on 2-D electrophoresis gels.
| Spot number | Expression patterna | Protein (species) accession numberb | Function classificationc | Mol. wt. gel/mol. (kDa) | pI gel/pI predictede | MASCOT scoref | Matched peptide g |
|---|---|---|---|---|---|---|---|
| 13 | F < H > M (AHP) | NAD(P)H-quinone oxidoreductase subunit K, chloroplastic (Glycine max) NDHK_SOYBN | Metabolism and energy | 21/24.1 | 5.7/6.82 | 25 | R.EIYENQMSSQR.E K.NSVISTTLNDLSNWSR.L |
| 18 | F < H > M (AHP) | Triosephosphate isomerase, cytosolic (Zea mays) gi|195605636 | Metabolism and energy | 28/27.2 | 5.7/5.53 | 303 | K.FFVGGNWK.C K.VIACVGETLEQR.E K.SQLRQEFHVAAQNCWVK.K K.VATPAQAQEVHASLRDWLK.T |
| 61 | F < H > M (AHP) | Maturase K (Thujaplicata) MATK_THUPL | Transcription and translation | 22.7/61.3 | 6.3/9.61 | 38 | K.GMDGWNHLR.S R.EDLHAIGHDHHLDR.S |
| 62 | F = H > M (HP) | Oil body-associated protein 2B/secreted protein (Zea mays) gi|226530017 | Defense and stress | 33.1/27.5 | 5.6/5.84 | 37 | R.LNQDVLQCAVYDSDKPSAR.L R.LIGVEYIVSDTIFEGLAPDEQR.L |
| 69 | F < H = M (HP) | Brittle 2 (Zea mays subsp. Mays) gi|23664291 | Metabolism and energy | 66/52.9 | 5.5/5.65 | 209 | R.FAPIYTQPR.H K.IYVLTQFNSASLNR.H K.AAANDSTYLNPQAHDSVLGIILGGGAGTR.L |
| 90 | F < H = M (D) | Globulin-1 S allele-like (Zea mays) gi|670440305 | Storage protein | 27/71.3 | 5.3/6.31 | 297 | K.GFETDVLR.L R.LGFGVKPEVVEAIK.S R.IYAIFTSEGINADDPSKPK.V |
| 105 | F = H > M (HP) | ATP synthase subunit beta, chloroplastic (Oltmannsiellopsis viridis) ATPB_OLTVI | Metabolism and energy | 15.1/51.6 | 6.2/4.70 | 41 | K.VINEDNLSESK.V R.MPSAVGYQPTLATEMGGLQER.I |
| 108 | F < H > M (AHP) | Glyoxalase family protein LOC100280456 (Zea mays) gi|226532762 | Defense and stress | 16/15.1 | 5.3/5.41 | 508 | K.AASFYDAAFGYTVR.R R.ETDELSGAVQLPDSSAAGR.G R.GSVEVCFAYADVDAAYKR.A R.KWAELESGATTIAFTPLHQR.E |
| 115 | F = H > M (HP) | Zein-beta (Zea mays) ZEB2_MAIZE | Storage protein | 20.4/20.2 | 6.8/7.48 | 82 | R.HQCSPAATPYGSPQCQALQQQCCHQIR.Q |
| 116 | F > H (Additive) | Myosin-9 (Arabidopsis thaliana) MYO9_ARATH | Cell cycle | 21.5/175.9 | 4.6/9.12 | 46 | K.FEIADIEPPPLIR.E R.MSQSFRGAPPGVNLAMINGAAGGGADTFR.Q |
| 122 | F > H (Additive) | NAD-dependent malic enzyme 62 kDa isoform, mitochondrial (Solanum tuberosum) MAOM_SOLTU | Metabolism and energy | 13.8/70.1 | 4.2/5.66 | 26 | R.LSSTLLKR.L K.IVVAGAGSAGIGVLNAARK.T |
| 137 | F < H (D) | Rab28 (Zea mays) gi|22460 | Defense and stress | 34.2/27.6 | 4.0/4.90 | 394 | R.IVTEFVAGQAVGQYLAR.D K.VTIGEALEATALAAGDAPVER.S R.LVADKPVESADALGVAGAENR.N |
| 154 | H < M (D) | Unknown (Zea mays) gi|224030527 | Storage protein | 23.3/65.3 | 5.1/6.39 | 478 | K.VFLAGADNVLQKLDR.V K.ALSFASKAEEVDEVLGSR.R R.LSPGTAFVVPAGHPFVAVASR.D K.ESGGHEEREQEEEEEREER.H R.SEEEEEESSEEQEEAGQGYHTIR.A R.RSEEEEEESSEEQEEAGQGYHTIR.A |
| 209 | H < M (Additive) | Cysteine protease ATG4 (Medicago truncatula) ATG4_MEDTR | Defense and stress | 66.3/54.0 | 6.7/6.69 | 40 | K.SSTEIVDNTQVPASSKAGSSDSK.F K.NEQGEQLLPMAIYVVSGDEDGER.G |
| 318 | F > H < M (BLP) | Nucleoredoxin 1 (Zea mays) gi|162459902 | Metabolism and energy | 76.5/64.0 | 4.8/4.8 | 100 | R.LEELQK.K K.VSGIPHLVILDAK.T K.MPWLAIPQGDIK.C R.CDECDFDLHPK.C K.MPWLAVPFSDSEGR.E K.MPWLAVPFSDSEGR.E R.GIPSLVAIGPTGQTVSR.D K.IRGIPSLVAIGPTGQTVSR.D K.SFEVVFASADRNEEAFNEYFAK.M |
a The expression pattern of differentially expressed protein spots in F1 hybrid (H) compared with parents (F - female, M - male) (AHP - above high parent, HP - high parent, LP - low parent, BLP - below low parent, D - different from additive, not belonging to any other class). b The accession number of each protein obtained via MASCOT software from the NCBInr and Swiss-Prot databases. c The function classification of each identified protein. d Molecular mass of proteins on gel and predicted molecular mass of proteins. e pI of proteins on gel and predicted pI of proteins. f Probability-based MASCOT score is defined as –10 × log (p), where p is the probability that the observed match is a random event. g Matching peptides unique to the identified proteins.
Table 2.
Mass spectrometric identification and annotation of proteins isolated from 24-h water-imbibed maize seeds that displayed differential accumulation in the hybrid DHM 117 compared to its parental inbreds (BML 6 and BML 7) after separation on 2-D electrophoresis gels.
| Spot number | Expression patterna | Protein (species) accession numberb | Function classificationc | Mol. wt. gel/mol. wt. predictedd (kDa) | pI gel/pI predictede | MASCOT scoref | Matched peptideg |
|---|---|---|---|---|---|---|---|
| 9 | F < H = M (HP) | Zein-beta (Zea mays) ZEB1_MAIZE | Storage protein | 17.8/19.9 | 7.3/8.15 | 221 | R.MMDVQSVAQQLQMMMQLER.A R.QQCCQQQMR.M |
| 26 | F = H < M (LP) | Vicilin-like embryo storage protein (Zea mays) gi|22284 | Storage protein | 22.8/66.6 | 5.3/6.23 | 354 | K.ALSFASKAEEVDEVLGSR.R R.SEEEEEESSEEQEEAGQGYHTIR.A R.RSEEEEEESSEEQEEAGQGYHTIR.A |
| 43 | F < H = M (HP) | Globulin-1 S allele-like (Zea mays) gi|670440305 | Storage protein | 31.6/71.3 | 5.4/6.31 | 287 | R.LGFGVKPEVVEAIK.S R.IYAIFTSEGINADDPSKPK.V R.VVAESEAGSVSAVDVADAAGTAYR.L |
| 58 | F = H < M (Additive) | Aldose reductase (Zea mays) gi|363543269 | Defense and stress | 37.8/35.9 | 6.9/6.47 | 146 | K.SGHTIPAVGLGTWR.A R.IKENIQVFGWEIPEEDFR.A R.IKENIQVFGWEIPEEDFR.A |
| 60 | F > H = M (LP) | Indole-3-acetaldehyde oxidase (Arabidopsis thaliana) ALDO2_ARATH | Hormone biosynthesis | 38/146.3 | 6.8/5.49 | 38 | K.LATHMEMIAAR.F K.VLYEAITLLGNVVVPEDGTSNPAYR.S |
| 64 | F > H = M (LP) | Type IV inositol polyphosphate 5-phosphatase 11 (Arabidopsis thaliana) IP5PB_ARATH | Defense and stress | 41/37.8 | 7.0/9.35 | 49 | K.TIEAVNSCSFSRK.A K.RDLTVWLGDLNYR.I |
| 136 | F > H = M (LP) | Chaperonin CPN60-1, mitochondrial (Zea mays) CH61_MAIZE | Defense and stress | 74.3/61.4 | 5.2/5.68 | 83 | K.APGFGENR.K R.NVVIEQSFGAPK.V |
| 154 | F = H < M (LP) | Sorbitol dehydrogenase homolog 1 (Zea mays) gi|926657619 | Metabolism and energy | 47.6/39.7 | 6.3/6.27 | 366 | R.EVDVVGVFR.Y R.VVVVDVDDHR.L R.VALEPGVSCWR.C K.AVGICGSDVHYLR.E K.IMPFKLPPVGPYDVR.V R.AGVGPETGVLVVGAGPIGLVSLLAAR.A |
| 172 | F > H < M (BLP) | Protein disulfide-isomerase (Zea mays) PDI_MAIZE | Metabolism and energy | 76/57.2 | 4.8/5.24 | 251 | K.FIDASTIPR.V K.NIQEYKGPR.E R.SDYDFGHTLHANHLPR.G K.LRSDYDFGHTLHANHLPR.G K.VDANEEKNRPLATKYEIQGFPTIK.I |
| 205 | F > H > M (D) | Globulin-1 S allele (Zea mays) GLB1_MAIZE | Storage protein | 65/65.4 | 7.1/6.63 | 304 | R.VAVLEANPR.S R.HGQDKGIIVR.A R.VLRPFDEVSR.L R.RSEEEEESSEEQEEVGQGYHTIR.A |
| 247 | F > H = M (LP) | Globulin-1 S allele (Zea mays) GLB1_MAIZE | Storage protein | 70/65.4 | 6.6/6.63 | 260 | R.RPYVFDRR.S R.VLRPFDEVSR.L K.IAYVPNGKGYAEIVCPHR.Q R.LSPGTAFVVPAGHPFVAVASR.D R.HASEGGHGPHWPLPPFGESR.G K.ILHTISVPGEFQFFFGPGGR.N |
| 279 | F < H = M (HP) | Oil body-associated protein 2A (Zea mays) gi|194693636 | Unknown | 30.5/27.8 | 7.1/6.90 | 286 | R.LIGVEYIVSR.K R.QVETHHYVSR.L K.GFAVDVVPHEMKR.H R.ADVEAPAEEHPGQADYWLR.H R.AARADVEAPAEEHPGQADYWLR.H |
| 319 | F = H < M (LP) | Glucose and ribitol dehydrogenase homolog (Oryza sativa subsp. Japonica) GRDH_ORYSJ | Metabolism and energy | 38.8/32.4 | 6.1/5.76 | 36 | R.GGAGAGGCSIINTSSINAYK.G |
| 321 | F > H < M (BLP) | Red chlorophyll catabolite reductase, chloroplastic (Arabidopsis thaliana) RCCR_ARATH | Defense and stress | 39.2/36.6 | 6.4/5.71 | 14 | K.EYYQDTALDSHR.Q |
| 377 | F < H (D) | Hydroxyproline-rich glycoprotein family protein (Zea mays) gi|226507242 | Metabolism and energy | 41.3/38.7 | 6.7/6.30 | 224 | K.LVPYNPGYQDESVLWTESR.D R.MVNNIYLNFDAFHGDKDHGGVR.D |
a The expression pattern of differentially expressed protein spots in F1 hybrid (H) compared with parents (F - female, M - male) (AHP - above high parent, HP - high parent, LP - low parent, BLP - below low parent, D - different from additive, not belonging to any other class). b The accession number of each protein obtained via MASCOT software from the NCBInr and Swiss-Prot databases. c The function classification of each identified protein. d Molecular mass of proteins on gel and predicted molecular mass of proteins. e pI of proteins on gel and predicted pI of proteins. f Probability-based MASCOT score is defined as –10 × log (p), where p is the probability that the observed match is a random event. g Matching peptides unique to the identified proteins.
Figure 6.
Functional classification of differentially expressed protein spots identified in the dry and 24-h water-imbibed seeds.
3.4. mRNA analysis of DEPs by qRT-PCR
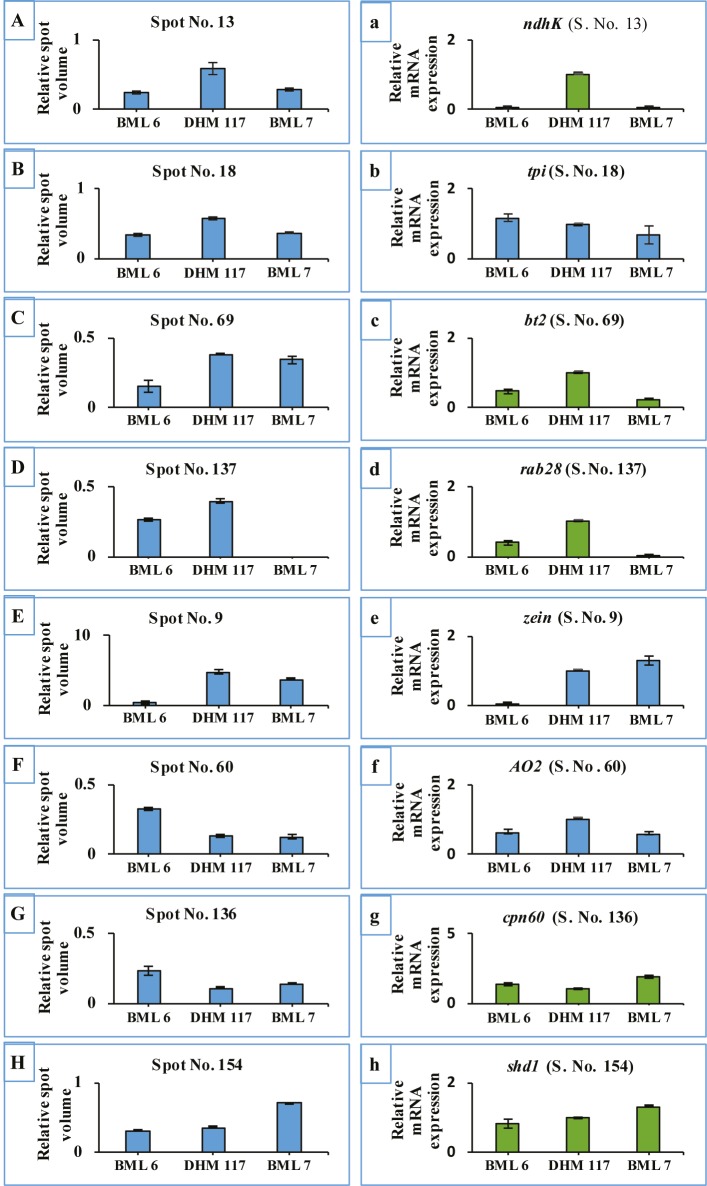
To further investigate whether the expression pattern of DEPs between the F1 hybrid and parents was correlated to the transcriptional level, qRT-PCR was performed with the categorized protein members, which have been reported to play important roles during stress, energy production, and storage in seeds. Of these, we randomly selected eight genes, brittle2 (spot 69/bt2), rab28 (spot 137/rab28), NAD(P)H-quinoneoxidoreductase subunit K (spot 13/ndhK), triose-phosphate isomerase (spot 18/ tpi), zein-beta (spot 9/zein), indole-3-acetaldehyde oxidase (spot 60/AO2), chaperonin CPN60-1 (spot 136/cpn60), and sorbitol dehydrogenase homolog1 (spot 154/sdh1), from dry and 24-h water-imbibed seeds, respectively (Figures 7 and 8; Table 3). We observed that nadK, rab28, zein, cpn60, and sdh1 displayed consistent expression patterns in the F1 hybrid and parental inbreds at transcriptional as well as translational levels. The expression pattern of tpi displayed additive expression in the F1 hybrid at the transcriptional level, which was different from the AHP expression at the translational level. Furthermore, the expression of bt2 and AO2 genes was significantly higher than that of both parents at the transcriptional level; however, these genes were expressed at levels similar to BML 7 and remained higher or lower than BML 6 at the translational level.
Figure 7.
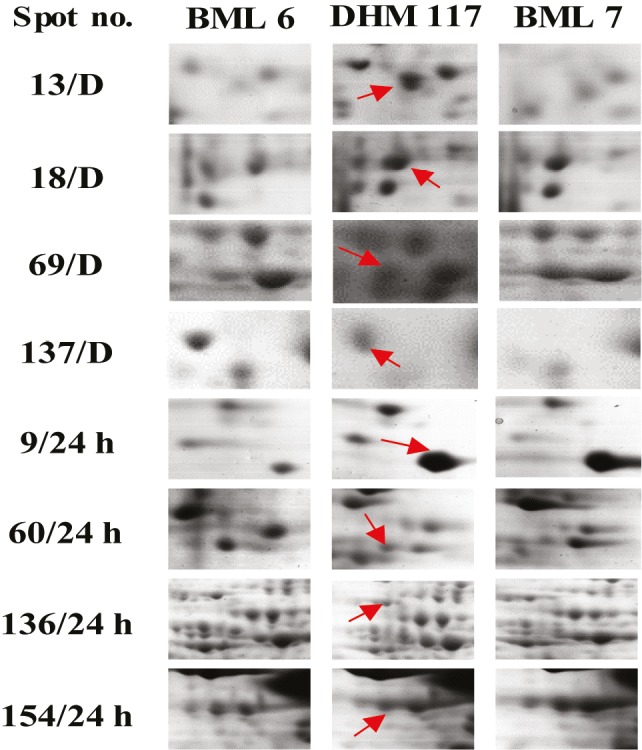
Close-up view of some differentially expressed proteins in 2-D electrophoresis gel maps of F1 hybrid and parents. The numbers of protein spots correspond to those detected in Figure 2 (D and 24 h represent dry and water-imbibed seeds, respectively).
Figure 8.
Relative expression level of differentially accumulated proteins (A to H) and corresponding RNA levels (a to h) of selected genes in dry and 24-h water-imbibed seeds of the F1 hybrid and its parents. Error bars indicate standard deviation of the mean.
Table 3.
List of qRT-PCR primer pairs used for validating the expression of genes at mRNA level.
| Spot number/sample | Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) | |
|---|---|---|---|---|
| 13/Dry | ndhK | GGTCGTTTGTCGTCTTGGC | GCTACATCATAGGCACATTGGG | |
| 18/Dry | tpi | TGGGTTTCTTGTCGGTGGAG | AGCTAAAAGTTGGGGGCAGG | |
| 69/Dry | bt2 | CATTGGAAAGAGGGTTCAGGC | TTGGAGCAAAACGGTCATAGAAG | |
| 137/Dry | rab28 | GCCATCGTTTTCGTTTCGTG | TCGTTCCTGTTGCGGTTCT | |
| 09/24 | h | zein | GTGTGGCTGGCTCTGTCA | CCCATCATCGTCGTCAGGC |
| 60/24 | h | AO2 | TAGACAAGGTGCGGGTCATC | TGTGCCAGTTTTAGCCTCCA |
| 136/24 | h | cpn60 | GAACTGCTTTGGTGGATGCTG | GATGTGTATGTGGGTGTGATGC |
| 154/24 | h | sdh1 | CAACCTCTAGGCGGGCAAG | TTCCAAAACCACGGCAAACTG |
| Control | GAPDH | GCCATCACTGCCACACAGAA | GGAACACGGAAGGACATACCA |
4. Discussion
4.1. DEPs may contribute to heterotic seed germination
Heterosis is a highly complex phenomenon that is influenced by trait, environment, and developmental stage (Romagnoli et al., 1990; Fu et al., 2011) . In this study, we did not observe any correlation between seed weight and germination; mature seeds of the male parent exhibited significantly higher seed weight than the F 1 hybrid and female parent but showed a slow seed germination rate as compared to the F1 hybrid. Guo et al. (2013) reported similar findings, where vigorous growth in the hybrid did not correlate with seed weight in maize. Thus, heterosis was manifested in the early seed germinating stage in the F1 hybrid used in the study.
In maize, there are only a few proteomic reports (Fu et al., 2011; Guo et al., 2013) related to heterotic seed germination. Thus, proteomic investigations were carried out in a highly heterotic F1 hybrid in comparison to its parental inbreds with dry seeds and water-imbibed seeds to obtain precise molecular information associated with heterotic seed germination. We have detected a higher number of protein spots in 24-h water-imbibed seeds in comparison to dry seeds of the F1 hybrid and its parental lines, which reflects an increase in translation of proteins during seed inhibition. Furthermore, a greater number of proteins were found to be differentially abundant between the F1 hybrid and its parental lines than between BML 6 and BML 7 in both dry and water-imbibed seeds and these differences in accumulation patterns might be contributing to the heterotic performance of F1 hybrid DHM 117. It has been reported previously that a higher rate of RNA and protein synthesis influences the heterosis in the growth of the embryonic axis during seed germination (Guo et al., 2013; Hu et al., 2013) .
Several proteomic studies on heterosis revealed that the fraction of nonadditive proteins in the hybrid varied depending on the genotype, tissue, and developmental stage. In this study, 53.4% and 58% of total proteins exhibited nonadditive patterns in dry and 24-h waterimbibed seeds of the hybrid DHM 117. In contrast, 47% and 34.5% of DEPs showed nonadditive expression in dry and 24-h imbibed embryos of maize hybrid Zong3/87-1 (Guo et al., 2013) , whereas in the third-leaf basal region, only 30.8% of DEPs displayed nonadditive patterns of accumulation in maize hybrid Mo17/B73 (Guo et al., 2014) . Marcon et al. (2010) reported that 49% of proteins accumulated differentially from the additivity in 3.5-dayold primary roots of maize hybrid. Thus, multiple molecular mechanisms might be leading to differential expression of proteins, which in turn might be contributing to heterosis.
Previous studies at the translational level showed additive, HP, and LP dominance and underdominance and overdominance in protein expression patterns in a global proteome comparison between hybrids and parents (Hoecker et al., 2008; Fu et al., 2011; Guo et al., 2013) ; the same was observed in our study. It has been suggested that additive patterns may dilute the detrimental effects of over- or underexpression of proteins by causing the total expression level of a protein to fall within an optimal range (Xing et al., 2016) . Among the nonadditively accumulated protein spots identified in this study, the LP pattern accounted for the largest class followed by below low parents (BLP) in water-imbibed seeds of the F1 hybrid.
However, no marked differences were noticed in the LP and BLP categories of nonadditive expression patterns in the dry seeds of the F1 hybrid. Hoecker et al. (2008) and Guo et al. (2013) reported similar expression patterns of nonadditive proteins in dry maize embryos and in primary roots, respectively. In total, eleven differentially accumulated proteins were overlapping with previously reported DEPs during heterosis (Supplementary Table 2).
Among them, the accumulation patterns of three DEPs, NAD(P)H-quinone oxidoreductase (spot no. 13; AHP), glyoxalase family protein (spot no. 108; AHP), and glucose and ribitol dehydrogenase homolog (spot no. 319; LP), were consistent with previous studies (Macron et al., 2010; Fu et al., 2011) . Other protein expression patterns varied at crop and organ levels. For example, triose-phosphate isomerase (spot no. 18), an important component of glycolysis, accumulated variably in maize and sorghumSudan grass hybrid as BLP or LP and HP, respectively, but in our study it was accumulated as AHP (Supplementary Table 2). Thus, a multitude of factors might be involved in the differential accumulation of proteins, which together lead to efficient processes in the F 1 hybrid, resulting in heterosis. These observations are in accordance with the hypothesis that heterosis is contributed by multiple, complex molecular mechanisms (Zhang et al., 2012) .
4.2. DEPs are associated with different cellular processes
To further elucidate the possible roles of differentially accumulated proteins in heterosis during germination, 30 DEPs were effectively identified in dry and 24-h water-imbibed seeds of F1 hybrid and parental inbreds. These DEP spots belong to several important cellular processes such as storage, metabolism and energy, defense and stress, transcription and translation, cell cycle, and hormone biosynthesis. During seed development, several proteins including globulin and vicilin have been stored in endosperm and mobilized after inhibition to initiate the seed germination (Han and Yang, 2015) . In the present study, we also identified several storage proteins such as globulin-1 S allele-like protein (spot nos. 90, 43, 205, 247), zein-beta (spot nos. 9, 115), and vicilin-like embryo storage protein (spot no. 26), and their accumulation in the F1 hybrid was greater than or equal to that of at least one parental inbred. In cereal seeds, proteolysis is an essential process for metabolism of seed proteins and germination (Szewinska et al., 2016) . In our study, we have identified cysteine protease ATG4 (spot no. 209) in the dry seed of the maize hybrid, which plays an important role in the hydrolysis of storage proteins. Seeds also contain several lipid bodies, which protect the lipid reserves in seeds during oxidation and hydrolysis until germination by interacting with specific structural proteins (Purkrtova et al., 2008) . The presence of HP expression of oil bodyassociated proteins (spot nos. 62 and 279) in the hybrid might be contributing to faster seed germination. This suggested that differentially abundant proteolytic proteins in the F1 hybrid could play an important role in mobilizing the storage proteins and might contribute to heterosis at the seed germination stage.
It is well known that ABA is the negative regulator of seed germination and promotes seed dormancy, whereas GA3 overcomes the effect of ABA by inducing germination (Yu et al., 2014) . A study on transgenic maize embryogenesis discovered that the amount of ABA-responsive protein Rab28 decreased after 1 day and became undetectable after the second day of inhibition (Niogret et al., 1996) . Rab28 was shown to have homology with late embryogenesis-abundant (LEA) D-34 proteins of cotton that are induced during stress and maintain the homeostatic environment of cells in dry seeds (Borrell et al., 2002) . In the present study, Rab28 (spot no. 137) protein showed higher expression in dry seeds of the F1 hybrid compared to BML 6 and was not detectable in BML 7. Thus, higher accumulation of Rab28 in the F 1 hybrid could be playing an important role in germination during heterosis even though the mechanism of its regulation and its precise role remain to be elucidated. Previous studies have demonstrated that overexpression of maize Rab28 (Amara et al., 2013) and Arabidopsis Atrab28 (Borrell et al., 2002) in transgenic plants resulted in rapid germination compared to wild-type. Moreover, indole-3-acetaldehyde, IAA, and synthetic auxins inhibited the germination of the dormant embryo in wheat and Arabidopsis whereas indole-3-acetaldehyde oxidase inhibitors decreased the inhibitory effects of IAA on embryos from dormant caryopses, further suggesting that IAA was involved in germination (Liu et al., 2013; Shuai et al., 2017) . In this study, the accumulation of indole-3-acetaldehyde oxidase (spot no. 60) remained lower in the F1 hybrid compared to BML 6 and BML 7. Thus, the decrease in abundance of indole-3-acetaldehyde oxidase protein could have influenced IAA biosynthesis, which is important for rapid seed germination in F1 hybrids. Recently, Chen et al. (2018) using proteomic analysis revealed the importance of auxin homeostasis on eighth internode length heterosis in maize.
Several proteomic studies revealed that metabolismrelated proteins, particularly those involved in the carbohydrate metabolic pathways, accumulated in mature dry seeds and remained stable or increased during germination (Fu et al., 2011; He and Yang, 2013; Han and Yang, 2015; Lee et al., 2016) . Due to limited oxygen availability during early seed germination, energy is primarily supplemented by glycolysis and fermentation followed by tricarboxylic acid (TCA) cycle, the gluconeogenesis and glyoxylate cycle, and the pentose phosphate pathway (He and Yang, 2013; Jin et al., 2014) . In the present study, we found that a protein triose-phosphate isomerase (TPI, spot no. 18), which catalyzes the reversible interconversion of dihydroxyacetone phosphate and glyceraldehyde-3-phosphate ( Kołodziejczyk et al., 2016 ) in glycolysis, is abundantly expressed in F1 hybrid seeds compared to parents. The products of glycolysis assimilate into the TCA cycle and generate energy as ATP and are reducing power as nicotinamide adenine dinucleotide (NADH) for the mitochondrial electron transport chain (van Dongen et al., 2011) . Remarkably, we have observed lower expression of the NAD-dependent malic enzyme 62-kDa isoform (spot no. 122) in dry seeds of the F1 hybrid than in BML 6 and this enzyme is known to catalyze the conversion of malate to oxaloacetate and reduce NAD+ into NADH, an important component of oxidative phosphorylation for ATP synthesis (Jin et al., 2014) . A high parental expression of the ATP synthase beta subunit (spot no. 105) was seen in F1 hybrid seeds in this study, which could be beneficial for catalyzing ATP production during germination (Mills and Richter, 1991; Ducos et al., 2001) . Thus, rapid seed germination of the F 1 hybrid could be due to higher accumulation of proteins involved in metabolism and energy production.
Water inhibition can also induce the oxidative stress and therefore several antioxidant enzymes like glutathione S-transferase (GST), catalase, and peroxidases could shield the cells from oxidative injury (Fu et al., 2011; He and Yang, 2013) . The glyoxalase family proteins catalyze the conversion of the methylglyoxal-glutathione conjugate to glutathione (Wu et al., 2011) and aldose reductase is used to detoxify the reactive aldehyde species (de Sousa et al., 2009) . Proteins related to oxidative stress tolerance, such as glyoxalase family protein (spot no. 108), were expressed more highly in the dry seeds of the F1 hybrid than the parental inbreds in this study. On the contrary, aldose reductase (spot no. 58) and chaperonin CPN60 (spot no. 136), a type of heat shock protein (HSP), displayed additive and LP expression, respectively, compared to parental inbreds in water-imbibed seeds of the F1 hybrid. Wu et al. (2011) reported that HSPs are highly correlated to the seed viability and start degradation during seed germination. Thus, differential accumulation of antioxidant and homeostasis-related proteins in dry and 24-h water-imbibed seeds might play an important role in maintaining the seed viability and protecting the cells from stress during seed inhibition.
4.3. Correlation between mRNA and protein levels during heterosis
It is well known from the literature that the relationship between mRNA and protein abundance is complex, being subject to a myriad of transcriptional, posttranscriptional, translational, and posttranslational determinants and regulations (Hu et al., 2013) . In the present study, 5 out of 8 selected genes exhibited similar expression patterns at mRNA and protein levels, whereas the expression pattern for the remaining genes did not correspond at mRNA and protein levels. Most notably, the expression of triosephosphate isomerase (tpi) was significantly higher than in both parents at the protein level but its expression was not significantly different from BML 6 at the transcript level. On the contrary, indole-3-acetaldehyde oxidase was expressed at lower levels in the F1 hybrid than in the female parent (BML 6) at the protein level but was more highly expressed than in both parents at the transcript level. The lack of correspondence between mRNA and protein abundances may be the result of varied protein synthesis and degradation, as suggested previously (Glickman and Ciechanover, 2002) .
DHM 117 displayed heterotic seed germination, which could be due to efficient regulation of molecular events associated with seed germination. The first step towards seed germination is always breaking the long dormant phase. Regulation for seed dormancy by hormones was induced in seeds so that they do not germinate during otherwise unfavorable conditions; however, for agricultural needs, a short dormancy period is desirable. Seed dormancy is regulated by hormones like ABA and auxins and it was interesting to note an upregulation of Rab28 (spot no. 137) at both protein and transcript levels. However, downregulation of indole-3-acetaldehyde oxidase (spot no. 60) was observed only at the protein level, which is needed for breaking seed dormancy and germination. DHM 117 also showed high levels of TPI (spot no. 18), which is much needed to provide energy for the growth and development of the embryo; we also noticed an upregulation of glyoxylate family protein (spot no. 108), which can protect the embryo from inhibitionassociated elevated ROS levels. This study revealed that a collection of proteins are upregulated to aid in seed germination as well as proteins like AO2 (spot no. 60) that are downregulated in order to save and divert energy required for seed germination.
In conclusion, our study led to the identification DEPs in dry and water-imbibed seeds of a maize F1 hybrid and its parental inbreds that exhibited additive and nonadditive expression patterns. The identified DEPs were involved in various cellular and metabolic processes; the higher accumulation of carbohydrate and energy metabolismrelated proteins in the F1 hybrid indicates that a higher energy supply is very important for heterosis during seed germination. Notably, the abundance of the ABAresponsive Rab28 protein was higher in dry seeds of the F1 hybrid in comparison to both parents, whereas indole-3acetaldehyde oxidase involved in IAA synthesis decreased in water-imbibed seeds of the F1 hybrid compared to the female parent. Thus, differential accumulation of these proteins in the F1 hybrid appears to be a part of the tight regulation of molecular events associated with heterotic seed germination.
Acknowledgments
The first author gratefully acknowledges the Department of Biotechnology (DBT), New Delhi, for providing a DBTJRF fellowship (DBT/JRF/14/AL/128, dated 20 June 2014) to carry out this work. We also thank the DBT-CREBB program of the School of Life Sciences and the DST-FIST Level-II (Phase-II) and UGC-SAP (DRS-1) programs of the Department of Plant Sciences, University of Hyderabad, for the infrastructural support. The authors are also thankful to Professor S Sokka Reddy, Institute of Biotechnology, Prof. Jayashankar Telangana State Agricultural University (PJTSAU), Hyderabad, and Dr MR Sudharshan, Principal Scientist, Maize Research Centre, PJTSAU, Hyderabad, for providing the seeds of the genotypes used in this study. We have received the help of Dr M Surender and Dr Deepti B Sagare of PJTSAU, Hyderabad, for SSR analysis, which is gratefully acknowledged. We would like to thank Monica Kannan, Proteomics Facility of the School of Life Sciences, for her assistance in mass spectrometry analysis. The first author also wishes to thank Sakshi Rampuria, Research Scholar, Department of Plant Sciences, University of Hyderabad, for editing the manuscript.
References
- Amara I , Capellades M , Ludevid MD , Pages M , Goday A ( 2013. ). Enhanced water stress tolerance of transgenic maize plants over-expressing LEA Rab28 gene . J Plant Physiol 170 : 864 - 873 . [DOI] [PubMed] [Google Scholar]
- Baranwal VK , Mikkilineni V , Zehr UB , Tyagi AK , Kapoor S ( 2012. ). Heterosis: emerging ideas about hybrid vigour . J Exp Bot 63 : 6309 - 6314 . [DOI] [PubMed] [Google Scholar]
- Borrell A , Cutanda MC , Lumbreras V , Pujal J , Goday A , CulianezMacia FA , Pages M ( 2002. ). Arabidopsis thaliana atrab28: a nuclear targeted protein related to germination and toxic cation tolerance . Plant Mol Biol 50 : 249 - 259 . [DOI] [PubMed] [Google Scholar]
- Chen Y Zhou Q Tian R Ma Z Zhao X Tang J Fu Z Proteomic analysis reveals that auxin homeostasis influences the eighth internode length heterosis in maize (Zea mays). Sci Rep. 2018;8:7159. doi: 10.1038/s41598-018-23874-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal D Mooney BP Newton KJ Specific changes in total and mitochondrial proteomes are associated with higher levels of heterosis in maize hybrids. Plant J. 2012;72:70–83. doi: 10.1111/j.1365-313X.2012.05056.x. [DOI] [PubMed] [Google Scholar]
- Dahal D Newton KJ Mooney BP Quantitative proteomics of Zea mays hybrids exhibiting different levels of heterosis. J Proteome Res. 2016;15:2445–2454. doi: 10.1021/acs.jproteome.5b01120. [DOI] [PubMed] [Google Scholar]
- de Sousa SM , Rosselli LK , Kiyota E , da Silva JC , Souza GH , Peroni LA , Stach-Machado DR , Eberlin MN , Souza AP , Koch KE et al. ( 2009. ). Structural and kinetic characterization of a maize aldose reductase . Plant Physiol Biochem 47 : 98 - 104 . [DOI] [PubMed] [Google Scholar]
- Ducos E , Touzet P , Boutry M ( 2001. ). The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes . Plant J 26 : 171 - 180 . [DOI] [PubMed] [Google Scholar]
- Faurobert M , Pelpoir E , Chaïb J ( 2007. ). Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues . In: Thiellement H , Zivy M , Damerval C , Méchin V , editors. Plant Proteomics: Methods and Protocols . Totowa, NJ, USA: Humana Press, pp. 9 - 14 . [DOI] [PubMed]
- Fu D , Xiao M , Hayward A , Fu Y , Liu G , Jiang G , Zhang H ( 2014. ). Utilization of crop heterosis: a review . Euphytica 197 : 161 - 173 . [Google Scholar]
- Fu D , Xiao M , Hayward A , Jiang G , Zhu L , Zhou Q , Li J , Zhang M ( 2015. ). What is crop heterosis: new insights into an old topic . J Appl Genet 56 : 1 - 13 . [DOI] [PubMed] [Google Scholar]
- Fu Z , Jin X , Ding D , Li Y , Tang J ( 2011. ). Proteomic analysis of heterosis during maize seed germination . Proteomics 11 : 1462 - 1472 . [DOI] [PubMed] [Google Scholar]
- Glickman , MH , Ciechanover A ( 2002. ). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction . Physiol Rev 82 : 373 - 428 . [DOI] [PubMed] [Google Scholar]
- Gomes MP , Garcia QS ( 2013. ). Reactive oxygen species and seed germination . Biologia 68 : 351 - 357 . [Google Scholar]
- Guo B , Chen Y , Li C , Wang T , Wang R , Wang B , Hu S , Du X , Xing H , Song X et al. ( 2014. ). Maize (Zea mays L.) seedling leaf nuclear proteome and differentially expressed proteins between a hybrid and its parental lines . Proteomics 14 : 1071 - 1087 . [DOI] [PubMed] [Google Scholar]
- Guo B , Chen Y , Zhang G , Xing J , Hu Z , Feng W , Yao Y , Peng H , Du J , Zhang Y et al. ( 2013. ). Comparative proteomic analysis of embryos between a maize hybrid and its parental lines during early stages of seed germination . PLoS One 8 : e65867 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C , Yang P ( 2015. ). Studies on the molecular mechanisms of seed germination . Proteomics 15 : 1671 - 1679 . [DOI] [PubMed] [Google Scholar]
- Han C , Yin X , He D , Yang P ( 2013. ). Analysis of proteome profile in germinating soybean seed, and its comparison with rice showing the styles of reserves mobilization in different crops . PLoS One 8 : e56947 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P , Lu X , Mi F , Dong J , Xue C , Li J , Han B , Zhang X ( 2016. ). Proteomic analysis of heterosis in the leaves of sorghumsudangrass hybrids . Acta Biochim Biophys Sin 48 : 161 - 173 . [DOI] [PubMed] [Google Scholar]
- He D , Yang P ( 2013. ). Proteomics of rice seed germination . Front Plant Sci 4 : 246 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker N , Lamkemeyer T , Sarholz B , Paschold A , Fladerer C , Madlung J , Wurster K , Stahl M , Piepho HP , Nordheim A et al. ( 2008. ). Analysis of nonadditive protein accumulation in young primary roots of a maize (Zea mays L.) F1-hybrid compared to its parental inbred lines . Proteomics 8 : 3882 - 3894 . [DOI] [PubMed] [Google Scholar]
- Hu G , Koh J , Yoo MJ , Grupp K , Chen S , Wendel JF ( 2013. ). Proteomic profiling of developing cotton bfiers from wild and domesticated Gossypium barbadense . New Phytol 200 : 570 - 582 . [DOI] [PubMed] [Google Scholar]
- Jin X , Fu Z , Ding D , Li W , Liu Z , Hu Y , Tang J ( 2014. ). Proteomic analysis of plumules and coleoptiles in maize between hybrids and their corresponding inbred lines . Acta Physiol Plant 36 : 355 - 370 . [Google Scholar]
- Klopfenstein TJ , Erickson GE , Berger LL ( 2013. ). Maize is a critically important source of food, feed, energy and forage in the USA . Field Crops Res 153 : 5 - 11 . [Google Scholar]
- Kołodziejczyk I , Dzitko K , Szewczyk R , Posmyk MM ( 2016. ). Exogenous melatonin expediently modifies proteome of maize (Zea mays L.) embryo during seed germination . Acta Physiol Plant 38 : 146 . [DOI] [PubMed] [Google Scholar]
- Kumar A , Bimolata W , Kannan M , Kirti PB , Qureshi IA , Ghazi IA ( 2015. ). Comparative proteomics reveals differential induction of both biotic and abiotic stress response associated proteins in rice during Xanthomonas oryzae pv. oryzae infection . Funct Integr Genomics 15 : 425 - 437 . [DOI] [PubMed] [Google Scholar]
- Lee SH , Gupta R , Kim YJ , Min CW , Kim SW , Seo WD , Kim ST ( 2016. ). Proteomic analysis indicates activation of reactive oxygen species signaling during seed germination and seedlings growth in Hordeum vulgare (barley) . J Proteins Proteomics 7 : 269 - 277 . [Google Scholar]
- Liu X , Zhang H , Zhao Y , Feng Z , Li Q , Yang HQ , Luan S , Li J , He ZH ( 2013. ). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis . P Natl Acad Sci USA 110 : 15485 - 15490 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ , Schmittgen TD ( 2001. ). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method . Methods 25 : 402 - 408 . [DOI] [PubMed] [Google Scholar]
- Marcon C , Schutzenmeister A , Schutz W , Madlung J , Piepho HP , Hochholdinger F ( 2010. ). Nonadditive protein accumulation patterns in maize (Zea mays L.) hybrids during embryo development . J Proteome Res 9 : 6511 - 6522 . [DOI] [PubMed] [Google Scholar]
- Mills DA , Richter ML ( 1991. ). Nucleotide binding to the isolated beta subunit of the chloroplast ATP synthase . J Biol Chem 266 : 7440 - 7444 . [PubMed] [Google Scholar]
- Niogret MF , Culianez-Macia FA , Goday A , Mar Alba M , Pages M ( 1996. ). Expression and cellular localization of rab28 mRNA and Rab28 protein during maize embryogenesis . Plant J 9 : 549 - 557 . [DOI] [PubMed] [Google Scholar]
- Nonogaki H ( 2014. ). Seed dormancy and germination-emerging mechanisms and new hypotheses . Front Plant Sci 5 : 233 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A , Jia Y , Marcon C , Lund S , Larson NB , Yeh CT , Ossowski S , Lanz C , Nettleton D , Schnable PS et al. ( 2012. ). Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents . Genome Res 22 : 2445 - 2454 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkrtova Z , Jolivet P , Miquel M , Chardot T ( 2008. ). Structure and function of seed lipid body-associated proteins . C R Biol 331 : 746 - 754 [DOI] [PubMed] [Google Scholar]
- Rockenbach MF , Correa CCG , Heringer AS , Freitas ILJ , SantaCatarina C , do Amaral-Junior AT , Silveira V ( 2018. ). Diefrentially abundant proteins associated with heterosis in the primary roots of popcorn . PLoS One 13 : e0197114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnoli S , Maddaloni M , Livini C , Motto M ( 1990. ). Relationship between gene expression and hybrid vigor in primary root tips of young maize (Zea mays L.) plantlets . Theor Appl Genet 80 : 769 - 775 . [DOI] [PubMed] [Google Scholar]
- Schoonheim PJ , Sinnige MP , Casaretto JA , Veiga H , Bunney TD , Quatrano RS , de Boer AH ( 2007. ). 14 -3 -3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination . Plant J 49 : 289 - 301 . [DOI] [PubMed] [Google Scholar]
- Schopfer P , Plachy C , Frahry G ( 2001. ). Release of reactive oxygen intermediates (superoxide radicals, hydrogen peroxide, and hydroxyl radicals) and peroxidase in germinating radish seeds controlled by light, gibberellin, and abscisic acid . Plant Physiol 125 : 1591 - 1602 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A , Wilm M , Vorm O , Mann M ( 1996. ). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels . Anal Chem 68 : 850 - 858 . [DOI] [PubMed] [Google Scholar]
- Shuai H , Meng Y , Luo X , Chen F , Zhou W , Dai Y , Qi Y , Du J , Yang F , Liu J et al. ( 2017. ). Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio . Sci Rep 7 : 12620 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X , Ni Z , Yao Y , Xie C , Li Z , Wu H , Zhang Y , Sun Q ( 2007. ). Wheat (Triticum aestivum L.) root proteome and differentially expressed root proteins between hybrid and parents . Proteomics 7 : 3538 - 3557 . [DOI] [PubMed] [Google Scholar]
- Stupar RM , Springer NM ( 2006. ). Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1-hybrid . Genetics 173 : 2199 - 2210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surender M , Suman K , Aparna M , Sokka Reddy S , Sudarshan MR ( 2015. ). DNA fingerprinting of DHM 121 maize hybrid using SSR markers . Res J Agric Sci 6 : 38 - 42 . [Google Scholar]
- Szewinska J , Siminska J , Bielawski W ( 2016. ). The roles of cysteine proteases and phytocystatins in development and germination of cereal seeds . J Plant Physiol 207 : 10 - 21 . [DOI] [PubMed] [Google Scholar]
- Vale EM , Reis RS , Santa-Catarina R , Pereira MG , Santa-Catarina C , Silveira V ( 2016. ). Comparative proteomic analysis of the heterosis phenomenon in papaya roots . Sci Hortic 209 : 178 - 186 . [Google Scholar]
- van Dongen JT , Gupta KJ , Ramirez-Aguilar SJ , Araujo WL , NunesNesi A , Fernie AR ( 2011. ). Regulation of respiration in plants: a role for alternative metabolic pathways . J Plant Physiol 168 : 1434 - 1443 . [DOI] [PubMed] [Google Scholar]
- Wang G , Zhang X , Wang F , Song R ( 2012. ). Isolation of high quality RNA from cereal seeds containing high levels of starch . Phytochem Anal 23 : 159 - 163 . [DOI] [PubMed] [Google Scholar]
- Wang H , Fang Y , Wang L , Zhu W , Ji H , Xu S , Sima Y ( 2015. ). Heterosis and differential gene expression in hybrids and parents in Bombyx mori by digital gene expression profiling . Sci Rep 5 : 8750 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X , Liu H , Wang W , Chen S , Hu X , Li C ( 2011. ). Proteomic analysis of seed viability in maize . Acta Physiol Plant 33 : 181 - 191 . [Google Scholar]
- Xing J , Sun Q , Ni Z ( 2016. ). Proteomic patterns associated with heterosis . Biochim Biophys Acta 8 : 908 - 915 . [DOI] [PubMed] [Google Scholar]
- Yu Y , Guo G , Lv D , Hu Y , Li J , Li X , Yan Y ( 2014. ). Transcriptome analysis during seed germination of elite Chinese bread wheat cultivar Jimai 20 . BMC Plant Biol 14 : 1471 - 2229 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C , Yin Y , Zhang A , Lu Q , Wen X , Zhu Z , Zhang L , Lu C ( 2012. ). Comparative proteomic study reveals dynamic proteome changes between superhybrid rice LYP9 and its parents at different developmental stages . J Plant Physiol 169 : 387 - 398 . [DOI] [PubMed] [Google Scholar]
- Zhang TF , Li B , Zhang DF , Jia GQ , Li ZY , Wang SC ( 2012. ). Genomewide transcriptional analysis of yield and heterosis-associated genes in maize (Zea mays L.) . J Integr Agric 11 : 1245 - 1256 . [Google Scholar]