Abstract
Background
KRAS and NRAS mutations are identified resistance mutations to anti-epidermal growth factor receptor monoclonal antibodies in patients with metastatic colorectal cancer. BRAF status is also routinely assessed for its poor prognosis value. In our institute, next-generation sequencing (NGS) is routinely used for gene-panel mutations detection including KRAS, NRAS and BRAF, but DNA quality is sometimes not sufficient for sequencing. In our routine practice, Idylla platform is used for the analysis of samples that don’t reach sufficient quality criteria for NGS assay.
Methods
In this study, data from mCRC samples analyzed from May 2017 to 2018 were retrospectively collected. All samples with a poor DNA quality for sequencing have been assessed using Idylla platform. First, KRAS Idylla assay cartridge has been used for the determination of KRAS mutational status. All KRAS wild-type samples have then been analyzed using NRAS-BRAF assay. Among 669 samples, 67 samples failed the DNA quality control and have been assessed on Idylla KRAS mutation test.
Results
Among 67 samples, 50 (75%) samples had a valid result with Idylla KRAS mutation test including 22 carrying a KRAS mutation. For 28 samples, NRAS and BRAF mutational statuses have been assessed using Idylla NRAS-BRAF mutation test. Among 28 samples, 27 (96%) had a valid result including 2 samples bearing a NRAS mutation and 3 samples bearing a BRAF mutation.
Conclusions
Our study shows that an integrated workflow using NGS and Idylla platform allows the determination of KRAS, NRAS and BRAF mutational statuses of 651/669 (97.3%) samples and retrieve 49/67 (73.1%)samples that don’t reach DNA quality requirements for NGS.
Introduction
Colorectal cancer (CRC) is the third most common cancer in men and the second in women worldwide [1]. Despite current early detection strategies for CRC, 20% of CRC are diagnosed at a metastatic stage [2,3].
Use of anti-epidermal growth factor receptor monoclonal antibodies (anti-EGFR mAbs) has improved overall outcome of patients with metastatic CRC (mCRC). Tumor KRAS and NRAS mutational statuses are a prerequisite for the prescription of anti-EGFR mAbs. Wild-type KRAS and NRAS statuses predict early response to anti-EGFR mAbs treatment whereas KRAS and NRAS hotspot mutations are associated with clinical resistance to anti-EGFR mAbs [4–6].
In mCRC context, BRAF mutational status is also commonly assessed because the presence of a V600E mutation is recognized as a poor prognosis factor [7]. According to these data, in mCRC context, research of KRAS, NRAS and BRAF somatic mutations has become a routine practice. Several sequencing or PCR-based assays are available for molecular sample characterization. According to guidelines, RAS analysis should include at least KRAS exons 2, 3 and 4 (codons 12, 13, 59, 61, 117 and 146) and NRAS exons 2, 3 and 4 (codons 12, 13, 59, 61 and 117). Turnaround time for RAS testing should be less than 7 working days from the time of receipt of the specimen by the testing laboratory to the time of issuing of the final report, for more than 90% of specimens. For prognostic assessment, tumor BRAF mutational status should be assessed alongside the assessment of tumor RAS mutational status [4].
Still according to these guidelines, next-generation sequencing (NGS) has been chosen for routine determination of KRAS, NRAS and BRAF mutations in patients with advanced stage of CRC in our institute. For approximately 10% of samples, DNA quality extracted from formalin-fixed paraffin embedded (FFPE) samples does not reach quality criteria and amplicon-based sequencing is not possible or leads to non-interpretable results. In our routine practice, Idylla platform (Biocartis, Mechelen, Belgium) is used as a second-line assay for samples that don’t reach sufficient quality criteria for NGS analysis.
In this study, we describe the integrated routine workflow used in our laboratory based on NGS testing and Idylla automated real-time PCR platform for low quality DNA samples for the determination of KRAS, NRAS and BRAF mutations for patients with mCRC.
Methods
Patients and samples
From May 2017 to May 2018, KRAS, NRAS and BRAF mutations have been assessed in a total of 669 FFPE samples (primary tumor or metastases) from patients with mCRC in our Institute. All samples were assessed for KRAS, NRAS and BRAF mutations in the routine management of their cancer. All patients give their consent for the detection of tumor mutations of KRAS, NRAS and BRAF genes. All data were anonymized prior to analysis for this study. This study has been approved by the ethical and scientific board of Institut de Cancérologie de Lorraine.
Workflow
Tumor specimens have been macrodissected after hematoxylin-eosin slide examination and tumor nuclei content determination by a senior pathologist. DNA has been extracted using QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany). After extraction, DNA concentration was measured using the Qubit dsDNA HS assay kit (which allows detection of concentrations between 0.01 and 100 ng/μL) (Qubit 3.0 Fluorometer, ThermoFisher Scientific Inc, Massachusetts, USA). TruSeq FFPE DNA Library Prep QC kit (Illumina, San Diego, USA) and qPCR using Cobas z480 (Roche Diagnostics, Meylan, France) were then used for DNA quality assessment. The PCR commercial kit is qPCR-based able to evaluate DNA fragmentation.
Two nanograms of DNA have been used for qPCR. DNA quality criteria is based on cycles of quantification (Cq) measurement. Cq is defined as the number of cycles needed to reach the threshold set for the assay. Delta-QC (ΔQC) have been calculated using LightCycler 480 Software W UDF 2.0.0 (Roche Diagnostics) by subtracting the Cq values of patient’s samples (= sample to test) from the Cq value of the kit’s internal control (= good quality DNA reference). Cq value will be low or high according to sample DNA quality which can be good or poor, respectively. Thus, a sample with good DNA quality will result in a low ΔQC value and a sample with degraded DNA will result in a high ΔQC value.
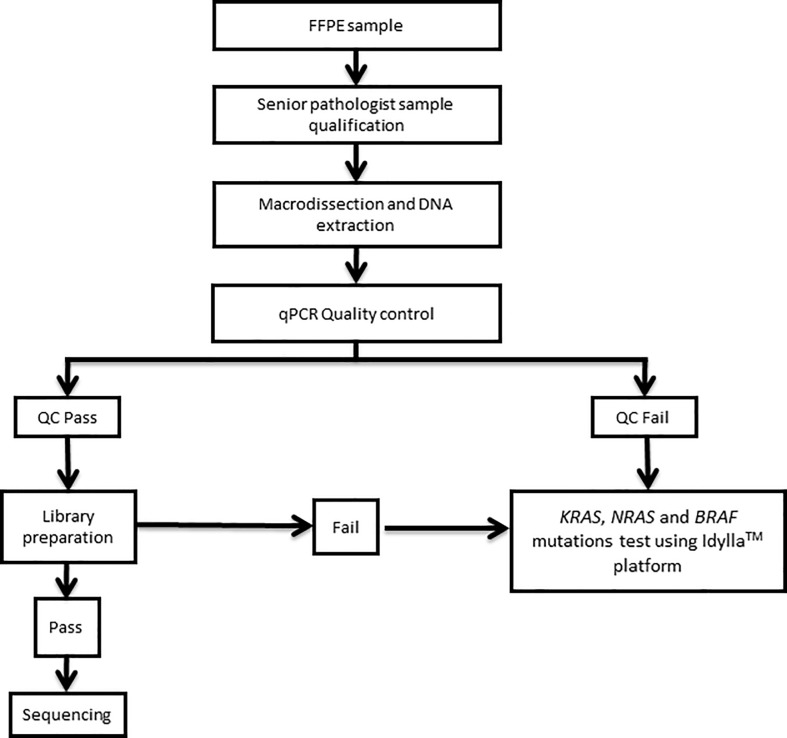
According to manufacturer’s protocol, DNA library were then prepared using TruSeq Custom Amplicon Library Preparation Kit v1.5 (Illumina, San Diego, CA, USA) for samples with ΔQC score lower than 6. Idylla platform was used for ΔQC score greater than 6 or for samples that failed at the library preparation step (Fig 1).
Fig 1. Routine workflow used in our laboratory.
DNA quality control is assessed for all samples. Samples that don’t reach quality criteria are assessed using Idylla assay.
Next generation sequencing assay
NGS library was prepared using the “Panel INCa” TruSeq Custom Amplicon Library Preparation Kit v1.5 (Illumina) that includes 16 genes of interest in theranostic, including KRAS (full exons 2, 3 and 4), NRAS (full exons 2, 3 and 4) and BRAF (full exons 11 and 15). The TruSeq Custom Amplicon Library Preparation Kit consists in two separate oligo pools (CATA and CATB). The two oligo pools were hybridized to DNA samples. The specific hybridized targets were ligated, extended and PCR amplified with adapters containing index with specific barcode sequences. Two complementary libraries were generated by targeting the forward and reverse DNA strands. A purification using AMPure XP beads was then performed on the PCR amplified amplicon libraries obtained for non-specific products and reaction components removal.
Libraries DNA concentration were quantified using Qubit 3.0 Fluorometer (ThermoFisher) and their quality was assessed on Fragment Analyzer (Agilent, Advanced Analytical, Ankeny, USA) using Standard Sensitivity NGS Fragment Analysis Kit (Advanced Analytical). PCR products sizes target was 260bp. All libraries were normalized to enable similar amplification and sequencing levels for each sample library within the same run. Sequencing was performed according to the manufacturer’s protocol. All libraries were pooled prior to sequencing on the MiSeq instrument (Illumina). After sequencing, data were treated with Sophia DDM software v.4.3 (Sophia genetics, Saint-Sulpice, Switzerland). Sequences were aligned using reference genome GRCh37/hg19. For patients with mCRC, KRAS, NRAS and BRAF mutational statuses were systematically reported to the clinician as well as relevant mutations in other genes for genomics guided clinical trials.
Idylla KRAS and NRAS-BRAF mutation test
The Idylla platform is a fully cartridge-based automated platform which uses microfluidics processing with all reagents on-board [8].
We used two different cartridges for this study. Idylla KRAS mutation test detects 21 mutations on the KRAS gene (exons 2, 3 and 4) and the Idylla NRAS-BRAF mutation test detects 18 mutations on the NRAS gene (exons 2, 3 and 4) and 5 on the BRAF gene (exon 15). Limit of detection (LOD) depends on each mutation is 1% for BRAF gene’s mutations, between 1 and 8.5% for mutations on the NRAS gene and between 1 and 5% for mutations on the KRAS gene. LOD per mutation are described in S1 and S2 Tables.
Briefly, FFPE tissue section was “sandwiched” in filter papers and introduced in the cartridge according to manufacturer’s protocol. The tissue area of the FFPE specimen should minimally be 50 mm2 when 5 μm FFPE tissue sections are used or 25 mm2 when 10 μm FFPE tissue sections are used. If tissue area with one section is less than required multiple FFPE tissue sections will be used. All samples in this study have been run in accordance with the manufacturer’s recommendations. After 130 min for Idylla KRAS mutation test and 110 min for Idylla NRAS-BRAF mutation test, final reports were directly available on the Idylla console. Results were presented as “no mutation detected”, “mutation detected in gene X (KRAS, NRAS or BRAF) codon XX” or “invalid”.
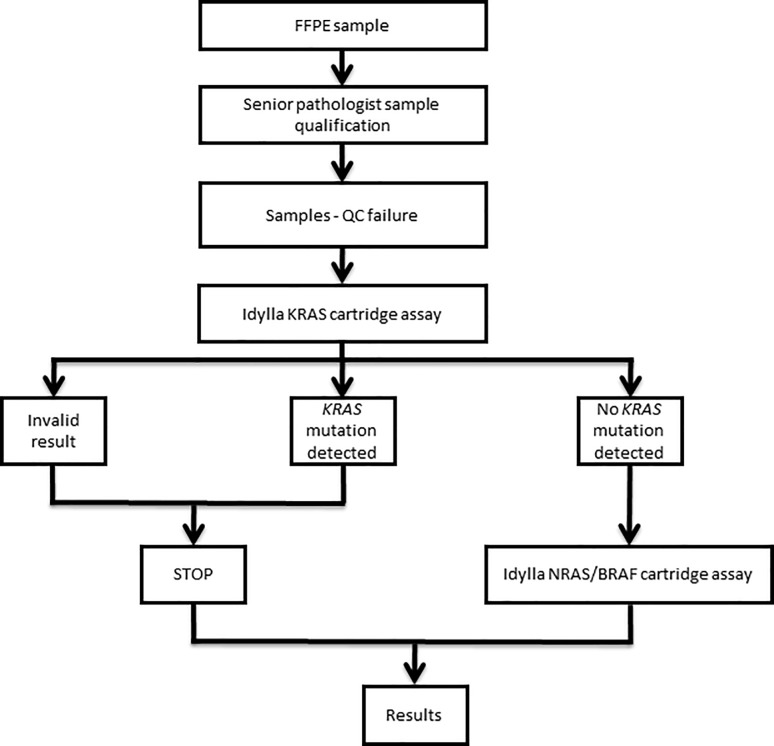
For each poor DNA quality sample, the required number of 5 or 10 μm FFPE tissue section was introduced in a KRAS cartridge. For « invalid » or « mutated » results, the process was stopped and no further test was assessed. For samples with no KRAS mutation detected, a Idylla NRAS-BRAF mutation test was used (Fig 2). A “post-analytic” slide corresponding to an extra FFPE slide after the slides introduced in the Idylla cartridges was systematically prepared for hematoxylin-eosin stain and re-evaluation of the tumor nuclei content by a pathologist to ensure the presence of tumor cells in the analyzed samples.
Fig 2. Decision algorithm used in our laboratory for the determination of KRAS, NRAS and BRAF mutations using the Idylla system.
Results
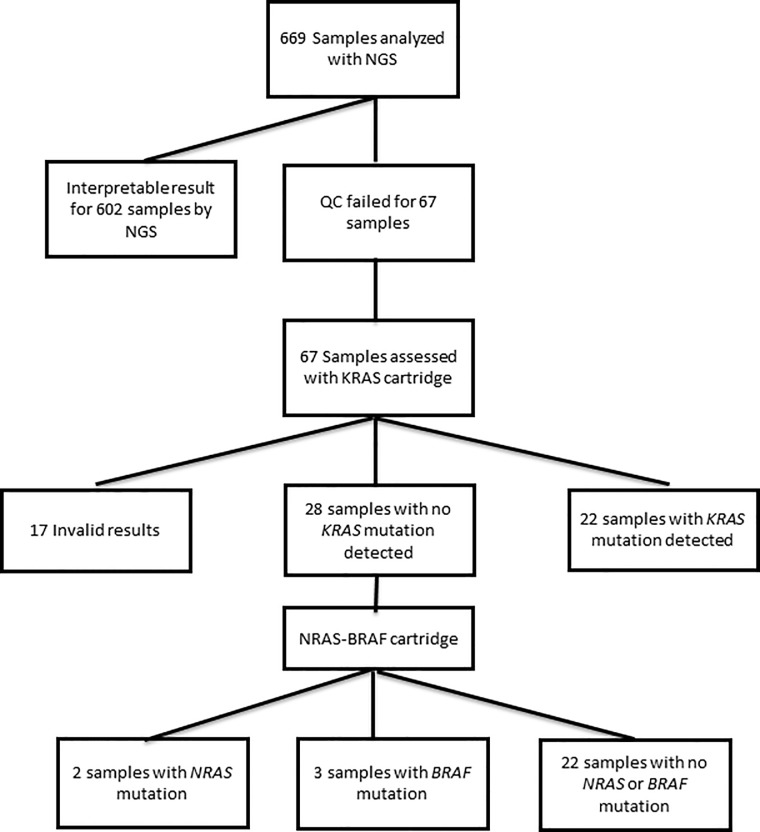
Among 669 samples analyzed in routine from May 2017 to May 2018, 67 samples failed to reach the quality requirement (ΔQC > 6, n=53) or reached the quality requirement (ΔQC ≤ 6, n=14) but did not allow library preparation or gave invalid results after DNA sequencing.
A total of 67 samples were thus tested using Idylla assay. Among 67 samples, 17 samples had an invalid result with Idylla KRAS mutation assay (25.4%) and 50 samples (74.6%) had interpretable results. For 50 patients, samples were successfully recovered using Idylla assay and an interpretable result has been addressed to the oncologist. In total, KRAS, NRAS or BRAF results were available for 652 samples (97.4%).
Among 67 samples, 14 have a ΔQC ≤ 6 (20.9%), 32 have a ΔQC between 6 and 10 (47.8%) and 21 have a ΔQC ≥ 10 (31.3%). Among the 14 samples with ΔQC ≤ 6, 13 samples (92.9%) were recovered by Idylla. Among 32 samples, 30 (93.6%) with a ΔQC between 6 and 10 have been recovered by Idylla. Finally, among the 21 samples which failed NGS with ΔQC > 10, 6 samples (28.6%) have been recovered by Idylla (Table 1).
Table 1. Samples recovered by Idylla according to ΔQC.
| ΔQC | Number of samples | Number of samples recovered with Idylla assay |
|---|---|---|
| ≤ 6 | 14 | 13 (92.9%) |
| > 6 and < 10 | 32 | 30 (93.7%) |
| ≥ 10 | 21 | 6 (28.6%) |
| Total | 67 | 49 (73.1%) |
Among the 50 samples with an interpretable result with Idylla KRAS mutation test, 22 (44%) had a KRAS mutation and 28 (56%) were wild-type for KRAS (Table 2). According to our standard operating procedure, these 28 KRAS wild-type samples have been tested with Idylla NRAS-BRAF mutation test. Among 28 samples, 2 samples had a NRAS mutation (7.1%), 3 (10.7%) a BRAF mutation and 22 (78.6%) samples were wild-type for NRAS and BRAF. One sample had an invalid result (3.6%). All mutations results are detailed in Table 3. Among 67 samples that were not suitable for NGS testing and tested with Idylla assay, 27 (40.3%) had a mutation with a direct clinical impact for the management of mCRC (Fig 3).
Table 2. KRAS, NRAS and BRAF results using Idylla assay.
| Mutation | Number of samples using Idylla testing (%) | |
|---|---|---|
| KRAS | Total | 22 (32.8%) |
| Codon 12 | 16 | |
| Codon 13 | 3 | |
| Codon 61 | 1 | |
| Codon 117 | 1 | |
| Codon 146 | 1 | |
| NRAS | Total | 2 (3.0%) |
| Codon 12 | 1 | |
| Codon 61 | 1 | |
| BRAF | Total | 3 (4.5%) |
| Codon 600 | 3 | |
| Wild-type | Total | 22 (32.8%) |
Table 3. Samples results including QC, ΔQC, DNA concentration (ng/μL) and mutation results for KRAS, NRAS and BRAF genes using the Idylla system.
| Sample | Percentage tumor cells content | QC | ΔQC | DNA concentration (ng/μl) |
Result Idylla KRAS | Result Idylla NRAS | Result Idylla BRAF |
|---|---|---|---|---|---|---|---|
| 1 | 60% | 34.9 | 16.3 | 7.8 | WT | WT | codon 600 mutation |
| 2 | 40% | 35.2 | 16.6 | 6.9 | WT | codon 12 mutation | WT |
| 3 | 30% | 23.2 | 4.5 | 0.7 | WT | WT | WT |
| 4 | 20% | 31.8 | 13.6 | 4.3 | Invalid | Not tested | Not tested |
| 5 | 40% | 26.2 | 5.9 | 0.9 | codon 117 mutation | WT | WT |
| 6 | 20% | 23.5 | 5.3 | 6.3 | codon 12 mutation | Not tested | Not tested |
| 7 | 50% | 29.8 | 9.5 | 3.4 | WT | WT | WT |
| 8 | 50% | 29.3 | 9.1 | 1.2 | WT | Invalid | Invalid |
| 9 | 60% | 27.4 | 8.1 | 25 | WT | WT | WT |
| 10 | 60% | 27 | 7.6 | 0.9 | codon 12 mutation | WT | WT |
| 11 | 80% | 34 | 16.3 | 0.1 | Invalid | Not tested | Not tested |
| 12 | 20% | 26 | 8.9 | 17.8 | codon 12 mutation | WT | WT |
| 13 | 60% | 26.4 | 8.7 | 5.9 | codon 12 mutation | Not tested | Not tested |
| 14 | 25% | 34.1 | 16.4 | 0.7 | Invalid | Not tested | Not tested |
| 15 | 40% | 28.1 | 10.9 | 0.3 | Invalid | Not tested | Not tested |
| 16 | 40% | 24.9 | 7.2 | 6.3 | codon 12 mutation | Not tested | Not tested |
| 17 | 60% | 33.3 | 15.7 | 0.6 | Invalid | Not tested | Not tested |
| 18 | 40% | 27.4 | 9.8 | 7.8 | WT | WT | WT |
| 19 | 70% | 20.9 | 3.7 | 1 | WT | WT | WT |
| 20 | 80% | 21.2 | 4 | 0.8 | codon 61 mutation | Not tested | Not tested |
| 21 | 30% | 30.5 | 11.7 | 2.5 | WT | WT | WT |
| 22 | 70% | 20.4 | 2.8 | 3.1 | codon 12 mutation | Not tested | Not tested |
| 23 | 40% | 32.1 | 13.3 | 0.1 | Invalid | Not tested | Not tested |
| 24 | 40% | 23.3 | 6.1 | 11.8 | WT | WT | WT |
| 25 | 40% | 21.4 | 2.6 | 2.7 | WT | WT | WT |
| 26 | 30% | 24.6 | 5.7 | 1.7 | WT | WT | WT |
| 27 | 80% | 19.3 | 1.7 | 0.4 | WT | WT | WT |
| 28 | 50% | 22.8 | 5.1 | 7.8 | WT | WT | WT |
| 29 | 40% | 30.1 | 12.4 | 0.4 | Invalid | Not tested | Not tested |
| 30 | 70% | 27.2 | 9.5 | 12.9 | WT | codon 61 mutation | WT |
| 31 | 70% | 23.3 | 6 | 14.6 | codon 12 mutation | Not tested | Not tested |
| 32 | 30% | 26.1 | 8.8 | out of range | Invalid | Not tested | Not tested |
| 33 | 10% | 23.9 | 7 | 29.8 | codon 12 mutation | Not tested | Not tested |
| 34 | 20% | 24.7 | 6.9 | 13 | WT | WT | codon 600 mutation |
| 35 | 85% | 21.6 | 4.7 | 3.9 | WT | WT | WT |
| 36 | 60% | 24.9 | 7.1 | 16.1 | WT | WT | codon 600 mutation |
| 37 | 60% | 24.4 | 6.1 | 11.5 | codon 12 mutation | Not tested | Not tested |
| 38 | 60% | 30.7 | 12.4 | 6.2 | WT | WT | WT |
| 39 | 50% | 24.6 | 6.8 | 2 | WT | WT | WT |
| 40 | 70% | 28.2 | 6.5 | 22 | codon 12 mutation | Not tested | Not tested |
| 41 | 70% | 33.6 | 11.9 | 1.3 | Invalid | Not tested | Not tested |
| 42 | 70% | 37.1 | 18.9 | 2.9 | Invalid | Not tested | Not tested |
| 43 | 60% | 33.1 | 13.8 | 8 | Invalid | Not tested | Not tested |
| 44 | 5% | 28.8 | 11.3 | 2.4 | Invalid | Not tested | Not tested |
| 45 | 70% | 25.5 | 6.3 | 35.7 | codon 12 mutation | Not tested | Not tested |
| 46 | 10% | 26.4 | 7 | 11.1 | WT | WT | WT |
| 47 | 80% | 26.7 | 6.6 | 27.7 | WT | WT | WT |
| 48 | < 10% | 25.4 | 5.3 | 7.3 | Invalid | Not tested | Not tested |
| 49 | 15% | 25.6 | 6.3 | 12.3 | codon 12 mutation | Not tested | Not tested |
| 50 | 30% | 28.4 | 9.9 | 1.9 | codon 12 mutation | Not tested | Not tested |
| 51 | 15% | 25.8 | 6.9 | 21.9 | WT | WT | WT |
| 52 | 80% | 25.9 | 7 | 7.5 | codon 13 mutation | Not tested | Not tested |
| 53 | 50% | 31 | 13.3 | 7.1 | Invalid | Not tested | Not tested |
| 54 | 90% | 23.6 | 6.2 | 21.3 | WT | WT | WT |
| 55 | 20% | 24.9 | 7.4 | 0.6 | WT | WT | WT |
| 56 | 10% | 26.2 | 8.7 | 44.3 | WT | WT | WT |
| 57 | 30% | 34.4 | 15.5 | 1.4 | Invalid | Not tested | Not tested |
| 58 | 40% | 27.7 | 8.9 | 3.5 | codon 13 mutation | Not tested | Not tested |
| 59 | 30% | 25.6 | 8 | 64.7 | codon 146 mutation | Not tested | Not tested |
| 60 | 50% | 29.7 | 12.1 | 46.2 | codon 13 mutation | Not tested | Not tested |
| 61 | 70% | 31.5 | 14 | 14.7 | codon 12 mutation | Not tested | Not tested |
| 62 | 20% | 25.3 | 7.9 | 10.5 | codon 12 mutation | Not tested | Not tested |
| 63 | 70% | 23.5 | 6.1 | 44.9 | codon 12 mutation | Not tested | Not tested |
| 64 | 30% | 27.6 | 10.2 | 1.1 | Invalid | Not tested | Not tested |
| 65 | 50% | 20.6 | 4 | 5.4 | WT | WT | WT |
| 66 | 20% | 29.3 | 12.7 | 2.8 | Invalid | Not tested | Not tested |
| 67 | 15% | 25.4 | 7.1 | 25.4 | WT | WT | WT |
Fig 3. A total of 669 mCRC samples have been analyzed.
53 mCRC samples failed QC for NGS assessment and 14 samples failed a valid sequencing result and have been analyzed with Idylla KRAS cartridge. No further analysis was conducted for the 17 invalid results and the 22 samples with a KRAS mutation. 28 wild-type KRAS samples have been analyzed with Idylla NRAS-BRAF mutation test.
Among the 602 samples with valid results with NGS, 12 (2%) had an uncommon mutational profile with non-hotspot mutations uncovered by the Idylla platform.
Discussion
In this study, we retrospectively collected data from 669 samples from patients with mCRC for KRAS, NRAS and BRAF mutational status assessment. DNA was of too poor quality for a NGS analysis for 10% of samples. The most common cause of DNA degradation in FFPE tissues is an inappropriate length of tissue fixation. In the large majority of the cases, the same poor DNA quality is found in a second block from the same lesion, because of the pre-analytical steps were similar. Moreover, asking a second block to the pathologist can improve delays for RAS and BRAF testing. In our experience, analyzing poor quality DNA (ΔQC > 6) using amplicon-based NGS assay leads to no library amplification or to results with a very high background noise which is not compatible with a good interpretation for samples with variant allele frequency (VAF) under 10% or more. The risk of false positive or false negative is too high when sequencing poor quality DNA, thus we chose a second assay which is less stringent on DNA quality to avoid a second biopsy to the patient. These samples have been analyzed with the Idylla platform and 75% of the samples have been retrieved. We assume that Idylla assay amplifies shorter amplicons than the library preparation kit we use and is then less influenced by DNA fragmentation than NGS assay. We tested this hypothesis by analyzing DNA fragments sizes from samples with a range of ΔQC and the results confirmed that a high ΔQC is associated with shorter DNA fragments (S1 Fig). Fourteen samples were found with a ΔQC ≤ 6 but did not allow library preparation or gave non interpretable results after sequencing. We assume that for these samples, ΔQC close to 6 combined with low DNA yield led to a library preparation failure and or high background noise. Using shorter amplicons for library preparation or using capture-based sequencing may address this issue.
Sample #8 had a valid result (WT) with KRAS cartridge and invalid result with NRAS-BRAF cartridge. The same quantity of FFPE sections have been used in the two cartridges, which suggests that DNA quality requirements may depends on the type of cartridge or a high DNA fragmentation in NRAS and BRAF loci.
Two strategies are commonly used for tumor driver mutations assessment: broad approach as NGS and targeted approach as PCR-based methods. NGS allows the exhaustive DNA analysis of regions of interest with no or few limitations. The targeted approach is only based on the detection of hotspot mutations, thus can’t detect rare mutations. Among the analyzed samples, 2% were found with a non-hotspot mutation on KRAS, NRAS or BRAF genes with NGS. By its design and its targeted approach the Idylla assay can’t detect these mutations. Whereas only mutations on codons 12, 13, 59, 61, 117 and 146 are validated resistance mutations to anti-EGFR mAbs, we have shown that non-hotspot KRAS or NRAS mutations may have an impact on resistance to anti-EGFR mAbs [9–11]. In the absence of more published data on rare mutations and resistance to anti-EGFR mAbs, targeted detection of KRAS, NRAS and BRAF mutation remains relevant and is recognized as the golden standard. In our experience, most of the sequencing prescriptions for patients with mCRC are used to choose the first-line therapy, thus use of NGS is only useful in a molecular tumoral board context.
DNA quality can sometimes make NGS analysis not possible; this assay also requires a minimal quantity of DNA to avoid false negative results. DNA quantity can sometimes be an issue with small tissue biopsies. Targeted assays often require less DNA than NGS and are feasible with DNA of poor quality. However, the development of capture-based NGS assays makes sequencing of DNA possible with few quantities of DNA and lower DNA quality [12]. However the Idylla system also has limitations since this assay requires a minimal amount of FFPE material (50 mm2 with 5μm tissue section and 25 mm2 with 10 μm tissue sections) with a tumor cell percentage superior to 10%.
NGS is known as an expensive assay and requires a highly-qualified team [13]. The analysis of raw sequencing data is highly-dependent upon bioinformatics and all detected variants need to be analyzed and biologically interpreted in the clinical context. These several steps mostly take from 3 to 5 days from DNA extraction to interpretation of the data. The targeted PCR assay we chose here is really easy to use and requires less than 5 minutes hands-on time. The results are directly available at the end of the analysis which requires 110 to 130 minutes depending on the test [8]. Idylla testing has been evaluated in several studies which have shown an excellent concordance with reference methods [14,15].
In our study, 97.3% of samples had an interpretable result using our integrated routine workflow. Avoiding a new biopsy which could be no possible nor ethical, we assume that this workflow allows to commence first-line therapy earlier for patients with mCRC.
We chose in our routine workflow to use a broad approach first and a targeted approach as a second choice. In some theranostics indications, it may sound relevant to firstly use a targeted approach and use NGS only for tumor molecular board purposes.
Indeed, turnaround time for gene panel analysis from sample reception to final results is in average 3 to 4 days whereas final results with Idylla system require only one day. Moreover, to optimize time and sequencing costs, most of routine laboratories use samples batches implying longer delays.
According to the French healthcare system, a targeted analysis of RAS and BRAF is evaluated to 556.20 euros (440.10 € for the analysis of the 6 exons of KRAS and NRAS and 116.10 € for the research of the V600 mutations of BRAF), whereas a gene panel with a size inferior to 20Kb is evaluated to 882.90 €.
This study focuses on the management of patients with mCRC, but KRAS, NRAS or BRAF mutations testing is also relevant in other cancers like metastatic melanoma or non-small cell lung cancer [16,17].
The Idylla system has been built to work with FFPE slides directly in the cartridge, but it has recently been showed that the detection of EGFR mutations was possible by adding DNA directly in the cartridge which can also be an advantage since DNA already extracted for the NGS assay can also be tested using Idylla without using more fixed-tissue [17,18]. For patients who require commencing a targeted therapy quickly, a two steps scheme using extracted DNA in Idylla first and then NGS, may be used, saving time and tissue.
In conclusion, our study shows that an integrated workflow that uses NGS and Idylla platform allows the determination of mutational status of KRAS, NRAS and BRAF genes of 97.4% of samples and can retrieve 75% of samples that don’t reach quality criteria for NGS testing.
Supporting information
The size of DNA fragments decreases when ΔQC increases.
(PDF)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors have no funding to report.
References
- 1.International Agency for Research on Cancer. GLOBOCAN 2012: estimated cancer incidence, mortality and prevalence worldwide in 2012. Accessed 8 June 2018.
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383(9927):1490–1502. 10.1016/S0140-6736(13)61649-9 [DOI] [PubMed] [Google Scholar]
- 3.Leufkens AM, van den Bosch MA, van Leeuwen MS, Siersema PD. Diagnostic accuracy of computed tomography for colon cancer staging: a systematic review. Scand J Gastroenterol 2011;46:887–94. 10.3109/00365521.2011.574732 [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol 2016;27:1386–1422. 10.1093/annonc/mdw235 [DOI] [PubMed] [Google Scholar]
- 5.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26(10):1626–1634. 10.1200/JCO.2007.14.7116 [DOI] [PubMed] [Google Scholar]
- 6.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66(8):3992–3995. 10.1158/0008-5472.CAN-06-0191 [DOI] [PubMed] [Google Scholar]
- 7.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol 2011;12(6):594–603. 10.1016/S1470-2045(10)70209-6 [DOI] [PubMed] [Google Scholar]
- 8.Harlé A, Salleron J, Franczak C, Dubois C, Filhine-Tressarieu P, Leroux A, et al. Detection of BRAF mutation using a Fully Automated Platform and Comparison with HighResolution Melting, Real-Time Allele Specific Amplification, Immunohistochemistry and NextGeneration Sequencing Assays, for patients with Metastatic Melanoma. PlosOne 2016;11(4):e0153576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369(11):1023–34. 10.1056/NEJMoa1305275 [DOI] [PubMed] [Google Scholar]
- 10.Shinozaki E, Yoshino T, Yamazaki K, Muro K, Yamaguchi K, Nishina T et al. Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for anti-EGFR monoclonal Antibodies by Comprehensive Cancer genomics (BREAC) study. Br J Cancer 2017;117(10):1450–1458. 10.1038/bjc.2017.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlé A, Filhine-Tresarrieu P, Husson M, Boidot R, Rouyer M, Dubois C, et al. Rare RAS Mutations in Metastatic Colorectal Cancer Detected During Routine RAS Genotyping Using Next Generation Sequencing. Target Oncol 2016;11(3):363–70. 10.1007/s11523-015-0404-7 [DOI] [PubMed] [Google Scholar]
- 12.Chung J, Son DS, Jeon HJ, Kim KM, Park G, Ryu GH, et al. The minimal amount of starting DNA for Agilent’s hybrid capture-based targeted massively parallel sequencing. Sci Rep 2016;6:26732 10.1038/srep26732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marino P, Touzani R, Perrier L, Rouleau E, Kossi DS, Zhaomin Z, et al. Cost of cancer diagnosis using next-generation sequencing targeted gene panels in routine practice: a nationwide French study. Eur J Hum Genet 2018;26(3):314–323. 10.1038/s41431-017-0081-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uguen A, Troncone G. A review on the Idylla platform: towards the assessment of actionable genomic alterations in one day. J Clin Pathol. 2018;71(9):757–762. 10.1136/jclinpath-2018-205189 [DOI] [PubMed] [Google Scholar]
- 15.Weyn C, Van Raemdonck S, Dendooven R, Maes V, Zwaenepoel K, Lambin S, et al. Clinical performance evaluation of a sensitive, rapid low-throughput test for KRAS mutation analysis using formalin-fixed, paraffin-embedded tissue samples. BMC Cancer. 2017;17:139 10.1186/s12885-017-3112-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011; 364(26):2507–16. 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Luca C, Rappa AG, Gragnano G, Malapelle U, Troncone G, Barberis M, et al. Idylla assay and next generation sequencing: an integrated EGFR mutational testing algorithm. J Clin Pathol 2018. [DOI] [PubMed] [Google Scholar]
- 18.De Luca C, Gragnano G, Pisapia P, Vigliar E, Malapelle U, Bellevicine C, et al. EGFR mutation detection on lung cancer cytological specimens by the novel fully automated PCR-based Idylla EGFR Mutation Assay. J Clin Pathol 2017;70(4):295–300. 10.1136/jclinpath-2016-203989 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The size of DNA fragments decreases when ΔQC increases.
(PDF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.