Abstract
Intestinal mucosa is the interface between the microbial content of the gut and the host’s milieu. The goal of this study was to modulate chicken intestinal microflora by in ovo stimulation with galactooligosaccharides (GOS) prebiotic and to demonstrate the molecular responses of the host. The animal trial was performed on meat-type chickens (Ross 308). GOS was delivered by in ovo injection performed into the air cell on day 12 of egg incubation. Analysis of microbial communities and mucosal gene expression was performed at slaughter (day 42 post-hatching). Chyme (for DNA isolation) and intestinal mucosa (for RNA isolation) from four distinct intestinal segments (duodenum, jejunum, ileum, and caecum) was sampled. The relative abundance of Bifidobacterium spp. and Lactobacillus spp. in DNA isolated from chyme samples was determined using qPCR. On the host side, the mRNA expression of 13 genes grouped into two panels was analysed with RT-qPCR. Panel (1) included genes related to intestinal innate immune responses (IL-1β, IL-10 and IL-12p40, AvBD1 and CATHL2). Panel (2) contained genes involved in intestinal barrier function (MUC6, CLDN1 and TJAP1) and nutrients sensing (FFAR2 and FFAR4, GLUT1, GLUT2 and GLUT5). GOS increased the relative abundance of Bifidobacterium in caecum (from 1.3% to 3.9%). Distinct effects of GOS on gene expression were manifested in jejunum and caecum. Cytokine genes (IL-1β, IL-10 and IL-12p40) were up-regulated in the jejunum and caecum of the GOS-treated group. Host defence peptides (AvBD1 and CATHL2) were up-regulated in the caecum of the GOS-treated group. Free fatty acid receptors (FFAR2 and FFAR4) were up-regulated in all three compartments of the intestine (except the duodenum). Glucose transporters were down-regulated in duodenum (GLUT2 and GLUT5) but up-regulated in the hindgut (GLUT1 and GLUT2). In conclusion, GOS delivered in ovo had a bifidogenic effect in adult chickens. It also modulated gene expression related to intestinal immune responses, gut barrier function, and nutrient sensing.
Introduction
Mucous tissue lines the gastrointestinal tract (GIT). It is composed of a vast surface area that forms an interface between luminal microflora and internal body structures. Mucosa is in constant cross-talk with the microbiome and reacts directly to signals from the gut environment [1]. The effects of such interaction on the host are immense: from intestinal health, improved digestion, metabolism regulation, and maturation of the immune system, to regulation of behaviour and body weight [2]. Composition of the microbiome can be altered by different environmental factors [3]. Some of them, including a low-fibre diet or use of antimicrobials or enteric infection agents, decrease the biodiversity and abundance of the beneficial microflora [3]. Others, such as prebiotics, probiotics, synbiotics, or postbiotics, exert positive effects on the composition of the gut microflora [3]. Every modification of the microbiome loops back to influence the health and metabolic status of the host. In this manner, the host and microbiome are interconnected in infinite symbiosis. On the host’s side, these inherent mechanisms are controlled by mucous tissue. For this reason, focus on the mucosa may help explain the mechanisms that follow microbiota modulation.
Intestinal mucosa is comprised of the epithelium, lamina propria, and smooth muscle. The outermost layer of the small intestine is lined with absorptive, columnar epithelial cells (enterocytes) with interspersed goblet cells that secrete mucins, and Paneth cells that secrete antimicrobials [4]. The mucosa forms intestinal villi and crypts, which take part in nutrient absorption and cellular turnover. Avian GIT reflects adaptation to type of food, environment, and motility [5]. The digestive system has to be flexible to accommodate a wide range of diets over the life cycle. Aside from digestion and absorption of ingested food, GIT plays a role in protecting internal tissues from luminal content. Such a barrier is physical, immunological, or microbial [6].
The immunological barrier of the intestine is formed by gut-associated lymphoid tissue (GALT), which includes bursa of Fabricius, caecal tonsils (double), Peyer’s patches (up to five), lymphoid aggregates (mostly in hindgut—urodeum and proctodeum) and intraepithelial T lymphocytes [7]. Innate and adaptive barrier mechanisms in the gut were reviewed by Kelsall (2008) [8]. Innate mechanisms include pattern recognition receptors (e.g., TLR, Toll-like receptors), mucins, cytokines and host defence peptides (HDP). Adaptive mechanisms develop in submucosal lamina propria, which is rich in all kinds of lymphatic cells, including dendritic cells, macrophages, and B lymphocytes. The latter produce secretory IgA that are released into lumen and neutralise antigens locally, without activation of inflammatory responses or the complement system. The immunological mechanisms are based on the subtle balance between immune tolerance and immune responses.
In avian species, GIT is relatively short, which results in the fast transfer time of chyme through the intestines [5]. It is much harder for the bacteria to colonise the proximal segments of the intestines. This is why most bacteria populations colonise crop, distal ileum, and caeca, where peristaltic movement is slower [9]. Stanley et al. (2014) reviewed the literature on spatial microbiome composition along the chicken GIT [10]. Currently, the most integrated report on chicken microbiome composition has been published by Wei et al. [11]. Based on 16s rRNA sequences from Sanger sequencing, chicken guts contain 70% Firmicutes (including 8% Lactobacillus), 12.3% Bacteroidetes, and 9.3% Proteobacteria. Bifidobacterium from Actinobacteria phyla was represented by about 1% of the sequences. The microbiome composition is strongly influenced by environmental rather than genetic factors [12]. Prebiotics, which are defined non-digestible oligosaccharides, are potent modulators of the intestinal microflora. Dietary prebiotics are metabolized only by particular intestinal bacteria, which in turn selectively stimulates their growth. The most common prebiotics include inulin, fructooligosaccharides (FOS), galactooligosaccharides (GOS), or mannanoligosaccharides (MOS) followed by novel prebiotics, such as xylooligosaacharides or isomaltooligosaccharides. For the detailed information on application of prebiotics in poultry we reference to some recent review papers [13–15].
Recent reports emphasize that early-life microbial stimulation has long-term effects on intestinal development and health by promoting maturation of the immune system and development of oral tolerance [16–18]. It also has been shown that microflora disturbances early in life by antimicrobials has long-lasting negative effects on the animal [19, 20]. For this reason, in ovo technologies have been adapted to administer a bioactive compound such as prebiotic, probiotic or synbiotic during embryonic development. In ovo technologies apply to early-stage (e.g., day 12 of egg incubation) or late-stage (e.g., day 17–18 of egg incubation) chicken embryos, reviewed by Roto et al. (2016) [21]. These methods are referred to by their authors as in ovo stimulation (early-stage embryos) [22] or in ovo feeding (late-stage embryos) [23–25]. We report effects of in ovo stimulation of the chicken microflora. This method is based on in ovo delivery of prebiotics or synbiotics on day 12 of egg incubation [26]. At this time-point, the chorioallantoic membrane is highly vascularized, which allows for a passive transfer of the small-weight oligosaccharides from the air cell to the blood vessels surrounding the embryo. The purpose of the early delivery of prebiotics is to boost growth of the indigenous microflora present in the egg, so that the microbiome can be developed along with the embryo. The in ovo stimulation has been patented [27] and validated at laboratory scale and in field trials [28–30].
Among many bioactive compounds tested for in ovo stimulation, GOS prebiotic demonstrated particularly beneficial properties in chickens, including an increase in the number of lactobacilli and bifidobacteria at hatching [28], long-term transcriptional modulation of the host [31], microstructure of the small intestine [32], immune system development [33–35], and improvement in broiler performance [30]. GOS was also used in chickens for dietary interventions to improve performance traits [36] as well as resistance against heat stress [37] and Salmonella colonisation [38, 39]. The effects of GOS delivered in ovo on the chicken were evaluated on multiple levels. The first sets of traits analysed in meat-type chickens was performance, measured by growth, efficiency of nutrition, and meat quality [29, 40]. A further step of looking into the effects of in ovo stimulation was the examining the development of intestinal tissue and microflora [41]. Analysis of morphology was followed by physiological modulation of nutrients and hormones [42]. Finally, the effects of GOS delivered in ovo on gene expression in spleen or caecal tonsils was demonstrated [31]. The next step was to study the effects of the molecular interaction between host and microbiome at their actual interface, which is intestinal mucosa.
This paper aims to provide insights into the mechanisms that drive beneficial effects of GOS stimulation in ovo in chicken. It focuses on both sides of microbiota-host interaction, i.e., selected microbial populations and intestinal mucosa physiology. For this purpose, the following effects of GOS delivered in ovo analyzed was analysed: (1) abundance of Bifidobacterium and Lactobacillus in the intestinal chyme as well as (2) the immune and physiological parameters of the intestinal mucosa measured by gene expression.
Materials and methods
GOS in ovo delivery
Prebiotic GOS was used for in ovo stimulation by in ovo delivery on day 12 of egg incubation. The GOS used in this study (trade name: Bi2tos, Clasado Biosciences Ltd., Jersey, UK) is manufactured by enzymatic transgalactosylation of the milk lactose by the whole cells of Bifidobacterium bifidum 41171 [43]. The GOS product obtained this way is a dry powder containing a mixture (wt:wt) of the following oligosaccharides: 45% lactose, 9.9% disaccharides [Gal (β 1–3)-Glc; Gal (β 1–3)- Gal; Gal (β 1–6)- Gal; Gal (α 1–6)- Gal], 23.1% trisaccharides [Gal (β 1–6)-Gal (β 1–4)- Glc; Gal (β 1–3)- Gal (β 1–4)- Glc], 11.55% tetrasaccharides [Gal (β 1–6)- Gal (β 1–6)- Gal (β 1–4)- Glc], and 10.45% pentasaccharides [Gal (β 1–6)- Gal (β 1–6)- Gal (β 1–6)- Gal (β 1–4)- Glc]. Injection dose was aseptically prepared by dissolving 3.5 mg of GOS mixture in 0.2 ml of physiological saline. The GOS/saline solution was manually deposited into an air chamber with a syringe and a needle. The in ovo injection was done in two steps: (1) the whole was punctured in the blunt end of the egg containing viable, 12-day-old embryo and (2) the 0.2 ml of GOS/saline solution was injected into the air chamber. The size of the needle used for puncturing the hole (0.9 mm) was bigger than the needle used for in ovo injection (0.45 mm). This way, enough space was left to let go air that was replaced by the solution. Also, only the outer egg membrane was punctured prior to injection. Precaution was taken not to break the semi-permeable inner egg membrane, which at this point (i.e., day 12 of eggs incubation) is highly vascularized and moist. Using blue dye for tracking, we determined that the substance (i.e., GOS) injected this way is gradually diffused through the inner egg membrane into the bloodstream of the embryo [22]. At the last step of the in ovo procedure, the hole punctured in the egg is sealed with a food-safe glue, which protects from water loss from the embryo. The egg tray is returned to the incubator within 20 minutes.
Animal trial
The trial was conducted on Ross 308 broiler chickens. Fertilized eggs (400 eggs) were incubated in standard conditions. On day 12 of incubation, the eggs were candled and viable embryos received an in ovo injection that contained either a biologically active compound (prebiotic) or physiological saline (mock injection). Experimental eggs (200 eggs) were injected in ovo with GOS. Control eggs (200 eggs) were mock-injected in ovo with 0.2 ml of physiological saline. All handling procedures of the eggs (punching hole, injecting liquid, sealing) were the same for GOS-injected and mock-injected eggs. After in ovo injection, incubation continued until hatching. One-day-old chicks were transported to the farm and distributed into pens (4 replicate pens/group, 10 birds/pen). The animals were reared in an open system, on a wooden litter with ad libitum access to feed and water. Table 1. shows the diet composition. On day 42 post-hatching randomly selected chickens (n = 10) were sacrificed by cervical dislocation and the intestinal samples were taken. The study was approved by the Local Ethics Committee for Animal Research (http://lke.utp.edu.pl) located at the Faculty of Animal Breeding and Biology, UTP University of Science and Technology in Bydgoszcz (study approval reference number 16/2014).
Table 1. Diet composition and nutritional value of the feed used for animal trial.
| Feeding phase | ||
|---|---|---|
| Ingredients (%) | Starter (1–21 days) | Grower (21–42 days) |
| Corn | 22 | 31.6 |
| Wheat | 19.5 | 15 |
| Soybean meal | 31.5 | 25 |
| Wheat middlings | 13 | 15 |
| Corn gluten | 10 | 10 |
| Soybean oil | 1.32 | 1.1 |
| Calcium Carbonate | 1.2 | 1 |
| Dicalcium Phosphate | 0.5 | 0.5 |
| NaCl | 0.2 | 0.2 |
| Sodium bicarbonate | 0.1 | 0.1 |
| Vitamin-mineral premix 1 1 | 0.3 | - |
| Vitamin-mineral premix 2 2 | - | 0.3 |
| Phytase | 0.1 | 0.1 |
| Coccidiostat | 0.1 | 0 |
| Color additives | 0.1 | 0.1 |
| Methionine | 0.08 | - |
| Calculated nutritional value of the diet (%) | ||
| Protein | 24.00 | 21.00 |
| Lipid | 4.50 | 4.50 |
| Crude fiber | 4.50 | 4.00 |
| Ash | 7.00 | 6.00 |
| Lysine, % | 1.10 | 1,00 |
| Methionine, % | 0.35 | 0.30 |
| Calcium, % | 1.30 | 1.10 |
| Available P, % | 0.70 | 0.60 |
| Sodium, % | 0.15 | 0.20 |
1Supplied per kilogram of diet: vitamin A, 12,000 IU; vitamin D3, 3,600 IU; vitamin E, 50.1 mg; vitamin B1, 3 mg; vitamin B12, 0.04 mg; vitamin B2, 6 mg; vitamin B6, 3.99 mg; CuSO4 5H2O (Cu, 10mg), 38.26mg; Ca(IO3)2 (I, 1.50mg), 2.31mg; FeCO3 (Fe, 45mg), 93.15mg; MnO (Mn, 36mg), 46.44mg; MnSO4 (Mn, 35mg),110.88mg; Na2SeO3 (Se, 0mg), 0.43mg; ZnO (Zn, 51mg), 63.24mg.
2Supplied per kilogram of diet: vitamin A, 10,000 IU; vitamin D3, 3,000 IU; vitamin E, 41.68 mg; vitamin B1, 2.90 mg; vitamin B12, 0.03 mg; vitamin B2, 5 mg; vitamin B6, 3.33 mg; CuSO4 5H2O (Cu, 8mg), 32.72mg; Ca(IO3)2 (I, 1.25mg), 1.93mg; Fe2O3 (Fe, 560mg), 800.8mg; FeCO3 (Fe, 38mg), 77.63mg; MnO (Mn, 30mg), 38.70mg; MnSO4 (Mn, 30mg), 92.40mg; Na2SeO3 (Se, 0mg), 0.36mg; ZnO (Zn, 43mg), 52.7mg.
During sampling the intestinal tract was excised and separated into four gut segments (duodenum, jejunum, ileum, and caecum). Each gut segment used for sampling contained approximately 5-cm-long fragment that was cut from the same sites in all animals. The duodenum was sampled in the middle of the duodenal loop (mid-duodenum). The jejunal segment was cut about 30–35 cm proximally from Meckel’s diverticulum (mid-jejunum). The ileal segment (distal ileum) was cut about 10–15 cm proximally to caeca. The caecum was sampled distal to caecum tonsil. Intestinal content was removed from each gut segment separately and frozen (-80°C) prior to bacterial DNA isolation. The gut segments were then rinsed in PBS and the mucosal layer was scraped with a glass slide. Mucosal scrapings were preserved in a DNA/RNA Stabilization Reagent (Fisher Molecular Biology, Terry Drive, PA, USA) prior to RNA isolation.
RNA and DNA isolation
Total bacterial DNA was isolated from the intestinal content. Approximately 100 mg of material was lysed and purified using a Genomic Mini AX Stool (A&A Biotechnology, Gdynia, Poland), according to manufacturer’s instruction. Next, isolated DNA was evaluated using the NanoDrop 2000 (Thermo Scientific Nanodrop Products, Wilmington, USA) and agarose gel electrophoresis. The evaluated DNA was diluted to a working concentration of 2 ng/μl and stored at 4°C prior to further analyses.
Total RNA was isolated from the mucosal scrapings sampled from the chicken intestines. About 100 mg of the mucosal scrapings were first homogenized in 1 ml of Trizol (Invitrogen, Carlsbad, USA) using a TissueRuptor homogenizer (Qiagen GmbH, Hilden, Germany). RNA was purified from the lysate using a Universal RNA Purification Kit (EURx, Gdańsk, Poland), according to manufacturer’s instruction. All RNA samples were evaluated using the NanoDrop 2000 (Thermo Scientific Nanodrop Products, Wilmington, USA) and agarose gel electrophoresis. Additionally, 10% of the RNA samples were evaluated on an Agilent 2100 Bioanalyzer using an Agilent RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA). RNA isolates were kept frozen at -20°C.
Bacteria quantification with qPCR
The relative abundances of Bifidobacterium spp. and Lactobacillus spp. in chyme samples from the duodenum, jejunum, ileum, and caecum were determined using quantitative PCR (qPCR) carried out on a LightCycler 480 II System (Roche-Diagnostics, Basel, Switzerland). A total reaction volume of 10 μl in a 384-well plate format contained 5 μl Maxima SYBR Green qPCR Master Mix (Thermo Scientific/Fermentas, Vilnius, Lithuania), 0.2 μM of each primer, specific to 16s rDNA of Bifidobacterium spp. (F: GCGTGCTTAACACATGCAAGTC, R: CACCCGTTTCCAGGAGCTATT) [44], Lactobacillus spp. (F: AGCAGTAGGGAATCTTCCA, R: CACCGCTACACATGGAG) [45, 46] or universal bacteria (F: ACTCCTACGGGAGGCAGCAGT, R: GTATTACCGCGGCTGCTGGCAC) [47] and 2 ng of bacterial DNA template. Each reaction was performed in four technical replicates. Thermal cycling consisted of initial denaturation at 95°C for 5 min, followed by 40 cycles of amplification: denaturation at 95°C for 15s, annealing at 58°C for 15s, and elongation at 72°C for 45s. Fluorescence was measured at the end of each extension step. After amplification, a melting curve was generated by increasing the temperature in small increments to 98°C and measuring the fluorescence of melting amplicons. Average Ct values of the four technical replicates obtained from the LightCycler 480 II System software were used for data analysis. Single Ct values differing by more than 0.3 were considered outliers. PCR efficiency for each primer pair was calculated in the LightCycler 480 II software based on the separate reaction of 5 dilutions (1x, 0.5x, 0.25x, 0.125x and 0.0625x) of pooled bacterial DNA template.
The relative abundances of the bacteria in the chyme were calculated as follows:
Relative Abundances [%] = (E universal)Ct universal / (E target)Ct target
Where, E universal is the efficiency of qPCR with primers for all bacteria, Ct universal is the Ct values for reaction with primers for all bacteria, E target is the efficiency of qPCR with primers specific for Bifidobacterium spp. or Lactobacillus spp., and Ct target is the Ct values for reaction with primers for Bifidobacterium spp. or Lactobacillus spp. [48].
Gene expression in intestinal mucosa
Gene expression analysis in the intestinal mucosa of chickens stimulated in ovo with GOS was performed for two gene panels. The full list and function of the genes are presented in Table 2. Panel (1) included genes related to intestinal innate immune responses, such as interleukins (IL-1β, IL-10 and IL-12p40) and HDP, defensins (AvBD1) and cathelicidins (CATHL2). Panel (2) contained genes involved in intestinal barrier function and nutrient sensing, including mucous compound (MUC6), tight junctions (TJ) proteins (CLDN1 and TJAP1), free fatty acids (FFA) receptors (FFAR2 and FFAR4) and glucose transporters (GLUT1, GLUT2 and GLUT5). Normalization of the expression levels of the target genes was performed with the geometric mean of two housekeeping genes: glucose-6-phosphate dehydrogenase (G6PDH) and beta actin (ACTB) [49]. Gene expression analysis was performed with two step reverse transcription quantitative PCR (RT-qPCR).
Table 2. Molecular function of the intestinal genes primer sequences used for RT-qPCR.
| Gene | Name | NCBI gene ID | Function in intestinal mucosa1 | Primer sequences 2 (5’ → 3’) |
Ref |
|---|---|---|---|---|---|
| Panel 1. Cytokine genes | |||||
| IL-1β | Interleukin 1 beta | 395196 | Pro-inflammatory cytokine produced by activated macrophages |
F: GGAGGTTTTTGAGCCCGTC R: TCGAAGATGTCGAAGGACTG |
this study |
| IL-10 | Interleukin 10 | 428264 | Anti-inflammatory cytokine, immunoregulator in the intestinal tract |
F: CATGCTGCTGGGCCTGAA R: CGTCTCCTTGATCTGCTTGATG |
[53] |
| IL-12p40 | Interleukin 12 subunit beta | 404671 | Cytokine with broad biological activities, activates T and NK cells, stimulates long-term immune responses |
F: TTGCCGAAGAGCACCAGCCG R: CGGTGTGCTCCAGGTCTTGGG |
[54] |
| Panel 2. Host defence peptide (HDP) genes | |||||
| AvBD1 | Avian beta-defensin 1 | 395841 | Host defence peptide involved in resistance of epithelia to microbial colonization |
F: AAACCATTGTCAGCCCTGTG R: TTCCTAGAGCCTGGGAGGAT |
this study |
| CATHL2 | Cathelicidin 2 | 420407 | Host defence peptide, inflammatory response regulator, functions in chemotaxis |
F: AGGAGAATGGGGTCATCAGG R: GGATCTTTCTCAGGAAGCGG |
this study |
| Panel 3. Barrier function genes | |||||
| MUC6 | Mucin 6 | 414878 | Forms insoluble mucin barrier that protects gut lumen and modulates mucus composition |
F: TTCAACATTCAGTTCCGCCG R: TTGATGACACCGACACTCCT |
this study |
| CLDN1 | Claudin 1 | 424910 | Component of tight junctions, regulates permeability of epithelia and water homeostasis |
F: TCTTCATCATTGCAGGTCTGTC R: AACGGGTGTGAAAGGGTCAT |
this study |
| TJAP1 | Tight junction associated protein 1 | 421455 | Component of tight junctions, takes part in vesicular trafficking |
F: AGGAAGCGATGAATCCCTGTT R: TCACTCAGATGCCAGATCCAA |
this study |
| Panel 4. Nutrient sensing genes | |||||
| FFAR2 | Free fatty acid receptor 2 | 100859369 | Receptor for short chain free fatty acids, regulates energy homeostasis through adipogenesis and controls intestinal immunity |
F: GCTCGACCCCTTCATCTTCT R: ACACATTGTGCCCCGAATTG |
this study |
| FFAR4 | Free fatty acid receptor 4 | 428963 | Receptor for medium and long chain free fatty acids (e.g., omega-3), represses macrophage-induced tissue inflammation |
F: AGTGTCACTGGTGAGGAGATT R: ACAGCAACAGCATAGGTCAC |
this study |
| GLUT1 | Glucose transporter 1 | 396130 | Basal, constitutive glucose transporter with broad substrate sensitivity |
F:AGATGACAGCTCGCCTGATG R:GTCTTCAATCACCTTCTGCGG |
this study |
| GLUT2 | Glucose transporter 2 | 396272 | Na(+)/glucose cotransporter in the small intestine, glucose sensor |
F:GGAGAAGCACCTCACAGGAA R:CAGGCTGTAACCGTACTGGA |
this study |
| GLUT5 | Glucose transporter 5 | 419438 | Fructose uptake by small intestine |
F:ACGGTTCCCAGAGCAAGTTA R:GTCTTGCATGTATGGGGCTG |
this study |
| 5. Reference genes | |||||
| ACTB | Actin, beta | 396526 | Ubiquitous cytoskeletal actin involved in cell motility, structure, integrity and intercellular signalingsignalling |
F: CACAGATCATGTTTGAGACCTT R: CATCACAATACCAGTGGTACG |
[49] |
| G6PDH | Glucose-6-Phosphate Dehydrogenase | 428188 | Cytosolic enzyme that produces NADPH in reductive biosynthetic reactions |
F: CGGGAACCAAATGCACTTCGT R: GGCTGCCGTAGAGGTATGGGA |
[49] |
1 gene function derived from GeneCards (http://www.genecards.org)
2 F–Forward primer, R–Reverse primer
For gene expression analysis, 1250 ng of total RNA of each sample was reversely transcribed to cDNA using Maxima First Strand cDNA Synthesis kit (Thermo Scientific/Fermentas, Vilnius, Lithuania). RT was performed at the volume of 10 μl, including 2 μl of 5x Reaction Mix and 1 μl of Maxima Enzyme Mix. Next, qPCR reactions were prepared at 10μL total volume in a 384-well plate format and contained: 5μl of Maxima SYBR Green qPCR Master Mix (Thermo Scientific/Fermentas, Vilnius, Lithuania), 1μM of forward and reverse primers, and 140 ng of cDNA. Oligonucleotide primers (Table 1) were designed using an NCBI/Primer-BLAST [50]. If it was possible, amplicon sequences spanned exon-exon junctions. RT-qPCR reactions were performed in the LightCycler 480 System (Roche-Diagnostics, Basel, Switzerland) and consisted of: initial denaturation at 95°C for 15 min, 40 cycles of amplification (denaturation at 95°C for 15s, annealing at 58°C for 20s, and elongation at 72°C for 20s) and melting curve. The annealing temperature was 58°C for all target genes except IL12p40, for which it was 65°C. Fluorescence was measured at the end of each elongation step. The melting curve was generated by increasing the temperature in small increments up to 98°C and measuring the fluorescence of the melting amplicon.
Relative gene expression was calculated with ΔΔCt algorithm and the amount of the target gene was calculated with formula 2–ΔΔCt [51]. A Multiexperiment Viewer (MeV) version 4.9 [52] was used to create a Hierarchical Cluster Tree based on fold change values. After loading data, results were clustered using the Hierarchical Clustering function with the standard option (Pearson Correlation) to prepare a heat map.
Statistical analysis
Statistical analysis was conducted with the SAS statistical software version 9.4 (SAS Institute, Cary, NC, USA). In both datasets (microbial communities and intestinal gene expression), the significance of the effects and their interaction was analysed with two-way ANOVA. The effects were: in ovo injected group (GOS vs. C) and intestinal segment (duodenum, jejunum, ileum, and caecum). An HSD Tukey post hoc test was used to determine differences in gene expression. Significance thresholds P < 0.05, 0.01 and 0.001 were used.
Results
Relative abundance of Lactobacillus spp. and Bifidobacterium spp.
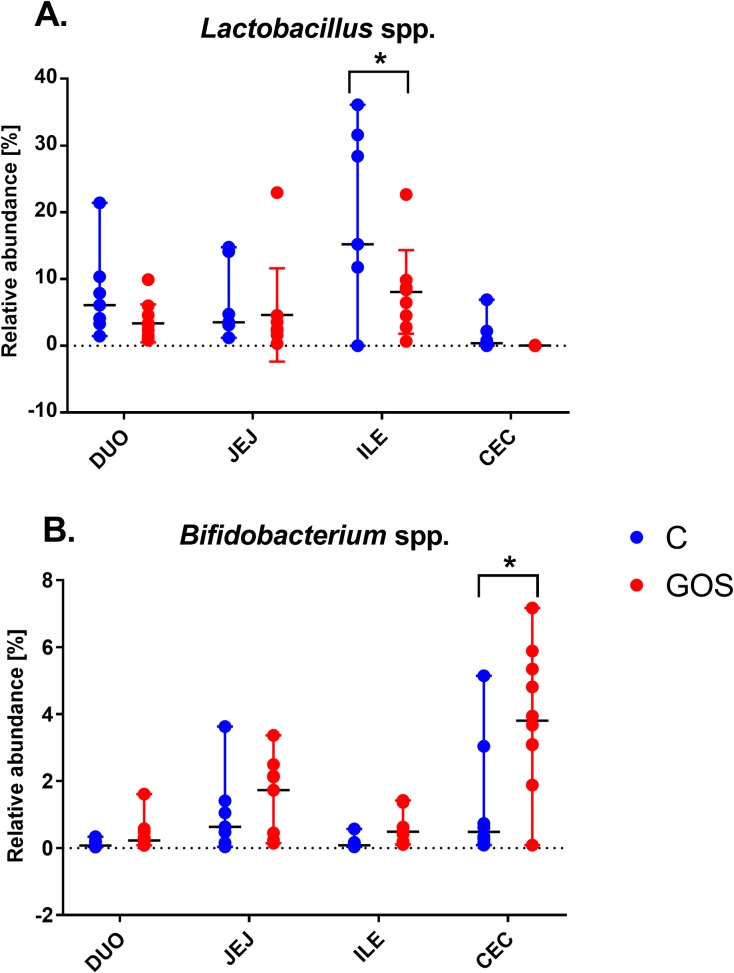
All the raw data obtained in this study have been made available in S1 File. Intestinal segment (P < 0.001) and prebiotic treatment in ovo (P < 0.01) had a significant effect on the relative abundance of microbial communities (Bifidobacterium and Lactobacillus) in the chyme. In Bifidobacterium spp. we found significant interaction between the intestinal section and prebiotic supplementation (P < 0.05). Fig 1 presents the relative abundance of (A) Bifidobacterium spp. and (B) Lactobacillus spp. in four sections of intestinal content (duodenum, jejunum, ileum, and caecum). Average values of Bifidobacterium ranged from 0.03% (in ileum) to 1.3% (in caecum) of C and from 0.4% (in ileum) to 3.9% (in caecum) of the GOS-injected group. The highest relative abundance of Bifidobacterium spp. (~4%) was found in caecum of GOS. It was significantly higher than caecum of C (P < 0.001) but also significantly higher than in other intestinal segments (P < 0.05).
Fig 1. The relative abundance of Bifidobacterium spp.
(A) and Lactobacillus spp. (B) in chyme from different sections of GIT in chickens supplemented in ovo with GOS prebiotic. GOS was delivered in ovo on day 12 of egg incubation. Samples of intestinal content were collected from 42-days-old chickens. Intestinal content was sampled from four distinct intestinal segments (duodenum, jejunum, ileum, and caecum) (n = 10). Bacteria quantification was done with qPCR based on bacterial DNA isolated from chyme. Statistical analysis was performed with two-way ANOVA with a Tukey HSD post hoc test. Significant differences found at P < 0.05.
There were more pronounced differences between treatment groups and intestinal section for Lactobacillus spp. There were higher values of Lactobacillus abundance in C vs. GOS. Median values of Lactobacillus spp. ranged from 0.40% (in caecum) to 15.20% (in ileum) of C and from 0.02% (in caecum) to 8.36% (in ileum) of GOS. GOS decreased Lactobacillus spp. along all GIT. The largest lactobacilli populations in this trial were found in ileum of C (P < 0.05).
Intestinal gene expression
Hierarchical clustering of gene expression profiles
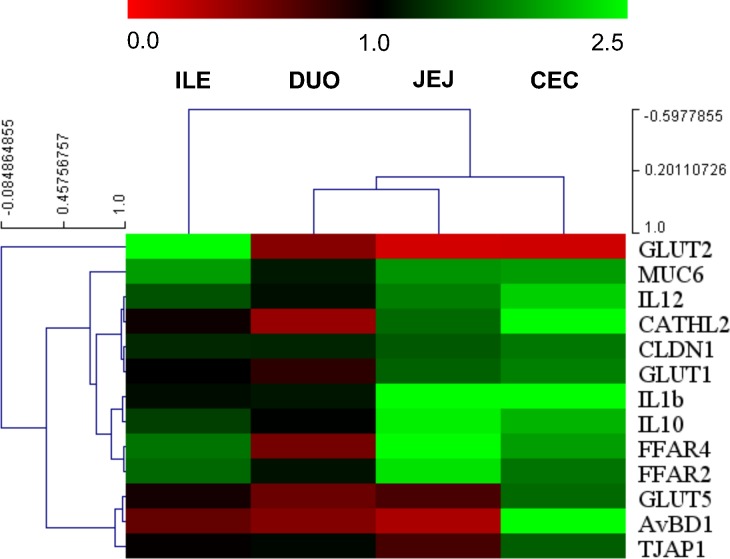
Hierarchical gene and sample clustering allowed determining the relationships between the samples and co-regulated genes. Based on correlation data of intestinal gene expression in GOS vs. C (Fig 2), we have determined that the pattern of mRNA expression was similar in jejunum and caecum. Three major gene clusters were identified, consisting of (1) IL-12p40, CATHL2, CLDN1 and GLUT1; (2) IL-1β, IL-10, FFAR2 and FFAR4; and (3) GLUT5, AvBD1 and TJAP1.
Fig 2. A heat map of hierarchically clustered gene expression in different segments of intestinal mucosa in chicken treated with GOS in ovo.
Intestinal segments: DUO–duodenum, JEJ–jejunum, ILE–ileum, and CEC–caecum. In ovo injection was carried out on day 12 of egg incubation. Intestinal samples (n = 10) collected from chickens on day 42 post-hatching. RT-qPCR data were generated with custom-designed primers used for amplification with SYBR green dye; Glucose-6-phosphate dehydrogenase (G6PDH) and beta-actin (ACTB) were used as reference genes; relative gene expression (fold change) calculated as 2–ΔΔCt. A Multiexperiment Viewer version 4.9 (MeV) was used for constructing a Hierarchical Cluster Tree based on fold change. Colours (red-black-green) show relative gene expression changes in GOS vs. C (red: down-regulated, green: up-regulated genes).
Effects of in ovo treatment and intestinal segment on gene expression
Effects of the in ovo treatment group and intestinal segment on mRNA gene expression in chicken intestinal mucosa were included in ANOVA (Table 3). The expression of every gene analysed depended on either variable (P < 0.05 or lower). Interaction was found significant for IL-1β, TJAP1, FFAR4, GLUT1, and GLUT2 (P < 0.05 or lower).
Table 3. Effects of experimental group, intestinal segment and their interaction on mRNA expression of immune, barrier function and nutrient sensing genes in chicken intestinal mucosa.
| Gene | Treatment 1 | Intestine 2 | Treatment x Intestine 3 |
|---|---|---|---|
| Panel 1. Cytokine genes | |||
| IL-1β | < 0.001 | < 0.001 | < 0.01 |
| IL-10 | < 0.001 | < 0.01 | ns |
| IL-12p40 | < 0.001 | < 0.001 | ns |
| Panel 2. Host defence peptide (HDP) genes | |||
| AvBD1 | ns | <0.001 | ns |
| CATHL2 | ns | <0.01 | ns |
| Panel 3. Barrier function genes | |||
| MUC6 | < 0.01 | < 0.001 | ns |
| CLDN1 | < 0.01 | < 0.001 | ns |
| TJAP1 | Ns | < 0.001 | <0.05 |
| Panel 4. Nutrient sensing genes | |||
| FFAR2 | < 0.001 | ns | ns |
| FFAR4 | < 0.05 | < 0.001 | < 0.01 |
| GLUT1 | < 0.05 | < 0.001 | < 0.05 |
| GLUT2 | ns | < 0.001 | < 0.01 |
| GLUT5 | ns | < 0.001 | ns |
Effects
1 in ovo delivery of GOS prebiotic (GOS vs C)
2 intestinal segment (duodenum, jejunum, ileum, or caecum) from which a chyme sample was collected and
3 the interaction between in ovo treatment and the intestinal segment on mRNA expression of immune and physiological genes in chicken mucosa. Gene expression analysis done with RT-qPCR. Significance of effects calculated with two-way ANOVA. Significance levels: P < 0.05, 0.01 or 0.001.
Relative expression of intestinal immune-related genes
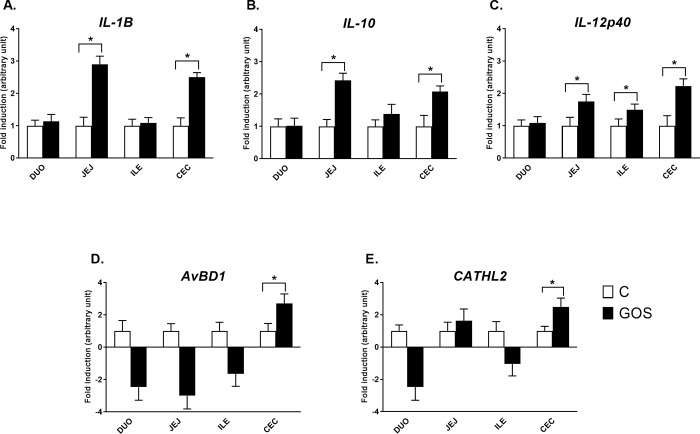
The relative expression of the intestinal innate immune response genes presented two patterns of gene expression regulation (Fig 3). Cytokine genes (IL-1β, IL-10 and IL-12p40) were up-regulated in jejunum and caecum of GOS (P < 0.05). The expression profile of those cytokines in duodenum and ileum did not deviate much from C, except IL-12p40, which was slightly up-regulated also in ileum (P < 0.05). HDP genes (AvBD1 and CATHL) were significantly up-regulated only in caecum of GOS (P < 0.05). In other intestinal segments (duodenum, jejunum, and ileum), those genes were down-regulated in GOS, but beyond statistical significance (P > 0.05). The fold change of the gene expression of significantly up-regulated immune genes in caecum ranged between 2.07 (IL-10) and 2.70 (AvBD1).
Fig 3. Relative mRNA expression of intestinal immune-related genes in different segments of intestinal mucosa in chickens injected in ovo with GOS.
The panel includes genes encoding cytokines, (A) IL-1β, (B) IL10 and (C) IL12p40; and host defence peptides, (D) AvBD1 and (E) CATHL. Intestinal segments: DUO–duodenum, JEJ–jejunum, ILE–ileum, and CEC–caecum. In ovo treatment groups: control (white bars), injected in ovo with physiological saline; GOS (black bars)–injected in ovo with galactooligosaccharides. In ovo injection was carried out on day 12 of egg incubation. Intestinal samples (n = 10) were collected from chickens on day 42 post-hatching. RT-qPCR data were generated with custom-designed primers used for amplification with SYBR green dye; Glucose-6-phosphate dehydrogenase (G6PDH) and beta-actin (ACTB) were used as reference genes; relative gene expression calculated as 2–ΔΔCt; Two-way ANOVA with post hoc HSD Tukey test was used to compare the groups. Asterisk indicates pair-wise significant differences (P < 0.05). The results of fold induction less than 1 have been transformed by the formula -1/fold induction.
Relative expression of barrier function and nutrient sensing genes
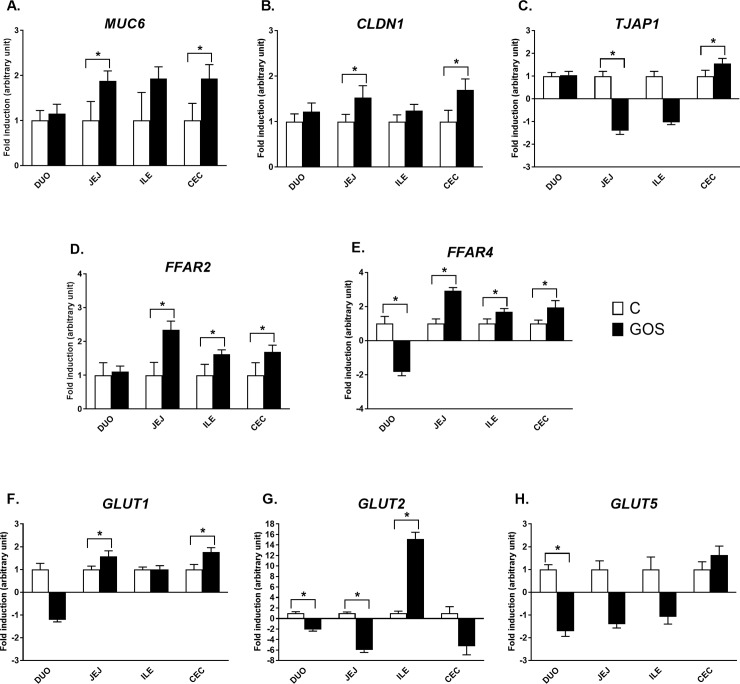
The mRNA expression of barrier function and nutrient sensing was varied and dependent mostly on intestinal segment (Fig 4). MUC6 and CLDN1 followed a similar pattern of gene expression regulation, with moderate (FC ~1.5–2.0) but significant up-regulation in jejunum and caecum of GOS (P < 0.05). TJAP1 had a distinct pattern of mRNA expression, also with moderate but significant down-regulation in jejunum (FC -1.4, P < 0.05) and up-regulation in caecum (FC 1.6, P < 0.05) of GOS. Free fatty acid receptors (FFAR2 and FFAR4) were up-regulated in jejunum, ileum, and caecum of GOS (P < 0.05). FFAR4 was significantly down-regulated in duodenum of GOS (P < 0.05). The most pronounced changes were determined in GLUT2, which was up-regulated in ileum of GOS (FC 15.2, P < 0.05), but down-regulated in duodenum (FC -2.1, P < 0.05), jejunum (FC -6, P < 0.05) and (suggestively) in caecum of GOS (FC -5.3, P > 0.05). GLUT1 was up-regulated in jejunum and caecum of GOS (P > 0.05), which seemed like a general pattern of expression regulation dependent on in ovo treatment. GLUT5 was significantly down-regulated only in duodenum of GOS (P < 0.05), with a slight but not significant fluctuation in mRNA expression in other intestinal segments.
Fig 4. Relative mRNA expression of barrier function and nutrient sensing genes in different segments of intestinal mucosa in chickens injected in ovo with GOS.
The panel includes genes encoding mucin, (A) MUC6; tight junctions components, (B) CLDN1 and (C) TJAP1; free fatty acid receptors, (D) FFAR2 and (E) FFAR4; and glucose transporters, (F) GLUT1, (G) GLUT2 and (H) GLUT5. Intestinal segments: DUO–duodenum, JEJ–jejunum, ILE–ileum and CEC–caecum. In ovo treatment groups: control (white bars)–injected in ovo with physiological saline; GOS (black bars)–injected in ovo with galactooligosaccharides. In ovo injection was carried out on day 12 of egg incubation. Intestinal samples (n = 10) were collected from chickens on day 42 post-hatching. RT-qPCR data were generated with custom-designed primers used for amplification with SYBR green dye; Glucose-6-phosphate dehydrogenase (G6PDH) and beta-actin (ACTB) were used as reference genes; relative gene expression calculated as 2–ΔΔCt; Two-way ANOVA with a post hoc HSD Tukey test was used to compare the groups. Asterisk indicates pair-wise significant differences (P < 0.05). The results of fold induction less than 1 have been transformed by the formula -1/fold induction.
Discussion
We have evaluated the impact of in ovo stimulation of chicken embryos with GOS prebiotic on the development of microbial populations along the GIT as well as the modulatory role in mucosal gene expression. We have demonstrated that a single injection of GOS on day 12 of egg incubation can efficiently and stably promote the growth of certain species of indigenous microflora, which has life-long consequences for the intestinal environment of broiler chickens. As such, these data provide insight into the spatial and functional interaction at the interface between intestinal microflora and mucosa.
Bifidogenic effect of GOS in chicken
The largest abundance of Bifidobacterium spp. was found in caecum, and of Lactobacillus spp., in ileum. This finding is supported by Bjerrum et al. [55], who identified Lactobacillus as the predominant species in chicken ileum, whereas caecum was inhabited by more diverse microbial communities, including Bifidobacterium spp. The primary and continuous source of Lactobacillus spp. in ileum is the crop, which is almost exclusively colonised by this genus (relative abundance within 97–98%) throughout the chicken lifespan [56]. From the crop, Lactobacillus spp. is shed down the GIT and inhabits the small intestine, becoming the predominant species, especially in the ileum. Intestinal microflora starts developing in perinatal period, but its most rapid growth and diversification increases with age, during environmental exposition of the host [57]. Caecal microflora are more numerous and diverse compared to that from the upper guts and its function reflects the large intestine in mammalian species [57]. It is colonised by obligately anaerobic bacteria, such as Clostridia, which successively exclude Lactobacillus spp. [56].
In ovo delivery of GOS prebiotic into the chicken embryos increased the relative abundance of Bifidobacterium spp. in the caecum and decreased the relative abundance of Lactobacillus spp. in the ileum. Competitive exclusion of Lactobacillus spp. can be attributed to bifidogenic effect of GOS prebiotic. GOS used in this study was obtained from lactose by β-galactosidase activity of Bifidobacterium bifidum NCIMB 41171 [43]. For this reason GOS specifically promotes growth of Bifidobacterium spp. [43]. Due to the complex carbohydrate structure of GOS, it passes the upper GIT without degradation [58]. The genome of Bifidobacterium spp. encodes carbohydrate-degrading enzymes with high affinity to GOS [59]. Jung et al. (2008) found that in-feed supplementation with high proportion of GOS (12%) significantly increased faecal lactobacilli and bifidobacteria counts [36]. GOS supplementation and Salmonella spp. challenge diverged microbiome diversity of the caecal content, but those differences disappeared by day 40 post-infection [60]. GOS delivered in ovo on day 12 of egg incubation increased the lactobacilli and bifidobacteria faecal counts of 1-day-old chicks [28]. Here we shown the effects of microbiota modulation with in ovo stimulation in adult broiler chickens.
Gene expression in jejunum and caecum
We have found that the administration of GOS had the most pronounced effects on gene expression regulation in jejunum and caecum. Both jejunum and caecum are of particular importance in avian GIT [61]. Most of the digestive and absorptive functions take place in jejunum. Jejunum is also the heaviest part of the small intestine. Even though nutrient absorption is a key physiological process, it happens quite rapidly—chyme passage time along jejunum is from 40 to 60 min.Due to availability of nutrients, jejunum is also target for both commensal bacteria and enteric pathogens. Shokker et al (2011) found correlation between Salmonella infection and jejunal gene expression [62]. Functions of jejunum in developing birds was characterized based on transcriptomic modulation; by day 21 post-hatching it was assigned to immune regulation [63]. These results are in line with this study in which we found immune-related gene expression that formed a jejunal cluster. Caecum is very distinct from jejunum in its function and physiology [9]. It takes part in electrolyte and water absorption but also allows for prolonged retention of chyme from two sources—jejunum and colon [9]. As such, it is caecum that is colonised by the majority of chicken intestinal microflora, which conveys numerous beneficial effects to the host [9]. Caecal gene expression been widely studied in chicken. Volf et al. (2017) compared germ-free, mono-associated and fully colonised chickens and determined that there was significant correlation between caecal microbiome and immunoglobulin expression [64]. Rychlik et al. (2014) reviewed the gene expression modulation in chicken caecal mucosa upon Salmonella challenge [65]. The authors confirmed the TLR pathway of bacterial recognition and pinpointed genes highly inducible in response to the enteric infection, such as metaloproteinases or cytokines. Based on transcriptomic caecal data, Higgins et al. (2011) inferred that probiotics clear Salmonella infection by inducing apoptosis in the caecal cells [66].
Cytokine genes
In ovo stimulation with GOS increased mRNA expression of IL-1β, IL-10 and IL-12p40 cytokine genes in jejunum and caecum. In gut health, IL-1β plays dichotomous role, exerting both classic pro-inflammatory as well as protective functions [67]. The transient and low expression of IL-1β mediates beneficial effects in the intestinal mucosa and manifests itself in faster healing of epithelia in case of damage. Intestinal IL1-β is expressed constitutively in lamina propria cells and secreted into intestinal lumen in a condition of intestinal health as well as disease [68]. The latter drastically increases IL-1β abundance, making this cytokine a key mediator of intestinal inflammation [68]. IL1-β expression is not only modulated by pathogenic bacteria but also by healthy gut microbiome [69]. Animals primed with healthy microflora mount higher abundance of IL-1β in the guts in comparison to animals with sterile guts [69]. Commensal microbiota inhabiting the guts stimulate macrophages residing in lamina propria to express pro-IL-1β, which is an inactive form of IL-1β [70]. Activation of pro-IL-1β by cleavage into IL-1β is executed by inflammasome [71]. On the other hand, presence of inflammasome guarantees maintaining microbiome and gut homeostasis [71]. In this study mRNA abundance of IL-1β was enhanced by in ovo delivered GOS and this effect was related to the intestinal fragment. Hereby we claim that the treatment primed gut mucosa towards immunostimulation.
In the literature regarding intestinal homeostasis, the synthesis of pro-inflammatory cytokine IL-12p40 is often compared to the level of anti-inflammatory cytokine IL-10 [72]. It allows determination of the IL-10/IL-12p40 ratio, which is indicative of the immunoregulatory (anti-inflammatory) vs. immunostimulatory (pro-inflammatory) function exerted by host-microbiome interaction in the guts [72]. Some probiotic strains are able to skew the IL-12p40/IL-10 ratio towards either of the cytokines, which can either boost immune response and activate the pro-inflammatory cascade or limit it and thus prevent the damage that excessive inflammation can do to the host. This model reflects cell polarization by luminal agents like probiotics [73]. We have previously demonstrated that in ovo stimulation with different prebiotics and synbiotics led to down-regulation of the immune-related gene expression in caecal tonsils [31, 74, 75]. Haghighi et al. (2008) also found multi-strain probiotic given to chicks on day of hatching reduced IL-12 gene expression in caecal tonsils of chickens challenged with Salmonella [76]. On the other hand, Kogut et al. (2013) demonstrated that feeding a peptide isolated from Brevibacillus texasporus probiotic to the chickens primed their immune responses to consecutive Salmonella challenge [77]. The dynamics between intestinal flora, environmental impact and immune status of the host is not completely understood. Maintaining the balance between activation and suppression of the immune responses upon microflora modulation will probably be a key point in the future research.
Barrier function genes
HDP belong to broad-spectrum and evolutionary-conserved innate immunity effector molecules, referred to in the past as antimicrobial proteins [78]. HDP expressed by intestinal mucosa take part in intestinal innate immunity and mucosal defence [79]. Intestinal HDP are bound with mucins, creating a firm immunological and mechanical barrier between the host and the intestinal antigens [79]. Their major function is antimicrobial activity, but HDP also play other roles, including immunomodulation, chemotaxis, or wound repair [80]. HDP gene expression depends on microbial modulation. Abkari et al. (2008) demonstrated that gene expression of β-defensins and cathelicidins was triggered by Salmonella challenge in cecal tonsils broiler chicks[81]. The same genes were suppressed to the negative control level by earlier oral supplementation with multi-strain probiotics. The authors attribute such modulatory effects of probiotics on HDP gene expression to competitive exclusion of Salmonella by probiotic strains [81]. Other authors also found relation between intestinal HDP gene expression microbiota composition, including and Salmonella [82] or E. coli [64] challenge[64]. [82]
Mucins contribute to maintaining gut-barrier function and protect from enteric pathogens. Mucins are secreted into intestinal lumen and form a protective layer, which is resistant to proteolysis [83]. Goblet cells produce mucins in reaction to microbial stimuli. Prebiotics can alter the composition of the intestinal microflora, which in turn stimulates goblet cells towards mucin production [84]. In this study, GOS delivered in ovo mediated increase in MUC6 expression in mucosa of jejunum and cecum. These results are in concordance with Bogucka et al. (2017), who demonstrated that GOS delivered in ovo increased the number of goblet cells in jejunum and ileum (caecum was not analysed) of 35-day-old broiler chickens [41]. Chicken mucin has confirmed in vitro cytotoxic activity against Salmonella [85]. Gut-barrier failure in chickens is associated with impaired expression of MUC2, a major intestinal gel-forming mucin [86]. Improved expression of MUC2 under LPS challenge was mediated by Bacillus-based probiotics [87]. Smirnov et al. (2006) also found in ovo feeding with carbohydrates (applied intraamniotically on day 17.5 of egg incubation) increased Goblet cells number and MUC2 expression in jejunum [88].
Integrity of intestinal mucosa is formed with epithelial cells connected with TJ proteins [89]. Intestinal permeability and its interaction with gut microbiota has been reviewed by Bischoff et al. [90]. Dysbiosis, defined as microbial imbalance, reduces intestinal integrity of the intestinal epithelia. When it happens and the host-microbiome barrier becomes faulty, intestinal antigens and lipopolysaccharides (LPS) leak into millieu of the body and cause inflammatory responses. Loosened TJ strands are major cause of the “leaky guts” [89]. Claudins constitute a key component of the TJ strand. They bind peripheral membrane proteins, including scaffold proteins, for example, zonula occludens protein 1 (ZO-1) [91]. Tight junction associated protein 1, encoded by the TJAP1 gene, is incorporated later in time into the TJ strand, after the claudin-based junction has been established [92]. Enteric pathogens such as Salmonella or Campylobacter often target integrity of the tight junctions [93]. On the other hand, prebiotics and probiotics are known to stimulate expression of different TJ genes [87], including claudins [94]. CLDN-1 encodes a barrier-forming claudin. Its increased expression leads to tightening the epithelial cells.
Nutrient sensing genes
Intestinal nutrient sensing takes part in regulating gut hormones release based on gut content. Proteins encoded by FFAR2 and FFAR4 regulate glucose and lipids metabolism [95]. In particular, they mediate the release of anorectic gut hormones (incretin hormones) from epithelia into the intestinal lumen [96]. One of those incretins, glucagon‐like peptide‐1 (GLP‐1), regulates energy uptake via the brain-gut axis [96]. GLP-1 is produced by the intestine right after nutrient ingestion to promote satiety and decrease further feed uptake [96]. Down-regulation of FFAR4 in duodenum of GOS suggests that GLP-1 activity was inhibited. Kolodziejski et al. [97] determined that GOS-based synbiotic delivered in ovo significantly reduced mRNA expression of GLP-1 in duodenal content and GLP-1 protein in blood serum of adult broiler chickens. Down-regulation of GLP-1 pathways via FFAR could be one of the mechanisms explaining increased body weight in chickens stimulated in ovo with GOS [98].
FFAR genes serve also as SCFA receptors. FFAR2 encodes GPR43, which is a receptor of SCFA, such as propionate, acetate and butyrate [99], end products of fibre fermentation by intestinal bacteria. FFAR4 encodes GPR120, which recognizes long-chain unsaturated fatty acids, including omega-3, which are usually of dietary origin [100]. Increase in both, FFAR2 and FFAR4 along the GIT (except duodenum) suggests increased FFA content. SCFA are not only energy source but they can also regulate inflammatory immune responses in the guts [101]. Activation of FFAR2 and FFAR4 by free fatty acids leads to increased cytokine production by intestinal epithelial cells [95, 101]. It was also demonstrated that SCFA activates chicken monocyte/macrophage cell line (HD11) [102]. By connecting metabolic and immune effects, FFAR genes are important biomarkers of the intestinal microflora activity.
Glucose transporters take part in the absorption of glucose through the membrane of epithelial cells in a passive way, i.e., down the gradient of glucose concentration. GLUT1 is a high-affinity constitutive glucose transporter. GLUT2 is a low-affinity intestinal glucose co-transporter. It has been manifested that expression of GLUT2 is activated by butyrate, which is one of the SCFA produced by intestinal microflora [103]. Also, studies on gnotobiotic mice showed that the presence of Clostridium ramosum activated GLUT2 expression in jejunum [104]. However, the exact mechanisms are still unknown. Kolodziejski et al. [97] found that the digestive ability of duodenum was improved by in ovo delivery of GOS-based synbiotic, which manifested itself by an increase in trypsin and lipase activity in duodenal chyme, followed by dampening 70% of the activity of the amylase (P < 0.05). Based on this finding we hypothesize that there was less glucose available in duodenum as a source of carbon, which could have inhibited expression GLUT2 and GLUT5 in GOS-stimulated group.
Conclusions
This paper provides insights into the mechanisms of the beneficial effects of the in ovo stimulation with GOS prebiotic in chickens. The effects were analysed on microbial and mucosal sites of the chicken GIT. Microbial stimulation with GOS delivered in ovo was manifested by increased abundance of Bifidobacterium spp. in caecum, and decreased abundance of Lactobacillus spp. in ileum. Mucosal gene expression was modulated mainly in jejunum and caecum. GOS delivered in ovo increased expression of the cytokine genes, barrier function genes and free fatty acid receptors. Varied expression of the glucose transporter genes indicated that GOS induced also modulation of energy metabolism. In conclusion, in ovo stimulation induced long-term, beneficial effects on the chicken gut homeostasis.
Supporting information
qPCR dataset for calculating (A) bacterial and (B) gene abundances.
(XLSX)
Acknowledgments
George Tzortzis from Clasado Biosciences Ltd. is kindly acknowledged for delivering GOS for the experiment.
Data Availability
All relevant data are within the paper and its Supporting Information files (S1 File).
Funding Statement
This study was supported by Ministry of Education, Universities and Research (MIUR) in Rome, Italy (OVOBIOTIC, grant number RBSI14WZCL), AS; and National Science Centre (NSC) in Cracow, Poland (grant number UMO-2013/11/B/NZ9/00783), AS. The study was published with support from the Statutory Research of the Department of Animal Biotechnology and Genetics, BS 10/2017. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Military Medical Research. 2017;4(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–70. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon M, Bird A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7(1):17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Current gastroenterology reports. 2010;12(5):319–30. 10.1007/s11894-010-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duke G. Alimentary canal: anatomy, regulation of feeding, and motility Avian physiology: Springer; 1986. p. 269–88. [Google Scholar]
- 6.Yegani M, Korver DR. Factors affecting intestinal health in poultry. Poultry Sci. 2008;87(10):2052–63. 10.3382/ps.2008-00091 WOS:000259403000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Befus AD, Johnston N, Leslie GA, Bienenstock J. Gut-associated lymphoid tissue in the chicken. I. Morphology, ontogeny, and some functional characteristics of Peyer's patches. J Immunol. 1980;125(6):2626–32. . [PubMed] [Google Scholar]
- 8.Kelsall B. Innate and adaptive mechanisms to control of pathological intestinal inflammation. The Journal of pathology. 2008;214(2):242–59. 10.1002/path.2286 [DOI] [PubMed] [Google Scholar]
- 9.Gabriel I, Lessire M, Mallet S, Guillot J. Microflora of the digestive tract: critical factors and consequences for poultry. World's poultry science journal. 2006;62(3):499–511. [Google Scholar]
- 10.Stanley D, Hughes RJ, Moore RJ. Microbiota of the chicken gastrointestinal tract: influence on health, productivity and disease. Appl Microbiol Biot. 2014;98(10):4301–10. [DOI] [PubMed] [Google Scholar]
- 11.Wei S, Morrison M, Yu Z. Bacterial census of poultry intestinal microbiome. Poult Sci. 2013;92(3):671–83. 10.3382/ps.2012-02822 . [DOI] [PubMed] [Google Scholar]
- 12.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- 13.Pourabedin M, Zhao X. Prebiotics and gut microbiota in chickens. FEMS microbiology letters. 2015;362(15). [DOI] [PubMed] [Google Scholar]
- 14.Micciche AC, Foley SL, Pavlidis HO, McIntyre DR, Ricke SC. A Review of Prebiotics Against Salmonella in Poultry: Current and Future Potential for Microbiome Research Applications. Frontiers in veterinary science. 2018;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricke SC. Focus: Nutrition and Food Science: Impact of Prebiotics on Poultry Production and Food Safety. The Yale journal of biology and medicine. 2018;91(2):151 [PMC free article] [PubMed] [Google Scholar]
- 16.Chistiakov DA, Bobryshev YV, Kozarov E, Sobenin IA, Orekhov AN. Intestinal mucosal tolerance and impact of gut microbiota to mucosal tolerance. Frontiers in microbiology. 2015;5:781 10.3389/fmicb.2014.00781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microbial ecology in health and disease. 2015;26(1):26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R, Nauta A, Ben Amor K, Knippels L, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Beneficial microbes. 2010;1(4):367–82. 10.3920/BM2010.0027 [DOI] [PubMed] [Google Scholar]
- 19.Simon K, Verwoolde MB, Zhang J, Smidt H, de Vries Reilingh G, Kemp B, et al. Long-term effects of early life microbiota disturbance on adaptive immunity in laying hens. Poult Sci. 2016;95(7):1543–54. 10.3382/ps/pew088 . [DOI] [PubMed] [Google Scholar]
- 20.Schokker D, Jansman AJ, Veninga G, de Bruin N, Vastenhouw SA, de Bree FM, et al. Perturbation of microbiota in one-day old broiler chickens with antibiotic for 24 hours negatively affects intestinal immune development. BMC Genomics. 2017;18(1):241 10.1186/s12864-017-3625-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roto SM, Kwon YM, Ricke SC. Applications of In Ovo Technique for the Optimal Development of the Gastrointestinal Tract and the Potential Influence on the Establishment of Its Microbiome in Poultry. Front Vet Sci. 2016;3:63 10.3389/fvets.2016.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siwek M, Slawinska A, K. S, Bogucka J, Dunislawska A, Bednarczyk M. Prebiotics and synbiotics—in ovo delivery for improved lifespan condition in chicken. BMC Veterinary Research. 2018;14 10.1186/s12917-018-1738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou T, Tako E. The In Ovo Feeding Administration (Gallus Gallus)—An Emerging In Vivo Approach to Assess Bioactive Compounds with Potential Nutritional Benefits. Nutrients. 2018;10(4):418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uni Z, Ferket P, Tako E, Kedar O. In ovo feeding improves energy status of late-term chicken embryos. Poultry Sci. 2005;84(5):764–70. [DOI] [PubMed] [Google Scholar]
- 25.Palermo-Neto J, Calefi A, Aloia T, Gomes A, Pinheiro M, Ribeiro A, et al. 63. Heat stress impairs performance parameters, immunity and increases Salmonella enteritidis migration to spleen of broilers chickens. Brain, Behavior, and Immunity. 2013;32:e18. [Google Scholar]
- 26.Villaluenga CM, Wardenska M, Pilarski R, Bednarczyk M, Gulewicz K. Utilization of the chicken embryo model for assessment of biological activity of different oligosaccharides. Folia Biol (Krakow). 2004;52(3–4):135–42. . [DOI] [PubMed] [Google Scholar]
- 27.Gulewicz K, Bednarczyk M, inventors; Mirosław Szykuła assignee. Method for stimulating favourable bacteria profile in hatched chicks (in Polish). Poland2008.
- 28.Bednarczyk M, Stadnicka K, Kozlowska I, Abiuso C, Tavaniello S, Dankowiakowska A, et al. Influence of different prebiotics and mode of their administration on broiler chicken performance. Animal. 2016:1–9. 10.1017/S1751731116000173 . [DOI] [PubMed] [Google Scholar]
- 29.Bednarczyk M, Urbanowski M, Gulewicz P, Kasperczyk K, Maiorano G, Szwaczkowski T. Field and in Vitro Study on Prebiotic Effect of Raffinose Family Oligosaccharides in Chickens. B Vet I Pulawy. 2011;55(3):465–9. WOS:000295554500019. [Google Scholar]
- 30.Maiorano G, Stadnicka K, Tavaniello S, Abiuso C, Bogucka J, Bednarczyk M. In ovo validation model to assess the efficacy of commercial prebiotics on broiler performance and oxidative stability of meat. Poultry Sci. 2017;96(2):511–8. [DOI] [PubMed] [Google Scholar]
- 31.Slawinska A, Plowiec A, Siwek M, Jaroszewski M, Bednarczyk M. Long-Term Transcriptomic Effects of Prebiotics and Synbiotics Delivered In Ovo in Broiler Chickens. PloS one. 2016;11(12):e0168899 10.1371/journal.pone.0168899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobolewska A, Bogucka J, Dankowiakowska A, Elminowska-Wenda G, Stadnicka K, Bednarczyk M. The impact of synbiotic administration through in ovo technology on the microstructure of a broiler chicken small intestine tissue on the 1 st and 42 nd day of rearing. Journal of animal science and biotechnology. 2017;8(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madej J, Bednarczyk M. Effect of in ovo-delivered prebiotics and synbiotics on the morphology and specific immune cell composition in the gut-associated lymphoid tissue. Poultry Sci. 2016;95(1):19–29. [DOI] [PubMed] [Google Scholar]
- 34.Madej JP, Stefaniak T, Bednarczyk M. Effect of in ovo-delivered prebiotics and synbiotics on lymphoid-organs' morphology in chickens. Poult Sci. 2015;94(6):1209–19. 10.3382/ps/pev076 . [DOI] [PubMed] [Google Scholar]
- 35.Slawinska A, Siwek M, Zylinska J, Bardowski J, Brzezinska J, Gulewicz KA, et al. Influence of synbiotics delivered in ovo on immune organs development and structure. Folia Biol (Krakow). 2014;62(3):277–85. . [DOI] [PubMed] [Google Scholar]
- 36.Jung S, Houde R, Baurhoo B, Zhao X, Lee B. Effects of galacto-oligosaccharides and a Bifidobacteria lactis-based probiotic strain on the growth performance and fecal microflora of broiler chickens. Poultry Sci. 2008;87(9):1694–9. [DOI] [PubMed] [Google Scholar]
- 37.Varasteh S, Braber S, Akbari P, Garssen J, Fink-Gremmels J. Differences in susceptibility to heat stress along the chicken intestine and the protective effects of galacto-oligosaccharides. PloS one. 2015;10(9):e0138975 10.1371/journal.pone.0138975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes R-A, Ali RA, Mendoza MA, Hassan HM, Koci MD. impact of Dietary galacto-Oligosaccharide (gOs) on chicken’s gut Microbiota, Mucosal gene expression, and Salmonella colonization. Frontiers in veterinary science. 2017;4 10.3389/fvets.2017.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azcarate-Peril MA, Butz N, Cadenas MB, Koci M, Ballou A, Mendoza M, et al. An attenuated Salmonella Typhimurium strain and galacto-oligosaccharides (GOS) accelerate clearance of Salmonella infection in poultry through modifications to the gut microbiome. Applied and environmental microbiology. 2017:AEM. 02526–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiorano G, Sobolewska A, Cianciullo D, Walasik K, Elminowska-Wenda G, Slawinska A, et al. Influence of in ovo prebiotic and synbiotic administration on meat quality of broiler chickens. Poultry Sci. 2012;91(11):2963–9. 10.3382/ps.2012-02208 WOS:000310421000033. [DOI] [PubMed] [Google Scholar]
- 41.Bogucka J, Dankowiakowska A, Elminowska-Wenda G, Sobolewska A, Jankowski J, Szpinda M, et al. Performance and small intestine morphology and ultrastructure of male broilers injected in ovo with bioactive substances. Ann Anim Sci. 2017;17(1):179–95. [Google Scholar]
- 42.Pruszynska-Oszmalek E, Kolodziejski PA, Stadnicka K, Sassek M, Chalupka D, Kuston B, et al. In ovo injection of prebiotics and synbiotics affects the digestive potency of the pancreas in growing chickens. Poultry Sci. 2015;94(8):1909–16. 10.3382/ps/pev162 WOS:000358186100024. [DOI] [PubMed] [Google Scholar]
- 43.Tzortzis G, Goulas AK, Gibson GR. Synthesis of prebiotic galactooligosaccharides using whole cells of a novel strain, Bifidobacterium bifidum NCIMB 41171. Appl Microbiol Biotechnol. 2005;68(3):412–6. 10.1007/s00253-005-1919-0 . [DOI] [PubMed] [Google Scholar]
- 44.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243(1):141–7. 10.1016/j.femsle.2004.11.052 . [DOI] [PubMed] [Google Scholar]
- 45.Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002;68(1):114–23. 10.1128/AEM.68.1.114-123.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter J, Tannock GW, Tilsala-Timisjarvi A, Rodtong S, Loach DM, Munro K, et al. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl Environ Microbiol. 2000;66(1):297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tannock GW, Tilsala-Timisjarvi A, Rodtong S, Ng J, Munro K, Alatossava T. Identification of Lactobacillus isolates from the gastrointestinal tract, silage, and yoghurt by 16S-23S rRNA gene intergenic spacer region sequence comparisons. Appl Environ Microbiol. 1999;65(9):4264–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen EG, Licht TR, Leser TD, Bahl MI. Dietary xylo-oligosaccharide stimulates intestinal bifidobacteria and lactobacilli but has limited effect on intestinal integrity in rats. BMC Res Notes. 2014;7:660 10.1186/1756-0500-7-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sevane N, Bialade F, Velasco S, Rebole A, Rodriguez ML, Ortiz LT, et al. Dietary inulin supplementation modifies significantly the liver transcriptomic profile of broiler chickens. PLoS One. 2014;9(6):e98942 10.1371/journal.pone.0098942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 52.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. 10.2144/03342mt01 . [DOI] [PubMed] [Google Scholar]
- 53.Rothwell L, Young JR, Zoorob R, Whittaker CA, Hesketh P, Archer A, et al. Cloning and characterization of chicken IL-10 and its role in the immune response to Eimeria maxima. J Immunol. 2004;173(4):2675–82. . [DOI] [PubMed] [Google Scholar]
- 54.Brisbin JT, Gong J, Parvizi P, Sharif S. Effects of lactobacilli on cytokine expression by chicken spleen and cecal tonsil cells. Clin Vaccine Immunol. 2010;17(9):1337–43. 10.1128/CVI.00143-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjerrum L, Engberg R, Leser TD, Jensen BB, Finster K, Pedersen K. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poultry Sci. 2006;85(7):1151–64. [DOI] [PubMed] [Google Scholar]
- 56.Ranjitkar S, Lawley B, Tannock G, Engberg RM. Bacterial succession in the broiler gastrointestinal tract. Applied and environmental microbiology. 2016;82(8):2399–410. 10.1128/AEM.02549-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiological reviews. 2010;90(3):859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 58.Roberfroid M. Prebiotics. 2003. [Google Scholar]
- 59.Pokusaeva K, Fitzgerald GF, Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes & nutrition. 2011;6(3):285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes R-A, Ali RA, Mendoza MA, Hassan HM, Koci MD. impact of Dietary galacto-Oligosaccharide (gOs) on chicken’s gut Microbiota, Mucosal gene expression, and Salmonella colonization. Frontiers in veterinary science. 2017;4:192 10.3389/fvets.2017.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svihus B. Function of the digestive system. J Appl Poultry Res. 2014;23(2):306–14. 10.3382/japr.2014-00937 WOS:000337352900022. [DOI] [Google Scholar]
- 62.Schokker D, Smits MA, Rebel JM, editors. Jejunal gene expression patterns correlate with severity of systemic infection in chicken. BMC proceedings; 2011: BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schokker D, Hoekman AJ, Smits MA, Rebel JM. Gene expression patterns associated with chicken jejunal development. Developmental & Comparative Immunology. 2009;33(11):1156–64. [DOI] [PubMed] [Google Scholar]
- 64.Volf J, Polansky O, Sekelova Z, Velge P, Schouler C, Kaspers B, et al. Gene expression in the chicken caecum is dependent on microbiota composition. Veterinary research. 2017;48(1):85 10.1186/s13567-017-0493-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rychlik I, Elsheimer-Matulova M, Kyrova K. Gene expression in the chicken caecum in response to infections with non-typhoid Salmonella. Veterinary research. 2014;45(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Higgins S, Wolfenden A, Tellez G, Hargis B, Porter T. Transcriptional profiling of cecal gene expression in probiotic-and Salmonella-challenged neonatal chicks. Poultry Sci. 2011;90(4):901–13. [DOI] [PubMed] [Google Scholar]
- 67.Lopetuso LR, Chowdhry S, Pizarro TT. Opposing Functions of Classic and Novel IL-1 Family Members in Gut Health and Disease. Front Immunol. 2013;4:181 10.3389/fimmu.2013.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Youngman KR, Simon PL, West GA, Cominelli F, Rachmilewitz D, Klein JS, et al. Localization of intestinal interleukin 1 activity and protein and gene expression to lamina propria cells. Gastroenterology. 1993;104(3):749–58. [DOI] [PubMed] [Google Scholar]
- 69.Khosravi A, Mazmanian SK. Disruption of the gut microbiome as a risk factor for microbial infections. Current opinion in microbiology. 2013;16(2):221–7. 10.1016/j.mib.2013.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamada N, Núñez G. Role of the gut microbiota in the development and function of lymphoid cells. The Journal of Immunology. 2013;190(4):1389–95. 10.4049/jimmunol.1203100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen GY. Regulation of the gut microbiome by inflammasomes. Free Radical Biology and Medicine. 2017;105:35–40. 10.1016/j.freeradbiomed.2016.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaji R, Kiyoshima-Shibata J, Tsujibe S, Nanno M, Shida K. Probiotic induction of interleukin-10 and interleukin-12 production by macrophages is modulated by co-stimulation with microbial components. J Dairy Sci. 2018. [DOI] [PubMed] [Google Scholar]
- 73.Barberi C, Campana S, De Pasquale C, Khorasgani MR, Ferlazzo G, Bonaccorsi I. T cell polarizing properties of probiotic bacteria. Immunology letters. 2015;168(2):337–42. 10.1016/j.imlet.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 74.Slawinska A, Siwek MZ, Bednarczyk MF. Effects of synbiotics injected in ovo on regulation of immune-related gene expression in adult chickens. Am J Vet Res. 2014;75(11):997–1003. 10.2460/ajvr.75.11.997 . [DOI] [PubMed] [Google Scholar]
- 75.Dunislawska A, Slawinska A, Stadnicka K, Bednarczyk M, Gulewicz P, Jozefiak D, et al. Synbiotics for broiler chickens—in vitro design and evaluation of the influence on host and selected microbiota populations following in ovo delivery. PloS one. 2017;12(1):e0168587 10.1371/journal.pone.0168587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haghighi HR, Abdul-Careem MF, Dara RA, Chambers JR, Sharif S. Cytokine gene expression in chicken cecal tonsils following treatment with probiotics and Salmonella infection. Veterinary microbiology. 2008;126(1–3):225–33. 10.1016/j.vetmic.2007.06.026 [DOI] [PubMed] [Google Scholar]
- 77.Kogut MH, Genovese KJ, He H, Swaggerty CL, Jiang Y. Modulation of chicken intestinal immune gene expression by small cationic peptides as feed additives during the first week post-hatch. Clinical and Vaccine Immunology. 2013:CVI. 00322–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sunkara L, Curtis A, Zhang G. Biology, expression, and regulation of host defense peptides: a minireview. Adv Anim Vet Sci. 2015;3(3s):9–20. [Google Scholar]
- 79.Robinson K, Deng Z, Hou Y, Zhang G. Regulation of the intestinal barrier function by host defense peptides. Frontiers in veterinary science. 2015;2:57 10.3389/fvets.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cuperus T, Coorens M, van Dijk A, Haagsman HP. Avian host defense peptides. Developmental & Comparative Immunology. 2013;41(3):352–69. [DOI] [PubMed] [Google Scholar]
- 81.Akbari MR, Haghighi HR, Chambers JR, Brisbin J, Read LR, Sharif S. Expression of antimicrobial peptides in cecal tonsils of chickens treated with probiotics and infected with Salmonella enterica serovar typhimurium. Clinical and Vaccine Immunology. 2008;15(11):1689–93. 10.1128/CVI.00242-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shao Y, Wang Z, Tian X, Guo Y, Zhang H. Yeast β-d-glucans induced antimicrobial peptide expressions against Salmonella infection in broiler chickens. International journal of biological macromolecules. 2016;85:573–84. 10.1016/j.ijbiomac.2016.01.031 [DOI] [PubMed] [Google Scholar]
- 83.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunological reviews. 2014;260(1):8–20. 10.1111/imr.12182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer–. The American journal of clinical nutrition. 2001;73(6):1131S–41S. 10.1093/ajcn/73.6.1131S [DOI] [PubMed] [Google Scholar]
- 85.Alemka A, Whelan S, Gough R, Clyne M, Gallagher ME, Carrington SD, et al. Purified chicken intestinal mucin attenuates Campylobacter jejuni pathogenicity in vitro. Journal of medical microbiology. 2010;59(8):898–903. [DOI] [PubMed] [Google Scholar]
- 86.Chen J, Tellez G, Richards JD, Escobar J. Identification of potential biomarkers for gut barrier failure in broiler chickens. Frontiers in veterinary science. 2015;2:14 10.3389/fvets.2015.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gadde UD, Oh S, Lee Y, Davis E, Zimmerman N, Rehberger T, et al. Dietary Bacillus subtilis-based direct-fed microbials alleviate LPS-induced intestinal immunological stress and improve intestinal barrier gene expression in commercial broiler chickens. Research in veterinary science. 2017;114:236–43. 10.1016/j.rvsc.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 88.Smirnov A, Tako E, Ferket PR, Uni Z. Mucin gene expression and mucin content in the chicken intestinal goblet cells are affected by in ovo feeding of carbohydrates. Poultry Sci. 2006;85(4):669–73. WOS:000236484300012. [DOI] [PubMed] [Google Scholar]
- 89.Madara JL. Loosening tight junctions. Lessons from the intestine. The Journal of clinical investigation. 1989;83(4):1089–94. 10.1172/JCI113987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke J-D, Serino M, et al. Intestinal permeability–a new target for disease prevention and therapy. BMC gastroenterology. 2014;14(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. The Journal of cell biology. 1986;103(3):755–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawabe H, Nakanishi H, Asada M, Fukuhara A, Morimoto K, Takeuchi M, et al. Pilt, a novel peripheral membrane protein at tight junctions in epithelial cells. J Biol Chem. 2001;276(51):48350–5. 10.1074/jbc.M107335200 [DOI] [PubMed] [Google Scholar]
- 93.Awad W, Hess C, Hess M. Enteric pathogens and their toxin-induced disruption of the intestinal barrier through alteration of tight junctions in chickens. Toxins. 2017;9(2):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rajput I, Li L, Xin X, Wu B, Juan Z, Cui Z, et al. Effect of Saccharomyces boulardii and Bacillus subtilis B10 on intestinal ultrastructure modulation and mucosal immunity development mechanism in broiler chickens. Poultry Sci. 2013;92(4):956–65. [DOI] [PubMed] [Google Scholar]
- 95.Alvarez-Curto E, Milligan G. Metabolism meets immunity: the role of free fatty acid receptors in the immune system. Biochemical pharmacology. 2016;114:3–13. 10.1016/j.bcp.2016.03.017 [DOI] [PubMed] [Google Scholar]
- 96.Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132(6):2131–57. 10.1053/j.gastro.2007.03.054 [DOI] [PubMed] [Google Scholar]
- 97.Kolodziejski PA, Sassek M, Chalupka D, Leciejewska N, Nogowski L, Mackowiak P, et al. GLP1 and GIP are involved in the action of synbiotics in broiler chickens. Journal of Animal Science and Biotechnology. 2018;9(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Slawinska A, Zampiga M, Sirri F, Meluzzi A, Tavaniello S, Maiorano G. Mitigating effects of galactooligosaccharides delivered in ovo on performance and welfare in broiler chickens under heat stress. Poultry Science (submitted). 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of lipid research. 2013;54(9):2325–40. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burns RN, Moniri NH. Agonism with the omega-3 fatty acids α-linolenic acid and docosahexaenoic acid mediates phosphorylation of both the short and long isoforms of the human GPR120 receptor. Biochemical and biophysical research communications. 2010;396(4):1030–5. 10.1016/j.bbrc.2010.05.057 [DOI] [PubMed] [Google Scholar]
- 101.Corrêa‐Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short‐chain fatty acids. Clinical & translational immunology. 2016;5(4):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sunkara LT, Achanta M, Schreiber NB, Bommineni YR, Dai G, Jiang W, et al. Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression. PLoS one. 2011;6(11):e27225 10.1371/journal.pone.0027225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mangian HF, Tappenden KA. Butyrate Increases GLUT2 mRNA Abundance by Initiating Transcription in Caco2‐BBe Cells. Journal of Parenteral and Enteral Nutrition. 2009;33(6):607–17. 10.1177/0148607109336599 [DOI] [PubMed] [Google Scholar]
- 104.Woting A, Pfeiffer N, Loh G, Klaus S, Blaut M. Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. MBio. 2014;5(5):e01530–14. 10.1128/mBio.01530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qPCR dataset for calculating (A) bacterial and (B) gene abundances.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files (S1 File).