Abstract
Background
Pediatric acute respiratory distress in tropical settings is very common. Bacterial pneumonia is a major contributor to morbidity and mortality rates and requires adequate diagnosis for correct treatment. A rapid test that could identify bacterial (vs other) infections would have great clinical utility.
Methods and Results
We performed RNA (RNA-seq) sequencing and analyzed the transcriptomes of 68 pediatric patients with well-characterized clinical phenotype to identify transcriptional features associated with each disease class. We refined the features to predictive models (support vector machine, elastic net) and validated those models in an independent test set of 37 patients (80%–85% accuracy).
Conclusions
We have identified sets of genes that are differentially expressed in pediatric patients with pneumonia syndrome attributable to different infections and requiring different therapeutic interventions. Findings of this study demonstrate that human transcription signatures in infected patients recapitulate the underlying biology and provide models for predicting a bacterial diagnosis to inform treatment.
Keywords: RNA-Seq, gene expression, biomarkers, machine learning, Pneumonia
Pediatric pneumonia syndrome, characterized by acute respiratory distress with fever, is associated with severe morbidity, mortality, and health service burden. Among children <5 years old, severe acute lower respiratory infection accounted for 15 million hospital admissions and 1.4 million deaths worldwide in 2010 [1]. Although the chief etiologic agents for pediatric pneumonia syndrome are viral and bacterial respiratory pathogens, syndromic overlap with severe malaria probably contributes to exceptionally high rates of apparent severe acute lower respiratory infection in malaria-endemic regions [1].
The significant clinical overlap between bacterial, viral, and malarial infections poses a special challenge for rapid diagnosis. Among children presenting with fever, multiple infections may be commonplace, amplifying the challenge of assigning a specific pathogenic cause for their illness [2]. Importantly, though the overlap of symptoms of severe malaria and severe pneumonia is common, co-occurrence of the diseases is not, emphasizing the importance of securing an etiologic diagnosis for appropriate clinical management. Correct identification of bacterial infection is of special importance, because it is often associated with a much higher case fatality rate and timely and appropriate antibiotic therapy may be life-saving [3].
Current approaches to diagnosis suffer a range of problems: profiles of clinical signs and symptoms provide some discrimination but not enough for accurate and actionable diagnosis even in expert hands, with still worse performance among low-skilled medical personnel; conventional clinical and laboratory tests involving specialized equipment may be unavailable or prohibitively expensive, often require expert interpretation, and still have marginal performance characteristics. Molecular diagnostics targeting the causal pathogens hold promise but likewise may have inadequate performance characteristics in pediatric populations prone to asymptomatic infection or colonization, especially when applied to accessible samples (blood, urine, or upper respiratory tract) rather than those from the site of infection (sputum), as is often necessary in children [4–12].
Even in advanced medical systems, the appropriate utilization of antimicrobial agents for acute respiratory syndromes is challenging, with therapy being withheld in a substantial proportion of antibiotic-appropriate infections and administered in antibiotic-inappropriate infections [13], leading to increased morbidity rates, health system inefficiencies, and antimicrobial resistance. This problem is amplified in resource-poor settings, where diagnostic testing is often limited and misapplied. Clinical data may not be captured, documented, or used to inform clinical decision-making in accordance with international guidelines, such as the World Health Organization (WHO) Integrated Management for Childhood Illness, which tend toward high sensitivity but low specificity and so have poor positive predictive value [14, 15]. A reliable and efficient diagnostic is urgently needed.
A compelling alternative is to leverage the fact that the underlying pathophysiologic mechanism and human “host” responses are very different among these disease classes. Classically, “active” viral infection produces a lymphoid response, bacterial infections produce a neutrophil response, and malaria is associated with monocyte/macrophage activation. Thus, tools that can assess these precise human responses may support development of more accurate tools to rapidly and appropriately administer treatment.
To characterize the underlying pathophysiologic mechanism in the molecular level, we developed a well-defined cohort of pediatric patients from a malaria-endemic area presenting with pneumonia syndrome attributable to a single entity—bacteria, malaria, or virus. We performed whole-transcriptome analyses of the peripheral blood human gene expression profiles to search for differentially expressed marker genes capable of discriminating between them. Using machine-learning algorithms, we built classifiers capable of discriminating between these specific causes of disease, or between bacterial and other causes of illness. Ultimately these marker panels may form the foundation for a transcript- or protein-based diagnostic test to help guide therapy for this vulnerable population.
METHODS
Study Population and Sample Collection
All children presenting to the outpatient clinic of the Manhiça District Hospital in Mozambique with symptoms of clinical pneumonia and fulfilling criteria for hospital admission between July 2010 and November 2012 were assessed for recruitment. Children <10 years of age who had documented fever at admission (>37.5°C axillary temperature) or a history of fever in the preceding 24 hours and met the WHO case definition for clinical pneumonia (increased respiratory rate and cough or difficulty in breathing) [16] were invited to participate in this study. Exclusion criteria included use of antimalarial drugs in the preceding 2 weeks, established or suspected pulmonary tuberculosis (based on a history of cough of >2 weeks in duration or history of direct contact with a patient with a tuberculosis diagnosis), ongoing participation in conflicting studies, and marked hypoxemia (oxyhemoglobin saturation ≤85%), to reduce the likelihood of Pneumocystis jirovecii infection.
Parents or guardians provided written informed consent after detailed explanation of the study objectives and procedures. Before the initiation of antimicrobial therapy, a nasopharyngeal aspirate was taken for polymerase chain reaction–based determination of respiratory viral infection, and venous blood was collected for blood culture, human immunodeficiency virus (HIV) testing, malaria diagnosis (by smear microscopy), and bacterial polymerase chain reaction. All children underwent anteroposterior chest radiography. Operators blinded to diagnosis and clinical status processed all laboratory tests and samples. All clinical, laboratory, and radiologic data were used to classify all patients enrolled in the study as having bacteria, viral, or malaria infection, either singly (used in this study) or mixed [17].
Ethics
This study was approved by the Mozambique National Bioethics committee (reference No. 262/CNBS/10), the Institutional Review Board of the Broad Institute/MIT Committee on the Use of Humans as Experimental Subjects (reference No. 1003003765), and the Barcelona Centre for International Health Research (reference No. 2010/5590). The data and software supporting the results of this article are freely available in the Zenodo repository (https://zenodo.org/record/45477).
RNA Sequencing
A complementary DNA library was first constructed (see Supplementary Notes). Pooled libraries were normalized to 2 nmol/L and denatured using 0.1N sodium hydroxide before sequencing. Flow cell cluster amplification and sequencing were performed according to the manufacturer's protocols, using either the HiSeq 2000 or HiSeq 2500 sequencing systems (Illumina). Each run consisted of a 76–base pair paired-end read with an 8-base index barcode read. Sequencing was performed in 96-well format in 2 batches.
Sequencing Analysis Pipeline
Unaligned reads were trimmed using the Trimmomatic preprocessing tool [18] to remove low-quality bases. RSEM software v 1.3.0 [19] was used to align reads to the human transcriptome and quantify gene expression. The reference genome assembly was hg19, using annotations from the University of California, Santa Cruz, Genome Browser [20]. Quality was evaluted with the RNA-sequenincg quality control (RNA-SeQC) module of GenePattern (http://archive.broadinstitute.org/cancer/cga/rna-seqc/) [12].
Modeling
We developed a classifier, performed cross-validation, created a final model, and evaluated the model with standard metrics; the details can be found in Supplementary Notes.
RESULTS
Patient Characteristics
A total of 105 enrolled patients met the criteria for a single specific cause of pneumonia syndrome and were included in this study (Supplementary File 1, Sample Distribution by Etiology). There were no significant differences in age, sex, size, nutritional status, fever, or respiratory rate between patients with bacterial, malarial, or viral infections (Supplementary File 2, Patient Demographics). Patients with malaria were significantly more anemic at admission. By definition, only patients with bacterial infection had positive blood cultures, and only patients with malaria had positive smears. Chest radiographic status was not an independent variable, because malarial patients were required to have clear radiographs and patients with viral infection were not allowed to have primary end-point pneumonia. (The only exception was sample 0314; it was originally classified as malaria but later reclassified as mixed bacteria/malaria. By the time that sample was reclassified the database was “locked.”) Of patients with viral infection, 45% had a nonconsolidative infiltrate or other radiographic abnormality. Of those with bacterial infection, 70% had primary end-point pneumonia. Patients with bacterial infection had higher leukocyte counts and were more likely to be HIV positive. They also had statistically significant longer hospital stays and higher in-hospital mortality rates. From a total of 105 samples, stratified random sampling was used to select a training set of 68 (and testing set of 37 samples while retaining the relative distribution of disease etiologic agents between training and test sets (Supplementary File 1).
Hierarchical Clustering
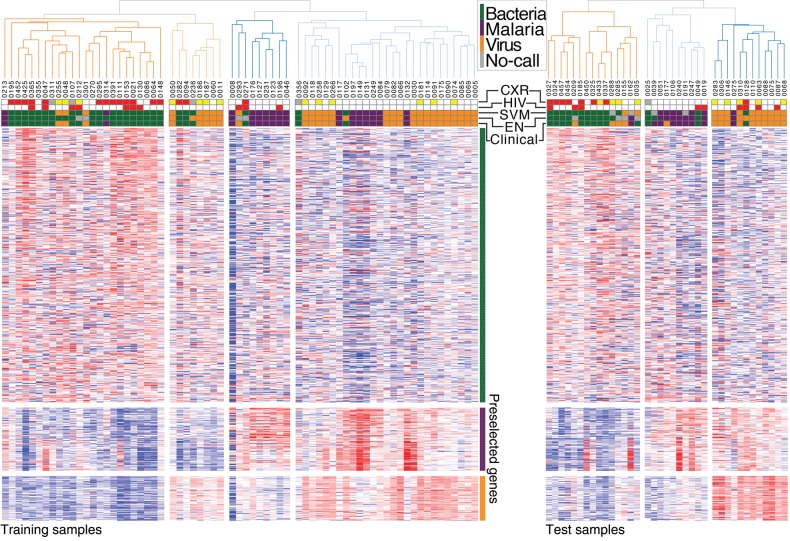
The cross-validation approach to marker selection yielded 600 total marker genes. Unsupervised hierarchical clustering of the training samples using these differentially expressed 600 genes (n = 600) is shown in Figure 1 [22], annotated with the clinical diagnosis, prediction using 3-class elastic net (EN) and support vector machine (SVM) models, HIV status, and radiographic pattern. The full list of marker genes is available in Supplementary File 3, and additional details on clustering are in Supplementary Notes.
Figure 1.
Hierarchical clustering using 600 preselected differentially expressed (DE) genes. Using the markers selected via the cross-validation procedure, we clustered the training samples (left) and testing samples (right) separately. The top row shows the clinical diagnosis, elastic net (EN) classification, support vector machine (SVM) classification, human immunodeficiency virus (HIV) status, and chest radiographic (CXR) diagnosis. Samples were separately row-normalized and clustered, displayed side by side for convenience. Expression was log-transformed and row-normalized by median-adjusted deviation. The distance metric was 1 minus the spearman correlation. HIV indicates whether HIV positive (red) or negative (white). CXR indicates chest radiographic diagnosis category: normal (white), other infiltrate/abnormality (yellow), primary end-point pneumonia (red), or data not available (gray).
Cross-Validation
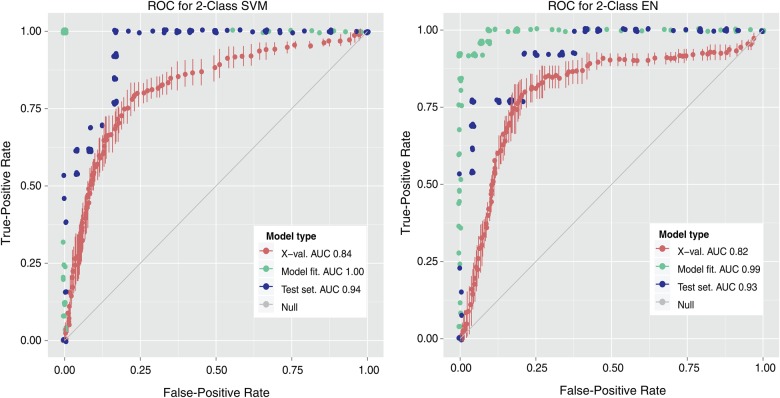
Figure 2 (red curve) shows the receiver-operating-characteristic (ROC) curves from cross-validation for both SVM (left) and EN (right) 2-class models. The performances of both classifiers are similar, with a cross-validation area under the curve (AUC) ROC of 0.84 using SVM and of 0.82 using EN. Despite biasing toward bacteria samples, false-positive rates remained acceptable, as measured by the high AUC-ROC. Quality performance across multiple algorithms provided substantial evidence that each etiology produced a unique transcriptomic signature.
Figure 2.
Receiver operating characteristic (ROC) curves for 2-class models. True-positive versus false-positive rates are shown for X-val, trained model, and test set, in support vector machine (SVM) (left) and elastic net (EN) (right) models. The area under the curve (AUC) for each data set is shown in the key. Abbreviation: X-val, cross-validation.
Independent Test Set
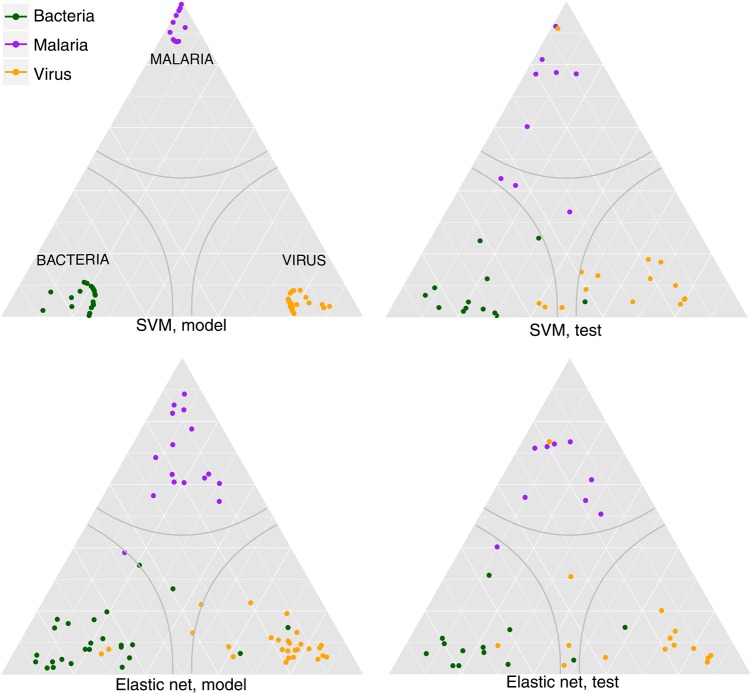
Confusion matrices for the training and test sets are shown for the 3-class EN model in Table 1 and the SVM model in Table 2. Because ROC curves do not aptly display the predictions of 3-class models, a ternary diagram displaying the predicted probabilities of each class is shown in Figure 3. EN has consistent performance between model and test sets (overall accuracy, 92.2% vs 87.8%), whereas SVM fits the model extremely well but has less consistent performance between model and test sets (overall accuracy, 100% vs 87.8%). A similar trend is apparent in 2-class accuracy statistics, shown in Table 3 (see Supplementary File 4 for the 2-class confusion matrices). The SVM models have perfect fit in the training set, and EN models have training accuracy of 90%–95%. This modest accuracy penalty occurs despite the fact that EN uses 5% of the markers used by SVM (29 vs 600 genes). This suggests that the full marker list, while large, has substantial redundancy.
Table 1.
Confusion Matrices for 3-Class Elastic Net
| Actual Diagnosis | Predicted Diagnosis for Trained (Test)
Model |
|||
|---|---|---|---|---|
| Bacterial Infection | Malaria | Viral Infetion | No-Call | |
| Bacterial infection | 21 (11) | 0 (0) | 2 (1) | 2 (1) |
| Malaria | 1 (1) | 15 (8) | 0 (0) | 0 (0) |
| Viral infection | 2 (1) | 0 (1) | 23 (11) | 2 (3) |
Table 2.
Confusion Matrices for 3-Class Support Vector Machine
| Actual Diagnosis | Predicted Diagnosis for Trained (Test)
Model |
|||
|---|---|---|---|---|
| Bacterial Infection | Malaria | Viral Infection | No-Call | |
| Bacterial infection | 25 (12) | 0 (0) | 0 (1) | 0 (0) |
| Malaria | 0 (0) | 16 (6) | 0 (0) | 0 (3) |
| Viral infection | 0 (2) | 0 (1) | 27 (11) | 0 (0) |
Figure 3.
Predicted probability ternary plot. The predicted probability of each classification is projected onto a triangle. Each corner represents a category having probability 1. There are 3 classes, and the probabilities for each class must sum to 1, hence there are only 2 degrees of freedom. The probability of a class at a given point is proportional to the distance along a line extending from the vertex to its opposing side. Grid lines represent constant weight for a class. For instance, the purely horizontal lines are lines of constant probability of malaria. Gray lines represent our confidence boundary; anything within that region has brier score <0.3 and is not called. Plots were generated using ggtern [41]. Abbreviation: SVM, support vector machine.
Table 3.
Performance Statistics of 2-Class Models for Trained Model and Test Set
| Performance | EN 2-Class |
SVM 2-Class |
||
|---|---|---|---|---|
| Trained Model | Test Set | Trained Model | Test Set | |
| Sensitivity, % | 95.8 | 100 | 100 | 100 |
| Specificity, % | 95.1 | 70.0 | 100 | 82.6 |
| PPV, % | 96.7 | 83.1 | 100 | 89.5 |
| No-call rate, % | 4.4 | 13.5 | 0 | 8.1 |
| AUC, % | 99.0 | 93.0 | 100 | 94.0 |
| Overall accuracy, % | 95.4 | 81.3 | 100 | 88.2 |
Abbreviations: AUC, area under the curve; EN, elastic net; PPV, positive predictive value; SVM, support vector machine.
There were 18 total HIV-positive samples in the collection. Combining errors and no-calls in both the training and test sets, there was no significant difference in classification performance between HIV-positive and HIV-negative samples (EN, P = .16; SVM, P = .55; Fisher exact test). Thus, the classifiers perform well irrespective of patients’ HIV status.
Gene Ontologic Analysis
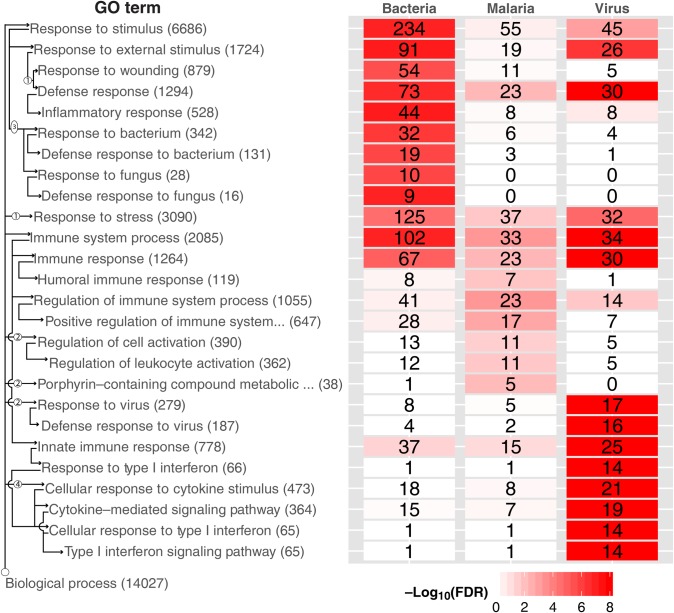
To explore the biological significance of our models, we performed gene ontology (GO) analysis on the 600 preselected marker genes. The top 10 biological process GO terms for each condition are displayed in Figure 4, and the complete lists are in Supplementary File 5. Enriched processes in bacterial genes included responses to bacteria and wounding. The major viral responses were type I interferon and cytokine signaling. Malarial responses, less well differentiated, included leukocyte activation and generic immune responses. The axis labels include arrows connecting parent/child terms. More detailed hierarchies are shown in Supplementary Figures 2–4, including the number of genes used by EN (as well as strongly correlated genes that might act as substitutes).
Figure 4.
Gene ontologic (GO) term enrichment heat map, displaying GO enrichment results using genes selected by cross-validation. The top 10 GO terms are shown by diagnostic category (regardless of significance), for a total of 26 terms across diagnostic categories. Four terms were represented in both bacterial and viral categories. Note that the bacterial and viral signatures top 10 GO terms were highly significant (by false discovery rate [FDR]), whereas the malaria signature was less significant. The top GO terms for malaria are all represented in the viral and bacterial GO term lists (just further down the list), suggesting that the malaria classification is at the exclusion of viral and bacterial classification. The total numbers of genes for each GO term are displayed in parentheses next to the title; the number of significant genes from each etiology for each term is displayed in each cell. Cells are colored by FDR. We illustrate the GO hierarchy via wiring diagram across the left side. Arrows indicate a “parent of” relationship, circled numbers indicate nodes not shown. For instance, there are 2 nodes between “Biological process” and “Response to virus”; the latter contains 279 genes total, of which 17 were found significant in viral samples, and is a parent of “Defense response to virus.”
DISCUSSION
We have shown that transcriptional signatures of host response can be used to classify the underlying etiology for pediatric patients in tropical settings presenting with a pneumonia syndrome. The idea that transcriptional signatures from the blood of ill patients might be used to parse the class of disease pathogen has been suggested in previous studies. Ramilo and colleagues [23] performed microarray analysis on messenger RNA isolated from peripheral blood mononuclear cells and used a k-nearest neighbors algorithm to classify pediatric patients with acute infections from influenza A virus, Escherichia coli, Staphylococcus aureus, or Streptococcus pneumoniae, associated with a wide range of clinical disease, including respiratory infections, localized abscesses, urinary tract infections, and meningitis. More recently, this group has leveraged whole-blood transcriptional profiles to characterize the host response to respiratory syncytial virus (RSV) lower respiratory tract infection in infants in contradistinction to influenza and human rhinovirus-associated response, and to establish biomarkers that might be use to assess disease severity [24]. An elegant series of articles from a related group [25, 26] has demonstrated the utility of an interferon-inducible transcriptional signature in blood to distinguish acute active tuberculosis disease from latent tuberculosis, other causes of bacterial pneumonia, and other pulmonary disease and to monitor response to tuberculosis therapy. The work presented here is, to our knowledge, the first application of this general approach using RNA sequencing (RNA-Seq) to identify biomarker candidates for pediatric pneumonia syndrome.
When hierarchical clustering is performed in the space of genes filtered for expression level and variance, but not selected for differential expression between classes, there is no significant segregation between samples by bacterial, malarial, and viral etiology. We attribute this to the large variance in gene expression across individuals relative to disease-specific host response, preventing unsupervised methods from detecting meaningful signal out of random variation. Nevertheless, supervised methods are able to extract reliable markers. Class segregation is quite good in the space of differentially expressed genes even before the development of classifiers (Figure 1). Of the 2 samples diagnosed as bacterial infection contained in rightmost, dominantly viral cluster, 1 sample, sample 0102, was erroneously diagnosed; its associated blood culture was positive for a contaminant rather than a pathogenic bacterial species, a determination not made until after the data set was locked. Of note, the EN model assigned it a viral etiology. The cluster representing an admixture of bacterial and viral samples is characterized by relative overexpression of both the bacterial markers, indicated by the green bar at the right edge of the figure, and the viral markers, indicated by the orange bar. It is well established that a substantial minority of children with severe clinical pneumonia will have viral and bacterial coinfection [2, 8, 27]. For the purposes of initial classifier development and testing, known coinfections were excluded from analysis. However, because the positive pathogenic bacterial blood cultures required to establish bacterial etiology are known to have good specificity but poor sensitivity [8, 10, 11], it is plausible that these samples represent undetected viral/bacterial coinfection. This cluster highlights the challenge of marker identification and classifier development in contexts where the “gold standard” diagnostics are themselves inadequate (see below).
Two different classification algorithms were equally effective in disease assignment in the independent test set, supporting the robustness of the response markers. Although up to 600 genes were used as markers with the SVM approach, accurate classification required no more than 29 with the more parsimonious elastic net (EN). The size of the differentially expressed set of genes is evidence of the substantial depth of the host response signature. Much of this information is probably redundant, however, because the 600-marker SVM model did not significantly outperform the 29-marker EN model with the test set. This redundancy is readily evident in the correlation structure of the differentially expressed genes (Supplementary Figure 5). This shows that some clusters of markers are relatively specific to particular disease causes, whereas others are shared across >1 cause, and it also demonstrates regions of high correlation within which individual markers carry essentially identical information. Even within the more selective EN model, there is clear evidence of that redundancy; the set of 29 markers is complemented by 118 surrogates that have highly correlated expression and could potentially be substituted in specific combinations for the original markers without degradation of model performance (Supplementary File 6). On average, there were 5 possibilities (the selected marker and 4 surrogates) for each of the 29 genes used by EN, with a range of 0–30 possibilities (complement component 3 had the most correlated genes). The addition of markers without addition of information content does not improve classification accuracy, though it may in some instances improve the robustness of classifiers (for instance if only subsets of markers are accurately measured or differential in each sample).
Although additional markers did not improve model performance, it is likely that model performance could be enhanced by access to a larger number of samples. Performance in 10-fold cross-validation consistently underperformed both training and test results (Figure 2), demonstrating that the withholding of 10% of the samples materially degraded model performance—and conversely, that better and more stable models would probably result from the inclusion of additional samples.
It is not strictly necessary for an effective disease marker to be directly related to the pathobiology of the disease, but it is probable that many or most of the best markers will have such a relationship. The biological relevance of the set of differentially expressed marker genes markers is shown in the plot of enrichment for GO terms, in Figure 4. The critical viral/bacterial distinction is particularly strong. Appropriately, bacterial markers are enriched in response/defense response to bacteria as well as in response to wounding and in inflammatory response, whereas viral markers are enriched in response/defense response to viruses and in interferon signaling. Both are enriched in the less specific categories of immune response and response to stimulus and stress. Malarial responses are less discrete, with GO term enrichment at the borderline of significance by the false discovery rate. For all 3 classes, the EN-selected markers are enriched (see EN ratios in Supplementary Figures 2–4) in the top GO terms and represented in processes that are biological relevant, whereas the SVM 600 gene model selected nonspecific GO terms (ie, response to bacteria vs response to fungi in Supplementary Figure 2).
Although informative markers are too numerous to explore individually, the reduced set represented in the EN model supports the biological relevance of the emerging candidates. Myosin-X, an unconventional myosin, is an actin-based motor protein known to be involved in filopodia formation and increasingly recognized to play a role in bacterial infection [28]. The importance of Myosin-X has been best established in infections with Shigella and Listeria [29], but many pathogenic bacteria, including Haemophilus influenzae and Klebsiella pneumoniae, are dependent on comparable actin cytoskeletal modulation to facilitate invasion of epithelial cells, intracellular and intercellular bacterial movement, and avoidance of immune responses [30].
Olfactomedin 4 (OLM4) is stored in the granules of mature neutrophils and modulates neutrophil-based killing and innate immunity against both gram-positive and gram-negative bacteria [31]. It may describe a distinct subpopulation representing about 30% of human neutrophils, but the functional significance of this subpopulation is as yet uncharacterized [32]. Although its function in human neutrophils is not established, OLM4 in mice negatively regulated the nucleotide-binding oligomerization domain 1, which broadly promotes neutrophil-based bactericidal activity [33] and inhibited the activity of cathepsin C, a protease involved in activation of other neutrophil proteases with roles in microbial killing [34]. Consistent with these activities, OLM4 knockout mice are less vulnerable to bacterial sepsis than wild type counterparts. Although in the current study OLM4 was a marker of bacterial infections, it has also been described as the most differentially expressed transcriptional marker of disease severity in children with RSV infection, with >40-fold higher expression in severe than in mild disease and a decrease after clinical recovery [35]. Whereas this raises the possibility that OLM4 is a biomarker of severe lung inflammation or injury rather than of bacterial pneumonia per se, the severity of RSV disease may also reflect bacterial coinfection, which was not tested for in this study but has been described to occur in 20%–40% of severe RSV bronchiolitis cases [36] and is established to be associated with severe pneumonia [2].
Viral markers are also biologically compelling. The secretion of interferons from cells infected with virus is a central hallmark of the host response. IFIT1, a marker of viral etiology, is a prototypical interferon-stimulated gene with antiviral activity. IFITs can specifically interact with a range of viral RNAs and proteins, thereby interfering with viral replication [37]. NEXN has also been recently described as an interferon-β response gene [38].
In this study, specific etiologies for pneumonia syndrome were determined by pathogen identification. An important alternative approach is the use of chest radiography. Radiographic end-point pneumonia has been recommended as the reference standard for bacterial pneumonia diagnosis [39] and is widely used in vaccine efficacy trials. The WHO radiologic end point may have underestimated the burden of pneumonia prevented in some pneumonia vaccination trials, consistent with its greater emphasis on specificity than on sensitivity. This has led WHO to reconvene a working group to try to establish a more sensitive end point for radiologically confirmed pneumonia [40]. The character of the chest radiograph in this study was an element in the diagnostic criteria and so cannot be analyzed as a variable independent of the diagnosis. No radiographic specifications were made with respect to bacterial cases, but viral cases were not allowed to have consolidative processes/radiologic end-point pneumonia (to decrease the likelihood of unidentified bacterial pneumonia), and malarial cases were required to have clear radiographs. Like the WHO radiographic criteria, the pathogenic bacteremia that was required in this study for bacterial classification emphasizes specificity over sensitivity. That only a subset of patients had radiographic end-point pneumonia may reflect the fact that we analyzed patients with a pneumonia syndrome, and not necessarily pneumonia per se; alternatively, it may highlight inadequate sensitivity of an anteroposterior chest radiograph for diagnosis of pneumonia. In either case, children with pathogenic bacteremia require antibiotics, and markers that predict bacterial sepsis may be clinically useful irrespective of the radiographic correlate.
Supplementary Material
Notes
Author Contributions. M. A. G., M. L., C. V., R. A., P. L. A., S. A. C., R. C. W., Q. B., J. P. M., D. A. M., and D. F. W. conceptualized and designed the study. J. S., M. L., R. A., S. A., and K. D. A. performed the research. C. V. and Q. B. prepared the study master database. J. S., Y. T., and J. P. M. analyzed the data. J. S., K. G. P., Q. B., J. P. M., D. A. M., and D. F. W. drafted the initial manuscript. M. A. G., M. L., C. V., R. A., K. D. A., P. L. A., S. A. C., R. C. W., Q. B., D. A. M., and D. F. W. contributed with data collection instruments. J. S., M. A. G., M. L., Y. T., Q. B., J. P. M., D. A. M., and D. F. W. contributed with interpretation of data. M. L., S. A., and K. D. A. coordinated and supervised sample and data collection at the study site, contributed with data acquisition, and supported data management activities. J. S., M. A. G., M. L., R. A., R. C. W., J. P. M., D. A. M., and D. F. W. revised the manuscript critically for important intellectual content. All authors reviewed and approved the final manuscript as submitted.
Disclaimer. The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the funding agencies.
Financial support. This work is supported by the Bill & Melinda Gates Foundation (grant OPP50092). Q. B. has a fellowship from the program Miguel Servet del Instituto de Salud Carlos III (ISCIII) (Plan Nacional de I+D+I 2008–2011; grant CP11/00269). L. M. has a fellowship from the program Río Hortega of the ISCIII (CM13/00260).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and all authors declare no conflict of interest.
References
- 1. Nair H, Simões EAF, Rudan I et al. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet 2013; 381:1380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. D'Acremont V, Kilowoko M, Kyungu E et al. Beyond malaria—causes of fever in outpatient Tanzanian children. N Engl J Med 2014; 370:809–17. [DOI] [PubMed] [Google Scholar]
- 3. Bassat Q, Machevo S, O'Callaghan-Gordo C et al. Distinguishing malaria from severe pneumonia among hospitalized children who fulfilled integrated management of childhood illness criteria for both diseases: a hospital-based study in Mozambique. Am J Trop Med Hyg 2011; 85:626–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Charkaluk M-L, Kalach N, Mvogo H et al. Assessment of a rapid urinary antigen detection by an immunochromatographic test for diagnosis of pneumococcal infection in children. Diagn Microbiol Infect Dis 2006; 55:89–94. [DOI] [PubMed] [Google Scholar]
- 5. Gwer S, Newton CRJC, Berkley JA. Over-diagnosis and co-morbidity of severe malaria in African children: a guide for clinicians. Am J Trop Med Hyg 2007; 77:6–13. [PMC free article] [PubMed] [Google Scholar]
- 6. Koram KA, Molyneux ME. When is malaria” malaria? the different burdens of malaria infection, malaria disease, and malaria-like illnesses. Am J Trop Med Hyg 2007; 77:1–5. [PubMed] [Google Scholar]
- 7. Gonsalves WI, Cornish N, Moore M, Chen A, Varman M. Effects of volume and site of blood draw on blood culture results. J Clin Microbiol 2009; 47:3482–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murdoch DR, O'Brien KL, Scott JAG et al. Breathing new life into pneumonia diagnostics. J Clin Microbiol 2009; 47:3405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Avni T, Mansur N, Leibovici L, Paul M. PCR using blood for diagnosis of invasive pneumococcal disease: systematic review and meta-analysis. J Clin Microbiol 2010; 48:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramirez M, Melo-Cristino J. Expanding the diagnosis of pediatric bacteremic pneumococcal pneumonia from blood cultures to molecular methods: advantages and caveats. Clin Infect Dis 2010; 51:1050–2. [DOI] [PubMed] [Google Scholar]
- 11. Resti M, Moriondo M, Cortimiglia M et al. Community-acquired bacteremic pneumococcal pneumonia in children: diagnosis and serotyping by real-time polymerase chain reaction using blood samples. Clin Infect Dis 2010; 51:1042–9. [DOI] [PubMed] [Google Scholar]
- 12. Bradley JS, Byington CL, Shah SS et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the pediatric infectious diseases society and the infectious diseases society of America. Clin Infect Dis 2011; 53:e25–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Donnelly JP, Baddley JW, Wang HE. Antibiotic utilization for acute respiratory tract infections in U.S. emergency departments. Antimicrob Agents Chemother 2014; 58:1451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. English M, Esamai F, Wasunna A et al. Assessment of inpatient paediatric care in first referral level hospitals in 13 districts in Kenya. Lancet 2004; 363:1948–53. [DOI] [PubMed] [Google Scholar]
- 15. Reyburn H, Mwakasungula E, Chonya S et al. Clinical assessment and treatment in paediatric wards in the north-east of the United Republic of Tanzania. Bull World Health Organ 2008; 86:123–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mulholland EK, Simoes EAF, Costales MOD, McGrath EJ, Manalac EM, Gove S. Standardized diagnosis of pneumonia in developing countries. Pediatr Infect Dis J 1992; 11:77–81. [DOI] [PubMed] [Google Scholar]
- 17. Valim C, Ahmad R, Lanaspa M et al. Responses to bacteria, virus and malaria distinguish the etiology of pediatric clinical pneumonia. Am J Respir Crit Care Med 2016; 193:448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. UCSC Genome Browser: Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12:996–1006. [DOI] [PMC free article] [PubMed]
- 21. DeLuca DS, Levin JZ, Sivachenko A et al. RNA-SeQC: RNA-Seq metrics for quality control and process optimization. Bioinformatics 2012; 28:1530–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gould J. GENE.E: Interact with GENE-E from R. R package version 1.8.0, 2013. http://www.broadinstitute.org/cancer/software/GENE-E. Accessed.
- 23. Ramilo O, Allman W, Chung W et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 2006; 109:2066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mejias A, Dimo B, Suarez NM et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berry MPR, Graham CM, McNab FW et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bloom CI, Graham CM, Berry MPR et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One 2013; 8:e70630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet 2011; 377:1264–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Courson DS, Cheney RE. Myosin-X and disease. Invited reviews: molecular motors 2015; 334:10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bishai EA, Sidhu GS, Li W et al. Myosin-X facilitates Shigella-induced membrane protrusions and cell-to-cell spread. Cell Microbiol 2013; 15:353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoshida S, Sasakawa C. Exploiting host microtubule dynamics: a new aspect of bacterial invasion. Trends Microbiol 2003; 11:139–43. [DOI] [PubMed] [Google Scholar]
- 31. Liu W, Yan M, Sugui JA et al. Olfm4 deletion enhances defense against Staphylococcus aureus in chronic granulomatous disease. J Clin Invest 2013; 123:3751–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amirbeagi F, Thulin P, Pullerits R et al. Olfactomedin-4 autoantibodies give unusual c-ANCA staining patterns with reactivity to a subpopulation of neutrophils. J Leukoc Biol 2015; 97:181–9. [DOI] [PubMed] [Google Scholar]
- 33. Liu W, Yan M, Liu Y et al. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc Natl Acad Sci USA 2010; 107:11056–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. doi: 10.4049/jimmunol.1103179. Liu W, Yan M, Liu Y, McLeish KR, Coleman WG Jr, Rodgers GP. Olfactomedin 4 inhibits cathepsin C-mediated protease activities, thereby modulating neutrophil killing of Staphylococcus aureus and Escherichia coli in mice. J Immunol 2012; 189:2460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brand HK, Ahout IML, de Ridder D et al. Olfactomedin 4 serves as a marker for disease severity in pediatric respiratory syncytial virus (RSV) infection. PLoS One 2015; 10:e0131927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thorburn K, Harigopal S, Reddy V, Taylor N, van Saene HKF. High incidence of pulmonary bacterial co-infection in children with severe respiratory syncytial virus (RSV) bronchiolitis. Thorax 2006; 61:611–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fensterl V, Sen GC. Interferon-induced IFIT proteins: their role in viral pathogenesis. J Virol 2015; 89:2462–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khsheibun R, Paperna T, Volkowich A, Lejbkowicz I, Avidan N, Miller A. Gene expression profiling of the response to interferon beta in Epstein-Barr-transformed and primary B cells of patients with multiple sclerosis. PLoS One 2014; 9:e102331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cherian T, Mulholland EK, Carlin JB et al. Standardized interpretation of paediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull World Health Organ 2005; 83:353–9. [PMC free article] [PubMed] [Google Scholar]
- 40. Mahomed N, Madhi S. Radiologic diagnosis of chest infection in children: WHO end-point consolidation. Pediatr Radiol 2014; 44:685–6. [DOI] [PubMed] [Google Scholar]
- 41. Hamilton N. ggtern: An Extension to ‘ggplot2’, for the Creation of Ternary Diagrams. R package version 2.1.5, 2016. https://CRAN.R-project.org/package=ggtern . Accessed .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.