Abstract
The FilmArray Respiratory Panel (FA-RP) is an FDA certified multiplex PCR that can detect 17 viruses and 3 bacteria responsible for upper respiratory tract infections, thus it is potentially useful to the assessment of the age-related prevalence of these pathogens.
In this observational study, we retrospectively analyzed the results of all the respiratory samples, which had been processed during 1 year-period (November 2015 to November 2016) with the FA-RP, in the Central Laboratories of Hygeia & Mitera General Hospitals of Athens, Greece. In order to have an age-related distribution, the following age groups were implemented: (<2), (≥2, <5), (≥5, <10), (≥10, <18), (≥18, <45), (≥45, <65), and (≥65) years old.
Among 656 respiratory samples tested, 362 (55%) were from male and 294 (45%) from female patients, while 356 (54.3%) were positive and 300 (45.7%) negative. In the first age-group (<2), 41/121 samples (33.9%) revealed human rhinovirus/enterovirus (HRV) and 16 (13.2%) adenovirus (Adv), followed by respiratory syncytial virus (RSV), coronavirus, human metapneumovirus (Hmpv), and parainfluenza viruses (PIV). In the age-group (≥2, <5), Adv predominated with 37/147 samples (25.2%), followed by HRV, RSV, coronavirus (all types), and influenza, Hmpv and PIV. In the age-group (≥5, <10), HRV was identified in 25/80 samples (31.3%), Adv in 18 (22.5%), influenza in 11 (13.8%), and Hmpv in 6 (7.5%). Influenza predominated in the age-group (≥10, <18), with 4/22 samples (18.2%), while in the remaining age-groups (≥18), HRV was the commonest isolated pathogen, 33/286 (11.5%), followed by influenza with 20 (7%) (influenza A H1-2009, 11/20).
In our patient series, HRV seemed to prevail in most age-groups, followed by Adv, although Influenza was the second most frequent pathogen isolated in the age-groups (≥18). Moreover, increasing age corresponded to increasing possibility of having a negative sample, indicating that FilmArray may be more useful before adolescence.
Keywords: age and respiratory infection, FilmArray, multiplex PCR, respiratory pathogen, respiratory virus
1. Introduction
Upper respiratory tract infections (URIs) can be a serious burden to the healthcare system.[1] The majority of URIs are of viral etiology, but definitive diagnosis can be difficult due to the overlapping clinical presentations of viral and bacterial infections, and the variable sensitivity, and lengthy turn-around time of viral culture.[1] The rapid and sensitive detection of respiratory viruses is essential for the early diagnosis and administration of appropriate antiviral therapy, as well as for the effective implementation of infection control measures.[2]
The FilmArray Respiratory Panel (FA-RP) assay is a fully automated, multiplex PCR system, which integrates nucleic acid extraction, nested amplification and detection in a reaction pouch preloaded with all reagents required for detection of 17 viruses and 3 bacteria.[3] It does not require advanced equipment or expertise in molecular diagnostics, making it a useful point-of-care test for acute respiratory infections,[3] and a valuable tool for the assessment of the age-related prevalence of the responsible pathogens.
2. Methods
In order to investigate the age-related prevalence of each respiratory pathogen, we retrospectively studied the results of all the samples, which had been processed during 1 year-period (November 2015 to November 2016) with the FA-RP, in the Central Laboratories of Hygeia & Mitera General Hospitals of Athens, Greece. Among 656 respiratory samples (Rs) tested, 362 (55%) were from male and 294 (45%) from female patients, while the median age of these 656 patients was 7 years old (0.1–92). In order to have an age-related distribution, the following age groups were implemented: (<2), (≥2, <5), (≥5, <10), (≥10, <18), (≥18, <45), (≥45, <65) and (≥65) years old. Ethical approval and/or patient consent were not necessary, because the paper does not report on primary research and all data analyzed were retrospectively collected as part of routine diagnosis.
2.1. BioFire FA-RP
Nasopharyngeal swab specimens were collected from patients with symptomatic respiratory tract infection and processed with FA-RP.[4] The assay detects nucleic acids of 17 respiratory viruses and 3 bacteria, including adenovirus (Adv), coronaviruses (HCoV: OC43, NL63, 229E and HKU1), human respiratory syncytial virus (HRSV or RSV), human metapneumovirus (Hmpv), influenza A virus (H1/2009, H1 and H3), influenza B virus, parainfluenza viruses (PIV, including PIV-1 to PIV-4), human rhinovirus/enterovirus (HRV), Bordetella pertussis, Chlamydophila pneumoniae, and Mycoplasma pneumoniae in a multiplex PCR.[4] The extraction, amplification, and detection steps take place in separate chambers of a self-contained, single-use pouch. The procedures were performed according to the manufacturer's instructions.
2.2. Statistical analysis
Chi-square tests and SPSS Statistics 17.0 (SPSS, Chicago, IL, USA) were used for statistical analysis, while P < .05 was considered statistically significant.
3. Results
3.1. Spectrum of respiratory pathogens detected
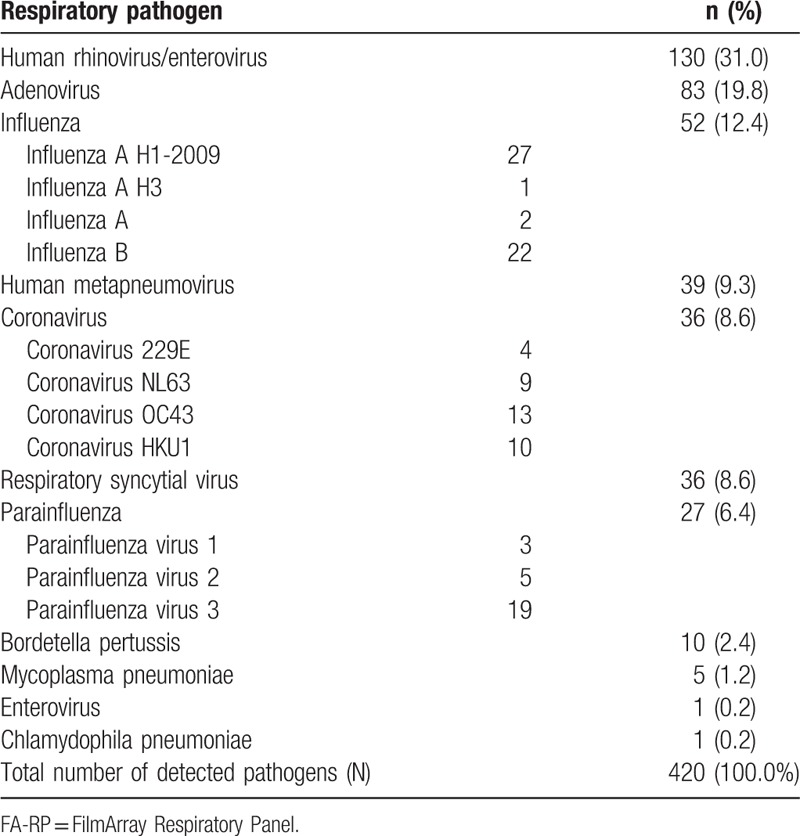
Among 656 respiratory samples tested, 356/656 (54.3%) were positive and 300/656 (45.7%) were negative. The total number of pathogens detected was larger than the positive samples, due to the frequent multiple isolations. Thus, 420 pathogens were detected, with HRV, Adv and influenza viruses (A and B) being the most usual isolated pathogens (Table 1).
Table 1.
The spectrum of pathogens detected by the FA-RP, irrespective of single or multiple identifications.
3.2. Age-related distribution of respiratory pathogens
In the first age-group (<2), 41 out of 121 samples (33.9%) revealed HRV, 16/121 (13.2%) Adv, 10/121 (8.3%) RSV, 10/121 (8.3%) coronavirus (229Ε, NL63, OC43, and ΗΚU1), 9 (7.4%) Hmpv, and 9 (7.4%) PIV (1, 2, and 3). In the age-group (≥2, <5), Adv predominated with 37 out of 147 samples (25.2%), followed by HRV (29/147, 19.7%), RSV (16/147, 16.9%), coronavirus (all types), and influenza (A and B), (14/147, 9.5%) each, Hmpv (12/147, 8.2%) and PIV (1, 2, and 3), 9 (6.1%). In the age-group (≥5, <10), HRV was identified in 25/80 samples (31.3%), Adv in 18/80 (22.5%), influenza (A and B) in 11 (13.8%), and Hmpv in 6/80 (7.5%). Influenza predominated in the age-group (≥10, <18), with 4/22 samples (18.2%), while in the remaining 3 age-groups (≥18), HRV was the commonest isolated pathogen 33/286 (11.5%), followed by Influenza with 20/286 (7%) (influenza A H1-2009, 11/20). The frequency of each pathogen in each respective age-group has been displayed in Figure 1. There was a statistically significant difference between all age groups, for HRV (P < .001), Adv (P < .001), influenza (ALL) (P = .02), RSV (P = .002), PIV-3 (P = .04), and coronavirus (ALL) (P = .02). Based on Figure 1, it is obvious that HRV prevailed in all age groups, but especially in the groups (<2) and (≥5, <10). Among Adv positive samples, there was a significant difference between the age group (≥2, <5) (most cases, 37/83 or 44.6%) and (≥45, <65) (no case) (P = .001), while most of the RSV positive samples (26/36, 72.2%) were noticed in ages under 5 years old. In regard with samples positive for influenza (A or B), the predominance of influenza in the age group (≥10, <18) was due to influenza B cases, while the majority of cases with influenza A H1-2009 were noticed in the ages under 10 years old (16/27, 59.3%). The latter was also true for Hmpv (27/39, 69.2%), while PIV-3, the main PIV type detected (Table 1) had a peak incidence in ages under 5 years old (15/19, 78.9%). Finally, most cases of coronavirus (all types included) were detected in children <10 years old (28 out of 35 cases, 80%), 10 of which (28.6%) were in children (<2), with 2 types of coronavirus being detected in a single sample.
Figure 1.
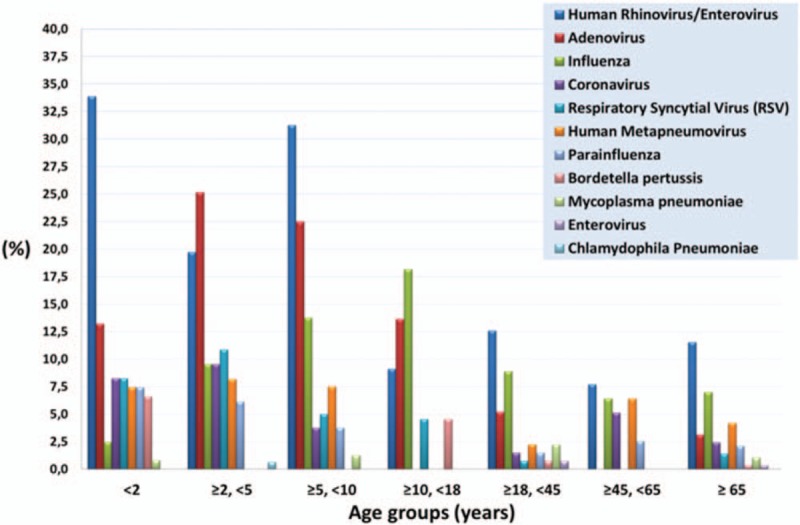
The frequency (%) of each pathogen's identification, among the total of samples in each separate age-group.
3.3. Multiple detections
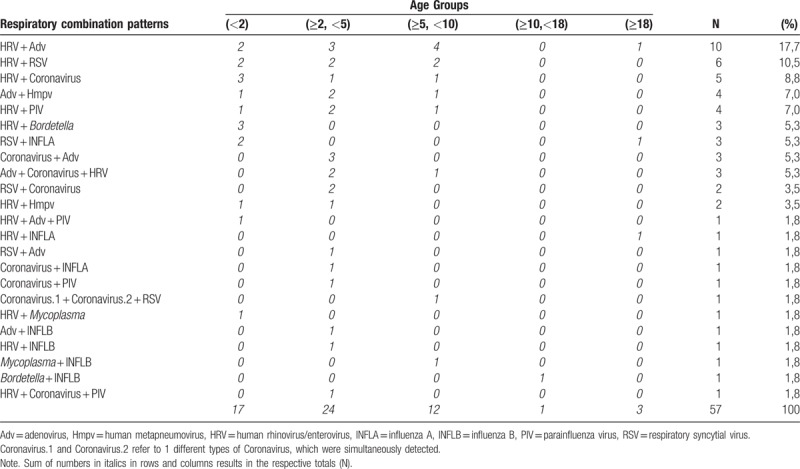
Among the total Rs, 57/656 (8.7%) revealed multiple isolations. Based on positive samples only, we had multiple pathogens in 57 out of 356 positive Rs. HRV participated in most multiple isolations, being the most frequent isolated pathogen (37/57, 64.9%) (Fig. 2). The second more frequently observed pathogens were PIV (1, 2, and 3), identified in 24 out of 57 samples (42.1%), and Adv identified in 23 out of 57 samples (40.4%), followed by RSV (15/57, 26.3%) (Fig. 2). It is remarkable that the combination HRV/Adv was the most commonly observed, with 10 cases out of 57 (17.5%), followed by HRV/RSV combination (6/57, 10.5%) (Table 2). Regarding the age-related distribution of multiple respiratory pathogens, it should be noted that most of them (41/57, 71.9%) concerned ages under 5 years old, while the combination HRV/Adv was mainly observed in the age group 2 to 10 (Table 2).
Figure 2.
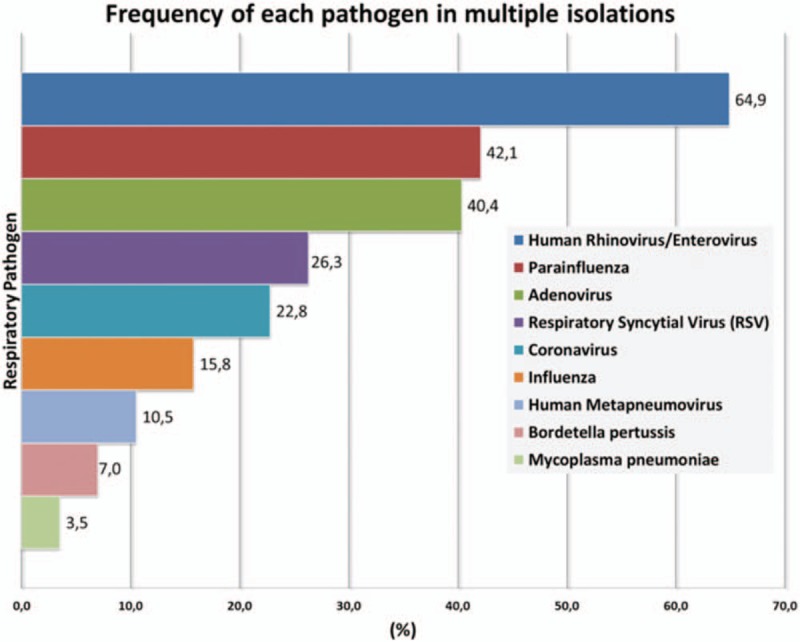
The frequency (%) of each pathogen in multiple isolations.
Table 2.
The age-related frequency of each respiratory combination pattern.
3.4. Negative samples
It was also remarkable that negative samples were mostly encountered in older ages. In particular, in the age group (<2), 33 out of 121 patients (27.3%) revealed no pathogen and the same was true for 42/147 (28.6%) patients in the age group (≥2, <5), 21/80 (26.3%) in the age group (≥5, <10), 12/22 (54.5%) in the age group (≥10, <18), 89/135 (65.9%) in the age group (≥18, <45), 56/78 (71.8%) in the age group (≥45, <65) and 47/73 (64.4%) in the age group ≥65 (Fig. 3).
Figure 3.
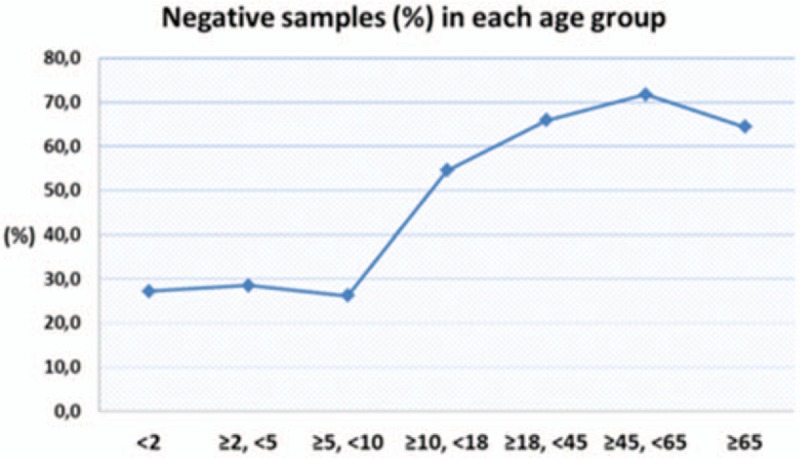
Negative samples were mostly encountered to older ages, with a peak incidence in the age group (≥45, <65).
4. Discussion
Respiratory infections are a common cause of pediatric morbidity[5] and the vast majority of acute URIs are caused by viruses.[6] Molecular point-of-care testing for respiratory viruses has the potential to improve the detection rate of respiratory viruses, improve the use of influenza antivirals and reduce unnecessary antibiotic use.[7] However, the age-related prevalence of these identifiable upper respiratory pathogens has not been clearly established. In this study, the routine clinical application of the FA-RP was indirectly used as a molecular point-of-care testing for the age-stratification of the respiratory viruses detected, in a tertiary hospital of Greece.
HRV was the most commonly identified virus in almost all age groups, but especially in the groups (<2) and (≥5, <10). The increasing availability of multiplex PCR panels allows rapid detection of HRV and provides the opportunity for timely treatment and early recognition of outbreaks.[8] The predominance of HRV in our patient series is in accordance with previously published data.[9–13] including data with patients under the age of 2.[14] HRV has been previously identified as the most common pathogen detected among symptomatic young children in a pediatric emergency department, with the majority of them requiring hospitalization.[15] The same was true in our series, where all cases were symptomatic and most cases had been hospitalized. Based on our results, HRV should certainly be included in the differential diagnosis of URIs in older patients, too.
In the case of Adv, our results indicate that the age group (≥2, <5) seem to be at the highest risk of getting infected. Until now, Adv has been considered as 1 of the 5 most frequent respiratory pathogens among children,[16–18] with infants being the population of the highest risk. In our study, Adv is referred for the first time as the most frequent responsible pathogen in the age group (≥2, <5) and second in priority among all the identified pathogens. These results may reflect a trend of increasing total incidence and higher-age peak incidence for Adv positive cases.
RSV is a regular winter visitor that is highly contagious among persons of all ages.[19] Although long recognized as the major viral pathogen of the lower respiratory tract of infants, it has also been implicated in severe lung disease in adults, especially the elderly.[20,21] This was not confirmed in our study, where most RSV positive samples were noticed in ages under 5 years old. However, it should be noticed that 4 out of the 5 positive cases which occurred in the ages ≥18 (data not shown), belonged to the age group (≥65), highlighting the relevance of this virus in all ages.[22]
As for samples positive for Influenza (A or B), we should underline the total prevalence of influenza A H1-2009, the high incidence of influenza A H1-2009 in ages under 10 years old and the preference of influenza B incidence in the middle ages. Influenza A and B are important causes of respiratory illness in all age groups.[23] Influenza has been referred as the most common etiologic agent (55.7%), especially among adults.[24] This was not true in our case, except for the age group (≥10, <18). However, influenza A has been previously considered as 1 of the 3 most frequent pathogens in the community setting.[16,18] Our data indicate that this applies totally to ages over 5 years.
Moreover, it has been proposed that the highest incidence of Hmpv infection is among children, as seropositivity for hMPV approaches 100% by 5 to 10 years of age.[25] In our series, although 70% of Hmpv positive cases were identified in ages under 10, we also had 12 cases after the age of 18 years old, underlying that a considerable number of Hmpv infections are diagnosed in adults and the elderly.[25]
Finally, PIV are single-stranded RNA viruses belonging to the paramyxovirus family that may cause of URIs and lower respiratory infections in infants, children and immunocompromised individuals.[26,27] It is considered that almost 90% of PIV-positive samples are from pediatric patients younger than 5 years old, with no infant under 1 month of age found positive.[26] These were true in our series, with PIV-3 being the most representative type. Also, HCoV are considered important respiratory pathogens, especially in hospitalized children under 2 years of age and in immunosuppressed patients.[28] Our data indicate that although 80% of cases belonged to the age group <10 years old, the peak incidence was met at the age group (≥2, <5).
The available reference data on the frequency of respiratory co-infections vary, although most reported percentages are higher than our results. In a study of 315 respiratory samples from children under 6 years of age, the FA-RP identified multiple co-infections (39%) with 2, 3, 4, and up to 5 different viruses.[29] Co-infection has also been found in 31%,[13] 36%,[9] 37%,[10] 42%,[30] and 51.8%[31] of positive respiratory samples. In our study, 9% of all our respiratory patients had multiple pathogens and 16% of all positive respiratory patients (almost 1 of 6 positive samples appeared with co-detections). There is no obvious explanation about the significance of lower frequency of co-detection in our series of patients. However, the predominance of HRV in our single and multiple detections is in accordance with previously published data.[32] Moreover, HRV/Adv and HRV/RSV combinations have been previously mentioned as the most frequent co-infection patterns.[30,33] Interestingly, it has been suggested that the presence of RSV reduces the probability of rhinovirus infection, but that, if a co-infection occurs, both viruses cause clinical symptoms.[34] Regarding the age-related distribution of multiple pathogens, our observation that patients under 5 years of age were more likely to have multiple detections, is almost in accordance with the eliminated previously published data.[33,35–37] Nevertheless, Tramuto et al have also noticed that most of their patients with multiple etiology were adults and elderly.[24]
In conclusion, understanding the viral etiology and age-specific incidence of URIs can help identify risk groups and probably inform vaccine delivery. In this study, HRV predominated in almost all ages, Adv in the age group (≥2, <5) and Influenza was second in the ages (≥18). Finally, it was remarkable that the negative samples were mostly attributed to older ages, with a peak incidence in the age group (≥45, <65), confirming that, as the age of the patient increases, the positivity rate for the PCR decreases proportionately,[32] a finding that needs further explanation.
Author contributions
Conceptualization: Nikolaos J. Tsagarakis.
Data curation: Nikolaos J. Tsagarakis, Anthi Sideri, Panagiotis Makridis, Argyro Triantafyllou.
Formal analysis: Nikolaos J. Tsagarakis, Argyro Triantafyllou.
Investigation: Nikolaos J. Tsagarakis, Anthi Sideri, Panagiotis Makridis.
Methodology: Nikolaos J. Tsagarakis, Panagiotis Makridis.
Software: Nikolaos J. Tsagarakis.
Supervision: Nikolaos J. Tsagarakis, Alexandra Stamoulakatou, Eleni Papadogeorgaki.
Validation: Nikolaos J. Tsagarakis, Panagiotis Makridis, Eleni Papadogeorgaki.
Visualization: Nikolaos J. Tsagarakis, Anthi Sideri, Panagiotis Makridis, Argyro Triantafyllou, Alexandra Stamoulakatou, Eleni Papadogeorgaki.
Writing – original draft: Nikolaos J. Tsagarakis.
Writing – review & editing: Nikolaos J. Tsagarakis, Eleni Papadogeorgaki.
Footnotes
Abbreviations: Adv = adenovirus, FA-RP = FilmArray Respiratory Panel, HCoV = coronaviruses, Hmpv = human metapneumovirus, HRSV, RSV = (human) respiratory syncytial virus, HRV = human rhinovirus/enterovirus, PIV = parainfluenza virus, Rs = respiratory samples, URIs = upper respiratory tract infections.
The authors report no conflicts of interest.
References
- [1].Layman CP, Gordon SM, Elegino-Steffens DU, et al. Rapid multiplex PCR assay to identify respiratory viral pathogens: moving forward diagnosing the common cold. Hawaii J Med Public Health 2013;72(9 suppl 4):24–6. [PMC free article] [PubMed] [Google Scholar]
- [2].Babady NE, Mead P, Stiles J, et al. Comparison of the Luminex xTAG RVP Fast assay and the Idaho Technology FilmArray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol 2012;50:2282–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Andersson ME, Olofsson S, Lindh M. Comparison of the FilmArray assay and in-house real-time PCR for detection of respiratory infection. Scand J Infect Dis 2014;46:897–901. [DOI] [PubMed] [Google Scholar]
- [4].Chen JH, Lam HY, Yip CC, et al. Clinical evaluation of the new high-throughput Luminex NxTAG respiratory pathogen panel assay for multiplex respiratory pathogen detection. J Clin Microbiol 2016;54:1820–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lim FJ, de Klerk N, Blyth CC, et al. Systematic review and meta-analysis of respiratory viral coinfections in children. Respirology 2016;21:648–55. [DOI] [PubMed] [Google Scholar]
- [6].Jain N, Lodha R, Kabra SK. Upper respiratory tract infections. Indian J Pediatr 2001;68:1135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Brendish NJ, Malachira AK, Clark TW. Molecular point-of-care testing for respiratory viruses versus routine clinical care in adults with acute respiratory illness presenting to secondary care: a pragmatic randomised controlled trial protocol (ResPOC). BMC Infect Dis 2017;17:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].To KKW, Yip CCY, Yuen KY. Rhinovirus—from bench to bedside. J Formos Med Assoc 2017;116:496–504. [DOI] [PubMed] [Google Scholar]
- [9].Bhuyan GS, Hossain MA, Sarker SK, et al. Bacterial and viral pathogen spectra of acute respiratory infections in under-5 children in hospital settings in Dhaka city. PLoS One 2017;12:e0174488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Finianos M, Issa R, Curran MD, et al. Etiology, seasonality, and clinical characterization of viral respiratory infections among hospitalized children in Beirut, Lebanon. J Med Virol 2016;88:1874–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Louie JK, Roy-Burman A, Guardia-Labar L, et al. Rhinovirus associated with severe lower respiratory tract infections in children. Pediatr Infect Dis J 2009;28:337–9. [DOI] [PubMed] [Google Scholar]
- [12].McGrath EJ, Thomas R, Asmar B, et al. Detection of respiratory coinfections in pediatric patients using a small volume polymerase chain reaction array respiratory panel: more evidence for combined droplet and contact isolation. Am J Infect Control 2013;41:868–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Morikawa S, Kohdera U, Hosaka T, et al. Seasonal variations of respiratory viruses and etiology of human rhinovirus infection in children. J Clin Virol 2015;73:14–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Anders KL, Nguyen HL, Nguyen NM, et al. Epidemiology and virology of acute respiratory infections during the first year of life: a birth cohort study in Vietnam. Pediatr Infect Dis J 2015;34:361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chu HY, Englund JA, Strelitz B, et al. Rhinovirus disease in children seeking care in a tertiary pediatric emergency department. J Pediatric Infect Dis Soc 2016;5:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Liu WK, Liu Q, Chen DH, et al. Epidemiology of acute respiratory infections in children in Guangzhou: a three-year study. PLoS One 2014;9:e96674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Malekshahi SS, Azad TM, Yavarian J, et al. Molecular detection of respiratory viruses in clinical specimens from children with acute respiratory disease in Iran. Pediatr Infect Dis J 2010;29:931–3. [DOI] [PubMed] [Google Scholar]
- [18].Wong-Chew RM, Espinoza MA, Taboada B, et al. Prevalence of respiratory virus in symptomatic children in private physician office settings in five communities of the state of Veracruz, Mexico. BMC Res Notes 2015;8:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hall CB. The nosocomial spread of respiratory syncytial viral infections. Annu Rev Med 1983;34:311–9. [DOI] [PubMed] [Google Scholar]
- [20].Hashem M, Hall CB. Respiratory syncytial virus in healthy adults: the cost of a cold. J Clin Virol 2003;27:14–21. [DOI] [PubMed] [Google Scholar]
- [21].Simoes EA. Respiratory syncytial virus infection. Lancet 1999;354:847–52. [DOI] [PubMed] [Google Scholar]
- [22].Gamino-Arroyo AE, Moreno-Espinosa S, Llamosas-Gallardo B, et al. Mexico Emerging Infectious Diseases Clinical Research Network (La Red). Epidemiology and clinical characteristics of respiratory syncytial virus infections among children and adults in Mexico. Influenza Other Respir Viruses 2017;11:48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clark NM, Lynch JP., III Influenza: epidemiology, clinical features, therapy, and prevention. Semin Respir Crit Care Med 2011;32:373–92. [DOI] [PubMed] [Google Scholar]
- [24].Tramuto F, Maida CM, Napoli G, et al. Burden and viral aetiology of influenza-like illness and acute respiratory infection in intensive care units. Microbes Infect 2016;18:270–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Principi N, Esposito S. Paediatric human metapneumovirus infection: epidemiology, prevention and therapy. J Clin Virol 2014;59:141–7. [DOI] [PubMed] [Google Scholar]
- [26].Liu WK, Liu Q, Chen DH, et al. Epidemiology and clinical presentation of the four human parainfluenza virus types. BMC Infect Dis 2013;13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pawelczyk M, Kowalski ML. The role of human parainfluenza virus infections in the immunopathology of the respiratory tract. Curr Allergy Asthma Rep 2017;17:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Trombetta H, Faggion HZ, Leotte J, et al. Human coronavirus and severe acute respiratory infection in Southern Brazil. Pathog Glob Health 2016;110:113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marcone DN, Carballal G, Ricarte C, et al. Respiratory viral diagnosis by using an automated system of multiplex PCR (FilmArray) compared to conventional methods. Rev Argent Microbiol 2015;47:29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Adam K, Pangesti KN, Setiawaty V. Multiple viral infection detected from influenza-like illness cases in Indonesia. Biomed Res Int 2017;2017:9541619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].O’Grady KF, Grimwood K, Sloots TP, et al. Prevalence, codetection and seasonal distribution of upper airway viruses and bacteria in children with acute respiratory illnesses with cough as a symptom. Clin Microbiol Infect 2016;22:527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Litwin CM, Bosley JG. Seasonality and prevalence of respiratory pathogens detected by multiplex PCR at a tertiary care medical center. Arch Virol 2014;159:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cebey-Lopez M, Herberg J, Pardo-Seco J, et al. Viral co-infections in pediatric patients hospitalized with lower tract acute respiratory infections. PLoS One 2015;10:e0136526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Karppinen S, Toivonen L, Schuez-Havupalo L, et al. Interference between respiratory syncytial virus and rhinovirus in respiratory tract infections in children. Clin Microbiol Infect 2016;22:208–16. [DOI] [PubMed] [Google Scholar]
- [35].Kono J, Jonduo MH, Omena M, et al. Viruses associated with influenza-like-illnesses in Papua New Guinea, 2010. J Med Virol 2014;86:899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu T, Li Z, Zhang S, et al. Viral Etiology of acute respiratory tract infections in hospitalized children and adults in Shandong Province, China. Virol J 2015;12:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Peng D, Zhao D, Liu J, et al. Multipathogen infections in hospitalized children with acute respiratory infections. Virol J 2009;6:155. [DOI] [PMC free article] [PubMed] [Google Scholar]


