Abstract
The vertebrate filamin family (A, B, and C) is part of the spectrin family of actin cross-linking proteins. Family members share high sequence similarity (>64%) and have both common and isoform-distinct functionalities. To identify the basis for isoform-specific functionality, we perform an evolutionary trace of chordate filamin at the granularity of single residues. Our trace methodology is constrained to focus on neofunctionality by requiring that one isoform remain the ancestral type, whereas at least one isoform has an accepted mutation. We call divergence meeting these characteristics “class-distinctive.” To obtain a temporal and spatial context for class-distinctive residues, we derive an all-atom model of full-length filamin A by homology modeling and joining individual domains. We map onto our model both conserved and class-distinctive residues along with the period (Teleostei, Amphibian, and Mammalian) in which they diverged. Our phylogenetic analysis suggests that filamins diverged from a common ancestral gene between urochordate and vertebrate lineages. Filamins also diverged the most just after gene duplication, in the Teleostei period, with filamin C remaining closest to ancestral filamin. At the residue level, domains with well-characterized interfaces, IgFLN 17 and IgFLN 21 (immunoglobulin, Ig), have diverged in potentially critical residues in their adhesion protein–binding interfaces, signifying that isoforms may bind or regulate ligand binding differentially. Similarly, isoform divergence in a region associated with F actin–binding regulation suggests that isoforms differentially regulate F-actin binding. In addition, we observe some class-distinctive residues in the vicinity of missense mutations that cause filamin A and B–associated skeletal disorders. Our analysis, utilizing both spatial and temporal granularity, has identified potentially important residues responsible for vertebrate filamin isoform–specific divergence—significantly in regions where few binding partners have been discovered to date— and suggests yet to be discovered filamin-binding partners and isoform-specific differential regulation with these binding partners.
Keywords: FLN filamin, Ig Immunoglobulin, ABD actin binding domain, CH calponin homology, ET evolultionary trace
Introduction
Gene duplication is a major source of genetic and functional diversity during evolution. After gene duplication, constraints on each gene are relaxed allowing for the accumulation of mutations in each gene (Wagner 2002). The high rate of accepted mutations after gene duplication (Lynch and Force 2000; Kondrashov et al. 2002) can result in development of neofunctionality in one gene while maintaining parental function in the other, or modifications to parental function in both genes such that the sum of function is equal to parental function (Lynch and Conery 2000). Amplifying this complexity, some proteins have several functional roles and therefore a multiplicity of sources of evolutionary constraints. For example, several proteins that physically maintain the cellular cytoskeleton also interact functionally with various noncytoskeletal proteins. Evolutionary biology seeks to use phylogenetic and structural information to reconstruct how protein families diverged and thus make nonbiased inferences about primary sequence changes that can influence functional changes among family members.
Of particular interest due to its involvement in numerous cellular processes is the filamin protein family, composed in vertebrate isoforms named A, B, and C (Stossel et al. 2001; Van der Flier and Sonnenberg 2001). Filamins participate in dynamical shaping and maintenance of the cytoskeleton and are involved in the physical stress response and mechanoprotection of cells (Glogauer et al. 1998; D'addario et al. 2001; Kainulainen et al. 2002). Filamins also have a role in cellular cytoskeletal anchoring by binding the tails of transmembrane cellular adhesion proteins such as GPIbα and integrins (Sharma et al. 1995; Loo et al. 1998; Takafuta et al. 1998; Xu et al. 1998; Feng et al. 2003; Travis et al. 2004). Filamins act as a protein scaffold binding over 70 proteins (Zhou, Boren, and Akyurek 2007) many of which are cytosolic effector proteins, transmembrane receptors, and ion channels. Most of the understanding about the role of filamin in cells has been derived from bottom-up studies in which filamin-binding partners have been initially identified through the yeast two-hybrid method. Functional studies of these filamin-binding proteins suggest that filamin A plays a more prominent role in cells compared with filamin B and that almost all the interactions with other proteins occur in the C-terminal region of filamin (Feng and Walsh 2004). In this study, we use an evolutionary-based technique to understand the primary sequence determinants that are responsible for functional difference among family members. This approach has advantages over the current bottom-up techniques in that it is not biased toward a particular isoform nor a particular region of an isoform. The basic technique we use, known as evolutionary trace (ET), addresses the following question: What are the sequence determinants associated with functional roles of filamin that after gene duplication are maintained across all isoforms, distributed between different isoforms, or newly evolved within an isoform?
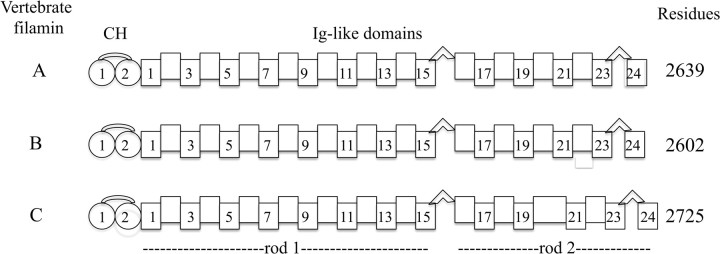
Structurally filamin isoforms are very similar to one another (fig. 1). Each is comprised of an N-terminal actin-binding domain (ABD) followed by a set of 24 repeated Ig-like domains (IgFLN; immunoglobulin, Ig) (Gorlin et al. 1990). The ABD consists of two tandem calponin homology (CH 1 and CH 2) domains containing three principal actin-binding sites (Nakamura et al. 2005). CH domains are all α-helical. Ig-like domains (∼100 residues) are ∼4 nm long and consist of seven β strands (A–G) arranged in two β-pleated sheets (Fucini et al. 1997). Two flexible hinge regions connect Ig-like domains 15 and 16, and 23 and 24. Short linkers connect the rest of the domains. Ig-like domains in the range 1–15 (rod 1) are devoid of known binding partners, whereas domains 16–24 (rod 2) have numerous binding partners. The 24th Ig-like domain is the dimerization domain. Although there is no high-resolution atomic structure of filamin, low-resolution electron micrographs (EMs) reveal a monomer of ∼80 nm (Hartwig and Stossel 1981; Nakamura et al. 2007), with the N-terminal Ig-like domains appearing in a linear conformation and the distal C-terminal Ig-like domains in a compact conformation (Nakamura et al. 2007).
FIG. 1.
Filamin family of proteins. Models of vertebrate filamin A-, B-, and C-domain architecture is composed of an ABD followed by 24 Ig-like domains. Ig-like domains are in two regions denoted by rod 1 (1–15) and rod 2 (16–24) showing their Ig-like and CHs; and regions rod 1 and rod 2. Large linker regions separate domains 15 from 16, and 23 from 24. All isoforms have the same domain architecture. The largest difference among isoforms is that filamin C domain 20 has a larger insert of ∼80 residues.
Experimental evidence and filamin-related developmental diseases suggest that isoforms have both overlapping and distinctive roles. Filamins A and B are ubiquitously expressed, and both are localized to the cortex and stress fibers (Sheen et al. 2002), whereas filamin C is expressed in smooth muscle and the heart and localized to the sarcomeric Z-line complex (Van der Ven et al. 2000). Disease mutations, many of which are caused by missense mutations, and filamin isoform depletion mouse models suggest that filamins A and B are critical for various aspects of skeletal, vasculature, cardio, and cerebral development (Sheen et al. 2002; Robertson 2004; Feng et al. 2006; Hart et al. 2006; Farrington-Rock et al. 2007; Lu et al. 2007; Zhou, Tian, and Sandzen 2007), whereas filamin C is critical for muscle and heart development (Goetsch et al. 2005; Dalkilic et al. 2006). At the cellular level, filamin A deficiency causes loss of motility and cell shape and promotes plasma membrane blebbing (Cunningham et al. 1992), whereas surface expression, localization, or turnover of many filamin-binding partners is disturbed. Many factors may contribute to isoform-specific properties. For example, the filamin C isoform unique histology contributes to functional differences between filamin C and the other isoforms. However, more subtle factors such as small coding differences can also lead to functional differences, for example, modifications to interactions with binding partners (Nakamura et al. 2009). Because the three filamins share at least 64% amino acid similarity, it is not surprising that isoforms have both overlapping function and localized sequence changes that lead to isoform functional variations.
One approach to identifying the underlying causes behind functional similarities and differences between protein isoforms is analyzing site-specific conservation and divergence between protein family members. Various techniques to derive functional significance from phylogenetic sequence constraints have been developed. These techniques are based on the observations that conserved function is correlated with conserved sequence within isoform families (Lichtarge et al. 1996; Gu 1999, 2006). Furthermore, clusters of family members that have conserved residues at specific sites but with different residue types (e.g., acidic vs. basic, polar vs. nonpolar) could have undergone a switch in function at or around these sites (Lichtarge et al. 1996; Gu 1999, 2006). Methods employed to distinguish functionally important sites in protein families include site-specific rate of evolution (Gu 1999), changes in residue type (e.g., acidic to basic) (Gu 2006), and structural thermodynamic properties (Dokholyan and Shakhnovich 2001).
To analyze functionally divergent features of vertebrate filamin isoforms, we perform a site-specific ET combined with a phylogenetic analysis and ancestral reconstruction to determine where and when isoforms diverge. Our trace methodology differs from that of Lichtarge et al. (1996) because we consider a site informative only if at least one isoform remains the ancestral type, while at least one other isoform diverges to become fixed to different type. We call this form of divergence “class-distinctive.” To isolate regional differences in evolutionary patterns, we then quantify ancestral and class-distinctive residues across domains. To visualize identified evolutionary divergent features in the context of a filamin structure, we develop an all-atom homology model of filamin A. Structurally, our model is very similar in size and features to low-resolution EMs. Finally, we map conserved and class-distinctive residues onto our filamin structure to observe in what time period (Teleostei, Amphibian, or Mammalian) and where in the context of a filamin structure different filamin isoforms have diverged. Phylogenetically, the filamins appear to diverge from a common single ancestral gene between chordate invertebrate and vertebrate lineages. Filamin isoforms diverge the most during the Teleostei period, whereas Filamin C diverges the least from the common ancestor to all three isoforms. From current reports, most filamin functionality is attributed to rod 2, but we do not observe a significant difference in quantity of either ancestral or class-distinctive sites between this region and the preceding region, rod 1. An examination of the well-characterized ABD and Ig-like domains 17 and 21 suggests isoform-specific regulation of ancestrally conserved function of actin and adhesion protein binding. Furthermore, we observe surface-exposed distinctive residues in the vicinity of missense mutations in both the ABD and some N-terminal Ig-like domains (e.g., 14) suggesting isoform distinct divergence involving disease-associated pathways. Thus, our results suggest that, although few binding partners have been localized to domains in rod 1, these domains may have played a role in the isoform-specific divergence and ancestral conservation and thus the function of filamin family members.
Materials and Methods
Modeling
An all-atom model of filamin A is created from homology models of individual filamin A domains. Most domains are joined by relative positioning pairs of domains onto a multidomain structure of ddFilamin (PDB ID 1wlh), and then loops are generated to connect domains.
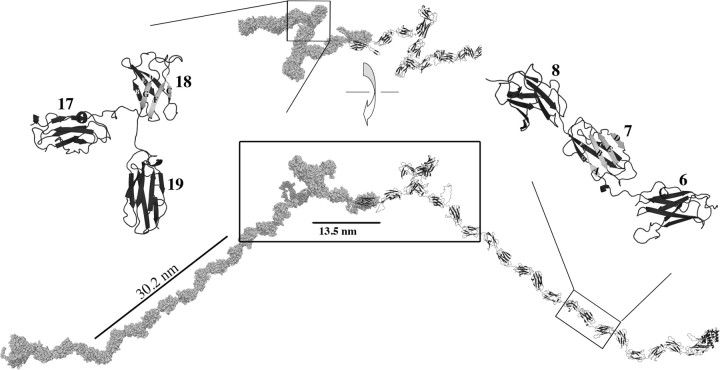
Homology modeling is performed using the Modeler module of the “InsightII” from Accelrys Inc. (www.accelrys.com). Homology modeling uses a template structure from a related protein to predict a structure for a target protein sequence. The homology models are evaluated for sequence–structure compatibility using the “Verify-3D” function of the Profiles-3D module from InsightII. Verify-3D scores residues based on how well they fit in their local environment. De novo loop generation is performed in “SYBYL” computational informatics software for molecular modeling from Tripos Inc. (www.tripos.com). To avoid residue clashes and approximate native structures, we relaxed pairs of linker-connected domains with “DMD/Eris/Medusa” (Yin et al. 2007a, 2007b; Ding et al. 2008). To create a filamin dimer (fig. 2), we superimpose each self-association domain (24) onto a structure of a dimer of IgFLNc24 (PDB ID 1v05) using “PyMOL.”
FIG. 2.
Atomic model of a filamin A dimer. This filamin dimer (space-filling model of a monomer left and cartoon model right) is produced by homology modeling individual domains, followed by their superimposition onto ddFilamin domains 4–5. Exception domains are 18 and 20. These domains have homology models lacking β strand A, resulting in a compact quaternary structure of C-terminal domains. The linear N-terminal domains have an extension of 30.2 nm. The length of last eight C-terminal domains equals to 15.3 nm. A 103° angle is observed between the filamin monomers.
Phylogenetic Analysis of the Filamin Family
Filamin protein sequences are obtained from a search of genomic databases. The general procedure is to obtain protein sequences using TBlastN with the query sequence of hsFilamin A on genomic databases. We only use sequences that have both the ABD and a large quantity of Ig-like domains (all but mosquito and sea urchin have the full complement of 24 Ig-like domains). The species included in this study are vertebrates: mammals (Homo sapiens, Mus musculus, Rattus norvegicus, and Canis familiaris), amphibians (Xenopus tropicalis), and teleosts (Danio rerio, Takifugu rubripes, and Tetraodon nigroviridis); urochordates: Ciona intestinalis and Ciona savignyi; and invertebrates: Strongylocentrotus purpuratus and Anopheles gambiae. Well-characterized sequences for H. sapiens filamin are obtained from NCBI (http://www.ncbi.nlm.nih.gov/) (hsFilamin A [NP_001447.2], B [NP_001448.2], and C [NP_001449.3]). Vertebrate and urochordate sequences are obtained from the Ensemble (release 41) database (http://www.ensemble.org), and invertebrates sequences are obtained from the NCBI database. Supplementary table S3, Supplementary Material online, lists the sequences we use in the study. To obtain a full-length Ciona filamin protein sequence with the full complement of 24 Ig-like domains, we use the consensus sequence from the following predicted proteins (ENSCSAVP00000017581, ENSCSAVP00000017582, ENSCSAVP00000017578, ENSCSAVP00000017579, ENSCSAVP00000017577, ENSCSAVP00000017583, ENSCSAVP00000017585, ENSCSAVP00000017580, ENSCSAVP00000017584, and ENSCINP00000006732).
Phylogenetic Tree Construction
Alignments are generated with the assistance of software alignment tools as well as by visual adjustment using both “JalView” (Clamp et al. 2004) and “Seaview” (Galtier et al. 1996). MrBayes (Huelsenbeck and Ronquist 2001) is employed to calculate all phylogenetic trees and for ancestral reconstruction. The outgroup chosen is a mosquito filamin from A. gambiae (AGAP004335-PA). We use the rate matrix amino acid model equalin, and fixed-rate model invgamma, 125,000 generations, sample frequency of 100, and three independent runs. A burn-in of 250 is used to create consensus trees as indicated by convergence of test runs. All but one branch possesses a posterior probability of partition of 1.00. This single exception occurs within the mammalian clade and possessed a posterior probability of (0.93). Tree-generation software “njplot” is employed to render trees (Perrière and Gouy 1996).
Evolutionary Trace
Determining isoform-specific divergence consists of ancestral reconstruction followed by a site-specific ET as detailed in supplementary figure S2, Supplementary Material online. Ancestors are reconstructed using MrBayes at the clade branch points as indicated in this figure. We used an amino acid rate model equalin with a fixed-rate model invgamma and either 50,000 or 10,000 generations, both of which converged well. To eliminate ancestral calls with high uncertainty, a consensus of 80% of the generations have to agree on an ancestral residue. We indicated with an X any residue in the ancestral sequence that did not meet these criteria. Using an alignment of these ancestral sequences, we performed an ET. At each site of this alignment, we monitor at periods Teleostei, Amphibian, or Mammalian, for a change in residue biochemical type that then becomes fixed until the human form. We additionally required that at least one isoform remained ancestral. Throughout this analysis, we use a reduced amino acid type set consisting of 16 residue types (S/T, I/L, K/R, D/E, and F/Y), 10 remaining single residues and a type for an insertion. We allow the skipping of at most one period to allow for period-specific divergence. We also account for divergence of Ciona by redefining the ancestral type, if all three Teleostei period ancestors are in agreement for residue type but disagreed with the common Ciona ancestor. The latter caveat occurred very infrequently.
We quantify ancestral and class-distinctive residues on a per-domain basis. Domain boundaries are derived from a multiple sequence alignment (MSA) of individual domains. Domain boundaries are listed in supplementary table S4, Supplementary Material online. Quantification is in absolute number of residues because most domains are of a similar size (mean 93 ± 8 with three outliers of greater than two standard deviations (SDs) from the mean, CH 1 (112), 18, and 20 (72)).
We also map distinctive residues and missense mutations onto our all-atom model of filamin A. Missense mutations are obtained from mutations listed along with filamin A (P21333) and filamin B (075369) in the UniProt database (http://www.uniprot.org).
Results
All-Atom Homology Model of a Filamin A Monomer
We construct an all-atom structural model of a human filamin A monomer using homology modeling (fig. 2). This monomer has an ABD, 24 Ig-like domains, linkers, and two hinge regions. In general, model construction consists of homology modeling individual domains, measurement of the quality of each model, positioning domains relative to one another, and generating linkers between domains (see Materials and Methods). We then construct a filamin A dimer model by superimposing the 24th Ig-like domain of each monomer onto the X-ray structure of the dimer of IgFLNc24 (PDB ID 1v05).
The protein-threading server “HHPRED” (Soding et al. 2005) is used to identify suitable templates for the ABD of filamin A. The top hit is to the ABD of α-actinin with an E value of 0. The architecture of filamin and the ABD of α-actinin are similar except for a longer negatively charged linker of filamin between calponin homology domain 1 (CH 1) and calponin homology domain 2 (CH 2) (supplementary fig. S1A, Supplementary Material online).
Homology Modeling of Individual Filamin A Domains
Filamin A Ig-like domains are modeled using templates composed of homologous Ig-like domains of human and Dictyostelium discoideum filamin (ddFilamin), except for the tridomain structure of domains 19–21 (PDB ID 2j3s). The MSA we use for template assignment is derived from a structural alignment of the human and ddFilamin Ig-like domains (supplementary fig. S1B, Supplementary Material online). A template for each target domain is chosen based on it having both a sequence and an insertion pattern that are similar to the query. Supplementary table S1, Supplementary Material online, contains the paired target/templates we use.
Homology models of filamin domains generally have high sequence–structure compatibility scores (supplementary table S1, Supplementary Material online). Quality analysis of each domain is performed using Verify-3d module of InsightII (Accelrys Inc.). This analysis measures the compatibility of modeled residues with their structural environment. A quality score of greater than 0.1 indicates a valid structural model with a correct fold (Luthy et al. 1992), whereas a higher score indicates a more accurate model. Typical experimental structures score around 1.0. The mean Verify-3d score we obtain for Ig-like domain models is 0.89 with SD 0.30. The score we obtain for the ABD is 0.75. The only model to produce a low score is IgFLNa18 (0.11). However, the homology model of IgFLNa18 has a high sequence identity (64% id) and a low root mean square deviation (RMSD) (0.23 Å) when compared with its IgFLNb18 template, suggesting that the IgFLNa18 model is correctly built based on the template. Furthermore, the structure of the template itself has a low-quality score of 0.56. Overall, the high mean quality score we obtain for the Ig-like domain models infers their high accuracy. Our homology model also shows a 1.1 Å RMSD, over 168 Cα atoms, with a recent crystal structure (PDB ID 3fer) of the filamin B ABD. The largest architectural difference between our model and the crystal structure is the positioning of a highly ancestral set of residues that lead to the neck region (linker between CH 2 and IgFLN 1). This may have the effect of modifying the relative position of the ABD and IgFLN 1.
Joining Models of Individual Filamin A Domains
We oriented domains relative to one another using the multiple-domain structure of ddIgFLN 4–6. Ig-like domains with short linkers (mean linker length 3 ± 1: supplementary table S2, Supplementary Material online) have an extended conformation (fig. 1, e.g., domains 6–8) (Nakamura et al. 2007) and have been proposed to have the same relative conformation as ddIgFLN 4 and 5 (PDB ID 1wlh) (Popowicz et al. 2004). Domains surrounding hinge regions are first oriented similar to that of short linkers, and then the distance between domains is extended to accommodate the larger linker. An adjustment to the orientation of domains around hinge 2, IgFLN 23 and IgFLN 24, is made based on visual inspection of a structure of these two domains produced by small angle X-ray light scattering (Sjekloca et al. 2007). After domain orientation, de novo loop generation is used to join domains.
During homology modeling, we identified several Ig-like domains that are potentially missing their N-terminal β strand. Ig-like domains are composed of two β sheets forming a β sandwich—one sheet with four (A–B–E–D) β strands and one with three (G–F–C) β strands. Our analysis of IgFLNb18 (PDB ID 2dmc) and IgFLNb20 (PDB ID 2e9i) revealed six-stranded Ig-like domains with most of β strand A forming a highly dynamic coil. In addition, the sequence of β strand A for these three domains shows low sequence identity with the canonical filamin β strand A sequence, a possible indication of a structural change. Thus, we adjusted our modeling such that Ig-like domains 18 and 20 lacked β strand A. The loss of β strand A affects the quaternary structure by placing the N- and C-termini adjacent to one another, thus inducing a compact multidomain structure (fig. 2, Ig domains 17–19). Because IgFLN 18 lacked strand A, we used ad hoc methods to determine its relative positioning to domain 17. IgFLN 18 and 19 share a short linker so we used ddFLN superimposition to derive their relative positioning. We modeled IgFLN 19–21 using the structure of IgFLN 19–21 (PDB ID 2j3s) (Lad et al. 2007). Both tri-domain models (17–19, 19–21) have an elbow-like architecture that, when incorporated into our structure, produce a zigzag conformation. In addition, we identify IgFLNa16 as possibly lacking β strand A. The structure of both IgFLNb16 (PDB ID 2ee9) and IgFLNc16 (PDIB ID 2d7n) reveal six-stranded Ig-like domains. However, sequence differences among β strand A of filamins A, B, and C suggest potential isoform-specific conformational differences. The sequence of β strand A IgFLNa16 has several residues that are conserved to canonical Ig-like domains (supplementary fig. S1B, Supplementary Material online). Therefore, we modeled IgFLNa16 with β strand A intact.
Comparison of All-Atom Homology Model of Filamin A to Structural Data
Our model of filamin A has similar dimensions and morphology to native filamin. The measured length of the model monomer is 82 nm, whereas the angle between the two monomers is 103°, similar to measured dimensions from early low-resolution EMs; Tyler et al. 1980; Hartwig and Stossel 1981). We conclude that the observed dimensions of filamin in EMs comes from two distinct regions, an N-terminal region from the ABD to hinge 1 made up of seven-stranded Ig-like domains and a compact region from hinge 1 to hinge 2 made up of both six- and seven-stranded Ig-like domains. In addition, the full range of conformational flexibility observed in EMs comes from hinge regions (Nakamura et al. 2007). Therefore, one model cannot show the full ensemble of conformations; molecular dynamic simulations on our all-atom filamin monomer would be required to sample these conformations.
Phylogenetic Analysis of the Filamin Family
We perform a Bayesian inference phylogenetic analysis on aligned invertebrate, invertebrate chordate, and vertebrate filamin sequences (fig. 3). All nodes of the consensus phylogenetic tree have at least a 90% level of confidence. Within the chordate filamin clade, we observe three monophyletic subclades, each corresponding to a different filamin isoform. The single ascidian sequence, from the urochordate genus, clusters outside these three vertebrate subclades, whereas invertebrates are even more distantly related. Therefore, two gene duplication events likely occur between the branching of urochordate and teleostei ancestors from the mammalian lineage, during early vertebrate evolution, to give rise to the three vertebrate filamin isoforms.
FIG. 3.
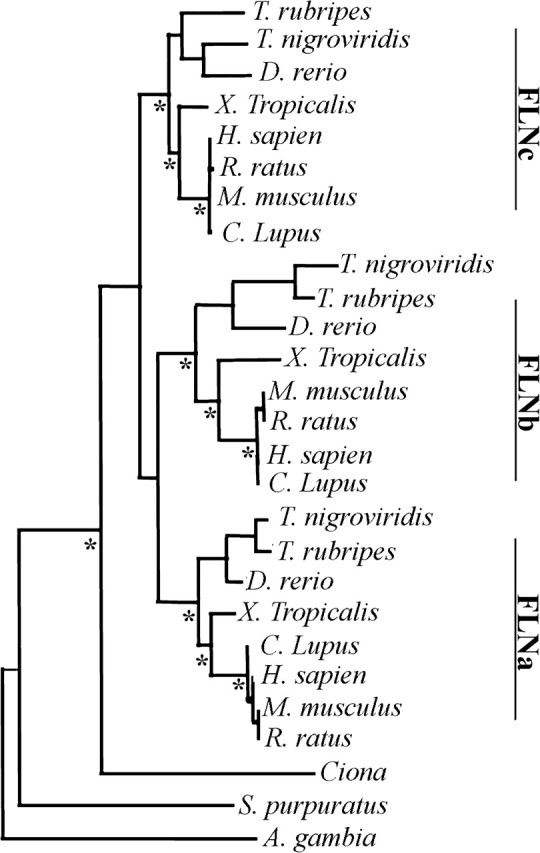
Phylogenetic analysis of the filamin family of proteins. Phylogenetic Bayesian analysis of aligned invertebrate and chordate filamin protein sequences suggests that gene duplication took place during early vertebrate evolution, prior to Teleosts, resulting in three filamin isoform genes. Furthermore, urochordate Ciona filamin is closely related to the chordate common ancestor of the three vertebrate filamin isoforms. The level of confidence in all branches is high. * denotes ancestral reconstructed branch points. (Abbreviations: Ciona family: consensus Ciona Intestinalis and Ciona Savignyi).
Determining the single invertebrate chordate filamin ancestor allows us to perform a vertebrate ET of the filamin family. If, at a particular site, a residue type is conserved across all isoforms including the Ciona ancestor, then maintenance of parental functionality in all isoforms is hypothesized to have occurred. We call these residues “ancestral.” If, however, at this site at least one isoform has maintained the residue type of the ancestral Ciona and at least one of the other isoforms became fixed with a new residue type, both maintenance of ancestral functionality and derivation of neofunctionality is assumed to have occurred. We call these isoform-specific divergent residues class-distinctive. All other modes of evolution are called “other.” We further annotate the class-distinctive residues by the period in which the divergence occurs, that is, as Teleostei, Amphibian, or Mammalian (supplementary fig. S2C, Supplementary Material online). Our method of isoform-specific divergence analysis is more restrictive than the ET method developed by Lichtarge et al. (1996), because we require maintenance of the ancestral type in at least one isoform for a site to be considered class distinctive.
Regional Analysis of Ancestral Conservation and Isoform-Specific Divergence
We analyzed how far and when each isoform functionally diverged from their common ancestor by quantifying both conserved and class-distinctive residues in the context of the complete protein. We find that filamin isoforms are 64% similar to one another and the common ancestor. We also find that 21% of the sites are class distinctive (fig. 4A). Of the class-distinctive sites, 78% diverged in the Teleostei period. Class-distinctive changes have few additions in the Amphibian period and a modest increment in the Mammalian period. The distribution of class-distinctive sites among isoforms is A 35%, B 43%, and C 22% (fig. 4B). Thus, we hypothesize that filamin B contains the most neofunctionality, and filamin C contains the most parental functionality.
FIG. 4.
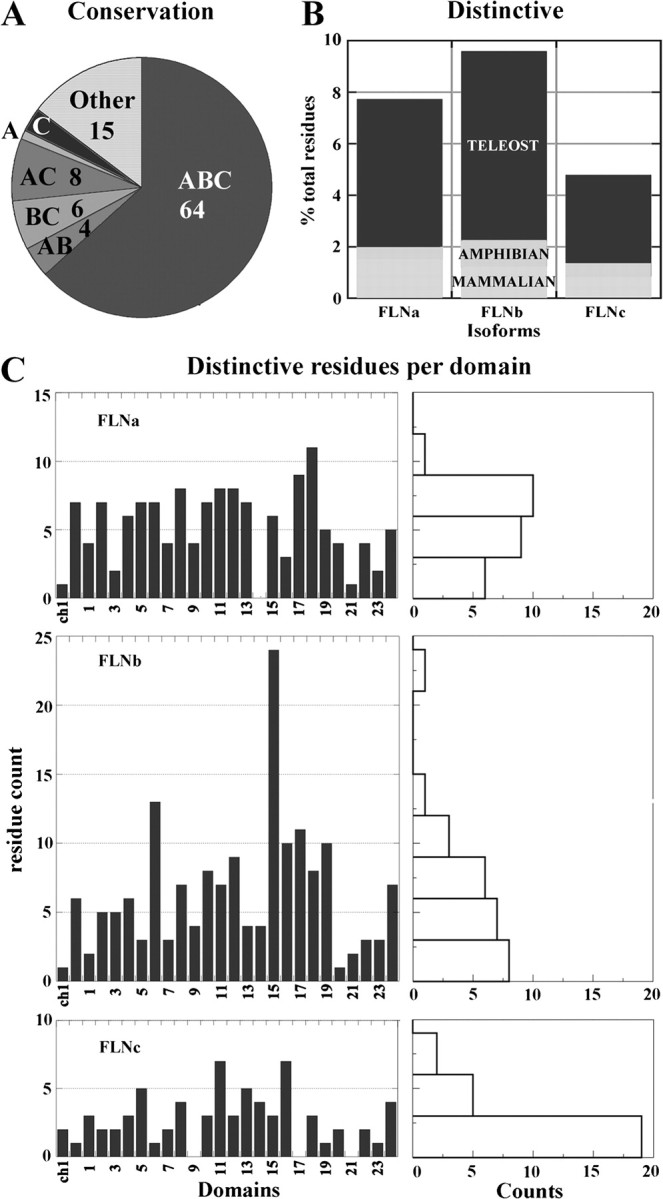
Quantification of ancestral conservation and distinctive divergence among filamin isoform. (A) Ancestral conservation analysis. An analysis of the similarity among isoforms derived from ET results. The cumulative count of sites that are ancestral in all three isoforms (ABC); are ancestral in exactly two isoforms and class-distinctive in a third (AB, BC, and AC); are ancestral in exactly one isoform, and distinctive class-distinctive in at least one isoform (A, B, and C). Most sites are fully conserved back to the urochordate common ancestor (64%). Filamin C retains the most ancestral residues. (B) Regional differences in isoform distinctive divergence. Cumulative count of distinctive class-distinctive residues distributed over periods. The most distinctively divergence occurred during the Teleostei period. Filamins A and B distinctively diverged the greatest and filamin C the least. (C) Distribution of Teleostei period distinctive class-distinctive residues across domains. Graph on the right is an accompanying histogram of each distribution. There is a nonuniform distribution in each isoform. The composition of many individual domains reflects the overall composition and is reflected in the mean per domain of A 7 ± 3, B 9 ± 5, and C 4 ± 2. Distinctive divergence attenuates proceeding C-terminal domains, A 20, B 19, and C 15. Domains having large counts are A 17, 18; B 6, 15; and C 11, 16. Domains having low counts are A CH 1, 14, 21; B CH 1, 20; and C CH 2, 6, 9, 17, 19, 21, 23.
To isolate regional differences in conservation of ancestral function among isoforms, we quantify ancestral sites within each domain. We observe a bimodal distribution in counts of ancestral residues (supplementary fig. S3 histogram, Supplementary Material online) with the first mode containing eight domains centered around ∼55% and the second mode containing 18 domains centered about ∼70%. Between domains 2–15, domains with lower conservation (mode 1) are interspersed with domains of higher conservation (mode 2). Most domains known to participate in protein–protein interactions (16–24) are contained within mode 2. CH 1 (actin binding) and the 21st Ig-like domain (integrin binding) have exceptionally high levels of conservation (>80%). Supplementary table S5, Supplementary Material online, summarizes the most notable findings. Surprisingly, a number of domains in rod 1 that currently have little experimental evidence of associated function also have as large a proportion of ancestral residues suggesting maintenance of ancestral function (e.g., Ig-like domains 3, 4, 9, 12, and 14).
To isolate regional differences in isoform divergence, we quantify class-distinctive residues within each domain (fig. 4C). We limit this analysis to the Teleostei period because only 22% of the class-distinctive residues follow this period. During the Teleostei period, class-distinctive divergence in the ABD is mostly limited to CH 2 (CH 2 (A 7, B 6, and C 1). CH 1 directly binds F-actin (Nakamura et al. 2005); thus, all isoforms may have a similar interface with F-actin. Hence, it is possible that isoforms differ in their regulation of actin binding because CH 2 has been proposed to play a regulatory role in binding F-actin (Robertson 2005; Sawyer et al. 2009).
We also observe the distribution of class-distinctive residues across Ig-like domains. All isoforms diverge in almost every Ig-like domain; thus, divergence through class-distinctive residues occurs ubiquitously but with variations in degree, rather than through modes such as localization to a limited number of specific domains. There are, however, isoform distinct patterns of divergence. First, in almost every domain, filamins A and B have more class-distinctive residues than filamin C. This pattern is consistent with our results of total counts per isoform. Second, there are two dissimilar regions in each isoform, region 1 having a count that fluctuates between low and high counts and region 2 having consistently low counts. These regions differ in location by isoform (A 1–20, 21–23; B 1–19, 20–23; and C 1–16, and 17–23). Although most known protein interactions occur in rod 2 (Feng and Walsh 2004), many of the domains in rod 1 have similar conservation patterns as the domains in rod 2.
Some individual domains have extreme counts of class-distinctive residues. In Filamin A, domains 14 and 21 (integrin binding) have very few class-distinctive modifications, whereas domains 17 and 18 have many class-distinctive residues (9 and 11, respectively). In filamin B, domain 20 has hardly diverged distinctively, whereas domains 6 and 15 have extreme counts of class-distinctive residues (13 and 24, respectively). In filamin C, domains 9, 17, and 21 have no class-distinctive residues, whereas domains 11 and 16 have a relatively large number of class-distinctive residues (both have 7). Supplementary table S5, Supplementary Material online, summarizes the most notable findings. We hypothesize that isoform-divergence patterns at this level can highlight either critical regions for conservation of ancestral functionality or isoform-specific divergence.
A Structural Mapping of Conservation and Divergence of Well-Characterized Domains
To increase our understanding of the pattern of divergences, we place ancestral, class-distinctive, and other residues in the spatial context of our model of filamin A. We also place class-distinctive residues in a temporal context according to the period (Teleostei, Amphibian, or Mammalian) in which they became fixed. In addition, we observe the relative positioning of ancestral and class-distinctive residues. A single accepted mutation in an ancestral background indicates a minor modification to ancestral function, whereas a cluster of accepted mutations can highlight neofunctionality. We also label residues involved in disease and note if they are in the vicinity of class-distinctive residues.
We first report our findings for well-characterized domains that have known binding partners. These include filamin A, IgFLN 21-β7 integrin (Kiema et al. 2006) (fig. 5A), IgFLN 17-GPIbα (Nakamura et al. 2006) (fig. 5B), and ABD (Nakamura et al. 2006) (fig. 5C). In addition, we examine the spatial distribution of class-distinctive residues for domains in the range 5–10 and 14–18 (supplementary figs. S7 and Supplementary Data, Supplementary Material online).
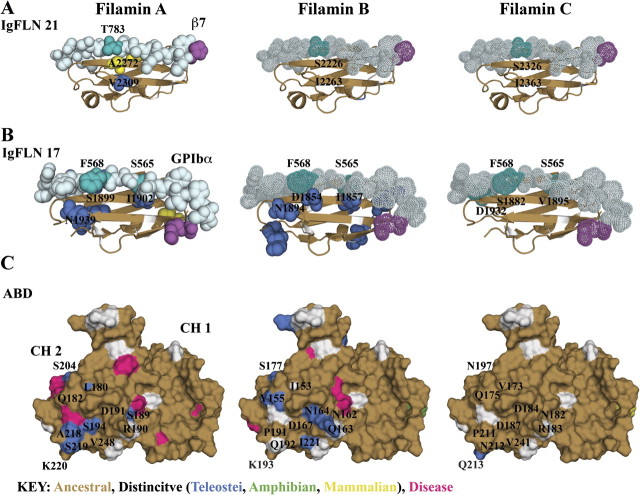
FIG. 5.
Analysis of class-distinctive residues in domains with well-characterized interfaces. Each column contains an all-atom model of filamin A, differentially colored based on isoform class-distinctive divergence (sand = ancestral, class-distinctive (blue = Teleostei, green = Amphibian, yellow = Mammalian), white = other). Ligands are colored in light cyan with residues adjacent to class-distinctive residues in dark cyan. Ligands are space filled for experimental and dotted for hypothetical complexes. The N-terminus of the ligand is colored magenta. (A) IgFLN 21 in complex with β7 integrin. There is one class-distinctive residue in the interface on filamin A. (B) IgFLN 17 in complex with GPIbα. There is one class-distinctive residue on filamin A and two on filamin B at the interface. (C) Surface model of the filamin A ABD, consisting of two calponin homology (CH) domains. CH 1 is highly ancestral, whereas CH 2 has a few class-distinctive residues that cluster around a few subdomains. Filamins A and B have more class-distinctive residues than C, and those are in the vicinity of colocalize near mutations that cause disease (pink).
A Structural Mapping of Conservation and Divergence of Adhesion Protein–Binding Domains
Isoforms may have evolved to differ in regulation of integrin binding. During the Mammalian period, filamin isoforms distinctively diverged in domain 21 at sites that are relevant to binding the developmentally critical binding partner, integrins. We find one class-distinctive substitution (fig. 5A) along β strand C in the well-characterized ligand-binding interface, an ancestral Ser/Thr in filamins B and C changed to an Ala at residue 2272 in filamin A during the Mammalian period. This class-distinctive site is localized to the end of the ligand-binding pocket and suggested to be critical for integrin binding based on several nuclear magnetic resonance perturbation (Kiema et al. 2006; Lad et al. 2007, 2008) studies. In addition, Kiema et al. showed in in vitro mutational studies that A2272 A2274/DK (a phosphomimic) diminished binding, whereas the substitutions AA/ST (ancestral residue type) did not (Kiema et al. 2006). These findings suggest the yet-to-be-confirmed possibility that the class-distinctive mutations to an Ala in filamin A disrupted the ability of this isoform to be posttranslationally regulated.
Isoforms may also have evolved to differ in regulation of binding GPIbα. Domain 17 has two Teleostei period distinct sites in the binding pocket with GPIbα (fig. 5B). On the N-terminal end of the interface, an ancestral Ser (filamin A S1899) to class-distinctive Asp substitution is observed in filamin B, whereas on the C-terminal end, an ancestral filamin C Val to class-distinctive Ile substitution is observed in filamins A and B. The N-terminal class-distinctive site aligns with the class-distinctive site in the integrin-binding interface. Evidence for its critical nature comes from the site-directed mutagenesis of the adjacent ligand residue (Cranmer et al. 2005). The Ser to Asp class-distinctive substitution at this interface suggests the possibility of isoform-specific regulation of interactions involving this domain. Distinctive sites on domains 17 and 21 also localize outside the well-characterized binding interface involving strands C and D (supplementary figs. S4A and Supplementary Data, Supplementary Material online), suggesting the possibility of other modes of binding to these domains in addition to the canonical interface involving these two strands.
We hypothesize that other domains having a pattern of distinctive residues on their β strands C and D similar to that of domains 17 and 21 (supplementary fig. S4C, Supplementary Material online) also possess a canonical interface. We find that rod 1 domain IgFLNc14 and rod 2 domains IgFLNa19 and IgFLNb19 have the closest match. Of these three domains, IgFLN 19 is most similar to IgFLNa21. This is yet another property suggesting binding to domains within rod 1 as well as rod 2. In addition, the substitutions of Ser/Thr residues in class-distinctive sites appears to be a recurrent pattern as all distinctive residues within these additional domains involve mutations to serine or threonine. There is some evidence that some ligands may bind multiple domains including domain 19, that is, migfilin and β7 integrins bind both domains 19 and 21 (Kiema et al. 2006; Nakamura et al. 2006). Therefore, it is possible that isoforms distinctively diverge at analogous sites because these locations are good targets to modulate binding of the canonical mode. Another possibility is that common modifications are the result of common selective pressure on each domain when multiple Ig-like domains bind the same ligand.
A Structural Mapping of Conservation and Divergence of the ABD
Of particular interest are class-distinctive sites implying that filamin isoforms may also differ in regulation of actin binding. The N-terminal ABD is composed of two CH domains connected by a larger linker (supplementary fig. S5A and Supplementary Data, Supplementary Material online). Quantification of class-distinctive residues suggests that isoforms have distinctively differentiated in CH 2 to a much greater extent than CH 1. The first CH domain is more conserved than the second and is involved in both directly binding F-actin and regulation of F-actin binding by interacting with calcium-activated calmodulin (Nakamura et al. 2005). The second CH domain has only been associated with regulating F-actin binding to date. Within CH 2, most class-distinctive residues are surface exposed and localize around helices B, C, E, and F (fig. 5C and supplementary fig. S5C, Supplementary Material online). The biochemical nature of class-distinctive residues differs by isoform, however, with transitioning from a more charged surface to a more uncharged polar surface appearing as a general trend. Distinctive changes in filamin A are primarily to serine residues that diverged from either large polar or charged residues. The filamin B class-distinctive residues transitioned from charged residues to large bulky residues. The only filamin C class-distinctive residue in CH 2 is a large polar residue that diverged from a charged residue. We also observe that gain-of-function missense mutations (Robertson et al. 2003; Sawyer et al. 2009) in this region, causing similar developmental skeletal disorders, localize in the vicinity of class-distinctive residues (residues colored pink in fig. 5C). Thus, we have isolated a particular region of CH 2 in which all isoforms have distinctly diverged from their ancestral form. Furthermore, we show that class-distinctive residues are in the vicinity of several missense mutations causing disease in both filamins A and B. Therefore, we propose that class-distinctive residues within this region may allow for filamin isoforms to differentially regulate F-actin binding.
A Structural Mapping of Conservation and Divergence of N-terminal Domains IgFLN 5–10
Loops between β-strands appear to play a prominent role in isoform divergence in N-terminal domains. Quantitative analysis of filamin class-distinctive residues implies that filamin isoforms diversified in central domains (supplementary fig. S6, Supplementary Material online). Many class-distinctive residues in IgFLN 5–10 are surface exposed, in loops between β strands, and within a residue environment that is mostly ancestral. IgFLNb6 was noted for large numbers of class-distinctive residues. These cluster to various regions of the domain (arrows in supplementary fig. S6A, Supplementary Material online, point to class-distinctive residue clusters). The largest cluster maps to a surface exposed loop between β strands C and D. This multiresidue substitution encompasses a negatively charged loop (784DARVLSEDEEDV795) in filamin B replacing an ancestral hydrophobic loop (APGVVGPAEADI) of filamins A and C. A motif scan (http://www.elm.eu.org/) suggests that filamin B potentially contains either a 14–3–3 or a casein kinase II (CK2) phosphorylation site, and filamins A and C potentially contain an SH3-binding motif.
Thus, in general, domains in the range 5–10 are highly conserved; however, we find class-distinctive residues in multiple domains where two isoforms utilized the same site for isoform-specific divergence, but accepted mutations were to different residue types. For example, in IgFLN 9 (supplementary fig. S6B, Supplementary Material online, e.g., IgFLNa9 and IgFLNb9, near center of the domain), two class-distinctive sites became fixed in filamin A, one during the Teleostei period (Q1115L, along β strand D) and the other during the Mammalian (P1087R, along β strand B) period. In filamin B, these sites became fixed during the Teleostei period (Q1115S, P1087E). These examples show that migration away from ancestral functionality occurred in both isoforms independently but at the same site.
We also see sites of mammalian divergence within domain 9 of filamins B and C, with a rare cluster of sites in IgFLNa9 (supplementary fig. S6B, Supplementary Material online). Thus, all three isoforms independently diverged in the Mammalian period within the same domain, although some sites in these domains diverged earlier in some isoforms. These sites suggest that critical functional changes were occurring in filamin during the Mammalian period involving domain 9.
A Structural Mapping of Conservation and Divergence of C-terminal Domains IgFLN 14–18
In domain IgFLNb15 class-distinctive residues clustered in two to three large groups, each surrounded by ancestral residues (supplementary fig. S7, Supplementary Material online). One of these clusters localizes to eight residues in the turn between β strands A and B that are encoded by a single exon (Chakarova et al. 2000). This region has been categorized previously as an alternative splice variant in filamin A (Maestrini et al. 1993; Patrosso et al. 1994) and a deletion in filamin B (Chakarova, et al. 2000). These previous classifications are consistent with our findings that filamin B distinctively diverged from ancestral filamins A and C by a deletion in this region. A motif scan suggests that this region may contain a GSK-3 phosphorylation site. These results imply that regions beyond the canonical interface involving strands C and D may be functionally important.
Our results show that class-distinctive sites may be correlated with the loss of β strand A in domain 18. Domain 18 from both filamins A and B have a large number of class-distinctive residues that localize to β strands A, B, and G. When we derived the homology model of a filamin A monomer, we modeled domain 18 as lacking β strand A. Because isoform divergent sites localize to this unique structural feature, each isoform may have a distinct intradomain architecture between domains 17, 18, and 19. The other domain we modeled lacking β strand A (domain 20) is highly ancestral in the analogous region (data not shown) suggesting that the architecture of 19, 20, and 21 predated gene duplication and is preserved from the common ancestor.
A Structural Mapping of Disease Mutations
Finally, we note that some class-distinctive residues occur near sites in which missense mutations in filamins A and B cause similar developmental skeletal disorders (supplementary figs. S6, S7, and S8A and S8B, Supplementary Material online). Almost all these disease mutations are on residues labeled ancestral. Sites where mutations exist are predominantly surface exposed on loops between β strands or near linkers between domains. Although disease mutations and distinctive sites are localized within one domain (e.g., IgFLNa10 and IgFLNb15 supplementary fig. S8A and S8B, Supplementary Material online), we also observe mutations and distinctive sites localized near each other while being on different domains (supplementary fig. S6B, intersection between IgFLNb7 and IgFLNb8, Supplementary Material online) and localized to conserved sites on different isoforms (e.g., IgFLNa10 and IgFLNb10). Thus, there appears to be a correlation with class-distinctive changes and etiology of disease. Disease mutations may interfere with protein–protein interactions or alter structural aspects of filamin domains.
Discussion
Structural Aspects of Filamin
Using phylogenetic analysis, an ET, and an all-atom structural model of filamin A, we analyze spatial and temporal patterns of filamin family divergence to identify elements involved in isoform class-distinctive and common function. Our structural model of filamin A is consistent with recent experimental data (Nakamura et al. 2007) that suggest filamin A has two distinct architectural regions, a linearly extended region followed by a more compact region.
Experimental evidence suggests that interdomain interactions in the compact region driven by β strand A play a role in modulating interactions with filamin-binding partners (Lad et al. 2007). We find that domains 18 and 20 have a similar ternary conformation lacking β strand A. Kesner et al. found that both domains are stable, independent of β strand A when pulled apart under low levels of force (Kesner et al. 2009); however, there are several class distinctive sites in β strand A of domain 18, whereas domain 20 is highly ancestral and lacks class distinctive sites in this strand. These data suggest that interdomain interactions involving β strand A of Ig-like domain 18 may have a biologically significant role similar to that of β strand A of Ig-like domain 20; however, the role of domain 18 may have arisen after gene duplication and could be distinct among isoforms. Therefore, we propose that some functional differences among isoforms may reflect differences in quaternary structure or difference may reflect coevolution of the interacting strands.
Phylogeny of Vertebrate Filamin
Our phylogenetic study of filamin suggests that vertebrate filamins (A, B, and C) originated from a single ancestral gene that existed prior to the advent of vertebrates and that this gene is also the progenitor of the single extant urochordate (tunicate) filamin gene. Evidence for a single filamin in tunicates comes from examining sequence databases of the Ciona genomes and to our knowledge is first reported here. Our analysis derives from the supposition that functionality of the progenitor to the single tunicate filamin was conserved and modified after the gene duplications leading to the three vertebrate isoforms.
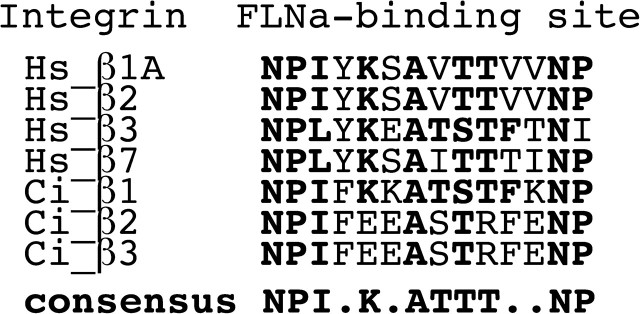
Assuming vertebrate filamin evolved from a single invertebrate chordate filamin, we hypothesize that the role of ancestral filamin involved integrin and actin binding because the filamin domains involved in binding integrins and actin have the highest content of ancestral residues of any domains, and all vertebrate filamin isoforms bind similar β1 integrin family members (Van der Flier and Sonnenberg 2001; Gontier et al. 2005). Furthermore, tunicates have been shown to have integrin-like transcripts (Ewan et al. 2005) including a filamin-binding sequence that is highly conserved with vertebrate integrins (fig. 6). Recently, the interaction of filamin with integrins has been suggested to play a critical role for cell-fate determination (Gehler et al. 2009), which could be critical for the evolution of various organs. Because all vertebrate filamin isoforms are critical to the development of the cardiovasculature system (Feng et al. 2006; Hart et al. 2006; Charitakis and Basson 2007; Zhou, Tian, and Sandzen 2007) and tunicates have a well-developed single chambered heart and vasculature (Davidson 2007) we hypothesize that filamin originated from a protein that had a role in cardiovasculature development.
FIG. 6.
Clustal alignment of β1 integrin family members in the filamin A binding region. Top four sequences are human (Hs), rest are from Ciona (Ci) (Ewan et al. 2005). Homologous residues in Ciona and human are in bold. The highest sequence similarity is between human β3 integrin and Cionaβ1 integrin (70% id).
Evolutionary Trace
Our ET of vertebrate filamin reveals ancestral conservation and isoform distinctive divergence down to the spatial granularity of an individual residue and temporal granularity to three periods—Teleostei, Amphibian, and Mammalian. We observe a nonuniform distribution of ancestrally conserved and class-distinctive residues that varies over periods, across domains, and among isoforms. These findings would suggest that transitions to an isoform distinctive residue with commensurate neofunctionality were governed both by spatial and temporal affects upon function. Most known protein interactions have been isolated to domains 16 and above. However, we do not see a clear delineation in conservation and divergence patterns between these C-terminal and N-terminal regions. In fact, we observe class-distinctive residues in the vicinity of missense mutations leading to disease in rod 1, suggesting relevance to these class distinctive changes. There may be a bias toward C-terminal interactions in part due to the primary method used to initially identify interactions with filamin, the yeast two-hybrid (Y2H) using binding partners as bait. Y2H libraries tend to be C-terminal biased based on the method used to derive them. In the few cases where other binding assays have been used, such as the Y2H method using filamin as “bait,” binding partners to the rod 1 region have been identified. These include actin-binding filamin A in the range 9–15 (Nakamura et al. 2007) and migfilin-binding filamin B domains 10–13 (Takafuta et al. 2003). Thus, from the distribution of ancestral and class-distinctive residues across domains, we propose that filamin has binding partners in rod 1 that have yet to be identified. Furthermore, the temporal distribution of class distinctive residues reveals a small number of Mammalian period distinctive residues in rod 1 that may have played a prominent role in filamin-isoform divergence during this period. An example discussed above was domain 9 from Filamin A. Because filamin A plays a critical role in heart development (Feng et al. 2006; Charitakis and Basson 2007), these sites may be associated with more recent evolutionary advances in vertebrates such as changes associated with the transition from a three to four chambered heart.
We also hypothesize that the class-distinctive sites may differentially regulate isoform interactions with binding partners. Evidence for this possibility is that significantly more class-distinctive changes to Ser/Thr residues are found in filamin A (21% or 2.3 SD) than B (11% or 1 SD) or C (13% or 0.76 SD). Commensurate with this, 26% of the in vivo S/T posttranslational modifications identified in UniProt (www.uniprot.org) records associated with filamin A are class distinctive (15, filamin A P21333; 14, filamin B O75369; and 2, filamin C Q14315), whereas none of the sites listed in UniProt records for filamin B or C are class distinctive. We also find evidence that class-distinctive sites may be involved in isoform-specific binding. In one example, we identify a class-distinctive residue on each of filamin-B and filamin-C domains 23 that was identified as critical for the distinct binding of filamin A to FILGAP (Nakamura et al. 2009).
As discussed above, we have also suggested that class-distinctive sites may be involved in regulation of actin binding because we observe several class-distinctive sites in ABD CH 2. This domain in filamin has been shown to be associated with actin-binding regulation based on gain-of-function disease mutations (Sawyer et al. 2009). This domain has also been associated with phospholipid binding in the homologous ABD of α-actinin (Full et al. 2007), and there are reports that suggest filamin is also regulated by lipid binding (Furuhashi et al. 1992). Isoform-specific regulation by lipids of actin binding may explain distinct cellular localization of filamins A and B in some cells, for example, proliferating chondrocytes (Krakow et al. 2004).
Recently, Kesner et al. reported the possibility that filamin A Ig domains have a heterogeneous set of stable conformations when forced to unfold under biologically significant levels of force (Kesner et al. 2009). The simplest explanation for the heterogeneity of unfolding is primary sequence differences among the filamin A Ig-like domains. Ig-like domains of different isoforms could have different forced unfolding properties because they also differ in primary sequence. Differences in behavior during forced unfolding could include variations in the stress required to induce conformational shifts and variations in the conformation of stable intermediates. Therefore, we posit that class-distinctive residues mediate isoform distinct responses to stress and may explain a potential mechanism for isoform distinct roles in cellular differentiation and mechanoprotection (Glogauer et al. 1998; D'Addario et al. 2003; Gehler et al. 2009).
In summary, our work has focused on understanding functions of filamin family members by analyzing site-specific divergence, using the ET method, spanning vertebrate developmental periods—Teleostei, Amphibian, and Mammalian. We find that isoforms diverge from one gene between invertebrate chordate and vertebrate lineages; that most divergence occurs in Teleostei; and that filamin C diverged the least. Our results also suggest that isoforms have diverged with regard to specificity for binding partners or regulatory function. In addition, similarities in the functional divergence we observe between rod 1 and rod 2 are not consistent with reported scaffold protein activity dominated by interactions domains in rod 2. Thus, our nonbiased identification of patterns of class-distinctive and ancestral residues in N-terminal domains should be used as a basis for further investigations.
Supplementary Material
Acknowledgments
This work is supported in part by the American Heart Association grant No. 0665361U and the National Institutes of Health grant R01GM080742.
References
- Chakarova C, Wehnert MS, Uhl K, Sakthivel S, Vosberg HP, van der Ven PF, Furst DO. Genomic structure and fine mapping of the two human filamin gene paralogues FLNB and FLNC and comparative analysis of the filamin gene family. Hum Genet. 2000;107:597–611. doi: 10.1007/s004390000414. [DOI] [PubMed] [Google Scholar]
- Charitakis K, Basson CT. Degenerating heart valves: fill them up with filamin? Circulation. 2007;115:2–4. doi: 10.1161/CIRCULATIONAHA.106.663237. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cranmer SL, Pikovski I, Mangin P, Thompson PE, Domagala T, Frazzetto M, Salem HH, Jackson SP. Identification of a unique filamin A binding region within the cytoplasmic domain of glycoprotein Ibalpha. Biochem J. 2005;387:849–858. doi: 10.1042/BJ20041836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 1992;255:325–327. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- D'Addario M, Arora PD, Ellen RP, McCulloch CA. Regulation of tension-induced mechanotranscriptional signals by the microtubule network in fibroblasts. J Biol Chem. 2003;278:53090–53097. doi: 10.1074/jbc.M309027200. [DOI] [PubMed] [Google Scholar]
- D'Addario M, Arora PD, Fan J, Ganss B, Ellen RP, McCulloch CA. Cytoprotection against mechanical forces delivered through beta 1 integrins requires induction of filamin A. J Biol Chem. 2001;276:31969–31977. doi: 10.1074/jbc.M102715200. [DOI] [PubMed] [Google Scholar]
- Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–6534. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2007;18:16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan NV, Shakhnovich EI. Understanding hierarchical protein evolution from first principles. J Mol Biol. 2001;312:289–307. doi: 10.1006/jmbi.2001.4949. [DOI] [PubMed] [Google Scholar]
- Ewan R, Huxley-Jones J, Mould AP, Humphries MJ, Robertson DL, Boot-Handford RP. The integrins of the urochordate Ciona intestinalis provide novel insights into the molecular evolution of the vertebrate integrin family. BMC Evol Biol. 2005;5:31. doi: 10.1186/1471-2148-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington-Rock C, Kirilova V, Dillard-Telm L, Borowsky AD, Chalk S, Rock MJ, Cohn DH, Krakow D. Disruption of the Flnb gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Hum Mol Genet. 2007;17:631–641. doi: 10.1093/hmg/ddm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Resendiz JC, Lu X, Kroll MH. Filamin A binding to the cytoplasmic tail of glycoprotein Ibalpha regulates von Willebrand factor-induced platelet activation. Blood. 2003;102:2122–2129. doi: 10.1182/blood-2002-12-3805. [DOI] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci USA. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA. The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol. 1997;4:223–230. doi: 10.1038/nsb0397-223. [DOI] [PubMed] [Google Scholar]
- Full SJ, Deinzer ML, Ho PS, Greenwood JA. Phosphoinositide binding regulates {alpha}-actinin CH2 domain structure: analysis by hydrogen/deuterium exchange mass spectrometry. Protein Sci. 2007;16:2597–2604. doi: 10.1110/ps.073146807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi K, Inagaki M, Hatano S, Fukami K, Takenawa T. Inositol phospholipid-induced suppression of F-actin-gelating activity of smooth muscle filamin. Biochem Biophys Res Commun. 1992;184:1261–1265. doi: 10.1016/s0006-291x(05)80018-x. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gehler S, Baldassarre M, Lad Y, Leight JL, Wozniak MA, Riching KM, Eliceiri KW, Weaver VM, Calderwood DA, Keely PJ. Filamin A-{beta}1 integrin complex tunes epithelial cell response to matrix tension. Mol Biol Cell. 2009;20:3224–3238. doi: 10.1091/mbc.E08-12-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem. 1998;273:1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- Goetsch SC, Martin CM, Embree LJ, Garry DJ. Myogenic progenitor cells express filamin C in developing and regenerating skeletal muscle. Stem Cells Dev. 2005;14:181–187. doi: 10.1089/scd.2005.14.181. [DOI] [PubMed] [Google Scholar]
- Gontier Y, Taivainen A, Fontao L, Sonnenberg A, van der Flier A, Carpen O, Faulkner G, Borradori L. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J Cell Sci. 2005;118:3739–3749. doi: 10.1242/jcs.02484. [DOI] [PubMed] [Google Scholar]
- Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Biol Evol. 2006;23:1937–1945. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Stossel TP. Structure of macrophage actin-binding protein molecules in solution and interacting with actin filaments. J Mol Biol. 1981;145:563–581. doi: 10.1016/0022-2836(81)90545-3. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kainulainen T, Pender A, D'Addario M, Feng Y, Lekic P, McCulloch CA. Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. J Biol Chem. 2002;277:21998–22009. doi: 10.1074/jbc.M200715200. [DOI] [PubMed] [Google Scholar]
- Kesner BA, Ding F, Temple BR, Dokholyan NV. N-terminal strands of filamin Ig domains act as a conformational switch under biological forces. Proteins. 2009;78:25–35. doi: 10.1002/prot.22479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakow D, Robertson SP, King LM, et al. (25 co-authors) Mutations in the gene encoding filamin B disrupt vertebral segmentation, joint formation and skeletogenesis. Nat Genet. 2004;36:405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- Lad Y, Jiang P, Ruskamo S, Harburger DS, Ylanne J, Campbell ID, Calderwood DA. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J Biol Chem. 2008;283:35154–35163. doi: 10.1074/jbc.M802592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad Y, Kiema T, Jiang P, Pentikainen OT, Coles CH, Campbell ID, Calderwood DA, Ylanne J. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. Embo J. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE. Evolutionarily conserved Galphabetagamma binding surfaces support a model of the G protein-receptor complex. Proc Natl Acad Sci USA. 1996;93:7507–7511. doi: 10.1073/pnas.93.15.7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo DT, Kanner SB, Aruffo A. Filamin binds to the cytoplasmic domain of the beta1-integrin. Identification of amino acids responsible for this interaction. J Biol Chem. 1998;273:23304–23312. doi: 10.1074/jbc.273.36.23304. [DOI] [PubMed] [Google Scholar]
- Lu J, Lian G, Lenkinski R, De Grand A, Vaid RR, Bryce T, Stasenko M, Boskey A, Walsh C, Sheen V. Filamin B mutations cause chondrocyte defects in skeletal development. Hum Mol Genet. 2007;16:1661–1675. doi: 10.1093/hmg/ddm114. [DOI] [PubMed] [Google Scholar]
- Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini E, Patrosso C, Mancini M, et al. (11 co-authors) Mapping of two genes encoding isoforms of the actin binding protein ABP-280, a dystrophin like protein, to Xq28 and to chromosome 7. Hum Mol Genet. 1993;2:761–766. doi: 10.1093/hmg/2.6.761. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Hartwig JH, Stossel TP, Szymanski PT. Ca2+ and calmodulin regulate the binding of filamin A to actin filaments. J Biol Chem. 2005;280:32426–32433. doi: 10.1074/jbc.M502203200. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Heikkinen O, Pentikainen OT, et al. (12 co-authors) Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS ONE. 2009;4:e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Pudas R, Heikkinen O, Permi P, Kilpelainen I, Munday AD, Hartwig JH, Stossel TP, Ylanne J. The structure of the GPIb-filamin A complex. Blood. 2006;107:1925–1932. doi: 10.1182/blood-2005-10-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrosso MC, Repetto M, Villa A, Milanesi L, Frattini A, Faranda S, Mancini M, Maestrini E, Toniolo D, Vezzoni P. The exon–intron organization of the human X-linked gene (FLN1) encoding actin-binding protein 280. Genomics. 1994;21:71–76. doi: 10.1006/geno.1994.1226. [DOI] [PubMed] [Google Scholar]
- Perrière G, Gouy M. An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Muller R, Noegel AA, Schleicher M, Huber R, Holak TA. Molecular structure of the rod domain of dictyostelium filamin. J Mol Biol. 2004;342:1637–1646. doi: 10.1016/j.jmb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Robertson SP. Molecular pathology of filamin A: diverse phenotypes, many functions. Clin Dysmorphol. 2004;13:123–131. doi: 10.1097/01.mcd.0000130235.95356.40. [DOI] [PubMed] [Google Scholar]
- Robertson SP. Filamin A: phenotypic diversity. Curr Opin Genet Dev. 2005;15:301–307. doi: 10.1016/j.gde.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, et al. (17 co-authors) Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the Filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: insights from the crystal structures of Filamin B actin binding domains. J Mol Biol. 2009;390:1030–1047. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Sharma CP, Ezzell RM, Arnaout MA. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J Immunol. 1995;154:3461–3470. [PubMed] [Google Scholar]
- Sheen VL, Feng Y, Graham D, Takafuta T, Shapiro SS, Walsh CA. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum Mol Genet. 2002;11:2845–2854. doi: 10.1093/hmg/11.23.2845. [DOI] [PubMed] [Google Scholar]
- Sjekloca L, Pudas R, Sjoblom B, Konarev P, Carugo O, Rybin V, Kiema TR, Svergun D, Ylanne J, Djinovic Carugo K. Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer. J Mol Biol. 2007;368:1011–1023. doi: 10.1016/j.jmb.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Takafuta T, Saeki M, Fujimoto TT, Fujimura K, Shapiro SS. A new member of the LIM protein family binds to filamin B and localizes at stress fibers. J Biol Chem. 2003;278:12175–12181. doi: 10.1074/jbc.M209339200. [DOI] [PubMed] [Google Scholar]
- Takafuta T, Wu G, Murphy GF, Shapiro SS. Human beta-filamin is a new protein that interacts with the cytoplasmic tail of glycoprotein Ibalpha. J Biol Chem. 1998;273:17531–17538. doi: 10.1074/jbc.273.28.17531. [DOI] [PubMed] [Google Scholar]
- Travis MA, van der Flier A, Kammerer RA, Mould AP, Sonnenberg A, Humphries MJ. Interaction of filamin A with the integrin beta 7 cytoplasmic domain: role of alternative splicing and phosphorylation. FEBS Lett. 2004;569:185–190. doi: 10.1016/j.febslet.2004.04.099. [DOI] [PubMed] [Google Scholar]
- Tyler JM, Anderson JM, Branton D. Structural comparison of several actin-binding macromolecules. J Cell Biol. 1980;85:489–495. doi: 10.1083/jcb.85.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- Van der Ven PF, Obermann WM, Lemke B, Gautel M, Weber K, Furst DO. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton. 2000;45:149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Wagner A. Asymmetric functional divergence of duplicate genes in yeast. Mol Biol Evol. 2002;19:1760–1768. doi: 10.1093/oxfordjournals.molbev.a003998. [DOI] [PubMed] [Google Scholar]
- Xu W, Xie Z, Chung DW, Davie EW. A novel human actin-binding protein homologue that binds to platelet glycoprotein Ibalpha. Blood. 1998;92:1268–1276. [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Modeling backbone flexibility improves protein stability estimation. Structure. 2007a;15:1567–1576. doi: 10.1016/j.str.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Meth. 2007b;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- Zhou X, Boren J, Akyurek LM. Filamins in cardiovascular development. Trends Cardiovasc Med. 2007;17:222–229. doi: 10.1016/j.tcm.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Zhou X, Tian F, Sandzen J, et al. (11 co-authors) Filamin B deficiency in mice results in skeletal malformations and impaired microvascular development. Proc Natl Acad Sci USA. 2007;104:3919–3924. doi: 10.1073/pnas.0608360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.