Abstract
Background:
Human telomerase reverse transcriptase (hTERT) plays an important role in cancer progression. Recently, several clinical studies investigated how the overexpression of hTERT predicts the poor prognosis of solid tumors. However, the results were inconclusive, partly because of the small numbers of patients included.
Method:
We systematically searched PubMed, Web of Science, and Embase to identify relevant studies until August 2017. Hazard ratios (HRs) with 95% confidence intervals (CIs) were used to evaluate the association of hTERT expression and survival outcomes.
Results:
A total of 27studies enrolling 2530 solid tumor patients were included in this meta-analysis. There were strong significant associations between hTERT overexpression and all endpoints: overall survival (OS) (HR = 1.50, 95% CI: 1.31–1.73, P = .00), disease-free survival (HR = 1.84, 95% CI: 1.38–2.46; P = .00), and recurrence-free survival (HR = 1.79, 95% CI: 1.07–2.99; P = .028). In the subgroup analysis, it was found that the overexpression of hTERT induced poor OS in lung cancer (HR = 1.51, 95% CI: 1.21–1.89; P = .00).
Conclusion:
Our comprehensive systematic review concluded that the overexpression of hTERT was associated with poor survival in human solid tumors. hTERT may be a valuable predictive biomarker for prognosis.
Keywords: cancer, hTERT, prognosis
1. Introduction
Cancer is a major public health problem worldwide. In China, it was predicted that there would be 4,292,000 newly diagnosed invasive cancer cases in 2015, which was almost 12,000 new cancer cases diagnosed on average each day.[1] It is very important to explore its pathogenesis and therapeutic methods. Increasing numbers of studies have demonstrated that the imbalance between oncogenes and tumor suppressor genes is a determinant factor that induces normal cell development into tumor tissues.[2–4] Therefore, it is meaningful to identify and characterize these genes that promote cancer progression and recognize the potential markers and specific targets for the prevention and individualized treatment of cancer.
Telomerase is a ribonucleoprotein enzyme that can extend the telomeres, which are composed of double-stranded tandem repeats of TTAGGG at the end of a chromosome.[5] In most somatic cells, the inactivated telomerase leads telomeres to shorten with each round of cell division, and cells lose the capacity to replicate when a critical length is reached.[6] However, in cancer cells, activated telomerase-induced cancer cells escaped from telomere shortening and ultimately promoted unlimited cancer cell proliferation and immortalization.[7] Telomerase consists of a catalytic protein unit, human telomerase reverse transcriptase (hTERT), and human telomerase RNA. hTERT is the rate-limiting component of telomerase, and the upregulated expression of hTERT represents a surrogate marker of increased telomerase activity in most cancers.[8,9]
It was hypothesized that the high expression of hTERT can predict the survival of cancer. A number of studies have demonstrated that the elevated expression of hTERT was associated with a poor prognosis in solid tumors such as gastric cancer, lung cancer, cervical and head cancer, glioblastoma, breast cancer, and ovarian cancer.[10–13] However, most studies that addressed hTERT expression were limited by small sample sizes. Furthermore, several studies failed to reveal the association between hTERT expression and cancer prognosis.[14–17] Therefore, this systematic review and quantitative meta-analysis sought to evaluate whether hTERT acts as a survival biomarker to predict cancer prognosis.
2. Materials and methods
2.1. Search strategy
This meta-analysis was conducted following the Preferred Reporting Items for systematic Reviews and Meta-Analyses guidelines.[18] A systematic literature search was performed in the electronic databases PubMed, Web of Science, and Embase (up to August1, 2017) with the followin g search terms: “hTERT," “human telomerase reverse transcriptase,” and “cancer or tumor or carcinoma or neoplasm." In addition, the reference lists of the original research articles and reviews were also manually searched. Two evaluators completed the comprehensive search independently, and differences were resolved through discussion.
2.2. Selection and exclusion criteria
The eligible studies met the following inclusion criteria: clinical trials exploring the association between hTERT expression and the prognosis of cancer patients; the survival endpoint index was overall survival (OS), disease-free survival (DFS), or recurrence-free survival (RFS); reverse transcription-polymerase chain action, immunohistochemistry, or in situ hybridization was used to assess hTERT mRNA or protein expression; using the hTERT cutoff value, research patients were divided into positive (+) and negative (–) groups for survival analysis; Kaplan-Meier analysis or Log-hazard ratio (HR) and its 95% confidence interval (CI) were reported or could be calculated by a survival curve.
The exclusion criteria of eligible studies were as follows: study methods for hTERT detection, survival follow-up, and statistical analysis were not mentioned or ambiguous; duplicate data or repeat analysis (if >1 study was published by the same medical center (we chose the study with sufficient data or larger sample size); non-solid cancer or non-human research; study with a total number of cases <20; insufficient data to calculate HR and its 95% CI.
Hao NB and Wang K independently performed an initial assessment by identifying the eligibility of abstracts from the identified studies according to the selection and exclusion criteria. The final selection decisions were made by reading the full text if the eligibility was unclear from the sole abstract. Disagreement was resolved by discussing quality assessment and data collection.
2.3. Data extraction
Data were extracted according to the standardized format, which included the necessary information: author/year, study location, cancer type, sample size, hTERT test method, survival outcome measure, follow-up period, methods of HR estimation, HR, and its 95% CI. As the data included in our study were extracted from articles, ethical approval and patients’ written consent were not needed in our study. HR is the definition of both time to the event and censoring, and it is recommended in prognostic research. Although the majority of eligible studies did not directly report HR and 95% CI, I2 value of 0% to 30%, 31% to 50%, 51% to 75%, and 75% to 100% represented insignificant, low, moderate, and considerable heterogeneity, respectively. A random-effect model was applied to the secondary analysis as I2 > 50%; otherwise, a fixed-effect model was used. Publication bias was evaluated by Begg funnel plot and Egger test. The statistical analysis was performed by STATA 14.0 (STATA Corporation, College Station, TX). A 2-tailed P value < .05 was considered statistically significant.
2.4. Quality assessment
The quality of the included studies was independently assessed using the Newcastle-Ottawa Scale (NOS score) by Hao and Wang. It is generally considered that an article evaluated in the range of 0 to 5 points is of low quality, and the data extracted from such a study should be interpreted with caution. A score >6 was considered to indicate high-quality articles.
2.5. Statistical analysis
The data collected from each eligible article was used to evaluate the relationships between hTERT overexpression and solid tumor prognosis by meta-analysis. Pooled HR with 95% CI was used to evaluate the outcomes. An observed HR >1 implied a poor prognosis for the group with hTERT overexpression. Conversely, an observed HR <1 implied a favorable prognosis for the group with hTERT overexpression. The heterogeneity between studies was evaluated using Cochran Q test and Higgins I2 statistic.[19]
3. Results
3.1. Study selection and characteristics
According to the search strategy, 2966 references were identified in total. However, after screening of these titles and abstracts, only 89 studies were eligible for a full-text review. Finally, 27 studies were included in our meta-analysis, all of which were published from 2000 to 2016.[10–12,14–17,20–38] The detailed process of selection and exclusion is shown in Figure 1.
Figure 1.
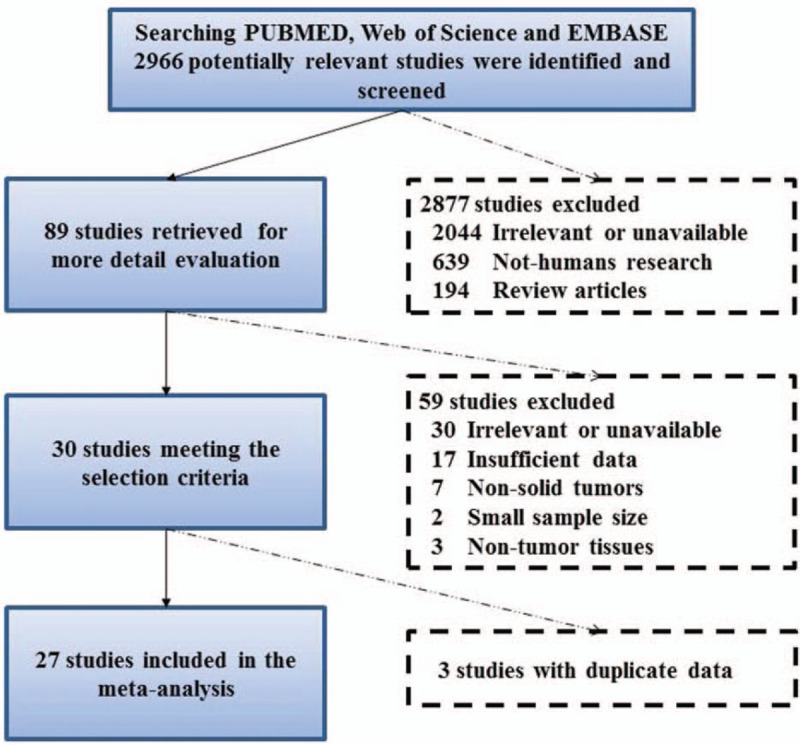
Flow diagram of dataset selection process.
The main characteristics of the 27 included studies are shown in Table 1. Twelve different types of cancer were investigated in this meta-analysis, including 9 cases of lung cancer, and 3 cases of digestive cancer, breast cancer, and ovarian cancer. All the studies were investigated in different countries, including China, the United States, France, and Australia. The sample sizes ranged from 34 to 201, and the follow-up period ranged from 18 to 264 months. All included studies were of high quality with an NOS score ≥7.
Table 1.
The basic characteristic of the included studies.
3.2. The prognostic significance of hTERT overexpression in OS, DFS and RFS of human cancer
Of all 27 studies, HRs for OS were available in 23 studies with 2071 patients.[10–12,14–16,21–30,34–38] As there was no statistical homogeneity in all the 23 datasets (I2 = 0.0%, P = .916), HR was calculated with the fixed-effects model. The forest plot results are shown in Figure 2, which indicated that the overexpression of hTERT induced poor OS (HR = 1.50, 95% CI: 1.31–1.73, P = .00).
Figure 2.
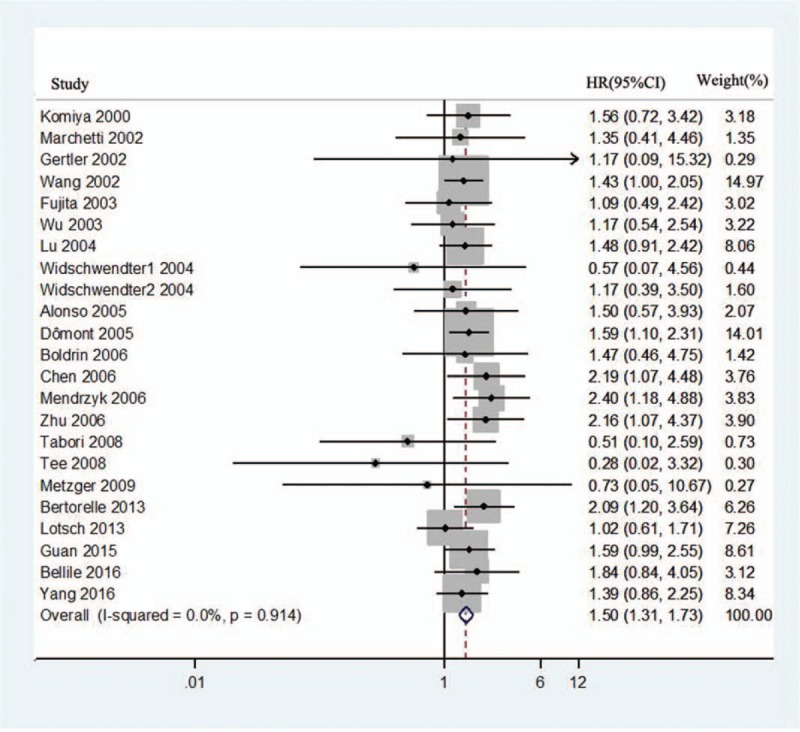
Forest plots for the correlation between human telomerase reverse transcriptase overexpression and overall survival in solid tumors. CI = confidence interval, HR = hazard ratio.
HRs for DFS were available in 4 studies with 413 patients.[17,23,28,37] Homogeneity was accepted (I2 = 47.6%, P = .126); thus, the fixed-effects model was chosen to calculate the HR. As shown in Figure 3, the pooled HR was 1.84 (95% CI: 1.38–2.46; P = .00), which indicated that hTERT overexpression was associated with poor DFS.
Figure 3.
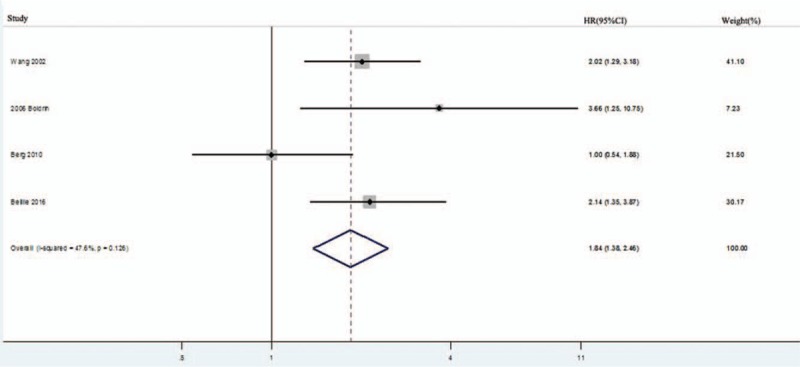
Forest plots for the correlation between human telomerase reverse transcriptase overexpression and disease-free survival in solid tumors. CI = confidence interval, HR = hazard ratio.
HRs for RFS were available in 4 studies with 374 patients.[20,30–31,33] Homogeneity was accepted (I2 = 0.0%, P = .882), and the fixed-effects model was chosen to calculate the HR. As shown in Figure 4, the pooled HR was 1.79 (95% CI: 1.07–2.99; P = .028), which indicated that hTERT overexpression was associated with poor RFS.
Figure 4.
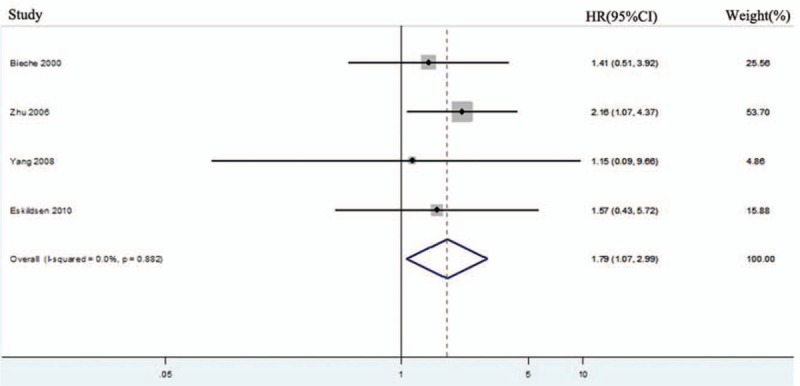
Forest plots for the correlation between human telomerase reverse transcriptase overexpression and recurrence-free survival in solid tumors. CI = confidence interval, HR = hazard ratio.
3.3. The prognostic significance of hTERT overexpression in OS of lung cancer
Among all 27 studies, 9 studies discussed the association between hTERT overexpression and lung cancer prognosis. Van den Berg et al[17] only reported hTERT overexpression with DFS. Finally, 8 studies with a total of 757 enrolled individuals dealt with the OS of hTERT overexpression in lung cancer.[10–11,21,23–25,30,32] As there was no statistical homogeneity in all 8 datasets (I2 = 0.0%, P = .928), HR was calculated with the fixed-effects model. The forest plot results are shown in Figure 5, which indicated that the overexpression of hTERT induced poor OS in lung cancer (HR = 1.51, 95% CI:1.21–1.89; P = .00).
Figure 5.
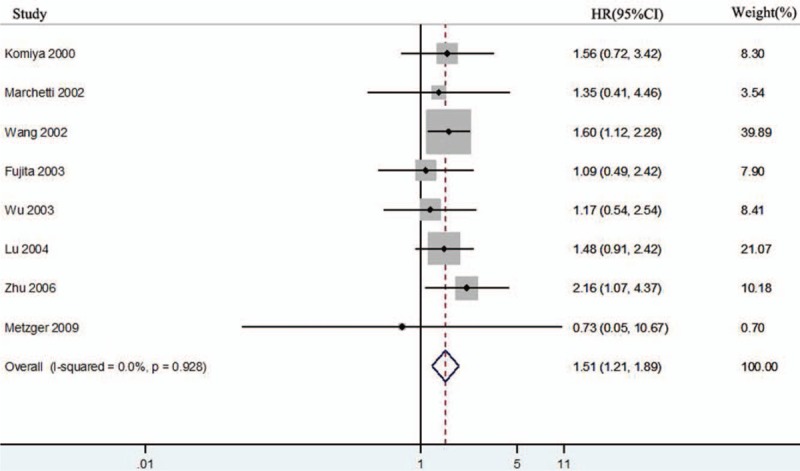
Forest plots for the correlation between human telomerase reverse transcriptase overexpression and overall survival in lung cancer. CI = confidence interval, HR = hazard ratio.
3.4. Sensitivity analysis and publication bias
Sensitivity analyses were performed to assess the influence of each individual study on the pooled HR. As shown in Figure 6, it was found that there was no significant change owing to the omission of any single article, demonstrating the statistical robustness of our results. Begg funnel plot and Egger test were utilized to evaluate the publication bias of the included studies. The shape of the funnel plot did not reveal any obvious asymmetry, and Egger tests revealed no publication bias regarding OS (data not show). These results still did not suggest any evidence of publication bias. Kaplan-Meier survival curves were read and extracted by Engauge Digitizer software as described by Tierney et al[39] to calculate HR and 95% CI data according to Parmar method.
Figure 6.
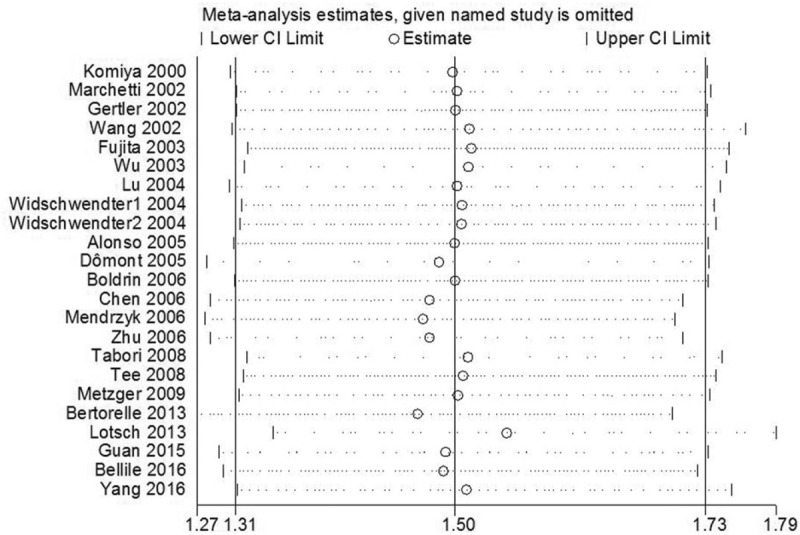
Sensitivity analysis on the relationships between human telomerase reverse transcriptase overexpression and OS in solid tumor patients. CI = confidence interval.
4. Discussion
hTERT fosters invasive cancer cell growth, which signifies the concerted activation of cell proliferation, angiogenesis, migration, and metastasis.[40–42] In recent years, research has focused on hTERT's maintenance of stability to promote cancer cell growth. Recent studies have found that hTERT does not depend on its telomere-lengthening function or its telomerase activity to promote cancer cell growth and metastasis.[43] Liu et al[44] reported that hTERT interacts with β-catenin and promotes the epithelial–mesenchymal transition and stem cell-like traits in cancer cells. Hu et al demonstrated that the hTERT-MDM2 complex enhances ITGB1 expression and ultimately promotes cancer cell metastasis. The high expression of hTERT is associated with TNM stage, lymphatic metastasis, and poor prognosis.[45] Moreover, our results demonstrated that the overexpression of hTERT was associated with poor OS and DFS/RFS of human cancer patients. Thus, hTERT could be a potential target gene for cancer therapy.
In this study, we conducted a meta-analysis of hTERT in different types of solid tumors. The results showed that hTERT overexpression acted as an unfavorable prognostic factor for solid tumors. Poor OS (HR = 1.50, 95% CI: 1.31–1.73, P = .00) was found in human cancers with hTERT overexpression. These results were consistent with most included studies. However, Tee et al[15] found that the activation of hTERT was not found to significantly predict the OS rate (HR = 0.28, 95% CI: 0.02–3.32) in early cervical cancer, as both the positive and negative expression of hTERT had practically the same prognostic value in our patients. It seems contradictory that overexpressed hTERT destabilizes cell proliferation, which in turn checks the deregulation of cancer cell growth.[46] Furthermore, Andreas et al reported that hTERT methylation but not hTERT overexpression can be an independent prognostic factor for cervical cancer.[14] For the RFS and DFS, the included studies demonstrated that hTERT was a poor prognostic biomarker. However, only 4 studies were included in the subgroup analysis, and these results were not conclusive. Further research is needed to explore the role of hTERT as a diagnostic marker for RFS and DFS in human cancers.
Lung cancer is one of the most common malignancies in the world and is the leading cause of cancer-related deaths. In the subgroup analysis, it was found that the overexpression of hTERT leads to a poor OS (HR = 1.51, 95% CI: 1.21–1.89; P = .00) in lung cancer. Among the included studies, Meterzger et al[32] found that the high expression of hTERT improved 5-year survival rates for non-small cell lung cancer, which was contradictory to the results of other studies. A possible reason was the limited number of telomerase-negative patients and total patients. Further research is still needed.
Aside from inspiring outcomes, some details need to be further refined. First, some of the included studies had a relatively small sample size, which may have caused a small-study effect. Second, the method of assessing hTERT expression was not consistent. Furthermore, the cutoff value for defining high and low hTERT expression varied in different studies, and it was difficult to obtain the same value. Finally, the follow-up period was not consistent in different studies, which may have influenced the final results, especially in the short follow-up period. Consequently, our estimation of the associations between hTERT overexpression and outcomes may have been overestimated.
In summary, our results showed that the overexpression of hTERT in solid tumor tissues was associated with poor survival (OS, DFS, and RFS). Meanwhile, subgroup analysis showed that hTERT overexpression also leads to the poor prognosis of lung cancer. Thus, hTERT might be a predictive biomarker for estimating the outcome of solid tumor patients. Furthermore, future studies related to specific tumor types and perspectives are needed to verify our results.
Author contributions
Conceptualization: Kai Wang, Ji Zhou, Ning-Bo Hao.
Data curation: Kai Wang, Rui-Ling Wang, Jian-Jun Liu, Ji Zhou, Ning-Bo Hao.
Formal analysis: Kai Wang, Rui-Ling Wang, Ji Zhou, Ning-Bo Hao.
Funding acquisition: Ning-Bo Hao.
Investigation: Kai Wang, Jian-Jun Liu, Xue Li, Wei-Jian Jiang.
Methodology: Xue Li, Wei-Jian Jiang, Ning-Bo Hao.
Project administration: Jian-Jun Liu, Wen-Wei Hu, Ning-Bo Hao.
Resources: Wen-Wei Hu.
Software: Kai Wang, Rui-Ling Wang, Xue Li, Ning-Bo Hao.
Writing – original draft: Kai Wang, Rui-Ling Wang, Ning-Bo Hao.
Writing – review & editing: Wei-Jian Jiang, Ning-Bo Hao.
Footnotes
Abbreviations: CI = confidence interval, DFS = disease-free survival, HR = Hazard ratios, hTERT = human telomerase reverse transcriptase, IHC = immunohistochemistry, ISH = in situ hybridization, OS = overall survival, RFS = recurrence-free survival, RT-PCR = Reverse transcription-polymerase chain action.
This work was supported by the fund of National Natural ScienceFoundation of China (No. 81502132) and the China Postdoctoral Science Foundation (2017M613374). The authors had no conflicts of interesting.
The authors report no conflicts of interest.
References
- [1].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [2].Hainaut P, Plymoth A. Targeting the hallmarks of cancer: towards a rational approach to next-generation cancer therapy. Curr Opin Oncol 2013;25:50–1. [DOI] [PubMed] [Google Scholar]
- [3].De Souza LM, Robertson BM, Robertson GP. Future of circulating tumor cells in the melanoma clinical and research laboratory settings. Cancer Lett 2017;392:60–70. [DOI] [PubMed] [Google Scholar]
- [4].Vicente-Duenas C, Romero-Camarero I, Cobaleda C, et al. Function of oncogenes in cancer development: a changing paradigm. EMBO J 2013;32:1502–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hahn WC, Stewart SA, Brooks MW, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med 1999;5:1164–70. [DOI] [PubMed] [Google Scholar]
- [6].Calado RT, Young NS. Telomere diseases. N Engl J Med 2009;361:2353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis 2010;31:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qian Y, Yang L, Cao S. Telomeres and telomerase in T cells of tumor immunity. Cell Immunol 2014;289:63–9. [DOI] [PubMed] [Google Scholar]
- [9].Janknecht R. On the road to immortality: hTERT upregulation in cancer cells. FEBS Lett 2004;564:9–13. [DOI] [PubMed] [Google Scholar]
- [10].Marchetti A, Pellegrini C, Buttitta F, et al. Prediction of survival in stage I lung carcinoma patients by telomerase function evaluation. Lab Invest 2002;82:729–36. [DOI] [PubMed] [Google Scholar]
- [11].Lu C, Soria JC, Tang X, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol 2004;22:4575–83. [DOI] [PubMed] [Google Scholar]
- [12].Chen HH, Yu CH, Wang JT, et al. Expression of human telomerase reverse transcriptase (hTERT) protein is significantly associated with the progression, recurrence and prognosis of oral squamous cell carcinoma in Taiwan. Oral Oncol 2007;43:122–9. [DOI] [PubMed] [Google Scholar]
- [13].Zachos I, Konstantinopoulos PA, Vandoros GP, et al. Predictive value of telomerase reverse transcriptase expression in patients with high risk superficial bladder cancer treated with adjuvant BCG immunotherapy. J Cancer Res Clin Oncol 2009;135:1169–75. [DOI] [PubMed] [Google Scholar]
- [14].Widschwendter A, Muller HM, Hubalek MM, et al. Methylation status and expression of human telomerase reverse transcriptase in ovarian and cervical cancer. Gynecol Oncol 2004;93:407–16. [DOI] [PubMed] [Google Scholar]
- [15].Tee YT, Wang PH, Yang SF, et al. Lymph node metastases, not human telomerase reverse transcriptase or p53 proteins, as the strongest prognostic factor for survival in early stage cervical cancer. J Obstet Gynaecol Res 2008;34:1002–9. [DOI] [PubMed] [Google Scholar]
- [16].Tabori U, Wong V, Ma J, et al. Telomere maintenance and dysfunction predict recurrence in paediatric ependymoma. Br J Cancer 2008;99:1129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Van den Berg RM, Brokx H, Vesin A, et al. Prognostic value of hTERT mRNA expression in surgical samples of lung cancer patients: the European Early Lung Cancer Project. Int J Oncol 2010;37:455–61. [DOI] [PubMed] [Google Scholar]
- [18].Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bieche I, Nogues C, Paradis V, et al. Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription-polymerase chain reaction assay. Clin Cancer Res 2000;6:452–9. [PubMed] [Google Scholar]
- [21].Komiya T, Kawase I, Nitta T, et al. Prognostic significance of hTERT expression in non-small cell lung cancer. Int J Oncol 2000;16:1173–7. [DOI] [PubMed] [Google Scholar]
- [22].Gertler R, Rosenberg R, Stricker D, et al. Prognostic potential of the telomerase subunit human telomerase reverse transcriptase in tumor tissue and nontumorous mucosa from patients with colorectal carcinoma. Cancer 2002;95:2103–11. [DOI] [PubMed] [Google Scholar]
- [23].Wang L, Soria JC, Kemp BL, et al. hTERT expression is a prognostic factor of survival in patients with stage I non-small cell lung cancer. Clin Cancer Res 2002;8:2883–9. [PubMed] [Google Scholar]
- [24].Fujita Y, Fujikane T, Fujiuchi S, et al. The diagnostic and prognostic relevance of human telomerase reverse transcriptase mRNA expression detected in situ in patients with nonsmall cell lung carcinoma. Cancer 2003;98:1008–13. [DOI] [PubMed] [Google Scholar]
- [25].Wu T. Loss of telomerase activity may be a potential favorable prognostic marker in lung carcinomas. Lung Cancer 2003;41:163–9. [DOI] [PubMed] [Google Scholar]
- [26].Alonso MM, Fueyo J, Shay JW, et al. Expression of Transcription Factor E2F1 and Telomerase in Glioblastomas: Mechanistic Linkage and Prognostic Significance. JNCI J Natl Cancer Inst 2005;97:1589–600. [DOI] [PubMed] [Google Scholar]
- [27].Domont J, Pawlik TM, Boige V, et al. Catalytic subunit of human telomerase reverse transcriptase is an independent predictor of survival in patients undergoing curative resection of hepatic colorectal metastases: a multicenter analysis. J Clin Oncol 2005;23:3086–93. [DOI] [PubMed] [Google Scholar]
- [28].Boldrini L, Pistolesi S, Gisfredi S, et al. Telomerase activity and hTERT mRNA expression in glial tumors. Int J Oncol 2006;28:1555–60. [DOI] [PubMed] [Google Scholar]
- [29].Mendrzyk F, Korshunov A, Benner A, et al. Identification of gains on 1q and epidermal growth factor receptor overexpression as independent prognostic markers in intracranial ependymoma. Clin Cancer Res 2006;12:2070–9. [DOI] [PubMed] [Google Scholar]
- [30].Zhu CQ, Cutz JC, Liu N, et al. Amplification of telomerase (hTERT) gene is a poor prognostic marker in non-small-cell lung cancer. Br J Cancer 2006;94:1452–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang CH, Hung WC, Wang SL, et al. Immunoexpression and prognostic role of hTERT and cyclin D1 in urothelial carcinoma. APMIS 2008;116:309–16. [DOI] [PubMed] [Google Scholar]
- [32].Metzger R, Vallbohmer D, Muller-Tidow C, et al. Increased human telomerase reverse transcriptase (hTERT) mRNA expression but not telomerase activity is related to survival in curatively resected non-small cell lung cancer. Anticancer Res 2009;29:1157–62. [PubMed] [Google Scholar]
- [33].Brems-Eskildsen AS, Zieger K, Toldbod H, et al. Prediction and diagnosis of bladder cancer recurrence based on urinary content of hTERT, SENP1, PPP1CA, and MCM5 transcripts. BMC Cancer 2010;10:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bertorelle R, Briarava M, Rampazzo E, et al. Telomerase is an independent prognostic marker of overall survival in patients with colorectal cancer. Br J Cancer 2013;108:278–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lotsch D, Ghanim B, Laaber M, et al. Prognostic significance of telomerase-associated parameters in glioblastoma: effect of patient age. Neuro Oncol 2013;15:423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Guan GG, Wang WB, Lei BX, et al. UBE2D3 is a positive prognostic factor and is negatively correlated with hTERT expression in esophageal cancer. Oncol Lett 2015;9:1567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gay-Bellile M, Romero P, Cayre A, et al. ERCC1 and telomere status in breast tumours treated with neoadjuvant chemotherapy and their association with patient prognosis. J Pathol Clin Res 2016;2:234–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang H, Zhang H, Zhong Y, et al. Concomitant underexpression of TGFBR2 and overexpression of hTERT are associated with poor prognosis in cervical cancer. Sci Rep 2017;7:41670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Saha D, Singh A, Hussain T, et al. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J Biol Chem 2017;292:15205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang C, Song C, Liu T, et al. KMT2A promotes melanoma cell growth by targeting hTERT signaling pathway. Cell Death Dis 2017;8:e2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu N, Ding D, Hao W, et al. hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res 2016;44:8693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Feng X, Xu X, Xiao X, et al. NMI inhibits cancer stem cell traits by downregulating hTERT in breast cancer. Cell Death Dis 2017;8:e2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu Z, Li Q, Li K, et al. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene 2013;32:4203–13. [DOI] [PubMed] [Google Scholar]
- [45].Hu C, Ni Z, Li BS, et al. hTERT promotes the invasion of gastric cancer cells by enhancing FOXO3a ubiquitination and subsequent ITGB1 upregulation. Gut 2017;66:31–42. [DOI] [PubMed] [Google Scholar]
- [46].Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer 2008;8:167–79. [DOI] [PubMed] [Google Scholar]


