Abstract
This study aims to investigate the role of miR-181a in multiple myeloma (MM). Fresh peripheral blood and bone marrows were collected. Expression of miR-181a, BCL-2 mRNA, and NOVA1 mRNA was detected by RT-qPCR. The correlation between miR-181a and clinical features of MM was further analyzed. miR-181a in serum and bone marrow mononuclear cells of MM patients were significantly higher. And, miR-181a level was significantly higher in MM Durie–Salmon stage III than that in stage I+II. miR-181a was positively correlated to Durie–Salmon staging, age, kidney injury, bone injury, β2-MG whereas negatively related to red blood cell, hemoglobin, and albumin. Additionally, BCL-2 and NOVA1 were predicted to be downstream targets of miR-181a. BCL-2 mRNA was significantly higher in the bone marrow mononuclear cells from MM patients. To sum up, the miR-181a expression is increased in peripheral blood and bone marrow of MM patients and is closely related to the clinical pathological indicators of MM.
Keywords: BCL-2, miR-181a, multiple myeloma, NOVA1, RT-qPCR
1. Introduction
Multiple myeloma (MM) is a malignant neoplasm characterized by abnormal proliferation of clonal malignant plasma cells in the bone marrow and increased monoclonal immunoglobulin.[1,2] Calcemia, renal insufficiency, anemia, and bone lesions are the 4 major clinical symptoms of MM.[3,4] So far, MM is still an incurable disease.[5] The pathogenesis of MM has been widely studied.[6] It has been shown that the abnormal expression of miRNA is involved in the occurrence and development of MM[7–10] and plays an important role in the transformation of monoclonal gammopathy of undetermined significance into MM.[11–13]
MicroRNAs (miRNAs) are small, endogenous, noncoding RNAs consisting of 20 to 25 nucleotides.[14,15] MiRNAs mainly bind with 3′ UTR of the target mRNAs to regulate the gene expression at the post-transcriptional level.[16] MiRNAs have been shown to be involved in the regulation of cell differentiation, proliferation, apoptosis, energy metabolism and tumor development.[17] MiRNAs exist not only in tissues but also in the body fluids such as blood, urine and cerebrospinal fluid, which are also known as circulating miRNAs.[18,19] Circulating miRNAs are stable and may be used as tumor markers.[20,21] MiR-181a belongs to miR-181 family and plays an important role in the process of cell differentiation and development.[22,23] It has been found that miR-181a can regulate gene expression by inhibiting or accelerating mRNA degradation and participates in tumor development.[24–26] Studies have shown that miR-181a may play a role in lymphocyte proliferation and differentiation into B cells.[24,27,28] At the same time, it has been reported that the expression of miR-181a is increased in MM.[29] However, the current research on the complex relationship between miR-181a and various clinical and pathological factors of MM disease is still lacking.
The aim of this study is to investigate the expression of miR-18a in MM. The relationship between miR-181a and the clinicopathological factors of MM was also analyzed. Additionally, we detected the target genes of miR-181a in order to reveal the mechanism of miR-18 in MM. Our study may provide a theoretical basis for studying the mechanism of miR-181a in the occurrence and development of MM.
2. Materials and methods
2.1. Clinical data
Patients with MM (n = 73), other hematological malignancies (n = 29), and benign hematological diseases (n = 6) hospitalized at the First Affiliated Hospital of Xi’an Jiaotong University from January 2016 to June 2017 were enrolled in this study. The diagnosis of MM followed the guidelines for the diagnosis and management of multiple myeloma in China (2015 revision).[30]
Inclusion criteria for MM: The ratio of bone marrow smear abnormal plasma cells was over 10% and/or plasmacytoma was confirmed by bone marrow biopsy; monoclonal protein M was detected from serum and/or urine. Abnormal clinical manifestations were observed.
Inclusion criteria for other hematological malignancies and benign hematological disease were that subjects were clearly diagnosed with other hematological malignancies and benign hematological disease. All cases were diagnosed by bone marrow smear or bone marrow biopsy, combined with the corresponding immunohistochemistry, cell morphological identification and flow cytometry diagnosis.
Exclusion criteria for MM: MM developed to plasma cell leukemia. MM patients complicated with other malignancies. Patients who had major organ failures. Patients who had immune system deficiency or other blood diseases. Patients with missing original data and incomplete records.
Exclusion criteria for other hematological malignancies: Patients with MM or MM history. Patients with other nonhematological malignancies. Patients who had major organ failures. Patients with missing original data and incomplete records. Exclusion criteria for benign hematological disease: Patients who were suffering from hematological malignancies. Patients with other nonhematological malignancies. Patients with major organ failures. Patients with missing original data and incomplete records.
The average age of MM patients was 61 ± 9 years old. The patients in the MM group were further divided into stage I+II (early) (n = 36) and stage III (n = 37) according to Durie–Salmon staging.[31] The diagnostic criteria of other hematological malignancies and benign hematological disease were based on the blood disease diagnosis and efficacy standards.[32] The 29 patients with other hematologic malignancies included 15 cases of acute myeloid leukemia, 7 cases of lymphoblastic leukemia, 6 cases of non-Hodgkin's lymphoma, and 1 case of hairy cell leukemi, with avearge age of 57 ± 11 years old. The 8 patients with benign hematological diseases with an avearge age of 56 ± 17 including 4 cases of iron deficiency anemia, 2 cases of megaloblastic anemia, 2 cases of idiopathic thrombocytopenic purpura. In the meanwhile, 30 cases of gender and age-matched normal subjects at the First Affiliated Hospital of Xi’an Jiaotong University were set as the control group.
Serum samples were obtained from 73 patients with MM, 29 patients with other hematological malignancies and 30 normal control individuals. The age and gender information of them is shown in Table 1. Bone marrow samples were collected from 19 patients with MM, 6 patients with other hematological malignancies and 8patients with benign hematological diseases, with the age and gender information of shown in Table 2. The other information of MM patients, including Durie–Salmon stage, bone injury, kidney injury, and immunophenotype, are listed in Table 3.
Table 1.
The gender and age information of patients for serum sample collection.

Table 2.
The gender and age information of patients for bone marrow sample collection.

Table 3.
Durie-Salmon staging distribution in patients with MM.
The study was approved by the ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU1AF2016LSK-46). Informed consent was obtained from each patient.
2.2. Extraction of bone marrow mononuclear cells
Totally, 4 mL of fresh bone marrow was collected. The bone marrow mononuclear cells were isolated within 4 hours of bone marrow collection using the cell separation kit (Virtue Pacific Biotechnology Co., Tianjin), according to the instruction. Briefly, fresh DMB water was added to the fresh bone marrow. The bone marrow mixture was then slowly added to the centrifuge tube with the cell separation liquid and was centrifuged at 1000 rpm for 20 minutes at 20 °C. The mononuclear cells were isolated from the middle layer.
2.3. qPCR
The expression of miR-181a, BCL-2 and NOVA-1 were analyzed by qPCR assays. The miRNA was extracted from the serum and bone marrow mononuclear cells using the miRcute serum/plasma miRNA isolation kit and miRcute miRNA isolation kit (Tiangen Co., Beijing) according to the instructions. The cDNA of miR-181a was generated using the miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen Co., Beijing). The sequence of miR-181a upstream primer was 5-AACATTCAACGCTGTCGGTGAGT-3. The downstream primer is provided by the miRcute Plus miRNA qPCR Detection Kit. The sequences of U6 primer were 5-CTCGCTTCGGCAGCACA-3 and 5-AACGCTTCACGAATTTGCGT-3. The PCR procedure was as follows: predenaturation at 95 °C for 15 minutes, followed by 45 cycles of 94 °C (20 seconds) and 60 °C (34 seconds). U6 was used as the internal control.
For BCL-2 and NOVA-1, mRNA was extracted from bone marrow mononuclear cells and reverse transcribed into cDNA using the FastKing RT Kit (Tiangen Co., Beijing) according to the instruction. The sequences of BCL-2 primers were 5-GGGAGGATTGTGGCCTTCTT-3 and 5-GGGCCAAACTGAGCAGAGTC-3. The sequences of NOVA-1 primers were 5-CCCTCCCCAGTGAGAACAAA-3 and 5-CCGCCATCATGTTTGCAGTT-3. GAPDH primer sequences were 5-AATGGGCAGCCGTTAGGAAA-3 and 5-GCGCCCAATACGACCAAATC-3. The PCR procedure included predenaturation at 95 °C for 15 minutes, followed by 45 cycles of 95 °C (10 seconds) and 60 °C (30 seconds). GAPDH was used as the internal control. Relative miR-181a and mRNA expression levels were determined by the 2-ΔCt method.
2.4. Bioinformatics softwares
The targets of miRNA were predicted by Targetscan (regRNA (http://regrna.mbc.nctu.edu.tw/php/prediction.php) and PicTar (http://pictar.mdc-berlin.de/).
2.5. Statistical analysis
The data were analyzed using the SPSS statistical package, version 17.0 (SPSS Inc., Chicago, IL) and the Graph Pad Prism version 5.0 (GraphPad Software, San Diego, CA). The measurement data with normal distribution were shown as mean ± standard deviation. The non-normal distributed measurement data were shown as median and interquartile spacing [M(P25-P75)]. Comparison between groups was performed using Kruskal–Wallis H (K) or Mann–Whitney U rank sum test. The comparison of rate was performed using χ2 test. The correlation analysis of parameters was performed using Spearman linear correlation analysis. P < .05 was considered as statistically significant.
3. Results
3.1. The expression of miR-181a in serum and bone marrow mononuclear cells
To detect the expression of miR-181a in the serum of MM, other hematological malignancies and normal control groups, we carried out qPCR assay. We found that the difference of miR-181a between the MM group and the normal control group was statistically significant (P < .05, Fig. 1A). There was also significant difference in miR-181a between the MM group and other hematologic malignancies (P < .05, Fig. 1A). However, there was no significant difference in miR-181a between the other hematologic malignancies and the normal controls (P > 0.05, Fig. 1A). We also tested the expression of miR-181a in Durie-Salmon staging of MM patients. Our result showed that the expression level of miR-181a in stage I+II and stage III was significantly different (P < .05; Fig. 1B). The qPCR was also used to determine the expression of miR-181a in the bone marrow mononuclear cells of patients with MM, other hematological malignancies and benign hematological diseases. We found that miR-181a level was significantly higher in MM patients than that in patients with other hematological malignancies and benign hematological diseases (P < .05, Fig. 1C). No significant difference was found between patients with other hematological malignancies and benign hematological diseases. The results indicate that miR-181a is highly expressed in the peripheral blood and bone marrow of MM, which is elevated with the increased staging level.
Figure 1.
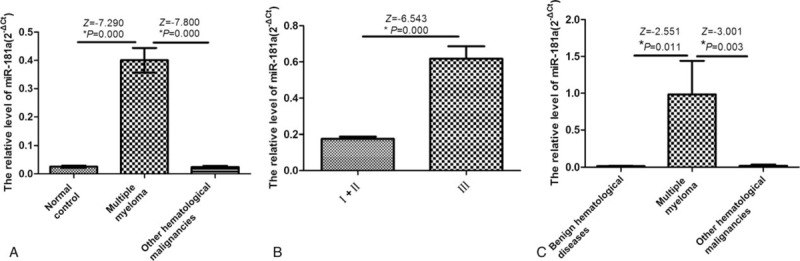
Analysis of miR-181a expression. The miR-181a expression was detected using qPCR. (A) miR-181a expression in patients with MM, other hematological malignancies and normal control individuals. (B) miR-181a expression in patients with MM Durie–Salmon stage I+II and stage III. (C) miR-181a expression in patients with MM, other hematological malignancies and benign hematological diseases. MM = multiple myeloma.
3.2. Correlation of miR-181a expression with clinical data of MM
We then carried out the Spearman correlation analysis to explore the relationship between the miR-181a expression and clinical indicators of MM. Our results showed that miR-181a was positively correlated with the Durie–Salmon stage, age, kidney injury and bone injury (Table 4). This result indicates that miR-181a could reflect the severity of the disease to some extent.
Table 4.
Correlation analysis of miR-181a expression with clinical index of MM patients.
![]()
3.3. Correlation of miR-181a expression with serum tumor burden indicators of MM
Spearman correlation analysis was also used to analyze the relationship between the miR-181a expression and serum tumor burden indicators of MM. The serum indicators of tumor burden in MM were shown in Table 5. Spearman correlation analysis showed that miR-181a was negatively correlated with red blood cell (RBC) (r = −0.423, P = .000) (Fig. 2A), hemoglobin (HMB) (r = −0.493, P = .000) (Fig. 2B) and albumin (ALB) (r = −0.449, P = .000) (Fig. 2C) whereas positively correlated with β2-microglobulin (β2-MG) (r = 0.633, P = .000) (Fig. 2D). Therefore, miR-181a is related to the pathogenesis of MM.
Table 5.
Tumor burden indicators in serum of MM patients.
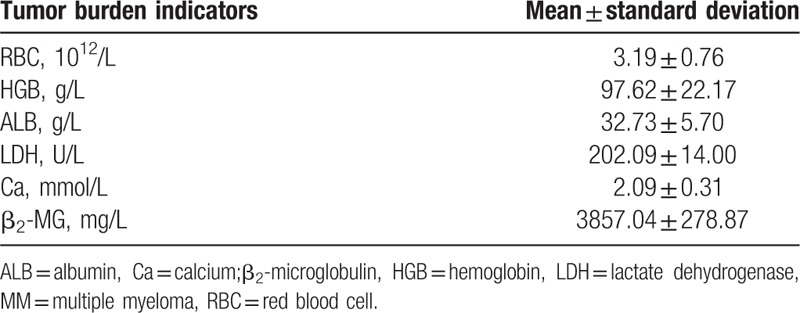
Figure 2.
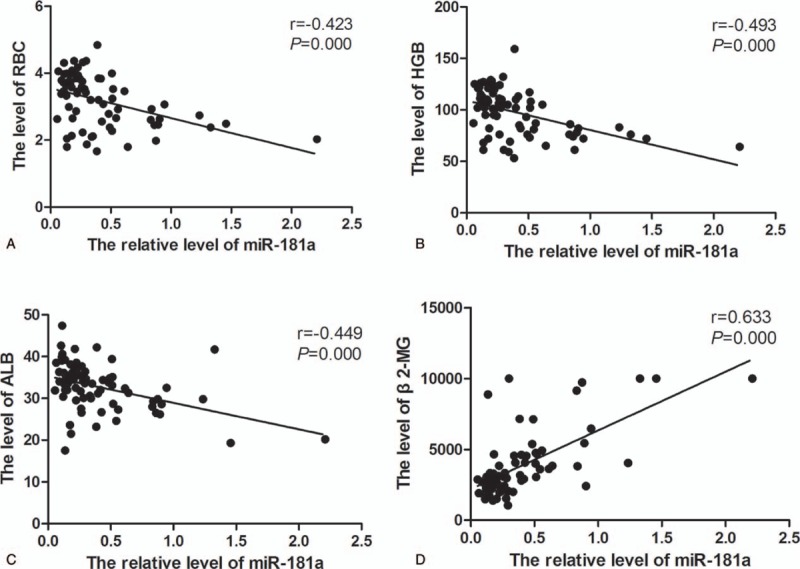
Correlation of miR-181a with tumor burden indicators. The spearman correlation analysis was used. (A) Correlation of miR-181a with RBC. (B) Correlation of miR-181a with HGB. (C) Correlation of miR-181a with ALB. (D) Correlation of miR-181a with β2-MG. ALB = albumin, HGB = hemoglobin, MM = multiple myeloma, RBC = red blood cell.
3.4. The expression of miR-181a target genes (BCL-2 and NOVA1) in the bone marrow mononuclear cells
To further study the function of miR-181a in MM, we used Targetscan and PicTar software to predict the target genes of miR-181a. The result showed that BCL-2 and NOVA1 were potential targets of miR-181a (Fig. 3A and B). Then we measured the expression of BCL-2 mRNA and NOVA1 mRNA in the bone marrow mononuclear cells by qPCR. We found that the expression of BCL-2 mRNA in MM patients and patients with other hematological malignancies was significantly increased compared with that in patients with benign hematological diseases (P < .05, Fig. 4A). However, the expression of BCL-2 between the patients with MM and other hematological malignancies showed no significant difference (P > .05). While the NOVA1 mRNA expression in the 3 groups showed no significant difference (P > .05, Fig. 4B). All these results showed that Bcl-2 is highly expressed in various hematological malignancies, including MM.
Figure 3.

Prediction of miR-181a targets using Targetscan and PicTar. (A) The base pairing between miR-181a and BCL-2. (B) The base pairing between miR-181a and NOVA1.
Figure 4.
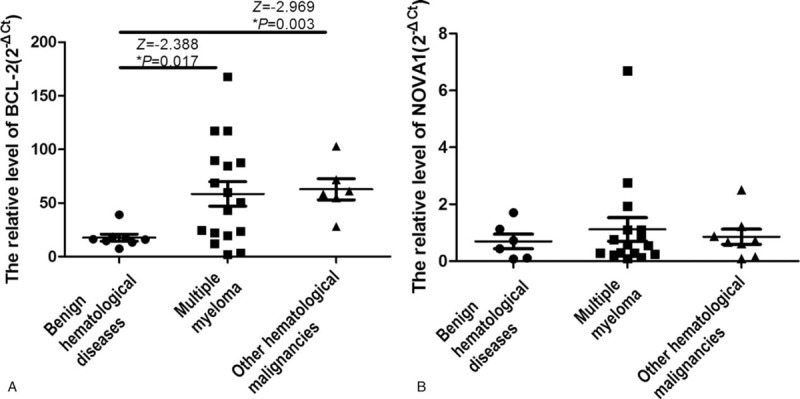
The mRNA expression of BCL-2 and NOVA1 in the bone marrow mononuclear cells. The mRNA expressions of BCL-2 and NOVA1 in the bone marrow mononuclear cells of patients with MM, other hematological malignancies and benign hematological diseases were detected using qPCR. (A) The expression of BCL-2 mRNA. (B) The expression of NOVA1 RNA.
3.5. Correlation of miR-181a, BCL-2 and NOVA1
We also studied the relationship between the expression of BCL-2 and NOVA1 and miR-181a using the Spearman correlation analysis. We found that there was a positive correlation between miR-181a and BCL-2 (r = 0.776, P = .001) (Fig. 5A). Moreover, BCL-2 was positively related to NOVA1 (r = 0.609, P = .012) (Fig. 5B). Thus, we speculate that the expression of miR-181a and the expression of BCL-2 are increased in MM patients, and NOVA1 may also be involved in the development of MM.
Figure 5.
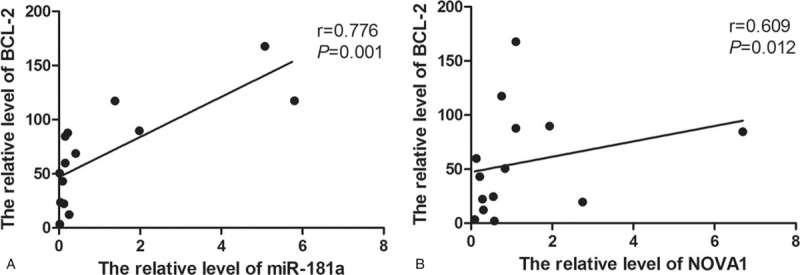
The relationship between miR-181a and BCL-2, and, between BCL-2 and NOVA1. The spearman correlation analysis was used. (A) The correlation between miR-181a and BCL-2. (B) The correlation between NOVA1 and BCL-2.
4. Discussion
MiRNA expression often significantly changes in tumors.[33–35] MiRNAs are not easily degraded by nucleic acid ribozymes in the blood and can maintain stable under adverse environments such as room temperature or repeated freeze-thaw.[36,37] Thus, miRNA may be used as useful marker for the diagnosis of diseases. Studies have detected the abnormal expression of miRNA in the occurrence and development of MM.[7–10] miRNA also plays an important role in the transformation of monoclonal gammopathy of undetermined significance into MM.[11–13] Additionally, Pichiorri et al[13] found that expression levels of miR-21, miR-32, miR-106b-25 clusters, miR-17–92 clusters, miR-181a, and miR-181b were abnormally expressed in MM cell lines.
In this study, the age distribution characteristics of selected MM patients were analyzed and the result showed that the onset age of MM was 51 to 60 years old, accounting for 47.9%. The patients of 61 to 70 years old accounted for 24.6%. Thus, the onset of MM was usually in the elderly, which is consistent with previous reports.[2,5,38] And, the older the patients were, the higher the staging level was, and the greater the chance of renal injury was. Our results also showed that miR-181a increased in the peripheral blood and bone marrow of MM patients, which is consistent with previous study.[13] In addition, Spearman correlation analysis showed that the relative expression level of miR-181a was positively correlated with the age, kidney injury, and bone injury in MM patients. The renal function and bone status of patients can intuitively reflect the severity of the disease.[39–42] Consistently, our further studies found that the expression of miR-181a was positively correlated with Durie–Salmon staging, suggesting that miR-181a could reflect the severity of MM disease to some extent.
We also studied the relationship between miR-181a and prognosis of disease. Anemia not only affects the prognosis of patients with MM, but also affects the quality of life.[38] IL-6 is a regulator of B cell differentiation into plasma cells.[43] The abnormal increase of IL-6 in patients with MM could reduce the ALB synthesis in liver.[44,45] β2-MG is a light chain of the complete histocompatibility antigen on the nuclear surface and is excreted by the kidneys.[46–48] Tumor cells have a strong ability to synthesize β2-MG, which directly reflects cell proliferation, tumor size, and renal impairment.[49,50] This paper analyzed the correlation between miR-181a and several tumor burden indicators. The results showed that miR-181a was positively correlated with β2-MG and negatively correlated with RBC, HGB and ALB. These results indicate that miR-181a is related to the progression of MM, which may have great value in the treatment and prognosis of MM.
It has been found that the miR-181 family can interact with different mRNAs via different binding sites of its target genes to inhibit their expression.[24–26] Studies have also shown that the expression and regulation of anti-apoptotic gene BCL-2 is one of the key factors that affect cell apoptosis.[51–53] The overexpression of BCL-2 is involved in the development of a variety of hematologic malignancies including MM, B-cell chronic lymphocytic leukemia, acute nonlymphocytic leukemia, and non-Hodgkin's lymphoma.[54–58] Furthermore, upregulation of BCL-2 gene expression has been shown to be associated with lifespan of MM cells and the drug resistance.[59,60] Consistently, our results showed that the expression of BCL-2 mRNA in patients with MM, other hematological malignancies and benign hematological diseases was significantly different. The elevated BCL-2 level was observed in patients with MM and other hematological malignancies. However, there was no significant difference between MM group and other hematologic malignancies group. We therefore suppose that elevated expression of miR-181a in MM and increased expression of BCL-2 may inhibit the apoptotic process in MM cells, and block the normal cell growth, which may be one of the important processes in the pathogenesis of MM.
NOVA1 was identified as one of the downstream targets of miR-181a in this study. The NOVA1 gene is a central nervous system-specific RNA-binding protein involved in alternative splicing of RNA after transcription. NOVA1 expressed ectopically in the breast, ovarian, and small-cell lung cancer[61–65] and NOVA1-associated antibodies are also found in the serum and or cerebrospinal fluid of these patients. However, the relationship between miR-181a and BCL-2, NOVA1 in MM is still unclear. Spearman correlation analysis in the current study showed that there was a positive correlation between miR-181a and BCL-2. The upregulation of miR-181a and BCL-2 and the positive correlation between them may inhibit cell apoptosis of tumor and the proliferation of normal cells. These are consistent with the low proliferative and pathological features of MM. The results of this study also showed that there was a positive correlation between BCL-2 and NOVA1. NOVA1 may be involved in the development of MM, but the specific role needs to be further studied.
This study has some limitations. First, this is a single center study. Second, the sample size is relatively small. Further multi-center study with large sample size is needed.
Overall, our results indicate that miR-181a is associated with MM and has a close relationship with clinical stage of MM. MiR-181a has a positive correlation with anti-apoptotic protein BCL-2. Furthermore, BCL-2 and NOVA1 might be the targets of miR-18a in MM. The miR-181a in peripheral blood may be used as a new marker for MM diagnosis and prognosis.
Author contributions
Conceptualization: Xuan Guo.
Data curation: Ni Liu, Lina Liu.
Formal analysis: Jinyu Yang.
Project administration: Ruili Yuan, Jing Peng.
Resources: Ni Liu.
Supervision: Xuan Guo.
Validation: Lina Liu.
Writing – original draft: Ruili Yuan.
Ruili Yuan: 0000-0002-1250-0403
Footnotes
Abbreviations: β2-MG = β2-microglobulin, ALB = albumin, HMB = hemoglobin, miRNAs = microRNAs, MM = multiple myeloma, RBC = red blood cell.
RY and NL contributed equally to this study.
This study is supported by Shaanxi Province International Science and Technology Cooperation and Exchange Program Project (2016KW-007).
The authors have no conflicts of interest to disclose.
References
- [1].Mimura N, Hideshima T, Anderson KC. Novel therapeutic strategies for multiple myeloma. Exp Hematol 2015;43:732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lu J, Lu J, Chen W, et al. Clinical features and treatment outcome in newly diagnosed Chinese patients with multiple myeloma: results of a multicenter analysis. Blood Cancer J 2014;4:e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Su YT, Xie XS, Sun H, et al. Clinical features of 46 multiple myeloma patients with different renal pathology. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2016;24:487–91. [DOI] [PubMed] [Google Scholar]
- [4].Talamo G, Farooq U, Zangari M, et al. Beyond the CRAB symptoms: a study of presenting clinical manifestations of multiple myeloma. Clin Lymphoma Myeloma Leuk 2010;10:464–8. [DOI] [PubMed] [Google Scholar]
- [5].Rollig C, Knop S, Bornhauser M. Multiple myeloma. Lancet 2015;385:2197–208. [DOI] [PubMed] [Google Scholar]
- [6].Neri P, Bahlis NJ, Lonial S. New strategies in multiple myeloma: immunotherapy as a novel approach to treat patients with multiple myeloma. Clin Cancer Res 2016;22:5959–65. [DOI] [PubMed] [Google Scholar]
- [7].Soley L, Falank C, Reagan MR. MicroRNA transfer between bone marrow adipose and multiple myeloma cells. Curr Osteoporos Rep 2017;15:162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bong IPN, Ng CC, Baharuddin P, et al. MicroRNA expression patterns and target prediction in multiple myeloma development and malignancy. Genes Genomics 2017;39:533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bi C, Chng WJ. MicroRNA: important player in the pathobiology of multiple myeloma. Biomed Res Int 2014;2014:521586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ahmad N, Haider S, Jagannathan S, et al. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia 2014;28:732–8. [DOI] [PubMed] [Google Scholar]
- [11].Hao M, Zang M, Wendlandt E, et al. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int J Cancer 2015;136:1835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Navarro A, Diaz T, Tovar N, et al. A serum microRNA signature associated with complete remission and progression after autologous stem-cell transplantation in patients with multiple myeloma. Oncotarget 2015;6:1874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pichiorri F, Suh SS, Ladetto M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA 2008;105:12885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang Y, Shen XJ, Zou Q, et al. Biological functions of microRNAs: a review. J Physiol Biochem 2011;67:129–39. [DOI] [PubMed] [Google Scholar]
- [15].Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis 2015;35:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Warf MB, Johnson WE, Bass BL. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA 2011;17:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis 2012;33:1126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li P, Teng F, Gao F, et al. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol 2015;35:433–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xu YN, Xiao CR, Huang YD, et al. Circulating serum microRNA as diagnostic biomarkers for multiple myeloma. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2017;25:471–5. [DOI] [PubMed] [Google Scholar]
- [20].Lin MS, Chen WC, Huang JX, et al. Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int J Clin Exp Med 2014;7:5226–34. [PMC free article] [PubMed] [Google Scholar]
- [21].Wang M, Gu H, Wang S, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep 2012;5:1514–20. [DOI] [PubMed] [Google Scholar]
- [22].Zhang P, Guo Z, Hu R, et al. Interaction between microRNA-181a and TNFAIP1 regulates pancreatic cancer proliferation and migration. Tumour Biol 2015;36:9693–701. [DOI] [PubMed] [Google Scholar]
- [23].Xu H, Zhu J, Hu C, et al. Inhibition of microRNA-181a may suppress proliferation and invasion and promote apoptosis of cervical cancer cells through the PTEN/Akt/FOXO1 pathway. J Physiol Biochem 2016;72:721–32. [DOI] [PubMed] [Google Scholar]
- [24].Nabhan M, Louka ML, Khairy E, et al. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene 2017;628:253–8. [DOI] [PubMed] [Google Scholar]
- [25].Cao Y, Zhao D, Li P, et al. MicroRNA-181a-5p impedes IL-17-induced nonsmall cell lung cancer proliferation and migration through targeting VCAM-1. Cell Physiol Biochem 2017;42:346–56. [DOI] [PubMed] [Google Scholar]
- [26].Li Z, Wang H, Xu Z, et al. Expression and mechanism of microRNA-181A on incidence and survival in late liver metastases of colorectal cancer. Oncol Rep 2016;35:1403–8. [DOI] [PubMed] [Google Scholar]
- [27].Verduci L, Azzalin G, Gioiosa S, et al. microRNA-181a enhances cell proliferation in acute lymphoblastic leukemia by targeting EGR1. Leuk Res 2015;39:479–85. [DOI] [PubMed] [Google Scholar]
- [28].Wang G, Zhao R, Zhao X, et al. MicroRNA-181a enhances the chemotherapeutic sensitivity of chronic myeloid leukemia to imatinib. Oncol Lett 2015;10:2835–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peng J, Thakur A, Zhang S, et al. Expressions of miR-181a and miR-20a in RPMI8226 cell line and their potential as biomarkers for multiple myeloma. Tumour Biol 2015;36:8545–52. [DOI] [PubMed] [Google Scholar]
- [30].Du J, Hou J. The guidelines for the diagnosis and management of multiple myeloma in China (2015 revision): interpretation of diagnosis. Zhonghua Nei Ke Za Zhi 2016;55:91–2. [DOI] [PubMed] [Google Scholar]
- [31].Kyle RA, Gertz MA, Witzig TE, et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin Proc 2003;78:21–33. [DOI] [PubMed] [Google Scholar]
- [32].Choudhry VP. Editorial: advances in diagnosis of hematological disorders. Indian J Pediatr 2013;80:754–5. [DOI] [PubMed] [Google Scholar]
- [33].Nana-Sinkam SP, Croce CM. Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 2013;93:98–104. [DOI] [PubMed] [Google Scholar]
- [34].Gao Y, Zhao H, Lu Y, et al. MicroRNAs as potential diagnostic biomarkers in renal cell carcinoma. Tumour Biol 2014;35:11041–50. [DOI] [PubMed] [Google Scholar]
- [35].Luo J, Zhao Q, Zhang W, et al. A novel panel of microRNAs provides a sensitive and specific tool for the diagnosis of breast cancer. Mol Med Rep 2014;10:785–91. [DOI] [PubMed] [Google Scholar]
- [36].Besse L, Sedlarikova L, Kryukov F, et al. Circulating serum microRNA-130a as a novel putative marker of extramedullary myeloma. PLoS One 2015;10:e0137294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kim K, Lee JH, Kim JS, et al. Clinical profiles of multiple myeloma in Asia—An Asian Myeloma Network study. Am J Hematol 2014;89:751–6. [DOI] [PubMed] [Google Scholar]
- [39].Engelhardt M, Kleber M, Udi J, et al. Current approaches in multiple myeloma and other cancer-related bone diseases. Dtsch Med Wochenschr 2012;137:1057–61. [DOI] [PubMed] [Google Scholar]
- [40].Gonsalves WI, Leung N, Rajkumar SV, et al. Improvement in renal function and its impact on survival in patients with newly diagnosed multiple myeloma. Blood Cancer J 2015;5:e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Khan R, Apewokin S, Grazziutti M, et al. Renal insufficiency retains adverse prognostic implications despite renal function improvement following total therapy for newly diagnosed multiple myeloma. Leukemia 2015;29:1195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kleber M, Udi J, Metzke B, et al. Challenging the current approaches to multiple myeloma- and other cancer-related bone diseases: from bisphosphonates to targeted therapy. Leuk Lymphoma 2012;53:1057–61. [DOI] [PubMed] [Google Scholar]
- [43].Perez-Chaparro PJ, Duarte PM, Shibli JA, et al. The current weight of evidence of the microbiologic profile associated with peri-implantitis: a systematic review. J Periodontol 2016;87:1295–304. [DOI] [PubMed] [Google Scholar]
- [44].Duan LJ, Li C, Yang RY. Values of detecting the levels of beta2-MG, TNF-alpha, CRP, IL-6 in the patients with multiple myeloma. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2015;23:1362–5. [DOI] [PubMed] [Google Scholar]
- [45].Matthes T, Manfroi B, Huard B. Revisiting IL-6 antagonism in multiple myeloma. Crit Rev Oncol/Hematol 2016;105:1–4. [DOI] [PubMed] [Google Scholar]
- [46].Argyropoulos CP, Chen SS, Ng YH, et al. Rediscovering Beta-2 microglobulin as a biomarker across the spectrum of kidney diseases. Front Med 2017;4:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li L, Dong M, Wang XG. The implication and significance of beta 2 microglobulin: a conservative multifunctional regulator. Chin Med J 2016;129:448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Monteiro MB, Thieme K, Santos-Bezerra DP, et al. Beta-2-microglobulin (B2 M) expression in the urinary sediment correlates with clinical markers of kidney disease in patients with type 1 diabetes. Metab Clin Exp 2016;65:816–24. [DOI] [PubMed] [Google Scholar]
- [49].Yoo C, Yoon DH, Kim S, et al. Serum beta-2 microglobulin as a prognostic biomarker in patients with mantle cell lymphoma. Hematol Oncol 2016;34:22–7. [DOI] [PubMed] [Google Scholar]
- [50].Fedele PL, Choy KW, Doery JC, et al. Inter-laboratory discordance of beta-2 microglobulin results: impact on the validity of the international staging system for multiple myeloma. Brit J Haematol 2014;166:951–3. [DOI] [PubMed] [Google Scholar]
- [51].Roufayel R. Regulation of stressed-induced cell death by the Bcl-2 family of apoptotic proteins. Mol Membr Biol 2016;33:89–99. [DOI] [PubMed] [Google Scholar]
- [52].Cory S, Roberts AW, Colman PM, et al. Targeting BCL-2-like proteins to kill cancer cells. Trends Cancer 2016;2:443–60. [DOI] [PubMed] [Google Scholar]
- [53].Kabbage M, Kessens R, Dickman MB. A plant Bcl-2-associated athanogene is proteolytically activated to confer fungal resistance. Microb Cell 2016;3:224–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cerella C, Gaigneaux A, Mazumder A, et al. Bcl-2 protein family expression pattern determines synergistic pro-apoptotic effects of BH3 mimetics with hemisynthetic cardiac glycoside UNBS1450 in acute myeloid leukemia. Leukemia 2017;31:755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kontro M, Kumar A, Majumder MM, et al. HOX gene expression predicts response to BCL-2 inhibition in acute myeloid leukemia. Leukemia 2017;31:301–9. [DOI] [PubMed] [Google Scholar]
- [56].Leonard JT, Rowley JS, Eide CA, et al. Targeting BCL-2 and ABL/LYN in Philadelphia chromosome-positive acute lymphoblastic leukemia. Sci Transl Med 2016;8:354ra114. [DOI] [PubMed] [Google Scholar]
- [57].Salem AH, Agarwal SK, Dunbar M, et al. Pharmacokinetics of venetoclax, a novel BCL-2 inhibitor, in patients with relapsed or refractory chronic lymphocytic leukemia or non-Hodgkin lymphoma. J Clin Pharmacol 2017;57:484–92. [DOI] [PubMed] [Google Scholar]
- [58].Li Y, Zhang B, Li W, et al. MiR-15a/16 regulates the growth of myeloma cells, angiogenesis and antitumor immunity by inhibiting Bcl-2, VEGF-A and IL-17 expression in multiple myeloma. Leuk Res 2016;49:73–9. [DOI] [PubMed] [Google Scholar]
- [59].Bajpai R, Matulis SM, Wei C, et al. Targeting glutamine metabolism in multiple myeloma enhances BIM binding to BCL-2 eliciting synthetic lethality to venetoclax. Oncogene 2016;35:3955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Matulis SM, Gupta VA, Nooka AK, et al. Dexamethasone treatment promotes Bcl-2 dependence in multiple myeloma resulting in sensitivity to venetoclax. Leukemia 2016;30:1086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Koukoui SD, Chaudhuri A. Neuroanatomical, molecular genetic, and behavioral correlates of fragile X syndrome. Brain Res Rev 2007;53:27–38. [DOI] [PubMed] [Google Scholar]
- [62].Kim EK, Yoon SO, Jung WY, et al. Implications of NOVA1 suppression within the microenvironment of gastric cancer: association with immune cell dysregulation. Gastric Cancer 2017;20:438–47. [DOI] [PubMed] [Google Scholar]
- [63].Shen B, Zhang Y, Yu S, et al. MicroRNA-339, an epigenetic modulating target is involved in human gastric carcinogenesis through targeting NOVA1. FEBS Lett 2015;589(20 pt B):3205–11. [DOI] [PubMed] [Google Scholar]
- [64].Xin Y, Li Z, Zheng H, et al. Neuro-oncological ventral antigen 1 (NOVA1): Implications in neurological diseases and cancers. Cell Prolif 2017;50: e12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhi F, Wang Q, Deng D, et al. MiR-181b-5p downregulates NOVA1 to suppress proliferation, migration and invasion and promote apoptosis in astrocytoma. PLoS One 2014;9:e109124. [DOI] [PMC free article] [PubMed] [Google Scholar]


