Abstract
Glucocorticoids are the primary anti-inflammatory therapy for asthma, but their effects are characterized by some interindividual variability that might have a genetic basis.
We aimed to determine the relationship between pulmonary function change and the variant of the glucocorticoid-induced transcript 1 (GLCCI1) gene in patients with asthma receiving long-term ICS treatment, the association of GLCCI1 genotypes and the level of GLCCI1 expression and cytokines production.
A total of 418 patients with asthma, including 25 individuals from 11 families with a history of asthma, were enrolled. The effects of single-nucleotide polymorphisms (SNPs) in GLCCI1 on changes in lung function in response to inhaled glucocorticoids were assessed. The expression levels of GLCCI1 mRNA and cytokines were also measured.
The SNP rs37973 in GLCCI1 was independently associated with changes in forced expiratory volume at one second (FEV1) and FEV1%pred. Individuals homozygous for the wild-type allele who had a percent FEV1 change greater than 5% were more common than individuals homozygous for the rare allele. When patients were stratified according to genotype, GLCCI1 expression was enhanced upon administration of low-dose dexamethasone among patients with the rs37973 A allele; however, GG homozygotes required high-dose dexamethasone to achieve enhanced GLCCI1 expression. Furthermore, the levels of some cytokines were significantly reduced after glucocorticoid treatment in individuals with the AA and AG genotypes.
The genetic variant rs37973 in GLCCI1 is associated with poorer clinical therapeutic response to inhaled glucocorticoids in a Chinese asthma population.
Keywords: asthma, GLCCI1, inhaled corticosteroid treatment, polymorphism, pulmonary function
1. Introduction
Asthma is a chronic disease that affects about 300 million people worldwide[1] and is characterized by airway inflammation and hyper-responsiveness, reversible airway obstruction, and airway remodeling. The efficacy of asthma treatment relies both on the control of environmental factors and pharmacological interventions.[2] Currently, many studies have shown that the response to inhaled corticosteroid treatment is remarkably variable.[3,4] To achieve the objective of personalized treatment for each patient, studies of the genetic influences on drug responsiveness are necessary.[5–7]
Inhaled corticosteroid (ICS) is the most commonly used and effective clinical medication for asthma therapy. ICS can alleviate the clinical symptoms of asthma, improve pulmonary function, and suppress airway inflammation.[8,9] The side effects of normal ICS use are modest, including hoarseness and oral ulcers,[10,11] whereas high-dose and sustained use of ICS is associated with more serious adverse effects.[12–15] However, many studies have found heterogeneity in the therapeutic responses to ICS among asthma patients who show poor improvements in the forced expiratory volume at one second (FEV1), even though they are highly compliant with medication use.[4,16] Thus, identifying those patients who do not benefit from ICS and modifying their treatment regimens to improve health outcomes would be meaningful. Increased FEV1, which is influenced by genetic factors, indicates the improvement of lung function. As the response to inhaled corticosteroid treatment in a given patient with asthma is highly reproducible,[17] and as FEV1 is a heritable trait,[18,19] it is plausible that genomic factors can determine drug responsiveness.[20,21] Therefore, we hypothesized that single nucleotide polymorphisms (SNPs) in genes might be associated with the therapeutic response to this class of asthmatic drugs.
Glucocorticoid induced transcript 1 (GLCCI1) is located on 7p21.3, induced by glucocorticoids, and might be an early marker for glucocorticoid-induced apoptosis.[22,23] A recently reported genome-wide association study identified one SNP, rs37972, located in the promoter of GLCCI1 that was associated with changes in lung function after ICS treatment. Further studies confirmed that another functional GLCCI1 promoter variant, rs37973, which is in complete linkage disequilibrium, was associated with a reduced response to ICS as measured over 4 to 8 weeks among 935 white non-Hispanic adults and children. In vitro experiments showed that both the rs37972 and rs37973 variants were associated with reduced GLCCI1 expression and that SNP rs37973 was significantly associated with reductions in luciferase reporter activity. Thus, the functional GLCCI1 variant rs37973 was associated with substantial reductions in the response to ICS in patients with asthma.
The present study aimed to investigate whether responses to glucocorticoid treatment were associated with the polymorphisms rs37973 in the GLCCI1 gene among adult patients, including a randomized asthma population and 25 individuals from 11 families after 24 weeks treatment with ICS in a Chinese population. Additionally, we sought to characterize the function of the rs37973 SNP by using dexamethasone stimulation in vitro cell approaches.
2. Methods
2.1. Subjects
This study included a total of 418 subjects, including 25 individuals from 11 families, who were recruited from an outpatient clinic at Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology from January 2011 to December 2012. At enrollment, 611 individuals were diagnosed as asthmatic first time based on clinical symptoms and positive results of bronchial provocation or dilation tests according to the GINA guidelines.[24] Participants were ruled out if they were current smokers or ex-smokers with a history more than 10 pack-years, had a course of oral or inhaled corticosteroids, or complicated with other respiratory diseases. At the first visit, 2 mL venous blood was obtained in Eathylene diamine tetraacetic acid blood collection tubes from those subjects and was then sent to a laboratory for genotyping. Additionally, a questionnaire asked about family history of asthma, basic demographic information, and the frequency of acute asthma attacks. Among them, 424 eligible asthmatics received inhaled fluticasone propionate/salmeterol combination (250/50 μg, twice daily) for the next 24 weeks consistently. Follow-up visits occurred at 4, 12, and 24 weeks, during which medication usage in the interval periods and asthma control tests were reviewed and lung function tests were performed using the same type of spirometer used at the first visit. All clinical data were recorded by the same physician, who was blinded to the genotype of patients. Additionally, 10 mL blood was drawn from those subjects who signed informed consent forms at the last visit for peripheral blood mononuclear cell (PBMC) isolation.
The study was approved by the Human Assurance Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology and informed consent was obtained from all study subjects.
2.2. SNP selection and genotyping
Rs37973, which were highly associated with changes in lung function in response to inhaled glucocorticoids and were located on chromosome 7p21, were selected for testing linkage to GLCCI1 based on the study of Tantisira et al. Genomic DNA was extracted from peripheral blood using an extraction kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. SNPs were genotyped using a TaqMan Genotyping master mix-based method on an ABI 7900 HT TaqMan sequence detection system (Applied Biosystems, Foster City, CA). Probes and primers were purchased from Applied Biosystems (Assay ID: rs37973, AHUACTL). The polymerase chain reaction amplification mixture (12.5 μL) contained 50 ng DNA, 300 nM of each specific probe, and 6.25 μL Taqman Universal PCR Master Mix (Applied Biosystems). Amplification was carried out under the following conditions: 95°C for 10 minutes followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were analyzed by using an Allelic Discrimination Program (Applied Biosystems).
2.3. Peripheral blood mononuclear cell culture
PBMCs were isolated from blood samples by gradient centrifugation using lymphoprep (Ficoll–Hypaque, TBD, China) and were centrifuged at 500g for 20 minutes. The gray mononuclear cell coats were carefully removed and then were washed 3 times with physiological saline. Cells were resuspended to 2 × 106 cells/mL with RPMI 1640 medium. Cell viability was determined using the Trypan Blue Dye exclusion test and was found to be greater than 90% in all experimental conditions. Cells from the same patient were seeded in 6-well plates (Corning, Corning, NY) and divided into 2 groups: the control group cultured with RPMI 1640 medium containing 10% heat-inactivated fetal calf serum and the asthma-like group cultured with RPMI 1640 medium containing 10% asthmatic serum (AS group). The asthmatic serum was obtained from different allergic asthmatic patients with serum IgE levels >1000 IU/mL and blended thoroughly to homogenize. Both groups were incubated with varying concentrations of dexamethasone (Sigma; 0, or 10−7–10−6 M) using dimethyl sulfoxide (DMSO) as a solvent at 37°C in the presence of 5% CO2 for 24 hours.
2.4. Quantitiative real-time polymerase chain reaction (PCR)
After 24 hours incubation, cells were harvested for RNA extraction. Total RNA was extracted using RNAiso Plus (Total RNA extraction reagent, TaKaRa, Shiga, Japan), and cDNA was generated from 500 ng RNA using a Prime Script RT Master Mix kit (TaKaRa). Real-time PCR was performed following the manufacturer's instructions using SYBR Premix Ex Taq (Takara) with an ABI Prism 7500 detection system. Data were analyzed using the 2-ΔΔCt method and were presented as arbitrary units.[25] The following oligonucleotide primers specific for genes were used in this study: GLCCI1, sense 5′-CGGAGGAGCAGCTCACCTGAG-3′ and antisense 5′-CGTGGCCACTGTCCTGTGAGGTA-3′; β-actin, sense 5′-AGCGAGCATCCCCCAAAGTT-3′ and antisense 5′-GGGCACGAAGGCTCATCATT-3′.
2.5. Measurements of cytokine production
Levels of IL-4, IL-5, IL-6, IL-8, IL-9, IL-10, IL-13, and INF-γ in the culture supernatants were quantified using enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer's protocol (NeoBioscience, China). The minimum detectable concentrations of the assays for IL-4, IL-5, IL-6, IL-8, IL-9, IL-10, IL-13, and IFN-γ were 8, 8, 8, 8, 24, 8, 15, and 8 pg/mL, respectively.
2.6. Statistical analysis
Continuous data were presented as means ± standard deviation (SD). Categorical data were presented as frequencies and percentages. Baseline values that were stratified by genotype were compared using Fisher's exact test (categorical variables) or an independent sample Student's t-test (continuous variables). All SNP markers were tested for deviation from Hardy–Weinberg equilibrium using a goodness-of-fit χ2 test with one degree of freedom. The changes in FEV1 and FEV1%pred in response to inhaled glucocorticoids were compared according to genotype using an independent sample Student's t-test because the data were normally distributed. SPSS software version 20.0 (SPSS Inc., Chicago, IL) was used for statistical evaluations of the above data. Probability (P) values of <0.05 were considered to indicate statistically significant differences.
3. Results
3.1. Subject characteristics
Demographic data and spirometry for the recruited subjects are shown in Table 1. Among the 611 enrolled asthmatics, 424 participants received ICS treatment consistently during the whole follow-up period. There were 6 individuals who lacked either baseline or endpoint FEV1 measurements. Thus, a total of 418 patients were analyzed. More females (59.09%) than males were recruited and the mean patient age was 44.4 years old. The mean body mass index (BMI) was 23.13. Participants only received short-actingβ-agonist therapy or no drug. Patients were stratified by genotype and tests for homogeneity did not identify statistically significant deviations for the key demographics.
Table 1.
The demographic characteristics of study subjects according to the rs37973 genotype of GLCCI1.
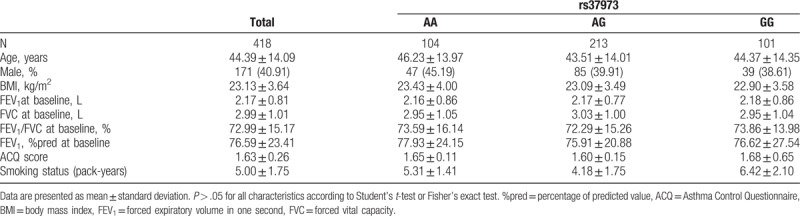
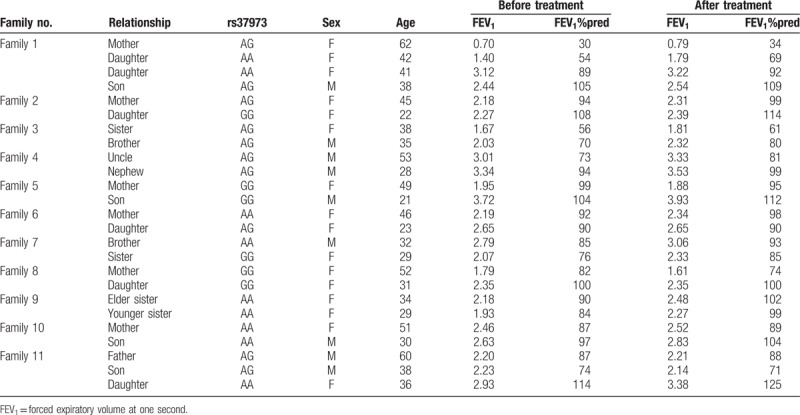
Among the 418 patients, there were 25 individuals who were from 11 families in which a first or second degree relative had a family history of asthma. These patients were treated with inhaled glucocorticoids, similarly to other individuals, and pulmonary function was tested before and after treatment (Table 2).
Table 2.
The demographic characteristics of 25 individuals from 11 families according to the rs37973 genotype of GLCCI1.
3.2. Genotype and allele frequencies
Among the 418 genotyped patients for rs37973, 104 (24.9%) were major AA homozygotes, 213 (51.0%) were AG heterozygotes, and 101 (24.2%) were minor GG homozygotes. None of the polymorphisms significantly deviated from Hardy–Weinberg equilibrium. Both SNPs had a call rate >95% and a genotyping accuracy >99%. The minor allele frequency for rs37973 was 0.496.
3.3. Association of genotypes and lung function changes
We examined changes in FEV1 and FEV1%pred according to genotype following a 24-week intervention (Fig. 1). We found that there was a strong effect of the rs37973 genotype on changes in FEV1 and FEV1%pred. Homozygotes for the wild-type allele of rs37973 were associated with a significant improvement in FEV1 (0.18 ± 0.28 L) compared to homozygous for the rare allele (0.09 ± 0.28 L, P < .05). For the FEV1%pred change (defined as FEV1%predtreatment– FEV1%predbaseline), homozygotes for the major allele had a FEV1%pred change of 9.82% ± 14.51% versus 6.33% ± 11.28% for homozygotes for the minor allele (P < .05).
Figure 1.
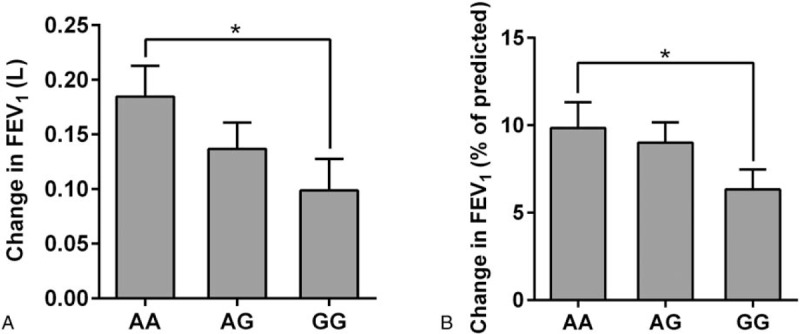
Association of rs37973 genotypes and changes in lung function after 24 weeks of inhaled corticosteroid therapy in 418 asthma patients. (A) The association between the rs37973 polymorphism and change in FEV1. (B) The association between the rs37973 polymorphism and change in FEV1 (% of predicted); ∗P < .05. FEV1 = forced expiratory volume in one second.
Furthermore, we analyzed the percent FEV1 change defined as (FEV1treatment – FEV1baseline) / FEV1baseline. There were great differences among different individuals after a 24-week ICS treatment. Although most individuals exhibited improved lung function, some did not benefit from ICS treatment. Homozygotes for the wild-type allele who had a percent FEV1 change greater than 5% were more common than were homozygotes of the rare allele (rs37973, AA 67.01% vs GG 49.49%, P < .05, Fig. 2).
Figure 2.
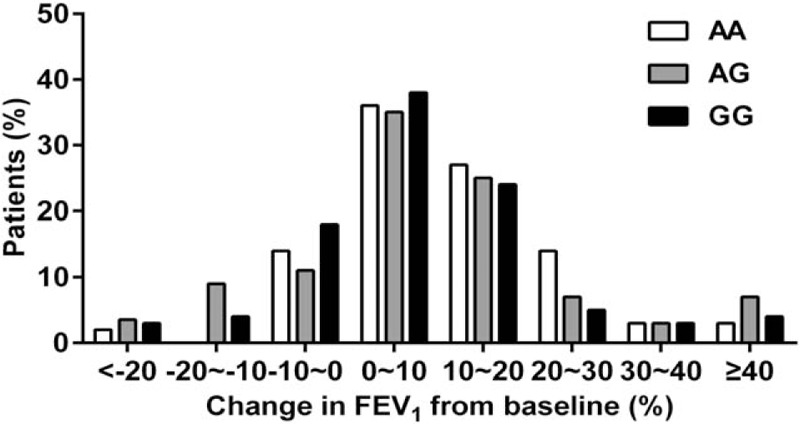
The distribution of FEV1 changes stratified by GLCCI1 rs37973 genotype in 418 patients with asthma after inhaled corticosteroid therapy.
3.4. Lung function changes in individuals with a family history of asthma
We analyzed FEV1 and FEV1%pred improvement in individuals who had a family history of asthma. There were 25 individuals from 11 families, including fathers or mothers, sons or daughters, and brothers or sisters. In all families, individuals who were homozygous carriers of the wild-type allele had an increased pulmonary function change (FEV1 and FEV1%pred) compared with those who were homozygous for the mutant allele or were heterozygous (Table 2).
Furthermore, the numbers of AA, AG, and GG individuals at rs37973 were 9, 10, and 6, respectively, in these families. FEV1 improved in response to inhaled glucocorticoids in homozygous carriers of the wild-type allele, as compared to individuals who were homozygous for the mutant allele or were heterozygous (P < .05), consistent with the initial association detected among all participants. The rare allele carriers at rs37973 (GG) were associated with less improvement in predicted FEV1% compared with wild-type allele homozygotes or heterozygous individuals (P < .05, Fig. 3).
Figure 3.
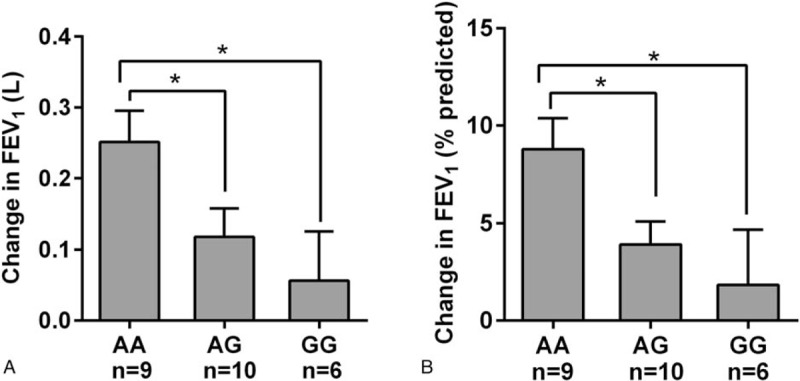
Changes in lung function stratified by genotype after 24 weeks of inhaled corticosteroid therapy in 25 subjects from 11 families with a history of asthma. (A) The association between the rs37973 polymorphism and change in FEV1. (B) The association between the rs37973 polymorphism and change in FEV1 (% of predicted); ∗P < .05. FEV1 = forced expiratory volume in one second.
3.5. Functional characterization
We hypothesized that rs37973 polymorphisms might influence GLCCI1 expression. At the final visit, 31 patients given written informed consent and provided blood for PBMC isolation and culture. By real-time PCR, mRNA levels of GLCCI1 in PBMCs, using β-actin as a reference gene, were measured and stratified by genotype (Fig. 4A). The expression of GLCCI1 was enhanced in the cells treated with 10−7 M dexamethasone compared to sham-treated cells for each genotype (AA, 1.72-fold; AG, 1.48-fold; GG, 1.19-fold). Individuals homozygous for the major allele or heterozygotes exhibited greater expression compared to those who were homozygous for the mutant allele.
Figure 4.
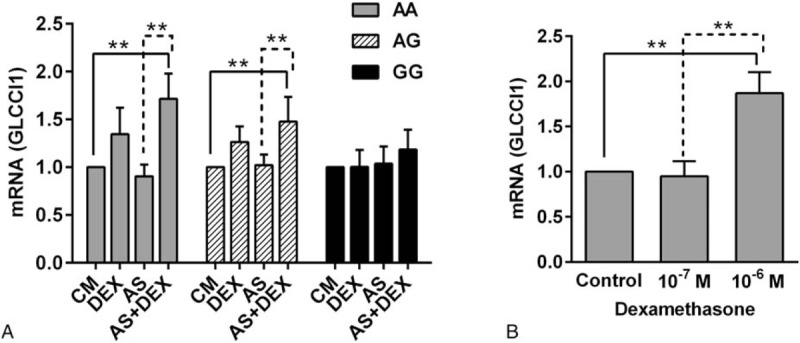
The expression of GLCCI1 mRNA induced by dexamethasone (DEX) in vitro mononuclear cells. (A) The expression of GLCCI1 mRNA stratified by the rs37973 genotype induced by 10−7 M dexamethasone with or without asthmatic serum (AS); (B) Elevated dexamethasone concentrations increased the expression of GLCCI1 in patients with the GG genotype (10−7 M vs. 10−6 M); ∗∗ P < .01.
However, GG homozygotes were less sensitive to glucocorticoid therapy, exhibited elevated concentrations of dexamethasone (10−6 M), and showed greater expression compared to 10−7 M dexamethasone-treated cells (1.869- vs 1.003-fold, P < .01, Fig. 4B). These findings demonstrated that individuals with a GG genotype required a high dose of glucocorticoids.
3.6. Cytokine production in individuals with different genotypes
Treatment with dexamethasone resulted in the inhibition of cytokine secretion. The concentrations of IL-6, IL-8, IL-10, and IL-13 were reduced after stimulation with dexamethasone, not only in the control group but also in the asthma-like group. Additionally, production of these cytokines was significantly reduced after dexamethasone stimulation in cells from AA and AG genotype patients (Fig. 5); however, there was a trend for less cytokine suppression induced by dexamethasone in PBMCs from GG genotype patients (P > .05). We also measured levels of other cytokines, including IL-4, IL-5, IL-9, and IFN-γ; however, the levels of these cytokines were too low to be detected.
Figure 5.
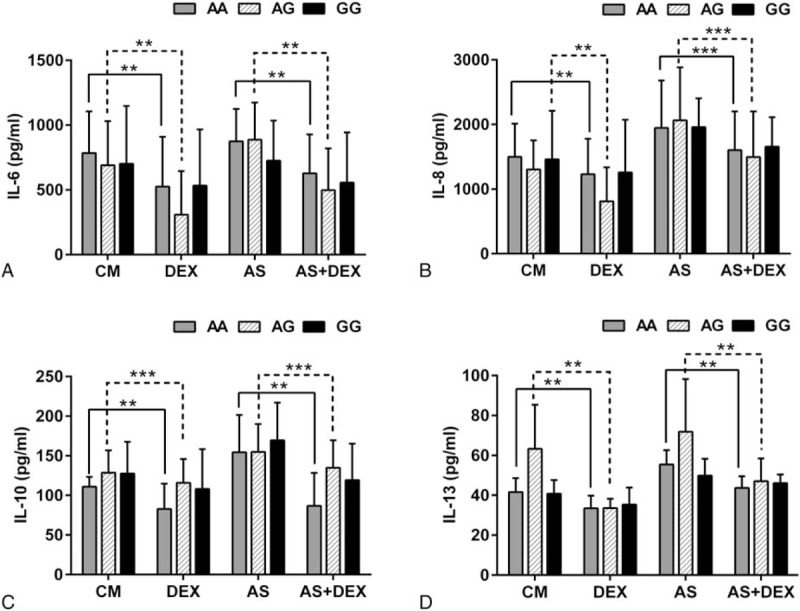
Cytokine production stimulated by 10−7 M dexamethasone (DEX) with or without asthmatic serum (AS). The concentrations of (A) IL-6, (B) IL-8, (C) IL-10 and (D) IL-13 stratified by GLCCI1 rs37973 genotype after cultured for 24 hours with dexamethasone. ∗∗P < .01; ∗∗∗P < .001. AS = asthmatic serum, DX = dexamethasone.
4. Discussion
Our findings demonstrated that a genetic variant in GLCCI1 rs37973 is associated with clinical responses to inhaled corticosteroids in a Han Chinese population. Homozygotes for the mutant allele showed a poorer improvement in FEV1 compared with homozygous for the wild-type allele. After induced by dexamethasone in vitro, the expression of GLCCI1 was higher, and the production of cytokines was significantly reduced in patients with the rs37973 A allele, including both wild-type homozygotes and heterozygotes.
The response to ICS, which represents the cornerstone for controlling and managing asthma, is characterized by high interindividual variability, which is likely to have a genetic basis. Genes are important for predicting the efficiency of ICS treatment and genetic variants predict patients who are non-responsive to ICS treatment and who should receive extra medical care.[26] A tailored treatment for each patient is beneficial for reducing cost and improving viability. Tantisira et al[27] showed that rs37972 and rs37973 were associated with changes in FEV1 after the administration of ICS in 3 of the 4 observed populations under an additive genetic model. Furthermore, rs37973 was in complete linkage disequilibrium with rs37972, and caused a reduced response to inhaled glucocorticoids in patients with asthma by causing changes in GLCCI1 expression. However, a recent study did not confirm that the rs37973 polymorphism influences treatment responses to inhaled corticosteroids in white subjects with asthma.[28] Cheong et al[29] examined the effect of GLCCI1 SNPs on steroid-responsiveness in nephrotic syndrome and found no clinically actionable effect. Another study showed that rs37972 was associated with the treatment effects of adjunct dexamethasone therapy in bacterial meningitis.[30] Hu et al[31] reported several SNPs in GLCCI1 were associated with ICS response in Chinese asthma patients. Our study verifies that GLCCI1 rs37973 polymorphisms are associated with corticosteroid treatment responses in a Chinese population with asthma.
Our study population included 11 families with a history of asthma, including a father or mother and their son, daughter, brothers, or sisters. Each member of the family had a similar or different genotype. However, subjects expressing the rs37973 A allele, including wild-type homozygotes and heterozygotes, exhibited improved pulmonary function after treatment with ICS. This finding is consistent with those for all participants, and subjects who were heterozygous also exhibited an exacerbated response to ICS. The genetic causes of asthma are not fully known, but many studies have suggested that asthma is genetically associated with ORMDL3, CHI3L1, PDE4D, and RAD50-IL13;[32–35] however, few genes have been identified that affect the response of an individual to treatment. In this present study, we chose a family-based design in an effort to increase our power to detect genetic associations, and patients with the rs37973 A allele showed a better response to treatment from these families, including parents and their first-degree relatives.
An important finding of our study is that GLCCI1 expression can be enhanced by dexamethasone (10−7 M) in patients with the rs37973 A allele, including both wild-type homozygotes and heterozygotes. However, GLCCI1 mRNA from homozygotes for the rare allele was not significantly enhanced by stimulation with low-dose dexamethasone (10−7 M). For PBMCs isolated from patients with a GG genotype, high-dose dexamethasone (10−6 M) significantly enhanced the mRNA levels of GLCCI1 compared with low-dose dexamethasone (10−7 M). Therefore, it seemed likely that patients with a GG genotype were relatively insensitive to glucocorticoid treatment, and that these patients needed comparatively higher doses of corticosteroids to control airway inflammation.
Chronic inflammation in airway is the core pathology of asthma, and glucocorticoids are the dominant agent used to treat asthma.[36] However, it has been recognized that glucocorticoids do not work well in 5–10% of all asthmatics, suggesting a reduced response to glucocorticoids.[37] Furthermore, some patients do not even respond to high doses of glucocorticoids.[38] The ultimate physiological responses to glucocorticoids are determined not only by the concentration of glucocorticoids, but also by differences between individuals in glucocorticoid sensitivity, which is influenced by multiple factors. The GC receptor gene (NR3C1) and corticotropin releasing hormone receptor 1 (CRHR1) have been found to be associated with reduced responses to glucocorticoids.[39,40] In our present study, levels of GLCCI1 expression induced by dexamethasone were associated with GLCCI1 genotypes, which suggested that genetic factors might determine glucocorticoid responsiveness by altering the expression of GLCCI1.
Another important finding of our study was that the levels of cytokines vary according to genotypes in response to glucocorticoids. The expression levels of some cytokines were significantly reduced after glucocorticoid treatment in subjects with the rs37973 A allele, including wild-type homozygotes and heterogyzotes. We also found reduced expression levels of cytokines in AS from stimulated PBMCs from these patients. The expression levels of these cytokines were significantly increased after AS treatment, and were reduced after the addition of glucocorticoids in wild-type homozygote and heterozygote individuals. However, subjects who were homozygous for the mutant allele showed no changes in response to low-dose glucocorticoid stimulation. Passive sensitization of human airway cells with serum from allergic patients provides an opportunity to study the interactions between allergic factors and cell behavior, immediate hypersensitivity,[41] or altered responsiveness to nonspecific agonists prior to antigen challenge.[42] In this present study, passively sensitized human PBMCs were used to study the response to treatment.
Asthma is characterized by variable degrees of chronic inflammation, and cytokines are immunomodulatory proteins that are important in regulating airway inflammation.[43] Cytokines are produced and released by a variety of cell types, including immune cells, which include macrophages, lymphocytes, and mast cells, and structural cells, which include endothelial cells, fibroblasts, and epithelial cells. The inflammatory process underlying asthma is coordinated by cytokines. Modulating this cytokine network with biological therapies represents a new paradigm for asthma treatment. In our present study, different genotypes were found to respond differently to glucocorticoid therapy. Our finding of differences in the expression of inflammatory cytokines after glucocorticoid treatment provides evidence that could guide the selection of different doses of glucocorticoids for therapy or other treatments.
There are also some limitations in our study. Because part of the patients refused to provide blood for PBMC isolation and culture in the last callback, only 31 independent experiments were conducted. Low sample size makes it difficult to obtain reliable statistical results. Although we observed homozygous for the major allele or heterozygotes exhibited greater GLCCI1 expression compared to those who were homozygous for the mutant allele, there was no significant difference. However, the study might be underpowered to detect this association and a larger study could confirm such an association. A prospective study would provide a stronger evidence for the relationship between rs37973 and response to ICS. Further study will focus on evaluation of the individual asthma therapy based on the rs37973 SNP.
In conclusion, the genetic variant rs37973 in GLCCI1 is associated with poorer clinical responses to corticosteroid therapy in a Chinese population, which resulted in reduced expression of GLCCI1 and altered production of cytokines.
Footnotes
Abbreviations: FEV1 = forced expiratory volume at one second, GLCCI1 = glucocorticoid-induced transcript 1, ICS = inhaled corticosteroid, SNP = single-nucleotide polymorphisms.
This study was supported by the National Natural Science Foundation of China (No. 81370145, 81370156, 81470227, and 81200029), The National Support Programme of the 12th five-year plan: clinical translational research on respiratory diseases (No. 2012BAI05B01), and the Natural Science Foundation of Hubei (No. 2014CFB971).
The authors have no conflicts of interest to disclose.
References
- [1].To T, Stanojevic S, Moores G, et al. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health 2012;12:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].London SJ, Romieu I. Gene by environment interaction in asthma. Ann Rev Public Health 2009;30:55–80. [DOI] [PubMed] [Google Scholar]
- [3].Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull 2000;56:1054–70. [DOI] [PubMed] [Google Scholar]
- [4].Szefler SJ, Martin RJ, King TS, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol 2002;109:410–8. [DOI] [PubMed] [Google Scholar]
- [5].Weiss ST. New approaches to personalized medicine for asthma: where are we? J Allergy Clin Immunol 2012;129:327–34. [DOI] [PubMed] [Google Scholar]
- [6].Weiss ST, McLeod HL, Flockhart DA, et al. Creating and evaluating genetic tests predictive of drug response. Nat Rev Drug Discov 2008;7:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ortega VE, Wechsler ME. Asthma pharmacogenetics: responding to the call for a personalized approach. Curr Opin Allergy Clin Immunol 2013;13:399–409. [DOI] [PubMed] [Google Scholar]
- [8].Naik SR, Wala SM. Inflammation, allergy and asthma, complex immune origin diseases: mechanisms and therapeutic agents. Recent Pat Inflamm Allergy Drug Discov 2013;7:62–95. [PubMed] [Google Scholar]
- [9].Busse WW. Inflammation in asthma: the cornerstone of the disease and target of therapy. J Allergy Clin Immunol 1998;102(4 pt 2):S17–22. [DOI] [PubMed] [Google Scholar]
- [10].Barnes PJ, Pedersen S, Busse WW. Efficacy and safety of inhaled corticosteroids. New developments. Am J Respir Crit Care Med 1998;157(3 pt 2):S1–53. [DOI] [PubMed] [Google Scholar]
- [11].Pinto CR, Almeida NR, Marques TS, et al. Local adverse effects associated with the use of inhaled corticosteroids in patients with moderate or severe asthma. J Bras Pneumol 2013;39:409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Saag KG. Glucocorticoid-induced osteoporosis. Endocrinol Metab Clin N Am 2003;32:135–57. vii. [DOI] [PubMed] [Google Scholar]
- [13].Hanania NA, Chapman KR, Sturtridge WC, et al. Dose-related decrease in bone density among asthmatic patients treated with inhaled corticosteroids. J Allergy Clin Immunol 1995;96(5 pt 1):571–9. [DOI] [PubMed] [Google Scholar]
- [14].Mathioudakis AG, Amanetopoulou SG, Gialmanidis IP, et al. Impact of long-term treatment with low-dose inhaled corticosteroids on the bone mineral density of chronic obstructive pulmonary disease patients: aggravating or beneficial? Respirology 2013;18:147–53. [DOI] [PubMed] [Google Scholar]
- [15].Bateman ED. Efficacy and safety of high-dose ciclesonide for the treatment of severe asthma. Expert Rev Respir Med 2013;7:339–48. [DOI] [PubMed] [Google Scholar]
- [16].Malmstrom K, Rodriguez-Gomez G, Guerra J, et al. Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. A randomized, controlled trial. Montelukast/Beclomethasone Study Group. Ann Intern Med 1999;130:487–95. [DOI] [PubMed] [Google Scholar]
- [17].Inglis GC, Ingram MC, Holloway CD, et al. Familial pattern of corticosteroids and their metabolism in adult human subjects—the Scottish Adult Twin Study. J Clin Endocrinol Metab 1999;84:4132–7. [DOI] [PubMed] [Google Scholar]
- [18].Wilk JB, Djousse L, Arnett DK, et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet Epidemiol 2000;19:81–94. [DOI] [PubMed] [Google Scholar]
- [19].Palmer LJ, Burton PR, James AL, et al. Familial aggregation and heritability of asthma-associated quantitative traits in a population-based sample of nuclear families. Eur J Hum Genet 2000;8:853–60. [DOI] [PubMed] [Google Scholar]
- [20].Maitland-van der Zee AH, Raaijmakers JA. Variation at GLCCI1 and FCER2: one step closer to personalized asthma treatment. Pharmacogenomics 2012;13:243–5. [DOI] [PubMed] [Google Scholar]
- [21].Martin RJ, Szefler SJ, King TS, et al. The Predicting Response to Inhaled Corticosteroid Efficacy (PRICE) trial. J Allergy Clin Immunol 2007;119:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chapman MS, Askew DJ, Kuscuoglu U, et al. Transcriptional control of steroid-regulated apoptosis in murine thymoma cells. Mol Endocrinol 1996;10:967–78. [DOI] [PubMed] [Google Scholar]
- [23].Chapman MS, Qu N, Pascoe S, et al. Isolation of differentially expressed sequence tags from steroid-responsive cells using mRNA differential display. Mol Cell Endocrinol 1995;108:R1–7. [DOI] [PubMed] [Google Scholar]
- [24].Masoli M, Fabian D, Holt S, et al. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy 2004;59:469–78. [DOI] [PubMed] [Google Scholar]
- [25].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- [26].Palmer LJ, Silverman ES, Weiss ST, et al. Pharmacogenetics of asthma. Am J Respir Crit Care Med 2002;165:861–6. [DOI] [PubMed] [Google Scholar]
- [27].Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med 2011;365:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hosking L, Bleecker E, Ghosh S, et al. GLCCI1 rs37973 does not influence treatment response to inhaled corticosteroids in white subjects with asthma. J Allergy Clin Immunol 2013;133:587–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cheong HI, Kang HG, Schlondorff J. GLCCI1 single nucleotide polymorphisms in pediatric nephrotic syndrome. Pediatr Nephrol 2012;27:1595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brouwer MC, van der Ende A, Baas F, et al. Genetic variation in GLCCI1 and dexamethasone in bacterial meningitis. J Infect 2012;65:465–7. [DOI] [PubMed] [Google Scholar]
- [31].Hu C, Xun Q, Li X, et al. GLCCI1 variation is associated with asthma susceptibility and inhaled corticosteroid response in a Chinese Han population. Arch Med Res 2016;47:118–25. [DOI] [PubMed] [Google Scholar]
- [32].Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–3. [DOI] [PubMed] [Google Scholar]
- [33].Ober C, Tan Z, Sun Y, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 2008;358:1682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Himes BE, Hunninghake GM, Baurley JW, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet 2009;84:581–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li X, Howard TD, Zheng SL, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. J Allergy Clin Immunol 2010;125:328–5.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pignatti PF. Trends in pharmacogenomics of drugs used in the treatment of asthma. Pharmacol Res 2004;49:343–9. [DOI] [PubMed] [Google Scholar]
- [37].Ito K, Chung KF, Adcock IM. Update on glucocorticoid action and resistance. J Allergy Clin Immunol 2006;117:522–43. [DOI] [PubMed] [Google Scholar]
- [38].McManus R. Mechanisms of steroid action and resistance in inflammation and disease. J Endocrinol 2003;178:1–4. [DOI] [PubMed] [Google Scholar]
- [39].Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet 2004;13:1353–9. [DOI] [PubMed] [Google Scholar]
- [40].Sousa AR, Lane SJ, Cidlowski JA, et al. Glucocorticoid resistance in asthma is associated with elevated in vivo expression of the glucocorticoid receptor beta-isoform. J Allergy Clin Immunol 2000;105:943–50. [DOI] [PubMed] [Google Scholar]
- [41].Ellis JL, Hubbard WC, Meeker S, et al. Ragweed antigen E and anti-IgE in human central versus peripheral isolated bronchi. Am J Respir Crit Care Med 1994;150:717–23. [DOI] [PubMed] [Google Scholar]
- [42].Mitchell RW, Ruhlmann E, Magnussen H, et al. Passive sensitization of human bronchi augments smooth muscle shortening velocity and capacity. Am J Physiol 1994;267(2 Pt 1):L218–222. [DOI] [PubMed] [Google Scholar]
- [43].Holgate ST. Innate and adaptive immune responses in asthma. Nat Med 2012;18:673–83. [DOI] [PubMed] [Google Scholar]


