Abstract
We report an intensiometric, near-infrared (NIR) fluorescent, genetically encoded calcium ion (Ca2+) indicator (GECI) with excitation and emission maxima at 678 nm and 704 nm, respectively. This GECI, designated NIR-GECO1, enables imaging of Ca2+ transients in cultured mammalian cells and brain tissue with sensitivity comparable to currently available visible-wavelength GECIs. We demonstrate that NIR-GECO1 opens up new vistas for multicolor Ca2+ imaging in combination with other optogenetic indicators and actuators.
Optically active, genetically encoded (optogenetic) proteins are near-ideal tools for recording and control of biological processes with high spatiotemporal resolution. However, the broad spectral profiles and limited range of colors of available optogenetic tools, such as fluorescent protein (FP)-based GECIs, limits the possibilities for multiplexing. Most genetically-encoded FPs fall into two classes: visibly fluorescent β-barrel FPs (β-FPs) that are homologues of the Aequorea green β-Fp1, and far-red to NIR fluorescent biliverdin (BV)-binding FPs (BV-FPs) derived from bacteriophytochromes (BphPs)2 or other BV-binding proteins3. β-FPs have emission peaks in the visible range (~450 nm to ~670 nm), and BV-FPs have emission peaks in the NIR (~670 nm to ~720 nm)4. While many GECIs and other indicators have been engineered from β-FPs, examples of BV-FP-based indicators are limited. Examples include BV-FPs as donors and acceptors in FRET-based indicators, and the use of split BV-FPs in protein complementation assays5.
To expand the range of GECI colors into the NIR, we have engineered an intensiometric GECI based on the monomeric BV-FP, mIFP6. We pursued a design with a Ca2+-binding domain (Calmodulin (CaM)-RS20), inserted into mIFP such that Ca2+ binding would modulate the BV chromophore environment and fluorescence intensity (Supplementary Note 1). We chose 4 potential insertion sites (between residues 9/10, 57/58, 138/139, and 170/176) based on inspection of the x-ray crystal structure of Deinococcus radiodurans BphP (PDB ID: 2O9B)7, which has 35% sequence identity with mIFP6. Only the replacement of residues 171–175 with CaM-RS20 yielded a protein with a Ca2+-dependent change in fluorescence in vitro (a 2-fold decrease) (Fig. 1a,b and Supplementary Fig. 1). To improve the indicator properties, we systematically optimized the insertion site (leading to deletion of mIFP residues 176 and 177) and the N- and C-terminal linkers (ultimately the sequences GAL and RRHD, respectively) connecting CaM-RS20 to mIFP.
Figure 1.
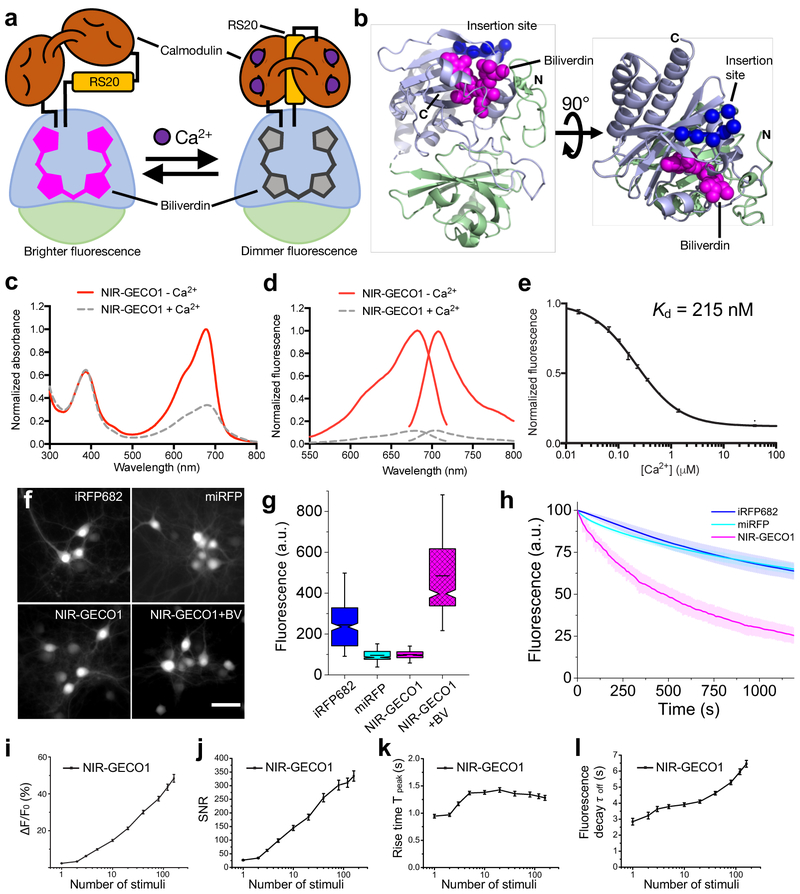
Structure and characterization of NIR-GECO1. (a) Schematic representation of NIR-GECO1 and its mechanism of response to Ca2+. The PAS domain is colored light green and the BV-binding GAF domain is colored light blue. RS20 is the CaM-binding peptide of smooth muscle myosin light chain kinase. (b) Orthogonal views of the structure of DrBphP (PDB ID: 2O9B)7, a close homologue of mIFP. The PAS and GAF domains are colored as in a, BV is shown as magenta spheres, and the Cα atoms of the 7 residues that were replaced with CaM-RS20 are shown as blue spheres. (c) Absorbance spectra in the presence (39 μM) and absence of Ca2+. (d) Fluorescence excitation and emission spectra in the presence (39 μM) and absence of Ca2+. (e) Fluorescence of NIR-GECO1 as a function of Ca2+ concentration. Center values are the mean and error bars are standard deviation. n = 3 independent experiments. (f) Representative wide-field fluorescence images (631/28 nm excitation (Ex) at 38 mW/mm2 and 664LP emission (Em)) of mouse neurons expressing iRFP682, miRFP, NIR-GECO1, and NIR-GECO1 supplemented with exogenous BV (25 μM) (n = 263, 326, 367, and 473 neurons for iRFP682, miRFP, NIR-GECO1 and NIR-GECO1 + BV, respectively, from 2 cultures). The dynamic ranges of these images have been normalized to facilitate visual comparison of protein localization. Fluorescence brightness quantification provided in g. Scale bar, 50 µm. (g) Relative fluorescence intensity for neurons shown in f. Box plots with notches are used. The narrow part of notch is the median; the top and bottom of the notch is the 95% confidence interval of the median; the horizontal line is the mean; the top and bottom horizontal lines are the 25% and 75% percentiles for the data; and the whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles. (h) Photobleaching curves for iRFP682, miRFP, and NIR-GECO1 (n = 84, 69, and 88 neurons, respectively, from 2 cultures; 631/28 nm Ex at 38 mW/mm2; solid lines represent mean value, shaded areas represent standard deviation). (i-l) NIR-GECO1 response amplitude (i), signal-to-noise ratio (SNR) (j), rise time (actually a fluorescence decrease) for Ca2+-binding (k), and decay time (actually a fluorescence increase) for Ca2+-dissociation (l), as a function of number of field stimulation-induced APs. Center values are the mean, and error bars are standard error of the mean (s.e.m). n = 55 neurons.
To facilitate iterative rounds of improvement based on fluorescence screening of randomly mutated variants in bacterial colonies, followed by functional tests in mammalian cells, we created a vector (pcDuEx2) for expression in both Escherichia coli and mammalian cells (Supplementary Fig. 2a). Following twelve rounds of library expression and screening (Supplementary Fig. 2b and 3), we designated our best variant as NIR genetically encoded Ca2+ indicator for optical imaging (NIR-GECO1; Supplementary Figs. 2c and 4). A parallel effort to engineer a GECI from the smURFP3 BV-FP was not successful (Supplementary Fig. 5). NIR-GECO1 has absorbance and emission peaks at 678 nm and 704 nm, respectively, and undergoes a 90% decrease in fluorescence intensity upon binding Ca2+ (Kd = 215 nM) (Fig. 1c-e and Supplementary Fig. 6). The fluorescent change and Kd are comparable to GCaMP3 (Fmax/Fmin = 13.6; Kd = 405 nM), which was the first broadly useful single FP-based GECI8. Key differences include the opposite directions of the responses to Ca2+ and NIR-GECO1’s lower Hill coefficient (n = 1.03). As an inverse response indicator, NIR-GECO1 is in its more brightly fluorescent form in resting cells (low Ca2+), and is therefore more susceptible to photobleaching under continuous illumination. In addition, excitation of resting cells above and below the imaging plane will contribute to an increased background signal. As expected when comparing a β-FP to a BV-FP, the Ca2+ bound state of GCaMP3 is ~6-fold brighter than the Ca2+-free state of NIR-GECO1 (Supplementary Table 1)8.
To evaluate the performance of NIR-GECO1 in cultured neurons, we compared intracellular fluorescence brightness and photostability to the spectrally similar BV-FPs, iRFP682 (Ref. 9) and miRFP10 (Supplementary Table 1). All three BV-FPs distributed evenly within the cytosol, dendrites, and nucleus of neurons, with no apparent puncta or localized accumulations (Fig. 1f). NIR-GECO1 baseline intracellular brightness was similar to miRFP and 2.5-fold lower than iRFP682 (Fig. 1g). Administration of 25 μM exogenous BV for 3 h resulted in ~5-fold increase in the NIR-GECO1 baseline fluorescence (Fig. 1g), indicating that ~80% of NIR-GECO1 is not bound to BV. Addition of BV also resulted in a slight increase in the mean value of the NIR-GECO1 fluorescence changes during spontaneous activity (16 ± 6% versus 20 ± 8% −ΔF/F0 for NIR-GECO1 and NIR-GECO1 + BV, respectively; mean ± standard deviation throughout; Supplementary Fig. 7a,b). This BV-free fraction is not fluorescent but presumably participates in contra-productive Ca2+ buffering. Co-expression of heme-oxygenase 1 (HO1) with NIR-GECO1 (Ref. 6), resulted in only a 1.4-fold enhancement of fluorescence intensity (Supplementary Fig. 7c,d). Under continuous wide-field illumination at 38 mW/mm2 (about 2–4 times higher than typically used for NIR-GECO1 imaging), the photobleaching rate of NIR-GECO1 was ~4-fold higher than miRFP and iRFP682 (Fig. 1h and Supplementary Table 1).
To characterize the fluorescence response of NIR-GECO1 to electric field stimulation-evoked action potentials (APs), field stimuli (50 V, 83 Hz, 1 ms) were delivered in trains of 1, 2, 3, 5, 10, 20, 40, 80, 120 and 160 to transfected neurons (Supplementary Fig. 8a). The resulting fluorescence changes, recorded from cell bodies, revealed that −ΔF/F0, signal-to-noise ratio (SNR), rise time, and decay time, all increased with the number of stimuli (Fig. 1i-l). Relative to GCaMP3, NIR-GECO1 has similar −ΔF/Fmin for 1–10 APs and a ~2-fold higher SNR, but these values are ~10-fold lower than those for GCaMP6s (Supplementary Fig. 9a-d). The near-linear stimulus-response over the range of ~2 to 40 stimuli is consistent with the near-unity Hill coefficient11. In cells, the rise and decay times of NIR-GECO1 appear substantially slower than GCaMP6s. This observation is inconsistent with the fast Ca2+-dissociation kinetics measured in vitro (koff = 1.93 s−1 for NIR-GECO1 vs. 1.08 s−1 for GCaMP6s; Supplementary Fig. 6b). With no targeting sequence attached, NIR-GECO1 distributes throughout the cytoplasm and nucleus. Measuring from the cell body, we found that nuclear-excluded NIR-GECO1 (NES-NIR-GECO1) exhibited similar kinetics to NIR-GECO1, ruling out slow Ca2+ diffusion in and out of the nucleus as an explanation for slower response kinetics (Supplementary Fig. 9c,d). When co-expressed in cultured neurons, NIR-GECO1 and GCaMP6s both report spontaneous oscillations in Ca2+ concentration with opposite fluorescence changes (Supplementary Fig. 9e,f).
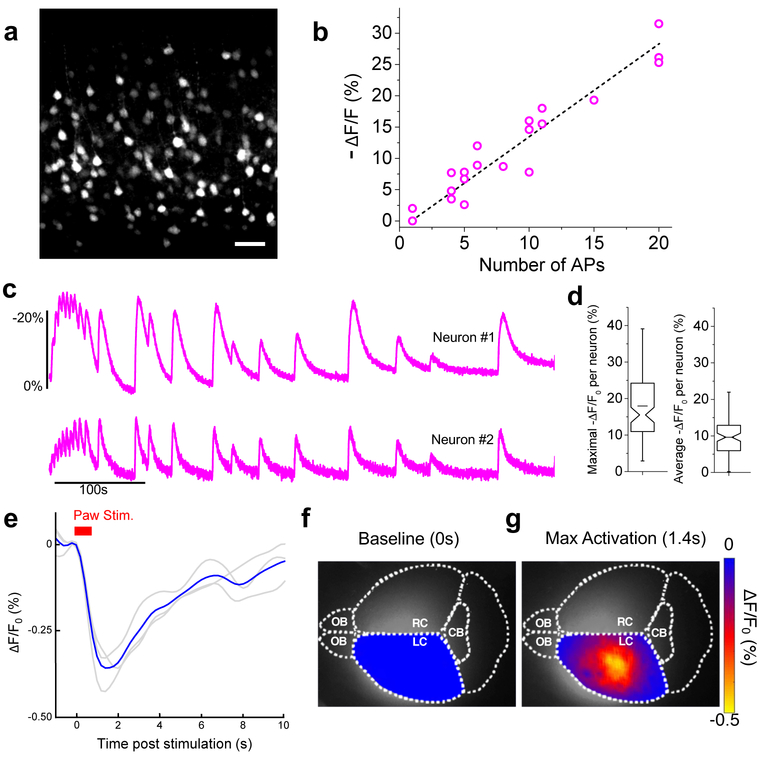
To evaluate in vivo expression of NIR-GECO1, the gene was expressed in layer 2/3 (L2/3) of mouse motor cortex via in utero electroporation (IUE). Imaging of brain slices revealed fluorescence through neuronal cell bodies and processes (Fig. 2a, Supplementary Fig. 8b) and no punctate structures. Stimulation of APs with whole-cell patch clamp electrophysiology gave −ΔF/F0s of 7.2 ± 2.8%, 13.4 ± 3.8%, and 27.6 ± 2.8% for 5, 10 and 20 APs, respectively (Fig. 2b, Supplementary Fig. 8c). Stimulation of neuronal activity with 4-aminopyridine resulted in mean maximal −ΔF/F0 of ~20% and mean averaged −ΔF/F0 of ~10% (Fig. 2c,d). To determine if NIR-GECO1 could be used for one-photon in vivo imaging, we injected adeno-associated virus (AAV), carrying the NIR-GECO1 gene (AAV2/9-hSyn1-NIR-GECO1), in the sensorimotor cortex of mice. Mesoscale fluorescence imaging through the intact skin (hair removed) and skull of anaesthetized mice, during two paradigms of paw stimuli, revealed transient stimuli- and NIR-GECO1-dependent fluorescence changes (decreases) of ~0.3% (Fig. 2e-g, Supplementary Figs. 10,11 and Supplementary Videos 1,2). Under similar conditions, GCaMP6s exhibited ~10-fold greater fluorescence changes (increases). We attribute the better performance of GCaMP6s to its inherently larger Ca2+-dependent fluorescence response (30× vs. 8× under identical conditions; see Supplementary Table 1), its higher Hill coefficient (2.4 vs. 1.0), and lower Kd (144 nM vs. 215 nM) that has been empirically optimized for neuronal activity imaging.
Figure 2.
Imaging of in vivo expressed NIR-GECO1. (a) Representative confocal image of live brain slice expressing NIR-GECO1 (641 nm Ex; 664LP Em; n = 4 slices from 2 mice at P11-22). Scale bar, 50 µm. (b) NIR-GECO1 fluorescence responses to AP trains evoked by current injections (n = 6 neurons from 4 mice at P11-22; dashed line indicates linear regression). Dashed line indicates linear regression. (c) Single-trial wide-field imaging of 4-aminopyridine (1 mM final concentration) evoked neuronal activity from the cell bodies of two representative neurons (631/28 nm Ex and 664LP Em; acquisition rate 20 Hz; n = 129 neurons from 2 slices from one mouse). (d) Maximal (left) and average (right) −ΔF/F0 for the experiment of c. Box plots are used as described in Fig. 1g. For experiments a-d, NIR-GECO1 was expressed in vivo by IUE at E15.5. (e-g) In vivo mesoscale imaging of foot shock responses in mouse sensorimotor cortex. Three mice (4 weeks old) were injected with AVV2/9-hSyn1-NIR-GECO1 in either the right or left side of the brain and imaged (671 nm Ex; 721/42 nm Em) 10 to 21 days later. (e) Response to a paw stimulation paradigm of 10 pulses in 700 ms (0.5 mA, 20 ms on and 50 ms off). Each grey line represents the averaged response of a mouse across 19 cycles and the blue line represents the mean response from all 3 mice (n = 3, i.e., 57 cycles). (f) Activation map of mouse brain, injected in left cortex, before stimulation. Diffuse fluorescence in the right cortex is attributed to diffusion of viral particles and light scattering. (g) Activation map of mouse brain at max activation 1.4 s post stimulation. Scale bar, 2 mm. OB, Olfactory Bulb; CB, Cerebellum; L/RC, Left or Right Cortex.
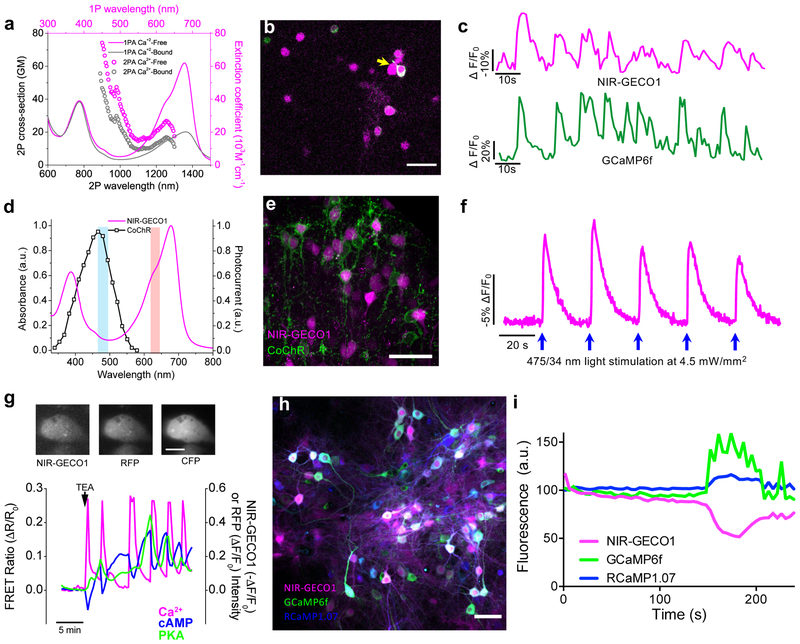
Due to its spectrally distinct fluorescence, NIR-GECO1 should be particularly useful for in vitro imaging in combination with optogenetic actuators and β-FP-based indicators. To explore such applications, we attempted two-photon imaging of NIR-GECO1 and GCaMP6f. NIR-GECO1 two-photon brightness at both 1250 nm and 880 nm excitation is sufficient to image neurons in culture and in mouse brain tissue ex vivo and in vivo (Fig. 3a,b and Supplementary Fig. 12). With 1250 nm excitation we observed neuronal activity-dependent changes in NIR-GECO1 fluorescence in cultured neurons, as confirmed by co-expression of GCaMP6f, with average −ΔF/F0 of 48 ± 28% (n = 37 neurons from one culture; Fig. 3c). With two-photon excitation at 880 nm (11.4 mW of total light power), both the intracellular brightness and photostability of NIR-GECO1 (t1/2 = 20 s) are slightly higher than miRFP but lower than iRFP682 (Supplementary Fig. 12a,b). However, when using 880 nm excitation, we did not observe characteristic fluorescence changes of NIR-GECO1 associated with neuronal Ca2+ dynamics in neurons either in culture or in live brain slices (Supplementary Fig. 12c). We have not succeeded in demonstrating in vivo imaging of neuronal activity using NIR-GECO1 with either 880 nm or 1250 nm two-photon excitation.
Figure 3. Spectral multiplexing of NIR-GECO1 with optogenetic indicators and actuators.
(a) One-photon (solid line; identical to Fig. 1c) and two-photon (open circles) absorption spectra of NIR-GECO1 in the presence and absence of Ca2+. Two-photon absorption spectra are presented versus laser wavelength used for excitation. GM, Goeppert-Mayer units. (b) Representative fluorescence image of cultured neurons expressing NIR-GECO1 (magenta) and GCaMP6f (green) acquired under two-photon excitation (imaging condition: NIR-GECO1 1250 nm Ex, 705/90 nm Em; GCaMP6f 920 nm Ex, 518/45 nm Em; n = 2 cultures). Scale bar, 50 µm. (c) Representative single-trial fluorescence recording of 4-aminopyridine (1 mM final concentration) evoked neuronal activity using NIR-GECO1 and GCaMP6f under imaging conditions as in b (n = 32 neurons from two cultures, yellow arrow indicates the neuron the fluorescence traces were obtained from; image acquisition rate 1 Hz). (d) CoChR action spectrum (black line; adapted from Ref. 12) and NIR-GECO1 absorbance spectrum (magenta line; identical to Fig. 1c with no free Ca2+) with wavelengths used for CoChR activation (475/34 nm; cyan bar) and NIR-GECO1 excitation (638/14 nm; orange bar). (e) Representative confocal images of neurons in L2/3 of motor cortex co-expressing NIR-GECO1 (magenta) and CoChR-mTagBFP2-Kv2.2motif (green) targeted by IUE at E15.5 (imaging conditions: NIR-GECO1 641 nm Ex, 664LP Em; CoChR-mTagBFP2-Kv2.2motif 405 nm Ex and 452/45 nm Em). Scale bar, 50 µm. (f) Single-trial wide-field imaging of NIR-GECO1 responses to CoChR activation (fluorescence excitation and activation as in d; 664LP Em; blue arrows, CoChR stimulation with 200 ms light pulses; image acquisition rate 5 Hz). Similar results were obtained using CheRiff20 (Supplementary Fig. 13d,e). (g) Top, representative fluorescence images of MIN6 β-cell coexpressing AKAR4 (left, 420/20 nm Ex and 475/40 nm Em for CFP and 535/25 nm Em for YFP), NIR-GECO1 (middle, 640/30 nm Ex and 700/75 nm Em), and Pink Flamindo16 (right, 555/25 nm Ex and 605/52 nm Em). Scale bar, 10 μm. Bottom, simultaneous visualization of Ca2+ (NIR-GECO1; −ΔF/F0, magenta line), cAMP (Pink Flamindo; ΔF/F0, blue line), and PKA (AKAR4; FRET emission ratio ΔR/R0, green line) in a MIN6 cell treated with 20 mM tetraethylammonium chloride (TEA) at t = 0 (arrow). Traces for 4 additional representative cells are provided in Supplementary Fig. 14. (h) Representative overlaid fluorescence image of dissociated neurons co-expressing NIR-GECO1, GCaMP6f, and RCaMP1.07. (i) Simultaneous detection of spontaneous neuronal activity reported by GCaMP6f, RCaMP1.07, and NIR-GECO1, in a single cell as in h. The percentage of responding cells (during a 3 minute imaging session) was 92% for GCaMP6f (n = 271 neurons), 79% for RCaMP1.07 (n = 178 neurons), and 59% for NIR-GECO1 (n = 331 neurons).
To explore the combined use of NIR-GECO1 with an optogenetic actuator, we prepared live brain slices expressing NIR-GECO1 and the high photocurrent channelrhodopsin CoChR12,13 (Fig. 3d). Activation of CoChR with cyan-colored light produced Ca2+ transients that were reliably reported by NIR-GECO1 (Fig. 3e,f), and there was no evidence of photophysical artifacts attributable to the illumination conditions (Supplementary Fig. 13a-c)14.
To demonstrate NIR-GECO1’s utility for use with β-FP-based indicators, we performed three-indicator (four-color) imaging using NIR-GECO1, the cyan and yellow β-FP based Protein Kinase A indicator AKAR415, and the red β-FP-based cAMP indicator Pink Flamindo16. Pharmacological stimulation of Ca2+ oscillations in MIN6 β-cells in vitro led to rapid and synchronous oscillations in Ca2+, cAMP, and PKA activity (Fig. 3g, Supplementary Fig. 14 and Supplementary Video 3). Co-expression of NIR-GECO1 with GCaMP6f17 and RCaMP1.07 (Ref. 18) enabled three color in vitro imaging of spontaneous neuronal activity (Fig. 3h,i and Supplementary Video 4).
We have demonstrated that NIR-GECO1 is a useful new addition to the GECI palette. As a first generation indicator, NIR-GECO1 falls short of the most extensively optimized β-FP-based GECIs in several critical performance parameters. Accordingly, NIR-GECO1 is not generally useful for in vivo imaging of neuronal activity. However, NIR-GECO1 does provide a robust inverse response to Ca2+ concentration changes in cultured cells, primary neurons, and in acute slices, roughly on par with GCaMP3. In addition, due to its highly red-shifted excitation maximum, it is the preferred Ca2+ indicator for pairing with blue light-activated optogenetic actuators, in order to minimize actuator activation during imaging19. Finally, it creates a multitude of new opportunities for multiparameter imaging in conjunction with multiple β-FP intensiometric or ratiometric FRET-based indicators.
As with many BV-FPs, NIR-GECO1 is substantially dimmer than state-of-the-art β-FP-derived GECIs such as GCaMP6s (10.7× brighter)17 and jRGECO1a (3× brighter)19. To enable general utility for in vivo imaging, future iterations of NIR-GECO1 should be optimized for brighter fluorescence (e.g., improved BV-binding efficiency could provide up to ~5× increase), increased affinity for Ca2+, increased photostability, and faster kinetics. We expect NIR-GECO1 to be just as amenable to further improvements as the GCaMP series, and for these advancements to be soon realized through protein engineering efforts.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available with the online version of this paper.
Methods
General methods and materials.
Synthetic DNA oligonucleotides were purchased from Integrated DNA Technologies. Q5 high-fidelity DNA polymerase (New England BioLabs) was used for routine PCR amplifications and Taq DNA polymerase (New England BioLabs) was used for error-prone PCR. QuikChange Mutagenesis Kit (Agilent Technologies) was used for site-directed mutagenesis. Restriction endonucleases, Rapid DNA ligation kits, and GeneJET miniprep kits were from Thermo Fisher Scientific. PCR products and products of restriction digests were purified using agarose gel electrophoresis and the GeneJET gel extraction kit (Thermo Fisher Scientific). All DNA sequences were confirmed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Reactions were analyzed at the University of Alberta Molecular Biology Service Unit. Absorbance measurements were made with a DU-800 UV-visible spectrophotometer (Beckman) and fluorescence spectra were recorded on a Safire2 platereader (Tecan).
Engineering of NIR-GECO1.
The gene encoding mIFP (a gift from Michael Davidson and Xiaokun Shu, Addgene plasmid #54620)6 was inserted between BamH1 and EcoR1 of a pBAD vector (Life Technologies) that expressed cyanobacteria Synechocystis HO-1 to convert endogenous heme in bacteria into BV, as previously described3,21.
The DNA sequence encoding CaM and RS20 (a peptide the corresponds to the CaM-binding peptide of smooth muscle myosin light chain kinase; VDSSRRKWNKAGHAVRAIGRLSS) portions of REX-GECO1 (Ref. 22), with mutations Q306D and M339F borrowed from jRGECO1a19, were genetically fused by overlap extension PCR using a DNA sequence that encodes for the flexible peptide linker GGGGS23.
For each site (X) of mIFP targeted for CaM-RS20 insertion, the full-length gene (encoding mIFP1 to X-CaM-RS20-mIFPX+1 to 320) was assembled by overlap extension PCR and then inserted into the pBAD vector. Variants were expressed in E. coli strain DH10B (ThermoFisher Scientific) in LB media supplemented with 100 μg/mL ampicillin and 0.0016% L-arabinose. Proteins were extracted using B-PER Bacterial Protein Extraction Reagent (ThermoFisher Scientific), and tested for fluorescence brightness and Ca2+-dependent response.
The most promising variant from was subjected to an iterative process of library generation and screening in E. coli. The pBAD vector was used in the first three rounds. From the fourth round, pcDuEx2 was used in order to enable expression in both E. coli and mammalian cells. Libraries were generated by error-prone PCR of the whole gene24 or site-directed mutagenesis using Quikchange (Agilent Technologies) and degenerate codons at the targeted positions.
For libraries generated by random mutagenesis, ~10,000 colonies were screened in a given round. For libraries generated by randomization of one or more codons, a number of colonies that was ~3-fold the theoretical number of gene variants were screened. For each round, the top 2% of colonies with high fluorescence intensity were picked, cultured, and tested on 396-well plates. Of those, approximately 25% of those picked variants were further screened in HeLa cell based on fluorescence. In a given round, screening was stopped when a substantially improved variant was identified. There were 12 rounds of screening before NIR-GECO1 was identified.
NIR-GECO1 expression vectors.
pcDuEx2 was constructed based on the pcDNA3.1 backbone. The Tac promoter and a gene sequence containing Kpn2I and XbaI sites was inserted immediately after the CMV promoter using overlap extension PCR. A DNA fragment containing the T7 promoter, the gene encoding NIR-GECO1, and the gene encoding HO-1, was amplified from the pBAD vector and inserted into the Kpn2I and XbaI sites.
For HeLa cell expression, pcDuEx2 vector was used. For expression in dissociated neurons, either an AAV2 vector or a lentivirus containing NIR-GECO1 were used. For AAV2 vector preparation, NIR-GECO1 was cloned from pcDuEx2 into BamHI and HindIII sites of AAV2 vector (a gift from Roger Tsien, Addgene plasmid #50970)25. To create lentivirus expressing NIR-GECO1, the gene for NIR-GECO1 or NIR-GECO1-T2A-HO1 was cloned into the BamHI and EcoRI sites of FCK lentivirus vector (Addgene plasmid #22217). HEK293FT cells at 80% confluency in 35-mm cell-culture dishes (Corning) were transfected with 1.5 μg FCK-CMV-NIR-GECO1 or FCK-CMV-NIR-GECO1-T2A-HO1, 1.0 μg psPAX2 (a gift from D. Trono, Addgene plasmid #12260), 0.5 μg pMD2.G (a gift from D. Trono, Addgene plasmid #12259), and 0.2 μg pAdvantage (Promega), with 9 μL Turbofect transfection reagent in 2 mL Opti-MEM medium (Thermo Fisher Scientific). Opti-MEM medium containing Turbofect and DNA mix were replaced with 2 mL complete cell-culture medium containing 110 mg/mL sodium pyruvate at 24 h post-transfection. At 48 h post-transfection, the virus-containing supernatant was collected, spun at 400g (relative centrifugal force (RCF)) for 5 min and filtered through a 0.45 μm PVDF Syringe Filter Unit (EMD Millipore) to get rid of pellet cellular debris. Dissociated neurons in 24-well plates were transduced with 2 mL virus-containing supernatant.
Protein purification and in vitro characterization.
The gene encoding NIR-GECO1, with a poly-histidine tag on the C-terminus, was expressed from the pBAD vector. Bacteria were lysed by cell disruptor (Constant Systems Ltd.), centrifuged at 15000g for 30 min, and proteins were purified by Ni-NTA affinity chromatography (Agarose Bead Technologies). The buffer was typically exchanged to 10 mM MOPS, 100 mM KCl (pH 7.2) with centrifugal concentrators (GE Healthcare Life Sciences). Extinction coefficients (EC) were determined by comparing the absorbance value at 683 nm to the absorbance value at the 391 nm, and assuming an EC of 39,900 M−1cm−1 at 391 nm (Refs. 2,6). For determination of quantum yields (QY or Φ), purified mIFP (QY = 0.08) was used as a standard. The concentration of NIR-GECO1 (Ca2+-free), NIR-GECO1 (Ca2+-saturated) and mIFP was adjusted to have absorbance of 0.2 to 0.6 at 650 nm. A series of dilutions, with absorbance ranging from 0.01 to 0.05, were prepared and integrated emission intensity vs. absorbance was plotted. QYs were determined from the slopes (S) of each line using the equation: Φprotein = Φstandard × (Sprotein/Sstandard). pH titrations were performed by diluting protein into buffers (pH from 2 and 11) containing 30 mM trisodium citrate, 30 mM sodium borate, and either 10 mM CaCl2 or 10 mM EGTA. Fluorescence intensities as a function of pH was then fitted by a sigmoidal binding function to determine the apparent pKa. Ca2+ titrations were carried out using EGTA-buffered Ca2+ solutions (Calcium Calibration Buffer Kit #1, Life Technologies). Buffers were prepared by mixing a CaEGTA buffer (30 mM MOPS, 100 mM KCl, 10 mM EGTA, 10 mM CaCl2) and an EGTA buffer (30 mM MOPS, 100 mM KCl, 10 mM EGTA) to give free Ca2+ concentrations ranging from 0 nM to 39 μM at 25 °C. Fluorescence intensities were plotted against Ca2+ concentrations and fitted by sigmoidal binding function to determine the Hill coefficient and Kd. To determine koff, a SX20 stopped-flow spectrometer (Applied Photophysics) was used. Briefly, proteins samples with 10 μM CaCl2 (30 mM MOPS, 100 mM KCl, pH 7.2) were rapidly mixed with 10 mM EGTA (30 mM MOPS, 100 mM KCl, pH 7.2) at room temperature, and absorption growth curve was measured and fitted by a single exponential equation.
Two-photon spectra and cross sections were measured using femtosecond excitation as described in Supplementary Note 2.
Animal care.
For experiments performed at Massachusetts Institute of Technology (MIT), all methods for animal care and use were approved by the MIT Committee on Animal Care and were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Four time pregnant Swiss Webster mice (Taconic) were used for this study, as were five C57BL/6 mice (Taconic), ages 4–12 weeks. Mice were used without regard to gender.
For experiments performed at Technical University of Munich, all animal in vivo experimentation was done in full compliance with the institutional guidelines of the Institute for Biological and Medical Imaging and with approval from the Government District of Upper Bavaria. A total of 9 mice were used for these experiments: three female FOXN1 nude mice that were injected with the NIR-GECO1 virus; three female Black6 (C57BL/6J) transgenic mice expressing GCaMP6s; and three mice (two female FOXN1 and one female Black6) that were injected with PBS as negative controls.
All experiments at University of Alberta for obtaining the cortical neurons were approved by the University of Alberta Animal Care and Use Committee and carried out in compliance with guidelines of the Canadian Council for Animal Care and the Society for Neuroscience’s Policies on the Use of Animals and Humans in Neuroscience Research.
For experiments at HHMI Janelia Research Campus, all surgical and experimental procedures were in accordance with protocols approved by the HHMI Janelia Research Campus Institutional Animal Care and Use Committee and Institutional Biosafety Committee.
Imaging of NIR-GECO1 in HeLa cells and dissociated neuron cultures.
HeLa cells (40 to 60% confluent) in 24-well glass bottom plates (Cellvis) were transfected with 0.5 µg of the NIR-GECO1-pcDuEx2 plasmid and 2 μL TurboFect (Thermo Fisher Scientific) in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco Fisher Scientific). Following 2 h incubation, the media was changed to DMEM supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 2 mM GlutaMax (Thermo Fisher Scientific), and 1% penicillin-streptomycin (Gibco). The cells were then incubated for 48 h at 37 °C in a CO2 incubator. Prior to imaging, culture medium was changed to Hanks’ Balanced Salt Solution (HBSS).
For dissociated hippocampal mouse neuron culture preparation, postnatal day 0 or 1 Swiss Webster mice (Taconic Biosciences, Albany, NY) were used as previously described10. Briefly, dissected hippocampal tissue was digested with 50 units of papain (Worthington Biochem) for 6–8 min at 37 °C, and the digestion was stopped by incubating with ovomucoid trypsin inhibitor (Worthington Biochem) for 4 min at 37 °C. Tissue was gently dissociated with Pasteur pipettes, and dissociated neurons were plated at a density of 20,000–30,000 per glass coverslip coated with Matrigel (BD Biosciences). Neurons were seeded in 100 µL plating medium containing MEM (Life Technologies), glucose (33 mM, Sigma), transferrin (0.01%, Sigma), Hepes (10mM, Sigma), Glutagro (2 mM, Corning), Insulin (0.13%, Millipore), B27 supplement (2%, Gibco), and heat inactivated FBS (7.5%, Corning). After cell adhesion, additional plating medium was added. AraC (0.002 mM, Sigma) was added when glia density was 50–70% of confluence. Neurons were grown at 37 °C and 5% CO2 in a humidified atmosphere. Cultured neurons were transduced at 4–5 days in vitro (DIV) by administering ~1010 viral particles of rAAV8-hSyn-iRFP682, rAAV8-hSyn-miRFP (both from Vector Core, University of North Carolina, Chapel Hill, NC) or rAAV9-hSyn-NIR-GECO1 (Department of Biochemistry and Microbiology, University of Laval, Quebec, Canada) per well (the rAAV genome titer was determined by dot blot). For co-expression of the GECIs, the rAAV8-hSyn-GCaMP6f, rAAV8-hSyn-RCaMP1.07 (both from Vector Core, University of North Carolina, Chapel Hill, NC), and rAAV9-hSyn-NIR-GECO1 viral particles were added in the 1:1:3 ratio, respectively. A biliverdin hydrochloride (Sigma-Aldrich) solution in DMSO (25 mM) was used as a 1000× stock (25 µM final concentration) for the experiments shown in Fig. 1g and Supplementary Fig. 7a,b. All measurements on neurons were taken after DIV 16.
For dissociated rat cortical neuron culture preparation, postnatal day 0 or 1 Sprague Dawley rats were used. Dissected cortices were digested in Papain Solution (50 units, Sigma) for 10 min at 37 °C followed by incubation with DNase (0.15 mg/ml, Sigma) for 5 min at 37 °C. After washing with FBS (Sigma) and removing supernatant, neurobasal B27 (Thermo Fisher Scientific) was added to tissue. Tissue was then gently dissociated with Pasteur pipettes, and dissociated neurons were plated at a density of ~1.5×105 on collagen-coated 24-well glass bottom dishes containing NbActiv4 culture medium (BrainBits LLC) supplemented with 2% FBS, penicillin-G potassium salt (50 units/mL), and streptomycin sulfate (50 mg/mL). Half of the culture media was replaced every 4 to 5 days. Neuronal cells were infected using the NIR-GECO1 lentivirus on day 8. Prior to imaging, the culture medium was changed to HBSS.
Wide-field fluorescence imaging of cultured neurons was performed using epifluorescence inverted microscope (Eclipse Ti-E, Nikon) equipped with a Photometrics QuantEM 512SC camera and a 75W Nikon xenon lamp or a Zyla5.5 sCMOS camera (Andor) and a SPECTRA X light engine (Lumencor). NIS-Elements Advanced Research (Nikon) was used for automated microscope and camera control. Cells were imaged with a 60× NA1.49 oil or 20× NA0.75 air objective lenses (Nikon) at room temperature. For dual-color imaging with GCaMP6s, NIR (650/60 nm Ex and 720/50 nm Em) and green (490/15 nm Ex and 525/50 nm Em) filter sets were rotated into the emission light path. Three-color Ca imaging with GCaMP6f and RCaMP1.07 was performed using an inverted Nikon Eclipse Ti microscope equipped with a spinning disk sCSUW1 confocal scanner unit (Yokogawa, Tokyo, Japan), 488, 561, and 642 nm solid state lasers, 525/25 nm, 579/34 nm, and 664LP emission filters, a 20× NA0.75 air objective lens (Nikon), and a 4.2 PLUS Zyla camera (Andor), controlled by NIS-Elements AR software. One cautionary note for confocal imaging is that gallium-arsenide-phosphide (GaAsP) photomultiplier tube (PMT) detectors have poor sensitivity at wavelengths greater than 700 nm.
Two-photon imaging as in Fig. 3b,c and Supplementary Fig. 12c was performed using an Olympus FVMPE-RS equipped with two lasers for fluorescence excitation. An InSight X3 laser (Spectra-Physics) tuned to 1250 nm at 8.0% transmissivity was used to excite NIR-GECO1, and a MaiTai HP Ti:Sapphire laser (Spectra-Physics) tuned to 920 nm at 17.4% transmissivity was used to excite GCaMP6f. The laser beams were focused by a 25× 1.05 NA water-immersion objective lens (Olympus). NIR-GECO1 emission was separated using a 660–750 nm filter, GCaMP6f emission was separated using a 495–540 nm filter, and signals were collected onto separate photomultiplier tubes. Imaging was performed at 2.0 µs/pixel sampling speed with one way galvano scanning. Raw scanner data was converted to an image z-stack using ImageJ (NIH).
Two-photon imaging for Supplementary Fig. 12a,b,d,e,f was performed using a two-photon laser scanning microscope (Ultima IV, Prairie Technologies, Sioux Falls, SD) with a mode-locked Ti-Sapphire laser (Mai-Tai, Spectra Physics) and a 16× NA0.8 water-immersion objective (CFI75 LWD 16; Nikon). For image acquisition, the laser was set to emit 880 nm at total light power 11.4 mW, and 535/50 and 731/137 nm emission filters (Semrock) were used. The microscope was operated using the ScanImage 3.8 software package26.
Electrophysiology and Ca2+ imaging in dissociated hippocampal neurons.
The genes encoding NIR-GECO1 and GCaMP6s were expressed under the control of synapsin promoter in cultured rat hippocampal neurons. Neurons were stimulated using a custom-built field stimulator using a stimulus isolator (A385, World Precision Instruments) with platinum wires. Field stimuli (50 V, 83 Hz, 1 ms) were delivered in trains of 1, 2, 3, 5, 10, 20, 40, 80, 120 and 160 field stimuli to the cultured neurons. Neurons were imaged using a Nikon Eclipse Ti2 inverted microscope equipped with a 40× objective (Nikon, 1.4 NA). A quad bandpass filter (set number: 89000, Chroma) was used along with a 480 nm LED (Spectra X light engine, Lumencor) or a 640 nm LED (Spectra X light engine, Lumencor) to image GCaMP6s and NIR-GECO1 respectively. Fluorescence was collected using a sCMOS camera (Orca-Flash4.0, Hamamatsu) at 34 Hz. For GCaMP6s, the response amplitude (ΔF/Fmin) was quantified as the change in fluorescence divided by baseline fluorescence over 0.5 s period preceding the stimulus. For NIR-GECO1, the response amplitude was quantified as the change in fluorescence divided by peak fluorescence during the stimulus (-ΔF/Fmin). Signal-to-noise ratio (SNR) was quantified as peak change in fluorescence over the standard deviation of the signal over the 0.5 s period preceding stimulation.
Multiplexed live-cell imaging with NIR-GECO1 in MIN6 β-cells.
MIN6 pancreatic β-cells were cultured in DMEM containing 4.5 g/L glucose, supplemented with 10% (v/v) FBS, 1% (v/v) Pen-Strep, and 50 μM β-mercaptoethanol, and maintained at 37 °C with a 5% CO2 atmosphere. Cells were plated onto 35-mm glass-bottomed dishes, grown to 40–60% confluence, and then transfected with 0.5 µg each of plasmids encoding AKAR415, Pink Flamindo16, and NIR-GECO1 using Lipofectamine 2000 (Invitrogen). After 48 h, cells were washed twice with HBSS (Gibco) and imaged in HBSS at 37 °C using a Zeiss AxioObserver Z1 inverted epifluorescence microscope (Carl Zeiss) equipped with a 40×/1.3 NA objective, a Lambda 10–2 filter-changer (Sutter Instruments), and a Photometrics Evolve 512 EMCCD (Photometrics, Tucson, AZ) controlled by METAFLUOR 7.7 software (Molecular Devices). Filters for cyan/yellow emission ratio were a 420DF20 excitation filter, a 450DRLP dichroic mirror, and two emission filters (475DF40 for CFP and 535DF25 for YFP). Filters for RFP were a 555DF25 excitation filter, a ZT568RDC dichroic mirror, and a 605DF52 emission filter. Filters for NIR-GECO1 were a 640DF30 excitation filter, a 700DF75 excitation filter, and a T660LPXR dichroic mirror. Exposure times ranged between 50 and 500 ms, with EM gain set from 10–50, and images were acquired every 20 s. Fluorescence intensities were corrected by background subtraction. The emission ratio change (R-R0) or fluorescence intensity change (F-F0) was divided by the initial ratio or intensity to obtain ΔR/R0 or ΔF/F0, with time zero defined as the time-point immediately preceding drug addition. Graphs were plotted using GraphPad Prism 7 (GraphPad Software).
In utero electroporation.
Embryonic day (E) 15.5 timed-pregnant female Swiss Webster (Taconic) mice were deeply anesthetized with 2% isoflurane. Uterine horns were exposed and periodically rinsed with warm sterile PBS. A plasmid encoding NIR-GECO1 or a mixture of plasmids encoding NIR-GECO1 and CoChR (pCAG-NIR-GECO1-WPRE, pCAG-CoChR-mTagBFP2-Kv2.2motif-WPRE; at total DNA concentration ~1–2 μg/μL) diluted with PBS were injected into the lateral ventricle of one cerebral hemisphere of an embryo. Five voltage pulses (50 V, 50 ms duration, 1 Hz) were delivered using round plate electrodes (ECM™ 830 electroporator, Harvard Apparatus). Injected embryos were placed back into the dam, and allowed to mature to delivery. The P0 pups were screened for corresponding fluorescence and negative pups were excluded for further experiments. All experimental manipulations were performed in accordance with protocols approved by the Massachusetts Institute of Technology Committee on Animal Care, following guidelines described in the US National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Acute brain slice preparation.
Acute brain slices were obtained from Swiss Webster (Taconic) mice at P11 to P22, using standard techniques. Mice were used without regard for sex. No statistical methods were used to estimate sample size for animal studies throughout. No randomization or blinding were used for animal studies throughout. Mice were anaesthetized by isoflurane inhalation, decapitated and cerebral hemispheres were quickly removed and placed in cold choline-based cutting solution consisting of (in mM): 110 choline chloride, 25 NaHCO3, 2.5 KCl, 7 MgCl2, 0.5 CaCl2, 1.25 NaH2PO4, 25 glucose, 11.6 ascorbic acid, and 3.1 pyruvic acid (339–341 mOsm/kg; pH 7.75 adjusted with NaOH) for 2 min, blocked and transferred into a slicing chamber containing ice-cold choline-based cutting solution. Coronal slices (300 μm thick) were cut with a Compresstome VF-300 slicing machine, transferred to a holding chamber with artificial cerebrospinal fluid (ACSF) containing (in mM) 125 NaCl, 2.5 KCl, 25 NaHCO3, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4 and 11 glucose (300–310 mOsm/kg; pH 7.35 adjusted with NaOH), and recovered for 10 min at 34 °C, followed by another 30 min at room temperature. Slices were subsequently maintained at room temperature until use. Both cutting solution and ACSF were constantly bubbled with 95% O2 and 5% CO2.
Concurrent electrophysiology and Ca2+ imaging in acute brain slice.
Slices were transferred to a recording chamber on an Olympus BX51WI upright microscope and superfused (2–3 mL/min) with ACSF at room temperature. Whole-cell patch clamp recordings were acquired via an Axopatch 700B amplifier (Molecular Devices) and Digidata 1440 digitizer (Molecular Devices). For recordings, borosilicate glass pipettes (Warner Instruments) with an outer diameter of 1.2 mm and a wall thickness of 0.255 mm were pulled to a resistance of 3–5 MΩ with a P-97 Flaming/Brown micropipette puller (Sutter Instruments) and filled with a solution containing 155 mM K-gluconate, 8 mM NaCl, 0.1 mM CaCl2, 0.6 mM MgCl2, 10 mM HEPES, 4 mM Mg-ATP, and 0.4 mM Na-GTP. The pipette solution pH was adjusted to 7.3 with KOH and the osmolarity was adjusted to 298 mOsm with sucrose. Cells were visualized through a 40× NA0.8 water-immersion objective with epifluorescence. Whole-cell current-clamp recordings were obtained from NIR-GECO1-positive neurons in layer 2/3 of motor cortex. Fluorescence was excited by a SPECTRA X light engine (Lumencor) with 638/14 nm excitation filter (Semrock), fluorescence was collected through the same objective, passed through a 664LP emission filter, and imaged onto an Orca-Flash4.0 V2 sCMOS camera (Hamamatsu) at 50 Hz acquisition frequency.
In vivo imaging of NIR-GECO1.
Methods for in vivo two-photon imaging to acquire the image shown in Supplementary Fig. 12f are provided as Supplementary Note 3. Methods for in vivo mesoscale imaging to acquire data and images shown in Fig. 2e-g and Supplementary Figs. 10 and 11, are provided as Supplementary Note 4.
Statistics and reproducibility.
All data are expressed as mean ± s.d or mean ± s.e.m, as specified in figure legends. Box plots with notches27 are used for Figs. 1g, 2d, and Supplementary Figs. 7b, 12a and 13c. In these plots, the narrow part of notch is the median; the top and bottom of the notch is the 95% confidence interval of the median; the horizontal line is the mean; the top and bottom horizontal lines are the 25% and 75% percentiles for the data; and the whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles. Sample sizes (n) are listed with each experiment. No samples were excluded from analysis and all experiments were reproducible. For experiments for which representative data is shown, the number of times each experiment was repeated independently with similar results is summarized in Supplementary Note 5. No randomization or blinding was used. All attempts at replication of the experiments were successful.
Reporting Summary.
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data Availability
The NIR-GECO1 gene sequence is available through GenBank with an accession code that is TBD (submission #MK134690). pDuEx2-NIR-GECO1 (plasmid #113680) and pAAV-hSyn-NES-NIR-GECO1 (plasmid #113683) are available via Addgene according to the terms of the Uniform Biological Material Transfer Agreement. Source data for all figures is available online.
Supplementary Material
Acknowledgements
The authors thank the University of Alberta Molecular Biology Services Unit, Y. Li, H. Zhou, A. Aggarwal, for technical support, A. Holt for providing access to the stopped-flow spectrophotometer, and M. Vanni, T. Murphy, A. Nimmerjahn, and S. Chen for preliminary AAV testing. We thank M-E. Paquet at the University of Laval Molecular Tools Platform, and the Janelia Research Campus (JRC) Virus core, for AAV production, We thank and V. Rancic and the JRC Histology group for preparing cultured neurons. We thank D. Park and H.J. Suk for help with characterization of NIR-GECO1 in brain slice and two-photon imaging. We thank Michael Reiss for assistance with the mouse handling, Dr. X. Luis Deán-Ben for help with the in vivo mesoscale data analysis, M. Davidson and X. Shu for the mIFP gene, and E. Rodriguez for the smURFP gene. Work in R.E.C.’s lab was supported by grants from NSERC (RGPIN 288338–2010), CIHR (MOP 123514 and FS 154310), Brain Canada, and NIH (UO1 NS090565). D.R. acknowledges support from the European Research Council (ERC-2015-CoG-682379). The work of D.R. and S.S was also supported by the NIH (R21-EY026382 and UF1-NS107680). E.S.B. was supported by John Doerr, the HHMI-Simons Faculty Scholars Program, the Open Philanthropy Project, Human Frontier Science Program (RGP0015/2016), U. S. Army Research Laboratory and the U.S. Army Research Office (W911NF1510548), U.S.-Israel Binational Science Foundation (2014509), and NIH (2R01-DA029639 and 1R01-GM104948).
Footnotes
Competing interests
The University of Alberta has non-exclusively licensed NIR-GECO1 to LumiSTAR Biotechnology.
Additional Information
Additional information Supplementary information is available for this paper at www.nature.com/.
References
- 1.Tsien RY Annu. Rev. Biochem 67, 509–544 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Shu X et al. Science 324, 804–807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodriguez EA et al. Nat. Methods 13, 763–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piatkevich KD et al. Biophys. J 113, 2299–2309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shcherbakova DM, Stepanenko OV, Turoverov KK & Verkhusha VV Trends Biotechnol. (2018). Doi: 10.1016/j.tibtech.2018.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu D et al. Nat. Methods 12, 763–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD & Forest KT J. Biol. Chem 282, 12298–12309 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Tian L et al. Nat. Methods 6, 875–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shcherbakova DM & Verkhusha VV Nat. Methods 10, 751–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piatkevich KD et al. Nat. Chem. Biol 14, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue M et al. Nat. Methods 12, 64–70 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Shemesh OA et al. Nat. Neurosci 20, 1796–1806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klapoetke NC et al. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J et al. ACS Chem. Neurosci 4, 963–972 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depry C, Allen MD & Zhang J Mol. Biosyst 7, 52–58 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Harada K et al. Sci. Rep 7, 7351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T-WW et al. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohkura M, Sasaki T, Kobayashi C, Ikegaya Y & Nakai J PLoS One 7, e39933 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dana H et al. Elife 5, e12727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochbaum DR et al. Nat. Methods 11, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gambetta GA & Lagarias JC Proc. Natl. Acad. Sci. U. S. A 98, 10566–10571 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J et al. Nat. Commun 5, 5262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heckman KL & Pease LR Nat. Protoc 2, 924–932 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Cirino PC, Mayer KM & Umeno D Directed Evolution Library Creation: Methods and Protocols 231, 3–9 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Lin JY et al. Neuron 79, 241–253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pologruto TA, Sabatini BL & Svoboda K BioMed Eng OnLine 2, 13 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krzywinski M & Altman N Nat. Methods 11, 119–120 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NIR-GECO1 gene sequence is available through GenBank with an accession code that is TBD (submission #MK134690). pDuEx2-NIR-GECO1 (plasmid #113680) and pAAV-hSyn-NES-NIR-GECO1 (plasmid #113683) are available via Addgene according to the terms of the Uniform Biological Material Transfer Agreement. Source data for all figures is available online.