Abstract
Background and Aims:
Long non-coding RNAs (lncRNAs) play a critical role in the regulation of many important cellular processes. However, the mechanisms by which lncRNAs regulate viral infection and host immune responses are not well understood. We sought to explore lncRNA regulation of HCV infection and interferon response.
Methods:
We performed RNAseq in Huh7.5.1 cells with or without IFNα treatment. CRISPR/Cas9 guide RNA (gRNA) was used to knock out selected genes. The promoter clones were constructed, and the activity of related interferon-stimulated genes (ISGs) were detected by the secrete-pair dual luminescence assay. We constructed the full-length and four deletion mutants of an interferon-induced lncRNA RP11–288L9.4 (lncRNA-IFI6) based on predicted secondary structure. Selected gene mRNAs and their proteins, together with HCV infection, in Huh7.5.1 cells and Primary Human Hepatocytes (PHHs) were monitored by qRT-PCR and Western blot.
Results:
We obtained 7901 lncRNAs from RNAseq. 1062 host-encoded lncRNAs were significantly differentially regulated by IFNα treatment. We found that lncRNA-IFI6 gRNA significantly inhibited HCV infection compared to negative gRNA control. The expression of the antiviral ISG IFI6 was significantly increased following lncRNA-IFI6 gRNA editing compared to negative gRNA control in JFH1-infected Huh7.5.1 cells and PHHs. We observed that lncRNA-IFI6 regulation of HCV was independent of JAK-STAT signaling. lncRNA-IFI6 negatively regulated IFI6 promoter function through histone modification. Overexpression of the truncated spatial domain or full-length lncRNA-IFI6 inhibited IFI6 expression and increased HCV replication.
Conclusion:
A novel lncRNA, lncRNA-IFI6, regulates antiviral innate immunity in the JFH1 HCV infection model. lncRNA-IFI6 regulates HCV infection independently of the JAK-STAT pathway. lncRNA-IFI6 exerts its regulatory function via promoter activation and histone modification of IFI6 through its spatial domain.
Keywords: lncRNA, RNAseq, ISGs, HCV, IFI6, antiviral immunity
Introduction
Coding genes only occupy around 2% of the human genome. The majority of the remaining transcripts are non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), extracellular RNAs (exRNAs) and piwi-interacting RNAs (piRNAs) (1, 2). Among these ncRNAs, lncRNAs are the most abundant in the genome of organisms (3). lncRNAs play multiple gene regulatory roles through modulation of transcription or via post-transcriptional mechanisms, and are involved in diverse physiological processes (4). One such family of genes that are regulated by lncRNAs are interferon-stimulated genes (ISGs). For example, bone marrow stromal cell antigen 2 (BST2) interferon-stimulated positive regulator (BISPR) is an RNA gene affiliated with the ncRNA class. Interferon (IFN) stimulated lncRNA BST2/BISPR regulates the expression of the antiviral factor BST2 to affect the potency of the antiviral response (5). In addition, the innate immune response-related lncRNA, negative regulator of antiviral response (NRAV), regulates the expression of multiple critical ISGs through interference with histone modification (6). Interestingly, several specific host lncRNAs are also involved in the regulation of viral infection. For example, the influenza A virus (IAV)-induced lncRNA VIN was observed to regulate replication and viral protein synthesis of IAV (7). lncRNA noncoding repressor of Nuclear Factor of Activated T cells (NRON) expressed in Jurkat cells has been shown to be regulated by HIV-1 infection, which in turn modulates HIV-1 replication in a Nuclear Factor of Activated T cells (NFAT)-dependent manner (8).
Hepatitis C virus (HCV) chronically infects approximately 71 million people worldwide, and is a leading cause of hepatocellular carcinoma (HCC) and liver cirrhosis (9). Prior to the development of direct-acting antiviral (DAA) therapy, IFN-based therapy (plus ribavirin) was the mainstay of therapy for acute and chronic HCV infection. Unfortunately, there is still a sizable population of HCV-infected persons non-responsive to IFN-based therapy in developing countries where DAA therapy is either not yet available or the cost of DAA therapy is prohibitive. Following HCV infection, the expression of hundreds of ISGs produces an antiviral state that restricts HCV spread within liver. Furthermore, pre-treatment levels of intrahepatic ISGs are closely related to polymorphisms in host IFNL3 genotype, and are also associated with IFN responsiveness (10–12). Although many ISGs exert antiviral functions in the context of HCV infection, the biological regulatory mechanisms of lncRNAs in the context of chronic HCV infection and the attendant ISG exhausted phenotype that is observed in chronic HCV infection have not been described. In addition, recent reports have suggested that DAA therapy for chronic HCV infection may be associated with reactivation of hepatitis B virus (HBV) infection in HCV/HBV co-infected individuals (13–15), provocation or induction of autoimmunity, and possibly a change in the incidence and pattern of HCC in individuals with advanced liver disease undergoing DAA therapy for chronic HCV infection. The mechanisms by which lncRNAs regulate innate immune responses and host-virus interactions during HCV treatment are still lacking.
We therefore studied the regulatory role of lncRNAs on ISG expression and type I IFN responses. We performed RNA sequencing (RNAseq) in Huh7.5.1 cells and found that IFN induced the expression of a large number of lncRNAs. As lncRNAs regulate gene expression in the nucleus through modulation of transcription or post-translational modification, siRNAs that target mRNA protein coding genes may not be the most effective strategy to knock down gene function (16). The CRISPR/Cas9 guide RNA (gRNA) system provides an effective tool to knock down non-coding RNAs (17, 18). We used gRNAs to study the regulatory roles of lncRNAs on ISGs and HCV replication. We obtained a total of 7901 lncRNAs from RNAseq of Huh7.5.1 cells with and without IFNα treatment. We identified a cluster of 1062 differentially expressed (>2-fold) lncRNAs that were involved in the regulation of the antiviral activity of IFN. We selected 10 differentially expressed lncRNAs for further characterization. We identified a novel lncRNA located within the IFI6 gene, subsequently named lncRNA-IFI6, as a negative regulator of IFI6. We found that lncRNA IFI6 plays a critical role in the containment of HCV through regulation of histone modification of the IFI6 promoter.
Methods
Cells and virus
Huh7.5.1, HepG2, LX2, TWNT4 and Huh7 cells were grown in DMEM (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Waltham, MA) and 1% penicillin/streptomycin (Invitrogen, Waltham, MA) at 37oC with 5% CO2. Huh7.5.1 cells were infected with genotype 2a JFH1 HCV at a multiplicity of infection of 0.2 (JFH1 cells) as previously described (1, 19, 20). Primary human hepatocytes (PHHs) were purchased from TRIANGLE Research Laboratories (Durham, NC), and cultured according to the manufacturer’s instructions. In brief, PHHs were seeded onto matrix-coated plates and cultured with Hepatocyte Plating Medium (Durham, NC). Cell viability was determined using a Cell Titer-Glo luminescent cell viability assay kit (Promega, Madison, WI) according to the manufacture`s protocol.
Long non-coding RNA high-throughput sequencing and data analysis
Total RNA from control Huh7.5.1 cells and IFNα-treated (30 IU/mL) Huh7.5.1 cells were used to prepare the lncRNA sequencing library. Oligo dT beads were used to select mRNA from the total RNA. The Illumina library preparation was performed using broadly designed indexed adapters following cDNA synthesis with RT primers. The samples were quantified by qPCR after enrichment, followed by lncRNA high-throughput sequencing performed at the Broad Institute (Cambridge, MA) on the Illumina HiSeq2000 platform using the Illumina Tru Seq™ Rapid SBS kit (#FC-402–4002, Illumina, San Diego, CA) according to the manufacturer’s instructions. The significantly differentially expressed lncRNAs for the IFNα treated sample vs. control sample were assessed by fold-change (see Bioinformatic analysis below), where ≥ 2.0 fold-change differential gene expression indicates up-regulation of lncRNA expression in that the lncRNA in IFNα-treated samples, while a fold-change value ≤ 0.5 indicates down-regulation of lncRNA gene expression in IFNα-treated samples. We then chose 5 up-regulated and 5 down-regulated IFN-induced lncRNAs from our RNAseq bioinformatic analysis for further validation.
Chromatin Immunoprecipitation (ChIP) assay
plncRNA-IFI6-overexpression, lncRNA-IFI6 gRNA, or negative control transfected Huh7.5.1 cells were selected for ChIP assays using the EZ-Magna ChIP A/G chromatin immunoprecipitation kit (Millipore, Burlington, MA) according to the manufacturer’s instructions. Briefly, cells were fixed with 1% formaldehyde for 10 minutes followed by quenching of the formaldehyde with 10x glycine. The cells were then lysed in lysis buffer. The nuclear fraction of cells was pelleted in nucleus lysis buffer. The chromatin was ultrasonically treated and immunoprecipitated with the following antibodies: anti-H3K4me3 (Cat# 17–614), anti-H3K27me3 (Cat# 07–449), anti-RNA Polymerase II (Cat# 05–623B) and IgG control (Cat# 12–371B) (Millipore, Burlington, MA). Elution of protein/DNA complexes and reverse cross-links of protein/DNA complexes to free DNA were performed using the ChIP Elution Buffer mixed with proteinase K, and the Magna Chip magnetic device. The DNA was purified for qRT-PCR to assess the promoter level of IFI6 and GAPDH expression.
Subcellular fractionation
Cytoplasmic and nuclear fractions of the cells were separated by using the SurePre™ Nuclear or Cytoplasmic RNA purification kit (Fisher Scientific, Waltham, MA), in accordance with the manufacturer’s instructions. In brief, Huh7.5.1 cells were treated with IFNα 30 IU/mL for 24 hours. After washing the cells with PBS, cells were lysed with ice-cold Lysis Solution, and the cell lysates were centrifuged at 13,000 RPM for 15 min at 4℃, after which the cytoplasmic RNA was located in the supernatant, and the nuclear RNA located within the pellets. The cytoplasmic RNA fraction and nuclear RNA fraction were individually bound to a column using a binding solution and washed three times. The cytoplasmic RNA and nuclear RNA were eluted in RNA elution solution.
5-Aza-2’-deoxycytidine (DAC) treatment
Cells overexpressing lncRNA-IFI6 gRNA, Neg gRNA, plncRNA-IFI6, or pEmpty were treated with or without 10 μM 5-Aza-2’-deoxycytidineDNA (DAC) (methyltransferase inhibitor decitabine) (Sigma, St Louis, MO) for 48 hours. Total RNA was isolated using the Total RNA kit (Qiagen, MD). The mRNA level of each sample was detected by qRT-PCR.
Secrete-Pair Dual Luminescence analysis (Detection of ISG promoter activity)
The Secrete-Pair Dual Luminescence Assay Kit for parallel bioluminescence assays of Gaussia luciferase (Gluc) and secreted Alkaline Phosphatase (SEAP) were provided by GeneCopoeia Inc (Rockville, MD). Cells overexpressing plncRNA-IFI6 and lncRNA-IFI6 gRNA were transfected with a pEZX-PG04-IFI6-promoter and pEZX-PG04-OAS3-promoter, respectively. Gluc and SEAP assays were performed to detect IFI6 and OAS3 promoter activity.
Statistical analysis
Data analyses were performed using a 2-tailed Student’s t-test. Data are expressed as mean ± standard deviation of at least three sample replicates, unless stated otherwise. In all analyses, * P < 0.05; ** P < 0.01; *** P < 0.001 for comparison of indicated treatments.
Notes: Additional Materials and Methods are available in the online methods supplement.
Results
lncRNA-IFI6 localization and expression
We obtained 7901 lncRNAs from RNAseq (Supplementary methods, STab 1). We identified 1062 lncRNAs that were significantly dysregulated greater than 2-fold following IFNα treatment (SFig 1A). We selected 5 upregulated and 5 downregulated lncRNAs for further qRT-PCR validation and characterization. We confirmed qRT-PCR results for these lncRNAs were consistent with the RNAseq data (SFig 1B-K, STab 1). lncRNA RP11–288L9.4 was the most upregulated gene following IFNα treatment. lncRNA RP11–288L9.4 is located on human chromosome 1 (p36, 11) (position: chr1: 27,669,468–27,670,276) and overlaps with the antisense strand of IFI6 within intron 1 (SFig 1L). We found that lncRNA RP11–288L9.4 expression in several human cell lines (Huh7.5.1, Huh7, LX2, TWNT4 and HepG2) and PHHs was highly induced following IFNα treatment (SFig 1M-R). These findings indicated that lncRNA RP11–288L9.4 levels are stable among our tested human hepatocytes and hepatic stellate cells. We subsequently named lncRNA RP11–288L9.4 as “lncRNA-IFI6” given its location within the IFI6 gene in the human genome.
lncRNA-IFI6 regulation of IFI6 expression and HCV infection
To further determine the regulatory effects of lncRNA-IFI6 on the antiviral response to HCV infection, we generated Huh7.5.1 cell lines stably expressing lncRNA-IFI6 gRNA by using the pSpCas9 BB-2A-Puro PX459 all-in-one system (SFig 2A) and Huh7.5.1 cells stably expressing full-length human lncRNA-IFI6. We found that lncRNA-IFI6 gRNA significantly inhibited lncRNA-IFI6 expression and IFNα-induced lncRNA-IFI6 expression in Huh7.5.1 and JFH1 cells (Fig 1A). lncRNA-IFI6 gRNA or overexpression of plncRNA-IFI6 did not significantly affect cell viability (SFig 2B-C). We confirmed that HCV RNA levels increased over time from 0 to 72 hours post inoculation with JFH1 virus, confirming HCV infection in JFH1-infected Huh7.5.1 cells. However, lncRNA-IFI6 gRNA significantly inhibited HCV RNA levels compared to negative (Neg) gRNA (SFig 3A).
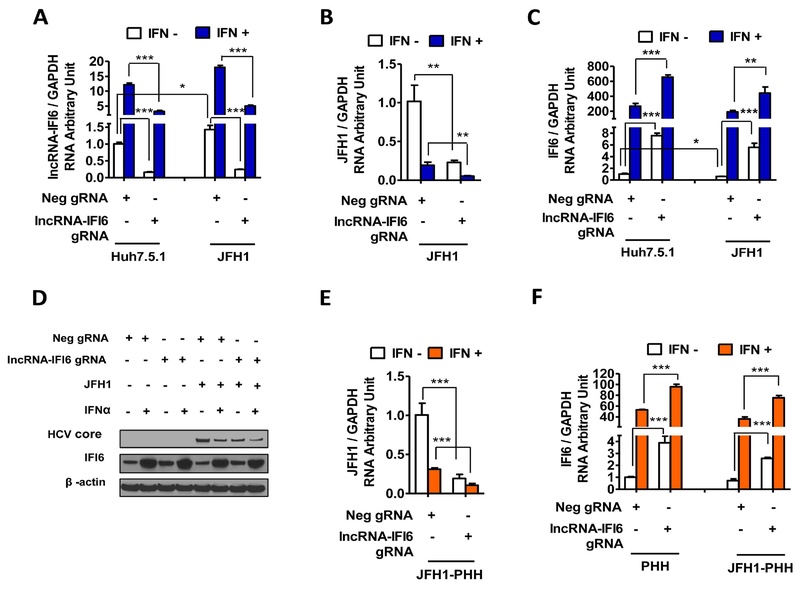
Figure 1. lncRNA-IFI6 regulation of IFI6 expression and HCV infection.
JFH1 HCV was inoculated in to cells for 48 hrs. At 24 hrs post infection, human IFNα was added at a final concentration of 30 IU/ mL. Total RNA or protein lysates were harvested at 48 hrs post infection. (A) lncRNA-IFI6 gRNA decreased lncRNA-IFI6 levels in Huh7.5.1 or JFH1 cells compared to Neg gRNA with or without IFNα, respectively. (B) lncRNA-IFI6 gRNA reduced HCV RNA levels in JFH1-infected Huh7.5.1 cells compared to Neg gRNA. IFNα plus lncRNA-IFI6 gRNA further decreased HCV RNA levels. (C) lncRNA-IFI6 gRNA significantly increased IFI6 mRNA expression in Huh7.5.1 and JFH1 cells compared to Neg gRNA with or without IFNα, respectively. (D) lncRNA-IFI6 gRNA increased IFI6 protein and reduced HCV core levels in Huh7.5.1 or JFH1 cells compared to Neg gRNA with or without IFNα, respectively. (E) lncRNA-IFI6 gRNA reduced HCV RNA levels in PHHs or JFH1-infected PHH cells compared to Neg gRNA. IFNα and lncRNA-IFI6 gRNA further decreased HCV RNA levels. (F) lncRNA-IFI6 gRNA significantly increased IFI6 mRNA expression in PHH and JFH1-infected PHH cells compared to Neg gRNA with or without IFNα, respectively.
We also found that both lncRNA-IFI6 gRNA and IFNα treatment inhibited HCV RNA levels (Fig 1B). IFNα plus lncRNA-IFI6 gRNA further reduced HCV infection (Fig 1B). Interestingly, we found that lncRNA-IFI6 specifically regulated IFI6 expression (Fig 1C, SFig 4A). In contrast, several classical ISGs including OAS3, IFIT1, MX1, IFIT5, ZAP, IRF7, ISG15 and ISG20 were not significantly affected by lncRNA-IFI6 gRNA (SFig 4B-I, SFig 5A-B). We found that lncRNA-IFI6 gRNA significantly enhanced IFI6 mRNA and protein expression levels compared to Neg gRNA in both Huh7.5.1 and JFH1 cells at 48 hours post HCV infection (Fig 1C-D). In contrast, overexpression of plncRNA-IFI6 significantly increased HCV RNA levels from 0, 12, 24, 36, 48 to 72 hours post HCV inoculation compared to pEmpty in JFH1-infected Huh7.5.1 cells (SFig 3B-C). Overexpression of plncRNA-IFI6 rescued IFNα-induced inhibition of HCV infection (SFig 3D). We also confirmed that overexpression of plncRNA-IFI6 inhibited IFI6 mRNA and protein levels compared to pEmpty (SFig 3E-F). IFNα rescued IFI6 mRNA and protein expression following overexpression of plncRNA-IFI6 in Huh7.5.1 and JFH1 cells (SFig 3E-F). As with lncRNA-IFI6 gRNA, we also found that classical ISGs were not significantly affected by overexpression of plncRNA-IFI6 (SFig 4J-R, SFig 5C-D). These findings indicate that IFI6 is specifically regulated by lncRNA-IFI6.
Because the Huh7.5.1 cell line is an immortalized cell line with deficiencies in the RIG-I pathway (21), we therefore validated our findings in PHHs. We confirmed that lncRNA-IFI6 gRNA inhibited lncRNA-IFI6 expression, and IFNα induced lncRNA-IFI6 expression in uninfected and JFH1-infected PHHs (SFig 3G). lncRNA-IFI6 gRNA also inhibited HCV infection and increased IFI6 expression in PHHs (Fig 1E-F). As expected, overexpression of plncRNA-IFI6 significantly promoted HCV infection and reduced IFI6 expression in PHHs (SFig 3H-J). In addition, we found that HCV infection increased lncRNA IFI6 (Fig 1A) and reduced IFI6 mRNA expressions (Fig 1C). We also found that higher HCV RNA levels are associated with lower IFN-induced IFI6 levels in liver biopsies from patients with chronic HCV infection (data not shown). These findings indicate that lncRNA-IFI6 is a specific regulator of the ISG IFI6 on HCV infection.
IFI6 regulates HCV infection
To confirm the effect of IFI6 on HCV infection, we performed siRNA knockdown and overexpression of IFI6 in JFH1-infected Huh7.5.1 cells. We found that siRNA knockdown or overexpression of IFI6 did not significantly affect cell viability (SFig 6A-B). Furthermore, IFI6 siRNA significantly increased HCV RNA and core protein levels in JFH1-infected Huh7.5.1 cells (SFig 6C-E). We also generated IFI6 knockout Huh7.5.1 cells by stably transfecting IFI6 gRNA. We found that IFI6 gRNA significantly increased HCV RNA and core protein levels in JFH1-infected Huh7.5.1 cells compared to Neg gRNA (Fig 2A-B). However, we observed partial impairment of anti-HCV activity with low dose IFNα treatment (30 IU/mL) with IFI6 knock down (Fig 2A-B, SFig 6C-E). These findings further confirm the anti-HCV effects of IFI6 (22–24), and that IFNα likely induces many other ISGs, such as IFITMs, RSAD2, ISG56, GBP-1, and CH25H that have been shown to have redundant effects against HCV replication (25–27). In contrast, overexpression of pIFI6 significantly decreased HCV RNA and protein levels in JFH1 cells (Fig 2C-E). These data further confirmed that IFI6 regulates HCV infection.
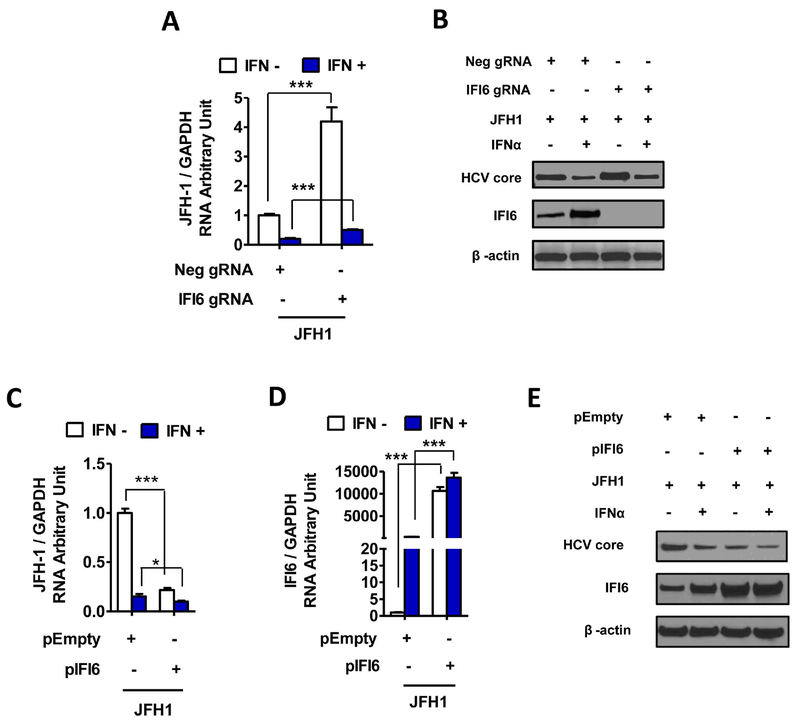
Figure 2. IFI6 regulates HCV infection.
Huh7.5.1 cells stably expressing specific IFI6 gRNA, Neg gRNA, pIFI6 and pEmpty were generated. Total RNA or protein of the cells was harvested at 48 hrs post JFH1 HCV (JFH1 at 0.2 MOI) infection, and 24 hrs post IFNα treatment. The selected gene mRNAs were assessed by qRT-PCR. The protein levels were monitored by Western blot. Data are shown as means ± standard deviation of three replicates. * P < 0.05; ** P < 0.01; *** P < 0.001. (A) IFI6 gRNA significantly increased HCV RNA level in JFH1 infected Huh7.5.1 cells compared to Neg gRNA with or without IFNα respectively. (B) IFI6 gRNA increased HCV core protein level compared to Neg gRNA. (C) Overexpression pIFI6 inhibited HCV RNA expression in JFH1 infected Huh7.5.1 cells compared to pEmpty with or without IFNα, respectively. (D) Overexpression of pIFI6 increased IFI6 mRNA levels compared to pEmpty. (E) Overexpression of pIFI6 increased IFI6 protein expression and reduced HCV core levels compared to pEmpty with or without IFNα, respectively.
IncRNA-IFI6 regulation of HCV infection is mediated by IFI6
To study the regulatory role of lncRNA-IFI6 and the association of IFI6 with lncRNA-IFI6, we performed siRNA-mediated knockdown of IFI6 in lncRNA-IFI6 gRNA stably transfected JFH1 cells, while an IFI6 overexpression plasmid was transiently transfected into lncRNA-IFI6 stably expressing JFH1 cells. Cell viability was not affected by these experimental conditions (SFig 6F-G). We demonstrated that RNAi-mediated knockdown of IFI6 rescues HCV infection in lncRNA-IFI6 gRNA stably expressing JFH1 cells (SFig 6H-J). Moreover, to avoid lncRNA-IFI6 gRNA off-target effects, we performed similar experiments in IFI6 knockout Huh7.5.1 cells by stably transfecting IFI6 gRNA. We confirmed that IFI6 knockout rescues HCV infection in lncRNA-IFI6 gRNA transfected JFH1 cells (Fig 3A-B). IFI6 knockdown or plncRNA-IFI6 overexpression significantly increased HCV mRNA and core protein levels (Fig 3C-D). We found that overexpression of IFI6 reversed the effects of plncRNA-IFI6 on increased HCV infection and inhibition IFI6 expression (Fig 3E-F, SFig 6K). We observed similar results in JFH1-infected PHHs, where siRNA of IFI6 rescued HCV infection in lncRNA-IFI6 gRNA-transfected JFH1-infected PHHs (SFig 6L-M). Furthermore, overexpression of IFI6 reversed the effect of plncRNA-IFI6 on HCV infection in JFH1-infected PHHs (SFig 6N-O). We found that lncRNA-IFI6 mRNA levels were not significantly affected by siRNA IFI6 knockdown or overexpression (SFig 6P-Q). These findings confirm that lncRNA-IFI6 regulation of HCV replication is mediated by IFI6.
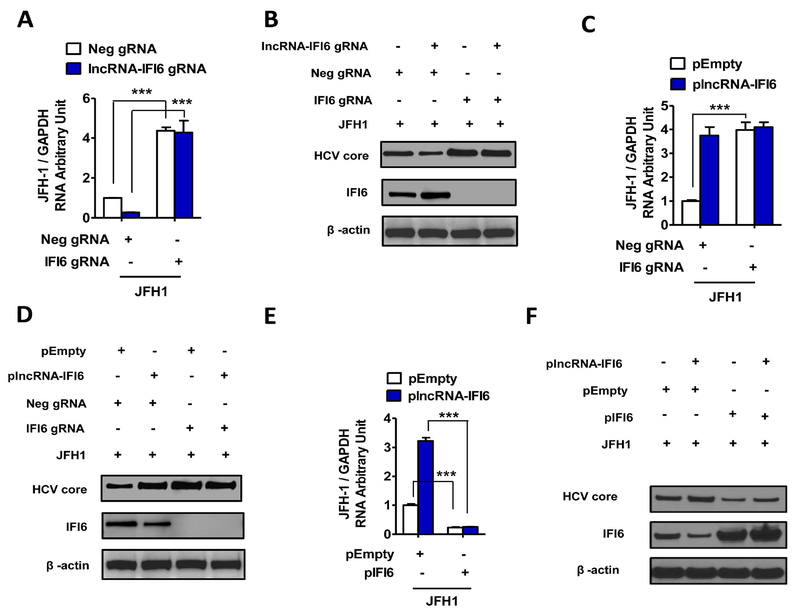
Figure 3. lncRNA-IFI6 regulates HCV infection through IFI6.
FI6 gRNA or Neg gRNA were transfected in to lncRNA-IFI6 gRNA stably transfected cells. pIFI6 or pEmpty were transfected to plncRNA-IFI6 overexpressing cells, respectively. Total RNA or protein of the cells was harvested at 72 hrs post vector transfection, 48 hrs post JFH1 HCV (JFH1 at 0.2 MOI) infection, and 24 hrs post IFNα treatment. (A) IFI6 gRNA rescued HCV RNA reduction mediated by lncRNA-IFI6 gRNA in JFH1 infected Huh7.5.1 cells compared to Neg gRNA. (B) IFI6 gRNA rescued HCV core protein reduction and depleted IFI6 protein expression in JFH1 infected Huh7.5.1 cells compared to Neg gRNA. (C) Overexpression of plncRNA-IFI6 or IFI6 gRNA increased JFH1 HCV RNA levels in JFH1 infected Huh7.5.1 cells compared to Neg gRNA (D) Overexpression of plncRNA-IFI6 or IFI6 gRNA increased JFH1 HCV core protein levels in JFH1 infected Huh7.5.1 cells compared to Neg gRNA. (E) Overexpression of pIFI6 inhibited plncRNA-IFI6 induction on HCV RNA levels compared to pEmpty in JFH1 cells. (F) Overexpression of pIFI6 increased IFI6 protein and decreased HCV core protein levels in cells overexpressing plncRNA-IFI6 compared to pEmpty.
lncRNA-IFI6 regulation of HCV infection through IFI6 is independent of JAK-STAT signaling
We found that lncRNA-IFI6 gRNA or overexpression did not significantly affect STAT1 mRNA and protein expression levels compared to Neg gRNA or pEmpty overexpression (SFig 7A-D). Moreover, STAT1, STAT2 or JAK1 siRNAs did not significantly affect lncRNA-IFI6 expression levels (SFig 7E), despite confirmed adequate STAT1, STAT2, JAK1 mRNA knockdown by their respective siRNAs compared to Neg siRNA control (SFig 7F-H). We found that siRNAs to STAT1, STAT2, JAK1 did not affect cell viability (SFig 7I). In addition, gRNA or overexpression of plncRNA-IFI6 did not significantly affect IFN-sensitive response element (ISRE)-directed signaling (SFig 8A-B), or cell viability (SFig 8C-D) in Huh7.5.1 or JFH1 cells.
Previous studies have shown that lncRNA may induce the production of certain cytokines (28). We therefore tested whether lncRNA-IFI6 regulates IFI6 through production of other cytokines. We found that IFI6 mRNA expression was not significantly different between Huh7.5.1 cells treated with cell culture supernatants from Huh7.5.1 cells stably overexpressing lncRNA-IFI6 gRNA and Neg gRNA control cells (SFig 8E-G). We confirmed that cell culture supernatant treatment did not affect Huh7.5.1 cell viability (SFig 8H-I). These results demonstrate that lncRNA-IFI6 regulation of IFI6 and HCV infection does not occur through the JAK-STAT and ISRE signaling pathways, or through other secreted cytokines.
lncRNA-IFI6 regulates initial transcription of IFI6
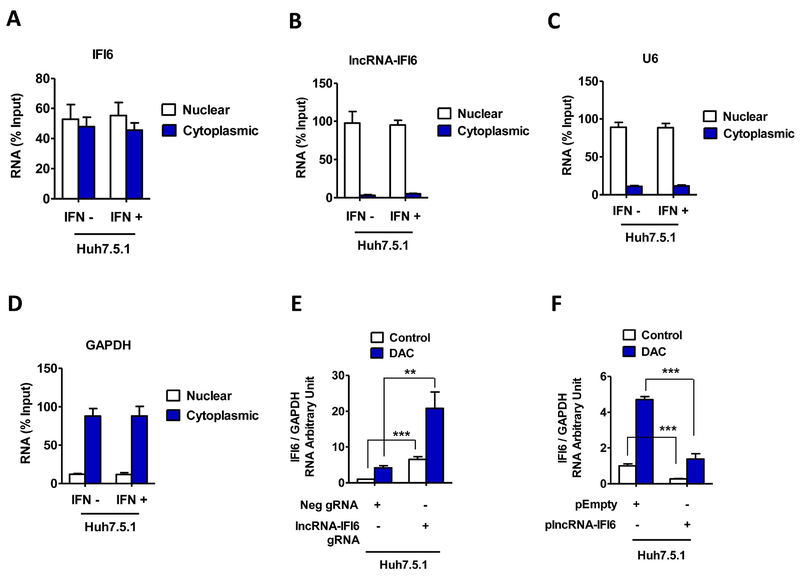
We performed subcellular fractionation and found that lncRNA-IFI6 mostly localizes in the nucleus of Huh7.5.1 cells (Fig 4A-D). lncRNAs have been shown to regulate the transcription processes of coding genes through the maintenance of DNA methylation (29). Our bioinformatic analysis found that a CpG island (304 base pair (bp) DNA) was present between exon 2 and intron 2 (4242–4545 bp) of the IFI6 gene (SFig 9A). We introduced the DNA methyltransferase inhibitor decitabine (DAC) in our cells and found that DAC treatment increased IFI6 mRNA levels in lncRNA-IFI6 gRNA or overexpressing lncRNA-IFI6 Huh7.5.1 cells; however, the expression of IFI6 was further increased in the presence of lncRNA-IFI6 gRNA. In contrast, overexpression of plncRNA-IFI6 still demonstrated inhibition of IFI6 (Fig 4E-F). Cell viability assays showed that DAC, lncRNA-IFI6 gRNA or overexpressed plncRNA-IFI6 did not affect cell viability (SFig 9B-C). These results indicate that lncRNA-IFI6 is not associated with IFI6 DNA methylation.
Figure 4. lncRNA-IFI6 is a nuclear transcript.
Huh7.5.1 cells were treated with IFNα (30 IU/ mL) for 24 hrs. Cytoplasmic and nuclear fractions of the cells were separated. The RNA levels of lncRNA-IFI6 and IFI6 were assessed by qRT-PCR in nuclear and cytoplasmic fractions from Huh7.5.1 cells. The total RNA was used as input control. Data are shown as percent (%) input. lncRNA-IFI6 gRNA, Neg gRNA, plncRNA-IFI6 and pEmpty stably overexpressing cells were treated with or without 10 μM 5-Aza-2’-deoxycytidineDNA (DAC) (methyltransferase inhibitor decitabine) for 48 hrs. Total RNA was isolated. IFI6 mRNA was assessed by qRT-PCR. (A) IFI6 mRNA was detected in both nuclear and cytoplasmic fractions in Huh7.5.1 cells. (B) lncRNA-IFI6 is a nuclear transcript. The majority of lncRNA-IFI6 was found in the nuclear fraction of Huh7.5.1 cells. (C) RNA levels of U6 (nuclear marker) were assessed by qRT-PCR in nuclear and cytoplasmic fractions from Huh7.5.1 cells. Total RNA was used as input control. Data are shown as percentage input. (D) RNA levels of GAPDH (cytoplasmic marker) were assessed by qRT-PCR in nuclear and cytoplasmic fractions from Huh7.5.1 cells. (E) lncRNA IFI6 gRNA or DNA methyltransferase inhibitor decitabine (DAC) (15 μM) treatment increased IFI6 mRNA levels in Huh7.5.1 cells. (F) Overexpression of plncRNA IFI6 blocked DAC treatment-induced IFI6 mRNA enhancement in Huh7.5.1 with or without IFNα, respectively.
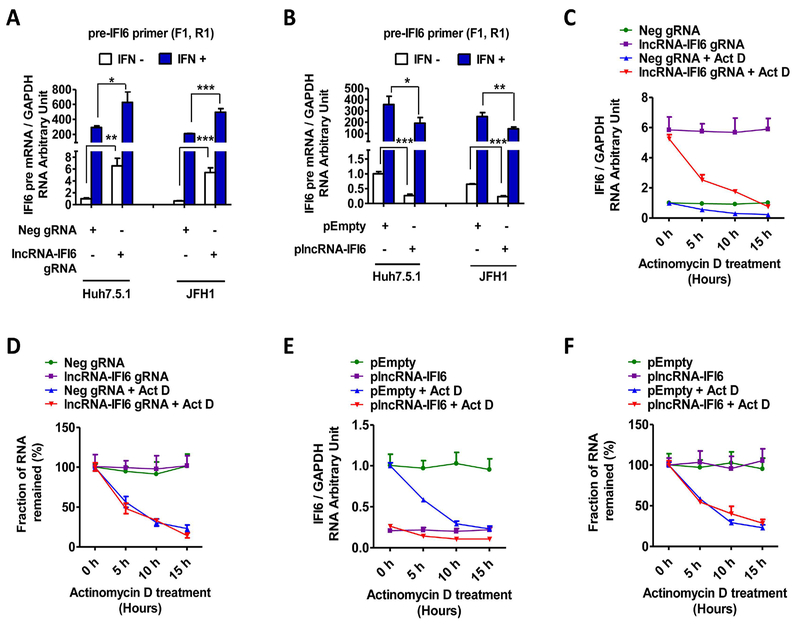
We designed specific primers to measure pre-mRNA, which represents the initial transcriptional level and mature IFI6 mRNA levels in Huh7.5.1 and JFH1 cells (STable 2). We found that lncRNA-IFI6 gRNA significantly increased IFI6 pre-mRNA levels compared to Neg gRNA control. As expected, overexpression of plncRNA-IFI6 significantly reduced IFI6 pre-mRNA levels in Huh7.5.1 cells (Fig 5A-B, SFig 9D-E). To further differentiate between initial transcription and downstream post-transcriptional modifications, including splicing and degradation in the nucleus, we introduced the transcription inhibitor actinomycin D (ActD) for comparison. We found that lncRNA-IFI6 gRNA or overexpression of plncRNA-IFI6 did not significantly affect IFI6 mRNA (Fig 5C-F) or IFI6 pre-mRNA (SFig 9F-I) half-life, indicating similar degradation rates between lncRNA-IFI6 gRNA treatment or overexpression of plncRNA-IFI6. These results suggest that the regulatory action of lncRNA-IFI6 on IFI6 occurs during initial transcription.
Figure 5. lncRNA-IFI6 regulates initial transcription of IFI6.
lncRNA-IFI6 gRNA or plncRNA-IFI6 stably transfected Huh7.5.1 cells were treated with or without actinomycin D (ActD) at 1 μg/mL. Total RNA was harvested at 0, 5, 10 and 15 hours post-treatment. The mRNA level of each sample was detected by qRT-PCR by using specific primers to measure precursor mRNA (pre-mRNA), which represents the initial transcriptional level, and mature IFI6 mRNA levels in Huh7.5.1 or JFH1 cells. (A) lncRNA-IFI6 gRNA significantly increased IFI6 pre-mRNA (pre-IFI6–1) levels compared to Neg gRNA in Huh7.5.1 or JFH1 cells with or without IFNα respectively. (B) Overexpression of plncRNA-IFI6 significantly reduced IFI6 pre-mRNA (pre-IFI6–1) levels compared to pEmpty in Huh7.5.1 or JFH1 cells with or without IFNα respectively. (C) ActD treatment inhibited IFI6 mRNA expression in lncRNA-IFI6 gRNA stably transfected Huh7.5.1 cells. The IFI6 mRNA values were normalized to GAPDH.(D) lncRNA-IFI6 gRNA did not significantly affect IFI6 mRNA half-life compared to Neg gRNA in Huh7.5.1 cells. The mRNA level at 0 hrs in each treatment in Fig 5C was set to 100% of the fraction value. The relative fraction values of each treatment at 5 hrs, 10 hrs or 15 hrs was obtained by normalized to the value at 0 hrs respectively. The IFI6 mRNA degradation half-life of Neg gRNA+ ActD is relatively equal to lncRNA-IFI6 gRNA+ ActD. (E)ActD treatment reduced IFI6 mRNA expression levels in plncRNA-IFI6 stably transfected Huh7.5.1 cells. The IFI6 mRNA values were normalized to GAPDH. (F) Overexpression of plncRNA-IFI6 did not significantly affect IFI6 mRNA half-life compared to pEmpty in Huh7.5.1 cells. The mRNA level at 0 hrs in each treatment in Fig 5E was set to 100% of the fraction value. The relative fraction values of each treatment at 5 hrs, 10 hrs or 15 hrs was obtained by normalized to the value at 0 hrs respectively. The IFI6 mRNA degradation half-life of lncRNA-IFI6 gRNA+ ActD is relatively equal to pEmpty+ ActD.
lncRNA-IFI6 regulates IFI6 through its promoter
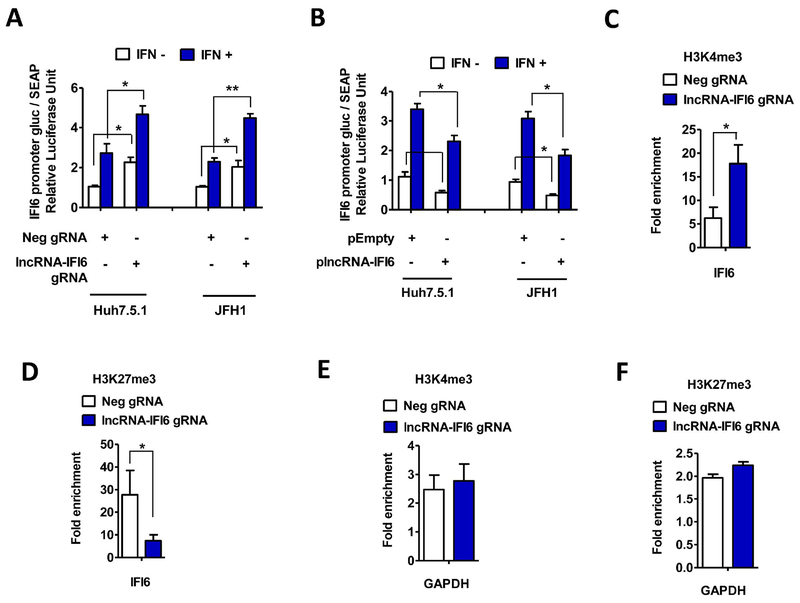
lncRNAs have also been shown to regulate coding gene transcription through modulation of promoter activity (6). To determine whether lncRNA-IFI6 regulates the promoter activity of IFI6, we constructed a pEZX-PG04-IFI6-promoter and pEZX-PG04-OAS3-promoter dual luminescence reporter vector. Because OAS3 is located in a different chromosome to IFI6 (chromosome 12 vs. chromosome 11, respectively) and is not affected by lncRNA-IFI6, we therefore selected the pEZX-PG04-OAS3-promoter vector as a control. We observed that lncRNA-IFI6 gRNA significantly increased IFI6 promoter activity compared to Neg gRNA control, while plncRNA-IFI6 overexpression significantly inhibited IFI6 promoter activity compared to the pEmpty control (Fig 6A-B). However, we found that lncRNA-IFI6 gRNA or plncRNA-IFI6 overexpression did not significantly affect OAS3 promotor activity (SFig 10A-B). We confirmed that lncRNA-IFI6 gRNA, plncRNA-IFI6 overexpression, or the Secrete-Pair Dual Luminescence Assay did not significantly affect cell viability (SFig 10C-F). These findings suggest that lncRNA-IFI6 is a specific negative regulator of IFI6 promoter activity. However, similar to a previous report (6), the GLuc and SEAP dual luciferase based promoter activity assay is less sensitive compared to PCR based mRNA assay in monitoring response to IFN treatment.
Figure 6. lncRNA-IFI6 regulates IFI6 through its promoter.
lncRNA-IFI6 gRNA, Neg gRNA, plncRNA-IFI6 and pEmpty stably over-expressing cells were co-transfected with pEZX-PG04-IFI6-promoter or pEZX-PG04-OAS3-promoter, respectively. Guassia luciferase (Gluc) assay and secreted Alkaline Phosphatase (SEAP) assay were performed to detect the promoter activity. plncRNA-IFI6 overexpression, pEmpty, lncRNA-IFI6 gRNA, or Neg gRNA transfected Huh7.5.1 cells were subjected to ChIP analysis. The relative amounts of IFI6 DNA immunoprecipitated by the H3K4me3 or H3K27me3 antibody were normalized to IFI6 DNA isolated by the control lgG. (A) lncRNA-IFI6 gRNA increased pEZX-PG04-IFI6-promoter induced Gaussia luciferase (Gluc) / alkaline phosphatase (SEAP) activity compared to Neg gRNA in Huh7.5.1 or JFH1 cells with or without IFNα, respectively. (B) Overexpression of plncRNA-IFI6 decreased pEZX-PG04-IFI6-promoter induced Gaussia luciferase (Gluc) / alkaline phosphatase (SEAP) activity compared to pEmpty in Huh7.5.1 or JFH1 cells with or without IFNα, respectively. (C) lncRNA-IFI6 gRNA significantly increased the enrichment of H3K4me3 at IFI6 transcription start sites. (D) lncRNA-IFI6 gRNA significantly reduced the enrichment of H3K27me3 of the IFI6 gene. (E) ChIP analysis of H3K4me3 levels at the GAPDH locus in lncRNA-IFI6 gRNA and Neg-gRNA control cells. (F) ChIP analysis of H3K27me3 levels at the GAPDH locus in lncRNA-IFI6 gRNA and Neg-gRNA control cells.
Another mechanism by which lncRNAs regulate gene transcription is through histone modification of transcription start sites (30). We performed chromatin immunoprecipitation (ChIP) to assess histone 3 lysine 4 trimethylation (H3K4me3, active mark) and histone 3 lysine 27 trimethylation (H3K27me3, repression signal) activity with lncRNA-IFI6 gRNA and plncRNA-IFI6 overexpression in Huh7.5.1 cells. We observed that lncRNA-IFI6 gRNA significantly increased the enrichment of H3K4me3 at IFI6 transcription start sites, but significantly reduced the enrichment of H3K27me3 at the IFI6 gene locus (Fig 6C-F). In contrast, the H3K4me3 enrichment at the IFI6 gene locus in plncRNA-IFI6 overexpression was significantly lower compared to pEmpty overexpression, while plncRNA-IFI6 overexpression significantly increased H3K27me3 enrichment at the IFI6 gene locus compared to pEmpty control in Huh7.5.1cells (SFig 10G-K). These results indicate that lncRNA-IFI6 affects the transcription of IFI6 through histone modification.
lncRNA-IFI6 exerts its regulatory function on IFI6 promoter activation and histone modification through its spatial domain
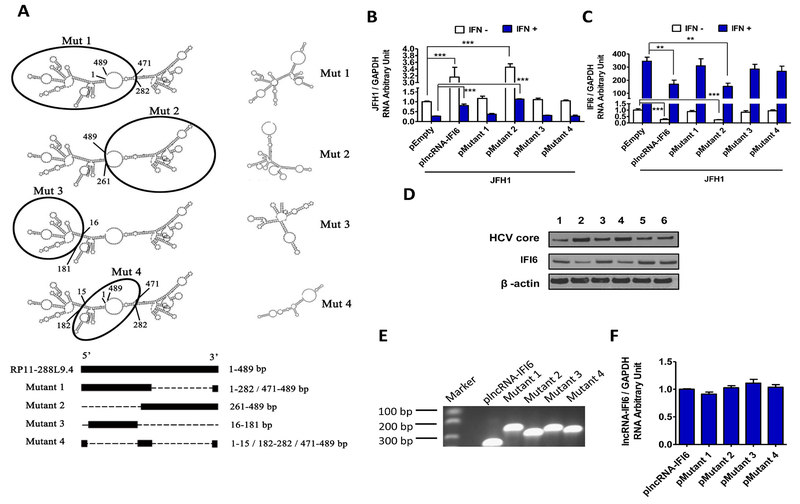
The secondary structure and domain of lncRNAs determine their regulatory function (31). To assess the specific functional structure of lncRNA-IFI6, we used the bioinformatic software, RNAfold and Vienna RNA, to predict the secondary structure of lncRNA-IFI6, which assigned four possible functional domains: mutant 1 (large left arm), mutant 2 (large right arm), mutant 3 (small left arm) and mutant 4 (central circle structure) (Fig 7A). We found that overexpression of full-length lncRNA-IFI6 or mutant 2 significantly increased HCV infection, and decreased IFI6 mRNA and protein levels in Huh7.5.1 cells. In contrast, mutant 1, 3 and 4 overexpression did not significantly affect HCV infection, IFI6 mRNA or IFI6 protein levels compared to the pEmpty in Huh7.5.1 and JFH1 cells (Fig 7B-D). We found that overexpression of full-length or each mutant lncRNA-IFI6 did not significantly affect cell viability (SFig 11A). We also confirmed that full-length and mutant 2 lncRNA-IFI6 overexpression exerted comparable levels of HCV enhancement and IFI6 mRNA inhibition in PHHs (SFig 11B-C). These results suggest that the large right arm structure of lncRNA-IFI6 (mutant 2) is the key functional domain.
Figure 7. lncRNA-IFI6 exerts its regulatory function on promoter activation and histone modification of IFI6 through its spatial domain.
Huh7.5.1 cells line stably expressing full-length plncRNA-IFI6, pMutant 1–4 and pEmpty were generated, respectively. JFH1 HCV was inoculated into the cells for 48 hrs. Each selected gene mRNA expression level was normalized to GAPDH mRNA yielding arbitrary units (fold-change). Protein levels were detected by Western blot. (A) The proposed lncRNA-IFI6 and 4 lncRNA mutations secondary structure based on the analysis results of bioinformatics softwares RNAfold and Vienna RNA. The black circles depict were the mutations are located. (B) Overexpression of wild type or mutant 2 lncRNA-IFI6 significantly increased HCV RNA levels compared to pEmpty in JFH1 cells with or without IFNα, respectively. (C) Overexpression of wild type or mutant 2 lncRNA-IFI6 significantly reduced IFI6 RNA levels compared to pEmpty in JFH1 cells with or without IFNα, respectively. (D) Overexpression of wild type or mutant 2 lncRNA-IFI6 significantly reduced IFI6 protein and increased HCV core protein levels compared to pEmpty in JFH1 cells. Lane 1: pEmpty, lane 2: plncRNA-IFI6, lane 3: pMutant 1, lane 4: pMutant 2, lane 5: pMutant 3, lane 6: pMutant 4. (E) PCR gel image of the similar lncRNA-IFI6 levels in Huh7.5.1 cells overexpressing full-length lncRNA-IFI6 or each mutant. (F) The relative equal amount of lncRNA-IFI6 levels in Huh7.5.1 cell overexpression with full-length lncRNA-IFI6 or each mutant. lncRNA-IFI6 level were measured by qRT-PCR.
Discussion
lncRNAs regulate a variety of biological processes through modulation of transcription and post-transcriptional mechanisms. Most lncRNAs are transcribed by RNA polymerase II, and show tissue and cell type specific expression (4, 16, 30, 32). lncRNAs have also been reported to be involved in epigenetic regulation. For example, lncRNA HOTAIR leads to altered histone H3 lysine 27 methylation through recruitment of Polycomb repressive complex 2 (PRC2) to a specific region of chromatin (33). In addition, lncRNA Kcnq1ot1 maintains the silencing of imprinted genes via modulation of DNA methylation (29). A recent report demonstrated that the immunoregulatory lncRNA-EPS plays a role as a repressor of inflammatory responses through restraining the expression of immune response genes (IRGs) (34). DDX5 and its associated lncRNA Rmrp was also reported to modulate T-helper 17 (TH17) cell effector functions (35). However, apart from the aforementioned studies, characterization of the immunoregulatory functions of most lncRNAs are largely lacking.
IFN-free combination DAA therapies are a highly effective therapy for chronic HCV infection (>95% cure rates), and are now widely used in clinical practice (36). Although DAAs directly inhibit specific HCV proteins involved in the HCV replication cycle and were not thought to be directly immunomodulatory, recent studies have demonstrated that innate immune profiles are altered during DAA therapy, including improvement in NK cell function (37, 38), as well as normalization of intrahepatic ISG expression in liver and restoration of intrahepatic type I IFN responses in HIV-1 patients with acute HCV infection (39) and in chronic HCV patients (39). In addition, DAA therapy has also been reported to partially improve adaptive immune cell function (37, 38, 40). Therefore, type I IFN and ISG responses are modulated and remain relevant during DAA therapy.
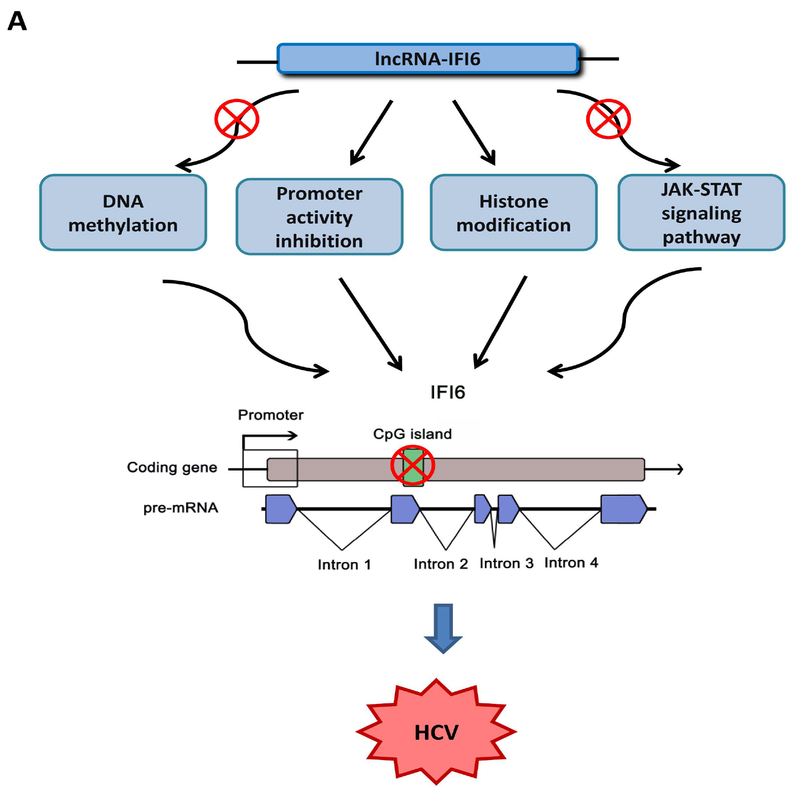
In order to study the regulatory mechanisms of lncRNAs on type I IFN response or ISGs in the context of HCV infection, we identified thousands of IFNα-induced lncRNAs, one of which was lncRNA-IFI6. We found that lncRNA-IFI6 regulated IFI6 with or without IFNα treatment. Type I interferons (IFNα/β) have been reported to induce over 300 ISGs. Many of these ISGs inhibit HCV replication (22, 23, 41, 42). However, some ISGs such as ISG15 and USP18 promote HCV replication (42–45). The expression of many lncRNAs is strongly dependent on the cell type and cellular state and is tightly controlled by various cellular signals. lncRNAs have been reported to play important roles in the regulation of the IFN response and ISG induction in HCV infection (27). For example, the lncRNA EGOT has been shown to increases HCV replication by suppressing the antiviral response through NF-κB pathway (46). HCV infection in turn activates PKR and induces NF-κB, EGOT and subsequently ISG15 transcription to block the RIG-I pathway, inhibiting the expression of several ISGs to enhance HCV replication (27, 46, 47). lncRNA-CMPK2/NRIR (Negative Regulator of the IFN Response) inhibits HCV replication through transcriptional downregulation of several ISGs in a Jak-Stat dependent manner. lncRNA NEAT1 controls Hantavirus infection through the RIG-I/IRF7 pathway (48). In this study, we found that IFN induces both lncRNA IFI6 and IFI6 expression, the latter of which exerts major inhibitory effects on HCV infection. lncRNA-IFI6 affected histone modification to regulate IFI6 through the enrichment of H3K4me3 (active mark) and H3K27me3 (repressive mark) to regulate HCV infection. We also determined that lncRNA-IFI6 regulated IFI6 independent of the JAK-STAT signaling pathway and DNA methylation (Fig 8A). The regulatory function of lncRNA can be determined by their specific secondary structure. For example, the tumor suppressing function of lncRNA MEG2 is determined by conservation of the secondary structure rather than its primary sequence (6, 40). Therefore, we designed 4 deletion mutants of lncRNA-IFI6 to investigate its functional structure, and found the large right arm structure of lncRNA-IFI6 (mutant 2) is the key functional domain. This study is the first to demonstrate that IFN-induced lncRNA-IFI6 regulates histone modification of the IFI6 promoter through its spatial domain (large right arm) to affect HCV infection.
Figure 8. Proposed model for lncRNA-IFI6 regulation of HCV infection.
lncRNA-IFI6 is an IFN-induced lncRNA, which regulates HCV infection through negative regulation of the antiviral gene, IFI6. lncRNA-IFI6 regulates IFI6 through histone modification and negatively regulates its promoter. lncRNA-IFI6 has no effect on DNA methylation and is independent of the JAK-STAT signaling pathway.
HCV replication has been shown to inhibit the expression of several ISGs including IFI6 in Huh7 HCV replicon cells (23). In this study, we confirmed that HCV infection reduced IFI6 and increased lncRNA IFI6 expression in JFH1-infected Huh7.5.1 cells. We found that lncRNA-IFI6 regulation of HCV infection through IFI6 is independent of Jak-Stat signaling. lncRNAs have been reported to play important roles in regulation of the IFN response and ISGs in HCV infection (27). We previously have demonstrated that HCV infection regulated ISGs through the Jak-Stat pathway (20, 21, 49, 50). We therefore speculate that HCV’s effects on lncRNA IFI6 and IFI6 expression are mediated through innate immune signaling pathways. These results further add to our understanding of the regulatory mechanisms of non-coding RNAs in the context of chronic HCV and the exhausted ISG phenotype, and will assist the development of applications that may prevent the establishment of persistent HCV infection, as well as potentially other viral infections.
Supplementary Material
Acknowledgements:
Funding:
This study was supported by the NIH AI069939 (RTC), AI082630 (RTC), DK098079 (RTC), DK108370 (RTC), China Scholarship Council (CSC) (XL, XD), Fundamental Research Funds for the Central Universities (Project Number: 2120134708)(XL), Doctoral Fund of Southwest University (Project Number: 22300501)(XL), Chongqing Social Undertakings and the People’s Livelihood Security Program (No. cstc2018jscx-msyb0797) (XL), Fundamental Research Funds for the Central Universities (XDJK2017A003) and NSFC (31772564).
We are grateful to these investigators for supplying the reagents listed here: Dr. Takaji Wakita (JFH1 HCV DNA construct); Dr. Francis Chisari (Huh7.5.1 cells).
Abbreviations:
- HCV
hepatitis C virus
- CRISPR
Clustered regularly interspaced short palindromic repeats
- IFN
interferon
- lncRNAs
long non-coding RNAs
- ISGs
Interferon-stimulated genes
- IFI6
Interferon alpha-inducible protein 6
Footnotes
Conflicts of interest:
The authors disclose no conflicts.
References
- 1.Duan X, Li S, Holmes JA, Tu Z, Li Y, Cai D, Liu X, et al. MicroRNA-130a regulates both HCV and HBV replication through a central metabolic pathway. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J-H, Li J-H, Jiang S, Zhou H, Qu L-H. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic acids research 2013;41:D177–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic acids research 2011;39:D146–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011;145:178–181. [DOI] [PubMed] [Google Scholar]
- 5.Barriocanal M, Carnero E, Segura V, Fortes P. Long non-coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor tetherin. Frontiers in immunology 2015;5:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell host & microbe 2014;16:616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winterling C, Koch M, Koeppel M, Garcia-Alcalde F, Karlas A, Meyer TF. Evidence for a crucial role of a host non-coding RNA in influenza A virus replication. RNA biology 2014;11:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON modulates HIV-1 replication in a NFAT-dependent manner and is differentially regulated by early and late viral proteins. Scientific reports 2015;5:8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. World Journal of Gastroenterology 2016;22:7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FLJ, Häussinger D, et al. Peginterferon Alfa-2a plus Ribavirin for Chronic Hepatitis C Virus Infection. New England Journal of Medicine 2002;347:975–982. [DOI] [PubMed] [Google Scholar]
- 11.Hadziyannis SJ, Sette H Jr, Morgan TR, et al. Peginterferon-α2a and ribavirin combination therapy in chronic hepatitis c: A randomized study of treatment duration and ribavirin dose. Annals of Internal Medicine 2004;140:346–355. [DOI] [PubMed] [Google Scholar]
- 12.Tai AW, Chung RT. Treatment failure in hepatitis C: Mechanisms of non-response. Journal of hepatology 2009;50:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes JA, Yu M-L, Chung RT. Hepatitis B reactivation during or after direct acting antiviral therapy – implication for susceptible individuals. Expert Opinion on Drug Safety 2017;16:651–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reig M, Mariño Z, Perelló C, Iñarrairaegui M, Ribeiro A, Lens S, Díaz A, et al. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy. Journal of hepatology 2016;65:719–726. [DOI] [PubMed] [Google Scholar]
- 15.Ravi S, Axley P, Jones D, Kodali S, Simpson H, McGuire BM, Singal AK. Unusually High Rates of Hepatocellular Carcinoma After Treatment With Direct-Acting Antiviral Therapy for Hepatitis C Related Cirrhosis. Gastroenterology;152:911–912. [DOI] [PubMed] [Google Scholar]
- 16.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends in molecular medicine 2014;20:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 2014;516:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho T-T, Zhou N, Huang J, Koirala P, Xu M, Fung R, Wu F, et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic acids research 2014:gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin W Tsai WL Shao RX, Wu G, Peng LF, Barlow LL, Chung WJ, et al. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology 2010;138:2509–2518, 2518 e2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin W Zhu C Hong J Zhao L, Jilg N, Fusco DN, Schaefer EA, et al. The spliceosome factor SART1 exerts its anti-HCV action through mRNA splicing. Journal of hepatology 2015;62:1024–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu C Xiao F Hong J, Wang K, Liu X, Cai D, Fusco DN, et al. EFTUD2 Is a Novel Innate Immune Regulator Restricting Hepatitis C Virus Infection through the RIG-I/MDA5 Pathway. J Virol 2015;89:6608–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, et al. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 2009;50:1727–1737. [DOI] [PubMed] [Google Scholar]
- 23.Itsui Y, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, Koyama T, Takeda Y, et al. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J Viral Hepat 2006;13:690–700. [DOI] [PubMed] [Google Scholar]
- 24.Meyer K, Kwon Y-C, Liu S, Hagedorn CH, Ray RB, Ray R. Interferon-α inducible protein 6 impairs EGFR activation by CD81 and inhibits hepatitis C virus infection. Scientific reports 2015;5:9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, Lemon SM. Innate immune responses in hepatitis C virus infection. Semin Immunopathol 2013;35:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metz P, Reuter A, Bender S, Bartenschlager R. Interferon-stimulated genes and their role in controlling hepatitis C virus. J Hepatol 2013;59:1331–1341. [DOI] [PubMed] [Google Scholar]
- 27.Valadkhan S, Fortes P. Regulation of the Interferon Response by lncRNAs in HCV Infection. Front Microbiol 2018;9:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei H, Wang S, Chen Q, Chen Y, Chi X, Zhang L, Huang S, et al. Suppression of interferon lambda signaling by SOCS-1 results in their excessive production during influenza virus infection. PLoS Pathog 2014;10:e1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammad F, Pandey GK, Mondal T, Enroth S, Redrup L, Gyllensten U, Kanduri C. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development 2012;139:2792–2803. [DOI] [PubMed] [Google Scholar]
- 30.Wang KCCHY. Molecular mechanisms of long noncoding RNAs. Molecular cell 2011;43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology 2013;20:300–307. [DOI] [PubMed] [Google Scholar]
- 32.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, et al. A long noncoding RNA mediates both activation and repression of immune response genes. science 2013;341:789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, et al. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 2016;165:1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, et al. DDX5 and its associated lncRNA Rmrp modulate Th17 cell effector functions. Nature 2015;528:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Burchill MA, Golden-Mason L, Wind-Rotolo M, R. Rosen H . Memory re-differentiation and reduced lymphocyte activation in chronic HCV-infected patients receiving direct-acting antivirals, 2015: n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 37.Serti E, Chepa-Lotrea X, Kim YJ, Keane M, Fryzek N, Liang TJ, Ghany M, et al. Successful Interferon-Free Therapy of Chronic Hepatitis C Virus Infection Normalizes Natural Killer Cell Function. Gastroenterology 2015;149:190–200. e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaan M, van Oord G, Kreefft K, Hou J, Hansen BE, Janssen HL, de Knegt RJ, et al. Immunological Analysis During Interferon-Free Therapy for Chronic Hepatitis C Virus Infection Reveals Modulation of the Natural Killer Cell Compartment. J Infect Dis 2016;213:216–223. [DOI] [PubMed] [Google Scholar]
- 39.Meissner EG, Wu D, Osinusi A, Bon D, Virtaneva K, Sturdevant D, Porcella S, et al. Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. The Journal of clinical investigation 2014;124:3352–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestell RG, Yu Z. Long and noncoding RNAs (lnc-RNAs) determine androgen receptor dependent gene expression in prostate cancer growth in vivo. Asian journal of andrology 2014;16:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 2001;69:912–920. [PubMed] [Google Scholar]
- 42.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014;32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Li S, McGilvray I. The ISG15/USP18 ubiquitin-like pathway (ISGylation system) in hepatitis C virus infection and resistance to interferon therapy. Int J Biochem Cell Biol 2011;43:1427–1431. [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Sun J, Meng L, Heathcote J, Edwards AM, McGilvray ID. ISG15, a ubiquitin-like interferon-stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. J Gen Virol 2010;91:382–388. [DOI] [PubMed] [Google Scholar]
- 45.Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, et al. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 2006;131:1584–1591. [DOI] [PubMed] [Google Scholar]
- 46.Carnero E, Barriocanal M, Prior C, Pablo Unfried J, Segura V, Guruceaga E, Enguita M, et al. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 2016;17:1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnaud N, Dabo S, Akazawa D, Fukasawa M, Shinkai-Ouchi F, Hugon J, Wakita T, et al. Hepatitis C virus reveals a novel early control in acute immune response. PLoS Pathog 2011;7:e1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma H, Han P, Ye W, Chen H, Zheng X, Cheng L, Zhang L, et al. The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. J Virol 2017;91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jilg N, Lin W, Hong J, Schaefer EA, Wolski D, Meixong J, Goto K, et al. Kinetic differences in the induction of interferon stimulated genes by interferon-alpha and interleukin 28B are altered by infection with hepatitis C virus. Hepatology 2014;59:1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin W, Choe WH, Hiasa Y, Kamegaya Y, Blackard JT, Schmidt EV, Chung RT. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology 2005;128:1034–1041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.