Abstract
Regulation of complex biological networks has proven to be a key bottleneck in synthetic biology. Interactions between the structurally flexible RNA and various other molecules in the form of riboswitches have shown a high-regulation specificity and efficiency and synthetic riboswitches have filled the toolbox of devices in many synthetic biology applications. Here we report the development of a novel, small molecule binding RNA aptamer, whose binding is dependent on light-induced change of conformation of its small molecule ligand. As ligand we chose an azobenzene because of its reliable photoswitchability and modified it with chloramphenicol for a better interaction with RNA. The synthesis of the ligand ‘azoCm’ was followed by extensive biophysical analysis regarding its stability and photoswitchability. RNA aptamers were identified after several cycles of in vitro selection and then studied regarding their binding specificity and affinity toward the ligand. We show the successful development of an RNA aptamer that selectively binds to only the trans photoisomer of azoCm with a KD of 545 nM. As the aptamer cannot bind to the irradiated ligand (λ = 365 nm), a light-selective RNA binding system is provided. Further studies may now result in the engineering of a reliable, light-responsible riboswitch.
INTRODUCTION
In recent years, light-controlled nucleotide tools have come into the focus of the biological and biochemical research (1–3). The development of new light sources has strengthened this progress. Light is an ideal tool that can be controlled spatially, temporally and in terms of its intensity without major technical effort. In biological systems, light can be used as an external, orthogonal trigger because (i) most of the cells do not respond to light (excepting photoreceptors), (ii) it does not provoke cell damage using an adequate wavelength and (iii) many cells and some organisms are at least partially translucent (1–3). The irradiation of biological samples can mostly be performed with a simple experimental setup and spatial and temporal control allows an exact location and timing of the desired effect. Biological regulation by light has often been carried out in transparent organisms such as Caenorhabditis elegans or zebra fish (4,5). For non-transparent organisms light can be used to address surface areas, like the skin. Furthermore, glass fibers can be used to facilitate deeper penetration and enable a possibility to treat internal organs. Lately, we could demonstrate that even ultraviolet (UV-) light can sufficiently penetrate the skin of mice (6).
Three technologies exist to couple a light signal to a biological effect: (i) Optogenetics makes use of endogenously expressible light-responsive proteins. (ii) Optochemical biology uses either photolabile groups installed in strategic places of molecules to temporarily block their activity (‘caging’). Alternatively, (iii) photoswitchable compounds can be used that exist in at least two photoisomeric forms, which can be interconverted for example with light of two different wavelengths. During the past decade, different photoswitchable molecules have been developed and investigated. Azobenzenes, spiropyrans and diarylethenes are some of the most prominent photoswitchable compounds that have been used for biological applications. Particularly, azobenzenes are reliable and well-investigated photoswitches to control biomolecules and have been used in various biological applications also referred to as ‘azalogization’ (7–11). However, while numerous light-directed manipulations of proteins (e.g. enzymes, ion channels etc.) have already been investigated, reports about light-responsive oligonucleotides are rare though they are interesting targets for several reasons. They are small, can be easily produced within cells or synthesized by solid-phase synthesis.
RNA is a versatile biomolecule capable of performing many different roles in the cell. The discovery of RNA elements mediating gene control, chemical reaction catalysis and signal transduction go far beyond their well-established role as the carrier of the genetic information and impressively demonstrate their functional versatility. Its ability to form complex three-dimensional structures that precisely present chemical moieties is imperative in enabling RNA to function as a biological catalyst, regulator or structural scaffold. Riboswitches are an excellent example of such RNA-based regulation. They are highly structured RNA elements mostly located in the 5′ UTR of many bacterial mRNAs as well as in the 3′ UTR in some plant mRNAs (12,13). The RNA forms a binding pocket that can recognize metabolites with high affinity and specificity. The ligand binding to this binding pocket, also called aptamer domain, is then read out by a second domain, the expression platform that can affect gene expression (12). In recent years, a number of synthetic riboswitches have been developed that are modeled on natural riboswitches (14,15). As binding domains, they often use aptamers that are not necessarily derived from natural riboswitches, but can also be produced de novo through a process of in vitro selection, also called SELEX (Systematic Evolution of Ligands by Exponential Enrichment) (16,17). With the help of this method, aptamers can be selected against almost any ligand of choice.
The overall intention of this work was to create a light-dependent synthetic riboswitch with structural changes only occurring upon binding to one photoisomer of a ligand. Whereas the riboswitch is expressed in the cell, the light-responsive ligand is introduced into the cell system and undergoes a structural change only upon irradiation. As a first step to engineer such a system, an aptamer has to be selected that responds in a light-dependent manner to a photoswitchable small molecule. Therefore the ligand needs to fulfill various requirements, such as high solubility under physiological conditions, favorable interactions with RNA (hydrogen bonds, stacking interactions, Coulomb attractions), sufficient cellular uptake and no toxicity to mention only a few. Furthermore, the photophysical requirements such as the ability to undergo structural change under physiological conditions, high switching amplitude for an optimal ‘ON’-‘OFF’ behavior, thermal stability (of the photoisomeric states) and no photobleaching should be guaranteed. Additionally, the photoswitches should not possess long tethers to allow an optimal hinge mechanism of the photoswitch. To achieve the ambitious goal of reversible light-controlled riboswitch regulation systems, as many as possible of the above prerequisites need to be fulfilled.
To our knowledge, up to now only three studies on SELEX methods against photochromic molecules have been published (18–20). In all cases, the aim was to direct the aptamer-binding to only one of the possible two photoisomeric states of the small molecule. Hayashi et al. performed a SELEX against the trans isomer of the photoswitchable peptide KRAzR (20,21). In surface plasmon resonance (SPR) studies, they demonstrated that individual aptamers bound KRAzRtrans with a dissociation constant in the high nanomolar to low micromolar range. This binding was tenfold worse for the UV-exposed KRAzRcis form, but could be completely restored by irradiation with visible light of 430 nm (conversion back to KRAzRtrans). Young and Deiters were also able to generate photoisomer-specific RNA aptamers, selecting for a spiropyran derivative (19). This allowed them to identify an aptamer, which exhibited a 14-fold discrimination between the closed spiropyran and open merocyanine form upon irradiation with UV light. However, the affinity was only at a KD value of ∼10 μM. Lee et al. obtained specific aptamers against the closed form of the dihydropyrene BDHP-COOH in their studies (18). The best candidate showed a 35-fold difference in its binding behavior between the respective photoisomers and a KD value of ∼2 μM.
Here, we present our approach toward generating RNA aptamers, which selectively bind to one isomer of a newly developed photoswitchable molecule in the nanomolar range, with 104 times better discrimination than the other photoisomer. These aptamers will extend the toolbox of synthetic biology compounds and provide now excellently suitable sensory domain for the engineering of light-responsive synthetic riboswitches.
MATERIALS AND METHODS
Biophysical characterization of the spectroscopic behavior of azoCm
The photophysical properties of the compound azoCm were determined in 1 × PBS buffer (phosphate-buffered saline), except for the stability measurements that were carried out in HeLa cell extracts and experiments in presence of RNA which were performed in the corresponding SELEX buffer. For the UV/vis-absorption measurements, the concentrations were kept below 100 μM and the samples were prepared in UV-permeable quartz glass cuvettes with 1 cm path length. Absorption spectra were recorded using a Specord S600 or S100 spectrometer (Analytik Jena). For irradiation experiments, ThorLabs light emitting diodes (LEDs) controlled by a ThorLabs DC4100-Driver were applied. UV-light for trans-cis isomerization was provided by a λ = 365 nm LED while cis-trans isomerization was induced using a λ = 420 nm LED (type M365L2 and M420L2, 300 mW).
The CD measurements were executed with a Jasco J-710 spectrometer under constant nitrogen flow of 1.5 l/min. Here, cuvettes with 1 mm path length were used and irradiation was also carried out with the same set of ThorLabs LEDs. In these experiments, the RNA concentration was 10 μM and the ligand azoCm was provided in 10-fold excess with a concentration of 100 μM.
SELEX—Pool design
We used three different pool designs. For the first SELEX, a previously described pool design was used (22). For the second SELEX a completely randomized RNA library was used. This library consisted of 74 nucleotides (nt) flanked by constant regions (5′ constant: 5′-GGA GCU CAG CCU UCA CUG C-3′/3′ constant: 5′-GGC ACC ACG GUC GGA UCC AC-3′) for amplification using the oligonucleotides Pool_fwd (5′-TCT AAT ACG ACT CAC TAT AGG AGC TCA GCC TTC ACT GC-3′, T7 promoter is underlined) and Pool_rev (5′-GTG GAT CCG ACC GTG GTG CC-3′). The third SELEX was carried out with a partially structured library containing a 15-nt long motif sequence (5′-GGC CUA CGG GAA AGG-3′) flanked by 30 random nt on the motifs 5′ end, and 5 random nt on its 3′ end (N30-motif-N5). The motif was partially randomized as well, with a 50% chance of a nucleotide exchange for the first two nt (GG), and a 2% chance for the remaining 13 nt. This partially structured library was flanked by constant regions (5′ constant: 5′-CCA AGC UAG AUC UAC CGG U-3′/3′ constant: 5′-AAA AUG GCU AGC AAA GGA GAA GAA CUU UUC ACU-3′) for amplification using the oligonucleotides Pool_fwd (5′-CCA AGT AAT ACG ACT CAC TAT AGG GCC AAG CTA GAT CTA CCG GT-3′) and Pool_rev (5′-AGT GAA AAG TTC TTC TCC TTT GCT AGC CAT TTT-3′).
Pool preparation
All pools were amplified using the following polymerase chain reaction (PCR) conditions: 10 mM Tris–Cl pH 9.0, 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.2 mM dNTPs (each), 30 nM pool template, 2 μM Pool_fwd, 2 μM Pool_rev, 50 U/ml Taq DNA Polymerase (NEB). 1015 pool template molecules were amplified in a 60 ml PCR reaction for only seven cycles to reduce PCR-induced bias. PCR efficiency was calculated according to Hall et al. (23).
After large-scale amplification, the DNA pool template was ethanol-precipitated and dissolved in MQ-H2O [de-ionized water purified with ion exchange resin and filtered through a Biofilter (ELGA) to remove possible RNAse contamination]. The purified DNA template was transcribed using T7 RNA polymerase as described previously (24). Afterward, the transcribed RNA was ethanol-precipitated, dissolved in formamide containing 25 mM ethylenediaminetetraacetic acid (EDTA) and loaded on a 6% denaturing polyacrylamide gel (8 M urea). The RNA was visualized by UV shadowing, sliced out and eluted overnight in 300 mM Na-acetate (pH 6.5). Hereafter, eluted RNA was ethanol-precipitated, the pellet was re-dissolved in a suitable amount of water and molarity was calculated.
Immobilization of the ligand azoCm
azoCm-amino was immobilized on Affi-Gel 10 (Bio-Rad). For this, the resin was pre-treated according to the manufacturer's instructions of anhydrous coupling. Afterward, the resin was mixed 3:1 with a 3 mM solution of azoCm in dimethyl sulfoxid (DMSO). The reaction was incubated for 4 h at room temperature (RT) on an H5600 rotator (Labnet). After two washes of the derivatized resin with DMSO, the remaining active groups were blocked by incubation with 0.1 M ethanolamine (MEA) for 1 h. After washing the resin twice with DMSO, it was resuspended 1:1 in 0.002% (w/v) NaN3 and 1 ml of the mix was distributed per SELEX column (bedvolume 1.2 ml). The columns were stored at 4°C in the dark.
Regular SELEX
1.2 × 1015 RNA molecules from the initial pool (1:1 mixture of completely randomized and pre-structured pool, see above (22)) were spiked with ∼250 kCPM of the 5′ 32P-labeled RNA pool in MQ-H2O to use in the first round. RNA folding was performed by heating the mixture to 95°C for 5 min and subsequently placed on ice water for additional 5 min. After this folding step, yeast tRNA was added to a final concentration of 1 mg/ml and the volume was adjusted to 1 column volume of resin (CV, 500 μl) with 1× binding buffer (40 mM Hepes pH 7.4, 125 mM KCl, 5 mM MgCl2, 5% DMSO). For depletion of RNAs able to bind the affinity matrix, the RNA library was first incubated for 30 min with 1 CV of a non-derivatized column (mock) during the first three rounds of SELEX. The mock column consisted of Affi-Gel 10 resin that had been treated with MEA only instead of azoCm. After negative selection, unbound RNAs were added to 1 CV azoCm-coupled resin and incubated for 30 min at RT. Next, the column was washed with 10 CV binding buffer and bound RNAs were eluted with either 4 CV 20 mM EDTA (round 1–5) or 4 CV 5 mM azoCm (round 6–9) in 1× binding buffer. To increase stringency, the number of washes was increased to 25 CV and an increased timeframe for the elution of the RNA from the column was used (round 8). Pre-elution steps were performed in round 8 and 9, adding an additional pre-elution step in round 9 (2 or 3 CV each). Details are described in Supplementary Table S1.
Light SELEX
During the second SELEX, 4.8 × 1014 RNA molecules from the initial pool (74 nt randomized region, see above) were spiked with ∼250 kCPM of 5′ 32P-labeled RNA pool in MQ-H2O to use in the first round. The folding, addition of yeast tRNA and adjustment of volume were carried out like in the first SELEX. Mock columns were used (round 1–4), RNA was eluted by either 20 mM EDTA (round 1) or 5 mM azoCm (round 2–9). Stringency was increased raising the number of washes from 10 CV to 20 CV or a decreasing binding time of the RNA pool to the column to 15 min, respectively (round 7 and 9). Two pre-elution steps (1 CV each) were performed starting from round 7. Starting with round 7, light-elution was carried out in parallel to the affinity SELEX. RNA was eluted by irradiating the column with light (λ = 365 nm, UVLED-365–1000-SMD, 2W (0.15 A, 13.5 V), 16 × 1 s) under exclusion of other light sources (distance from column to light source = 10 cm), while washing the column with 6 CV of 1× SELEX buffer (10 mM Tris–HCl pH 7.6, 250 mM NaCl, 5 mM MgCl2). Details are described in Supplementary Table S1.
Doped SELEX
For the third SELEX, 1.2 × 1015 RNA molecules from the initial pool (motif doped pool, see pool design) were spiked with ∼250 kCPM of 5′ 32P-labeled RNA pool in MQ-H2O to use in the first round. The folding, addition of yeast tRNA and adjustment of volume were carried out like in the first SELEX. The columns with immobilized RNA were washed with 10 CV of 1× SELEX buffer, before RNA was eluted with 1 mM azoCm in 1× SELEX buffer (round 1–5). Details are described in Supplementary Table S1.
SELEX cycle
In all SELEX experiments and after each round, eluted RNA was ethanol-precipitated with Na-acetate pH 6.5 in the presence of 15 μg GlycoBlue™ Coprecipitant (Ambion) and washed twice with 70% (v/v) ethanol. The air-dried pellets were dissolved in a total volume of 50 μl MQ-H2O and reverse-transcribed and amplified (RT-PCR). For RT-PCR, 50 μl eluted RNA was mixed with 1× PCR buffer (10 mM Tris–Cl pH 9.0, 50 mM KCl, 0.1% Triton X-100), 1× first strand buffer (Invitrogen), 2 mM dithiothreitol (DTT, Roche), 1 μM Pool_fwd, 1 μM Pool_rev, 1.5 mM MgCl2 and 0.3 mM dNTPs (each). The reaction was heated to 65°C for 5 min and then quickly placed on ice. After that, 5 U Taq DNA Polymerase (NEB) and 200 U SuperScript™ II (Thermo Fisher Scientific) were added to the reaction and RNA was reverse-transcribed and amplified (54°C for 10 min followed by 6 to 10 cycles of 95°C for 1 min, 58°C for 1 min and 72°C for 1 min). Product formation was monitored on a 3% agarose gel. For the following rounds (all except round 1), RNA was transcribed as follows: 10 μl of RT-PCR product was mixed with 40 mM Tris-Cl (pH 8.0), 5 mM DTT, 2.5 mM NTPs (each), 15 mM MgCl2, 100 U T7 RNA Polymerase (NEB), 40 U ribonuclease inhibitor (moloX) and 33 nM 32P-α-UTP (Hartmann analytics) in a total volume of 100 μl. Transcription was carried out at 37°C for 1 h. Afterward, RNA was precipitated with NH4-acetate/ethanol, washed twice with 70% EtOH and the pellet was dissolved in a suitable amount of water. A total of 500 kCPM RNA was folded, diluted in 1× binding buffer and subsequently loaded onto the column for the next round of SELEX.
NGS library preparation and data analysis
4-mer barcodes to assign each sequence to the specific round after sequencing were introduced by PCR (sequences available upon request). Forward and reverse oligonucleotides hybridized at the 5′ and 3′ constant regions, respectively, thus the sequence of the T7 polymerase promoter was removed. After amplification, the samples were Gel-purified (Zymoclean Gel DNA Recovery Kit, Zymo Research) and mixed in equimolar amounts for Illumina sequencing reaction (GenXPro GmbH, Frankfurt, Germany).
In vitro transcription of RNA
For in vitro analysis (binding studies and ITC measurements), RNA was transcribed from hybridized oligonucleotide containing a T7 promoter sequence in front of the aptamer (sequences are available upon request). After ethanol precipitation, the DNA template was used for in vitro transcription with T7 RNA polymerase (NEB) as done during the SELEX cycle. The RNA was gel purified and molarity was determined by spectrophotometric measurement using NanoDrop 1000 Spectrophotometer (Thermo Scientific).
Binding studies
The elution capacity of RNA aptamers by different solutions was tested using column binding assays. The aptamers were radioactively body labeled and after folding (in a total volume of 500 μl, in 1× SELEX buffer with a final concentration of 1 mg/ml yeast tRNA) applied on the azobenzene-derivatized columns. After incubation for 30 min at RT, the columns were washed 4× with 1 ml 1× SELEX buffer each, followed by two washing steps of 500 μl each. The binding RNA molecules were tried to be eluted with different solutions (cis- or trans-azoCm, UV-light and buffer or just buffer). Each aptamer measurement set was normalized to the RNA eluted by azoCm trans (100%).
ITC measurements
The 10 μM RNA were folded using the SELEX folding program (heating to 95°C for 5 min and subsequent cooling on ice water for 5 min) and diluted in 350 μl 1× SELEX buffer. Ligand was adjusted to a 100 or 200 μM solution in 1× SELEX buffer. ITC experiments were carried out with either an MicroCal iTC200 or a MicroCal PEAQ iTC200, with the ligand solution being titrated to the RNA. After thermal equilibration at 25°C, an initial 150 s delay and one initial 0.4 μl injection, 18 serial injections of 2.0 μl at intervals of 150 s and at a stirring speed of 750 rpm were performed. Raw data were recorded as power (μcal/s) over time (min). The temperature associated with each titration peak was integrated and plotted against the corresponding molar ratio of ligand and RNA. The dissociation constant (KD) was extracted from a curve fit of the corrected data calculated using the Origin 7.0 ITC Software or the MicroCal PEAQ-ITC Analysis Software 1.1.0.1262. Measurements were repeated twice.
Toxicity tests
To test toxicity against Escherichia coli DH5a, Bacillus subtilis (strain 168) and Saccharomyces cerevisiae (strain RS453α), cells were plated densely on LB- or SD-URA-Agar plates. Sterile filter discs (diameter of 6 mm) were placed on the agar, allowing space between each other. Ligand solutions with different concentrations (0, 0.1, 0.5, 1, 2 and 5 mM, 25 μl each) were pipetted on the disks to allow radial diffusion into the agar. The cultures were grown over night at 37°C or 30°C until they formed a cell layer. Analysis of potential inhibition zone sizes around the filter discs allowed judgement whether the ligand was toxic and at which concentration toxic effects occurred.
To test toxic effects on human cells, 2 × 103 HeLa cells per well were seeded in a 96-well plate in 100 μl media (D-MEM). After incubation (37°C) for 24 h, 2 μl of 500 μM, 5 mM or 50 mM ligand solution in DMSO were added to the cells to reach concentrations of 10 μM, 100 μM or 1 mM ligand in the media, respectively. Controls consisted of adding DMSO to a final concentration of 2% (negative control) and 10 μM Taxol (positive control) to wells with cells. After another 24 h of incubation, cells were washed twice with warm 1× PBS. After adding 110 μl of a 1:10 WST-1:media mix (white D-MEM) per well, the cells were incubated again and samples were taken at intervals for analysis. Using a TECAN 200 pro reader, viable cells were quantified. Samples were measured in triplicates, shown are the averages and standard deviations of two independent measurements.
NMR measurements of compounds
Nuclear magnetic resonance (NMR) spectra for the characterization of the compounds were recorded on a Bruker AVIII-HD 500 MHz instrument equipped with a N2 cooled cryogenic probe head using d6-DMSO as solvent. High resolution mass spectrometry (HRMS) spectra were recorded using a Thermo Scientific MALDI LTQ Orbitrap. For flash chromatography silica gel 60 by Macherey-Nagel was used. Thin layer chromatography (TLC) analyses were performed on aluminum plates coated with silica gel 60 F 254 (Merck).
RESULTS AND DISCUSSION
Synthesis of a light-switchable azobenzene compound
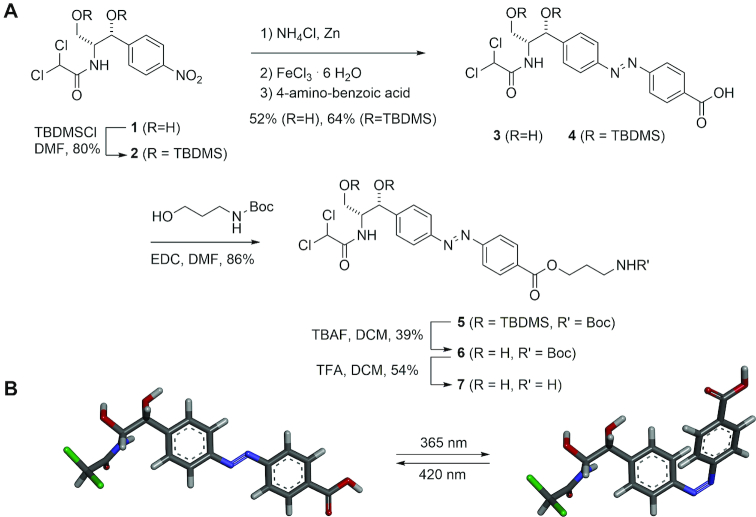
We chose azobenzene as the core of our light-switchable ligand because it is a very reliable photoswitch. However, azobenzene itself offers only poor interactions with RNA. Therefore, it has to be modified. The modification should have functional groups that can be easily recognized by RNA and do not have too many internal degrees of freedom in order to maximize the effect of photoswitching whereby this modification and hence also the RNA anchor points are moved through space. Due to previous good experience with the generation of aptamers against chloramphenicol (22) and its low toxicity, we chose the latter as binding motif. The synthesis of such a designed photoswitchable compound named azoCm (compound (3)) started from chloramphenicol (1) and followed Trauner's ‘azalogization’ strategy conceptually by converting the nitrophenyl group into an azobenzene derivative (9–11). To this end, chloramphenicol (1) was reduced with ammonium chloride and zinc to form the hydroxylamine which was oxidized with iron (III)-hexahydrate to form the nitroso derivative that was further reacted with 4-amino benzoic acid to form azoCm (3) (Figure 1). To arrive at the aminopropyl-modified azoCm-amino (7) we first protected the hydroxyls of chloramphenicol (1) with TBDMS groups (→2) and performed the same azo coupling (→4). After ester formation with N-Boc-protected aminopropanol using EDC as coupling agent (→5) and deprotection, aminopropylated azoCm-amino (7) was obtained. Details about the synthesis of asoCm and its intermediates can be found in Supplementary Figures S1–6.
Figure 1.
Synthesis of the light-switchable azobenzene compound azoCm. (A) Synthesis of azoCm (3) and the aminopropyl-derivative (7). (B) 3D model of the cis-trans conversion of azoCm. Details about the synthesis of asoCm and its intermediates can be found in Supplementary Figures S1–6.
Characterization of spectroscopic behavior of azoCm
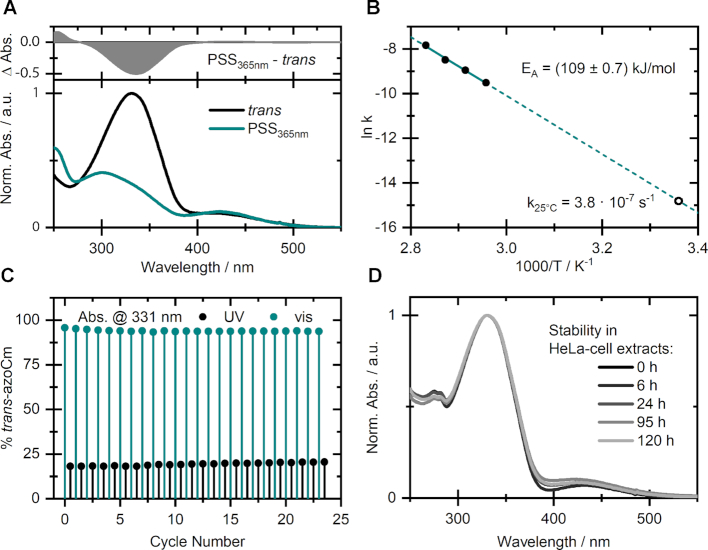
Prior to the application of azoCm as a photochromic ligand for selected RNA aptamers, the structurally modified azobenzene compound was characterized regarding its photophysical properties in aqueous media. The UV/vis spectrum of the thermally stable and favored trans-isomer of azoCm (Figure 2A) exhibits a pronounced ππ*-band around 330 nm and a minor nπ*-band around 420 nm, as typical for this class of photoswitches. Upon irradiation with 365 nm a photostationary state (PSS365nm) containing about 80% of the cis-isomer can be accumulated. The reverse isomerization can be induced through 420 nm irradiation, resulting in a nearly quantitative conversion to the trans-isomer (containing less than 5% cis).
Figure 2.
Spectroscopic behavior of azoCm. (A) UV/vis-spectra of trans-azoCm (black) and the PSS (cyan) obtained upon irradiation with λ = 365 nm of azoCm in 1xPBS buffer. (B) Arrhenius plot based on thermal cis to trans relaxation studies with determined activation energy (filled circles) and extrapolated rate (open circle) at 25°C. (C) Photofatigue resistance study monitoring the absorption at 331 nm throughout alternating irradiation with 420 and 365 nm. (D) Stability in HeLa cell extracts of a sample that was switched forth and back once prior to being stored in the dark for the given time.
Thermal relaxation studies of pre-illuminated samples at different temperatures allowed for determination of the thermal recovery rate k25°C = 3.8 × 10−7 s−1 for the cis to trans isomerization. This reveals an enormously high stability of the cis-isomer e.g. compared to the unsubstituted azobenzene (25) and corresponds to a lifetime of ∼27 days for the respective conformation. Relying on the Arrhenius-equation, the energy barrier for this isomerization is calculated to be EA = (109 ± 0.7) kJ/mol (Figure 2B). Additionally, the compound showed virtually no decomposition due to photo- or hydrolysis throughout a photofatigue experiment over multiple alternating switching cycles (Figure 2C) and no additional absorption bands could be detected afterward. Moreover, the highly beneficial switching amplitude of roughly 75% is maintained throughout these multiple cycles. In view of a biological application of the compound, the outstanding stability of azoCm was also confirmed in HeLa-cell extracts, where the sample showed virtually no significant changes in its absorption spectrum after 120 h (Figure 2D). Thus, the compound can be used for days without considerable decomposition. Beside its outstanding photochemical and biological stability, we investigated the toxicity of the compound. We performed toxicity tests for bacteria (E. coli, B. subtilis), yeast (S. cerevisiae) and human cell lines (HeLa cells) and showed that azoCm has no significant influence on cell growth (Supplementary Figure S7).
Selection of azoCm-binding aptamers
The characterization showed that azoCm exhibits stable switching behavior even under cellular conditions and is therefore a very good candidate to start an aptamer selection. For the selection process, the aminopropyl derivative of azoCm containing an additional amino group (7) was immobilized on activated resin. For better access of the RNA library to the azoCm moiety a C10 linker was used. Two selections were started in parallel with two established library compositions, one with a randomized region of N74 and a smaller library (N64) being a 1:1 mixture of both a completely randomized region and two shorter randomized regions (N26) separated by a preformed stem loop (Supplementary Figure S8). It was discussed that preformed stem-loops provide favorable conditions for aptamer selection by acting as nucleation sites for RNA structure formation (26–29). We had no a priori knowledge of the nature of the aptamer we were exploring, including both completely randomized regions of different size and a preformed stem loop gave us the full scope to unrestrainedly select for the best fit. Importantly, both library designs were already successfully applied for the selection of small molecule-binding aptamers (22,30).
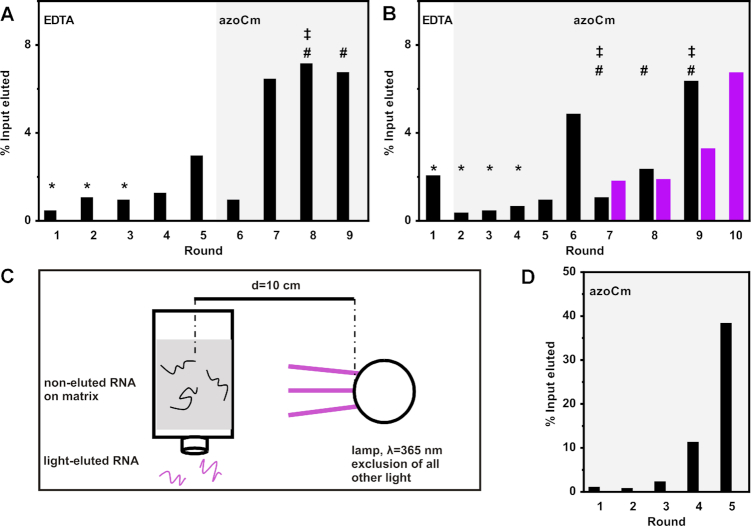
A first selection was carried out using the N64 1:1 RNA library with a starting diversity of 1.2 × 1015 RNA molecules. In the first five rounds of selection, we eluted unspecifically with EDTA to ensure elution of every RNA molecule neglecting their binding properties. This approach was chosen to guarantee that the first enrichment of the pool introduces no bias toward low affinity aptamers because of the mild selection conditions. Negative selection steps during the first three rounds were carried out to remove RNA molecules that specifically recognize the resin or the linker region on which azoCm was immobilized. After an initial enrichment observed in round 5 (Figure 3A), a specific elution with 5 mM azoCm was performed in round 6. Stringency was increased in round 8 by raising the number of washing steps and longer elution times. Despite increased stringency, larger amounts of RNA were eluted from the column. In consequence, we decided to implement a pre-elution step each in rounds 8 and 9 to eliminate RNA species with fast Koff rates (31). All selection rounds are summarized in Figure 3A and Supplementary Table S1.
Figure 3.
In vitro selections for azoCm binding aptamers. (A) Shown is the fraction of loaded RNA that could be eluted from azoCm-derivatized columns after each selection round. RNA was eluted by either 20 mM EDTA (round 1–5) or 5 mM azoCm (round 6–9). In the first three rounds, a negative selection was performed (*). In round 8, stringency was increased by raising the number of column washes from 10 to 25 and an increased timeframe for the elution of the RNA from the column, respectively (‡). Pre-elution steps were performed in round 8 and 9, adding an additional pre-elution step per round (#). Details are given in Supplementary Table S1. (B) Shown is the fraction of loaded RNA that could be eluted from azoCm-derivatized columns after each selection round. RNA was eluted by either 20 mM EDTA (round 1) or 5 mM azoCm (round 2–9) for regular SELEX (black bars). In the first four rounds, a negative selection was performed (*). In round 7 and 9, stringency was increased by doubling the number of column washes or a decreased binding time of the RNA pool to the column, respectively (‡). Two pre-elution steps were performed starting from round 7 (#). Starting with round 7, light elution was carried out in parallel to the regular SELEX (violet bars). For RNA elution, the column was irradiated with light (λ = 365 nm) under exclusion of other light sources, while washing the column with SELEX buffer. Details are given in Supplementary Table S1. (C) Set-up of light elution of RNA. The column was irradiated with UV-light (λ = 365 nm) under exclusion of other light sources, while washing the column with SELEX buffer to achieve elution of aptamers which lose binding affinity when the ligand is switched from trans to cis. The UV lamp was placed 10 cm from the column middle. (D) Doped SELEX. Shown is the fraction of loaded RNA that could be eluted from azoCm-derivatized columns after each selection round. RNA was eluted with 1 mM azoCm (round 1–5).
In parallel, we performed a second selection approach with the N74 RNA library with a starting diversity of 4.8 × 1014 RNA molecules. Unspecific elution was reduced to only the first round, while negative selection was performed during the first four rounds. Starting from round two, specific elution was performed using a 5 mM azoCm solution. After a first enrichment in round 6, stringency was increased in rounds 7 and 9 by doubling the number of column washes or a decreased binding time of the RNA pool to the column, respectively. Additionally, two pre-elution steps were performed starting from round 7. All selection rounds are summarized in Figure 3B and Supplementary Table S1.
We also implemented a light elution step. From this we hoped for specific enrichment of light-dependent aptamers. Since the second selection (from the N74 library) led to a faster enrichment, we decided in favor of this. We split the RNA pool transcribed from round 6 to carry out a light elution parallel to the regular round 7. While the column was treated and washed using the same conditions as in the regular SELEX, the RNA-loaded column was then placed in a closed box, shutting out all external light sources (Figure 3C). Inside this box, the column was irradiated with UV-light of 365 nm inducing azoCm to change from its trans- to cis-conformation. After irradiation, the column was washed with buffer to specifically elute those RNA aptamers that were detached from the ligand due to its conformational change. Amplification of these RNAs should result in an enrichment of light-responsive aptamers and were used for the next round of light SELEX. The data are included in Figure 3B as violet bars. A significant enrichment of aptamers that have been specifically eluted after irradiation with UV-light was clearly visible in round 10 of the light SELEX.
For a first glimpse into the selection progress, we sequenced 87 candidates (27 of round 9 of N64 1:1, 20 of round 9 of the regular SELEX (N74-R) and 40 of round 10 of the light SELEX (N74-L)). We obtained 27 unique sequences (11, 4 and 12, respectively, for the three different selections, shown in Supplementary Table S2) and analyzed their binding capacities by their interaction with azoCm derivatized columns. Only 6 out of 27 tested candidates could be specifically eluted from the column by 5 mM azoCm (Supplementary Figure S9). The remaining 21 candidates showed less than 15% RNA eluted and were not considered as binders. Interestingly, when comparing the sequences of the six candidates using the program MEME (http://meme-suite.org/tools/meme V5.0.2 (32)) we identified a 13-nt long motif in three of them (one from the N64 1:1 SELEX and two in the light branch of the N74 SELEX, displayed in Figure 4C).
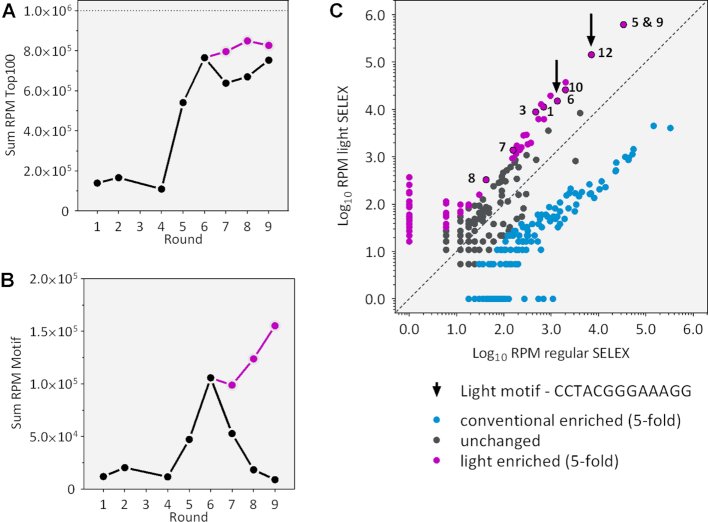
Figure 4.
Deep sequencing analysis of all rounds of selection. (A) Enrichment of the 100 most enriched sequences in each round. Shown is the sum of the RPM in black for traditional SELEX and in violet for light SELEX. (B) Occurrence of the identified light motif within each selection round clearly shows an enrichment in the light SELEX compared to the regular SELEX. (C) Scatter plot of the families found in the regular and the light SELEX. Application of light clearly sets some families apart (at least 5-fold enriched). Analysed families are marked with numbers.
Deep sequencing uncovers the enrichment of a ‘light motif’
We performed deep sequencing to analyse whether and to what extent our identified sequence motif was enriched in the course of the different selections rounds. Therefore, we compared the regular with the light branch of the N74 SELEX (for financial reasons we limited ourselves to the N74 SELEX). By Illumina sequencing, we obtained a total number of 820 000 reads for all investigated rounds, of which 90.1% could be sorted according to their corresponding barcoding. Identical sequences were summed up and the total read count was normalized for each round to reads per million (RPM). To show the enrichment process we computed the sum of RPM of the 100 most enriched sequences (Figure 4A). For the most abundant sequences we see a clear exponential enrichment in the first six rounds and then a transition into saturation when the overall sum of the Top 100 did not change any more, in neither of both branches. We also tracked the enrichment of the 13 nt motif we identified. It was enriched until round six. Then, in the light branch of the SELEX, the motif was further enriched, whereas in the regular branch, the motif gradually decreased over the rounds (Figure 4B). Due to its clear enrichment in the light branch, we have now called it ‘light motif’. It seems as if the increase of stringency we took in the regular SELEX to select for aptamers with a slow Koff rate lead to the disfavoring of aptamers containing the motif.
We then clustered the sequences for each round into families by comparing their sequence identities by Levenshtein distance (Lv dist) (33). We have previously evaluated that an Lv dist of five is ideal to split the sequence distribution into useful families (22). The distribution of families enriched in the regular and the light-branch, respectively, are displayed in the scatter plot in Figure 4C. It shows a clear separation of the two populations of families (comparing the blue with the violet dots). The detailed analysis of the families enriched by light SELEX revealed 131 unique sequence families, the most prominent ones marked with numbers in Figure 4C. Interestingly, only two of them carry the identified ‘light motif’ (marked with an arrow), but these are highly enriched and make up 14.4% of all sequences.
Motif doped SELEX
The deep sequencing analysis revealed the importance of the light motif. In order to generate highly specific aptamers of a smaller size able to selectively and reversibly bind to only the trans-isomer of azoCm we generated a new pool with the light motif included but flanked by random sequences. This should enable us to identify flanking sequences that present the light motif in the best possible way. As the motif occurred always in the 3′ end of the randomized sequence, we designed a pool consisting of 30 randomized nts followed by two G′s (often found by deep sequencing) and the 13 nt motif. Afterward, five more randomized nucleotides completed a total of 50 nt that represent the new, doped aptamer pool. In order to find potentially better variants of the motif, a 6% mutation rate was allowed for the 13 nt of the motif. As it was unclear whether the two G’s in front of the motif were a necessary or a helpful addition to the motif, they were added but were allowed to mutate with 50% probability (Supplementary Figure S8D). A fast enrichment took place when using this doped pool, reaching 38% RNA elution within five rounds of selection even with reduced ligand concentration (1 mM instead of 5 mM).
We sequenced 96 individual aptamers of rounds 2–5. Interestingly, within these sequences, we found barely any sequences more than twice. There were no dominant aptamer sequences, not even in round 5 with its exceptionally strong enrichment. This suggests that the motif indeed is the core binding region on which the ligand binding solely depends. While over the course of the doped SELEX no single aptamer could be found to dominate the pool, a re-enrichment of the original motif occurred. For 7 out of 13 positions, no single mutation was allowed, for the remaining positions, a mutation rate of only 1–2% occurred (with 6% mutation rate allowed for the complete motif, Supplementary Figure S8D). The analysis of the motif revealed as well that the first G’s upstream of the motif are not essential for the binding. With the allowed nucleotide exchange rate of 50%, no preference for the first nucleotide have been identified. The position of the second G however showed a migration from a dominance of a G on this position in round 2 (80% probability) toward a preference of purine bases in general (A, 40% and G, 56%). But it seems like the sequences of the motif-adjacent nucleotides are not important for binding.
The analysis shows that the 13 nt long motif is essential for the binding of our light-dependent ligand. However it was unexpected that no specific sequence enrichment of the surrounding sequences could be found with the doped SELEX. We therefore considered whether the light motif is embedded in a structural motif. To test this, we used LocARNA (www.rna.informatik.uni-freiburg.de/LocARNA/V 1.9.1 (34,35)) to search for a secondary structural motif within the 96 aptamers sequences. The analysis resulted in a small structural motif consisting of a three-way-junction with a 4-nt long closing stem P1 (see Figure 5). The two Gs of the upper two base pairs of P1 are formed by the last two nts of the light sequence motif. Sequence and length of P2 are fully variable whereas P3 is defined by the motif. It consists of only two GC base pairs closed by an apical UNCG tetraloop that are connected to P1 by three As facing a CU. To prove the predicted secondary structure we performed in-line probing of the aptamer 42 (Supplementary Figure S10). The probing pattern supports the existence of P1 and P2 whereas it proposes that the short 2 nt long P2 is not formed for this candidate. However it clearly shows an involvement of the light motif in ligand binding. We observe a strong reduction of the cleavage for the nucleotide GGGAAAG (nt 6–12 of the motif), whereas the first 5 nucleotides are clearly accessible (showing that the small P2 helix is unlikely to be formed as such) and cleavage tends to increase. In sum, it can be stated that the light motif represents the binding pocket for the ligand that is held in place by the two helices P1 and P2.
Figure 5.
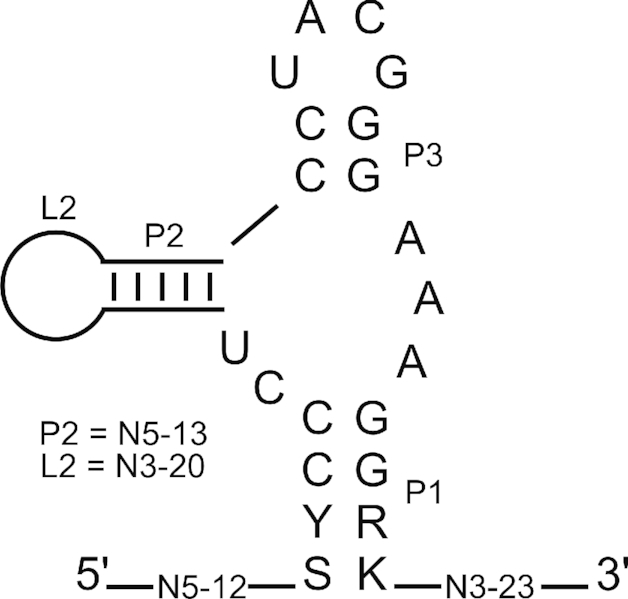
Structure motif. LocARNA analysis was performed to identify a common structural motif using 96 sequences from round 5 of the motif doped SELEX (34,35). Conserved nucleotides are shown in capital letters, variable sizes of the stem P2 and the corresponding loop L are displayed.
Biochemical characterization of light-responsive aptamers
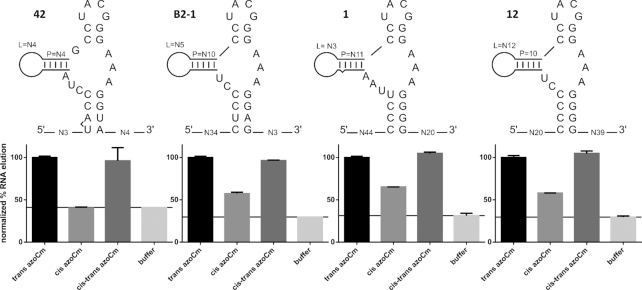
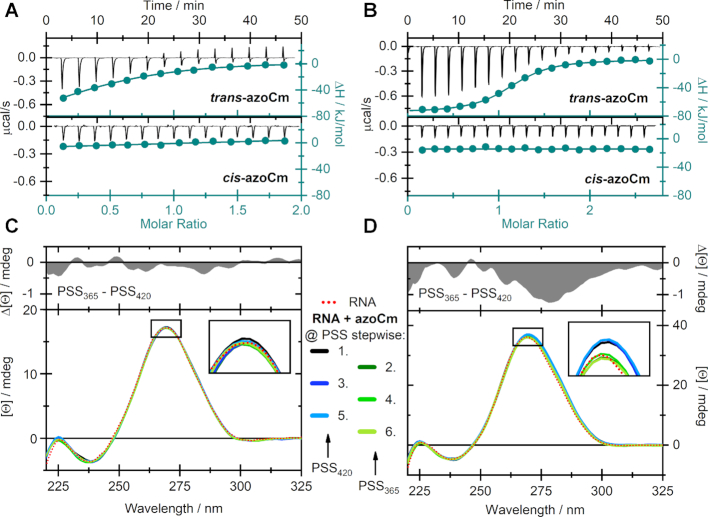
Four individual aptamers that all fold in the secondary structural motif predicted with LocARNA (Figure 5) were tested in column binding assays for their specific binding to the photoisomers of azoCm (candidate 42 from first affinity SELEX, candidates 1 and 12 from the light SELEX and candidate B2–1 from the motif doped SELEX). Aptamer RNA was radioactively labeled, folded and immobilized on azoCm-derivatized columns using the same conditions as during the SELEX. Immobilized RNA was eluted from the columns using azoCm in trans, cis and cis-trans switched conformation, or using 1× SELEX buffer. The fraction of RNA eluted was normalized (elution with trans azoCm was set to 100%). As shown in Figure 6, aptamer 42 shows the most prominent reduction when comparing elution of the trans-ligand to the cis-ligand (-60%), reaching buffer elution level (-59%). B2–1 shows a 43% reduction, aptamer 1 shows 35% reduction and aptamer 12 shows 42% reduction. To determine the binding affinities of the two best aptamers 42 and B2–1 to both the trans- and cis-azoCm photoisomer, ITC measurements were performed with pre-illuminated azoCm to accumulate the corresponding photoisomer. As depicted in Figure 7A and B, the KD values of 42 and B2–1 binding to trans-azoCm were determined to be 1.9 μM and 545 nM, respectively (Supplementary Table S3). On the contrary, the ITC experiments with cis-azoCm showed no binding curve at all. Thus, an exact KD could not be determined, but considering the total amount of ligand that was titrated to the aptamers, it is reasonable to give an estimate for the lower boundary of the binding strength to the cis-isomer of at least mM. This discrimination is extremely high and with three and four orders of magnitude, respectively, much better than previously selected aptamers against light-switchable compounds.
Figure 6.
Aptamer binding studies. Above: Secondary structures of aptamer candidates 42, B2–1, 1 and 12 derived from common LocARNA analysis. Below: Radioactively labeled aptamer RNA was folded and immobilized on azoCm derivatized columns using the same conditions as during SELEX. Four individual elution steps with 50 μM azoCm (in the trans, cis and cis-trans switched conformation) or buffer were performed. The percentage of eluted RNA versus total RNA was calculated. Each aptamers azoCm trans value was normalized to 100%. Buffer elution values are marked with a line in each graph.
Figure 7.
Biochemical characterization of light-responsive aptamers. ITC measurements of (A) 42 and (B) B2–1 with both the trans- (upper panels) and cis-isomer (lower panels) of azoCm. Measurements were performed using MicroCal iTC200 (aptamer 42) or MicroCal PEAQ iTC200 (aptamer B2–1). CD-monitored alternating photoswitching experiments of azoCm in presence of (C) 42 and (D) B2–1 at 40°C. The red dotted line represents the spectra of the pure RNA-aptamers and the solid colored lines indicate the spectra in the PSS while illuminated with either 420 or 365 nm. The upper panels display difference spectra of the aptamers at both PSS. All CD spectra were corrected for the contribution of the corresponding azoCm photoisomers.
In order to provide evidence, that the aptamers not only binds to trans-azoCm specifically but also releases the ligand upon irradiation, we conducted CD experiments to monitor conformational changes of the RNA upon photoisomerization of azoCm (Figure 7C and D). Generally, the spectral shape of both aptamers can be ascribed to hairpin structures (36–38), exhibiting a pronounced positive CD signal around 270 nm and a minor negative signal at 240 nm. Concerning the ligand azoCm, only the cis-isomer shows a significant CD signal (Supplementary Figure S11). Throughout the depicted experiments, solutions of each RNA construct in presence of 10-fold excess of ligand, were alternatingly illuminated with 420 and 365 nm. As in the case of aptamer 42 no serious spectral variation was detected, we assume that a reversible release of cis-azoCm is not feasible once the trans-isomer is bound (Figure 7C). Yet, in the case of B2–1 a small decrease of the signal at 270 nm of ∼3% was observed upon switching azoCm from trans to cis reversibly, which indicates a dissociation of the ligand accompanied by slight changes in the tertiary structure of the aptamer.
CONCLUSION
In summary, we have developed an RNA aptamer against an azobenzene-derived ligand, which can only bind to the trans-isomer of azoCm but not to its cis-photoisomer. The ligand itself is non-toxic, stable in biological medium and can be photoswitched multiple times without significant degradation and with high photoswitching amplitude. High-quantum yields, low-thermal rates of conversion and spectrally addressable photoisomers make the ligand ideal for an application in a biological context. In addition azoCm retains its switching properties in presence of RNA. A light SELEX protocol based on liberation of RNA after trans- to cis-conversion of azoCm was able to enrich a sequence motif which we had also found in a regular SELEX approach before. However, the RNA found with the latter approach did not perform any liberation of the ligand upon photoisomerization. By next-generation sequencing, we were able to show the enrichment of the photoisomerization-responsive RNA motif only in the light SELEX but not in the regular one performed in parallel. The 13-nt sequence motif identified in the light SELEX is embedded in a conserved a three-way-junction. With a KD of 545 nM, the affinity of aptamer B2–1 to the trans-photoisomer of azoCm is relatively high while the KD toward the cis-photoisomer is at least 1 mM. By CD spectroscopy, we could show that in multiple switching cycles the bound trans-azoCm was liberated and sequestered, repeatedly. All of these properties make this system of RNA aptamer and photoswitchable ligand ideal for many interesting applications, including the further development of a light-sensitive riboswitch.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Britta Schreiber for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB902 A2/A6/A7 to J.W., A.H., B.S.]. Funding for open access charge: Deutsche Forschungsgemeinschaft [SFB902 A2/A6/A7].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ankenbruck N., Courtney T., Naro Y., Deiters A.. Optochemical control of biological processes in cells and animals. Angew. Chem. Int. Ed. Engl. 2018; 57:2768–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brieke C., Rohrbach F., Gottschalk A., Mayer G., Heckel A.. Light-controlled tools. Angew. Chem. Int. Ed. Engl. 2012; 51:8446–8476. [DOI] [PubMed] [Google Scholar]
- 3. Klan P., Solomek T., Bochet C.G., Blanc A., Givens R., Rubina M., Popik V., Kostikov A., Wirz J.. Photoremovable protecting groups in chemistry and biology: reaction mechanisms and efficacy. Chem. Rev. 2013; 113:119–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Griepenburg J.C., Rapp T.L., Carroll P.J., Eberwine J., Dmochowski I.J.. Ruthenium-Caged antisense morpholinos for regulating gene expression in zebrafish embryos. Chem. Sci. 2015; 6:2342–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Husson S.J., Gottschalk A., Leifer A.M.. Optogenetic manipulation of neural activity in C. elegans: from synapse to circuits and behaviour. Biol. Cell. 2013; 105:235–250. [DOI] [PubMed] [Google Scholar]
- 6. Lucas T., Schafer F., Muller P., Eming S.A., Heckel A., Dimmeler S.. Light-inducible antimiR-92a as a therapeutic strategy to promote skin repair in healing-impaired diabetic mice. Nat. Commun. 2017; 8:15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kusebauch U., Cadamuro S.A., Musiol H.-J., Lenz M.O., Wachtveitl J., Moroder L., Renner C.. Photo-controlled folding and unfolding of a collagen triple helix. Angew. Chem. Int. Ed. Engl. 2006; 45:7015–7018. [DOI] [PubMed] [Google Scholar]
- 8. Spörlein S., Carstens H., Satzger H., Renner C., Behrendt R., Moroder L., Tavan P., Zinth W., Wachtveitl J.. Ultrafast spectroscopy reveals sub-nanosecond peptide conformational dynamics and validates molecular dynamics simulation. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7998–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stein M., Middendorp S.J., Carta V., Pejo E., Raines D.E., Forman S.A., Sigel E., Trauner D.. Azo-propofols: photochromic potentiators of GABA(A) receptors. Angew. Chem. Int. Ed. Engl. 2012; 51:10500–10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schonberger M., Trauner D.. A photochromic agonist for mu-opioid receptors. Angew. Chem. Int. Ed. Engl. 2014; 53:3264–3267. [DOI] [PubMed] [Google Scholar]
- 11. Broichhagen J., Frank J.A., Trauner D.. A roadmap to success in photopharmacology. Acc. Chem. Res. 2015; 48:1947–1960. [DOI] [PubMed] [Google Scholar]
- 12. McCown P.J., Corbino K.A., Stav S., Sherlock M.E., Breaker R.R.. Riboswitch diversity and distribution. RNA. 2017; 23:995–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wachter A., Tunc-Ozdemir M., Grove B.C., Green P.J., Shintani D.K., Breaker R.R.. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007; 19:3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berens C., Groher F., Suess B.. RNA aptamers as genetic control devices: the potential of riboswitches as synthetic elements for regulating gene expression. Biotechnol. J. 2015; 10:246–257. [DOI] [PubMed] [Google Scholar]
- 15. Berens C., Suess B.. Riboswitch engineering—making the all-important second and third steps. Curr. Opin. Biotechnol. 2015; 31:10–15. [DOI] [PubMed] [Google Scholar]
- 16. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 17. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 18. Lee H.W., Robinson S.G., Bandyopadhyay S., Mitchell R.H., Sen D.. Reversible photo-regulation of a hammerhead ribozyme using a diffusible effector. J. Mol. Biol. 2007; 371:1163–1173. [DOI] [PubMed] [Google Scholar]
- 19. Young D.D., Deiters A.. Light-regulated RNA-small molecule interactions. Chembiochem. 2008; 9:1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayashi G., Hagihara M., Nakatani K.. RNA aptamers that reversibly bind photoresponsive azobenzene-containing peptides. Chemistry. 2009; 15:424–432. [DOI] [PubMed] [Google Scholar]
- 21. Hayashi G., Hagihara M., Dohno C., Nakatani K.. Reversible regulation of binding between a photoresponsive peptide and its RNA aptamer. Nucleic Acids Symp. Ser. (Oxf). 2007; 93–94. [DOI] [PubMed] [Google Scholar]
- 22. Groher F., Bofill-Bosch C., Schneider C., Braun J., Jager S., Geißler K., Hamacher K., Suess B.. Riboswitching with ciprofloxacin—development and characterization of a novel RNA regulator. Nucleic Acids Res. 2018; 46:2121–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hall B., Micheletti J.M., Satya P., Ogle K., Pollard J., Ellington A.D.. Design, synthesis, and amplification of DNA pools for in vitro selection. Curr. Protoc. Mol. Biol. 2009; doi:10.1002/0471142727.mb2402s88. [DOI] [PubMed] [Google Scholar]
- 24. Groher F., Suess B.. In vitro selection of antibiotic-binding aptamers. Methods. 2016; 106:42–50. [DOI] [PubMed] [Google Scholar]
- 25. Zimmerman G., Chow L.-Y., Paik U.-J.. The photochemical isomerization of azobenzene1. J. Am. Chem. Soc. 1958; 80:3528–3531. [Google Scholar]
- 26. Davis J.H., Szostak J.W.. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11616–11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nutiu R., Li Y.. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. Engl. 2005; 44:1061–1065. [DOI] [PubMed] [Google Scholar]
- 28. Uhlenbeck O.C. Tetraloops and RNA folding. Nature. 1990; 346:613–614. [DOI] [PubMed] [Google Scholar]
- 29. Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berens C., Thain A., Schroeder R.. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001; 9:2549–2556. [DOI] [PubMed] [Google Scholar]
- 31. McKeague M., McConnell E.M., Cruz-Toledo J., Bernard E.D., Pach A., Mastronardi E., Zhang X., Beking M., Francis T., Giamberardino A. et al. Analysis of in vitro aptamer selection parameters. J. Mol. Evol. 2015; 81:150–161. [DOI] [PubMed] [Google Scholar]
- 32. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 33. Levenshtein V. Binary codes capable of correcting deletions, insertions and reversals. Soviet Phys. Doklady. 1966; 10:707–710. [Google Scholar]
- 34. Smith C., Heyne S., Richter A.S., Will S., Backofen R.. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010; 38:W373–W377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Will S., Joshi T., Hofacker I.L., Stadler P.F., Backofen R.. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012; 18:900–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uhlenbeck O.C., Borer P.N., Dengler B., Tinoco I. Jr.. Stability of RNA hairpin loops: A 6 -C m -U 6. J. Mol. Biol. 1973; 73:483–496. [DOI] [PubMed] [Google Scholar]
- 37. Williams D.J., Hall K.B.. Thermodynamic comparison of the salt dependence of natural RNA hairpins and RNA hairpins with non-nucleotide spacers. Biochemistry. 1996; 35:14665–14670. [DOI] [PubMed] [Google Scholar]
- 38. Burke D.H., Hoffman D.C., Brown A., Hansen M., Pardi A., Gold L.. RNA aptamers to the peptidyl transferase inhibitor chloramphenicol. Chem. Biol. 1997; 4:833–843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.