Abstract
Posttranslational modifications of the Forkhead family transcription factor, FOXO1, have been known to have important regulatory implications in its diverse activities. Several types of modifications of FOXO1, including acetylation, phosphorylation, and ubiquitination, have been reported. However, lysine methylation of FOXO1 has not yet been identified. Here, we reported that FOXO1 is methylated by G9a at K273 residue in vitro and in vivo. Methylation of FOXO1 by G9a increased interaction between FOXO1 and a specific E3 ligase, SKP2, and decreased FOXO1 protein stability. In addition, G9a expression was increased by insulin and resulted in insulin-mediated FOXO1 degradation by K273 methylation. Tissue array analysis indicated that G9a was overexpressed and FOXO1 levels decreased in human colon cancer. Cell proliferation assays revealed that G9a-mediated FOXO1 methylation increased colon cancer cell proliferation. Fluorescence-activated cell sorting (FACS) analysis indicated that apoptosis rates were higher in the presence of FOXO1 than in FOXO1 knock-out cells. Furthermore, we found that G9a protein levels were elevated and FOXO1 protein levels were decreased in human colon cancer patients tissue samples. Here, we report that G9a specific inhibitor, BIX-01294, can regulate cell proliferation and apoptosis by inhibiting G9a-mediated FOXO1 methylation.
INTRODUCTION
Forkhead box O (FOXO) family members, FOXO1 (FKHR), FOXO3 (FKHRL1), FOXO4 (AFX) and FOXO6, are multifunctional transcription factors that regulate the transcription of target genes that play critical roles such as tumor suppression, energy metabolism, lifespan extension, apoptosis, and oxidative stress resistance (1–4). Recent studies have shown that by regulating pro-apoptotic genes such as BIM, FasL and TRAIL, FOXOs acts as tumor suppressors resulting in the inhibition of the growth of several tumors including prostate, breast, glioblastoma, and colon cancers (5–9). The expression of FOXO3 is significantly decreased in colon cancer when compared to normal tissue, which correlates to the pathological stage (10).
Many studies have reported that the transcriptional activity of FOXO1 is regulated by posttranslational modifications (PTMs) such as phosphorylation, acetylation, and ubiquitination (4,11–14). For example, the FOXO family is acetylated by p300, CBP and PCAF, and deacetylated by histone deacetylases (HDACs) and Sirtuins (SIRTs) family. FOXO acetylation results in the regulation of transcriptional activity by inhibiting the DNA binding ability or increasing protein stability (12,15). FOXO1 stability is regulated via proteasomal degradation by an E3 ligase, SKP2 and FOXO3 stability is regulated by MDM2 via ERK-mediated phosphorylation (16,17).
G9a (EHMT2) is constituted by the Su(var)3–9, Enhancer-of-zeste, Trithorax (SET) domain and exhibits histone methyltransferase (HMTase) activity. It forms heterodimers with G9a-like protein (GLP; also known as EHMT1) and together, they catalyze H3K9-me1 and H3K9-me2, which are transcriptional repressive histone markers (18,19). Peptide methylation arrays have revealed that G9a also methylates non-histone proteins such as CDYL1, WIZ, and ACINUS (20). In addition, G9a methylates p53 at K373 and regulates apoptosis by preserving p53 in an inactive form (21). Pontin is methylated by G9a/GLP methyltransferase in hypoxic conditions and it potentiates HIF-1α-mediated activation by increasing the recruitment of p300 to a subset of HIF-1α target promoters (22). Moreover, C/EBPβ is methylated by G9a at K39 resulting in the repression of transcriptional activity (23). Reptin, a chromatin-remodeling factor, is methylated by G9a at K67 in hypoxic conditions. Methylated Reptin by G9a binds to the promoters of a subset of hypoxia-responsive genes and negatively regulates their transcription (24).
In this study, we tried to identify specific HMTase responsible for FOXO1 and association with colon cancer. We discovered that G9a interacts with and methylates FOXO1 at K273 in vitro and in vivo. Methylation of FOXO1 by G9a increases the interaction between FOXO1 and SKP2, which is a specific E3 ligase for FOXO1. We observed that G9a was upregulated by insulin signaling, which resulted in decreased FOXO1 protein stability via the G9a-mediated methylation of FOXO1. In addition, we observed that G9a was overexpressed and FOXO1 was underexpressed in human colon cancer tissues when compared to normal tissues. Moreover, attenuated FOXO1 stability via G9a-mediated methylation increased cell proliferation, which was inhibited by the G9a inhibitor, BIX-01294, by decreasing FOXO1 methylation. Consistent with our cell line works, G9a protein levels were increased and those of FOXO1 protein were reduced in human colon cancer patient tissue samples and it is associated with the tumor–node–metastasis (TNM) grade. Collectively, the data suggested that G9a-mediated FOXO1 methylation promoted colon cancer, the inhibition of which could be a potential therapeutic target.
MATERIALS AND METHODS
Plasmid constructs
The p3XFlag-CMV10-FOXO1 and pGEX4T1-FOXO1 deletion constructs were designed as previously described (25). The pGEX4T1-FOXO Δ2, pGEX4T1-FOXO1 Δ2 N-terminal fragment, pGEX4T1-FOXO1 Δ2 C-terminal fragment, pGEX4T1-FOXO1 Δ2-K245R, K248R, K262R, K265R, K272R, K273R, K274R, 2KR (K272R, K273R), p3XFlag-CMV10-FOXO1 K273R, pcDNA3.0-Flag-G9a, Flag-G9a-685C, Flag-G9a-936C, pEGFP-G9a, pEGFP-G9a ΔSET, pGEX4T1-G9a SET and HA-Ub were used in each experiment.
Cell culture and transient transfection
293T cells, shG9a stable 293T cells, MEF WT, and MEF G9a knock-out cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco), and HCT116, DLD-1, and SW480 cells were grown in RPMI 1640 medium (Gibco) containing 10% heat-inactivated fetal bovine serum (Welgene) and 0.05% penicillin-streptomycin (Welgene) at 37°C in 5% CO2. HCT116 cells and 293T cells were transfected with the indicated DNA constructs using polyethylenimine (PEI) (Polyscience).
Reagents
BIX-01294 was purchased from Santa Cruz Biotechnology. MG132 (20 μM) and cycloheximide (100 μM) were purchased from Enzo Life Science.
RNA interference
This procedure was previously described (26). Briefly, DNA oligonucleotides encoding G9a shRNA (5′-CACACATTCCTGACCAGAGAT-3′) were subcloned into pLKO.1-puro (Addgene) lentiviral vector according to standard procedures. To produce the viral particles, 293T cells were cotransfected with plasmids encoding VSV-G, NL-BH and the shRNAs. Two days after transfection, the media containing the viruses were collected and used to infect cells in the presence of polybrene (8 μg/ml).
Histone methyltransferase (HMTase) assay
GST-FOXO1 WT, GST-FOXO1 deletion constructs, and/or GST-FOXO1 point mutants were incubated overnight at 30°C with GST-G9a in the presence of 40 nCi of S-adenosyl-[methyl-14C]-l-methionine [14C-SAM] (Perkin Elmer) in HMTase assay buffer (50 mM Tris–HCl [pH 8.5], 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 1.25 M sucrose). Reaction products were separated by SDS-PAGE and analyzed by a phosphorimager.
Antibodies
Antibodies against FKHR (FOXO1; sc-11350, sc-3.74427), H3 (sc-8654), β-actin (sc-47778), GFP (sc-9996), HA (sc-805), Ub (sc-166553; Santa Cruz Biotechnology), β-tubulin (T4026), Flag (F3165; SIGMA), EZH2 (3147S; Cell signaling), Methyl-lysine (Me-K; ab-7375; Abcam), H3K9-me2 (07-441) and G9a (07-551; Millipore) were used for western blotting and immunoprecipitation (IP) analysis.
Immunoprecipitation (IP)
To determine the level of FOXO1 methylation in vivo and for the FOXO1 and G9a binding assays, transfected cells were lysed in RIPA buffer (50 mM Tris–HCl [pH 8.0], 150 mM NaCl, 0.1% SDS, 0.5% SDC, 1% NP-40, 1× protease inhibitor cocktail, and 1 mM EDTA) and immunoprecipitated overnight at 4°C with indicated antibodies in IP buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM PMSF and 1× protease inhibitor cocktail). Protein A/G agarose beads (GenDEPOT) were added for 2 h with agitation at 4°C. Bound proteins were eluted and analyzed by immunoblotting with indicated antibodies.
GST-pull-down assay
Cell lysates, ectopically expressing pEGFP-G9a in 293T cells, were incubated with either GST-FOXO1 or GST-FOXO1 deletion mutants in TNT reaction buffer (50 mM Tris–HCl [pH 7.6], 150 mM NaCl, 0.1% Triton X-100). Next, the protein complexes were washed three times with TNT washing buffer (50 mM Tris–HCl [pH 7.6], 300 mM NaCl, 0.5% Triton X-100). Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies.
LTQ-orbitrap mass spectrometry
Samples were separated by SDS-PAGE and isolated via gel cutting. After an overnight trypsin or chymotrypsin digestion, the eluted peptides were separated using a C18 column with a linear gradient (A: 100% H2O, 0.1% formic acid, B: 100% ACN) at a flow rate of 300 nl/min. Typically, 2 μl of sample was injected. Mass spectrometry was performed with a dual-mass spectrometer (LTQ Orbitrap Velos; Thermo Scientific) coupled to a nano-LC system (EASY nLC; Thermo Scientific). This method consisted of a cycle combining one full MS scan (mass range: 150–2000 m/z). Proteins were identified by searching the MS/MS spectra using SEQUEST.
Subcellular fractionation
Preparation of nuclear and cytosolic fractions was carried out by lysing cells for 10 min on ice using buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 1× protease inhibitor cocktail, and 0.4% NP-40), followed by centrifugation at 15,000 × g for 3 min. Supernatants were retained as cytosolic fractions, whereas the pellets were subjected to further lysis in buffer B (20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, 0.5 mM PMSF and 1× protease inhibitor cocktail). The pelleted material was then resuspended by pipetting. After a 2 h agitation at 4°C, lysates were centrifuged at 15 000 × g, and the resulting supernatants were collected as nuclear fractions.
Reverse transcription and real-time PCR
Total RNA was isolated from transfected cells using RNAiso Plus (TaKaRa). After synthesis, cDNA was quantified and then subjected to FOXO1, G9a, and HNF4a mRNA expression analysis. The following PCR primers were used: FOXO1 forward, 5′-TACGAGTGGATGGTCAAGAG-3′; reverse, 5′-ATGAACTTGCTGTGTAGGGAC-3′, G9a forward, 5′-GAGCGAGGGTTTGAGGAGTT-3′; reverse, 5′-TGATGCGGTCAATCTTGGGT-3′, HNF4a forward, 5′-ACAGATGTCCACCCCTGAGA-3′, reverse 5′-AGAGGGGCTTGACGATTGTG-3′. The following FOXO1 PCR primers were used: Disassociation curves were performed after each PCR run to ensure that a single product of the appropriate length was amplified. Mean threshold cycle (CT) and standard error values were calculated from individual CT values obtained from triplicate reactions per stage. The normalized mean CT value (ΔCT) was estimated by subtracting the mean CT of β-actin. The value ΔΔCT was calculated as the difference between control ΔCT and values obtained for each sample. The n-fold change in gene expression, when compared to a control, was calculated as 2-ΔΔCT.
Chromatin immunoprecipitation (ChIP) assay
ChIP analyses were performed as described in the literature (1). Briefly, HCT116 cells were transfected with the indicated plasmids and harvested after incubation for 48 h. The cells were cross-linked by addition of 1% formaldehyde to the medium and incubation for 10 min at 37°C, followed by addition of 125 mM glycine and incubation for 5 min at room temperature. The cells were then lysed in SDS lysis buffer, and the samples were sonicated and immunoprecipitated using the indicated antibodies. The immunoprecipitates were eluted and reverse cross-linked. The DNA fragments were then purified and PCR amplified for quantification. The following PCR primers were used G9a forward, 5′-GCAGAACTGAGGCCAGGAAT-3′; reverse, 5′-AAGGAGCCGCTACATGCTTT-3′, Disassociation curves were generated after each PCR run to ensure amplification of a single product of the appropriate length. The mean threshold cycle (CT) and standard error values were calculated from individual CT values, obtained from duplicate reactions per stage. The normalized mean CT value was estimated as ΔCT by subtracting the mean CT of the input from that of HNF4a.
In vivo ubiquitination assay
Cells were transfected with indicated plasmids using PEI and harvested 48 h later. MG132 (Enzo Life Science; 20 μM) was added to cells 6 h before lysis in modified RIPA buffer (10 mM Tris–HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.025% SDS, 1× protease inhibitor cocktail) as described previously (27). Ubiquitinated protein was immunoprecipitated overnight at 4°C with anti-HA antibody in IP buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM PMSF and 1 × protease inhibitor cocktail). Protein A/G agarose beads (GenDEPOT) were added for 2 h with agitation at 4°C. Bound proteins were eluted and analyzed by immunoblotting with anti-Flag antibody.
Fluorescence-activated cell sorting (FACS) analysis
HCT116 and FOXO1 KO HCT116 cells were treated with BIX-01294 for 24 h. Immediately before FACS analysis, the cells were treated with RNase A (20 mg/ml) and stained with Annexin V-FITC (BD bioscience) and propidium iodide (PI) (BD bioscience) for 30 min. Cells were then subjected to FACS analysis using a BD Accuri™ C6 Plus Flow Cytometer (BD bioscience).
CRISPR/Cas9 KO system
A guide sequence (5′-GCGCGAGCTCAATGACCGGC-3′) targeting the first exon of FOXO1 was selected from the CRISPR design web site (http://crispr.mit.edu). Two complementary oligos containing the FOXO1 guide sequence and BsmBI ligation adapters were synthesized. Each oligo was phosphorylated and annealed using T4 polynucleotide kinase (New England Biolabs). The annealed oligo was ligated by T4 DNA ligase (Enzynomcis) to lentiCRISPRv2 vector. The lentiCRISPRv2 or lentiCRISPRv2-gRNA FOXO1 construct was transfected by PEI in HCT116 cells. After transfection for 48 h, selection was performed with 500 ng/ml of puromycin (Sigma) for 3 days. Selected cells by puromycin were seeded a single cell. FOXO1 knock-out was confirmed by western blotting and sequencing.
Tissue array
Formalin-fixed, paraffin-embedded tissue array slides containing colon cancer and normal tissues were purchased from US BIOMAX. Briefly, after deparaffinization in xylene and rehydration in graded ethanol, endogenous peroxidase activity was blocked by incubating with 3% hydrogen peroxide for 10 min. Next, tissue sections were heated in 100 mM citrate buffer (pH 6.0) for 10 min to retrieve antigens and then preincubated with normal horse serum for 20 min at room temperature. Anti-FOXO1 and anti-G9a antibodies (diluted 1:100) were used as the primary antibodies. The specimens were subsequently incubated with biotinylated anti-rabbit secondary antibody (Vectastain Laboratories) and streptavidin–horseradish peroxidase (Zymed Laboratories Inc.). DAB (3,3-diaminobenzidine; Vectastain Laboratories) was used as a chromogen, and eosin was used for counterstaining.
Soft agar assay
For the soft agar colony formation assay, 5 × 103 cells were plated in a six-well culture dish as a suspension in 1.5 mL of 2 × RPMI containing 10% FBS and 0.35% agar on a layer of 1.5 ml of the same medium containing 0.5% agar. The cells were treated with 1 μM BIX-01294. The plates were incubated for 2–3 weeks until colonies formed.
Colon cancer patient tissue samples
This study utilized tissue samples derived from human colon or rectum (n = 35; n = 12 for TNM stage I, n = 12 for TNM stage II, n = 11 for TNM Stage III and n = 12). Normal tissues were extracted from adjacent sections of cancer tissues (n = 6). All patient tissue samples were obtained under Institutional Review Board (IRB)–approved protocols. Written informed consent was obtained from all patients. All tissue samples were deidentified, and protected health information was reviewed according to the Health Insurance Portability and Accountability Act guidelines. The biospecimens and data used for this study were provided by the Biobank of Chonnam National University Hwasun Hospital, a member of the Korea Biobank Network. Tissue samples were homogenized and lysed in RIPA buffer (50 mM Tris–HCl [pH 8.0], 150 mM NaCl, 0.1% SDS, 0.5% SDC, 1% NP-40, 1× protease inhibitor cocktail, and 1 mM EDTA). After a 2 h agitation at 4°C, samples were centrifuged at 20 000 × g, and the resulting supernatants were collected.
Statistical analysis
Data are expressed as mean ± SD of three or more independent experiments. Statistical significance (P < 0.05) was calculated using Microsoft Excel. Differences between groups were evaluated by a Student's t-test.
RESULTS
The methylation of FOXO1 by G9a
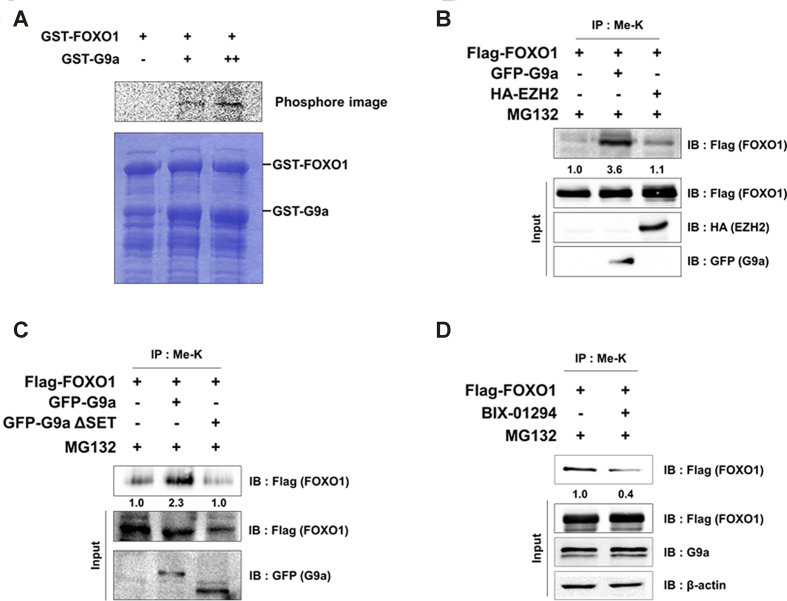
Although PTMs of FOXO1 such as phosphorylation, acetylation, and ubiquitination has been discovered (28–30), the only known instance of FOXO1 methylation until now was the arginine methylation by PRMT1, which blocks FOXO1 phosphorylation and results in nuclear accumulation (31). Lysine methylation of proteins plays an important role in the transcriptional regulation and protein stability of transcription factors such as p53, E2F1, androgen receptor (AR), and Reptin (24,32–34). Therefore, we hypothesized that lysine methylation of FOXO1 is also important to its function. To identify the HMTase specific for FOXO1, we initially screened several HMTases including G9a for FOXO1 methylation activity. We performed the HMTase assay using purified FOXO1 along with HMTases including G9a, SMYD2, EZH2, SET8 and WHSC1. Among the tested HMTases, only G9a could methylate FOXO1 (Supplementary Figure S1A). To further confirm FOXO1 methylation by G9a, we conducted the HMTase assay using purified GST-FOXO1 and GST-G9a SET domain. Again, FOXO1 was methylated by G9a in a dose-dependent manner (Figure 1A). To determine whether G9a methylated FOXO1 in vivo, we checked the methylation level of FOXO1 via IP assay in HCT116 cells. We found an increase of FOXO1 methylation levels in G9a-overexpressed cells, but not EZH2-overexpressed cells (Figure 1B). When G9a wild type (G9a WT) was overexpressed, G9a WT increased FOXO1 methylation however, SET domain deleted G9a (G9a ΔSET) fail to increase FOXO1 methylation (Figure 1C). To further investigate whether endogenous methylation of FOXO1 was mediated by G9a, we treated with BIX-01294 and measured FOXO1 methylation levels in colon cancer cell lines. As expected, FOXO1 methylation levels were decreased in BIX-01294 treated HCT116, DLD-1 and SW480 cells, which indicated that G9a catalyzed the methylation of FOXO1 in vivo (Figure 1D, Supplementary Figure S1B and S1C). This concentration of BIX-01294 is enough to inhibit G9a activity toward H3K9-me2 (Supplementary Figure S1D). Altogether, these results suggest that FOXO1 is methylated by G9a, both in vitro and in vivo.
Figure 1.
FOXO1 is methylated by G9a. (A) Purified GST-FOXO1 was incubated overnight at 30°C with increasing amounts of GST-G9a SET. Reaction mixtures were separated by SDS-PAGE and analyzed via a phosphorimager. (B and C) HCT116 cells were transfected with the indicated plasmids and immunoprecipitated with anti-methyl lysine antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag, anti-HA, and anti-GFP antibodies. Methyl lysine levels were normalized by input of FOXO1. (D) HCT116 cells were transfected with the indicated plasmids and immunoprecipitated with anti-methyl lysine antibodies. After 24 h of transfection, the cells were treated with 5 μM BIX-01294 for 24 h. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag, anti-G9a and anti-β-actin antibodies. Methyl lysine levels were normalized by input of FOXO1.
G9a-mediated methylation of FOXO1 at lysine 273 in vitro and in vivo
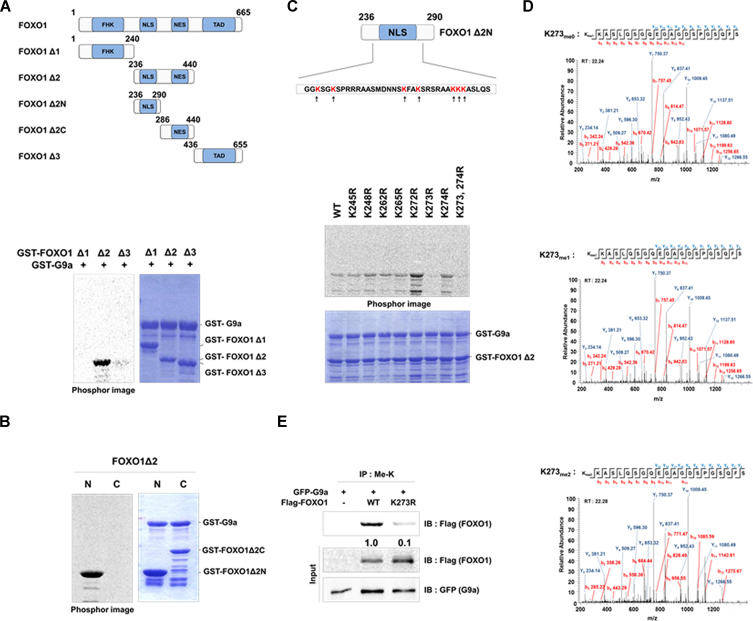
To precisely identify the methylation site(s) of FOXO1, we conducted the HMTase assay using FOXO1 deletion constructs FOXO1 Δ1, Δ2, and Δ3 (Figure 2A, upper panel). Among the deletion constructs tested, only FOXO1 Δ2, which spanned amino acids 236–440, was methylated by G9a (Figure 2A, lower panel). We further generated FOXO1 Δ2 N- and C-terminus fragments to perform the HMTase assay (Figure 2A, upper panel). Among the two FOXO1 Δ2 deletion constructs, only the FOXO1 Δ2 N-terminus fragment was methylated by G9a (Figure 2B). There are seven lysine residues within the FOXO1 Δ2 N-terminus fragment (Figure 2C, upper panel). Each lysine residue was individually mutated to arginine and FOXO1 point mutants were tested for G9a-mediated methylation (Figure 2C, lower panel). Among the tested FOXO1 Δ2 point mutants, G9a-mediated methylation was abrogated in the FOXO1 K273R mutant, thereby indicating FOXO1 K273 as the methylation site targeted by G9a (Figure 2C, lower panel). Additionally, we performed the LTQ-orbitrap mass spectrometry analysis of the FOXO1 Δ2 fragment, which indicated that G9a carried out both, mono- and di-methylation of FOXO1 K273 (Figure 2D). To confirm this data in vivo, we compared the methylation levels between FOXO1 WT and FOXO1 K273R. Methylation of FOXO1 K273R was lower when compared to that of FOXO1 WT (Figure 2E). In addition, previous results demonstrated that endogenous FOXO1 methylation was observed by IP assay in DLD-1 and SW480 cells (Supplementary Figure S1B and C). To further identify FOXO1 methylation in vivo, we conducted the LTQ-orbitrap mass spectrometry analysis in FOXO1 transfected HCT116 cells. As expected, FOXO1 methylation at K273 was detected in vivo (Supplementary Figure S2A). FOXO1 at K273 is a conserved residue in the human FOXO family and is evolutionarily preserved between Mus musculus, Sus scrofa, Xenopus laevis, and Danio rerio indicating the functional importance of the residue (Supplementary Figure S2B). Altogether, these data suggest that G9a carries out mono- and di-methylation of FOXO1 at K273 in vitro and in vivo.
Figure 2.
G9a methylates FOXO1 at K273. (A) The figure represents the domains of the FOXO1 deletion constructs (upper panel). Purified GST-FOXO1 deletion mutants were incubated overnight at 30°C with GST-G9a SET. Reaction mixtures were separated by SDS-PAGE and analyzed via a phosphorimager (lower panel). (B) Purified GST-FOXO1 deletion mutants were incubated overnight at 30°C with GST-G9a SET. Reaction mixtures were separated by SDS-PAGE and analyzed via a phosphorimager. (N: nucleus, C: cytoplasm) (C) Lysine residues of the FOXO1 Δ2 N-terminus protein are indicated (upper panel). Purified GST-FOXO1 Δ2 or GST-FOXO1 Δ2 point mutants were incubated overnight at 30°C with GST-G9a SET. Reaction mixtures were separated by SDS-PAGE and analyzed via a phosphorimager (lower panel). (D) The LTQ-orbitrap mass spectrometry analysis of the FOXO1 Δ2 protein identified mono- and di- methylation of the K273 residue. (E) HCT116 cells were transfected with the indicated plasmids and immunoprecipitated with anti-methyl lysine antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag and anti-GFP antibodies.
Interaction between FOXO1 and G9a
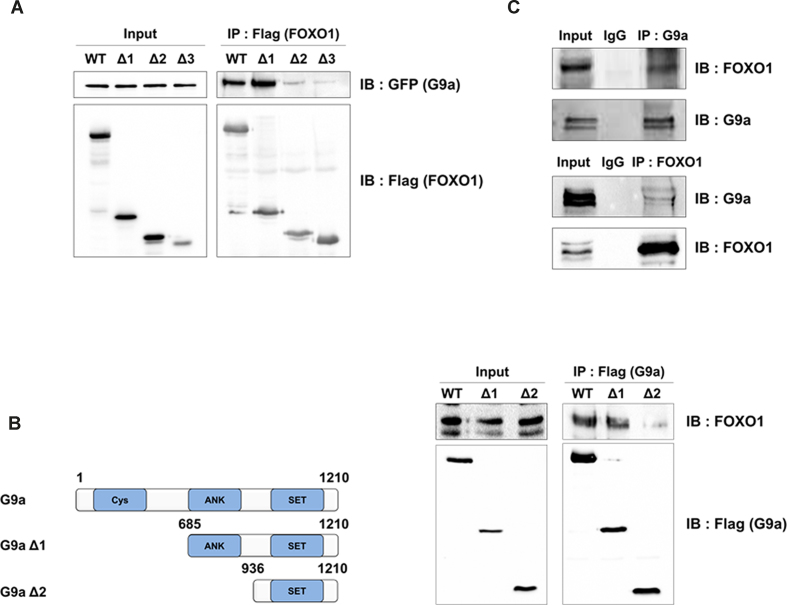
Next, we examined the interaction between FOXO1 and G9a to determine the mechanism of G9a-mediated methylation. To test the interaction between the two proteins in vivo, we performed an IP assay with anti-Flag antibodies (FOXO1) in 293T cells that ectopically expressed FOXO1 WT or FOXO1 deletion constructs. G9a strongly interacted with FOXO1 WT and FOXO1 Δ1 (both of which contained the Forkhead domain) and showed weak interaction with FOXO1 Δ2 and Δ3 (both of which lacked the Forkhead domain; Figure 3A). The Forkhead domain is known as a DNA binding domain and has been suggested to play a role in protein-protein interaction (35). To identify the G9a domain involved in this interaction, we performed another IP assay using G9a deletion constructs. When G9a WT and deletion constructs were immunoprecipitated with anti-Flag antibodies (G9a), we observed that FOXO1 interacted with G9a WT and G9a Δ1 (which contained the ANK and SET domains), but weakly interacted with G9a Δ2 (Figure 3B). Next, to determine whether these proteins interacted endogenously, we performed a co-IP assay by using antibodies against G9a and FOXO1. The results showed that endogenous G9a could interact with endogenous FOXO1 (Figure 3C). Furthermore, we performed IP assay to determine where G9a and FOXO1 interacted. After HCT116 cells were separated into the cytoplasm and nuclear fractions, the cell lysates were co-immunoprecipitated with FOXO1. We observed that FOXO1 interacted with G9a in the nucleus (Supplementary Figure S3). Collectively, the data demonstrated that the Forkhead domain of FOXO1 interacted with the ANK domain of G9a in the nucleus in vivo.
Figure 3.
FOXO1 interacts with G9a. (A) 293T cells were transfected with the indicated plasmids and immunoprecipitated with anti-Flag antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag or anti-GFP antibodies. (B) Schematic diagram of G9a deletion constructs (left panel). 293T cells were transfected with the indicated plasmids and immunoprecipitated with anti-Flag antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag or anti-FOXO1 antibodies (right panel). (C) Endogenous interactions between FOXO1 and G9a were examined in 293T cells. 293T cell lysates were immunoprecipitated with anti-FOXO1 or anti-G9a antibodies and associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-G9a or anti-FOXO1 antibodies.
G9a-mediated FOXO1 methylation decreased FOXO1 stability
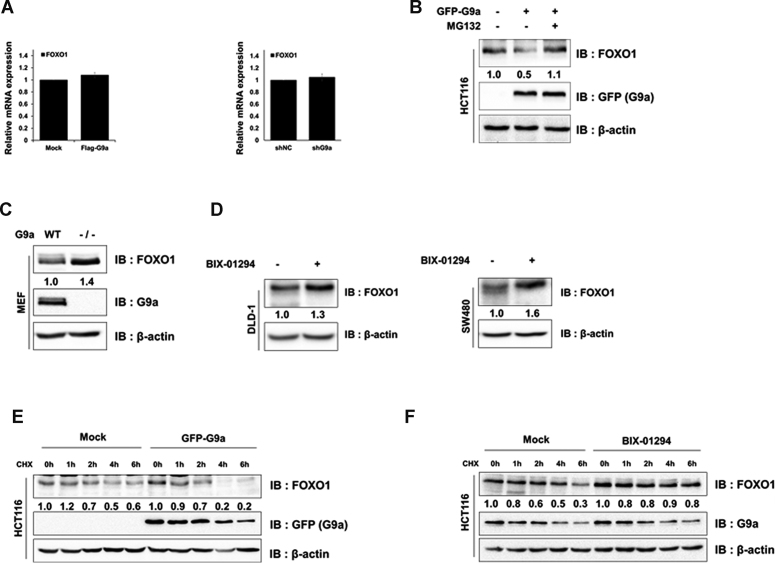
Since previous studies reported that FOXO1 phosphorylation regulated its subcellular localization (4,12), we analyzed whether G9a-mediated methylation altered this feature. Cell fractionation and IP analysis indicated that FOXO1 methylation did not seem to affect FOXO1 subcellular localization (Supplementary Figure S4A). Since another report demonstrated that FOXO3 protein stability was regulated by SET9-mediated methylation (36), we hypothesized that G9a-mediated FOXO1 methylation attenuates FOXO1 stability. First, we checked the FOXO1 mRNA and protein levels in G9a-overexpressed cells. Even though G9a does not regulate FOXO1 transcription (Figure 4A), protein expression patterns were regulated in G9a overexpressed or depleted 293T cells (Supplementary Figures S4B and C). Next, we decided to test FOXO1 stability via G9a-mediated methylation in colon cancer cells. Interestingly, FOXO1 protein levels were decreased when G9a was overexpressed in HCT116 cells (Figure 4B). Reduction of FOXO1 protein levels by G9a were markedly recovered by the proteasome inhibitor MG132, suggesting that the FOXO1 K273 methylation possibly regulated the stability of FOXO1 via proteasomal degradation (Figure 4B and Supplementary Figure S4D). In addition, we confirmed that the decrease of FOXO1 stability was dependent on the G9a SET domain in HCT116 and MEF cells (Supplementary Figures S4E and S4F). The hypothesis was further supported by increased FOXO1 protein levels in MEF G9a null cells (Figure 4C). Moreover, FOXO1 protein level was elevated in BIX-01294 treated DLD-1 and SW480 cells (Figure 4D). Next, we measured FOXO1 stability in various cell lines in the presence of a protein synthesis inhibitor, cycloheximide (CHX). FOXO1 stability was attenuated when G9a was overexpressed in HCT116 and 293T cells (Figure 4E and Supplementary Figure S4G), while it was maintained for up to 4 h in G9a knock-down cells, which supports the reduced FOXO1 stability via G9a-mediated methylation (Supplementary Figure S4H). Again, FOXO1 stability was enhanced in HCT116 and 293T cells treated with BIX-01294 (Figure 4F and Supplementary Figure S4I). Also, we obtained consistent results in DLD-1 cells (Supplementary Figure S4J). Therefore, these results suggest that G9a-mediated methylation attenuates FOXO1 protein stability via proteasomal degradation.
Figure 4.
Methylation of FOXO1 by G9a induces proteasomal degradation. (A) The mRNA levels of FOXO1 in transfected with GFP-G9a HCT116 or shG9a stable 293T cells, were analyzed by qRT-PCR and normalized by β-actin. (B) HCT116 cells transfected with GFP-G9a or empty vector were lysed, immunoblotted with anti-FOXO1, anti-GFP and anti-β-actin antibodies, and treated with 20 μM of MG132 for 6 h. (C) MEF WT and MEF G9a (–/–) cells were lysed and immunoblotted with anti-FOXO1, anti-G9a, and anti-β-actin antibodies. (D) DLD-1 (left panel) and SW480 (right panel) cells were lysed and immunoblotted with anti-FOXO1, and anti-β-actin antibodies. (E and F) HCT116 cells transfected with GFP-G9a or empty vector (E) and BIX-01294 treated cells (F) were subjected to CHX (100 μg/ml), and harvested at the indicated time points. Cells were lysed and immunoblotted with anti-GFP, anti-FOXO1, anti-G9a, and anti-β-actin antibodies.
G9a-mediated methylation induced poly-ubiquitination of FOXO1
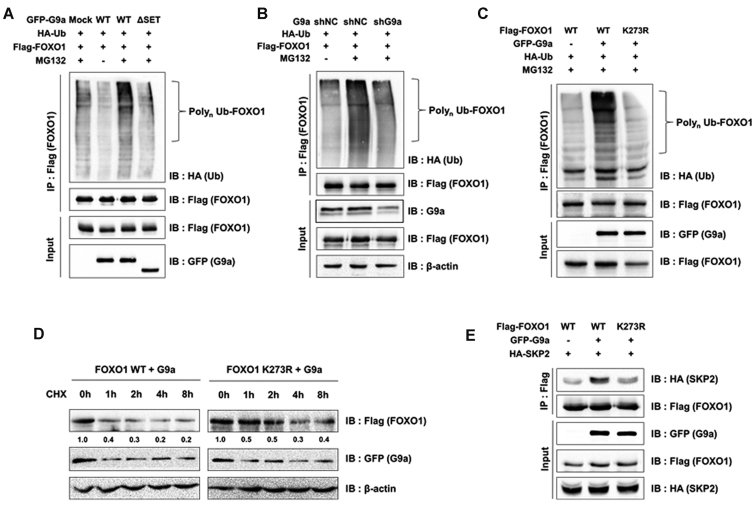
Next, we studied the mechanism of methylated FOXO1 degradation. It is interesting that increased FOXO1 poly-ubiquitination was observed when G9a was overexpressed (Figure 5A). G9a ΔSET did not induce FOXO1 poly-ubiquitination, indicating that ubiquitination of FOXO1 was dependent on the G9a HMTase activity (Figure 5A). Consistent results were obtained in shG9a stable 293T cells (Figure 5B). Therefore, it was reasonable to speculate that FOXO1 methylation at K273 probably affected the poly-ubiquitination of FOXO1. To test this, we conducted ubiquitination assays using FOXO1 K273R mutant. The results showed that FOXO1 poly-ubiquitination was inhibited when FOXO1 K273R was overexpressed in 293T and HCT116 cells (Figure 5C and Supplementary Figure S5A). The poly-ubiquitination of FOXO1 was decreased in G9a knocked out MEF cells compared to G9a WT MEF cells (Supplementary Figure S5B). Consistent with poly-ubiquitination data, protein stability of FOXO1 K273R was increased despite of G9a overexpression (Figure 5D). A previous study demonstrated that SKP2, an oncogenic subunit of the Skp1/Cul1/F-box protein ubiquitin complex, promotes the degradation of FOXO1 via proteasomal degradation (17). Therefore, we further questioned whether E3 ubiquitin ligase SKP2 might be involved in G9a-mediated FOXO1 degradation. We reasoned that FOXO1 methylation by G9a might enhance the interaction between FOXO1 and SKP2 and therefore, tested this possibility. The interaction between FOXO1 and SKP2 substantially increased when G9a was overexpressed, while the FOXO1 K273R mutant attenuated this interaction (Figure 5E). Cumulatively, the data indicated that FOXO1 methylation by G9a promoted poly-ubiquitination of FOXO1 and its proteasomal degradation via increased interaction with SKP2.
Figure 5.
FOXO1 methylation increases interaction with SKP2. (A) 293T cells were transfected with the indicated plasmids. After treatment with MG132 (20 μM) for 6 h, cells were lysed and immunoprecipitated with anti-Flag antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-HA, anti-Flag, and anti-GFP antibodies. (B) shG9a stable 293T cells were transfectedf with the indicated plasmids. After treatment with MG132 (20 μM) for 6 h, cells were lysed and immunoprecipitated with anti-Flag antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-HA, anti-Flag, anti-β-actin, and anti-G9a antibodies. (C) 293T cells were transfected with the indicated plasmids. After treatment with MG132 (20 μM) for 6 h, cells were lysed and immunoprecipitated with anti-Flag antibody. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-HA, anti-Flag, and anti-GFP antibodies. (D) 293T cells transfected with the indicated plasmids were subjected to CHX (100 μg/ml) and harvested at the indicated time points. Cells were lysed and immunoblotted with anti-Flag, anti-GFP and anti-β-actin antibodies. (E) 293T cells were transfected with the indicated plasmids and immunoprecipitated by anti-Flag antibody. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag, anti-HA, and anti-GFP antibodies.
Insulin enhanced G9a-mediated FOXO1 methylation
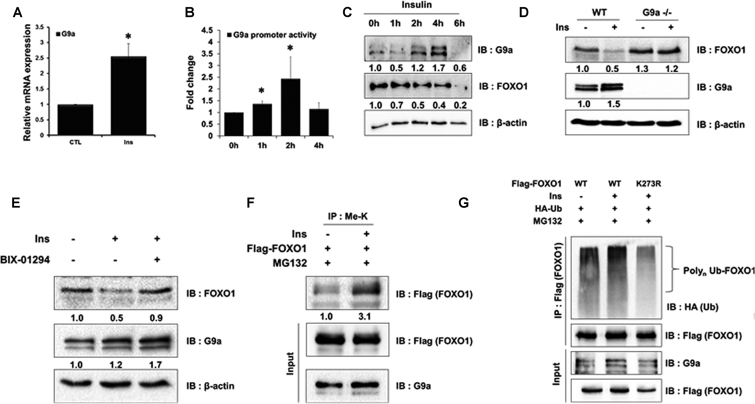
Several studies have associated hyperinsulinemia with colon and prostate cancers (37–39). For instance, the top 25% of C-peptide, which is an indicator of hyperinsulinemia, increased a 3-fold higher risk of colon cancer (40,41). Moreover, a previous study also reported that FOXO1 was degraded in insulin-treated cells (42). Therefore, we hypothesized that G9a-mediated FOXO1 degradation led to tumorigenesis via insulin activity. First, we tested whether G9a was induced by insulin. When HCT116 cells were treated with insulin, G9a transcription and promoter activity were increased (Figure 6A and 6B). Consistent with the transcription data, G9a protein levels were increased by insulin (Figure 6C, Supplementary Figure S7A and S7B). Next, we tested the effect of insulin signaling in G9a knock-out MEF cells. G9a protein levels were increased and FOXO1 protein levels were decreased in the presence of insulin in wild-type MEF cells. However, FOXO1 protein levels were not changed in G9a null cells despite of insulin treatment (Figure 6D). Furthermore, insulin-mediated reduced FOXO1 protein levels were restored in BIX-01294 treated HCT116 cells (Figure 6E). Next, we tested whether FOXO1 degradation by insulin was associated with G9a-mediated FOXO1 methylation. To our surprise, FOXO1 methylation levels were increased upon treatment with insulin (Figure 6F). These data indicate that insulin-mediated FOXO1 degradation was induced by G9a-mediated FOXO1 methylation. Next, we tested whether insulin treatment increased poly-ubiquitination of FOXO1. As expected, FOXO1 poly-ubiquitination levels were increased in insulin-treated cells. Again, FOXO1 K273R mutants did not experience increased poly-ubiquitination (Figure 6G). Altogether, the data suggested that FOXO1 degradation by insulin signaling required G9a-mediated FOXO1 methylation.
Figure 6.
FOXO1 methylation by G9a and insulin-induced increase in G9a expression. (A) HCT116 cells were analyzed by real-time qPCR to examine the mRNA expression levels of G9a. Cells were untreated or treated with insulin (Ins; 600 nM) for 6 h. Results are shown as means ± SD; n = 3. *P < 0.05, (B) HCT116 cells were co-transfected with the G9a promoter-luc construct, treated with insulin, and harvested at the indicated time points. Cell extracts were then assayed for luciferase activity, which was normalized to β-galactosidase. Results are shown as means ± SE; n = 3. *P < 0.05. (C) HCT116 cells were treated with insulin (600 nM) and harvested at the indicated time points. Cells were lysed and immunoblotted with anti-FOXO1, anti-G9a, and anti-β-actin antibodies. (D) MEF WT and MEF G9a null cells were treated with insulin (600 nM) for 6 h. Cells were lysed and immunoblotted with anti-FOXO1, anti-G9a, and anti-β-actin antibodies. (E) HCT116 cells were treated with insulin (600 nM) for 6 h and BIX-01294 (5 μM) for 24 h. Cells were lysed and immunoblotted with anti-FOXO1, anti-G9a, and anti-β-actin antibodies. (F) HCT116 cells were transfected with the indicated plasmids and immunoprecipitated with anti-methyl lysine antibodies. Associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-Flag and anti-G9a. (G) HCT116 cells were transfected with the indicated plasmids. After treatment with MG132 (20 μM) and insulin (600 nM) for 6 h, associated proteins were eluted, resolved by SDS-PAGE, and immunoblotted with anti-HA, anti-Flag and anti-G9a antibodies.
HNF4a-mediated induction of G9a expression by insulin
Next, we investigated the mechanism of G9a induction by insulin. We searched binding proteins on the G9a promoter using cisRED program (http://www.cisred.org/human9). We found several binding motifs for proteins including HNF4, HNF3, STAT5A, MRF-2 and PAX-6 on G9a promoter. Previous report suggests that FOXO1 inhibits DNA binding ability of HNF4 via interacting between FOXO1 and HNF4. However, the interaction is dissociated when insulin was treated and resulted in increase of HNF4a activity (43). Additionally, microarray data (GSE69639) showed that HNF4a was increased 2.4-fold in HT29 human colon cancer cell when insulin was treated (44). Therefore, we hypothesized that induction of HNF4a expression by insulin may regulate G9a expression. To test this hypothesis, we first measured HNF4a expression levels by qPCR in insulin treated HCT116 cells. Consistent with the microarray data, HNF4a was increased in HCT116 cells by insulin (Supplementary Figure S7A). Next, we tested whether G9a mRNA and protein expression levels were increased in HNF4a transiently transfected HCT116 cells. Indeed, the results showed that HNF4a induced both mRNA and protein of G9a (Supplementary Figure S7B). To further confirm these results, we measured G9a promoter activity in HNF4a overexpressed cells. G9a promoter activity was elevated by HNF4a in a dose dependent manner (Supplementary Figure S7C). We next determined whether HNF4a is directly recruited to G9a promoter by ChIP assay. The result showed that highly recruited HNF4a at G9a promoter when HNF4a was overexpressed (Supplementary Figure S7D). These data suggest that HNF4a increases G9a expression via recruitment to G9a promoter when insulin was treated.
G9a-mediated FOXO1 methylation promotes colon cancer cell proliferation
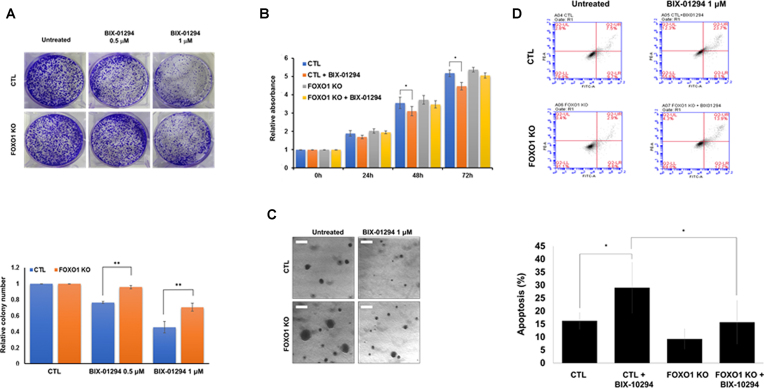
Previous reports showed that the decreased transcriptional activity of FOXO1 promoted tumorigenesis in various cancers such as breast, lung, and prostate cancer (5,7,17). G9a also promoted tumorigenesis and was overexpressed in different types of cancer (21,45,46). In addition, abnormal induction of insulin promoted aberrant crypt foci, which are the earliest changes seen in the colon leading to cancer (47,48). Therefore, we tested whether G9a regulated cell proliferation and apoptosis by negatively regulating FOXO1 stability. To test this, we generated FOXO1 knock-out (KO) HCT116 cells via the CRISPR/Cas9 system (Supplementary Figure S8), and conducted colony formation assay with or without BIX-01294 treatment. The number of colonies decreased in BIX-01294 treated control cells; however, we observed that the decrease in the number of colonies in the FOXO1 KO cells was less than that of control cells (Figure 7A). To further confirm these observations, we conducted MTT cell proliferation assay. Consistent with the colony formation assay result, control cells showed decreased cell proliferation in BIX-01294-treated G9a inactive cells when compared to FOXO1 KO cells (Figure 7B). Likewise, FACS analysis also showed that apoptosis was further induced in BIX-01294-treated HCT116 control cells (from 16.3% to 29%) than in FOXO1 KO cells (from 9.3% to 15.7%) (Figure 7C). Moreover, we measured anchorage independent cell growth via soft agar assay. Consistent with other proliferation assays, the result showed that BIX-01294 increasingly inhibited cell growth in control cells than in FOXO1 KO cells (Figure 7D).
Figure 7.
G9a-mediated FOXO1 methylation regulates colon cancer proliferation. (A) HCT116 cells (5 × 103) were treated with BIX-01294 (1 μM). After 5 days, colony numbers were measured by Quantity One software (colony counting; sensitivity: 5, averaging: 5) (B) Cell viability was determined via MTT assay. HCT116 cells and HCT116 FOXO1 KO cells were either treated or untreated with BIX-01294 (1 μM) for 24 h. Results are shown as means ± SD; n = 3. **, P < 0.01. (C) Apoptotic cells were measured by FACS analysis. Cells were stained with PI and Annexin V for 15 min at room temperature. Cells were then subjected to FACS analysis using a BD Accuri™ C6 Plus Flow Cytometer. Results are shown as means ± SD; n = 3 (lower panel). (D) Representative images of cells through the soft agar assay in HCT116 WT and FOXO1 KO HCT116 cells. Cells were treated BIX-01294 (1 μM) or not.
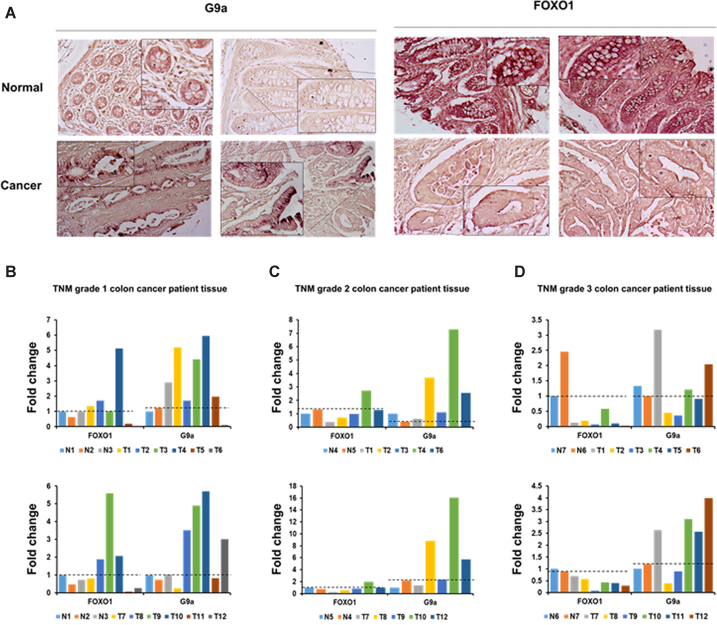
Since we identified the role of G9a-mediated FOXO1 methylation in colon cancer cell lines, we next checked G9a and FOXO1 expression levels in human colon cancer patients tissue samples. The analysis of the Human Protein Atlas Database (http://www.proteinatlas.org) indicated that G9a was upregulated and FOXO1 was downregulated in colon cancer (Supplementary Figure S9). To further confirm this, we examined FOXO1 and G9a expression in a large cohort of human colon cancer samples using immunohistochemical analysis of tissue microarrays. As expected, G9a was overexpressed in colon cancer crypt cells when compared to normal cells (Figure 8A, left panel). On the contrary, FOXO1 levels were lower in human colon cancer crypt cells than in normal cells (Figure 8A, right panel). These data supported our findings that showed G9a negatively regulating FOXO1 expression in human colon cancer cell lines.
Figure 8.
FOXO1 protein levels were decreased by G9a in colon cancer patients tissue. (A) Formalin-fixed tissue array slides were used in immunohistochemistry experiments. Tissue array slides from colon cancer and normal tissues were used for immuno-histochemical 3,3-diaminobenzidine (DAB) staining of FOXO1 or G9a. (B–D) Colon cancer patients tissue were lysed and immunoblotted with anti-FOXO1 anti-G9a and anti-β-actin antibodies. (B) TNM grade 1, (C) TNM grade 2 and (D) TNM grade 3. FOXO1 and G9a protein levels were normalized by β-actin. The dotted line indicates the degree of expression of normal tissue.
Furthermore, we investigated whether FOXO1 expression was dysregulated by G9a thorough measuring the levels of FOXO1 and G9a protein levels in 35 colon cancer patients tissue samples. We observed that 23 colon patients showed elevated G9a protein expression (74.2%). Importantly, over half of the G9a overexpressed patients had decreased protein expression of FOXO1 (53.8%). Especially, FOXO1 expression negatively correlated with different tumor grades (TNM grade 1; 22.2%, TNM grade 2; 50%, TNM grade 3; 100%), indicating that downregulation of FOXO1 plays an important role in colon cancer progression (Figure 8B–D). Altogether, the data suggested that G9a-mediated FOXO1 methylation attenuates stability of FOXO1, resulting in promoting cancer cell proliferation and colon tumorigenesis.
DISCUSSION
Posttranslational modification of proteins is important for various cellular processes such as cell growth regulation and development. However, not many studies have been reported regarding the methylation of the FOXO protein family. Recently, it was observed that FOXO1 arginine methylation by PRMT1 inhibited FOXO1 phosphorylation and resulted in nuclear accumulation under oxidative stress (31). Another report suggested that FOXO3 methylation at K271 by SET9 resulted in the decrease of FOXO3 stability, but an increase in its transcriptional activity (36). Additionally, FOXO3 was also methylated at K270 by SET9, resulting in suppression of FOXO3 transcriptional activity (49).
In the present study, we first showed that FOXO1 was methylated at K273 by G9a in vitro. In addition, FOXO1 methylation was also detected in vivo by mass spectrometry analysis (Supplementary Figure S2A) and IP assay using K273R mutant (Figure 2E). Importantly, this site is well-preserved evolutionarily in the human FOXO family and different species (Supplementary Figure S2). The methylation of FOXO1 did not completely disappear in FOXO1 K273R point mutant, suggesting the possibility that other methylation target sites by SET9 such as K270 and K271 may exist (Figure 2E). The other FOXO1 modifications such as acetylation, phosphorylation, and other methylations still need to be studied for possible crosstalk with K273 methylation.
Many reports suggest that epigenetic modifications regulate protein stability. For instance, methylation of PCNA at K248 by SETD8 enhances the stability of PCNA by inhibiting poly-ubiquitination and increasing the interaction between PCNA and FEN1 (50). SETD7 methylates p53 at K372, enhancing p53 stability and its transcriptional activity (34). Similarly, we observed that G9a-mediated FOXO1 methylation attenuated FOXO1 protein stability (Figure 4). In addition, G9a-mediated FOXO1 methylation enhanced the interaction between SKP2 and FOXO1, resulting in increased poly-ubiquitination of FOXO1 and its subsequent degradation (Figure 5).
Furthermore, we showed that insulin induced G9a expression and promoted FOXO1 methylation, resulting in FOXO1 degradation via G9a HMTase activity (Figure 6). Consistent with our suggested mechanism, previous studies revealed that various cancers such as colon, prostate, and breast cancers were promoted by abnormal production of insulin (37,39,51). Crypt foci (the early stage of colon cancer), in particular, has been reported to be stimulated by insulin (47,48). Moreover, insulin promoted phosphorylation dependent poly-ubiquitination of FOXO1, resulting in FOXO1 degradation (42). Another report revealed that the interaction between FOXO1 and G9a increased in insulin treated HepG2 cells (52). Insulin also regulated FOXO1 localization from the nucleus to the cytoplasm (29). However, since this regulation is reversible, nucleus-accumulated FOXO1 could be methylated by G9a, which was upregulated by insulin.
Consistent with previous studies, we also detected differences in FOXO1 and G9a expression between normal and colon cancer tissue samples. Tissue array analysis showed that G9a was upregulated, while FOXO1 was downregulated in human colon cancer crypt cells (Figure 8A). Moreover, the Human Protein Atlas database showed the same results (Supplementary Figure S9). It is likely that the increase in crypt foci and colon cancer risk via insulin activity might be associated with the induction of G9a, which results in the degradation of FOXO1 via G9a-mediated methylation. We also observed a robust effect of FOXO1 in colon cancer cell proliferation but not FOXO1 KO colon cancer cells, which strongly suggests the role of G9a-mediated FOXO1 methylation in colon cancer growth. Human Protein Atlas database showed that lower expressed FOXO1 cancer patients of survival rate is shorter than higher expressed FOXO1 cancer patients in rectum adenocarcinoma (READ), renal and liver cancers. G9a was increased in many cancer types including colon, bladder, liver, lung and leukemia according to Oncomine database. In addition, there are reports that G9a promotes cancer cell growth, invasion and metastasis in lung and colon cancer (45,46,53). We further confirmed the expression levels of FOXO1 and G9a using human colon cancer patient samples. Many patients were overexpressed G9a and downregulated FOXO1. Expression in majority of the patients, the amount of protein of FOXO1 was decreased by G9a, and this pattern was found to be more pronounced with higher TNM grade. Together, these data suggest that G9a-mediated FOXO1 degradation may play an important role in progression of colon cancers.
There are reports that G9a promotes cell proliferation and cell cycle exit during myogenic differentiation (54). Additionally, G9a promotes tumor cell growth and invasion by increasing H3K9-me2 activity at the CASP1 promoter (55). G9a could regulate cell proliferation and apoptosis by regulating H3K9-me1/2, however we obtained more dramatic effect in FOXO1 WT cells and that of KO cells (demonstrated in the colony formation and MTT assays) (Figure 7). These results indicated the possibility of a mechanism for FOXO1 methylation as well as H3K9-me1/2 catalyzed silencing of specific target genes’ expression. In this study, we demonstrated that insulin-induced G9a expression via induction of HNF4a resulted in FOXO1 methylation. G9a attenuated the stability of methylated FOXO1 via enhanced interaction between FOXO1 and SKP2. Degradation of FOXO1 via G9a-mediated methylation increased cell proliferation and decreased apoptosis in colon cancer cells.
In summary, we have uncovered a mechanism involving G9a-mediated FOXO1 methylation via insulin signaling lead to the development of colon cancer. G9a-mediated FOXO1 methylation attenuates FOXO1 protein stability. Finally, our results demonstrated that FOXO1 methylation is important for cell proliferation and colony formation in colon cancer cells. Our studies suggest that G9a inhibitor, BIX-01294, may have therapeutic potential in colon cancer by inhibiting FOXO1 degradation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Sung Hee Baek (Seoul National University) for the pcDNA3.0-Flag-G9a; Dr A. Tarakhovsky (Rockefeller University) for G9a KO MEFs; and Dr Martin J. Walsh (Mount Sinai School of Medicine) for pEGFP-hG9a and pEGFP-hG9SET clones. We thank National Center for Inter-University Research Facilities (NCIRF) for assistance with LTQ-orbitrap analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT & Future Planning [NRF-2016R1A4A1008035, NRF-2017R1A2B4004407 and NRF-2018R1A6A3A11050656]; Chung-Ang University Research Grants in 2017.
Conflict of interest statement. None declared.
REFERENCES
- 1. Salih D.A., Brunet A.. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 2008; 20:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Daitoku H., Fukamizu A.. FOXO transcription factors in the regulatory networks of longevity. J. Biochem. 2007; 141:769–774. [DOI] [PubMed] [Google Scholar]
- 3. Essers M.A., Weijzen S., de Vries-Smits A.M., Saarloos I., de Ruiter N.D., Bos J.L., Burgering B.M.. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004; 23:4802–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S., Anderson M.J., Arden K.C., Blenis J., Greenberg M.E.. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999; 96:857–868. [DOI] [PubMed] [Google Scholar]
- 5. Greer E.L., Brunet A.. FOXO transcription factors in ageing and cancer. Acta Physiol. (Oxf.). 2008; 192:19–28. [DOI] [PubMed] [Google Scholar]
- 6. Dijkers P.F., Medema R.H., Lammers J.W., Koenderman L., Coffer P.J.. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000; 10:1201–1204. [DOI] [PubMed] [Google Scholar]
- 7. Hu M.C., Lee D.F., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H. et al. . IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004; 117:225–237. [DOI] [PubMed] [Google Scholar]
- 8. Seoane J., Le H.V., Shen L., Anderson S.A., Massague J.. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004; 117:211–223. [DOI] [PubMed] [Google Scholar]
- 9. Modur V., Nagarajan R., Evers B.M., Milbrandt J.. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J. Biol. Chem. 2002; 277:47928–47937. [DOI] [PubMed] [Google Scholar]
- 10. Bullock M.D., Bruce A., Sreekumar R., Curtis N., Cheung T., Reading I., Primrose J.N., Ottensmeier C., Packham G.K., Thomas G. et al. . FOXO3 expression during colorectal cancer progression: biomarker potential reflects a tumour suppressor role. Br. J. Cancer. 2013; 109:387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Horst A., de Vries-Smits A.M., Brenkman A.B., van Triest M.H., van den Broek N., Colland F., Maurice M.M., Burgering B.M.. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006; 8:1064–1073. [DOI] [PubMed] [Google Scholar]
- 12. Matsuzaki H., Daitoku H., Hatta M., Aoyama H., Yoshimochi K., Fukamizu A.. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:11278–11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A.. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10042–10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y. et al. . Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004; 303:2011–2015. [DOI] [PubMed] [Google Scholar]
- 15. Perrot V., Rechler M.M.. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol. Endocrinol. 2005; 19:2283–2298. [DOI] [PubMed] [Google Scholar]
- 16. Yang J.Y., Zong C.S., Xia W., Yamaguchi H., Ding Q., Xie X., Lang J.Y., Lai C.C., Chang C.J., Huang W.C. et al. . ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 2008; 10:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang H., Regan K.M., Wang F., Wang D., Smith D.I., van Deursen J.M., Tindall D.J.. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peters A.H., Kubicek S., Mechtler K., O'Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y. et al. . Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003; 12:1577–1589. [DOI] [PubMed] [Google Scholar]
- 19. Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y.. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005; 19:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rathert P., Dhayalan A., Murakami M., Zhang X., Tamas R., Jurkowska R., Komatsu Y., Shinkai Y., Cheng X., Jeltsch A.. Protein lysine methyltransferase G9a acts on non-histone targets. Nat. Chem. Biol. 2008; 4:344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang J., Dorsey J., Chuikov S., Perez-Burgos L., Zhang X., Jenuwein T., Reinberg D., Berger S.L.. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J. Biol. Chem. 2010; 285:9636–9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J.S., Kim Y., Bhin J., Shin H.J., Nam H.J., Lee S.H., Yoon J.B., Binda O., Gozani O., Hwang D. et al. . Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13510–13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pless O., Kowenz-Leutz E., Knoblich M., Lausen J., Beyermann M., Walsh M.J., Leutz A.. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. J. Biol. Chem. 2008; 283:26357–26363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee J.S., Kim Y., Kim I.S., Kim B., Choi H.J., Lee J.M., Shin H.J., Kim J.H., Kim J.Y., Seo S.B. et al. . Negative regulation of hypoxic responses via induced Reptin methylation. Mol. Cell. 2010; 39:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chae Y.C., Kim K.B., Kang J.Y., Kim S.R., Jung H.S., Seo S.B.. Inhibition of FoxO1 acetylation by INHAT subunit SET/TAF-Ibeta induces p21 transcription. FEBS Lett. 2014; 588:2867–2873. [DOI] [PubMed] [Google Scholar]
- 26. Kim K.B., Son H.J., Choi S., Hahm J.Y., Jung H., Baek H.J., Kook H., Hahn Y., Kook H., Seo S.B.. H3K9 methyltransferase G9a negatively regulates UHRF1 transcription during leukemia cell differentiation. Nucleic Acids Res. 2015; 43:3509–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nie J., Liu L., Wu M., Xing G., He S., Yin Y., Tian C., He F., Zhang L.. HECT ubiquitin ligase Smurf1 targets the tumor suppressor ING2 for ubiquitination and degradation. FEBS Lett. 2010; 584:3005–3012. [DOI] [PubMed] [Google Scholar]
- 28. Zhao Y., Wang Y., Zhu W.G.. Applications of post-translational modifications of FoxO family proteins in biological functions. J. Mol. Cell Biol. 2011; 3:276–282. [DOI] [PubMed] [Google Scholar]
- 29. Daitoku H., Sakamaki J., Fukamizu A.. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim. Biophys. Acta. 2011; 1813:1954–1960. [DOI] [PubMed] [Google Scholar]
- 30. Calnan D.R., Brunet A.. The FoxO code. Oncogene. 2008; 27:2276–2288. [DOI] [PubMed] [Google Scholar]
- 31. Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., Fukamizu A.. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell. 2008; 32:221–231. [DOI] [PubMed] [Google Scholar]
- 32. Gaughan L., Stockley J., Wang N., McCracken S.R., Treumann A., Armstrong K., Shaheen F., Watt K., McEwan I.J., Wang C. et al. . Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2011; 39:1266–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kontaki H., Talianidis I.. Lysine methylation regulates E2F1-induced cell death. Mol. Cell. 2010; 39:152–160. [DOI] [PubMed] [Google Scholar]
- 34. Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J. et al. . Regulation of p53 activity through lysine methylation. Nature. 2004; 432:353–360. [DOI] [PubMed] [Google Scholar]
- 35. Puigserver P., Rhee J., Donovan J., Walkey C.J., Yoon J.C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D. et al. . Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003; 423:550–555. [DOI] [PubMed] [Google Scholar]
- 36. Calnan D.R., Webb A.E., White J.L., Stowe T.R., Goswami T., Shi X., Espejo A., Bedford M.T., Gozani O., Gygi S.P. et al. . Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY). 2012; 4:462–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Giovannucci E. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am. J. Clin. Nutr. 2007; 86:s836–s842. [DOI] [PubMed] [Google Scholar]
- 38. Saydah S.H., Platz E.A., Rifai N., Pollak M.N., Brancati F.L., Helzlsouer K.J.. Association of markers of insulin and glucose control with subsequent colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev. 2003; 12:412–418. [PubMed] [Google Scholar]
- 39. Venkateswaran V., Haddad A.Q., Fleshner N.E., Fan R., Sugar L.M., Nam R., Klotz L.H., Pollak M.. Association of diet-induced hyperinsulinemia with accelerated growth of prostate cancer (LNCaP) xenografts. J. Natl. Cancer Inst. 2007; 99:1793–1800. [DOI] [PubMed] [Google Scholar]
- 40. Kaaks R., Toniolo P., Akhmedkhanov A., Lukanova A., Biessy C., Dechaud H., Rinaldi S., Zeleniuch-Jacquotte A., Shore R.E., Riboli E.. Serum C-peptide, insulin-like growth factor (IGF)-I, IGF-binding proteins, and colorectal cancer risk in women. J. Natl. Cancer Inst. 2000; 92:1592–1600. [DOI] [PubMed] [Google Scholar]
- 41. Bonser A.M., Garcia-Webb P.. C-peptide measurement: methods and clinical utility. Crit. Rev. Clin. Lab. Sci. 1984; 19:297–352. [DOI] [PubMed] [Google Scholar]
- 42. Aoki M., Jiang H., Vogt P.K.. Proteasomal degradation of the FoxO1 transcriptional regulator in cells transformed by the P3k and Akt oncoproteins. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:13613–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirota K., Daitoku H., Matsuzaki H., Araya N., Yamagata K., Asada S., Sugaya T., Fukamizu A.. Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J. Biol. Chem. 2003; 278:13056–13060. [DOI] [PubMed] [Google Scholar]
- 44. Chitforoushzadeh Z., Ye Z., Sheng Z., LaRue S., Fry R.C., Lauffenburger D.A., Janes K.A.. TNF-insulin crosstalk at the transcription factor GATA6 is revealed by a model that links signaling and transcriptomic data tensors. Sci. Signal. 2016; 9:ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding J., Li T., Wang X., Zhao E., Choi J.H., Yang L., Zha Y., Dong Z., Huang S., Asara J.M. et al. . The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab. 2013; 18:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen M.W., Hua K.T., Kao H.J., Chi C.C., Wei L.H., Johansson G., Shiah S.G., Chen P.S., Jeng Y.M., Cheng T.Y. et al. . H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010; 70:7830–7840. [DOI] [PubMed] [Google Scholar]
- 47. Koohestani N., Tran T.T., Lee W., Wolever T.M., Bruce W.R.. Insulin resistance and promotion of aberrant crypt foci in the colons of rats on a high-fat diet. Nutr. Cancer. 1997; 29:69–76. [DOI] [PubMed] [Google Scholar]
- 48. Corpet D.E., Jacquinet C., Peiffer G., Tache S.. Insulin injections promote the growth of aberrant crypt foci in the colon of rats. Nutr. Cancer. 1997; 27:316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie Q., Hao Y., Tao L., Peng S., Rao C., Chen H., You H., Dong M.Q., Yuan Z.. Lysine methylation of FOXO3 regulates oxidative stress-induced neuronal cell death. EMBO Rep. 2012; 13:371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takawa M., Cho H.S., Hayami S., Toyokawa G., Kogure M., Yamane Y., Iwai Y., Maejima K., Ueda K., Masuda A. et al. . Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012; 72:3217–3227. [DOI] [PubMed] [Google Scholar]
- 51. Wysocki P.J., Wierusz-Wysocka B.. Obesity, hyperinsulinemia and breast cancer: novel targets and a novel role for metformin. Expert Rev. Mol. Diagn. 2010; 10:509–519. [DOI] [PubMed] [Google Scholar]
- 52. Arai T., Kano F., Murata M.. Translocation of forkhead box O1 to the nuclear periphery induces histone modifications that regulate transcriptional repression of PCK1 in HepG2 cells. Genes Cells. 2015; 20:340–357. [DOI] [PubMed] [Google Scholar]
- 53. Zhang J., He P., Xi Y., Geng M., Chen Y., Ding J.. Down-regulation of G9a triggers DNA damage response and inhibits colorectal cancer cells proliferation. Oncotarget. 2015; 6:2917–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rao V.K., Ow J.R., Shankar S.R., Bharathy N., Manikandan J., Wang Y., Taneja R.. G9a promotes proliferation and inhibits cell cycle exit during myogenic differentiation. Nucleic Acids Res. 2016; 44:8129–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang T., Zhang P., Li W., Zhao T., Zhang Z., Chen S., Yang Y., Feng Y., Li F., Shirley Liu X. et al. . G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 2017; 8:e2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.