Abstract
Single-cell RNA sequencing (scRNA-seq) methods generate sparse gene expression profiles for thousands of single cells in a single experiment. The information in these profiles is sufficient to classify cell types by distinct expression patterns but the high complexity of scRNA-seq libraries often prevents full characterization of transcriptomes from individual cells. To extract more focused gene expression information from scRNA-seq libraries, we developed a strategy to physically recover the DNA molecules comprising transcriptome subsets, enabling deeper interrogation of the isolated molecules by another round of DNA sequencing. We applied the method in cell-centric and gene-centric modes to isolate cDNA fragments from scRNA-seq libraries. First, we resampled the transcriptomes of rare, single megakaryocytes from a complex mixture of lymphocytes and analyzed them in a second round of DNA sequencing, yielding up to 20-fold greater sequencing depth per cell and increasing the number of genes detected per cell from a median of 1313 to 2002. We similarly isolated mRNAs from targeted T cells to improve the reconstruction of their VDJ-rearranged immune receptor mRNAs. Second, we isolated CD3D mRNA fragments expressed across cells in a scRNA-seq library prepared from a clonal T cell line, increasing the number of cells with detected CD3D expression from 59.7% to 100%. Transcriptome resampling is a general approach to recover targeted gene expression information from single-cell RNA sequencing libraries that enhances the utility of these costly experiments, and may be applicable to the targeted recovery of molecules from other single-cell assays.
INTRODUCTION
New methods that measure mRNA abundance in hundreds to thousands of single cells have been used to understand gene expression heterogeneity in tissues (1–4). But these single-cell RNA-seq experiments have a tradeoff: instead of surveying gene expression at great depth, they generate a sparse gene expression profile for each cell in a population. This information is often sufficient to identify cell types in a population, but provides only a glimpse of genes expressed in a given cell (5). Moreover, mRNAs in each cell are captured stochastically, leading to false negatives in identification of expressed genes in many cells (6).
Single-cell RNA-seq experiments can identify rare cell populations that have distinct gene expression profiles. Previous studies have identified retinal precursors (2,7), hematopoietic stem cells (8), rare immune cells (9), and novel lung cell types (10) in complex populations, where these cell types represent a small fraction of the cell mixture. Historically, the information known about a cell lineage is correlated with its abundance and thus these rare cell types often contain new information for uncharacterized cell types. Whereas scRNA-seq methods can identify these rare cell populations, they provide only a glimpse of the RNA expression patterns in rare cells because of the detection bias for highly expressed RNAs. Moreover, because the mRNAs from these rare cells represent a small fraction of the total library, increasing the sequencing depth is not an efficient way to learn more about these cells. More complete analysis of their expression might identify e.g., cell surface markers that could be used to isolate these rare cell populations.
Recently an approach termed DART-seq was developed that enables acquisition of both global and targeted gene expression information in a single experiment (BioRxiv: https://doi.org/10.1101/328328). In DART-seq, gene-specific probes are ligated to oligo-dT terminated Drop-seq beads (2), enabling both oligo-dT-primed and site-specific cDNA synthesis during reverse transcription. This approach is valuable if the mRNAs of interest are known a priori to provide increased coverage for specific mRNAs. Additionally a method to enrich cell barcodes of interest from pooled single cell libraries was developed that uses hemi-specific multiplexed PCR to selectively resequence individual cells (11), which could be useful to more deeply investigate cell specific gene expression patterns.
Here, we developed ‘transcriptome resampling’ to address limitations of single-cell RNA sequencing. Many single-cell RNA sequencing platforms have been developed (Supplementary Table S1) and all of them incorporate a unique DNA sequence into mRNAs derived from a single cell. We reasoned that this sequence could serve as a molecular handle to isolate RNAs derived from a cell of interest, and that these isolated RNAs could be resequenced to higher depth to interrogate the transcriptional profile of targeted cells. Moreover, this same principle could be applied to isolate RNAs by their unique sequences, enabling their detection in a second round of DNA sequencing. By physically isolating RNA-derived cDNA fragments, we find that transcriptome resampling can more deeply interrogate RNAs in specific cells, or can be used to determine whether specific mRNAs are expressed across cells in a mixture.
MATERIALS AND METHODS
Cell isolation and single cell RNA-seq generation
For the mouse/human experiment, NIH:3T3 and 293T cells were grown in standard media and mixed at a 1:1 ratio prior to capture. For the mouse xenograft experiment, an estrogen receptor positive patient derived xenograft primary cell line (UCD65 (12)) was labeled with luc-eGFP using lentivirus (13). Cells were xenografted into NOD/SCID/IL2rg−/− mice via intracardiac injection to generate disseminated metastases or injected into the mammary pad to generate a primary tumor. Cells were isolated from the primary cell line, a primary fat pad tumor, a brain metastasis and a bone metastases. Contaminating murine bone marrow cells copurified with human tumor cells isolated from the bone metastasis. Single cells were then captured using the iCell8 system from WaferGen Bio-Systems.
For the Jurkat libraries, Jurkat cells were thawed from a frozen vial, grown for one passage in RPMI-1640 + 10% FBS media and diluted to 500 cells/μl for capture on the 10X Genomics Chromium controller. For the megakaryocyte experiment, blood from healthy controls between ages 15 and 55 was collected in heparinized tubes and peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation using Ficoll to obtain the buffy coat.
Wafergen libraries were prepared according to the manufacturer instructions. 5′ and 3′ gene expression libraries were prepared on the 10× Genomics Chromium controller and libraries were prepared according to manufacturer protocols. TCR enrichment libraries were generated using the 10X Genomics 5′ library construction and VDJ enrichment kit.
PCR primer design and synthesis
For 10× Genomics cell barcode pulldown libraries, 5′ biotinylated oligos with 4 bp of consensus sequence followed by the 16 bp barcode with LNA bases added every third base. These 6 LNA base pairs allow for increased binding specificity of the barcode sequence. For 10× Genomics gene pulldown libraries, 5′ biotinylated oligo probes were designed against selected CD3D using primer3 (14). For LNA probes, LNA bases were added every 4th base. Regions to target were selected based on inspection of read coverage of the 5′ UTR of the CD3D gene (see Figure 4H for genome browser tracks). LNA molecules were purchased from either Exiqon or Qiagen. For Wafergen libraries, 5′ biotinylated oligos with 17 bp of consensus sequence followed by the 11 bp barcode were designed containing three phosphorothioate bonds on the three terminal 3′ positions to prevent degradation. All oligonucleotides used in this study are described in Supplementary Table S2.
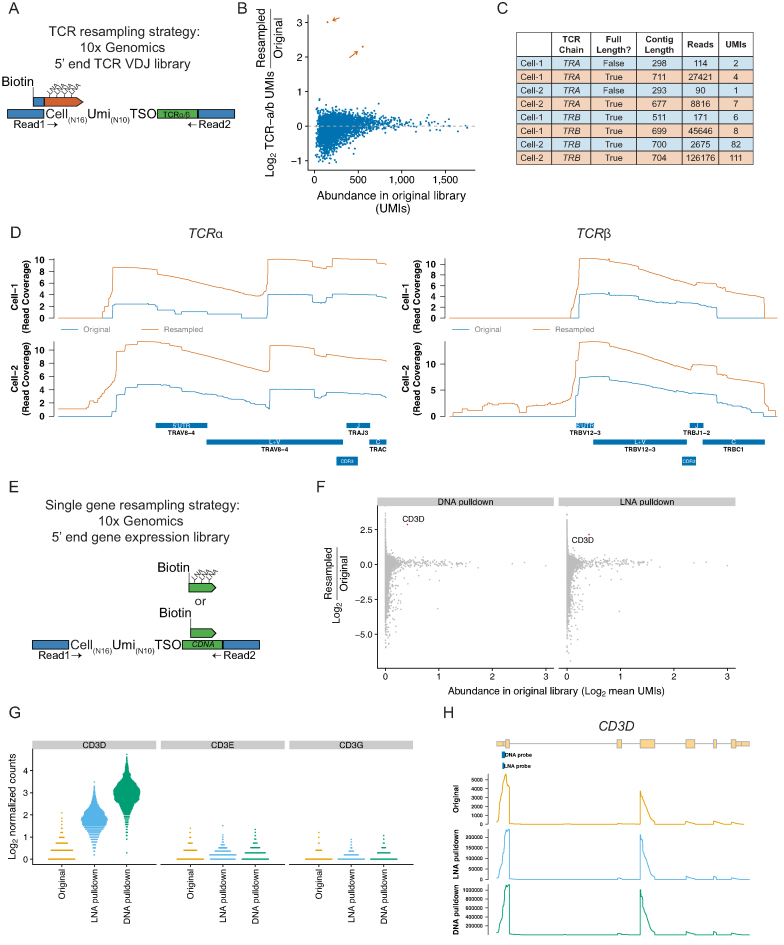
Figure 4.
Resampling individual mRNAs and TCR sequences. (A) Schematic of hybridization method applied to resampling single cell VDJ sequences from a 5′ end VDJ enrichment library. (B) Enrichment of UMIs derived from the TCR alpha or TCR beta chains for the targeted cells (resampled cells; n = 2, shown in orange). (C) Summary statistics of the TCR assemblies from the original (blue) or resampled libraries (orange). *Note that UMIs with less than 10 reads were excluded from the assembly and are not included in the displayed UMI and read counts. (D) Read coverage (Log scale) across the consensus Jurkat TCR-alpha and TCR-beta sequences in the original library (blue) or the resampled library (orange). (E) Schematic of hybridization method applied to resampling a single mRNA from a 5′ end gene expression library. Hybridization probes were designed with LNA nucleotides or with only DNA. (F) Average fold-change of normalized UMI counts across all cells after resampling with a DNA only oligo or an LNA oligo targeting CD3D. X-axis indicates the normalized expression as average of the expression values in the original and resampled libraries. (G) CD3 expression per cell for each CD3 chain. (H) Read coverage across CD3D in the original or resampled libraries.
Library preparation and sequencing
10× Genomics Illumina libraries at 2 nM concentration were diluted 20-fold and PCR amplified with Truseq Pcr F and R oligo for 14 cycles with 1 unit of Phusion. PCR amplification was confirmed by gel electrophoresis and the remaining sample was Ampure XP purified at a ratio of 1.8× bead/PCR volume. Samples were eluted in 10 μl and molarity was determined using a Qubit. 10 μl of a 100 nM library was mixed with 10 pmol LNA molecule in annealing buffer (10 mM Tris 8.0, 50 mM NaCl) and heated to 98° for 10 min followed by slow cooling to 59–64°C depending on the LNA and held overnight at the annealing temperature. Annealed DNA was added in equal volume to washed Dynabead M270 streptavidin beads at the annealing temperature. Beads were incubated for 15 minutes with gentle shaking. Beads were washed five times in binding buffer at the annealing temperature. Hybridized molecules were eluted in 20 μl of elution solution buffer (50 mM NaCl, 0.1 mM EDTA) by heating for 10 min at 100°C and centrifuging briefly to pellet the streptavidin-bound LNA molecules (which can inhibit PCR). The elution was performed two times to ensure all the DNA was removed from the LNA. 10μl of streptavidin purified input DNA was added to PCR using primers Truseq PCR F and R for 8 cycles. PCR products were ampure purified, analyzed by Tapestation D1000, quantified by the Qubit, and sequenced on an Illumina MiSeq or Nova Seq 6000.
Wafergen Illumina libraries at 2 nM concentration were diluted 20-fold and PCR amplified with 1 unit of Phusion and the cell specific biotinylated primer and Nextera read 2 sequencing primer to specifically amplify the sequences from the selected cells for 14 cycles. PCR amplification was confirmed by gel electrophoresis and the remaining sample was purified with Ampure XP beads at a ratio of 1.8× bead/PCR volume. Samples were eluted and added in equal volume to prepared Dynabead M270 streptavidin beads. Beads were incubated for 15 min at room temperature with gentle rotation. Beads were washed 5× in bind and wash buffer and eluted in 20 μl of water. 10 μl of streptavidin-purified input DNA was added to PCR using PEPCRPrimer 1.0 and Nextera N703 for 9 cycles. PCR products were AMPure purified, confirmed on the Agilent TapeStation D1000, quantified by Qubit and submitted for sequencing.
Data analysis
We provide our data analysis pipeline and custom scripts in a github repository (https://github.com/rnabioco/scrna-subsets). Single cell RNA-Seq libraries were preprocessed to append the read 1 sequence to the paired read 2 read id followed by quality trimming and poly(A) tail removal from read 2 using cutadapt (v. 1.8.3) (15). Reads were next aligned with STAR (16) to either the human genome assembly (Gencode GRCh38) for the PBMC experiments or a genome with both human (GRCh38) and mouse (GRCm38) sequences for all other experiments. Sequence headers in the human/mouse combined genome were prefixed with either an ‘H_’ or ‘M_’ to designate human or mouse references respectively.
Following alignment, BAM files were processed to extract the cell barcode and UMI sequences into tags (CN and BX) within the BAM file. The cell barcode was error corrected against a list of cell barcodes, either as known well barcodes (Wafergen experiments), or generated from the original 10x Genomics single cell libraries processed with the 10× Genomics software cellranger (v. 2.1.1). Cell barcodes within an edit distance of 1 of the known barcodes were considered valid cell barcodes and corrected to the known barcode. Alignments not designated as multi-mapping that overlapped distinct exonic features were tagged in the BAM file (subread v. 1.6.0) (17). Gencode v25 annotations was used for human data, and a union reference containing Gencode v25 and mouse v11 was used for human/mouse datasets. UMIs per gene were enumerated per cell using umi-tools (v 0.5.3) (18) using the directional method to disambiguate similar UMI sequences.
tSNE projections were generated using the Seurat R package (19). Briefly, PCA analysis was performed on scaled, log-transformed, library-size-normalized UMI matrices using variable gene sets. PCA was used to reduced the dimensionality and tSNE projections were generated with a perplexity of 30. Graph-based clustering was performed to identify clusters using the first 15 principal components. Markers per cluster were identified using a Wilcoxon rank sum test. K-nearest neighbors were identified using the top 20 PCA dimensions using the RANN R package.
RESULTS
A strategy to recover individual transcriptomes from scRNA-seq libraries
The cDNA molecules in single-cell RNA sequencing libraries contain unique oligonucleotide barcodes that are used to associate a sequencing read to its cell of origin (Figure 1A and B). Because these barcodes are typically long (11–19 continuous or discontinuous base pairs, depending on the platform, Supplementary Table S1) and are present in every molecule in the library, we reasoned that these sequences could be used as sites for oligonucleotide hybridization, enabling selective recovery of specific molecules from a pool of molecules containing many different barcodes.
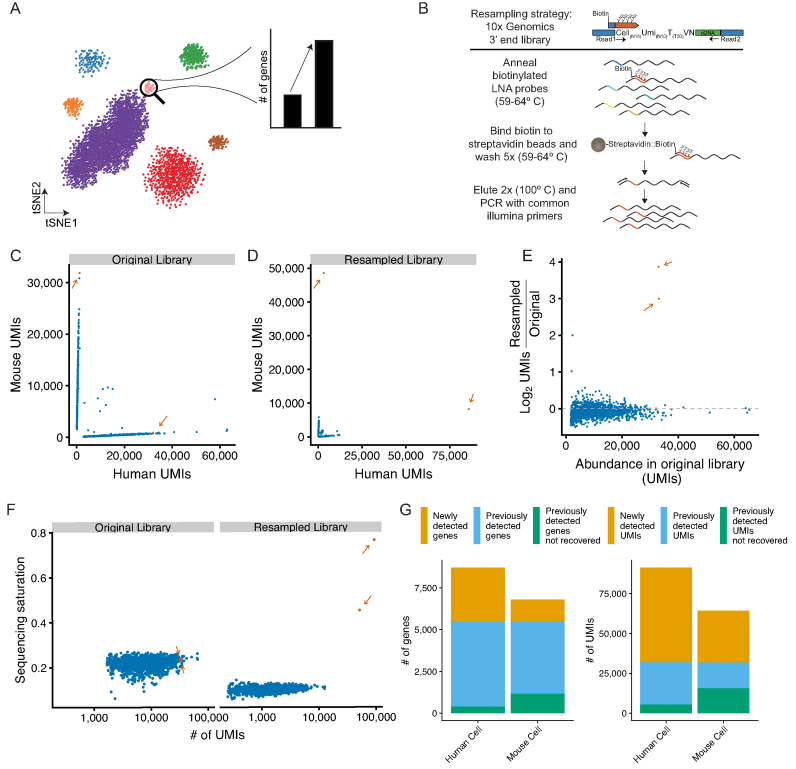
Figure 1.
Resampling specific cell transcriptomes from pooled single-cell RNA-seq libraries. (A) Resampling single cell libraries from rare cell populations to enable deeper characterization of a targeted cell type. (B) Schematic of Locked Nucleic Acid (LNA) hybridization-based approach to enrich mRNAs from targeted cells from 10X Genomics single-cell mRNA-seq libraries. (C) Species specificity of cells recovered from a 10X Genomics 3′ end gene expression scRNA-seq library containing a 1:1 mix of mouse (NIH-3T3) and human (293T) cells. Orange dots and arrows (n = 2) indicate cells selected for resampling; blue dots (n = 1505) are untargeted cells. (D) Species specificity of cells from the resampled library. Colors are are the same as in C. (E) Enrichment of targeted libraries after resampling. The y-axis plots the log2 enrichment of UMIs normalized by the size of the entire scRNA-seq library. Colors are the same as in C. (F) Sequencing saturation, as defined by 1 minus the ratio of the number of UMIs to the number of reads, per cell for resampled and untargeted cells in the original scRNA-seq library or after resampling. Colors are the same as in C. (G) Number of genes (left) or UMIs (right) in the resampled cells that are either newly detected by resampling (orange), previously detected in the original library (blue), or previously detected in the original library but not found after resampling (green).
As proof-of-principle, we resampled a mouse and a human cell transcriptome from a mixed-species single cell RNA-seq library. We generated a 3′ end single cell library on the 10X Genomics Chromium platform with a 1:1 mixture of mouse NIH-3T3 fibroblasts and human 293T cell lines (Supplementary Figure S1). After sequencing and analysis of this library, we selected a single mouse cell and a single human cell and designed oligonucleotide probes to target their unique cell barcodes (16 nt) and a short common region (4 nt) 5′ of the cell barcode for each cell library (Figure 1B, Supplementary Table S1). To increase the specificity of hybridization, we incorporated locked nucleic acid (LNA) nucleotides at six sites in the oligo, increasing the Tm to 74°C. Biotin was added to the 5′ end of the oligo to enable purification of hybridized DNA using streptavidin beads. These probes were hybridized with PCR amplified library DNA. Following streptavidin purification and elution, the enriched libraries were reamplified with PCR primers containing common Illumina sequences and sequenced.
After resampling, the targeted cell barcodes were the most abundant barcodes detected in the sequencing libraries (Figure 1C and D). The number of UMIs for the resampled cells increased by 1.56-fold and 2.85-fold for mouse and human, respectively, while non-targeted cells were largely depleted at the expense of the resampled sequences (Figure 1E). A useful measure of complexity in single-cell mRNA sequencing libraries is ‘saturation’, which is an estimate of the number of single mRNAs (as measured by UMI counts) captured from each cell in the experiment. The original single cell RNA-seq libraries were sequenced to an average saturation (i.e. the proportion of UMIs observed for a given cell at a given sequencing depth) of 21.75% ± 2.4, and after resampling the saturation for the selected cells increased to 45.44% (mouse) and 76.63% (human) (Figure 1F). Finally, we examined the number of both genes and UMIs recovered in each resampled cell and found largely the same genes and UMIs previously observed in each cell (Figure 1G and Supplementary Figure S2A).
The novel UMIs recovered in the resampled cells had diverse sequence content compared to previously detected UMIs from the same gene, indicating that the novel UMIs were not simply due to resampling artifacts (Supplementary Figure S3A). UMIs recovered in the resampled transcriptomes were largely assigned to the expected species, although the species purity decreased by 6.06% (human cell) and 2.23% (mouse cell) upon resampling, likely reflecting increased detection of free RNA molecules present in each droplet (Supplementary Figure S4A). In addition, increased sequencing depth correlates to decreased species purity in our original scRNA-seq library as well as in publicly available datasets (Supplementary Figure S4B, C). Overall these data demonstrate the feasibility of resampling individual cell transcriptomes using LNA-based hybridization and that the rate of misassignment of reads to resampled cells is low relative to the gains achieved by resampling.
Isolation and resequencing of rare cell transcriptomes
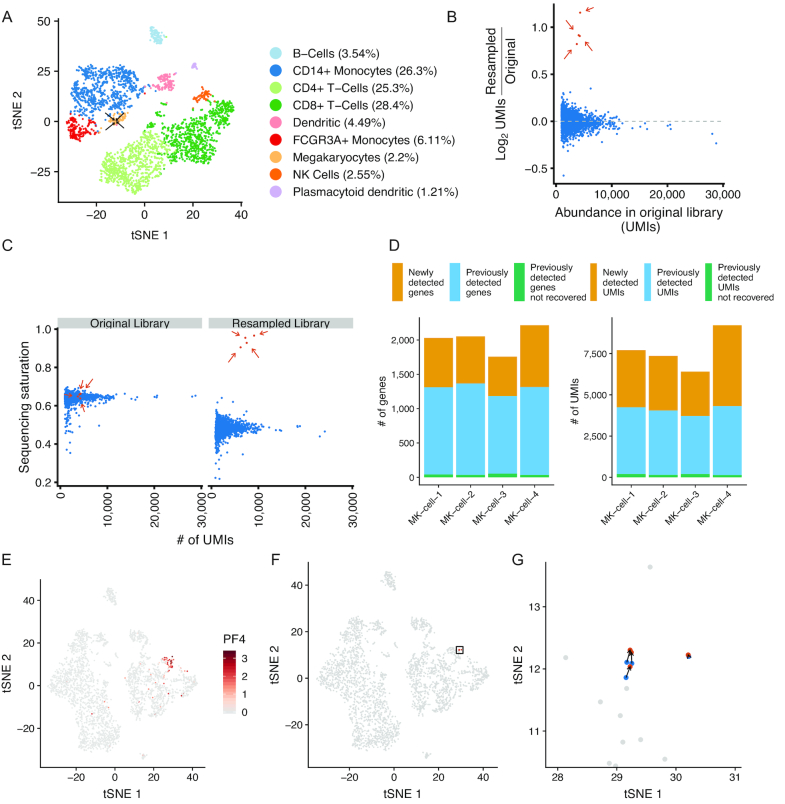
We next sought to more fully characterize transcriptomes derived from rare cells in a complex cell population. We generated a 10× Genomics 3′ end scRNA-seq library from a sample of peripheral blood mononuclear cells (PBMCs) taken from a healthy human adult (Figure 2A). Megakaryocytes are present at ∼0.1% in the bone marrow (21), have been found to populate the lung (22), and are rare in a typical PBMC sample from a healthy person. We found that megakaryocytes represented 2.2% of the PBMCs as judged by expression of the megakaryocyte marker PF4 (Supplementary Figure S5). We selected four megakaryocytes for resampling to further characterize the transcriptomes of these cells. After hybridization and resequencing, the barcodes for selected cells were enriched over non-targeted cell barcodes (Figure 2B), were driven to a higher sequencing saturation (65.21% ± 1.1 in original, 93.86% ± 2.8 in resampled, Figure 2C), had increased numbers of cell-derived reads (typically an order of magnitude), and resulted in the detection of an additional 677.25 ± 143.1 genes per cell (Figure 2D). To assess the specificity of cell type identification after resampling, we supplemented the original scRNA-seq dataset with the resampled cell transcriptomes. The resampled transcriptomes maintained the megakaryocyte identity as based on t-SNE visualization (Figure 2E–G), and the nearest neighbor for each resampled cell in PCA space was the original cell transcriptome selected for resampling (Supplementary Figure S6). From these data we conclude that transcriptome resampling is an efficient method to efficiently isolate and more deeply analyze the transcriptome of targeted cell types.
Figure 2.
Resampling rare megakaryocytes from peripheral blood mononuclear cells. (A) tSNE projection of a sample of peripheral blood mononuclear cells (PBMCs; n = 3194 cells) with cells labeled by their inferred cell type. Arrows indicate megakaryocytes selected for resampling (n = 4 resampled megakaryocytes; n = 69 total megakaryocytes). (B) Enrichment of UMIs in targeted cells following resampling. Y-axis indicates log2 enrichment of UMIs normalized by library size of the entire resampled scRNA-seq library. Orange dots and arrows (n = 4) indicate cells selected for resampling; blue dots (n = 3190) are untargeted cells. (C) Sequencing saturation of the targeted cells in the original library and after resampling. Colors are the same as in B. (D) Number of genes or UMIs in the resampled megakaryocyte (MK) cells that are either newly detected in the resampled library (orange), previously detected in the original library (blue), or previously detected in the original library but not found after resampling (green). (E) tSNE projection of the original scRNA-seq dataset supplemented with the resampled cells. Cells are colored by the expression (natural log) of the megakaryocyte marker PF4. (F) tSNE with the original cell transcriptomes (blue), resampled transcriptomes (orange), and non-targeted cells (gray). The rectangle indicates the location of cells highlighted in panel G. (G) tSNE projection of resampled cells in the region highlighted in panel F.
Identification of novel marker genes from resampling data
One goal of scRNA-seq experiments is to define novel marker genes that characterize a cell population. To address the utility of transcriptome resampling for this application, we examined the gene expression patterns of the resampled megakaryocyte transcriptomes. Genes most strongly enriched in the resampled libraries were more lowly expressed in the original libraries, and also had reduced levels of sequencing saturation (Figure 3A). In addition to detecting more genes, each resampled cell also contained more marker genes of megakaryocytes defined from the megakaryocyte cluster in the original library (Figure 3B).
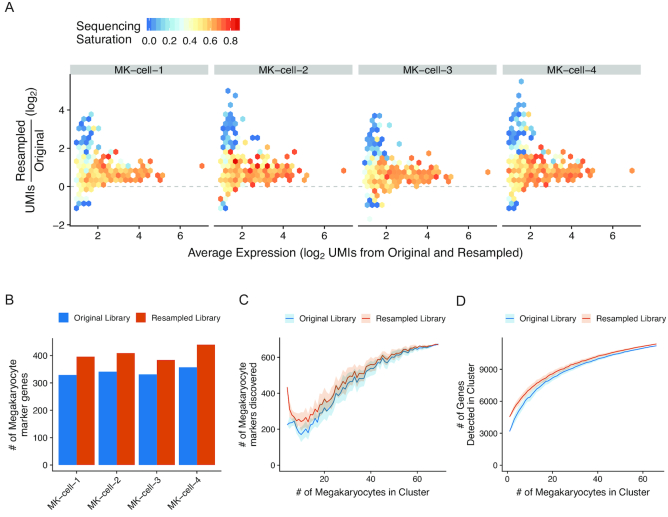
Figure 3.
Enrichment of megakaryocyte markers in resampled cells. (A) Relationship between sequencing saturation and enrichment of genes in the resampled libraries. X-axis indicates the normalized expression as the average of the expression values in the original and resampled libraries. Y-axis indicates UMI enrichment. Genes are binned into hexagons colored by their sequencing saturation (1 – UMIs/reads) calculated from the original library values (scale bar from 0 to 1.0). For genes not detected in the original library, the average sequencing saturation from all megakaryocytes is displayed. (B) The number of genes recovered in each resampled cell that were defined as megakaryocyte markers in the original scRNA-seq dataset. Orange dots are for the resampled libraries; blue is from the original. (C) Markers for megakaryocytes were calculated using the resampled cells (first set of dots), using either the UMI counts from the original library (blue dots) or the UMI counts from the resampled library (orange dots). The same procedure was repeated with increasing numbers of randomly selected non-resampled megakaryocytes supplementing the resampled cells. The random selection was repeated 10 times and the average is shown with the standard deviation displayed in the shaded areas. (D) Number of unique genes detected in a hypothetical megakaryocyte cluster containing the cells selected for resampling from the resampled library (orange line) or the original library (blue line) with increasing numbers of randomly selected non-targeted megakaryocytes supplementing the resampled cells. The random selection was repeated 100 times and the average is shown with the standard deviation displayed in the shaded areas.
A key parameter to motivate a resampling experiment is to determine the appropriate number of cells to resample to gain additional insight into the gene expression profile of a particular cell population. In our megakaryocyte resampling experiment, we resampled 4 of the 69 (∼5%) megakaryocytes from the dataset. However, with only ∼5% of the megakaryocytes resampled, we did not find an increase in the number of genes that are differentially expressed (i.e. marker genes) between the megakaryocyte cluster and other clusters in the dataset (674 marker genes in the original library; 669 in the resampled). The resampled cells have lower normalized gene expression values (Supplementary Figure S7) due to increased gene diversity, particularly for highly expressed genes, resulting in small decreases in the number of novel markers detected in the resampled data.
We next investigated the relationship between the number of resampled cells in a cell population and the ability to detect new cell-specific markers. We computed differentially expressed genes between the four megakaryocyte cells selected for resampling and all other non-megakaryocyte cells in the PBMC dataset, and then iteratively supplemented the four cells with additional megakaryocyte cells until all of the megakaryocytes were present in the cluster (Figure 3C). At small cluster sizes, the resampled cells provided additional power to detect novel markers, but the relative increase in markers detected decreased with increasing cluster size.
Finally, we examined the relationship between the number of resampled cells and the overall number of genes recovered in the megakaryocyte cluster after resampling. Supplementing the cluster with the resampled libraries increases the overall number of genes recovered; however, at higher cells numbers the relative increase in genes detected is smaller (Figure 3D). Overall, we increased the total number of detectable genes in the megakaryocyte cluster from 11,209 to 11,377. These results demonstrate that the resampled libraries allow for increased recovery of marker genes in a cell population, but the contribution of any specific resampled cell will be influenced by the initial size of the cell population sampled and the increase in the number of recovered genes in resampled cells.
Specificity of cell barcode targeting
We considered whether hybridization-based isolation of cell-specific transcriptomes might lead to stochastic enrichment of other, non-targeted cells, so we examined the level of cross-reactivity between hybridization probes and non-targeted cell barcodes. For both the human/mouse cell and megakaryocyte experiments, we compared the level of enrichment for targeted and non-targeted barcodes with their propensity for cross-hybridization using a Smith-Waterman alignment-based score and their Hamming distance (Supplementary Figure S8). For the targeted human and mouse cells, we found that the cognate cell barcode was the highest alignment score and lowest Hamming distance to the hybridization probe (Supplementary Figure S8A). We also found two non-targeted cell barcodes with elevated enrichments above two-fold and we currently cannot explain why they are enriched. It is possible that this apparent enrichment is due to stochasticity in the low numbers of UMIs recovered for these cells. Alternatively, it is possible that these barcodes may have been enriched due to hybridization to intervening sequence in the cDNA, which is invisible in the experiment because we collected short reads from either end of the amplicon (Supplementary Figure S8A). We performed a similar analysis for targeted megakaryocytes and found that for all four cell barcodes targeted by hybridization, the cognate barcodes had the highest alignment scores and lowest Hamming distances of any recovered barcodes (Supplementary Figure S8B), and we did not find any enrichment for non-targeted barcodes.
Enhanced recovery of TCR mRNAs from single cells
Another common scRNA-seq application is profiling VDJ-rearranged B and T-cell receptor sequences. We applied LNA-based hybridization method to resample individual cells to recover additional VDJ rearranged receptor sequences. A TCR receptor enrichment library was prepared from the 5′ end Jurkat cell library using targeted PCR with primers specific for the TCRA and TCRB mRNAs (Figure 4A). We selected two cells for which the TCRB chain was assembled, but in which the TCRA chain had not been successfully assembled. After resampling we detected enrichment for the targeted cells (Figure 4B), and were able to assemble full-length TCRA chain in these cells after resampling (Figure 4C and Supplementary Figure S9). The resampled cells had additional read coverage that spanned the consensus Jurkat TCRA and TCRB chains (Figure 4D). These results demonstrate that resampling can be applied to recover single cell VDJ-rearranged TCR sequences from libraries that did not yield fully assembled TCR receptor genes.
Isolation and resequencing of targeted mRNA subsets
The hybridization-based resampling method can in principle be applied to isolate arbitrary oligonucleotide sequences in a single cell RNA-seq library. A common challenge with single cell RNA-seq libraries is the low cellular detection rate of low or moderately expressed genes due to stochastic capture of mRNA molecules from cells. These ‘gene dropout’ events (i.e. false negative mRNA identifications) can prevent the identification of cells expressing key marker genes or trangenes designed to identify cell populations. We therefore examined the suitability of the resampling method to enrich specific mRNA sequences and enable interrogation of the expression of specific mRNAs across all cells.
As a proof of principle, we generated a non-saturated 5′ end gene expression library on the 10x Genomics platform from Jurkat cells (an immortalized human T cell line). An initial round of sequencing showed that the mRNA for delta chain of the CD3 T-Cell coreceptor (CD3D) was only detectable in 59.7% of cells despite being highly expressed in this cell line (23). To increase the cellular detection rate of the CD3D mRNA, we designed hybridization probes that target the 5′ end of CD3D mRNA (Figure 4). We considered several points in the design of probes to the CD3D mRNA. When designing probes against an mRNA sequence there are fewer sequence constraints than with cell barcodes with fixed position and length, and therefore longer DNA-only probes might be used to increase the probe Tm. Both LNA and DNA hybridization probes directed to the 5′ ends of CD3D mRNA were designed and used to isolate CD3D fragments from 5′ gene expression libraries. We first identified a region with high read coverage in the original library near the annotated CD3D transcription start site. We then used a oligonucleotide probe design tool (Primer3) to pick candidate ∼25 nt sequences for LNA probes; we also used these sequences a basis for DNA hybridization probes, but included another 10–15 bases on each side of the LNA probe candidate sequences. Next, we used BLAST to rule out probes with poor E-scores or long stretches of off-target complementarity in human DNA. Finally, we selected probes with GC content >50% (especially at their 3′ ends), Tm values between 75 and 85°C, and a low propensity form secondary structures at the predicted Tm. In addition to an LNA containing probe (20 nt, Tm = 71°C), we also generated a DNA only probe (40 nt, Tm = 66°C) to determine whether LNAs would be necessary for specific hybridization (Figure 4E).
After resampling with the LNA and DNA probes we observed specific enrichment of the CD3D mRNA (Figure 4F and G), increasing the cellular detection rate of CD3D from 59.7% to 100% of cells. In addition, other CD3-associated mRNAs (CD3E and CDEG) were not enriched after resampling (Figure 4G). After resampling, the read coverage profiles across the CD3D mRNA remained similar, indicating minimal bias in the read coverage introduced by resampling (Figure 4H). These results demonstrate the utility of using LNA and DNA hybridization probe for resampling specific mRNA species.
Recovery of individual transcriptomes by targeted PCR
We additionally tested a PCR-based strategy to recover targeted single cell transcriptomes. The 10x Genomics platform uses cellular barcodes that are at a minimum Hamming distance 2 apart, which cannot be reliably distinguished by standard PCR approaches. In contrast, other platforms such as the Wafergen iCell8 system, have more diverse barcodes (Hamming distance of 3), with limited numbers of barcodes detected per experiment (1000s of detectable barcodes versus 100,000s of barcodes in 10× Genomics or DropSeq libraries). The Wafergen iCell8 libraries contain an 11 nucleotide cell barcode (Supplementary Figure S10A).
To investigate the utility of a PCR-based approach, we designed PCR primers to anneal to cell barcodes and recover the transcriptomes of single cells from a scRNA-seq library. We tested three PCR approaches to recover single cell transcriptomes. The first strategy used standard DNA primers, the second strategy incorporated a 5′ biotin to enable stringent purification of PCR products, and the final strategy additionally incorporated phosphorothioate linkages into the terminal three 3′ nucleotides to prevent 3′-to-5′ exonucleolytic cleavage by the proofreading Phusion DNA polymerase (see Supplementary Table S2 for primer design).
To test these PCR strategies, we selected either 10 (standard and biotinylated approach) or 5 cells (phosphorothioate approach) that spanned ∼100-fold in sequencing coverage and were derived from either human cells (from a breast cancer tumor xenograft) or mouse cells (host mouse bone marrow derived cells). We performed two rounds of PCR with low cycle numbers to enrich the libraries. We performed 14 cycles of amplification in individual reactions, then either pooled the resulting PCR products for the standard approach, or for the biotinylated and phosphorothioate approach purified the PCR products using AMPure purification followed by streptavidin magnetic beads to remove unamplified library material. Lastly, we performed a second round of PCR to incorporate library indexes and sequences required for flow cell clustering. We observed 7.7-, 8.6- and 19.7-fold enrichment for the targeted barcodes in the resampled sequencing libraries, for the standard, biotinylated, and biotinylated with phosphorothioate approach respectively (Supplementary Figure S10C–E). Both the standard PCR and biotinylated purification approach were enriched for many non-targeted sequences. In contrast, barcodes were specifically enriched over non-targeted barcodes by the third PCR approach. These results indicate that incorporation of phosphorothioates is necessary to achieve specific amplification, and suggests that 3′-to-5′ exonucleolytic activity of the Phusion polymerase is responsible for the observed non-specific amplification.
A caveat of using a direct PCR resampling approach is the potential for rebarcoding of similar barcode sequences. Non-specific hybridization to similar barcode sequences would result in the amplification of non-targeted cell barcode sequences. These off-target sequences would be misclassified as a resampled cell due to the PCR primer sequence rebarcoding the original off-target cell barcode. To investigate the extent of rebarcoding in these libraries we assessed the proportion of UMIs that were found in multiple cells in the original library and the resampled library. In the LNA-based hybridization method, the fraction of UMIs that were detected in multiple cells in the original library did not appreciably increase in the resampled libraries (0.76% ± 0.26 shared in original library, 1.80% ± 0.47 shared in resampled libraries) (Supplementary Figure S3B), indicating that the novel UMIs recovered after resampling were not likely derived from other cells. In contrast, resampling with the PCR method resulted in increases in the proportion of UMIs that were found in multiple cells (0.39% ± 0.26 to 3.52% ± 1.75 for the phosphorothioate approach) (Supplementary Figure S10G). Additionally the percentage of UMIs assigned to the correct species was lowered after resampling with the PCR approach (for the phosphorothioate approach, 91.26% ± 8.8 in the original library compared to 76.66% ± 27.5 for the resampled library) (Supplementary Figure S10H). These results demonstrate that a direct PCR based resampling approach can result in undesired off-target rebarcoding, which for the purposes of examining single cell gene expression profiles makes the PCR approach less desirable that the LNA-based approach.
DISCUSSION
Here, we demonstrate how resampling of individual transcriptomes from scRNA-seq libraries provides richer information for selected cell and mRNA populations. One consideration in the design of probes for the resampling approach is the design and structure of barcode information in single-cell mRNA sequencing libraries. We leveraged the contiguity of the cell barcode in 10× Genomics and Wafergen libraries to design probes and primers that can effectively target molecules by hybridization. Such an approach could also be used for other platforms where a contiguous cell barcode is synthesized on each bead (e.g. Drop-seq, Supplementary Table S1) (2). The 10× Genomics Chromium platform uses a small set of fixed cell barcodes (737,280 in version ‘737K-august-2016.txt’) and each of these have on average ∼13 sequences within Hamming distance of 2. For 10× Genomics libraries, we opted for the hybridization approach as it is less likely to lead to ‘recoding’ of cells in the library. Moreover, because cell barcodes are short in 10× Genomics libraries (16 nt), we used locked nucleic acid probes that target only the barcode region.
A recent study used hemi-specific primers to amplify cell barcodes of interest from 10× Genomics scRNA-seq libraries (11). This study showed enrichment for the targeted barcodes of interest (up to ∼100-fold), and the recovered libraries had similar expression profiles to the original expression. However, we found that PCR resampling is subject to artifacts because primers designed against one barcode might misprime on and amplify a related barcode, causing inclusion of mRNA from an unrelated cell in the resampled transcriptome (Supplementary Figure S10). In addition, primers that mishybridize to a template are subject to 3′-5′ exonuclease activity by proofreading DNA polymerases (e.g. Phusion) during PCR, causing amplification of non-targeted barcodes and reducing recovery of targeted barcodes. Terminal phosphorothioate linkages effectively mitigate this exonuclease activity (Supplementary Figure S10E), improving specific recovery of targeted barcodes. Addition of 3′ phosphorothioate linkages is therefore strongly recommended if a PCR approach is used for resampling.
Before embarking on a resampling experiment one must ensure that a single-cell mRNA sequencing library is sufficiently complex as to yield additional information after targeted resampling and resequencing. Libraries with high overall saturation observed in the first round of sequencing are unlikely to benefit from resampling because most UMIs have already been observed. We selected libraries with less than ∼66% saturation to maximize the information gained from specific cells, although libraries with higher saturation levels could still provide novel UMI or gene discovery, the relative enrichment will be diminished as the saturation increases. New methods that increase cell numbers in single-cell experiments (BioRxiv: https://doi.org/10.1101/237693 and https://doi.org/10.1101/315333) will benefit from transcriptome resampling because as cell numbers increase, DNA sequencing becomes limiting and fewer reads are recovered per-cell from these more complex libraries. Transcriptome resampling may enable an initial low-depth examination of many cells followed by more targeted analysis of defined populations. As an example, we increased sequencing coverage for four megakaryocytes (out of a library with 3194 detected cells; Figure 2A) between 6- and 20-fold after resampling. The current recommended maximum for captured cells in the 10X Genomics workflow is 10,000 cells; therefore if a cell were resampled from this larger population, we would expect and 18- to 60-fold increase in its sequencing coverage, assuming the same sequencing depth and efficiency of hybridization for cell-specific probes.
Both LNA and DNA probes performed well for CD3D mRNA, increasing the percentage of cells with detected expression from 59.7% to 100%. We also found a DNA probe that targeted the same region of CD3D provided enhanced recovery of UMIs associated with CD3D mRNAs (Figure 3C). We attribute the superior recovery of DNA compared to the LNA probe to the increased length of the DNA probe (40nt DNA versus 20nt LNA), as the Tm of the DNA probe (66°C) was not appreciably different from the LNA probe (71°C). DNA probes are less expensive than LNA probes (∼$60 per probe for biotinylated DNA versus ∼$200 per LNA probe) and may be more cost effective when targeting multiple mRNAs. In theory a PCR approach could also be used to target specific genes for resampling, however we instead used a hybridization approach which can be scaled to target 10s or 100s of target mRNAs with minimal modification to the method. We envision pooling subsets of mRNA-specific probes to more fully characterize gene expression programs in cells from specific contexts (e.g. interferon-stimulated (24) or stress-response (25) expression programs).
In principle, transcriptome resampling might be used to query other features of mRNA expression and processing. For example, full-length mRNAs might be studied in greater detail with single-molecule sequencing approaches by isolating molecules from libraries or library preparation steps that contain full-length cDNA (e.g., Smart-Seq2 (26) or the full-length cDNA generated during the 10× Genomics library preparation). Indeed in a recent study, T-cell and B-cell receptor mRNAs were isolated via hybridization from full-length cDNA from a 10× Genomics library and subjected to single-molecule sequencing, enabling characterization of immune repertoires in parallel with gene expression (BioRxiv: https://doi.org/10.1101/424945). However, probe-based mRNA isolation is subject to some caveats. Several biological processes create variation at mRNA 3′ ends including alternative polyadenylation and 3′ UTR splicing. In addition, 3′ end libraries have a large degree of internal mispriming at genomically-encoded poly(A) stretches (27), potentially rendering a large proportion of the cDNA from a given mRNA unable to be captured using a 3′ end targeted probe. It is possible that 5′ gene expression libraries have a more homogeneous representation of a given mRNA 5′ end, enabling the design of hybridization probes that target a majority of mRNA isoforms.
We anticipate that the recovery of individual transcriptomes will facilitate characterization of rare cell populations identified in scRNA-seq experiments. However, the structure of DNA barcodes in a scRNA-seq library impacts the generality of the resampling approach. We applied resampling to libraries wherein cell barcodes are encoded by a contiguous region of DNA. As such, a single hybridization probe can specifically recover information for an individual cell. Other library designs employ discontinuous cell bar codes (e.g. sci-RNA-seq (4)); here the information needed to associate an mRNA with a single cell is present at different sites in the molecule (i.e. a linker sequence in addition to the two library indices). In this case, enrichment for a portion of the cell barcode would likely provide additional information for the cell of interest, but would also enrich for other unrelated cells because some cell barcode information is distant to the site of hybridization.
Resampling could also be used to recover molecules from other types of complex single cell libraries. Single-cell ATAC-seq and DNA-seq have been used to probe chromatin accessibility and copy number variation in individual cells (28–30). Because the amplicons in these libraries have structure similar to scRNA-seq libraries with a cell barcode and UMI, one could resample cells with interesting chromatin properties from these mixed populations. Detection of regulatory regions and transcription factor footprints is highly dependent on read coverage (31) and deeper sequencing of recovered libraries could provide more insight into gene regulation than can be gained from the mixed population, possibly enabling interrogation of how promoters and enhancers are correlated in accessibility in single-cell ATAC experiments or providing increased depth of coverage for targeted domains in single-cell Hi-C (32).
DATA AVAILABILITY
DNA sequencing data are available from NCBI GEO under accession GSE119428. A reproducible software pipeline (including Snakemake (20) pipeline and R Markdown documents) is available at https://github.com/rnabioco/scrna-subsets. Processed data is available from zenodo at https://doi.org/10.5281/zenodo.1405578.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Srinivas Ramachandran for comments on the manuscript, and Katrina Diener and Todd Woessner for expert technical assistance. Single-cell RNA sequencing libraries and Illumina sequencing was performed in the University of Colorado Cancer Center Genomics Shared Resource (P30 CA046934).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
RNA Bioscience Initiative at the University of Colorado School of Medicine and the National Institutes of Health [R35 GM119550 to J.R.H.]. Funding for open access charge: Grant funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. et al. . Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. et al. . Highly parallel Genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W.. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015; 161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J. et al. . Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017; 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pollen A.A., Nowakowski T.J., Shuga J., Wang X., Leyrat A.A., Lui J.H., Li N., Szpankowski L., Fowler B., Chen P. et al. . Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014; 32:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bacher R., Kendziorski C.. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016; 17:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M. et al. . Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016; 166:1308–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olsson A., Venkatasubramanian M., Chaudhri V.K., Aronow B.J., Salomonis N., Singh H., Grimes H.L.. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016; 537:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu Y., Tsang J.C.H., Wang C., Clare S., Wang J., Chen X., Brandt C., Kane L., Campos L.S., Lu L. et al. . Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its developmental pathway. Nature. 2016; 539:102–106. [DOI] [PubMed] [Google Scholar]
- 10. Plasschaert L.W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., Jaffe A.B.. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018; 560:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ranu N., Villani A.-C., Hacohen N., Blainey P.C.. Targeting individual cells by barcode in pooled sequence libraries. Nucleic Acids Res. 2018; doi:10.1093/nar/gky856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kabos P., Finlay-Schultz J., Li C., Kline E., Finlayson C., Wisell J., Manuel C.A., Edgerton S.M., Harrell J.C., Elias A. et al. . Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res. Treat. 2012; 135:415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanna C., Kwok L., Finlay-Schultz J., Sartorius C.A., Cittelly D.M.. Labeling of breast cancer patient-derived xenografts with traceable reporters for tumor growth and metastasis studies. J. Vis. Exp. 2016; 117:e54944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G.. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011; 17:10–12. [Google Scholar]
- 16. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 18. Smith T., Heger A., Sudbery I.. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017; 27:491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Köster J., Rahmann S.. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 21. Nakeff A., Maat B.. Separation of megakaryocytes from mouse bone marrow by velocity sedimentation. Blood. 1974; 43:591–595. [PubMed] [Google Scholar]
- 22. Lefrançais E., Ortiz-Muñoz G., Caudrillier A., Mallavia B., Liu F., Sayah D.M., Thornton E.E., Headley M.B., David T., Coughlin S.R. et al. . The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017; 544:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. et al. . The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider W.M., Chevillotte M.D., Rice C.M.. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014; 32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Nadal E., Ammerer G., Posas F.. Controlling gene expression in response to stress. Nat. Rev. Genet. 2011; 12:833–845. [DOI] [PubMed] [Google Scholar]
- 26. Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R.. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013; 10:1096–1098. [DOI] [PubMed] [Google Scholar]
- 27. Shepard P.J., Choi E.-A., Lu J., Flanagan L.A., Hertel K.J., Shi Y.. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011; 17:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cai X., Evrony G.D., Lehmann H.S., Elhosary P.C., Mehta B.K., Poduri A., Walsh C.A.. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2015; 10:645. [DOI] [PubMed] [Google Scholar]
- 29. Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J.. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015; 348:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J.. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015; 523:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. et al. . ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012; 22:1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ramani V., Deng X., Qiu R., Gunderson K.L., Steemers F.J., Disteche C.M., Noble W.S., Duan Z., Shendure J.. Massively multiplex single-cell Hi-C. Nat. Methods. 2017; 14:263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequencing data are available from NCBI GEO under accession GSE119428. A reproducible software pipeline (including Snakemake (20) pipeline and R Markdown documents) is available at https://github.com/rnabioco/scrna-subsets. Processed data is available from zenodo at https://doi.org/10.5281/zenodo.1405578.