Abstract
Changes in gene regulation are important for phenotypic and in particular morphological evolution. However, it remains challenging to identify the transcription factors (TFs) that contribute to differences in gene regulation and thus to phenotypic differences between species. Here, we present TFforge (Transcription Factor forward genomics), a computational method to identify TFs that are involved in the loss of phenotypic traits. TFforge screens an input set of regulatory genomic regions to detect TFs that exhibit a significant binding site divergence signature in species that lost a particular phenotypic trait. Using simulated data of modular and pleiotropic regulatory elements, we show that TFforge can identify the correct TFs for many different evolutionary scenarios. We applied TFforge to available eye regulatory elements to screen for TFs that exhibit a significant binding site decay signature in subterranean mammals. This screen identified interacting and co-binding eye-related TFs, and thus provides new insights into which TFs likely contribute to eye degeneration in these species. TFforge has broad applicability to identify the TFs that contribute to phenotypic changes between species, and thus can help to unravel the gene-regulatory differences that underlie phenotypic evolution.
INTRODUCTION
Morphological differences are a hallmark of phenotypic diversity between species. It is assumed that changes in morphology largely involve changes in the expression pattern of genes that play key roles in development (1–3). Such expression changes are often due to differences in cis-regulatory elements (CREs) such as promoters and distal enhancers that control the expression level and pattern of a gene. Cis-regulatory activity is determined by transcription factors (TFs) that bind to a CRE and activate or repress transcription. To understand how differences in morphology and other phenotypes evolved, it is necessary to identify functional differences in CREs. However, despite the availability of numerous sequenced genomes and functional genomics approaches that uncover CREs active in specific tissues, it remains challenging to detect the TFs and CREs that contribute to phenotypic differences between species.
To detect CREs that are associated with phenotypic differences, we recently extended the general Forward Genomics framework (4) and developed a computational method called Regulatory Element forward genomics (REforge) (5). This approach focuses on phenotypes that are lost in independent lineages and screens for regulatory elements that exhibit TF binding site (TFBS) divergence in species that lost the given phenotype. We expect CREs that are only necessary for a single phenotypic trait to evolve neutrally upon loss of this trait. Neutral evolution will lead to a gradual decay of important TFBSs, eventually leading to loss of regulatory activity in trait-loss species. In contrast, these CREs typically evolve under selection to preserve regulatory activity in species that possess the trait. This selective pressure will often preserve binding sites for important transcriptional regulators within the CRE sequence. Over time, this difference between selection and neutral evolution is expected to result in a preferential maintenance of TFBSs in trait-preserving lineages and a preferential decay of these binding sites in trait-loss lineages. Given a set of motifs of relevant TFs, REforge uses this characteristic divergence signature to screen genome-wide for CREs that exhibit preferential decay of TFBSs in the independent trait-loss lineages.
The success of REforge crucially depends on prior knowledge about TFs that are relevant for the given phenotype. While functional annotations such as gene expression patterns or knockout phenotypes in model organisms (6) can be used to select potentially relevant TFs, it is in general unknown if these TFs actually contribute to the phenotypic change. Furthermore, this annotation-based TF selection strategy is limited to the small subset of phenotypes for which relevant TFs are known. Finally, while many computational methods exist to find motifs that are enriched in a given set of DNA sequences, TFBSs that are overrepresented in one set relative to another set, or TFBSs that are evolutionarily-conserved (7–10), no computational method exists to detect TFs that preferentially lost binding sites in trait-loss species and thus may contribute to this phenotypic change.
To computationally detect TFs that are associated with phenotypic differences, we developed a new method called TFforge (Transcription Factor forward genomics). In contrast to REforge, TFforge jointly considers a set of CREs and screens a library of motifs to infer which TFs exhibit a widespread binding site decay signature in the trait-loss lineages. We validated TFforge on synthetic data obtained by simulating regulatory element evolution. We further applied TFforge to the phenotype ‘eye degeneration in subterranean mammals’, which provided novel insights into which TFs are involved in this repeated trait loss. By identifying transcriptional regulators, TFforge will help to understand the changes that contribute to gene regulatory differences and ultimately phenotypic changes between species.
MATERIALS AND METHODS
Overview of TFforge
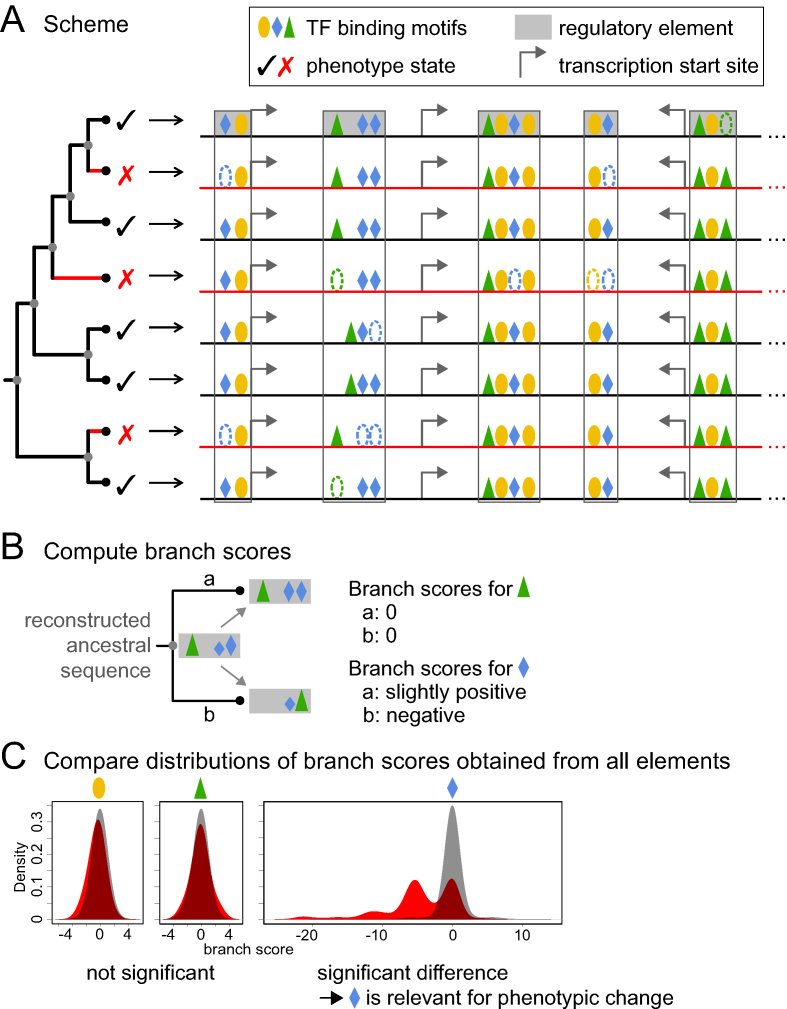
The main idea behind TFforge is illustrated in Figure 1A. If a TF is important for gene regulation in a certain cell type or tissue in trait-preserving species, then its binding sites should be largely conserved. If a TF becomes less important for this cell type or tissue in trait-loss species, it is expected that neutral mutations weaken or destroy many binding sites of this TF over time.
Figure 1.
Illustration of the TFforge principle. (A) Illustration of TFBS differences across a phylogeny of species. In contrast to the green-triangle and orange-circle TF, the blue-diamond TF preferentially lost binding sites on the branches (red) leading to the trait-loss species (red cross). Dashed ovals illustrate TFBS loss. Cases where the presence but not the position of TFBSs is conserved illustrate TFBS turnover. Black and gray dots in the phylogenetic tree represent extant species and reconstructed ancestors, respectively. A grey filled box represents a tissue-specific or non-tissue-specific CRE experimentally determined for the first (reference) species in the tree. The open box represents orthologous sequence of other species that aligns to this CRE. (B) Illustration of branch scores. For each TF motif separately, TFforge scores binding sites in all extant and reconstructed ancestral sequences and computes the score difference between the start and end of a branch, which captures TFBS differences that occurred on each branch. The height of the symbol represents binding site strength. (C) Comparing branch score distributions. After computing all branch scores for all regulatory elements, TFforge compares the score distributions between the two groups of branches (red versus black in panel A). If there is no preferential loss of TFBSs on the branches leading to trait-loss species, both distributions will be similar, as shown for the orange-circle and green-triangle TF. In case of preferential loss of binding sites on the trait-loss branches, as illustrated for the blue-diamond TF, there should be a significant difference between both distributions. Bimodality may arise from branches along which TFBSs were conserved (score of ∼0) or lost (negative score).
TFforge requires as input (i) a library of Position Weight Matrices (PWMs) that represent the TF binding motifs, (ii) a phylogenetic tree, (iii) a set of CREs and their orthologous sequences in a set of species (fasta format) and (iv) a binary classification that assigns each species in the tree to either group A or group B. Without loss of generality, we assume in the following that group A species have lost a certain phenotype which is present in group B species and test if the branches associated with group A (trait-loss) species have a tendency to lose or weaken TFBSs compared to the branches associated with group B (trait-preserving) species. However, the general framework is flexible as branches can be assigned to more than two arbitrary groups and the opposite direction of TFBS divergence (gain or strengthening of binding sites) can be tested as well.
Given a set of CREs that are active in the tissues relevant for the phenotype and a library of TF binding motifs, TFforge considers one TF at the time and determines if this TF has a tendency to lose TFBSs in the trait-loss species in comparison to all other species. CREs that are active in the relevant tissues can be obtained from high-throughput functional genomics approaches like ATAC-seq, DNaseI-seq, or ChIP-seq for histone marks or transcription factors (11–13). It is sufficient if the CRE annotation is provided for only one species (for example human or mouse) that serves as a reference species in the comparative framework, since TFforge only uses sequences of other species that are orthologous to the given CREs. TFforge can be applied to both tissue-specific CREs and CREs that are also active in other tissues (pleiotropic CREs, see below).
Computing sequence scores
Given a CRE, we first reconstruct all ancestral sequences in the phylogenetic tree with Maximum Likelihood, using a multiple alignment of the CRE sequences of all species. TFforge uses a given TF motif to estimate the binding affinity of this TF to the sequences that represent either extant or ancestral species in the tree. To this end, we use the Hidden Markov model (HMM)-based method Stubb (14,15) (version 2.1). Stubb uses the Forward Algorithm to compute the probability that the sequence was generated by an HMM that emits either TFBSs, sampled from the given motif, or background sequence. Stubb then computes the probability that the sequence was generated by a second HMM that only emits background sequence without TFBSs. The Stubb score is the log-likelihood ratio of both probabilities, capturing how likely the sequence was generated by the motif-emitting HMM. Since the Forward Algorithm considers each possible path through the HMM, weighted by its probability, Stubb does not require fixed thresholds for TFBSs but rather integrates the contribution of both weak and strong binding sites proportionally to their strength. This avoids a main drawback of threshold-based approaches that ignore TFBSs just below the threshold and consider all TFBSs above it as equal regardless of their strength. Furthermore, Stubb makes no assumption about the absolute position of a TFBS within the CRE sequence. Consequently, if mutations destroy a TFBS at one place and create an equivalent TFBS elsewhere in the CRE sequence (TFBS turnover, (16,17)), the Stubb score will be largely the same.
TFforge starts by scoring the sequence of the common ancestor, for which we let Stubb optimize the transition probability from the background into the motif state with expectation maximization. To avoid fluctuations in transition probability estimation that can influence the comparability of Stubb scores, TFforge then uses the same transition probabilities to score the sequences of extant or ancestral species that descends from the common ancestor. While the emission probabilities of the motif state are determined by the PWM, the emission probabilities of the background HMM state are estimated from a given background sequence. To make Stubb scores comparable between species, we generated a fixed set of random sequences with different GC-contents and use the pre-defined random sequence that matches the GC-content of the real sequence as the input background sequence. Finally, TFforge converts Stubb scores into ‘sequence scores’ by shuffling the bases in each sequence 10-times and subtracting the average Stubb score of these 10 randomized sequences from the real Stubb score. In contrast to Stubb scores, these sequence scores are on average zero for random sequences that contain TF binding sites only by chance. TFforge uses this property to exclude uninformative branches, as described below.
Computing branch scores
Since species are phylogenetically related, the sequence scores cannot be directly compared between species. Therefore, TFforge adopts the Forward Genomics branch method (18) and computes ‘branch scores’ that capture differences in TFBSs for every branch in the phylogenetic tree. TFforge traverses the phylogenetic tree from root to leaves and computes for every branch the score difference between the end and the start node (Figure 1B). These branch scores are positive if existing TFBSs were strengthened or if new TFBSs were gained. Weakening or loss of TFBSs results in negative branch scores. A branch score of ∼0 indicates that TFBSs remained largely the same; however, TFBS turnover is allowed as there is no constraint that TFBSs must occur at the same positions. Alternatively, a branch score of ∼0 can also arise if both the start and end node of the branch have sequence scores of ≤0, indicating that in comparison to randomized sequences no significant TFBS is present. Since such branches are uninformative, we excluded branch scores for which both start and end node have sequence scores ≤0 for computing the significance (below). Since every branch is phylogenetically independent, branch scores can be directly compared and no further correction for phylogenetic relatedness is necessary.
Computing the significance of the TF motif
Given the list of group A and B species, TFforge employs Dollo parsimony to assign all internal tree nodes to either group A (trait-loss) or B (trait-preserving). Then, we assign each branch to group A or B, depending on the group assignment of the end node of the branch. If binding sites of a TF preferentially evolve neutrally on the group A branches, the respective branch scores should be lower than the scores of group B branches. To test this, TFforge pools the group A and B branch scores from the entire set of CREs and computes the significance P-value of a positive Pearson correlation between the branch scores and the group assignment. This P-value is used to identify the TFs that are significantly associated with the given phenotypic difference. Using simulated data (below), we tested the power of a t-test, Wilcoxon-rank sum test and other methods, and found that Pearson correlation performed best for our data (Supplementary Figure S1). By considering each TF motif, one at the time, TFforge outputs a list of TFs ranked by their P-value. Finally, P-values are adjusted for multiple testing by the Benjamini-Hochberg procedure. While TFforge considers a single TF motif at the time, it combines the branch scores obtained for a set of CREs, which typically provides larger sample sizes and thus statistical power.
Construction of TF motif library
In order to compile a library of TF binding motifs, we integrated motifs from three widely used databases. First, we downloaded all PWMs in forward complement orientation from UniPROBE (19), a database containing TF motifs obtained with protein-binding microarrays. We kept all motifs for which the UniProt or Swiss-Prot ID of the TF could be converted to an Ensembl gene ID. Second, we obtained motifs from TRANSFAC Pro 2014.3 (20), a database for eukaryotic TFs and their binding motifs. We focused on motifs of vertebrate TFs and required that each frequency matrix is either based on at least 20 sequences or that the motif was derived from 3D structures. Third, we downloaded the collection of non-redundant vertebrate TFs from JASPAR (21). All frequency matrices were converted into probability matrices. We removed all unspecific motifs by requiring that a motif has an information content of at least six bits and removed motifs for which we could not determine the Ensembl gene ID of the TF. This resulted in a total of 2197 motifs, which we converted into Stubb's weight matrix input file format.
In order to cluster highly-similar motifs or redundant motifs for the same TF, we computed pairwise similarity scores with Tomtom (22) (parameters: ‘-thresh 1’, ‘-dist ed’). Motifs with a pairwise similarity score ≤0.0001 were clustered together, resulting in 614 clusters. We then selected the PWM that is most similar to all other motifs within a cluster as the cluster representative. The motifs of the 614 clusters representatives constitute our motif library (Supplementary Table S1). While clustering very similar motifs reduces the runtime of TFforge by avoiding scoring redundant motifs repeatedly, clustering is an optional step.
Creating simulated CRE datasets
To test TFforge, we first generated a synthetic dataset of CREs where the TFs whose binding sites evolve under purifying selection along group B branches and neutrally along group A branches are fully known. To this end, we made use of GEMSTAT and PEBCRES (23,24) to simulate the evolution of regulatory elements. GEMSTAT predicts regulatory activity from the sequence of a CRE using the binding preference of TFs and information of TF expression level and activator/repressor strength. PEBCRES evolves a CRE using a discrete-time Wright-Fisher model with a fixed size population. In each generation, PEBCRES introduces random mutations into the CRE sequences and samples sequences with replacement for the next generation. The probability of sampling a sequence is proportional to its fitness, which in turn is proportional to how well the predicted CRE activity matches a chosen ideal activity. The ideal activity is a user-defined fixed expression profile. A maximum fitness of 1 is reached if predicted and ideal activity are equal. We set PEBCRES mutagenesis parameters to mutation_rate 1e-04, substitution_probability 0.95, insertion_probability 0.5, and tandem_repeat_probability 0.2 and simulated a population of 50 sequences.
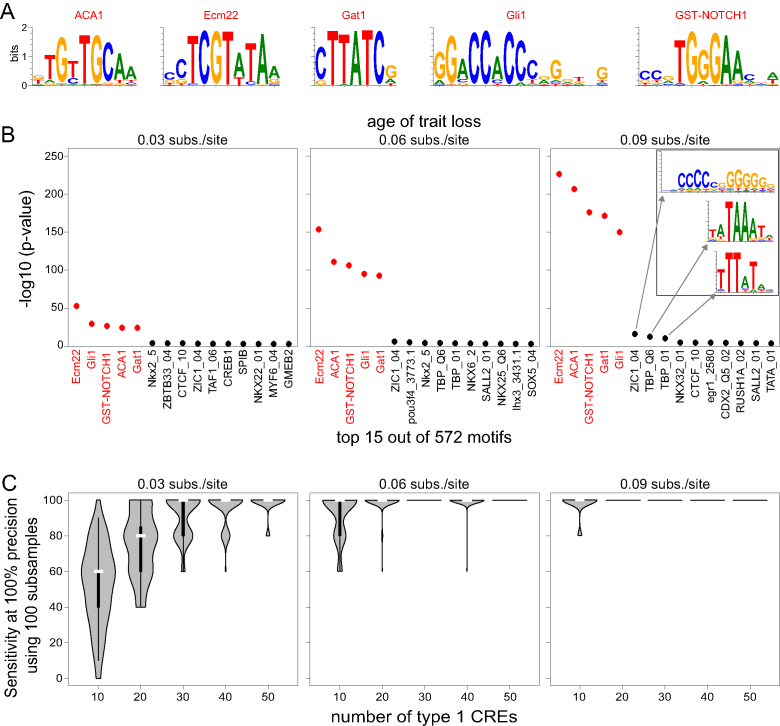
First, we simulated modular (non-pleiotropic) CREs that have an ideal activity of 100% expression level in a single tissue. In this simulation, CRE activity is controlled by five foreground TFs (Figure 2A), which have equal concentration levels in this tissue and are activators of equal strength. These five TFs were selected from all UniPROBE motifs and are sufficiently different from each other. The start point for the simulation of a CRE’s evolution is the sequence of the common ancestor. To this end, we randomly generated a 200 bp sequence, in which we implanted five non-overlapping binding sites for randomly selected foreground TFs at random positions. We discarded all ancestral CRE sequences with a start fitness of <0.85. Then we evolved the CRE sequence along every branch in the 20-species phylogeny. The PEBCRES parameter ‘num_generations’ was set such that the total number of mutations expected on a branch is equal to the branch length (e.g. 100 generations at a mutation rate of 1e-04 correspond to 0.01 substitutions per site). After obtaining the evolved population of 50 sequences at an internal node, we independently evolved this population along the two descending branches. For every internal node and every extant species in the tree, we selected the sequence with median fitness out of the 50 simulated sequences as the single representative sequence to compute sequence and branch scores.
Figure 2.
Application of TFforge to simulated data. (A) Motifs of the five randomly-selected foreground TFs. (B) The plots show the top-ranked 15 TF motifs for three trait-loss ages (corresponding to neutral evolution for 0.03/0.06/0.09 substitutions per site). Red font indicates motifs for foreground TFs that control the activity of 100 simulated type 1 CREs that evolve neutrally after trait loss. The inset on the right side shows the top 3 background motifs. Despite belonging to different motif clusters, these background motifs partially resemble foreground motifs (ZIC1 has some similarity to GST-Notch and Gli1, the two TBP motifs to Gat1). This suggests that predicted binding sites for these background TFs may overlap suboptimal binding sites of some of the foreground TFs, which provides an explanation why TFforge detects these motifs at ranks 6 to 8. Importantly, the significance of these motifs is substantially lower than the significance of the five foreground motifs. (C) Performance of TFforge on 100 subsamples of type 1 CREs of various sizes. Violin plots show the distribution of the sensitivity at a precision of 100%, which corresponds to the number of foreground TF motifs that have a higher significance than the most significant background TF motif. The vertical black bar inside a violin plot spans the first and third quartile, the white bar indicates the median.
We assigned three independent species as trait-loss species and the remaining 17 as trait-preserving species (Supplementary Figure S2). Trait-preserving branches evolved under selection to preserve the ideal regulatory activity by setting the PEBCRES selection parameters to D_max = 1, selectionExp = 2, selectionScale = 100, and selectionCoeff = 0.1. Branches leading to a trait-loss species were split into two parts. The first (upstream) part evolved under purifying selection until the simulated trait loss event occurred. After trait loss, the second (terminal) part of the branch evolved neutrally by setting selectionCoeff = 0, which removes the influence of fitness during the Wright–Fisher selection step. We simulated three different trait-loss scenarios where the events correspond to a final branch length part of 0.09, 0.06 or 0.03 substitutions per site (Supplementary Figure S2). For each of the three trait-loss time points, we simulated the evolution of a total of 1000 of such CREs, called type 1 CREs in the following.
To test TFforge on this single-tissue scenario, we obtained background TF motifs that are irrelevant for the activity of the simulated CREs by extracting the 567 motifs from our library that are sufficiently distinct (Tomtom similarity score ≥ 0.01) from any of the five foreground TFs. Then, we determined if TFforge is able to detect the five foreground TF motifs given a set of 572 motifs that contains 99.1% background motifs.
We explored robustness of TFforge to detect foreground TF motifs given an input CRE dataset that not only contains type 1 CREs. To this end, we simulated two additional types of CREs. Type 2 CREs are created using an identical simulation setting as type 1 CREs, but they evolve under selection in all 20 species. Type 3 CREs evolve as type 2 CREs, but they are active in another single tissue and regulatory activity of these CREs is controlled by five different activator TFs.
To assess the influence of ancestral sequence reconstruction on the performance, we aligned the sequences of extant species with PRANK (25) (parameters ‘-once -gaprate = 0.05 -gapext = 0.2 -termgap -showanc’) using the phylogenetic tree as input (Supplementary Figure S2). PRANK also reconstructs all ancestral sequences, which we then used instead of the known ancestral sequences to compute branch scores.
Second, we simulated pleiotropic CREs that have regulatory activity in two tissues. To this end, we redefined the ideal regulatory activity of a CRE as 100% expression in two tissues. In the second tissue, five different TFs are expressed at an equal level and these TFs have an equal activating strength. For pleiotropic CREs, we assume that after trait loss expression in the first tissue is no longer under purifying selection, but expression in the second tissue still is. Therefore, after trait loss, we changed the ideal regulatory activity from 100% expression in both tissues to 100% expression in the second tissue only. Since the CRE is still under selection to maintain expression in the second tissue, the regulatory input required for expression in this tissue remains under selection, which limits the overall sequence divergence of the CRE in the trait-loss species.
Application of TFforge to real data
We used a multiple genome alignment of 29 species with the mouse mm10 assembly as the reference, generated by lastz, axtChain, chainNet and Multiz (26–28), as described previously (18). To obtain conserved non-coding elements, we excluded coding exon regions from evolutionarily conserved elements detected by PhastCons (29) and GERP (30).
Crx ChIP-seq data from adult mouse retina tissue was taken from reference (31). We only considered peaks that have a quality score of ≥45 and that were detected in both replicates. For each peak, we obtained the region ±100 bp around the center position and retained those regions that overlap with at least 100 bp conserved non-coding elements (CNEs). This resulted in 1075 genomic regions of which the majority (769, 72%) does not overlap promoter regions (300 bp upstream of the transcription start site). Nrl ChIP-seq peaks from adult mouse retina tissue were kindly provided by Anand Swaroop (32). We restricted the analysis to the central 200 bp regions and filtered peaks for CNE overlap as done for Crx peaks. This resulted in 500 peaks, 401 (80%) of which do not overlap promoter regions. Lens-specific Pax6 ChIP-seq data was obtained from reference (33) by selecting lens peaks that do not overlap forebrain Pax6 peaks provided in the same study. Filtering the central 200 bp region of each lens-specific peak for a minimal CNE overlap of 100 bp resulted in 929 regions.
Given a CNE that overlaps these TF-bound regions, we reconstructed all ancestral sequences with PRANK (25) (parameters ‘-keep -showtree -showanc -prunetree -seed = 10’) using the species phylogeny (Supplementary Figure S3) as input and applied TFforge to all placental mammals.
RESULTS
Proof of concept
To test TFforge, we first used synthetic CRE datasets and determined if TFforge is able to detect the motifs of the five randomly-selected foreground TFs from 567 background TFs that are not used in the simulation. We generated three sets of 100 CREs that differ in the age of the trait loss by setting the final (neutral) part of the trait-loss branches to 0.09, 0.06 or 0.03 substitutions per site. As shown in Figure 2B, TFforge is able to detect significant binding site losses for all five foreground TFs for the three trait-loss ages. The P-values decrease with an increasing age of the trait loss, which is expected since an increased number of neutral mutations should lead to an increased amount of TFBS loss. In contrast, the 567 background TFs, whose binding sites should only occur by chance, do not show a pronounced binding site loss for the three trait-loss ages.
We next explored how random variation in the simulation affects the TFforge performance and how many CREs are necessary to detect significant binding site loss for the five foreground TFs. To this end, we first generated 900 additional CREs for each of the three trait-loss ages. From the total of 1,000 CREs, we then generated 100 subsamples comprising 50, 40, 30, 20 and 10 CREs each. Since only 0.87% (5 of 572) of the motifs correspond to foreground TFs, we compared sensitivity (percent of correctly detected foreground TFs) versus precision (percent of foreground TFs among all detected TFs) in the following. As shown in Figure 2C, for subsamples of 50 CREs, TFforge achieves a median sensitivity of 100% at a fixed precision of 100%, showing that 50 simulated CRE are sufficient to distinguish the five foreground from all 567 background motifs. For subsamples of less than 50, the sensitivity decreases; however, TFforge is often still able to correctly identify four (80% sensitivity) or three (60% sensitivity) of the five foreground TFs at 100% precision. Overall, this simulated data serves as a proof of concept that TFforge can identify TFs that have a tendency to lose binding sites on trait-loss branches.
Testing TFforge on more realistic evolutionary scenarios
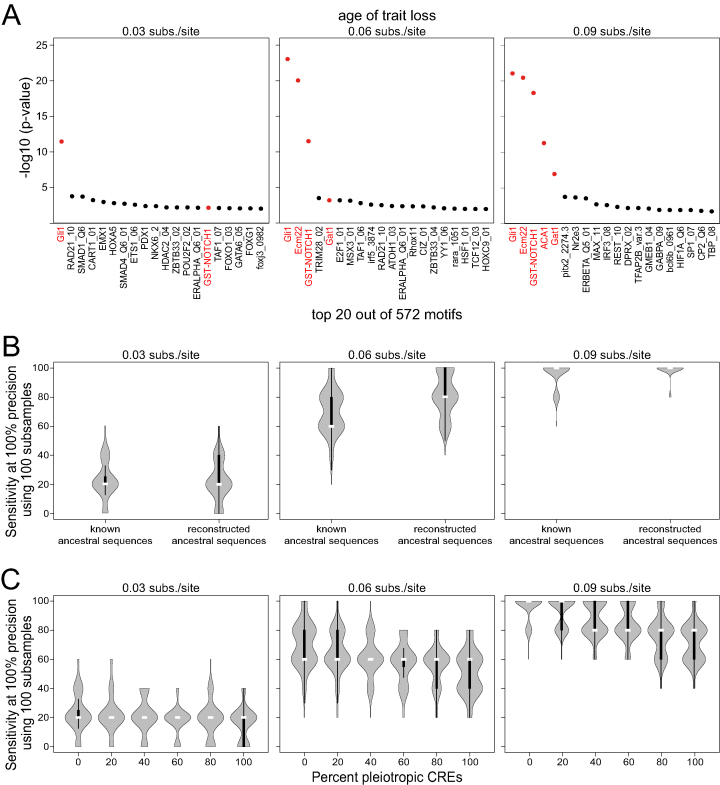
Up to now, all considered CREs of type 1 evolved without selection to preserve a regulatory activity on the trait-loss branches. For a real data set, consisting of CREs that are active in a selected tissue, it is unlikely that all CREs evolve neutrally in the trait-loss lineages. Therefore, we tested the performance of TFforge on simulated data that additionally includes CREs that still evolve under purifying selection in the trait-loss species. To this end, we generated two additional CRE sets. The regulatory activity of type 2 CREs is controlled by the same five foreground TFs, while the regulatory activity of type 3 CREs is controlled by five different TFs. In contrast to type 1 CREs, type 2 and 3 CREs evolve under purifying selection on every branch in the tree. Based on our observations that a minimum number of ∼50 type 1 CREs is required to reliably detect foreground TFs, we applied TFforge to a combined dataset comprising 50 type 1, 150 type 2 and 150 type 3 CREs. As shown in Figure 3A, having 86% type 2 and 3 CREs makes it harder to identify the five foreground TFs. However, TFforge still achieved a median sensitivity of 20%, 60% or 100% at a high precision of 100% for the three trait-loss ages of 0.03, 0.06 or 0.09 substitutions per site, respectively. Different numbers of type 2 and type 3 CREs give similar results (Supplementary Figure S4A). This shows that it is possible to identify the foreground TF motifs also under conditions where <15% of the input CREs are expected to exhibit preferential binding site loss in the trait-loss species.
Figure 3.
Performance of TFforge on simulated datasets containing different background CREs or pleiotropic CREs. (A) Plots show the 20 top-ranked TF motifs for three trait-loss ages on a combined dataset that consists of 50 type 1, 150 type 2 and 150 type 3 CREs. Foreground TF motifs are shown in red. (B) Performance of TFforge on 100 subsamples of datasets where the ancestral sequences were either known or were reconstructed from an alignment of extant sequences. Each dataset consists of a total of 50 type 1, 150 type 2 and 150 type 3 CREs. Violin plots show the distribution of the sensitivity at a precision of 100%, which corresponds to the number of foreground TF motifs that have a higher significance than the most significant background TF motif. The vertical black bar inside a violin plot spans the first and third quartile, the white bar indicates the median. (C) Performance of TFforge on 100 subsamples of datasets where different percentages of modular type 1 CREs were replaced with pleiotropic type 1 CREs. Each dataset consists of a total of 50 type 1, 150 type 2 and 150 type 3 CREs.
Next, we investigated the influence of computationally reconstructing ancestral sequences on the performance of TFforge. To this end, we compared the sensitivity at a precision of 100% for simulated CREs where the ancestral sequences were either known or were computationally reconstructed. As shown in Figure 3B and Supplementary Figure S4B, computationally reconstructing ancestors does not impair the performance, suggesting that TFforge is robust towards ancestral reconstruction uncertainty.
Another key assumption has been so far that CREs are modular and control gene expression only at a specific time and tissue. However, it is known that some CREs are pleiotropic and control expression at multiple time points or in different tissues, for example, many enhancers drive gene expression in both the developing limbs and genitals (34). After the loss of a trait that results in the absence of selection to maintain enhancer activity in one of these tissues, purifying selection would still preserve the regulatory activity in the other tissues. Therefore, we tested the performance of TFforge on simulated pleiotropic CREs that have regulatory activity in two tissues, controlled by two sets of five TFs. We adapted the simulation such that after trait loss purifying selection acts exclusively on regulatory activity in the second tissue. We combined various percentages of modular and pleiotropic CREs to obtain a total of 50 type 1 CREs and added 150 type 2 and 150 type 3 CREs. As shown in Figure 3C, replacing various percentages of modular CREs by pleiotropic CREs has only a minor effect on the ability of TFforge to identify the five foreground TFs. We conclude that the ability of TFforge to identify TFs that preferentially lose binding sites on trait-loss branches is largely unaffected by the presence of pleiotropic CREs that do not evolve entirely neutrally after trait loss.
Application of TFforge to real regulatory data
To validate TFforge on real data, we applied it to identify TFs that are likely involved in the degeneration of eyes in the blind mole-rat, naked mole-rat, star-nosed mole, and cape golden mole (Supplementary Figure S3). These four independently evolved subterranean mammals have poor vision or are completely blind and possess degenerated retinas and lenses (35–37). Furthermore, the genome of these mammals has been sequenced (38,39) and ChIP-seq datasets exist that provide genomic regions bound by eye-related TFs in the retina of mouse, which we used as the reference species in this analysis.
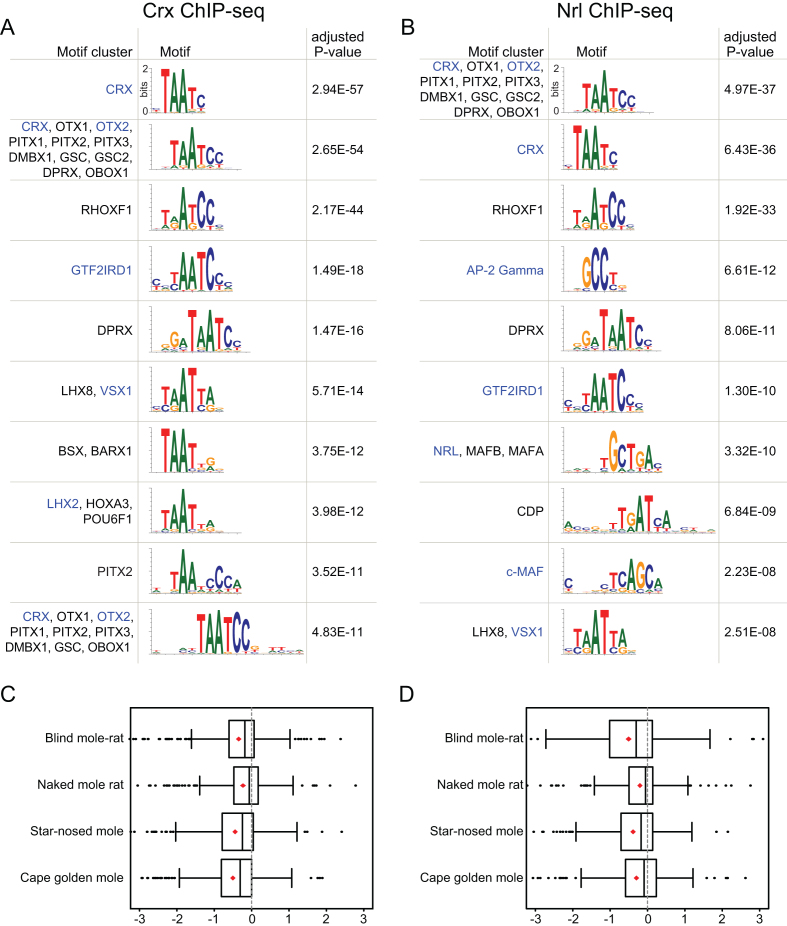
We first focused on genomic regions bound by Crx (cone-rod homeobox), a TF that is required for photoreceptor development (31). We applied TFforge to all Crx-bound regions that overlap CNEs and screened our library of 614 similarity-clustered TF motifs for preferential binding site loss on the branches leading to the four subterranean mammals. This screen identified the Crx motif as the most significant out of all TF motifs (Figure 4A, Supplementary Table S2), suggesting that subterranean mammals have lost a substantial number of Crx binding sites. Many other top-ranked motifs are similar to the Crx motif; however, they also highlight other TFs that have roles in the eye and interact with Crx. For example, the motif at rank 2 describes the binding preference of other homeobox TFs, among them Otx2 that is also required for photoreceptor development. Interestingly, Otx2 directly interacts with Crx and regulates Crx expression (40,41). Consistent with previous observations that Otx2 co-binds regulatory elements of Crx target genes (42), we found that 91% of the analysed Crx-bound CNEs overlap Otx2 ChIP-seq data obtained from the mouse retina (43). The motif at rank 4 is Gtf2ird1, a TF that directly interacts with Crx and regulates gene expression in rod and cone photoreceptors (44). This suggests that not only Crx but also co-factor binding sites preferentially evolve neutrally in Crx-bound regions in subterranean mammals.
Figure 4.
TFforge identifies TFs associated with eye degeneration in subterranean mammals. (A, B) Ten top-ranked motif clusters for Crx-bound (A) and Nrl-bound (B) regions in mouse retina. TFs that are Crx/Nrl co-factors or that have a role in eye development and function are shown in blue font. (C, D) Boxplots show the distribution of branch scores of the Crx (C) and Nrl (D) motif for the branches leading to the four subterranean mammals. Red diamonds indicate the average. For visual clarity, a few outlier data points outside the branch score range [−3,3] are not shown.
Next, we used TFforge to analyse CNEs bound by Nrl (neural retina leucine zipper), a TF that is necessary for the development of rod photoreceptors (45). TFforge identified the Nrl motif at rank 7 with an adjusted P-value of 3e-10 (Figure 4B, Supplementary Table S3), consistent with a substantial loss of Nrl binding sites in subterranean mammals. Interestingly, our screen identified many of the same TFs that show preferential binding site loss in subterranean mammals in Crx bound CNEs (Crx, Otx2, Gtf2ird1; Figure 4B). This is likely explained by the fact that Nrl interacts, co-binds regulatory regions and co-regulates many photoreceptor genes with these TFs (32,44,46,47). This is further supported by our observation that 60% and 78% of the analysed Nrl-bound CNEs overlap Crx and Otx2 ChIP-seq data, respectively. In addition, the analysis of Crx- and Nrl-bound CNEs also identified other TF motifs that have a role in specific cell types within the eye (Figure 4A, B). For example, TFforge detected the motif of an AP-2 transcription factor (encoded by Tfap2c) that is specifically expressed in retinal amacrine cells (48). The top-ranked motifs also include Vsx1 (visual system homeobox 1), a TF required for cone bipolar cell development that binds the opsin locus control region (49–51), c-Maf, a TF necessary for lens development (52), and Lhx2, a factor necessary for Mueller glia cell development (53). Finally, applying TFforge to lens-specific Pax6 ChIP-seq data (Supplementary Table S4) highlights a preferential loss of binding sites of additional TFs, such as N-myc, a TF required for fiber cell differentiation during lens development (54), Isl1, a direct target gene of Pax6 in the lens that is involved in retinal ganglion cell differentiation (55,56), and NeuroD, which is required for photoreceptor survival (57).
Given that we used a set of Crx- or Nrl-bound genomic regions as input, it might not be surprising to detect the Crx or Nrl motif. However, TFforge is different from tools that detect motif enrichment, as our scoring procedure does not count binding site occurrences but contrasts the conservation pattern of such binding sites on two sets of branches in the phylogeny. To illustrate this difference, we applied TFforge to the same set of genomic regions but selected other mammals as ‘trait-loss species’. Specifically, we considered all 4809 combinations of four placental mammals that do not contain sister species and do not include subterranean mammals. None of these 4809 combinations results in P-values for the Crx and Nrl motifs that are lower than the P-values obtained for the four subterranean mammals (Supplementary Figure S5A, B). This indicates that the subterranean mammals collectively have the strongest tendency to lose Crx and Nrl binding sites in genomic regions bound by these TFs in retina tissue. We further computed the average branch scores for the four individual branches leading to these species (Figure 4C, D). This revealed differences in the amount of binding site loss between the subterranean mammals with the cape golden mole and blind mole rat showing the strongest tendency to lose Crx and Nrl binding sites, respectively, while the naked mole rat has lost the fewest binding sites for both TFs.
DISCUSSION
TFforge is a new method to identify motifs of TFs that preferentially lose binding sites in species that lost a given phenotype. Using datasets obtained by simulating the evolution of cis-regulatory elements under selection and under neutrality on different branches, we showed that TFforge is able to identify the correct motifs for a variety of scenarios and parameters. We found that it is not necessary to assume that CREs evolve entirely neutrally, as TFforge could also identify many correct motifs when applied to pleiotropic CREs that control expression in different tissues and that still evolve under purifying selection to preserve the regulatory activity in some of the tissues. This is likely important as the degree of pleiotropy of CREs is not well characterized. For example, a recent study found that many enhancers control expression in both the developing limbs and genitals (34), suggesting that pleiotropic CREs might be more common than previously thought. While simulating modular or pleiotropic CREs certainly represents a simplification of regulatory element evolution, it allowed us to establish a proof of principle for TFforge and to probe the limitations of the method.
To gain insights into the TFs that are likely involved in the degeneration of eyes in subterranean mammals, we applied TFforge to eye TF ChIP-seq datasets obtained from mouse retina tissue. Screening our library of 614 motifs, TFforge detected a significant binding site decay signature in subterranean mammals of the TFs that were immunoprecipitated (Crx and Nrl) as well as motifs of other TFs that interact or co-bind with Crx and Nrl and are also involved in eye development. Importantly, systematic hypothesis tests involving non-subterranean mammals showed that the four subterranean mammals collectively have the most significant tendency for losing binding sites of the immunoprecipitated TFs, showing that TFforge not simply detects overrepresentation of a motif but rather specific divergence patterns of the respective binding sites.
TFforge will help to identify TFs that are involved in a given phenotypic change, and thus reveal the transcriptional regulators that contribute to phenotypic evolution. Given the rapid increase in the number of sequenced genomes and advances in functional genomics that make it possible to comprehensively identify relevant CREs, TFforge has broad applicability to identify TFs that are involved in phenotypes that differ between the sequenced species. By utilizing the ever-growing list of TF binding motifs to detect such TFs based on a large-scale binding site decay signature, our approach is complementary to predicting phenotype-relevant TFs based on functional annotations. Thus, TFforge will also be a valuable upstream step of REforge, which can then detect associations between individual CREs and phenotypic differences between species and thus provide insights into the genomic basis of nature's phenotypic diversity.
DATA AVAILABILITY
All simulated CREs and the analysed Crx and Nrl regions are available at https://bds.mpi-cbg.de/hillerlab/TFforge/. The TFforge source code is available at https://github.com/hillerlab/TFforge.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Henrike Indrischek for helpful comments on the manuscript and the Computer Service Facilities of the MPI-CBG and MPI-PKS for their support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society; ELBE PhD Project Funding. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wray G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007; 8:206–216. [DOI] [PubMed] [Google Scholar]
- 2. Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008; 134:25–36. [DOI] [PubMed] [Google Scholar]
- 3. Wittkopp P.J., Kalay G.. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011; 13:59–69. [DOI] [PubMed] [Google Scholar]
- 4. Hiller M., Schaar B.T., Indjeian V.B., Kingsley D.M., Hagey L.R., Bejerano G.. A “forward genomics” approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2012; 2:817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Langer B.E., Roscito J.G., Hiller M.. REforge associates transcription factor binding site divergence in regulatory elements with phenotypic differences between species. Mol. Biol. Evol. 2018; doi:10.1093/molbev/msy187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H. et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016; 537:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 8. Smith A.D., Sumazin P., Zhang M.Q.. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. PNAS. 2005; 102:1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sinha S., Blanchette M., Tompa M.. PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics. 2004; 5:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siddharthan R. PhyloGibbs-MP: module prediction and discriminative motif-finding by Gibbs sampling. PLoS Comput. Biol. 2008; 4:e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Consortium Encode Project. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B. et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017; 14:959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sinha S., Liang Y., Siggia E.. Stubb: a program for discovery and analysis of cis-regulatory modules. Nucleic Acids Res. 2006; 34:W555–W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinha S., van Nimwegen E., Siggia E.D.. Stubb: A probabilistic method to detect regulatory modules. Bioinformatics. 2003; 19:i292–i301. [DOI] [PubMed] [Google Scholar]
- 16. Huang W., Nevins J.R., Ohler U.. Phylogenetic simulation of promoter evolution: estimation and modeling of binding site turnover events and assessment of their impact on alignment tools. Genome Biol. 2007; 8:R225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otto W., Stadler P.F., Lopez-Giraldez F., Townsend J.P., Lynch V.J., Wagner G.P.. Measuring transcription factor-binding site turnover: a maximum likelihood approach using phylogenies. Genome Biol. Evol. 2009; 1:85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prudent X., Parra G., Schwede P., Roscito J.G., Hiller M.. Controlling for phylogenetic relatedness and evolutionary rates improves the discovery of associations between species' phenotypic and genomic differences. Mol. Biol. Evol. 2016; 33:2135–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hume M.A., Barrera L.A., Gisselbrecht S.S., Bulyk M.L.. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015; 43:D117–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006; 34:D108–D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014; 42:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He X., Samee M.A., Blatti C., Sinha S.. Thermodynamics-based models of transcriptional regulation by enhancers: the roles of synergistic activation, cooperative binding and short-range repression. PLoS Comput. Biol. 2010; 6:e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duque T., Samee M.A., Kazemian M., Pham H.N., Brodsky M.H., Sinha S.. Simulations of enhancer evolution provide mechanistic insights into gene regulation. Mol. Biol. Evol. 2014; 31:184–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loytynoja A., Goldman N.. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008; 320:1632–1635. [DOI] [PubMed] [Google Scholar]
- 26. Harris R.S. Improved pairwise alignment of genomic DNA. The Pennsylvania State University. 2007; Ph.D. Thesis. [Google Scholar]
- 27. Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D.. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. PNAS. 2003; 100:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D. et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004; 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S.. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010; 6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corbo J.C., Lawrence K.A., Karlstetter M., Myers C.A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L.G. et al. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 2010; 20:1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hao H., Kim D.S., Klocke B., Johnson K.R., Cui K., Gotoh N., Zang C., Gregorski J., Gieser L., Peng W. et al. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012; 8:e1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun J., Rockowitz S., Xie Q., Ashery-Padan R., Zheng D., Cvekl A.. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015; 43:6827–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Infante C.R., Mihala A.G., Park S., Wang J.S., Johnson K.K., Lauderdale J.D., Menke D.B.. Shared enhancer activity in the limbs and phallus and functional divergence of a Limb-Genital cis-Regulatory element in snakes. Dev Cell. 2015; 35:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanyal S., Jansen H.G., de Grip W.J., Nevo E., de Jong W.W.. The eye of the blind mole rat, Spalax ehrenbergi. Rudiment with hidden function. Invest. Ophthalmol. Vis. Sci. 1990; 31:1398–1404. [PubMed] [Google Scholar]
- 36. Hetling J.R., Baig-Silva M.S., Comer C.M., Pardue M.T., Samaan D.Y., Qtaishat N.M., Pepperberg D.R., Park T.J.. Features of visual function in the naked mole-rat Heterocephalus glaber. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2005; 191:317–330. [DOI] [PubMed] [Google Scholar]
- 37. Nemec P., Cvekova P., Benada O., Wielkopolska E., Olkowicz S., Turlejski K., Burda H., Bennett N.C., Peichl L.. The visual system in subterranean African mole-rats (Rodentia, Bathyergidae): retina, subcortical visual nuclei and primary visual cortex. Brain Res Bull. 2008; 75:356–364. [DOI] [PubMed] [Google Scholar]
- 38. Fang X., Nevo E., Han L., Levanon E.Y., Zhao J., Avivi A., Larkin D., Jiang X., Feranchuk S., Zhu Y. et al. Genome-wide adaptive complexes to underground stresses in blind mole rats Spalax. Nat. Commun. 2014; 5:3966. [DOI] [PubMed] [Google Scholar]
- 39. Kim E.B., Fang X., Fushan A.A., Huang Z., Lobanov A.V., Han L., Marino S.M., Sun X., Turanov A.A., Yang P. et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011; 479:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., Furukawa T.. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 2003; 6:1255–1263. [DOI] [PubMed] [Google Scholar]
- 41. Fant B., Samuel A., Audebert S., Couzon A., El Nagar S., Billon N., Lamonerie T.. Comprehensive interactome of Otx2 in the adult mouse neural retina. Genesis. 2015; 53:685–694. [DOI] [PubMed] [Google Scholar]
- 42. Peng G.H., Chen S.. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis. Neurosci. 2005; 22:575–586. [DOI] [PubMed] [Google Scholar]
- 43. Samuel A., Housset M., Fant B., Lamonerie T.. Otx2 ChIP-seq reveals unique and redundant functions in the mature mouse retina. PLoS One. 2014; 9:e89110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Masuda T., Zhang X., Berlinicke C., Wan J., Yerrabelli A., Conner E.A., Kjellstrom S., Bush R., Thorgeirsson S.S., Swaroop A. et al. The transcription factor GTF2IRD1 regulates the topology and function of photoreceptors by modulating photoreceptor gene expression across the retina. J. Neurosci. 2014; 34:15356–15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mears A.J., Kondo M., Swain P.K., Takada Y., Bush R.A., Saunders T.L., Sieving P.A., Swaroop A.. Nrl is required for rod photoreceptor development. Nat. Genet. 2001; 29:447–452. [DOI] [PubMed] [Google Scholar]
- 46. Mitton K.P., Swain P.K., Chen S., Xu S., Zack D.J., Swaroop A.. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J. Biol. Chem. 2000; 275:29794–29799. [DOI] [PubMed] [Google Scholar]
- 47. Hsiau T.H., Diaconu C., Myers C.A., Lee J., Cepko C.L., Corbo J.C.. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One. 2007; 2:e643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bassett E.A., Korol A., Deschamps P.A., Buettner R., Wallace V.A., Williams T., West-Mays J.A.. Overlapping expression patterns and redundant roles for AP-2 transcription factors in the developing mammalian retina. Dev. Dyn. 2012; 241:814–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chow R.L., Volgyi B., Szilard R.K., Ng D., McKerlie C., Bloomfield S.A., Birch D.G., McInnes R.R.. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. PNAS. 2004; 101:1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ohtoshi A., Wang S.W., Maeda H., Saszik S.M., Frishman L.J., Klein W.H., Behringer R.R.. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr. Biol.: CB. 2004; 14:530–536. [DOI] [PubMed] [Google Scholar]
- 51. Hayashi T., Huang J., Deeb S.S.. RINX(VSX1), a novel homeobox gene expressed in the inner nuclear layer of the adult retina. Genomics. 2000; 67:128–139. [DOI] [PubMed] [Google Scholar]
- 52. Kim J.I., Li T., Ho I.C., Grusby M.J., Glimcher L.H.. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. PNAS. 1999; 96:3781–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Melo J., Zibetti C., Clark B.S., Hwang W., Miranda-Angulo A.L., Qian J., Blackshaw S.. Lhx2 is an essential factor for retinal gliogenesis and notch signaling. J. Neurosci. 2016; 36:2391–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cavalheiro G.R., Matos-Rodrigues G.E., Zhao Y., Gomes A.L., Anand D., Predes D., de Lima S., Abreu J.G., Zheng D., Lachke S.A. et al. N-myc regulates growth and fiber cell differentiation in lens development. Dev. Biol. 2017; 429:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xie Q., Yang Y., Huang J., Ninkovic J., Walcher T., Wolf L., Vitenzon A., Zheng D., Gotz M., Beebe D.C. et al. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS One. 2013; 8:e54507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pan L., Deng M., Xie X., Gan L.. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008; 135:1981–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pennesi M.E., Cho J.H., Yang Z., Wu S.H., Zhang J., Wu S.M., Tsai M.J.. BETA2/NeuroD1 null mice: a new model for transcription factor-dependent photoreceptor degeneration. J. Neurosci. 2003; 23:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All simulated CREs and the analysed Crx and Nrl regions are available at https://bds.mpi-cbg.de/hillerlab/TFforge/. The TFforge source code is available at https://github.com/hillerlab/TFforge.