Abstract
Telomeres are nucleoprotein structures at the ends of linear chromosomes and present an essential feature for genome integrity. Vertebrate telomeres usually consist of hexameric TTAGGG repeats, however, in cells that use the alternative lengthening of telomeres (ALT) mechanism, variant repeat sequences are interspersed throughout telomeres. Previously, it was shown that NR2C/F transcription factors bind to TCAGGG variant repeats and contribute to telomere maintenance in ALT cells. While specific binders to other variant repeat sequences have been lacking to date, we here identify ZBTB10 as the first TTGGGG-binding protein and demonstrate direct binding via the two zinc fingers with affinity in the nanomolar range. Concomitantly, ZBTB10 co-localizes with a subset of telomeres in ALT-positive U2OS cells and interacts with TRF2/RAP1 via the N-terminal region of TRF2. Our data establishes ZBTB10 as a novel variant repeat binding protein at ALT telomeres.
INTRODUCTION
Telomeres are nucleoprotein structures at the end of linear chromosomes that in vertebrates consist of double-stranded (ds) TTAGGG repeats. They are constitutively bound by the shelterin complex, which consists of six proteins: the telomere repeat binding factors TRF1 and TRF2, the TRF2-interacting protein RAP1, the telomeric ssDNA-binding protein POT1 with its direct interactor TPP1 as well as the bridging factor TIN2 (1). The shelterin complex is important for telomere end protection by preventing recognition of the chromosome end as a DNA double strand break (DSB) and protecting it from illegitimate DNA repair activities (2). Within the complex, TRF2 plays an important role in end protection by inducing and stabilizing t-loop structures (3–5). Thereby, chromosomal ends are safeguarded from nucleolytic degradation and protected from the DNA damage response and repair activities such as non-homologous end joining. TRF2 and RAP1 together repress homology directed repair by preventing telomeric localization of PARP1 and SLX4 (6). TRF2 has furthermore been shown to repress ATM kinase activity at telomeres (7,8). Moreover, TRF2 recruits various factors such as RTEL1 during S-phase to enable t-loop unwinding (9) or ORC to stimulate pre-replication complex assembly at telomeres (10).
During each cell division telomeres progressively shorten, which is counteracted by the ribonucleoprotein telomerase in the majority of cancers (11). In the absence of telomerase, replicative immortality can be achieved by alternative lengthening of telomeres (ALT) in 10–15% of cancers (12). ALT is based on homology directed repair (13,14), leading to long heterogeneous telomeres and the interspersion of variant repeats such as TCAGGG, TGAGGG and TTGGGG that are otherwise confined to subtelomeres (15,16). As a result, NR2C/F nuclear receptors, which have been shown to preferentially bind TCAGGG repeats, are recruited along the interspersed telomeric sequence and contribute to the ALT phenotype (i.e. ALT-associated PML bodies, C-circles, telomere sister chromatid exchanges) (16). Moreover, NR2C/F receptors have also been described as bridging proteins leading to targeted telomeric insertion throughout chromosomes in ALT cells (17). However, proteins specifically binding to other telomeric variant repeat sequences have not been described yet.
Zinc finger and BTB domain containing proteins offer a large surface for protein-protein interactions, are able to form homo- and heterodimers and have been suggested to play a role in regulating gene expression (18). Recently the family member ZBTB48, also known as TZAP, has been described as a telomeric protein involved in telomere length regulation (19,20). In a phylointeractomics screen across 16 vertebrate species, we found multiple ZBTB proteins including ZBTB10 as a binder of TTAGGG repeats (21). Here, we show that ZBTB10 preferably binds to the TTGGGG variant repeat, partially localizes to telomeres in the ALT cancer cell line U2OS and interacts with the TRF2/RAP1 heterodimeric complex irrespective of the telomere maintenance mechanism.
MATERIALS AND METHODS
Cell culture and plasmids
All cell lines (HEK293, HeLa E1 (22), U2OS, GM847 and SAOS2) were cultivated in 4.5 g/l glucose, 1 mM sodium pyruvate Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco), 2 mM glutamine (Life Technologies), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) in a humidified incubator at 5% CO2 and 37°C.
The ZBTB10 clone was obtained from Bioscience (Q96DT7-isoform 3; ENST00000610895.2) and full-length TRF2 (Q15554; ENST00000254942.7) was amplified from HEK293 cDNA (First strand synthesis Kit; Thermo Scientific). Deletion constructs of both proteins were obtained by PCR amplification using specific primers (Supplementary Table S1). Constructs were confirmed by Sanger sequencing (GATC) and sequences were LR recombined into Gateway-compatible expression vectors: pDest-pcDNA3.1 with N-terminal FLAG tag or pcDNA-Dest47 with C-terminal GFP tag.
Transfections
Plasmid transfection in HEK293 cells was performed using linear polyethylenimine (PEI, MW 25 000; Polysciences). 450 000 cells/ml were seeded one day before in 10 cm dishes and transfected with 12 μg DNA and 48 μl PEI diluted in DMEM. U2OS cells were transfected using linear PEI HCl MAX (MW 4000; Polysciences) or with the Amaxa Cell Line Nucleofector Kit V (Lonza) according to manufacturer's instructions. SAOS2, GM847 and HeLa E1 cells were transfected using Lipofectamine 2000 (Thermo) diluted in Opti-MEM (Life Technologies). Stable cell lines were generated by selection in 0.8 mg/ml G418 (Sigma) for one week, enriched by sorting GFP-positive cells (Becton Dickinson FACSAria III SORP) and further cultured in 0.4 mg/ml G418.
DNA pulldown
Twenty five microgram of forward and reverse oligonucleotides of telomeric, variant repeat or control sequences (Supplementary Table S1) were denatured at 80°C and annealed by cooling. Double-stranded oligonucleotides were then polymerized using 50 U T4 polynucleotidekinase (Thermo Scientific) and 80 U T4 DNA ligase (NEB), biotinylated with desthiobiotin-dATP (Jena Bioscience) by Klenow fragment (Thermo Scientific) and purified using G-50 columns (GE Healthcare). Chemically synthesized DNA was immobilized on 0.5 mg streptavidin-coated magnetic beads (Dynabeads MyOne Streptavidin C1, Invitrogen) for 15 min at RT and incubated with 400–800 μg protein lysates diluted in PBB buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 0.5% IGEPAL CA-630, 1 μM Pepstatin, 1 μg/ml Leupeptin and 0.5 mM PMSF) for 1.5 h on a rotation wheel at 4°C. Sheared salmon sperm DNA (16.7 μg; Ambion) was added as a competitor. After three washes with PBB buffer, bound proteins were eluted in LDS sample buffer supplemented with 0.1 M DTT, boiled for 10 min at 70°C and separated on a 4–12% NuPAGE Novex Bis–Tris precast gel (Life Technologies) for 50 min at 180 V in 1× MES buffer.
Protein purification
ZBTB10-2Znf (aa 540–606) and ZBTB10-2Znf-C (aa 540–682) were cloned into a modified pET28a vector with a SUMO protein fused at the N-terminus after a His6 tag. Proteins were expressed in Escherichia coli BL21 (DE3) cells (Stratagene). After induction for 24 h with 0.1 mM IPTG at 16°C, the cells were harvested and then lysed by sonication. Proteins were purified by Ni-NTA agarose beads (Qiagen), followed by Ulp1 cleavage to remove the His-SUMO tag and size-exclusion chromatography on a Hiload 16/60 Superdex 75 column (GE Healthcare) in a buffer containing 20 mM Tris–HCl (pH 7.5) and 1 M NaCl. Purified proteins were dialyzed with Buffer A (20 mM Tris–HCl pH 7.5, 150 mM NaCl) and concentrated for subsequent analysis.
Fluorescence polarization assay
The protein stock was diluted in 1/2 series in Buffer A to the lowest desired concentration. The protein dilution series was then incubated with 50 nM FAM-labeled probe in Buffer A (200 μl final volume) for 30 min at RT. Fluorescence polarization was measured using a SpectraMax M5 plate reader (Molecular Devices). Curves were fit individually using the equation 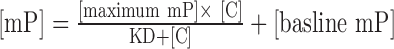 , where mP is millipolarization, and [C] is protein concentration. KD values and the fitting figures were derived by fitting the experimental data (two experimental replicates) to the equation using a fitting script written in python.
, where mP is millipolarization, and [C] is protein concentration. KD values and the fitting figures were derived by fitting the experimental data (two experimental replicates) to the equation using a fitting script written in python.
Nuclear and chromatin extract preparation
Cells were harvested, washed in PBS and incubated for 10 min on ice in five pellet volumes of cold buffer A (10 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl). Cells were pelleted, resuspended in two volumes of cold buffer A supplemented with 0.2% IGEPAL CA-630 and complete protease inhibitor mix (Roche) and homogenized with 30 strokes (type B pestle) in a dounce homogenizer. The cytoplasmic fraction was collected after centrifugation for 15 min at 3900 rpm at 4°C. After a PBS wash, nuclei were resuspended in two volumes of buffer C (420 mM NaCl, 20 mM HEPES–KOH pH 7.9, 20% glycerol, 2 mM MgCl2, 0.2 mM EDTA pH 8, 0.1% IGEPAL CA-630, 0.5 mM DTT and complete protease inhibitor mix (Roche)), incubated for 1 h on a rotating wheel at 4°C and centrifuged at 14 800 rpm for 1 h at 4°C to collect the nuclear extract. The chromatin pellet was incubated for 30 min on a rotating wheel at 4°C in buffer C supplemented with 0.05% SDS, sonicated for 15 cycles (30 s on and 45 s off) and further incubated for 1 h on a rotating wheel at 4°C. The extract was cleared by centrifugation at 14 500 rpm for 1 h at 4°C.
GFP pulldown
Ten microliters slurry of GFP-M Nanotrap (Chromotek) magnetic beads were used per immunoprecipitation (IP) and equilibrated in wash buffer (150 mM NaCl, 10 mM Tris–HCl pH 7.5, 0.5 mM EDTA). Cells were lysed in wash buffer supplemented with 0.5% IGEPAL CA-630 and 0.1% sodium deoxycholate, 1 μM Pepstatin, 1 μg/ml Leupeptin and 0.5 mM PMSF for 30 min on ice. Cleared supernatants were either first incubated with 500 U Sm-nuclease (in-house) for 30 min on ice or directly incubated with equilibrated GFP beads for 2 h on a rotation wheel at 4°C. After three washes on ice, proteins were eluted in LDS sample buffer supplemented with 0.1 M DTT, boiled for 10 min at 95°C and separated on a 4–12% NuPAGE Novex Bis–Tris precast gel (Life Technologies) for 10 min at 180 V in 1× MOPS buffer.
Western blot
Separated proteins were transferred from a SDS gel to a nitrocellulose membrane (Amersham) at 300 mA for 1 h. The membrane was blocked in PBS containing 5% (w/v) non-fat milk (Sigma) and 0.1% Tween-20 (PBST) for 1 h at RT and incubated with primary antibodies for 1–2 h at RT or overnight at 4°C. Detailed information about antibodies can be found in Supplementary Table S1. Antibodies were diluted in PBST containing 5% BSA and 0.02% sodium azide. After three washes in PBST, the membrane was incubated for 1 h at RT with secondary antibody. Chemiluminescence detection was performed using SuperSignal West Pico solution (Pierce). The SeeBlue Plus2 Pre-stained Protein Standard (Thermo Scientific) was used as a marker.
MS sample preparation, data acquisition and data analysis
In-gel digestion was performed as previously described (23). Tryptic peptides were desalted on StageTips and loaded on an in-house packed C18 column (25 cm long, 75 μm inner diameter) for reverse-phase chromatography. The EASY-nLC 1000 system (Thermo Scientific) was mounted to a Q Exactive Plus mass spectrometer (Thermo Scientific) and peptides were eluted from the column in an optimized 90 min (pulldown) or 5 h (proteome) gradient from 2 to 40% MS grade acetonitrile/0.1% formic acid solution at a flow rate of 200 nl/min. The mass spectrometer was used in a data-dependent acquisition mode with one MS full scan and up to 10 MS/MS scans using HCD fragmentation. All raw files were processed with MaxQuant (version 1.5.2.8) and searched against the human Uniprot database (81 194 entries). Carbamidomethylation (Cys) was set as fixed modification, while oxidation (Met) and protein N-acetylation were considered as variable modifications. For enzyme specificity, trypsin was selected with a maximum of two miscleavages. LFQ quantification (without fast LFQ) using at least two LFQ ratio counts and the match between run option were activated. Data analysis was performed in R using existing libraries (knitr, reshape2, dplyr, ggplot2, ggrepel) and in-house scripts. Protein groups reported by MaxQuant were filtered removing known contaminants, protein groups only identified by site and those marked as reverse hits. Missing values were imputed at the lower end of LFQ values using random values from a beta distribution fitted at 0.1–1.5%. For statistical analysis, P-values were calculated using the Welch's t-test. Enrichment values in the volcano plots represent the median difference of log2 transformed LFQ intensities between ZBTB10-GFP and GFP. For the definition of enriched protein groups in all GFP pulldowns we applied an identical cut-off (S0 = 1.6, c = 0.8 and P = 0.05).
Immunofluorescence (IF) staining
One day before, cells were seeded on a 96-well imaging plate (Perkin Elmer) or on glass slides (#1216691, Marienfeld). For synchronization at the G2/M border, cells were incubated with 10 μM RO-3306 (SML0569#; Sigma) inhibitor for 20 h and after two washes in medium released into cell cycle progression for 4.5 h to reach G1 phase. Cells were briefly washed with PBS and fixed in 10% formalin solution (Sigma) for 10 min at RT followed by two washes with 30 mM glycine–PBS. Permeabilization was performed for 5 min at 4°C in 0.5% Triton X-100 in PBS. After two more washes in 30 mM glycine–PBS, cells were blocked for 15 min at RT in blocking solution (0.2% fish skin gelatin in PBS). Primary antibodies were diluted in blocking solution and incubated for 1 h at RT or overnight at 4°C. Detailed information regarding antibodies can be found in Supplementary Table S1. After three washes in blocking buffer for 3 min each, cells were incubated for about 45 min with secondary antibodies in the dark at RT. After three more washes, slides were briefly rinsed in distilled water and mounted using DAPI Prolong Diamond Antifade Reagent (Invitrogen). In case of imaging plates, cells were incubated for 10 min in 10 μg/ml Hoechst 33342 (Life Technologies) followed by PBS washes and stored in PBS at 4°C. Imaging was performed using the 63× water magnification and the spinning disk confocal mode of the high-content screening microscope Opera Phenix (Perkin Elmer) or the 63× oil magnification of the inverted TCS SP5 confocal microscope (Leica). Raw images were used for quantification of co-localization events using the maximum projection of acquired z-stacks. Images were deconvolved using the Huygens Essentials software.
Single molecule localization microscopy (SMLM)
2D SMLM was performed on a SR GSD microscope set up (Leica Microsystems CMS GmbH) based on an inverse widefield microscope DMI AF6000 equipped with an oil immersion objective (160×/1.43 plan apochromatic, HCX PL APO, TIRF). The temporal isolation of single molecules was obtained by fluorescence activation and deactivation (on and off switching) of organic dyes by laser excitation in an appropriate buffer system, which supports reversible photoswitching. The method is known as ground state depletion and individual molecule return (GSDIM) (24) or direct stochastically optical reconstruction microscopy (dSTORM) (25). U2OS cells were seeded on imaging dishes (#81156, ibidi) overnight and IF staining was performed as described using the anti-GFP Atto488 nanobody and anti-TRF2 antibody coupled to rabbit Alexa-647 (F(ab)2). Detailed information regarding antibodies can be found in Supplementary Table S1. Before imaging, fixed cells were submersed with freshly prepared photoswitching buffer containing the oxygen scavenging system Pyranose-Oxidase-Catalase in combination with a thiol as reducer to provide stable pH conditions during measurement time. The buffer was adapted from (26): pyranose-oxidase (3 U/ml), catalase (90 U/ml), glucose 100 mg/ml (10% w/v), 5 mM cysteamin/β-mercaptoethylamine hydrochloride, Tris buffer (20 mM Tris–HCl pH 7.5, 50 mM NaCl), 2 mM cyclooctatetraene and 20% glycerol. Lasers (488 nm/300 mW, 642 nm/500 mW) were used for excitation and photoactivation, and a 405 nm laser (30 mW) was used for backpumping. TRF2-Alexa647 was imaged before ZBTB10-Atto488 to prevent photobleaching of the far-red dye by the 488 nm laser. Bleed through was controlled by single fluorophore stained samples. Fluorescent emission was filtered (488 HP-T, dichroic 405/488 nm, em 505–595 nm; 647 HP-T, dichroic 405/642nm, em 660–760 nm) and imaged on an EMCCD camera with a pixel size of 100 nm (iXon3 ultra 897, Andor Technologies). Sequential two-color SMLM imaging was performed in TIRF mode ca. 200 nm above the surface of the imaging dish. First, corresponding widefield images of ZBTB10-GFP positive cells and controls were created in the 488 nm and the 647 nm channel. On average 15 000 frames of a 180 × 180 pixel (18 × 18 μm) camera sub-region were recorded with a camera frame rate of 50 Hz (20 ms). The laser intensity for activation was progressively increased during acquisition to keep the amount of emitting fluorophores constant (0.5–2 kW/cm2) and ca. 5% of the 405 nm laser was used for backpumping. Chromatic aberrations were controlled by a ‘sandwich method’, imaging TetraSpeck microspheres (0.1 μm, 1000× dilution in PBS, pre-adsorbed on the surface of the imaging dish; Thermo Scientific) before and after the biological sample. The offset (shift between channels 647 and 488) was obtained by autocorrelation (chromatic shift correction, Huygens software, SVI), and the correction matrix was transferred to the respective sample. Fluorescence image acquisition and processing was performed with the LAS AF 4.0 software (Leica microsystems), Fiji, and ThunderSTORM (27). The post-processing procedure was done in ThunderSTORM: uncertainty 2–25, merging maximum distance of 20 nm in xy, drift correction. The shift between the 642 and 488 nm channel was corrected by cross correlation of the TetraSpeck beads (chromatic aberration corrector, Huygens Essential, SVI). Distances between TRF1/TRF2 and ZBTB10/TRF2 protein pairs were measured between the intensity maxima (peak-to-peak distance) in the reconstructed two-color SMLM images.
Generation of knockout cells
ZBTB10 knockouts (KOs) were generated using three different guide RNAs targeting the second exon of the ZBTB10 gene (Supplementary Table S1). Guide oligonucleotides carrying the PAM sequence were cloned into the pX459 V2 vector containing Cas9 and the sgRNA scaffold and validated by Sanger sequencing (GATC). One day after transfection, HEK293 and U2OS cells were selected in 750 ng/ml puromycin. After successful selection gDNA was extracted using the QIAamp DNA Mini Blood Kit (Qiagen) according to manufacturer's instructions and a ∼1100 bp long target region around the cut locus was amplified by PCR. Cas9 activity was tested in a T7 endonuclease assay. In a first annealing step mismatched DNA was allowed to form, which was visualized by agarose gel electrophoresis after T7 endonuclease digest for 30 min at 37°C. After confirming Cas9 activity, single cell sorting was performed (Becton Dickinson FACSAria III SORP) and cells were expanded to screen for ZBTB10 KO clones by Western blot using an anti-ZBTB10 antibody (Supplementary Table S1).
RESULTS
ZBTB10 binds telomeric and telomeric variant repeats directly via its zinc fingers
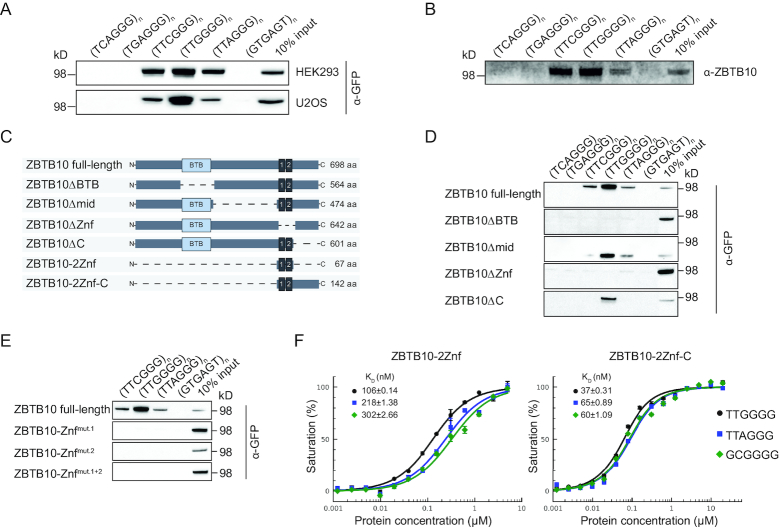
In our previous phylointeractomics screen for telomere-associated proteins across 16 vertebrate species, we identified the zinc finger and BTB domain-containing protein 10 (ZBTB10) as a binder to the telomeric TTAGGG sequence in six different species ranging from fish to mammals (21). To verify the ability of human ZBTB10 to bind to telomeric repeats and to compare specificity with variant repeat sequences, we performed DNA pulldown assays with whole cell lysates from telomerase-positive HEK293 and telomerase-negative U2OS cell lines stably expressing a C-terminal GFP-tagged ZBTB10 (Figure 1A, Supplementary Table S1). Indeed, while human ZBTB10 was able to bind to TTAGGG repeats, we also observed binding to TTCGGG and TTGGGG variant repeats. In fact, ZBTB10 showed strongest enrichment at TTGGGG variant repeats. To investigate whether ZBTB10’s binding ability is due to a direct interaction with DNA, we tested a recombinantly expressed His-tagged ZBTB10 in DNA pulldowns. The recombinant His6-ZBTB10 showed the same binding behavior (Figure 1B), demonstrating that recognition of the DNA sequence is a function of the protein and not mediated by auxiliary factors. To identify the DNA-binding domain, we generated four deletion constructs: ΔBTB (aa Δ164–297), Δmid (aa Δ306–529), ΔZnf (aa Δ540–595; two classical Cys2–His2 zinc fingers) and ΔC (aa Δ602–698) (Figure 1C, Supplementary Table S1). Cells stably expressing ZBTB10 fusion proteins did not show any growth defect and no change in cell cycle distribution (Supplementary Figure S1A). While ZBTB10Δmid and ZBTB10ΔC were still able to bind telomeric variant repeats, the loss of either the BTB domain or the zinc finger region abolished binding to any tested sequence (Figure 1D). To further substantiate the importance of both zinc fingers for DNA binding, we introduced a single point mutation in either one of the two zinc fingers (C551A or C579A) or in both. DNA pulldown assays revealed that both zinc fingers need to be intact for binding to telomeric DNA (Figure 1E). As we observed different enrichment of ZBTB10 to the distinct repeat sequences, we next quantified binding affinities by fluorescence polarization. Here, we determined the affinity of ZBTB10 to its predicted DNA-binding motif GCGGGG, the telomeric sequence TTAGGG and the variant repeat TTGGGG. To this end, we purified a minimal construct consisting of both zinc fingers (ZBTB10-2Znf: aa 540–606, Figure 1C) and a longer construct containing the C-terminus (ZBTB10-2Znf-C: aa 540–682, Figure 1C). We decided to include the longer construct as we found a putative atypical C2HR finger C-terminally adjacent to the two canonical C2H2 fingers, which might also be important for DNA binding. In agreement with the DNA pulldown data, ZBTB10-2Znf showed strongest binding to the TTGGGG probe with 106 nM, a two-fold higher affinity in comparison to 218 nM for the TTAGGG probe and to 302 nM for the GCGGGG motif (Figure 1F). Noteworthy, the longer ZBTB10-2Znf-C construct showed increased specificity for all tested motifs exhibiting strongest affinity for TTGGGG with a KD value of 37 nM, again twice as high as for the other tested motifs (Figure 1F). We thereby quantitatively confirmed the preferred binding of ZBTB10 to the TTGGGG variant repeat sequence and established that the two classical zinc fingers combined are necessary and sufficient for DNA binding, while the C-terminal region further increases binding affinity.
Figure 1.
ZBTB10 preferably binds the TTGGGG variant repeat sequence. (A) Binding of stably expressed ZBTB10-GFP from HEK293 and U2OS cells to telomeric and telomeric variant repeats. Biotinylated concatenated DNA oligonucleotides were immobilized on streptavidin magnetic beads and bound proteins were analyzed by Western blot. A scrambled GTGAGT repeat sequence was used as control bait. (B) DNA pulldowns with extracts containing recombinantly expressed His6-ZBTB10 to telomeric and telomeric variant repeats. (C) Schematic representation of constructed ZBTB10 deletion variants together with their respective length in amino acids (aa). (D) Binding of ZBTB10-GFP variants in DNA pulldown experiments stably expressed in U2OS cells. (E) DNA pulldowns of ZBTB10 zinc finger mutants zinc finger 1 (C551A) and zinc finger 2 (C579A) stably expressed in U2OS cells. (F) Fluorescence polarization assays of the minimal ZBTB10 construct consisting of both zinc fingers (ZBTB10-2Znf) and the longer construct containing both C2H2 fingers and the C-terminus (ZBTB10-2Znf-C). Binding affinities to the telomeric variant repeat TTGGGG (black circle), the telomeric repeat TTAGGG (blue square) and the predicted binding motif GCGGGG (green diamond) were determined.
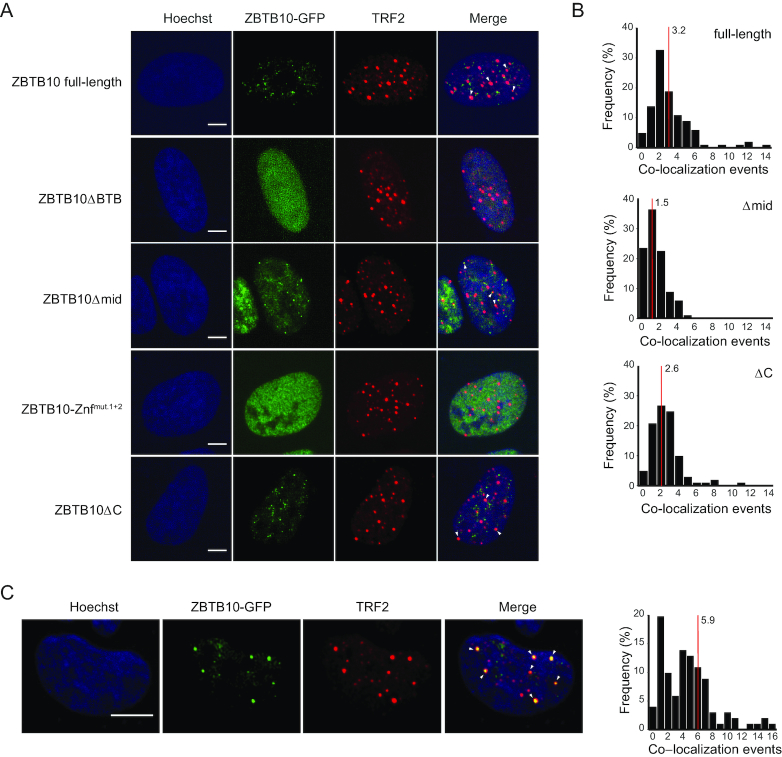
ZBTB10 localizes to a subset of telomeres in ALT cells
To investigate whether ZBTB10 localizes to telomeres in vivo, we analyzed U2OS cells stably expressing ZBTB10-GFP. First, we assessed whether the expression level of the ZBTB10-GFP fusion is similar to the endogenous level and compared expression of ZBTB10 (WT), stably expressed (SE) and transiently overexpressed (OE) ZBTB10-GFP in U2OS cells by label-free quantitative mass spectrometry. In contrast to a >100-fold overexpression when transiently transfected, the expression level of the stably expressed protein were within 10-fold of the endogenous protein level (Supplementary Figure S1B). Using these stable ZBTB10-GFP cells, we observed a punctuated pattern of ZBTB10 with an average of 17 foci per nucleus (Figure 2A, Supplementary Figure S1C). To examine whether ZBTB10 foci co-localize with telomeres, we performed immunofluorescence staining of endogenous TRF2 as a bona fide marker for telomeres in unsynchronized U2OS ZBTB10-GFP cells. For full-length ZBTB10 we quantified on average three foci to co-localize with TRF2 showing that 19% of ZBTB10 foci are overlapping with telomeres (Figure 2A and B). To test whether we observe a difference in co-localization of ZBTB10 and TRF2 between ALT and telomerase-positive cells, we analyzed U2OS, GM847 and SAOS2 cells overexpressing ZBTB10-GFP as well as HeLa E1 and HEK293 cells. On average, co-localization events are slightly higher in the three ALT cell lines compared to the two telomerase-positive cell lines (Supplementary Figure S2A and B). Moreover, it is interesting to note that we quantified up to 20 co-localization events in ALT cells, whereas we found a maximum of five events per nucleus in HeLa E1 or HEK293 cells (Supplementary Figure S2A and B). When we analyzed co-localization of ZBTB10 deletion constructs and TRF2, we find that distinct foci are completely lost upon removal of the dimerization domain (ZBTB10ΔBTB) or mutation of both zinc fingers (ZBTB10ΔZnfmut.1+2) and thus, co-localization with TRF2 cannot be determined (Figure 2A). In accordance with our in vitro DNA-binding data, deletion of the middle region (ZBTB10Δmid) and the C-terminus (ZBTB10ΔC) only moderately affected the number of ZBTB10 and TRF2 co-localization events (Figure 2A and B). The co-localization of full-length ZBTB10-GFP with TRF2 was increased to on average six overlapping foci in G1 cells that were synchronized at the G2/M border and released into cell cycle for 4.5 h (Figure 2C). To further study ZBTB10 nuclear localization during cell cycle progression, we performed live-cell imaging for 40 h in the stable U2OS cell line (Supplementary Figure S2C and Supplementary Video). We observed dynamic changes in foci number, intensity and size, which did not correlate with a specific cell cycle phase. However, we monitored absence of foci during mitosis and foci formation in both daughter cells after cell division.
Figure 2.
Cellular localization of ZBTB10 in U2OS cells. (A) Immunofluorescence staining of TRF2 in U2OS cells stably expressing the different GFP-tagged ZBTB10 deletion variants. DNA was stained with Hoechst. Scale bars represent 5 μm. (B) Histogram of co-localization events between TRF2 and ZBTB10 full-length (n = 103 cells), ZBTB10Δmid (n = 101 cells) or ZBTB10ΔC (n = 100 cells). The average number of co-localization events per cell is indicated with a red line. Due to the diffuse distribution of ZBTB10ΔBTB and ZBTB10ΔZnfmut.1+2 co-localization events could not be determined. (C) Immunofluorescence staining of TRF2 in G1-synchronized U2OS cells stably expressing full-length ZBTB10-GFP. DNA was stained with Hoechst. Scale bars represent 5 μm. Quantification of co-localization events between TRF2 and ZBTB10 (n = 101 cells). The average number of co-localization events is marked with a red line.
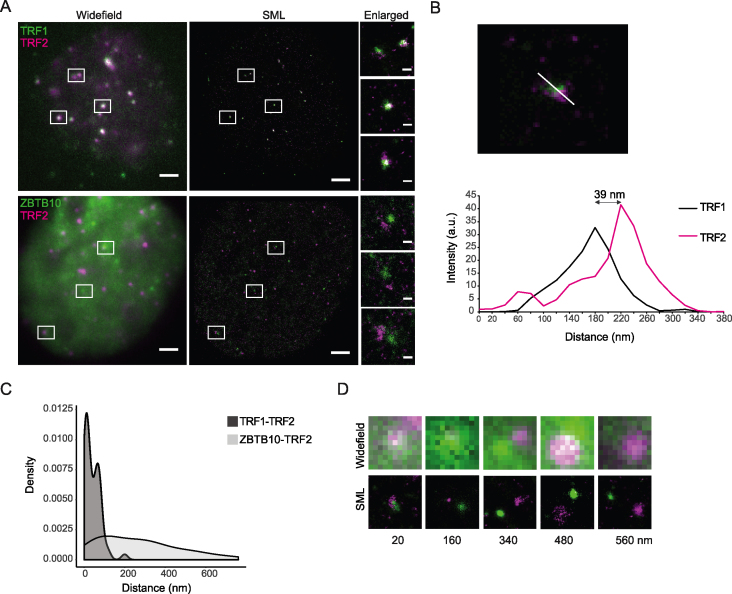
As conventional microscopy has limited capacity in further resolving the localization, we performed super-resolution microscopy based on single molecule localization. For the TRF1/TRF2 complex, serving as a positive control, we measured distances ranging from 1 to 200 nm with an average of 41 nm (Figure 3A–C). In comparison, ZBTB10/TRF2 pairs revealed a higher diversity in measured distances ranging from 14 to 760 nm with an average of 267 nm (Figure 3A, C and D). The observed distances between ZBTB10/TRF2 in comparison to TRF1/TRF2 pairs indicate that a fraction of ZBTB10 (measured distances < 200 nm) co-localizes with telomeres.
Figure 3.
Super-resolution microscopy (GSDIM/dSTORM) and nanometer resolved intermolecular two-color distance analysis of ZBTB10-GFP stably expressed in U2OS cells in relation to TRF2. (A) Dual-color immunostaining of TRF1 (green)/TRF2 (magenta) or ZBTB10-GFP (green)/TRF2 (magenta) demonstrate the different interrelation between the respective protein pairs. Widefield images (left) and corresponding single molecule localizations (SML) rendered as Normalized Gaussian distribution with a localization precision of 15–20  6 nm. Scale bars represent 2 μm and 200 nm in enlarged SMLM images. (B) Intermolecular distances were measured in the reconstructed two-color images by drawing an intensity line profile (in arbitrary units) crossing corresponding protein clusters in the respective channels and recording the distances between the intensity maxima of Atto488 and Alexa647. In this example, the measured distance is 39 nm. (C) Density plot distribution of peak-to-peak distances measured between TRF1/TRF2 pairs (n = 58) with on average 41 nm and ZBTB10/TRF2 (n = 67) pairs with on average 267 nm in reconstructed SMLM images. Image reconstruction and shift correction with a localization precision of 30–40 nm. (D) Examples of selected ZBTB10 (green)/TRF2 (magenta) pairs with measured peak-to-peak distances demonstrate the capabilities of SML (lower panel) to further differentiate between co-localization and co-distribution events compared to conventional light microscopy (upper panel). Images are 1 μm × 1 μm.
6 nm. Scale bars represent 2 μm and 200 nm in enlarged SMLM images. (B) Intermolecular distances were measured in the reconstructed two-color images by drawing an intensity line profile (in arbitrary units) crossing corresponding protein clusters in the respective channels and recording the distances between the intensity maxima of Atto488 and Alexa647. In this example, the measured distance is 39 nm. (C) Density plot distribution of peak-to-peak distances measured between TRF1/TRF2 pairs (n = 58) with on average 41 nm and ZBTB10/TRF2 (n = 67) pairs with on average 267 nm in reconstructed SMLM images. Image reconstruction and shift correction with a localization precision of 30–40 nm. (D) Examples of selected ZBTB10 (green)/TRF2 (magenta) pairs with measured peak-to-peak distances demonstrate the capabilities of SML (lower panel) to further differentiate between co-localization and co-distribution events compared to conventional light microscopy (upper panel). Images are 1 μm × 1 μm.
Depletion of ZBTB10 showed only few changes on transcript level
To study the effect of the putative transcription factor ZBTB10 on gene expression, we generated CRISPR/Cas9 knockout (KO) cells using three different guide RNAs targeting the second exon of ZBTB10 in two cell lines: HEK293 and U2OS (Supplementary Figure S3A). KO clones were validated by Western blot (Supplementary Figure S3B) and sequencing of genomic DNA (Supplementary Figure S3C). To compare transcript levels between WT and KO, we performed RNA-seq analysis using at least three cell clones per cell line (Supplementary Figure S3D, Supplementary Tables S2 and S3). Differential expression analysis was performed for 16,182 genes in HEK293 and 17 936 genes in U2OS cells. When employing a threshold of log2(fold change) >1 and –log10(ad. P-value) >2, we detected only a small number of differentially regulated genes: 39 up- and 17 downregulated genes in HEK293 cells as well as five up- and two downregulated in U2OS cells. Our data therefore shows very few cell line-specific transcriptional changes upon ZBTB10 knockout and further bioinformatics analysis did not reveal any functionally enriched category.
ZBTB10 is not involved in telomere homeostasis
To probe a role of ZBTB10 in telomere homeostasis, we performed telomerase activity measurements using the quantitative telomeric repeat amplification protocol in telomerase-positive HEK293 cells, C-circle assay in telomerase-negative U2OS cells and telomere restriction fragment analysis in both cell lines. Using lysates of the five biological HEK293 WT and KO clones we did not detect differences in telomerase activity in three independent experiments (Supplementary Figure S4A). Additionally, for HEK293 WT clones we measured an average telomere restriction fragment length of 3.5 kb in comparison to a slightly shorter telomere fragment length of 2.8 kb in HEK293 KO clones that did not reach statistical significance (Supplementary Figure S4B). Similarly, no difference in telomere restriction fragment length was observed in U2OS WT and KO cells after culturing them for five months (Supplementary Figure S4C). Concomitantly, quantitative telomeric fluorescence in situ hybridization experiments to assess telomere length in WT and KO clones of HEK293 and U2OS cells did not reveal changes (Supplementary Figure S4D and E). To analyze possible ALT phenotypes, we quantified extrachromosomal telomeric DNA by C-circle assay (Supplementary Figure S4F) and counted ALT-associated promyelocytic leukaemia bodies (APBs) (Supplementary Figure S4G). For both, we did not observe any significant changes between WT and KO U2OS cells. These data suggest that ZBTB10 is not involved in the assayed telomeric functions.
ZBTB10 interacts with TRF2 and RAP1
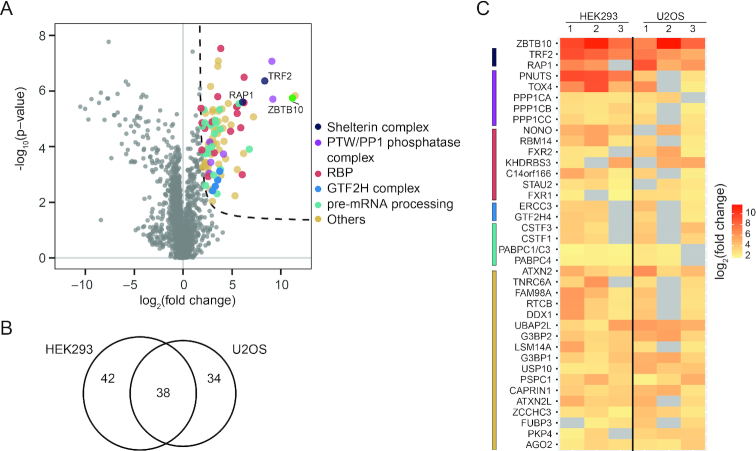
We then performed GFP pulldowns coupled to quantitative label-free proteomics in HEK293 and U2OS stable ZBTB10-GFP cells to gain insights into the molecular function of ZBTB10 (Figure 4A, Supplementary Figure S5A and B, Supplementary Table S4). We reproducibly detected (in at least two of three experiments per cell line) 42 proteins enriched in HEK293, 34 proteins enriched in U2OS and 38 commonly enriched proteins delineating a set of cell line-specific and common interaction partners (Figure 4B and C). Performing functional annotation clustering on the 37 proteins putatively interacting with ZBTB10, 27 of them formed a network with five high confidence clusters (EASE score >3) that share either annotated protein domains, specific gene ontology descriptors or KEGG pathway terms (Supplementary Figure S5C, Supplementary Table S5). Four of these clusters are associated with RNA-related terms, originating from several proteins involved in cleavage and polyadenylation of pre-mRNA 3′ ends, splicing regulation and RNA binding. In addition to RNA-associated proteins, we found the core transcription factor II H complex (GTF2H), which plays a role in nucleotide excision repair of DNA and when bound to the CAK complex, it functions in RNA polymerase II-dependent transcription (28). Moreover, we identified several subunits of the PTW/PP1 phosphatase complex, which plays a role in the control of chromatin structure, RNA processing, and cell cycle progression during the transition from mitosis to interphase (29). Our interaction data indicates that ZBTB10 functions in several cellular processes and might be involved in post-transcriptional activities via interaction with a diverse set of proteins. In addition to these factors, we also co-purified the shelterin complex members TRF2 and RAP1 with very high fold enrichments (Figure 4C). To note, none of the other shelterin subunits were detected in any of our experiments implying that ZBTB10 only interacts with the TRF2/RAP1 heterodimer (30). As observed in telomerase-positive HEK293 and telomerase-negative U2OS cells, the interaction is independent of the telomere maintenance mechanism. To investigate whether loss of ZBTB10 influences TRF2 levels, we compared protein abundance by Western blot and IF staining but could not detect differences in HEK293 or U2OS cells (Supplementary Figure S6A and B). To nevertheless exclude putative effects on telomere end protection, we analyzed chromosomal rearrangements on metaphase chromosomes, changes in the occurrence of telomere-induced foci (TIF) and ATM kinase activation (Supplementary Figure S6C–G). In none of these assays, we could detect a difference between ZBTB10 KO and WT U2OS cells. Overall, we show that ZBTB10 does not influence TRF2 protein stability or its role in end protection.
Figure 4.
Identification of ZBTB10 interaction partners. (A) Volcano plot of a representative GFP pulldown from HEK293 cells stably expressing ZBTB10-GFP using quantitative mass spectrometry. GFP transfected cells served as control and each experiment was performed in quadruplicates. Proteins co-purifying with ZBTB10 are determined by S0>1.6, c = 0.8 and p < 0.05 (dotted line) and colored according to functional description or complex membership (see legend). (B) Venn diagram to show overlap of proteins found enriched in at least two out of three independent experiments from U2OS and HEK293 cells. (C) Heat map illustrating enrichment of the 37 common interactors and ZBTB10. Interaction partners are grouped according to their functional description or complex membership: shelterin complex (dark blue), PTW/PP1 phosphatase complex (purple), RBPs (red), GTF2H complex (blue), pre-mRNA processing (green) and others (beige). Grey squares represent no enrichment in the respective pulldown.
ZBTB10 interacts with TRF2 through its N-terminal domain
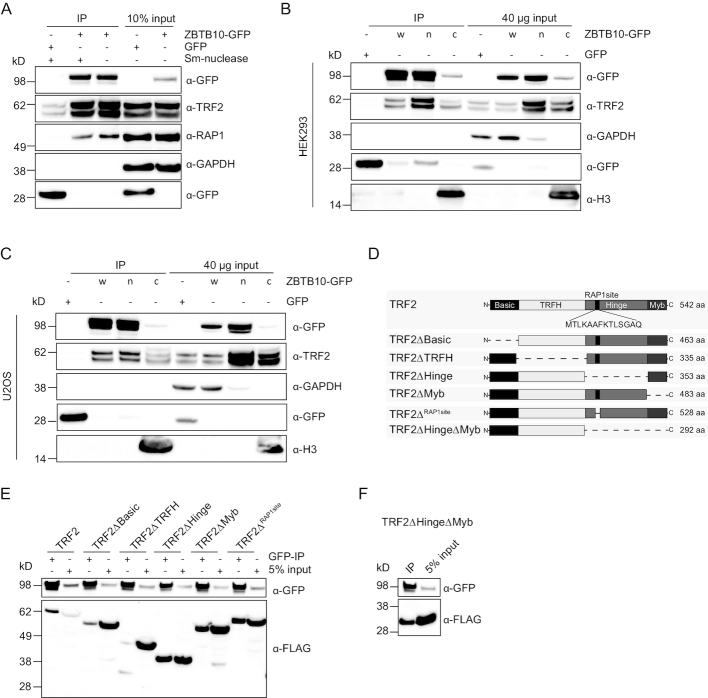
Prior to mapping the interaction between ZBTB10 and the TRF2/RAP1 complex, we validated TRF2 and RAP1 enrichment in the ZBTB10-GFP IP by immunostaining and show that the interaction is not mediated by RNA or DNA binding as Sm-nuclease treatment did not abrogate TRF2/RAP1 enrichment (Figure 5A, Supplementary Figure S7A). To test if the ZBTB10-GFP/TRF2 interaction occurs in the nucleoplasm or in the context of chromatin, we prepared nuclear and chromatin extracts from HEK293 and U2OS cells stably expressing ZBTB10-GFP. While TRF2 is equally abundant in both subcellular fractions, the amount of chromatin-bound ZBTB10 is 6× less in HEK293 cells and even 34× less in U2OS cells (Figure 5B and C). GFP pulldowns of nuclear and chromatin extracts showed similar stoichiometry of TRF2 and ZBTB10 (Figure 5B and C) revealing that their interaction is also stable in nucleoplasmic extracts and does not require telomeric chromatin. To determine the interaction site of TRF2 required for binding to ZBTB10, we generated various deletion constructs of TRF2: ΔBasic (aa Δ1–81; N-terminal domain), ΔTRFH (aa Δ83–290; the dimerization domain), ΔHinge (aa Δ293–482) and ΔMyb (aa Δ483–542; the DNA-binding domain) (Figure 5D) (31). We also constructed a TRF2ΔRAP1site (aa Δ328-341) (32) construct to evaluate whether RAP1 binding to TRF2 was required for the interaction with ZBTB10. Except for TRF2ΔBasic and TRF2ΔTRFH, all TRF2 variants co-immunoprecipitated with ZBTB10-GFP (Figure 5E) revealing that the interaction between ZBTB10 and TRF2 is mediated via the Basic and TRFH domains of TRF2. Indeed, a minimal construct of the N-terminal region of TRF2 missing the Hinge and Myb domain (TRF2ΔHingeΔMyb; aa 1–292) was sufficient to be co-IPed by ZBTB10-GFP (Figure 5F). These results establish that the interaction of ZBTB10 with TRF2 does not require the DNA-binding domain of TRF2 and is independent of RAP1.
Figure 5.
Mapping of the interaction site between ZBTB10 and TRF2. (A) Western blot validation of endogenous TRF2 and RAP1 co-purifying with ZBTB10 from stable HEK293 cells with and without Sm-nuclease treatment. GFP transfected cells served as control pulldown and GAPDH was used as a loading control. (B) GFP pulldowns using 1 mg whole lysate (w), 400 μg nuclear (n) and 400 μg chromatin (c) extracts of HEK293 cells stably expressing ZBTB10-GFP. GFP transfected cells served as control pulldown. Histone H3 served as a marker protein for the chromatin fraction. (C) GFP pulldowns using 1 mg whole lysate (w), 400 μg nuclear (n) and 400 μg chromatin (c) extracts of U2OS cells stably expressing ZBTB10-GFP. GFP transfected cells served as control pulldown. Histone H3 served as a marker protein for the chromatin fraction. (D) Schematic overview of constructed TRF2 deletions with protein length in amino acids (aa). (E) GFP pulldowns from HEK293 cells stably expressing ZBTB10-GFP in combination with different transiently overexpressed FLAG-TRF2 variants pinpoint the N-terminal region of TRF2 as the interaction site. (F) Co-immunoprecipitation of ZBTB10-GFP with the minimal FLAG-TRF2ΔHingeΔMyb construct from stable HEK293 cells.
DISCUSSION
Our previous phylointeractomics screen suggested that ZBTB10 can associate with the telomeric repeat sequence. Indeed, we here show that the dsTTAGGG motif is recognized by the ZBTB10 minimum DNA-binding fragment with similar affinity (KD = 218 nM) as the recently described telomeric protein ZBTB48/TZAP (KD = 170 nM) (33). Unlike ZBTB48/TZAP that surprisingly uses a single zinc finger (Znf11) aided by an adjacent C-terminal arm to bind telomeric DNA (19,33), we demonstrate that the two classical Cys2His2 zinc fingers of ZBTB10 combined are necessary and sufficient for binding. Moreover, when we investigated the binding ability of ZBTB10 at telomeric variant repeats, we found that ZBTB10 exhibited a two-fold stronger binding to the TTGGGG motif (KD = 106 nM) than to the TTAGGG motif, differentiating it from ZBTB48/TZAP, which has less affinity for this telomeric variant repeat (KD = 380 nM) (33). We further reveal that the additional atypical zinc finger in the C-terminal region strongly enhances ZBTB10 binding affinity to telomeric DNA with a KD value of 37 nM for the TTGGGG motif. In the same way, a cooperative binding mode for Zif268 has been reported by addition of a third zinc finger that strongly increased binding affinity (34). We thus identified ZBTB10 as a new telomere-binding protein with a preference for the TTGGGG motif. Similarly to ZBTB10, the nuclear receptor NR2C2 has been characterized to preferably bind the TCAGGG sequence with a two-fold higher binding affinity (KD = 28 nM) compared to the telomeric motif (KD = 58 nM) (16). While variant repeat sequences are restricted to the proximal 2 kb of telomeres in telomerase-positive cells, they are more frequently occurring in the longer heterogeneous telomeres of ALT cells due to homology-directed telomere maintenance (16). Concomitantly with the in vitro affinity of ZBTB10 to telomeric repeats, we observe co-localization of ZBTB10 and telomeres. There seems to be a subset of ALT cells that have more than five co-localization events compared to the telomerase-positive cell lines.
Our additional experiments using HEK293 and U2OS ZBTB10 knockout cells demonstrated that absence of ZBTB10 has no effect on telomere homeostasis in telomerase-positive or ALT cells for multiple hallmarks such as telomere length, C-circle abundance and APB frequency. In our phylointeractomics screen, we identified several TTAGGG-associated proteins and we cannot exclude a redundant function of ZBTB10 with any of them. However, using quantitative interactomics, we identify TRF2 and RAP1 as interaction partners of ZBTB10 and we show that ZBTB10 interacts with the N-terminal region of TRF2 in a DNA and RNA-independent manner. The interactome further revealed an interaction with the GTF2H complex and the PTW/PP1 phosphatase complex. Interestingly, the regulatory subunit of this complex, PNUTS, was highly enriched in our pulldowns and has previously been reported as an interactor of TRF2 and shown to co-localize with a subset of telomeres (35). In a recent quantitative proximity ligation assay to decipher the DNA repair network, ZBTB10 showed between 1.50- and 1.74-fold enrichment with all three APEX2-tagged DNA repair proteins: 53BP1, MDC1 and BRCA1 (36). Previously, ZBTB10 has also been found in a TAP-MS screen to possibly interact with the BRCT-domain of 53BP1 (37). Together with the interaction of PNUTS that influences 53BP1-mediated DNA repair (38), it is tempting to speculate that ZBTB10 in cooperation with the TRF2/RAP1 complex is involved in DNA damage repair at telomeres, especially as ZBTB10 has been shown to localize to sites of damaged chromatin after UV laser microirradiation (39).
DATA AVAILABILITY
The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE (40) partner repository with the dataset identifier PXD010672. The sequencing data has been deposited in the NCBI Gene Expression Omnibus database with the accession number GSE117214.
Supplementary Material
ACKNOWLEDGEMENTS
Assistance by the following IMB facilities is gratefully acknowledged: Microscopy Core Facility and the use of its Opera Phenix (INST 247/845-1 FUGG), Cytometry Core Facility, Proteomics Core Facility, Genomics Core Facility and Media Lab. We further thank Joachim Lingner for providing the HeLa E1 cell line.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Bu2996/1-2]; Research in the Kappei laboratory was supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative and by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education's AcRF Tier 3 grants [MOE2014-T3-1-006]; Research in the Shi laboratory was supported by The Strategic Priority Research Program (Pilot study) ‘Biological basis of aging and therapeutic strategies’ of the Chinese Academy of Sciences [XDPB10]. Funding for open access charge: Deutsche Forschungsgemeinschaft [Bu2996/1-2].
Conflict of interest statement. None declared.
REFERENCES
- 1. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 2. Maciejowski J., de Lange T.. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017; 18:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doksani Y., Wu J.Y., de Lange T., Zhuang X.. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013; 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 5. van Steensel B., Smogorzewska A., de Lange T.. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998; 92:401–413. [DOI] [PubMed] [Google Scholar]
- 6. Rai R., Chen Y., Lei M., Chang S.. TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat. Commun. 2016; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karlseder J., Hoke K., Mirzoeva O.K., Bakkenist C., Kastan M.B., Petrini J.H.J., De Lange T.. The telomeric protein TRF2 binds the ATM Kinase and Can Inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004; 2:1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denchi E.L., de Lange T.. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007; 448:1068–1071. [DOI] [PubMed] [Google Scholar]
- 9. Sarek G., Vannier J., Panier S., Petrini J.H.J., Boulton S.J.. TRF2 recruits RTEL1 to telomeres in S phase to promote T-Loop unwinding. Mol. Cell. 2015; 57:622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Higa M., Kushiyama T., Kurashige S., Kohmon D., Enokitani K., Iwahori S., Sugimoto N., Yoshida K., Fujita M.. TRF2 recruits ORC through TRFH domain dimerization. BBA - Mol. Cell Res. 2017; 1864:191–201. [DOI] [PubMed] [Google Scholar]
- 11. Kim N.W., Piatyszek K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W., Prowse M.A.. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994; 226:2011–2015. [DOI] [PubMed] [Google Scholar]
- 12. Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R.. Telomere elongation in immortal human cells without detectable telomerase activity. Embo. J. 1995; 14:4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dilley R.L., Verma P., Cho N.W., Winters H.D., Wondisford A.R., Greenberg R.A.. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016; 539:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sobinoff A.P., Pickett H.A.. Alternative lengthening of Telomeres: DNA repair pathways converge. Trends Genet. 2017; 33:921–932. [DOI] [PubMed] [Google Scholar]
- 15. Lee M., Hills M., Conomos D., Stutz M.D., Dagg R.A., Lau L.M.S., Reddel R.R., Pickett H.A.. Telomere extension by telomerase and ALT generates variant repeats by mechanistically distinct processes. Nucleic Acids Res. 2014; 42:1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conomos D., Stutz M.D., Hills M., Neumann A.A., Bryan T.M., Reddel R.R., Pickett H.A.. Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol. 2012; 199:893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marzec P., Armenise C., Pérot G., Roumelioti F., Basyuk E., Gagos S., Chibon F., Déjardin J.. Nuclear-Receptor-Mediated telomere insertion leads to genome instability in ALT cancers. Cell. 2015; 160:913–927. [DOI] [PubMed] [Google Scholar]
- 18. Perez-Torrado R., Yamada D., Defossez P.A.. Born to bind: The BTB protein-protein interaction domain. BioEssays. 2006; 28:1194–1202. [DOI] [PubMed] [Google Scholar]
- 19. Jahn A., Rane G., Paszkowski‐Rogacz M., Sayols S., Bluhm A., Han C., Draškovič I., Londoño‐Vallejo J.A., Kumar A.P., Buchholz F. et al. ZBTB48 is both a vertebrate telomere‐binding protein and a transcriptional activator. EMBO Rep. 2017; 18:929–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J.S.Z., Fusté J.M., Simavorian T., Bartocci C., Tsai J., Karlseder J., Denchi E.L.. TZAP: A telomere-associated protein involved in telomere length control. Science. 2017; 355:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kappei D., Scheibe M., Paszkowski-Rogacz M., Bluhm A., Gossmann T.I., Dietz S., Dejung M., Herlyn H., Buchholz F., Mann M. et al. Phylointeractomics reconstructs functional evolution of protein binding. Nat. Commun. 2017; 8:14334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cristofari G., Lingner J.. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006; 25:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shevchenko A., Tomas H., Havliš J., Olsen J. V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2007; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 24. Fölling J., Bossi M., Bock H., Medda R., Wurm C.A., Hein B., Jakobs S., Eggeling C., Hell S.W.. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat. Methods. 2008; 5:943–945. [DOI] [PubMed] [Google Scholar]
- 25. Van De Linde S., Löschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., Sauer M.. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011; 6:991–1009. [DOI] [PubMed] [Google Scholar]
- 26. Swoboda M., Henig J., Cheng H.M., Brugger D., Haltrich D., Plumeré N., Schlierf M.. Enzymatic oxygen scavenging for photostability without ph drop in single-molecule experiments. ACS Nano. 2012; 6:6364–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ovesný M., Křížek P., Borkovec J., Švindrych Z., Hagen G.M.. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014; 30:2389–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Compe E., Egly J.-M.. Nucleotide excision repair and transcriptional regulation: TFIIH and beyond. Annu. Rev. Biochem. 2016; 85:265–290. [DOI] [PubMed] [Google Scholar]
- 29. Lee J.H., You J., Dobrota E., Skalnik D.G.. Identification and characterization of a novel human PP1 phosphatase complex. J. Biol. Chem. 2010; 285:24466–24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bae N.S., Baumann P.. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell. 2007; 26:323–334. [DOI] [PubMed] [Google Scholar]
- 31. Arnoult N., Karlseder J.. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015; 22:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okamoto K., Bartocci C., Ouzounov I., Diedrich J.K., Iii J.R.Y., Denchi E.L.. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013; 494:502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao Y., Zhang G., He C., Mei Y., Shi Y., Li F.. The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA. Cell Res. 2018; 28:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J., Kim J., Seok C.. Cooperativity and specificity of Cys 2 His 2 zinc finger protein–DNA interactions: a molecular dynamics simulation study. 2010; [DOI] [PubMed] [Google Scholar]
- 35. Kim H., Lee O.-H., Xin H., Chen L.-Y., Qin J., Chae H.K., Lin S.-Y., Safari A., Liu D., Songyang Z.. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 2009; 16:372–379. [DOI] [PubMed] [Google Scholar]
- 36. Gupta R., Somyajit K., Narita T., Maskey E., Stanlie A., Kremer M., Typas D., Lammers M., Mailand N., Nussenzweig A. et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell. 2018; 173:972–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Woods N.T., Mesquita R.D., Sweet M., Carvalho M.A., Li X., Liu Y., Nguyen H., Thomas C.E., Iversen E.S. Jr, Marsillac S. et al. Charting the landscape of tandem BRCT domain-mediated protein interactions. Sci. Signal. 2012; 5:rs6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Landsverk H.B., Mora-Bermúdez F., Landsverk O.J.B., Hasvold G., Naderi S., Bakke O., Ellenberg J., Collas P., Syljuåsen R.G., Küntziger T.. The protein phosphatase 1 regulator PNUTS is a new component of the DNA damage response. EMBO Rep. 2010; 11:868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izhar L., Adamson B., Ciccia A., Lewis J., Pontano-vaites L., Liang A.C., Westbrook T.F., Harper J.W., Stephen J.. A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors. Cell Rep. 2015; 11:1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE (40) partner repository with the dataset identifier PXD010672. The sequencing data has been deposited in the NCBI Gene Expression Omnibus database with the accession number GSE117214.