Abstract
The research field of systems biology has greatly advanced and, as a result, the concept of network pharmacology has been developed. This advancement, in turn, has shifted the paradigm from a “one-target, one-drug” mode to a “network-target, multiple-component-therapeutics” mode. Network pharmacology is more effective for establishing a “compound-protein/gene-disease” network and revealing the regulation principles of small molecules in a high-throughput manner. This approach makes it very powerful for the analysis of drug combinations, especially Traditional Chinese Medicine (TCM) preparations. In this work, we first summarized the databases and tools currently used for TCM research. Second, we focused on several representative applications of network pharmacology for TCM research, including studies on TCM compatibility, TCM target prediction, and TCM network toxicology research. Third, we compared the general statistics of several current TCM databases and evaluated and compared the search results of these databases based on 10 famous herbs. In summary, network pharmacology is a rational approach for TCM studies, and with the development of TCM research, powerful and comprehensive TCM databases have emerged but need further improvements. Additionally, given that several diseases could be treated by TCMs, with the mediation of gut microbiota, future studies should focus on both the microbiome and TCMs to better understand and treat microbiome-related diseases.
Keywords: network pharmacology, Traditional Chinese Medicine, database, assessment and comparison, completeness
Opportunities and Challenges for the Modernization of Traditional Chinese Medicine
Traditional Chinese Medicine
Traditional Chinese Medicine (TCM) is one of the greatest treasures of Chinese culture, has a long history of use in East and Southeast Asia, and has been widely used since ancient civilization. Through continuous development and innovation, physicians have selected the essence and discarded the dross of TCM. As a result, TCM has become one of the main forms of alternative medicine in East Asia, North America, and Europe. TCMs use therapeutic herbs to treat diseases according to the combinatorial principle of “King, Vassal, Assistant, and Delivery servant” based on a patient’s syndrome and to restore balance of life and body functions (Yi and Chang, 2004; Qiu, 2007). Each prescribed combination of these herbs is referred to as a TCM preparation, or TCM prescription, for example the LiuWeiDiHuangWan (LWDHW) pill. Traditionally, the discovery of most new drugs is focused on identifying or designing a pharmacologically effective agent that specifically interacts with a single target. During the past 30 years, such an approach has generated highly successful drugs. However, drugs acting on individual molecular targets usually exert unsatisfying therapeutic effects or have toxicity when used to treat certain diseases, such as diabetes, inflammation, and cancer (Kola and Landis, 2004). Given the major bottleneck in drug discovery, drug research, and development have gradually shifted from a “one-target, one-drug” mode to a “network-target, multiple-component-therapeutics” mode (Li, 2011; Li et al., 2011, 2014b). To treat disease, the holistic philosophy of TCM shares key concepts with the new mode of drug discovery. Therefore, making full use of TCM is of great importance for drug research. In recent years, TCM has gradually garnered interest thanks to its low toxicity and therapeutic effects (Chan, 1995; Corson and Crews, 2007; Qiu, 2007). For example, Ganoderma lucidum, termed “LingZhi” in China, has been used as a health-preserving and therapeutic agent. Previous studies have shown that it possesses medicinal properties, including anti-cancer, anti-diabetic, anti-hepatotoxic, and immunomodulatory effects (Paterson, 2006; Boh et al., 2007, Boh, 2013; Ma et al., 2015). Panax ginseng is another TCM that has been used therapeutically for more than 2,000 years to treat such diseases as cardiovascular disease, Alzheimer’s disease, and diabetes (Lee et al., 2008; Kim, 2012; Yuan et al., 2012; Chan et al., 2013). Unlike modern drugs discovered by targeting a specific protein, however, the understanding of the molecular basis of TCM remains limited, and research into modern TCM theory has lagged, which has slowed down the development of novel TCMs (Corson and Crews, 2007). TCM studies must keep pace with the rapid development of modern science to remain relevant. Therefore, a scientific and effective assessment system needs to be established, which will be key for studying and making full use of TCMs.
Network Pharmacology: An Appropriate Approach for Modern TCM Research
Given the rapid progress in bioinformatics, systems biology, and polypharmacology, network-based drug discovery is considered to be a promising approach for cost-effective drug development. Systems biology examines biological systems by systematically perturbing them; monitoring the gene, protein, and informational pathway responses; integrating these data; and, ultimately, formulating mathematical models to describe the structure of the system and its response to individual perturbations (Ideker et al., 2001). Based on a systems biology approach, the concept of network pharmacology was first proposed by Li et al. (2014b). Because network pharmacology can provide a full or partial understanding of the principles of network theory and systems biology, it has been considered the next paradigm in drug discovery (Hopkins, 2008). Furthermore, the network pharmacology approach has been used to study “compound-proteins/genes-disease” pathways, which are capable of describing complexities among biological systems, drugs, and diseases from a network perspective, sharing a similar holistic philosophy as TCM. Applications of systems biology methods to determine the pharmacological action, mechanism of action, and safety of TCMs are invaluable for modern research and development of TCM. Thus, a new interdisciplinary method termed TCM network pharmacology has been proposed (Li and Zhang, 2013; Liang et al., 2014a; Li et al., 2014b), which has initiated a new research paradigm for transforming TCM from an experience-based to evidence-based medicine. In this work, we first summarized the currently widely used databases and tools for TCM network pharmacology research. Second, we concentrated on the different applications of network pharmacology to TCM research, including TCM recipes, target prediction, and network toxicology. Third, we compared different TCM databases based on their basic properties and search results (Figure 1).
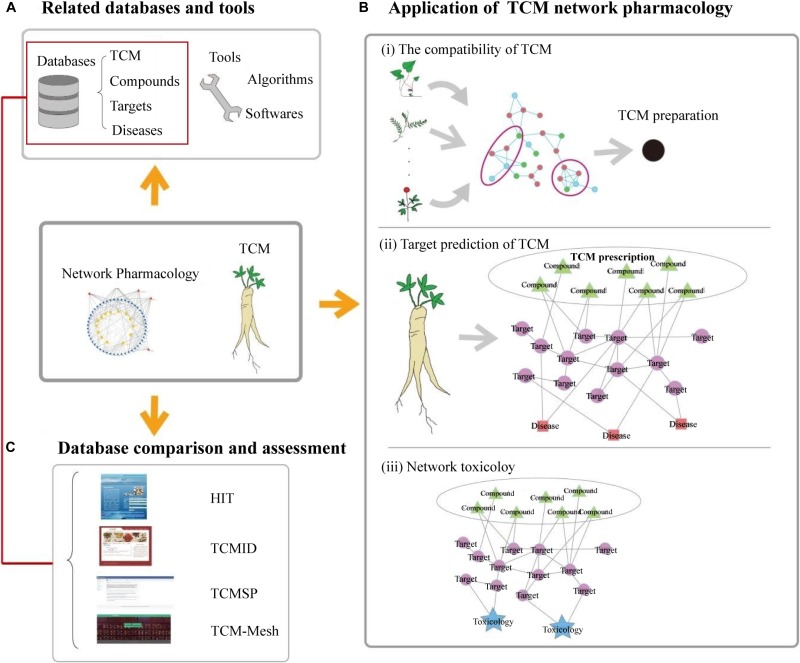
FIGURE 1.
Overall schema of the databases, tools, and applications of TCM network pharmacology analysis. (A) Related databases (TCM, compounds, targets, and disease databases) and tools (algorithms and software) used for TCM network pharmacology research. (B) The application of network pharmacology to TCM research: (i) the compatibility of TCM; (ii) target prediction of TCM; and (iii) network toxicology. (C) Comparison among several TCM databases based on the general statistics and search results.
Tools and Databases for TCM Network Pharmacology Research
With the rapid development of research, network pharmacology has become a new approach for drug mechanism research and drug development. In recent years, a variety of related databases and tools have provided crucial support for TCM network pharmacology research. Commonly used databases for TCM network pharmacology research include TCM databases [TCM database@Taiwan (Chen, 2011), HIT (Ye et al., 2011), TCMSP (Ru et al., 2014), and TCMID (Xue et al., 2013)), compound and drug information databases (Drugbank (Wishart et al., 2006), STITCH (Kuhn et al., 2014), ChEMBL (Gaulton et al., 2012), and PubChem (Wang et al., 2009)], target interaction databases [STRING (Szklarczyk et al., 2015), HPRD (Peri et al., 2003), MINT (Zanzoni et al., 2002), IntAct (Kerrien et al., 2012), Reactome (D’Eustachio, 2009), and HAPPI (Chen et al., 2009)], and gene-disease association databases [OMIM (Hamosh et al., 2002) and GAD (Becker et al., 2004; Table 1)]. Recently, we also proposed TCM-Mesh (Zhang et al., 2017), a more comprehensive TCM database that embodies the core idea of TCM network pharmacology. Based on these biological databases and clinical trial results, researchers can analyze the “herbs-compounds-proteins/genes-diseases” interaction network from the perspective of systems biology, which will provide an understanding of the effects of herbs on diseases. Furthermore, network pharmacology algorithms and tools are particularly vital in mining these databases for knowledge. For example, the Random Walk (Chen et al., 2012) algorithm is a commonly used network clustering algorithm, which starts from a random node (drug, target, or disease) and calculates the similarity of this node and its adjacent node to construct a “drug-target-disease” network. Moreover, the PRINCE (Vanunu et al., 2010) algorithm is used to prioritize disease genes and infer protein complex associations, based on formulating constraints on the prioritization function which is related with its smoothness over the network and usage of prior information. In addition to data acquisition and analysis, visualization is a key element of network pharmacology that makes the network more intuitive. Cytoscape (Shannon et al., 2003) is an open-source platform suitable for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles, and other state data. Pajek (Dohleman, 2006) is another powerful network analysis tool that is used to examine various complex non-linear networks.
Table 1.
Public databases, algorithms, and software related to TCM network pharmacology.
| Type | Name | Description | Website for database or tool | Reference | |
|---|---|---|---|---|---|
| Databases | TCM-related databases | TCM-Mesh | An integration of database and a data-mining system for network pharmacology analysis of TCM preparations | http://mesh.tcm.microbioinformatics.org | Zhang et al., 2017 |
| TCM database@Taiwan | The world’s largest and most comprehensive free small molecular database on TCM for virtual screening | http://tcm.cmu.edu.tw | Chen, 2011 | ||
| HIT | A comprehensive and fully curated database to complement available resources on protein targets for FDA-approved drugs as well as the promising precursor compounds | http://lifecenter.sgst.cn/hit | Ye et al., 2011 | ||
| TCMSP | A unique systems pharmacology platform of TCMs that captures the relationships among drugs, targets, and diseases | http://lsp.nwu.edu.cn/tcmsp.php | Ru et al., 2014 | ||
| TCMID | A comprehensive database that provides information and bridges the gap between TCM and modern life sciences | http://www.megabionet.org/tcmid | Xue et al., 2013 | ||
| Drug-related databases | A unique bioinformatics and cheminformatics resource that combines detailed drug data with comprehensive drug target information | https://www.drugbank.ca | Wishart et al., 2006 | ||
| STITCH | A database of known and predicted interactions between chemicals and proteins | http://stitch.embl.de | Kuhn et al., 2014 | ||
| ChEMBL | An Open Data database containing binding, functional, and ADMET information for a large number of drug-like bioactive compounds | https://www.ebi.ac.uk/chembl | Gaulton et al., 2012 | ||
| PubChem | A public information system for analyzing the bioactivities of small molecules | https://pubchem.ncbi.nlm.nih.gov | Wang et al., 2009 | ||
| Target-related databases | STRING | A database of known and predicted protein-protein interactions | https://string-db.org | Szklarczyk et al., 2015 | |
| HPRD | An object database that integrates a wealth of information relevant to the function of human proteins in health and disease | http://www.hprd.org | Peri et al., 2003 | ||
| MINT | A database that focuses on experimentally verified protein-protein interactions mined from the scientific literature by expert curators | https://mint.bio.uniroma2.it | Zanzoni et al., 2002 | ||
| IntAct | A freely available, open-source database system and analysis tool for molecular interaction data | https://www.ebi.ac.uk/intact | Kerrien et al., 2012 | ||
| Reactome | A free, open-source, curated, and peer-reviewed pathway database | https://reactome.org | D’Eustachio, 2009 | ||
| HAPPI | An online database of comprehensive human annotated and predicted protein interactions | http://discovery.informatics.uab.edu/HAPPI | Chen et al., 2009 | ||
| Disease-related databases | OMIM | A comprehensive, authoritative compendium of human genes and genetic phenotypes that is freely available and updated daily | https://www.omim.org | Hamosh et al., 2002 | |
| GAD | A database of genetic association data from complex diseases and disorders | https://geneticassociationdb.nih.gov | Becker et al., 2004 | ||
| Algorithms | Random walk | An algorithm that predicts potential drug-target interactions on a large scale under the hypothesis that similar drugs often target similar target proteins and the framework of Random Walk | https://www.rdocumentation.org/packages/diffusr/versions/0.1.4/topics/random.walk | Chen et al., 2012 | |
| PRINCE | A global, network-based method for prioritizing disease genes and inferring protein complex associations | https://github.com/fosterlab/PrInCE | Vanunu et al., 2010 | ||
| Software | Cytoscape | A software environment for integrated models of biomolecular interaction networks | https://cytoscape.org | Shannon et al., 2003 | |
| Pajek | A tool for complex network analysis | http://mrvar.fdv.uni-lj.si/pajek | Dohleman, 2006 | ||
These public databases are categorized into four types: TCM-related, drug-related, target-related, and disease-related. Publicly available tools and visualization software use data from these databases. Websites and references are provided for all of these resources.
Network Pharmacology Research and TCM
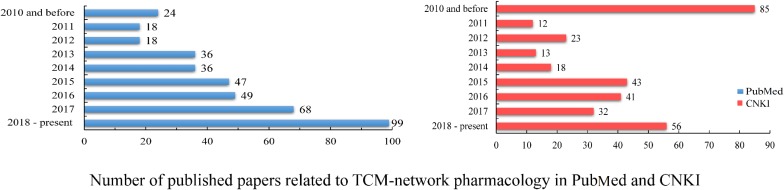
In recent years, an increasing number of studies have focused on the area of TCM network pharmacology. According to statistics in PubMed and the China National Knowledge Infrastructure (CNKI) databases, the number of published papers (Figure 2) has increased over time. These statistics demonstrate the increasing interest in the application of network pharmacology to TCM.
FIGURE 2.
Published papers identified from PubMed and CNKI databases. Papers in PubMed were searched by Title/Abstract containing the following keywords: “network pharmacology,” “Chinese,” and “medicine.” Papers in CNKI were searched with the following key words: “network pharmacology” and “Chinese herb,” excluding all conference papers. The analysis time was from 2010 and before, to the end of 2018.
To date, network pharmacology has been applied to studies of many traditional Chinese herbs and herbal prescriptions. As listed in Table 2, researchers have conducted many network-based computational and experimental studies to detect effective substances and determine the mechanisms of herbal formulas against many diseases.
Table 2.
TCM network pharmacology studies in international journals from 2011 to 2017.
| Herb/Herbal prescriptiona | Related disease/effect | Year | Reference |
|---|---|---|---|
| Aconiti Lateralis Radix | Multi-targets | 2011 | Wu et al., 2011 |
| Compound DanShen formula | Cardiovascular disease | 2012 | Li et al., 2012 |
| Chinese herbal Radix Curcumae formula | Cardiovascular disease | 2013 | Tao et al., 2013 |
| QingLuoYin | Rheumatoid arthritis | 2013 | Zhang et al., 2013 |
| TaoHongSiWu decoction | Osteoarthritis | 2013 | Zheng et al., 2013 |
| Eucommia ulmoides Oliv. | Hypertension | 2014 | Li et al., 2014d |
| GeGenQinLian decoction | Type 2 diabetes | 2014 | Li et al., 2014a |
| BuShenHuoXue formula | Chronic kidney disease | 2014 | Shi et al., 2014 |
| LiuWeiDiHuang pill | Colon cancer and esophagitis | 2014 | Li et al., 2014b |
| QiShenYiQi formula | Myocardial infarction | 2014 | Li et al., 2014c |
| QiShenYiQi formula | Acute myocardial ischemia | 2014 | Wu et al., 2014 |
| Dragon’s blood tablet | Colitis | 2014 | Xu et al., 2014 |
| ZhiZiDaHuang decoction | Alcoholic liver disease | 2015 | An and Feng, 2015 |
| Rhubarb | Renal interstitial fibrosis | 2015 | Xiang et al., 2015 |
| SiMiaoWan | Gout | 2015 | Zhao et al., 2015 |
| ErXian decoction | Menopause-related symptoms | 2015 | Wang S. et al., 2015 |
| QiGuiTongFeng tablet | Gout | 2015 | Ke et al., 2015 |
| Salvia miltiorrhiza | Cardiovascular disease | 2015 | He et al., 2015 |
| SiNiSan | Depression | 2015 | Wang H.H. et al., 2015 |
| ShengMai preparations | Cardio-cerebral ischemic diseases | 2015 | Li F. et al., 2015 |
| Dendrobium nobile | Type 2 diabetes | 2015 | Li M.M. et al., 2015 |
| MaHuangFuZiXiXin decoction | Allergic rhinitis | 2015 | Tang et al., 2015 |
| XiaoYaoSan | Depression | 2015 | Gao et al., 2015 |
| Eriobotrya japonica - Fritillaria usuriensis dropping pills | Pulmonary diseases | 2016 | Tao et al., 2016 |
| XiaoYao powder | Anovulatory infertility | 2016 | Liu et al., 2016 |
| Euphorbia kansui and Glycyrrhiza | Hepatocellular carcinoma ascites | 2016 | Zhang et al., 2016 |
| QingFeiXiaoYanWan | Acute lung inflammation | 2016 | Hou et al., 2016 |
| YinChenHaoTang | Severe acute pancreatitis | 2016 | Xiang et al., 2016 |
| YinHuangQingFei capsule | Chronic bronchitis | 2016 | Yu et al., 2017 |
| YangHe decoction | Breast cancer | 2017 | Zeng and Yang, 2017 |
| Fructus Aurantii Immaturus | Anti-coagulation Gastrointestinal motility regulation activities | 2017 | Tan et al., 2017 |
| SheXiangBaoXin pill | Cardiovascular disease | 2017 | Fang et al., 2017 |
| Radix salviae miltiorrhizae | Hepatoprotective effect | 2017 | Hong et al., 2017 |
aThe biological ingredients of herbal prescription were listed in Supplementary Table S1.
Compatibility of TCM Ingredients
Traditional Chinese Medicine preparation (in other words, TCM prescription) is the main form of TCM, and it is produced under the guidance of TCM syndrome differentiation and treatment, which is highly scientific. The traditional “King, Vassal, Assistant, and Delivery servant” combination rule of TCM herbal formulas contains the rich and profound scientific connotation of TCM theory. Thus, the ability to explain the compatibility of TCM is one of the most challenging tasks for TCM research. Li et al. (2010) proposed a method named the Distance-Based Mutual Information Model (DMIM) to identify useful relationships that exist among herbs in numerous herbal formulas. DMIM combines mutual information entropy and “between-herb-distance” to score herb interactions and construct an herb network. It achieves a good balance among the herb’s frequency, independence, and distance in herbal formulas when used to retrieve herb pairs. They used 3,865 collateral-related herbal formulas to construct an herb network and illustrated the traditional herbal pairing and compatibility. LWDHW was used as an example to identify possible compatibility mechanisms, and it was found that LWDHW-treated disease shows high phenotype similarity and that certain “co-modules” are enriched in cancer pathways and neuro-endocrine-immune pathways, which may be the mechanism of action by which the same LWDHW formula treats different diseases. Zhang et al. (2008, 2017) focused on the five flavors of Chinese medicinal properties and constructed Bayesian network models of bitterness, pungent flavor, and sugariness. The five flavors of some TCMs were predicted by these established models, providing crucial support for research on TCM compatibility.
Target Prediction of TCM
Understanding the molecular basis of TCM is crucial for the modernization of TCM, which not only would provide full theoretical support for TCM research but also would increase acceptance of TCM worldwide. In recent years, researchers have done a lot of work to study the molecular mechanisms of TCMs. We proposed the TCM-Mesh system (Zhang et al., 2017), which was designed as an integration of a database and a data-mining system for network pharmacology analysis of TCM preparations. We used TCM-Mesh to identify candidate targets for ginseng and LWDHW based on existing clues (e.g., molecular structures, bioactivity) from curated databases along with a pharmacology approach. The results showed that dammarane-type tetracyclic triterpenoids from ginseng (i.e., ginsenoside Rg1, ginsenoside Re, and ginsenoside Rb1) might be used to treat Alzheimer’s disease, hypertension, and atherosclerosis by targeting tumor necrosis factor (TNF), nitric oxide synthase 3 (NOS3), and AKT serine/threonine kinase 1 (AKT1). Chemical constituents from LWDHW (i.e., loganin, paeonol) can be used to treat diabetes, diabetic nephropathy, and diabetic retinopathy by targeting intercellular adhesion molecule 1 and connective tissue growth factor. Li et al. (Liang et al., 2014b) established a novel pharmacology approach based on a network pharmacology approach to analyze the traditional herbal formulas for LWDHW. The authors found that the compounds of LWDHW may play an important role in esophagitis and colon cancer by regulating the expression of C-C chemokine receptor 2 (CCR2), estrogen receptor 1 (ESR1), peroxisome proliferator-activated receptor gamma (PPARG), and retinoic acid receptor alpha (RARA). They also selected a XinAn medical family’s anti-rheumatoid arthritis (RA) herbal formula “QingLuoYin” (QLY) as an example (Zhang et al., 2013), which consisted of four herbs: KuShen (Sophora flavescens), QingFengTeng (Sinomenium acutum), HuangBai (Phellodendron chinensis), and BiXie (Dioscorea collettii). Some anti-angiogenic and anti-inflammatory active ingredients, such as kurarinone, matrine, sinomenine, berberine, and diosgenin, were identified among the 235 ingredients of QLY. In addition, the synergistic effects of the major ingredients were identified in QLY, such as matrine and sinomenine, which may have been derived from the feedback loop and compensatory mechanisms by targeting TNF- and vascular endothelial growth factor-induced signaling pathways involved in RA. According to the molecular structures of proteins and the corresponding targets of 1,401 drugs approved by the U.S. Food and Drug Administration, Wu et al. (2011) constructed a target prediction model by using the “Random Forest” algorithm. The authors used the compounds of “fuzi” (Aconiti Lateralis Radix Praeparata) to predict the targets and construct a multi-target network. The study found that the 22 compounds of “fuzi” have been used to predict several targets that reflect the characteristics of TCM. In addition, Li et al. (An and Feng, 2015) studied the antioxidant effects of ZhiZiDaHuang Decoction (ZZDHT) based on a network pharmacology approach. The authors found that ZZDHT may exert antioxidant effects to regulate reactive oxygen species, thereby treating alcoholic liver disease by targeting cytochrome P450 2E1 (CYP2E1), xanthine dehydrogenase (XDH), nitric oxide synthase 2 (NOS2), and prostaglandin-endoperoxide synthase 2 (PTGS2).
Network Toxicology
Network toxicology is based on an understanding of the “toxicity-(side effect)-gene-target-drug” interaction network, which utilizes network analysis to speculate and estimate the toxicity and side effects of drugs. Liu et al. (2012) proposed that technologies that feature rapid preparation, high-throughput screening, toxic components exclusion, and biochip along with the drug-target network are of great importance to TCM research on active ingredient screening, toxic components exclusion, and molecular mechanism, which could increase the safety of TCMs. Fan et al. (2011) proposed the concept and framework of network toxicology of TCM. The related tools and technologies were briefly introduced, and the prospects for network toxicology of TCM were forecasted. In their work, they used network pharmacology methods to reconstruct the network of “compound-protein/gene-toxicity” to identify toxic substances and predict the toxic side effects of known compounds, which provides valuable information for understanding the toxic mechanisms. Currently, the related databases for the study of network toxicology includes CTD (Davis et al., 2011), TOXNET (Wexler, 2001), and the National Toxicology Program. Furthermore, additional foresting toxicity software, such as TOPKAT, HazardExpert, and DEREK (Wolfgang and Johnson, 2002), are available for TCM network toxicology studies.
Pros and Cons of Current TCM Databases: Benchmarking
Comparisons Among TCM Databases
As discussed, various databases are used for TCM analysis. However, network analysis of TCM is limited by several aspects of the currently available databases (Figure 3, current statistics on various databases can be obtained from the homepage of each database).
FIGURE 3.
Comparisons among the four TCM databases. The comparison contains the paper information, different input types, general statistics, and limitations of several TCM databases.
For example, HIT is a comprehensive TCM database that supports many different input formats. It is limited, however, in that the total information from HIT for TCM analysis is not sufficient, as it only contains about 586 compounds and 1,301 targets. Thus, the data that can be collected from the HIT are limited. TCMSP is a TCM database that captures the relationships among drugs, targets, and diseases. It allows users to use many input formats; however, TCMSP should be more concise as the database has a large amount of redundancies when searching for herbs, which must be trimmed to be improved. TCMID is another comprehensive database that provides information and bridges the gap between TCM and modern life science. It collects TCM-related information from the TCM@Taiwan database and the literature, including prescription of TCM, and also offers a module for network visualization. Although TCMID is available for multiple inputs, including compound name, STITCH database ID, and CAS number, when searching for ingredients, it is not sufficiently comprehensive, and many compounds are searched without a related network in TCMID. The newly launched TCM-Mesh is a more comprehensive database that not only contains the data on network pharmacology, including the “herb-compound-target-disease” network, but also includes data on TCM side effects and toxicity. As a result, TCM-Mesh offers a more holistic perspective for those conducting TCM research. Problems in TCM-Mesh remain to be resolved, however; for example, the web service is limited and many functions need to be improved, as the CAS number query is not supported. In addition, detailed descriptions about the compounds, genes, and diseases are lacking.
Detailed Information on Herbs and Prescriptions for Assessment
We used prescriptions for LWDHW and ZZDHT to assess TCM-Mesh and randomly selected 10 well-known herbs to test different TCM databases for systematic assessment (Table 3).
Table 3.
Detailed information on the data sets of herbs and prescriptions.
| Herb or prescription | Index of assessment | Source |
|---|---|---|
| LWDHW | Targets | Liang et al., 2014b |
| ZZDHT | An and Feng, 2015 | |
| Ganoderma lucidum | (i) Number of ingredients of herbs found in the database | |
| Ginseng | (ii) Number of ingredient-target found in the database | Famous Chinese herbs |
| Codonopsis pilosula | (iii) Number of target-disease association found in the database | |
| Astragalus membranaceus | ||
| Chinese yam | ||
| Pseudo-ginseng | ||
| Polygonum multiflorum | ||
| Radix Angelicae dahuricae | ||
| Coptis chinensis | ||
| Cordyceps sinensis | ||
Assessment Using LWDHW as an Example
As mentioned earlier, Li et al. (2010) found that the compounds of LWDHW may play an indispensable role in esophagitis and colon cancer by regulating the expression of CCR2, ESR1, PPARG, and RARA. To validate the effects of LWDHW on esophagitis and colon cancer, we used TCM-Mesh. We found 11 candidate targets (including PPARG) for colon cancer and four potential targets for esophagitis (Table 4), all of which were validated by the literature.
Table 4.
Candidate targets of LWDHW for colon cancer and esophagitis validated by TCM-Mesh.
| Disease | Target | Related compounds (herbs) | Reference |
|---|---|---|---|
| Colon cancer | ACP1 (acid phosphatase 1) | Adenine (Poria cocos) | Spina et al., 2008 |
| EGFR (epidermal growth factor receptor) | Capsaicin (Poria cocos), Cholesterol (Chinese yam) | Press et al., 2008 | |
| GSTM1 (glutathione S-transferase mu 1) | Methyleugenol (cornel) | Huang et al., 2006 | |
| IL10 (interleukin 10) | Dopamine (Chinese yam) | Cacev et al., 2008 | |
| IL1B (IL-10b protein precursor) | Capsaicin (Poria cocos) | Lurje et al., 2009 | |
| IL1RN (interleukin 1 receptor antagonist) | Palmitate (Poria cocos, Chinese yam, cornel) | Lurje et al., 2009 | |
| PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha) | Palmitate (Poria cocos, Chinese yam, cornel) | Ogino et al., 2009 | |
| PIK3R1 (phosphoinositide-3-kinase regulatory subunit 1) | Palmitate (Poria cocos, Chinese yam, cornel) | Li et al., 2008 | |
| PPARG (peroxisome proliferator activated receptor gamma) | Capsaicin (Poria cocos) | Slattery et al., 2006 | |
| RXRA (retinoid X receptor alpha) | Palmitate (Poria cocos, Chinese yam, cornel) | Egan et al., 2010 | |
| SLC22A3 (solute carrier family 22 member 3) | Choline (Poria cocos), dopamine (Chinese yam) | Cui et al., 2011 | |
| Esophagitis | CYP2C19 (cytochrome P450 family 2 subfamily C member 19) | Capsaicin (Poria cocos), dopamine (Chinese yam), ursolic acid (cornel) | Kawamura et al., 2007 |
| GSTP1 (glutathione S-transferase pi 1) | Methyleugenol (cornel) | Kala et al., 2007 | |
| IL1B (interleukin 1 beta) | Capsaicin (Poria cocos) | Koivurova et al., 2003 | |
| IL1RN (interleukin 1 receptor antagonist) | Palmitate (Poria cocos, Chinese yam, cornel) | Koivurova et al., 2003 | |
Assessment Using ZZDHT as an Example
As discussed in Section 3.2, “Target prediction of TCM,” ZZDHT decoction has been applied to treat alcoholic liver disease by targeting CYP2E1, XDH, NOS2, and PTGS2. We used TCM-Mesh to validate the effects of ZZDHT, leading to the identification of 12 candidate targets (including CYP2E1) for alcoholic liver disease (Table 5), all of which were supported by the literature.
Table 5.
Candidate targets of ZZDHT for alcoholic liver disease validated by TCM-Mesh.
| Disease | Target | Related chemical composition (herbs) | References |
|---|---|---|---|
| Alcoholic liver disease | ADH1B (alcohol dehydrogenase 1B (class I), beta polypeptide) | Zinc (Cape jasmine) | Lorenzo et al., 2006 |
| ADH1C (alcohol dehydrogenase 1C (class I), gamma polypeptide) | Zinc (Cape jasmine) | Khan et al., 2010 | |
| ALDH2 (aldehyde dehydrogenase 2 family member) | Manganese (Cape jasmine) | Yu et al., 2002 | |
| CYP1A1 (cytochrome P450 family 1 subfamily A member 1) | Apigenin, genistein, quercetin (Cape jasmine), naringin, nobiletin, sinensetin, tangeretin, progesterone (rheum officinale)- | Burim et al., 2004 | |
| CYP2E1 (cytochrome P450 family 2 subfamily E member 1) | Calcium (Cape jasmine), Geniposide (Cape jasmine) | Khan et al., 2009 | |
| GSTM1 (glutathione S-transferase mu 1) | Zinc (Cape jasmine) | Ladero et al., 2005 | |
| HFE (homeostatic iron regulator) | Calcium (Cape jasmine) | Ropero et al., 2001 | |
| IL10 (interleukin 10) | Beryllium (Cape jasmine) | Gleeson et al., 2008 | |
| IL2 (interleukin 2) | Beryllium (Cape jasmine) | Marcos et al., 2008 | |
| IL6 (interleukin 6) | Chromium (Cape jasmine) | Gleeson et al., 2008 | |
| PPARG (peroxisome proliferator activated receptor gamma) | Genistein (Cape jasmine) | Marcos et al., 2009 | |
| XRCC1 (X-ray repair cross-complementing 1) | Chromium (Cape jasmine) | Rossit et al., 2002 | |
Comparison of Search Results Among the TCM Databases
To further compare the databases, we selected 10 well-known herbs to randomly evaluate the TCM databases: G. lucidum, ginseng, Codonopsis pilosula, Astragalus membranaceus, Chinese yam, pseudo-ginseng, Polygonum multiflorum, Radix Angelicae dahuricae, Coptis chinensis, and Cordyceps sinensis. We used each herb to search the different databases and compared the search results (Table 6).
Table 6.
Search results of 10 well-known TCMs from three TCM databases.
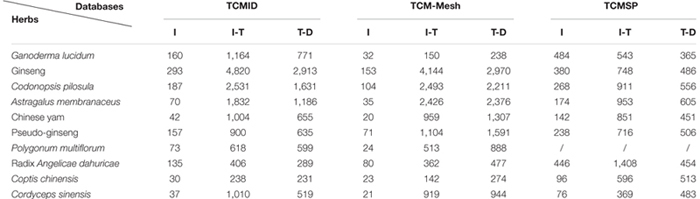 |
I, Ingredients of herb; I-T, Ingredient-target associations; T-D, Target-disease associations.
We compared the results from different databases and found that although the compound data recorded in TCM-Mesh were abundant, the data on the ingredients of the herb were limited. For each herb, the number of ingredients obtained from TCM-Mesh was lower than the number obtained from TCMID and TCMSP. We also found that the number of ingredients collected from TCMSP was much higher than that collected from the other two databases, in accordance with the fact that the database had a large amount of redundancies when searching for herbs. Additionally, we discovered higher completeness of TCM-Mesh in the ingredient-target associations and target-disease associations. For many herbs, the search results produced more hits in ingredient-target associations and target-disease associations in TCM-Mesh, although fewer ingredients were collected. Furthermore, a search for P. multiflorum in TCMSP produced no hits, revealing the limited herb data in TCMSP. The TCM database must keep pace with the development of TCM research to support the modernization of TCM. Therefore, a more comprehensive and complete database with powerful web services is necessary.
Discussion and Conclusion
Research on TCM has a long history in China, illustrating the enormous potential and opportunities for new drug innovation. The inception of TCM network pharmacology offers researchers a new chance to gain systematic insights into TCM, as the research strategy of network pharmacology is in accordance with the holistic understanding of the effects of TCM on disease. This understanding may lead to a new direction for the research of pharmacological mechanisms and safety evaluation of TCM. In addition, related methods and studies must keep pace with the rapid developments in TCM research and must continue to be powerful.
Unification and Integration: A More Powerful Database System
In recent years, several databases have been proposed, many of which share similar functions although they have different data sources. For most databases, the patterns of input differ from database to database. For example, the herb “Dried Tangerine Peel” in TCMSP must be searched by “chenpi” or “Citrus reticulata,” whereas in TCMID, it must be searched by “chen pi” or “Citri reticulatae pericarpium.” In addition, many compounds and proteins have different aliases in different databases (e.g., compounds: chemical name, CID number, STITCH ID, CAS number, PubChem CID, EC number, UNII; proteins: protein name, gene symbol, node ID, target ID, target drugbank ID, uniprot ID). The uniform format of input and output not only would make the current databases more concise, but also would enable researchers to take full advantage of the database resources. Thus, a standard transforming platform is needed. Such a platform should fully understand the data structure and content of various databases. Furthermore, it should support the translation among different formats of herbs, compounds, or proteins to bridge the gap between current databases for TCM research. Currently, the output of previous databases can be directly used as the input for another database. Most databases have associated web services; thus, the transforming platform that could transfer data from one database to another, can be presented to users as web services that are linked to different databases or to a browser plug-in. In addition to the unification of databases, the integration of diverse databases is also of great importance. Currently, more than 10,000 herbs used in more than 100,000 herbal formulas have been recorded in TCM (Qiu, 2007), posing a huge challenge for researchers who want to better understand the efficacy and the safety of the TCM. It is difficult for a single database to resolve all these problems as a result of the incompleteness of data. Each database has its own advantages regarding the specificity of its data and its algorithm. Therefore, the full integration of available resources will facilitate the production of an even more powerful and comprehensive database system, which undoubtedly will more effectively promote the modernization of TCM.
The Content of Compounds: A Crucial Factor
TCM aims to restore whole-body balance in patients by using a herbal formula (Li et al., 2007), and herbal prescriptions are usually composed of two or more medical herbs in a certain proportion. For example, LWDHW consists of six herbs (Zhao et al., 2007; Xie et al., 2008): Rehmannia glutinosa libosch., Cornus officinalis Sieb., Dioscorea oppositifolia L., Paeonia ostii, Alisma orientale Juz, and Poria cocos (Schw.) Wolf, with a dose proportion of 8:4:4:3:3:3 (Wu et al., 2007; Wang et al., 2010). For current studies on TCM formulas, researchers focus on the chemical composition rather than on the proportion of each compound of TCM. Therefore, the influence of the compound content on the effect of the TCM is often ignored. The weight of the content of each compound should be taken into consideration in future studies to better understand the pharmacological effects of the TCM.
TCM-Gut Microbiota Network Pharmacology: A New Frontier
The significant involvement of the gut microbiota in human health and disease suggests that manipulation of commensal microbial composition through combinations of probiotics, antibiotics, and prebiotics could be a novel therapeutic approach. Previous studies have shown that many TCMs can be used as agents to prevent gut dysbiosis (Chang et al., 2015; Zhou et al., 2016). Therefore, a more comprehensive database about TCM and gut microbiota is needed, which should not only include information on the interaction between the TCM and the gut microbiota but also integrate respective advantages of TCM databases and microbiome databases. This integration will be conducive to accelerating the internationalization of TCM and will allow researchers to fully understand the efficacy of TCM from a holistic perspective. Research in the area of TCM network pharmacology is still in its infancy and remains to be developed. However, with data accumulation on TCM and clinical research and interaction of various methods of analysis and experiment, researchers can obtain more substantive and authentic information. This information is conducive to the modernization and internationalization of TCM and may offer critical technological support for drug development, clinical diagnosis, and personalized medicine.
Author Contributions
KN and HB conceived and proposed the idea. KN, HB, and RZ designed the work. KN, RZ, and XZ contributed to the interpretation of data for the work. KN, HB, and RZ drafted the work. KN, HB, XZ, and RZ revised it critically for important intellectual content. All authors read and approved the final manuscript to be published, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was partially supported by National Science Foundation of China grants 81573702, 81774008, 31871334, and 31671374, and the Ministry of Science and Technology (High-Tech) grant (No. 2018YFC0910502).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.00123/full#supplementary-material
References
- An L., Feng F. (2015). Network pharmacology-based antioxidant effect study of zhi-zi-da-huang decoction for alcoholic liver disease. Evid. Based. Complement. Alternat. Med. 2015:492470. 10.1155/2015/492470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. G., Barnes K. C., Bright T. J., Wang S. A. (2004). The genetic association database. Nat. Genet. 36 431–432. 10.1038/ng0504-431 [DOI] [PubMed] [Google Scholar]
- Boh B. (2013). Ganoderma lucidum: a potential for biotechnological production of anti-cancer and immunomodulatory drugs. Recent Pat. Anticancer Drug Discov. 8 255–287. 10.2174/1574891X113089990036 [DOI] [PubMed] [Google Scholar]
- Boh B., Berovic M., Zhang J., Zhi-Bin L. (2007). Ganoderma lucidum and its pharmaceutically active compounds. Biotechnol. Annu. Rev. 13 265–301. 10.1016/s1387-2656(07)13010-6 [DOI] [PubMed] [Google Scholar]
- Burim R. V., Canalle R., Martinelli Ade L., Takahashi C. S. (2004). Polymorphisms in glutathione S-transferases GSTM1, GSTT1 and GSTP1 and cytochromes P450 CYP2E1 and CYP1A1 and susceptibility to cirrhosis or pancreatitis in alcoholics. Mutagenesis 19 291–298. 10.1093/mutage/geh034 [DOI] [PubMed] [Google Scholar]
- Cacev T., Radosević S., Krizanac S., Kapitanović S. (2008). Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis 29 1572–1580. 10.1093/carcin/bgn164 [DOI] [PubMed] [Google Scholar]
- Chan G. H., Law B. Y., Chu J. M., Yue K. K., Jiang Z. H., Lau C. W., et al. (2013). Ginseng extracts restore high-glucose induced vascular dysfunctions by altering triglyceride metabolism and downregulation of atherosclerosis-related genes. Evid. Based. Complement. Alternat. Med. 2013:797310. 10.1155/2013/797310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. (1995). Progress in traditional Chinese medicine. Trends Pharmacol. Sci. 16 182–187. 10.1016/S0165-6147(00)89019-7 [DOI] [PubMed] [Google Scholar]
- Chang C. J., Lin C. S., Lu C. C., Martel J., Ko Y. F., Ojcius D. M., et al. (2015). Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 6:7489. 10.1038/ncomms8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y. (2011). TCM Database@Taiwan: the world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One 6:e15939. 10.1371/journal.pone.0015939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Y., Mamidipalli S., Huan T. (2009). HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC Genomics 10:S16. 10.1186/1471-2164-10-S1-S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu M. X., Yan G. Y. (2012). Drug–target interaction prediction by random walk on the heterogeneous network. Mol. Biosyst. 8 1970–1978. 10.1039/c2mb00002d [DOI] [PubMed] [Google Scholar]
- Corson T. W., Crews C. M. (2007). Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell 130 769–774. 10.1016/j.cell.2007.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui R., Okada Y., Jang S. G., Ku J. L., Park J. G., Kamatani Y., et al. (2011). Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut 60 799–805. 10.1136/gut.2010.215947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. P., Murphy C. G., Johnson R., Lay J. M., Lennonhopkins K., Saracenirichards C., et al. (2011). The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 39 1067–1072. 10.1093/nar/gkq813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Eustachio P. (2009). Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 37 D619–D622. 10.1093/nar/gkn863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohleman B. S. (2006). Exploratory social network analysis with Pajek. Psychometrika 71 605–606. 10.1007/s11336-005-1410-y [DOI] [Google Scholar]
- Egan J. B., Thompson P. A., Ashbeck E. L., Conti D. V., Duggan D., Hibler E., et al. (2010). Genetic polymorphisms in vitamin D receptor VDR/RXRA influence the likelihood of colon adenoma recurrence. Cancer Res. 70 1496–1504. 10.1158/0008-5472.CAN-09-3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Zhao X., Jin Y., Shen X., Liu C. (2011). Network toxicology and its application to traditional Chinese medicine. Zhongguo Zhong Yao Za Zhi 36 2920–2922. [PubMed] [Google Scholar]
- Fang H. Y., Zeng H. W., Lin L. M., Chen X., Shen X. N., Fu P., et al. (2017). A network-based method for mechanistic investigation of Shexiang Baoxin Pill’s treatment of cardiovascular diseases. Sci. Rep. 7:43632. 10.1038/srep43632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Gao L., Gao X. X., Zhou Y. Z., Qin X. M., Tian J. S. (2015). An exploration in the action targets for antidepressant bioactive components of Xiaoyaosan based on network pharmacology. Yao Xue Xue Bao 50 1589–1595. [PubMed] [Google Scholar]
- Gaulton A., Bellis L. J., Bento A. P., Chambers J., Davies M., Hersey A., et al. (2012). ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40 D1100–D1107. 10.1093/nar/gkr777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson D., Bradley M. P., Jones J., Peck R. J., Bond S. K., Teare M. D., et al. (2008). Cytokine gene polymorphisms in heavy drinkers with and without decompensated liver disease: a case-control study. Am. J. Gastroenterol. 103 3039–3046. 10.1111/j.1572-0241.2008.02150.x [DOI] [PubMed] [Google Scholar]
- Hamosh A., Scott A. F., Amberger J., Bocchini C., Valle D., McKusick V. A. (2002). Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 30 52–55. 10.1093/nar/30.1.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. B., Zhang B. X., Wang H. H., Wang Y., Qiao Y. J. (2015). Study on mechanism of Salvia miltiorrhiza treating cardiovascular disease through auxiliary mechanism elucidation system for Chinese medicine. Zhongguo Zhong Yao Za Zhi 40 3713–3717. [PubMed] [Google Scholar]
- Hong M., Li S., Wang N., Tan H. Y., Cheung F., Feng Y. (2017). A biomedical investigation of the hepatoprotective effect of Radix salviae miltiorrhizae and network pharmacology-based prediction of the active compounds and molecular targets. Int. J. Mol. Sci. 18:E620. 10.3390/ijms18030620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins A. L. (2008). Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4 682–690. 10.1038/nchembio.118 [DOI] [PubMed] [Google Scholar]
- Hou Y., Nie Y., Cheng B., Tao J., Ma X., Jiang M., et al. (2016). Qingfei Xiaoyan Wan, a traditional Chinese medicine formula, ameliorates Pseudomonas aeruginosa-induced acute lung inflammation by regulation of PI3K/AKT and Ras/MAPK pathways. Acta Pharm. Sin B 6 212–221. 10.1016/j.apsb.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Sandler R. S., Millikan R. C., Schroeder J. C., North K. E., Hu J. (2006). GSTM1 and GSTT1 polymorphisms, cigarette smoking, and risk of colon cancer: a population-based case-control study in North Carolina (United States). Cancer Causes Control 17 385–394. 10.1007/s10552-005-0424-1 [DOI] [PubMed] [Google Scholar]
- Ideker T., Galitski T., Hood L. (2001). A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2 343–372. 10.1146/annurev.genom.2.1.343 [DOI] [PubMed] [Google Scholar]
- Kala Z., Dolina J., Marek F., Izakovicova Holla L. (2007). Polymorphisms of glutathione S-transferase M1, T1 and P1 in patients with reflux esophagitis and Barrett’s esophagus. J. Hum. Genet. 52 527–534. 10.1007/s10038-007-0148-z [DOI] [PubMed] [Google Scholar]
- Kawamura M., Ohara S., Koike T., Iijima K., Suzuki H., Kayaba S., et al. (2007). Cytochrome P450 2C19 polymorphism influences the preventive effect of lansoprazole on the recurrence of erosive reflux esophagitis. J. Gastroenterol. Hepatol. 22 222–226. 10.1111/j.1440-1746.2006.04419.x [DOI] [PubMed] [Google Scholar]
- Ke Z. P., Zhang X. Z., Ding Y., Cao L., Li N., Ding G., et al. (2015). Study on effective substance basis and molecular mechanism of Qigui Tongfeng tablet using network pharmacology method. Zhongguo Zhong Yao Za Zhi 40 2837–2842. [PubMed] [Google Scholar]
- Kerrien S., Aranda B., Breuza L., Bridge A., Broackescarter F., Chen C., et al. (2012). The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40 D841–D846. 10.1093/nar/gkr1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. J., Husain Q., Choudhuri G., Parmar D. (2010). Association of polymorphism in alcohol dehydrogenase and interaction with other genetic risk factors with alcoholic liver cirrhosis. Drug Alcohol Depend. 109 190–197. 10.1016/j.drugalcdep.2010.01.010 [DOI] [PubMed] [Google Scholar]
- Khan A. J., Ruwali M., Choudhuri G., Mathur N., Husain Q., Parmar D. (2009). Polymorphism in cytochrome P450 2E1 and interaction with other genetic risk factors and susceptibility to alcoholic liver cirrhosis. Mutat. Res. 664 55–63. 10.1016/j.mrfmmm.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Kim J. H. (2012). Cardiovascular diseases and Panax ginseng: a review on molecular mechanisms and medical applications. J. Ginseng. Res. 36 16–26. 10.5142/jgr.2012.36.1.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivurova O. P., Karhukorpi J. M., Joensuu E. T., Koistinen P. O., Valtonen J. M., Karttunen T. J., et al. (2003). IL-1 RN 2/2 genotype and simultaneous carriage of genotypes IL-1 RN 2/2 and IL-1beta-511 T/T associated with oesophagitis in Helicobacter pylori-negative patients. Scand. J. Gastroenterol. 38 1217–1222. 10.1080/00365520310006504 [DOI] [PubMed] [Google Scholar]
- Kola I., Landis J. (2004). Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 3 711–715. 10.1038/nrd1470 [DOI] [PubMed] [Google Scholar]
- Kuhn M., Szklarczyk D., Pletscher-Frankild S., Blicher T. H., von Mering C., Jensen L. J., et al. (2014). STITCH 4: integration of protein-chemical interactions with user data. Nucleic Acids Res. 42 D401–D407. 10.1093/nar/gkt1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladero J. M., Martinez C., Garcia-Martin E., Fernandez-Arquero M., Lopez-Alonso G., de la Concha E. G., et al. (2005). Polymorphisms of the glutathione S-transferases mu-1 (GSTM1) and theta-1 (GSTT1) and the risk of advanced alcoholic liver disease. Scand. J. Gastroenterol. 40 348–353. 10.1080/00365520510012109 [DOI] [PubMed] [Google Scholar]
- Lee S. T., Chu K., Sim J. Y., Heo J. H., Kim M. (2008). Panax ginseng enhances cognitive performance in Alzheimer disease. Alzheimer Dis. Assoc. Disord. 22 222–226. 10.1097/WAD.0b013e31816c92e6 [DOI] [PubMed] [Google Scholar]
- Li F., Lv Y. N., Tan Y. S., Shen K., Zhai K. F., Chen H. L., et al. (2015). An integrated pathway interaction network for the combination of four effective compounds from ShengMai preparations in the treatment of cardio-cerebral ischemic diseases. Acta Pharmacol. Sin. 36 1337–1348. 10.1038/aps.2015.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhao L., Zhang B., Jiang Y., Wang X., Guo Y., et al. (2014a). A network pharmacology approach to determine active compounds and action mechanisms of ge-gen-qin-lian decoction for treatment of type 2 diabetes. Evid. Based. Complement. Alternat. Med. 2014:495840. 10.1155/2014/495840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Plummer S. J., Thompson C. L., Tucker T. C., Casey G. (2008). Association between phosphatidylinositol 3-Kinase regulatory subunit p85α Met326Ile genetic polymorphism and colon cancer Risk. Clin. Cancer Res. 14 633–637. 10.1158/1078-0432.CCR-07-1211 [DOI] [PubMed] [Google Scholar]
- Li M. M., Zhang B. X., He S. B., Zheng R., Zhang Y. L., Wang Y. (2015). Elucidating hypoglycemic mechanism of Dendrobium nobile through auxiliary elucidation system for traditional Chinese medicine mechanism. Zhongguo Zhong Yao Za Zhi 40 3709–3712. [PubMed] [Google Scholar]
- Li S. (2011). Network target: a starting point for traditional Chinese medicine network pharmacology. Zhongguo Zhong Yao Za Zhi 36 2017–2020. [PubMed] [Google Scholar]
- Li S., Fan T. P., Jia W., Lu A., Zhang W. (2014b). Network pharmacology in traditional Chinese medicine. Evid. Based. Complement. Alternat. Med. 2014:138460. 10.1155/2014/138460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang B. (2013). Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 11 110–120. 10.1016/s1875-5364(13)60037-0 [DOI] [PubMed] [Google Scholar]
- Li S., Zhang B., Jiang D., Wei Y., Zhang N. (2010). Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinformatics 11:S6. 10.1186/1471-2105-11-S11-S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang B., Zhang N. (2011). Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst. Biol. 5(Suppl. 1):S10. 10.1186/1752-0509-5-s1-s10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Z. Q., Wu L. J., Zhang X. G., Li Y. D., Wang Y. Y. (2007). Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst. Biol. 1 51–60. 10.1049/iet-syb:20060032 [DOI] [PubMed] [Google Scholar]
- Li X., Wu L., Liu W., Jin Y., Chen Q., Wang L., et al. (2014c). A network pharmacology study of Chinese medicine QiShenYiQi to reveal its underlying multi-compound, multi-target, multi-pathway mode of action. PLoS One 9:e95004. 10.1371/journal.pone.0095004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xu X., Wang J., Yu H., Wang X., Yang H., et al. (2012). A system-level investigation into the mechanisms of Chinese traditional medicine: compound Danshen formula for cardiovascular disease treatment. PLoS One 7:e43918. 10.1371/journal.pone.0043918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Han C., Wang J., Xiao W., Wang Z., Zhang J., et al. (2014d). Investigation into the mechanism of Eucommia ulmoides Oliv. based on a systems pharmacology approach. J. Ethnopharmacol. 151 452–460. 10.1016/j.jep.2013.10.067 [DOI] [PubMed] [Google Scholar]
- Liang X., Li H., Li S. (2014a). A novel network pharmacology approach to analyse traditional herbal formulae: the Liu-Wei-Di-Huang pill as a case study. Mol. Biosyst. 10 1014–1022. 10.1039/C3MB70507B [DOI] [PubMed] [Google Scholar]
- Liang X., Li H., Li S. (2014b). A novel network pharmacology approach to analyse traditional herbal formulae: the Liu-Wei-Di-Huang pill as a case study. Mol. Biosyst. 10 1014–1022. 10.1039/c3mb70507b [DOI] [PubMed] [Google Scholar]
- Liu H., Zeng L., Yang K. (2016). A network pharmacology approach to explore the pharmacological mechanism of xiaoyao powder on anovulatory infertility. Evid. Based. Complement. Alternat. Med. 2016:2960372. 10.1155/2016/2960372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zhang Z., Fang L., Jiang Y., Yin X. (2012). Application of network pharmacology and high through-put technology on active compounds screening from traditional Chinese medicine. Zhongguo Zhong Yao Za Zhi 37 134–137. [PubMed] [Google Scholar]
- Lorenzo A., Auguet T., Vidal F., Broch M., Olona M., Gutiérrez C., et al. (2006). Polymorphisms of alcohol-metabolizing enzymes and the risk for alcoholism and alcoholic liver disease in Caucasian Spanish women. Drug Alcohol Depend. 84 195–200. 10.1016/j.drugalcdep.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Lurje G., Hendifar A. E., Schultheis A. M., Pohl A., Husain H., Yang D., et al. (2009). Polymorphisms in interleukin 1 beta and interleukin 1 receptor antagonist associated with tumor recurrence in stage II colon cancer. Pharmacogenet. Genomics 19 95–102. 10.1097/FPC.0b013e32831a9ad1 [DOI] [PubMed] [Google Scholar]
- Ma H. T., Hsieh J. F., Chen S. T. (2015). Anti-diabetic effects of Ganoderma lucidum. Phytochemistry 114 109–113. 10.1016/j.phytochem.2015.02.017 [DOI] [PubMed] [Google Scholar]
- Marcos M., Pastor I., Gonzalez-Sarmiento R., Laso F. J. (2008). A new genetic variant involved in genetic susceptibility to alcoholic liver cirrhosis: -330T > G polymorphism of the interleukin-2 gene. Eur. J. Gastroenterol. Hepatol. 20 855–859. 10.1097/MEG.0b013e3282fd0db1 [DOI] [PubMed] [Google Scholar]
- Marcos M., Pastor I., Gonzalez-Sarmiento R., Laso F. J. (2009). A functional polymorphism of the NFKB1 gene increases the risk for alcoholic liver cirrhosis in patients with alcohol dependence. Alcohol. Clin. Exp. Res. 33 1857–1862. 10.1111/j.1530-0277.2009.01023.x [DOI] [PubMed] [Google Scholar]
- Ogino S., Nosho K., Kirkner G. J., Shima K., Irahara N., Kure S., et al. (2009). PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J. Clin. Oncol. 27 1477–1484. 10.1200/JCO.2008.18.6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson R. R. (2006). Ganoderma - a therapeutic fungal biofactory. Phytochemistry 67 1985–2001. 10.1016/j.phytochem.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Peri S., Navarro J. D., Amanchy R., Kristiansen T. Z., Jonnalagadda C. K., Surendranath V., et al. (2003). Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 13 2363–2371. 10.1101/gr.1680803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press O. A., Zhang W., Gordon M. A., Yang D., Lurje G., Iqbal S., et al. (2008). Gender-related survival differences associated with EGFR polymorphisms in metastatic colon cancer. Cancer Res. 68 3037–3042. 10.1158/0008-5472.CAN-07-2718 [DOI] [PubMed] [Google Scholar]
- Qiu J. (2007). Traditional medicine: a culture in the balance. Nature 448 126–128. 10.1038/448126a [DOI] [PubMed] [Google Scholar]
- Ropero Gradilla P., Villegas Martínez A., Fernández Arquero M., García-Agúndez J. A., González Fernández F. A., Benítez Rodríguez J., et al. (2001). C282Y and H63D mutations of HFE gene in patients with advanced alcoholic liver disease. Rev. Esp. Enferm. Dig. 93 156–163. [PubMed] [Google Scholar]
- Rossit A. R., Cabral I. R., Hackel C., da Silva R., Froes N. D., Abdel-Rahman S. Z. (2002). Polymorphisms in the DNA repair gene XRCC1 and susceptibility to alcoholic liver cirrhosis in older Southeastern Brazilians. Cancer Lett. 180 173–182. 10.1016/S0304-3835(02)00029-0 [DOI] [PubMed] [Google Scholar]
- Ru J., Li P., Wang J., Zhou W., Li B., Huang C., et al. (2014). TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6:13. 10.1186/1758-2946-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S. H., Cai Y. P., Cai X. J., Zheng X. Y., Cao D. S., Ye F. Q., et al. (2014). A network pharmacology approach to understanding the mechanisms of action of traditional medicine: bushenhuoxue formula for treatment of chronic kidney disease. PLoS One 9:e89123. 10.1371/journal.pone.0089123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M. L., Curtin K., Wolff R., Ma K. N., Sweeney C., Murtaugh M., et al. (2006). PPARγ and colon and rectal cancer: associations with specific tumor mutations, aspirin, ibuprofen and insulin-related genes (United States). Cancer Causes Control 17 239–249. 10.1007/s10552-005-0411-6 [DOI] [PubMed] [Google Scholar]
- Spina C., Bottini P. S., Gloria-Bottini F. (2008). ACP1 genetic polymorphism and colon cancer. Cancer Genet. Cytogenet. 186 61–62. 10.1016/j.cancergencyto.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43 D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Li Y., Wang Y., Zhang Z., Wang T., Zhou Q., et al. (2017). Anti-coagulative and gastrointestinal motility regulative activities of Fructus Aurantii immaturus and its effective fractions. Biomed. Pharmacother. 90 244–252. 10.1016/j.biopha.2017.03.060 [DOI] [PubMed] [Google Scholar]
- Tang F., Tang Q., Tian Y., Fan Q., Huang Y., Tan X. (2015). Network pharmacology-based prediction of the active ingredients and potential targets of Mahuang Fuzi Xixin decoction for application to allergic rhinitis. J. Ethnopharmacol. 176 402–412. 10.1016/j.jep.2015.10.040 [DOI] [PubMed] [Google Scholar]
- Tao J., Hou Y., Ma X., Liu D., Tong Y., Zhou H., et al. (2016). An integrated global chemomics and system biology approach to analyze the mechanisms of the traditional Chinese medicinal preparation Eriobotrya japonica - Fritillaria usuriensis dropping pills for pulmonary diseases. BMC Complement. Altern. Med. 16:4. 10.1186/s12906-015-0983-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W., Xu X., Wang X., Li B., Wang Y., Li Y., et al. (2013). Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J. Ethnopharmacol. 145 1–10. 10.1016/j.jep.2012.09.051 [DOI] [PubMed] [Google Scholar]
- Vanunu O., Magger O., Ruppin E., Shlomi T., Sharan R. (2010). Associating genes and protein complexes with disease via network propagation. PLoS Comput. Biol. 6:e1000641. 10.1371/journal.pcbi.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. H., Zhang B. X., Ye X. T., He S. B., Zhang Y. L., Wang Y. (2015). Study on mechanism for anti-depression efficacy of Sini San through auxiliary mechanism elucidation system for Chinese medicine. Zhongguo Zhong Yao Za Zhi 40 3723–3728. [PubMed] [Google Scholar]
- Wang P., Sun H., Lv H., Sun W., Yuan Y., Han Y., et al. (2010). Thyroxine and reserpine-induced changes in metabolic profiles of rat urine and the therapeutic effect of Liu Wei Di Huang Wan detected by UPLC-HDMS. J. Pharm. Biomed. Anal. 53 631–645. 10.1016/j.jpba.2010.04.032 [DOI] [PubMed] [Google Scholar]
- Wang S., Tong Y., Ng T. B., Lao L., Lam J. K., Zhang K. Y., et al. (2015). Network pharmacological identification of active compounds and potential actions of Erxian decoction in alleviating menopause-related symptoms. Chin. Med. 10:19. 10.1186/s13020-015-0051-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiao J., Suzek T. O., Jian Z., Wang J., Bryant S. H. (2009). PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 37 W623–W633. 10.1093/nar/gkp456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler P. (2001). TOXNET: an evolving web resource for toxicology and environmental health information. Toxicology 157 3–10. 10.1016/S0300-483X(00)00337-1 [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Knox C., Guo A. C., Shrivastava S., Hassanali M., Stothard P., et al. (2006). DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34 D668–D672. 10.1093/nar/gkj067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang G. H., Johnson D. E. (2002). Web resources for drug toxicity. Toxicology 173 67–74. 10.1016/S0300-483X(02)00022-7 [DOI] [PubMed] [Google Scholar]
- Wu C. R., Lin L. W., Wang W. H., Hsieh M. T. (2007). The ameliorating effects of LiuWei Dihuang Wang on cycloheximide-induced impairment of passive avoidance performance in rats. J. Ethnopharmacol. 113 79–84. 10.1016/j.jep.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Wu L., Gao X., Wang L., Liu Q., Fan X., Wang Y., et al. (2011). Prediction of multi-target of Aconiti Lateralis Radix Praeparata and its network pharmacology. Zhongguo Zhong Yao Za Zhi 36 2907–2910. [PubMed] [Google Scholar]
- Wu L., Wang Y., Li Z., Zhang B., Cheng Y., Fan X. (2014). Identifying roles of “Jun-Chen-Zuo-Shi” component herbs of QiShenYiQi formula in treating acute myocardial ischemia by network pharmacology. Chin. Med. 9:24. 10.1186/1749-8546-9-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H., Wang G., Qu J., Xia S., Tao X., Qi B., et al. (2016). Yin-Chen-Hao tang attenuates severe acute pancreatitis in rat: an experimental verification of in silico network target prediction. Front. Pharmacol. 7:378. 10.3389/fphar.2016.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z., Sun H., Cai X., Chen D., Zheng X. (2015). The study on the material basis and the mechanism for anti-renal interstitial fibrosis efficacy of rhubarb through integration of metabonomics and network pharmacology. Mol. Biosyst. 11 1067–1078. 10.1039/c4mb00573b [DOI] [PubMed] [Google Scholar]
- Xie B., Gong T., Tang M., Mi D., Zhang X., Liu J., et al. (2008). An approach based on HPLC-fingerprint and chemometrics to quality consistency evaluation of Liuwei Dihuang Pills produced by different manufacturers. J. Pharm. Biomed. Anal. 48 1261–1266. 10.1016/j.jpba.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Xu H., Zhang Y., Lei Y., Gao X., Zhai H., Lin N., et al. (2014). A systems biology-based approach to uncovering the molecular mechanisms underlying the effects of dragon’s blood tablet in colitis, involving the integration of chemical analysis, ADME prediction, and network pharmacology. PLoS One 9:e101432. 10.1371/journal.pone.0101432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R., Fang Z., Zhang M., Yi Z., Wen C., Shi T. (2013). TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 41 D1089–D1095. 10.1093/nar/gks1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Ye L., Kang H., Zhang D., Tao L., Tang K., et al. (2011). HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 39 D1055–D1059. 10.1093/nar/gkq1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y. D., Chang I. M. (2004). An overview of traditional Chinese herbal formulae and a proposal of a new code system for expressing the formula title. Evid. Based. Complement. Alternat. Med. 1 125–132. 10.1093/ecam/neh019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Li Y., Chen W., Yue M. (2002). Genotype of ethanol metabolizing enzyme genes by oligonucleotide microarray in alcoholic liver disease in Chinese people. Chin. Med. J. 115 1085–1087. [PubMed] [Google Scholar]
- Yu G., Zhang Y., Ren W., Ling D., Li J., Geng Y., et al. (2017). Network pharmacology-based identification of key pharmacological pathways of Yin–Huang–Qing–Fei capsule acting on chronic bronchitis. Int. J. Chron. Obstruct. Pulmon. Dis. 12 85–94. 10.2147/COPD.S121079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H. D., Kim J. T., Kim S. H., Chung S. H. (2012). Ginseng and diabetes: the evidences from in vitro, animal and human studies. J. Ginseng. Res. 36 27–39. 10.5142/jgr.2012.36.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. (2002). MINT: a Molecular INTeraction database. FEBS Lett. 513 135–140. 10.1016/S0014-5793(01)03293-8 [DOI] [PubMed] [Google Scholar]
- Zeng L., Yang K. (2017). Exploring the pharmacological mechanism of Yanghe Decoction on HER2-positive breast cancer by a network pharmacology approach. J. Ethnopharmacol. 199 68–85. 10.1016/j.jep.2017.01.045 [DOI] [PubMed] [Google Scholar]
- Zhang B., Wang X., Li S. (2013). An integrative platform of TCM network pharmacology and its application on a herbal formula, Qing-Luo-Yin. Evid. Based. Complement. Alternat. Med. 2013:456747. 10.1155/2013/456747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li J., Wang Y. (2008). Bayesian belief network and its application in predicting the five flavors of Chinese medicinal components. Mod. Tradit. Chin. Med. Mater. Mater. World Sci. Technol. 10 114–117. 10.11842/wst.2008.5 [DOI] [Google Scholar]
- Zhang R. Z., Yu S. J., Bai H., Ning K. (2017). TCM-Mesh: the database and analytical system for network pharmacology analysis for TCM preparations. Sci. Rep. 7:2821. 10.1038/s41598-017-03039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lin Y., Zhao H., Guo Q., Yan C., Lin N. (2016). Revealing the effects of the herbal pair of euphorbia kansui and glycyrrhiza on hepatocellular carcinoma ascites with integrating network target analysis and experimental validation. Int. J. Biol. Sci. 12 594–606. 10.7150/ijbs.14151 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao F., Guochun L., Yang Y., Shi L., Xu L., Yin L. (2015). A network pharmacology approach to determine active ingredients and rationality of herb combinations of Modified-Simiaowan for treatment of gout. J. Ethnopharmacol. 168 1–16. 10.1016/j.jep.2015.03.035 [DOI] [PubMed] [Google Scholar]
- Zhao X., Wang Y., Sun Y. (2007). Quantitative and qualitative determination of Liuwei Dihuang tablets by HPLC-UV-MS-MS. J. Chromatogr. Sci. 45 549–552. 10.1093/chromsci/45.8.549 [DOI] [PubMed] [Google Scholar]
- Zheng C. S., Xu X. J., Ye H. Z., Wu G. W., Li X. H., Xu H. F., et al. (2013). Network pharmacology-based prediction of the multi-target capabilities of the compounds in Taohong Siwu decoction, and their application in osteoarthritis. Exp. Ther. Med. 6 125–132. 10.3892/etm.2013.1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. S., Xu J., Zhu H., Wu J., Xu J. D., Yan R., et al. (2016). Gut microbiota-involved mechanisms in enhancing systemic exposure of ginsenosides by coexisting polysaccharides in ginseng decoction. Sci. Rep. 6:22474. 10.1038/srep22474 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.