Abstract
During blood feeding haematophagous arthropods inject into their hosts a cocktail of salivary proteins whose main role is to counteract host haemostasis, inflammation and immunity. However, animal body fluids are known to also carry miRNAs. To get insights into saliva and salivary gland miRNA repertoires of the African malaria vector Anopheles coluzzii we used small RNA-Seq and identified 214 miRNAs, including tissue-enriched, sex-biased and putative novel anopheline miRNAs. Noteworthy, miRNAs were asymmetrically distributed between saliva and salivary glands, suggesting that selected miRNAs may be preferentially directed toward mosquito saliva. The evolutionary conservation of a subset of saliva miRNAs in Anopheles and Aedes mosquitoes, and in the tick Ixodes ricinus, supports the idea of a non-random occurrence pointing to their possible physiological role in blood feeding by arthropods. Strikingly, eleven of the most abundant An. coluzzi saliva miRNAs mimicked human miRNAs. Prediction analysis and search for experimentally validated targets indicated that miRNAs from An. coluzzii saliva may act on host mRNAs involved in immune and inflammatory responses. Overall, this study raises the intriguing hypothesis that miRNAs injected into vertebrates with vector saliva may contribute to host manipulation with possible implication for vector-host interaction and pathogen transmission.
Introduction
Mosquitoes are vectors of parasitic and arboviral diseases of great importance to human health. Malaria, which is transmitted by Anopheles mosquitoes, affected >200 million people with ~450 thousands deaths in 20161 and dengue, transmitted by Aedes mosquitoes, may be responsible for >100 million symptomatic infections per year2. Most vector-borne pathogens, as malaria parasites and dengue viruses, are transmitted to vertebrates through hematophagous arthropod saliva during the blood meal. Saliva of blood feeding arthropods (BFA) is a complex cocktail including hundreds of salivary proteins and its role in hematophagy is pretty well known3–6. As far as mosquitoes are concerned, transcriptomic4,7–15, genomic16–18 and proteomic studies19–22 allowed to clarify that mosquito saliva carries ~100–150 salivary proteins whose main role is to facilitate blood feeding by counterbalancing host responses to tissue injury, namely haemostasis, inflammation and immunity23. Moreover, in virtue of its immunomodulatory properties, mosquito saliva generates at the biting site a local environment that may affect pathogen transmission24–30.
MicroRNAs (miRNAs) are small non-coding RNAs of ~22 nt in length with a relevant role in post-transcriptional gene regulation. Typically, primary miRNA transcripts (pri-miRNAs) are first processed to hairpins of ~80 nt in length (pre-miRNAs) and then into the mature miRNA duplex31,32. One strand of the duplex, named the guide strand, is preferentially loaded into the miRNA-induced silencing complex (miRISC) and drives it to the target mRNA promoting its degradation or translational inhibition31,33,34. Target recognition mainly involves imperfect base pairing between the mRNA 3′UTR and the miRNA, with the seed region of the miRNA (nucleotides 2 to 8) playing a crucial role in target selection31,33,35. miRNAs are essentially found in all animal cell types where they exhibit tissue-specific expression patterns and, as part of complex networks, contribute to the regulation of practically every aspect of cell life, from cell growth and differentiation to apoptosis, development and immunity31,36. miRNAs are not only present within cells but also extracellularly. They have been found in all human body fluids37–39, as well as in the saliva of disease vectors as Aedes mosquitoes and Ixodes ticks40,41. Extracellular miRNAs in body fluids may be either in complex with proteins, as Argonaute (Ago) family members or High Density Lipoproteins, or may be carried within exosomal microvesicles42–44. The role of extracellular miRNAs is still debated44,45 but there is clear evidence that miRNAs enclosed within exosomes may play roles in cell-cell communication46–48. In this scenario miRNAs carried by exosomes may enter the target cells by direct fusion to plasma membranes or receptor-mediated endocytosis, whereas the vesicle-free miRNAs bound to Ago proteins may find their way through gap junction channels or some other yet unknown mechanism45,49.
miRNAs typically target endogenous genes, nevertheless, it is known that viral-encoded miRNAs target host mRNAs within infected cells50. In addition, exosomal miRNAs from parasitic nematodes may target host genes associated with immunity and inflammation51,52. Overall, these observations raise the fascinating hypothesis that miRNAs in mosquito saliva, perhaps encapsulated within exosomes, are injected into vertebrate hosts during blood feeding and may represent additional players in vector-pathogen-host interactions, contributing to manipulation of host inflammatory and immune responses.
Anopheline miRNAs have been studied in different experimental conditions in the malaria vectors Anopheles gambiae53–56, An. coluzzii57,58, An. stephensi59–62, An. funestus63, An. anthropophagus64 and An. sinensis65,66. However, so far no information has been obtained on the presence of miRNAs in the saliva of Anopheles species. To get insights into anopheline saliva miRNA composition and verify whether saliva-enriched miRNAs may have the capacity to manipulate host responses, we carried out a small RNA-Seq study on adult female salivary glands and saliva of the African malaria vector An. coluzzii using adult males and females as reference.
Results
Deep sequencing of small RNAs from Anopheles coluzzii
Anopheles coluzzii saliva (S), salivary glands (G), adult males (M) and females (F) were collected as described in the method section. Samples were in triplicate; for the saliva sample a pilot study including a duplicate was previously performed and, therefore, a total of five saliva replicates were analysed in this study (Supplementary Fig. S1A). Overall, 14 small RNA libraries were constructed and used for Illumina high-throughput sequencing, yielding a total of ~180 million reads (S = 38.80, G = 42.85, F = 48.65, M = 49.57). After quality filtering, adapter trimming and size selection (≥14 nt) approximately 126 million reads were retained for mapping to the An. gambiae genome (PEST strain, AgamP4 assembly), a choice motivated by its more complete assembly and annotation and the very close evolutionary relationships between An. gambiae and An. coluzzii (see methods). Approximately 84 million reads aligned to AgamP4 (S = 7.75; G = 21.06; F = 28.77; M = 26.46) and were mapped to rRNAs and to a list of 273 miRNA precursors (Supplementary File S1) and other ncRNAs (Table 1). Counts mapping to hairpins and other ncRNAs were used to investigate the linear relationships between replicates of the different samples. The correlation was very high for G, F and M (Pearson’s coefficients in the range 0.88–0.99) and moderate to high for the five S replicates (0.41–0.92, Supplementary Fig. S1B). Variation between libraries was also evaluated calculating distances based on fold change and biological coefficient of variation. Samples G, F and M formed three distinct clusters with little variation among replicates, whereas the five saliva libraries appeared more heterogeneous, but still clustering together and independently from the other samples (Supplementary Fig. S1C).
Table 1.
Summary of deep sequencing of small RNAs from An. coluzzii.
| sample | raw reads | filtered reads | AgamP4 | rRNAs | ncRNAs |
|---|---|---|---|---|---|
| S | 38.807 | 27.875 | 7.758 | 5.295 | 0.193 |
| G | 42.856 | 27.458 | 21.069 | 13.262 | 1.880 |
| F | 48.652 | 36.762 | 28.771 | 9.023 | 6.756 |
| M | 49.570 | 33.875 | 26.466 | 11.854 | 8.729 |
| total | 179.885 | 125.970 | 84.065 | 39.436 | 17.559 |
Numbers indicate million reads. S, saliva; G, female salivary glands; F, adult females; M, adult males. Filtered reads, reads remaining after adapter removal and size selection (≥14). Reads mapping to the An. gambiae genome (AgamP4), ribosomal RNAs (rRNAs) and to a list of 273 miRNA precursors plus other non-coding RNAs (ncRNAs) are indicated.
Size distribution and mapping
Reads mapping to the An. gambiae genome, excluding those representing ribosomal RNAs, were analysed for their size distribution. The G, F and M samples showed a peak at 22 nt, which is typical of mature miRNAs and was prominent in the M sample, less pronounced in F and smaller in the G sample (Fig. 1, left panels). The F sample also showed a secondary peak in the range 25–29 nt likely representing piRNAs; a similar pattern was reported in a previous study on An. gambiae ovaries54. No peak of ~22 nt was visible in the S sample, with most of the reads falling in the 14–16 nt range, which is indicative of degradation that most likely occurred because of the elaborated saliva collection procedure. Nevertheless, ~0.23 million reads from the S sample mapped in the 20–24 nt range.
Figure 1.

Features of small RNA sequenced from the four Anopheles coluzzii samples. The bar plots on the left show the frequency and size distribution of reads 14–33 nt in length mapping to the An. gambiae genome (AgamP4) and subtracted of those mapping to rRNAs. The pie charts on the right summarize the results of read alignment to rRNAs, miRNA precursors, tRNAs, other non-coding RNAs (including snoRNAs, snRNAs, Metazoa SRP, arthropod 7SK and RNase P) and to An. gambiae transcripts and repeats. Unannotated, reads mapping to AgamP4 but no to the other classes. Total, number of reads mapping to AgamP4. S, saliva; G, salivary glands; F, adult females; M, adult males.
The result of reads alignment to rRNAs, miRNA precursors, tRNAs, other non-coding RNAs and to An. gambiae transcripts and repeats is reported in Fig. 1 (right panels). The relative abundance of rRNAs was higher in the G and S samples, likely due to partial degradation of large rRNAs that occurred during gland dissection and saliva collection. Reads mapping to miRNA precursors (hairpins) was high for the M, F and G samples (1.2 to 6.7 million reads) but rather small for the S sample (40,305 reads) (Fig. 1, Table 2). The proportion of reads mapping to other RNA types or to unannotated regions of the genome were comparable in the different samples, whereas those mapping to repeats were approximately 3 to 4 times higher in F (21.8%). The reason of this difference is unclear, however a similarly high frequency of reads mapping to repeats (>30%) was previously reported in adult An. gambiae females53.
Table 2.
Reads mapping to precursor and mature miRNAs.
| sample | AgamP4 | hairpins (%) | mature (%) |
|---|---|---|---|
| S | 7,758,653 | 40,305 (0.52) | 28,475 (0.37) |
| G | 21,069,482 | 1,244,151 (5.90) | 1,188,871 (5.64) |
| F | 28,771,027 | 5,628,344 (19.56) | 5,514,013 (19.17) |
| M | 26,466,637 | 6,766,708 (25.57) | 6,624,225 (25.03) |
Reads mapping to miRNA precursors were re-mapped to a collection of 438 mature (5p + 3p) miRNAs (Supplementary File S1). Altogether 28,475 reads from the saliva sample mapped to mature miRNAs, whereas 1.19, 5.51 and 6.62 million reads from the G, F and M samples, respectively, represented mature miRNAs (Table 2).
miRNAs found in saliva, salivary glands, adult males and females
Overall, setting as a threshold for inclusion (i) the presence of counts in at least two replicates (three for the S sample) and (ii) a mean count per million (CPM) ≥ 3 in at least one of the four samples, we found in silico evidence for the expression of 214 mature miRNAs. Among these 178 were An. coluzzii orthologues of previously known An. gambiae miRNAs (miRBase53–55), and 87 of them matched 50 of the 57 An. coluzzii miRNA precursors previously identified by bioinformatics predictions67 and available in VectorBase68. The remaining 36 represent novel An. coluzzii and An. gambiae miRNAs. A list of these 214 miRNAs is reported in Supplementary File S2, where miRNAs were named using the prefix aco- followed by an identification code. For miRNAs previously described in An. gambiae we used the same identification; for those predicted by miRDeep* we used miR-N followed by a number; for those predicted by MapMi we kept as identification the name of the query miRNA. We then determined the subsets of miRNAs expressed in each sample and found 77 miRNAs in the saliva sample, with 66 supported by more than 500 mean CPM. As expected, a larger number of miRNAs was found in the other samples, with G, F and M including 147, 196 and 171 miRNAs, respectively (Supplementary File S2). Seventy-three miRNAs were common among the four samples and 51, instead, were unique to G, M or F samples (Fig. 2, Supplementary File S2).
Figure 2.
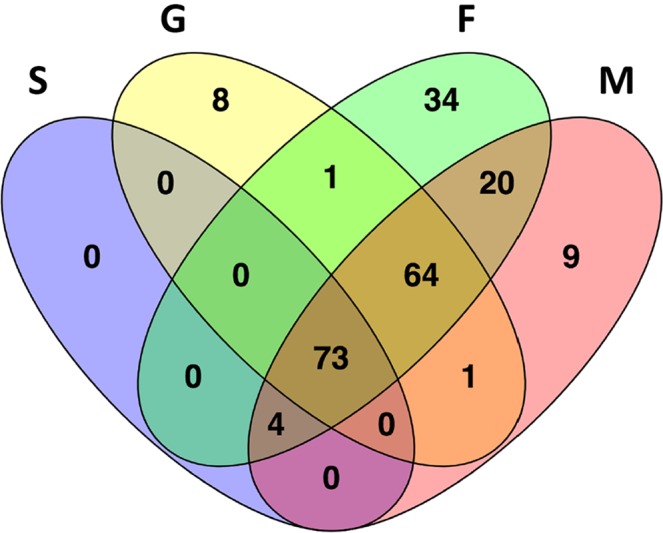
Distribution of the 214 mature miRNAs in the four samples analysed. The Venn diagram depicts the degree of overlap of the 214 miRNAs in the four samples: S (purple), G (yellow), F (green) and M (pink).
Novel miRNAs
In the attempt to identify novel miRNAs, two complementary prediction tools were employed to search the AgamP4 assembly: miRDeep*69 and MapMi70, which exploit small RNA-Seq data and known miRNAs from other species, respectively. This way 99 putative mature miRNAs were predicted (Supplementary File S1). Overall, after mapping and applying the threshold for inclusion, 36 miRNAs appeared as bona fide An. gambiae and An. coluzzii novel miRNAs (Supplementary File S3). A large majority (30/36) was represented in at least six of the fourteen libraries and most of them (31/36) were of low abundance (Fig. 3). This is in agreement with previous studies on An. gambiae where the majority of novel miRNAs were found expressed at very low levels58. The remaining five miRNAs, instead, were well supported by counts (2,761-39,588) and CPM (103-58,697): these include two miRNAs matching the Aedes aegypti miR-71 stem-loop (MI0013440) and 3 completely novel miRNAs named aco-miR-N96, aco-miR-N56, aco-miR-N951. The secondary structure of the hairpins encoding these three abundant novel miRNAs from An. coluzzii are shown in Supplementary Figure S2.
Figure 3.
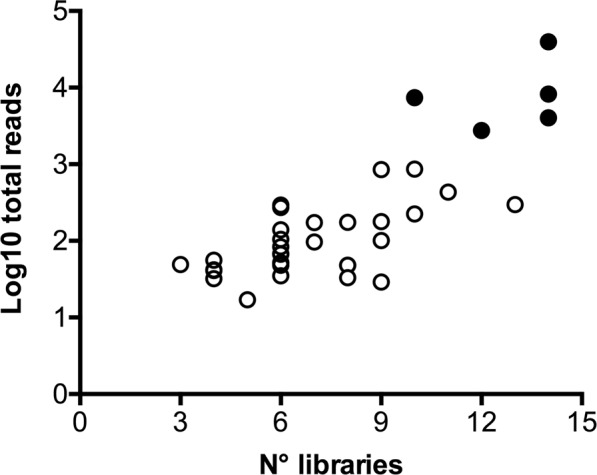
Abundance of the thirty-six novel miRNAs in the fourteen libraries. For each of the putative 36 novel miRNAs from An. coluzzii the number of total reads and the total number of libraries are reported. The five most abundant miRNAs, represented by >1000 reads, are shown with filled circles.
Homologues of these 36 putative novel miRNAs were searched in the genomes of several anopheline and a few culicine mosquitoes as well as of other BFA and of the non-blood feeding Diptera Drosophila melanogaster and Musca domestica. The four miRNAs predicted by MapMi as well as aco-miR-N636 appeared widely conserved in mosquito species and also in a few other blood feeders (Supplementary Fig. S3). With the exception of aco-miR-N645, restricted to An. gambiae and An. coluzzii, all other miRNAs were found among members of the An. gambiae species complex but only occasionally in other species. Only three miRNAs showed a somehow wider distribution among mosquitoes, with aco-miR-N1044 exclusively found in anophelines and aco-miR-N149 and aco-miR-N1306 also found in culicine mosquitoes.
A subset of nine miRNAs of low (CPM < 20), medium (20 ≤ CPM < 100) and high (CPM ≥ 100) abundance in the F sample, were selected for validation by the Stem-loop Reverse-Transcription Polymerase Chain Reaction (slRT-PCR) technique71. Using small RNA from adult An. coluzzii females as template, clean amplifications of all selected miRNAs but one was obtained by slRT-PCR. The only exception was the least abundant aco-miR-N135 (7 CPM) that yielded an unspecific amplification product, most likely because of a not optimal primer design. An inverse correlation was found when mean CPM values were compared to Ct values as determined by the real time PCR amplification (Spearman r -0.833, p 0.015; Supplementary Fig. S4). Overall, these observations suggest that the large majority of the putative novel miRNAs identified here are indeed real and expressed in An. coluzzi.
Differential expression analysis
At first, differential expression (DE) analysis of mature miRNAs was performed on all four samples (S, G, F and M). This yielded a subset of miRNAs upregulated in the saliva sample; however, while ~40% of these miRNAs were well supported by counts, the remaining appeared differentially expressed despite the low or very low number of mapping reads. We assumed this was most likely due to the large difference in the number of reads mapping to miRNAs in the four samples. Therefore, to avoid any possible bias and make the downstream analysis more robust and reliable, we decided to (i) use the reads from the S sample just for assembling a catalogue of salivary miRNAs from An. coluzzii and (ii) repeat the DE analysis using only the G, F and M samples, which carried a comparable number of mapping reads. The mature miRNAs expression heatmap and the cluster analysis confirmed the overall quality of replicates, highlighting groups of miRNAs with specific profile signatures (Supplementary Fig. S5). Sample-specific miRNA enrichment was evaluated by pairwise comparisons between the three samples: fold change (FC) and false discovery rates (FDR) were calculated to provide statistical validation (Supplementary File S4). Using as threshold parameters FC > 2 and FDR < 0.05 we could identify subsets of miRNAs specifically enriched in adult female salivary glands, in adult females and in adult males. More in detail, the comparison G vs F highlighted 38 miRNAs enriched in female salivary glands and 103 significantly more abundant in adult females (Fig. 4, top panel), whereas the comparison G vs M pointed out the differential enrichment of 125 miRNAs, 41 upregulated in female salivary glands and 84 in adult males (Fig. 4, central panel). Finally, the comparison F vs M identified 68 sex-biased miRNAs that may play roles in sexual dimorphism: among these, 50 were more abundant in females and 18 in males (Fig. 4, bottom panel). The numbers of miRNAs found upregulated in the three pairwise comparisons according to different false discovery rates (FDR < 0.05, FDR < 0.01 and FDR < 0.001) are reported in Supplementary Table S1. Matching the two subsets of miRNAs upregulated in female salivary glands in the pairwise comparisons to F (38) and M (41) revealed that 30 miRNAs were common. These may be considered with good confidence as miRNAs specifically enriched in female salivary glands and, therefore, likely to play sex- and/or tissue-specific roles in salivary gland physiology and blood feeding.
Figure 4.
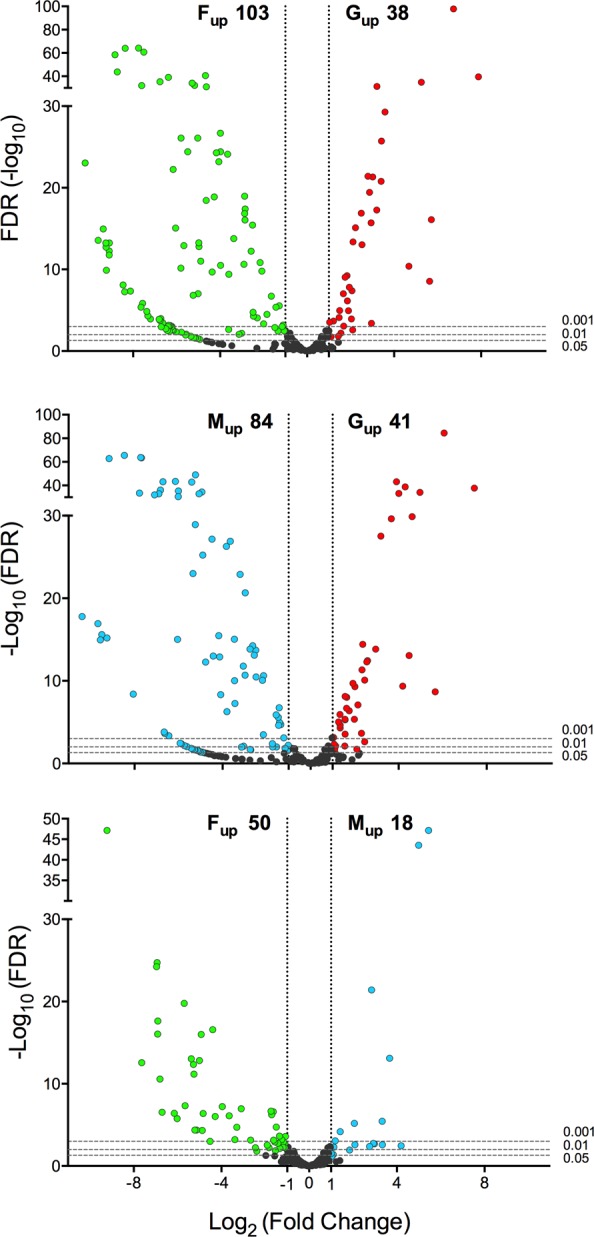
Differential abundance of miRNAs in Anopheles coluzzii salivary glands, adult females and adult males. The volcano plots show the differential abundance of miRNAs in the pairwise comparisons G-F (top), G-M (middle) and M-F (bottom). The log2 of fold change (FC) versus the negative log10 of false discovery rate (FDR) as calculated by the Fisher’s exact test are reported. Vertical dotted lines mark FC = 2, horizontal dashed lines mark FDR threshold equal to 0.05, 0.01 and 0.001. miRNAs with a FC > 2 and FDR < 0.05 in the different pairwise comparisons were considered as differentially expressed and are shown in red (upregulated in female salivary glands), green (upregulated in adult females) and blue (upregulated in adult males).
Asymmetric distribution of miRNAs in saliva and salivary glands of Anopheles coluzzii
As mentioned above, the S sample was not included in DE analysis to avoid introducing any bias due to the low number of reads mapping to a group of miRNAs in the saliva sample. Nevertheless, we wondered whether miRNAs were or not symmetrically distributed between salivary glands and saliva. Therefore, we selected the 30 most abundant miRNAs from the S (CPM range 3,098-161,759) and G (CPM range 3,828-175,456) samples and, comparing the two lists, we found an overlap restricted to 21 miRNAs. When the S/G CPM ratio was calculated for the 39 miRNAs from the combined lists we found an asymmetric miRNA distribution between salivary glands and saliva. Indeed, 9 miRNAs had an S/G ratio > 4.00 and, therefore, appeared to be more abundant in saliva, whereas other 11 had an S/G ratio < 0.25 and were more represented in the salivary glands. The remaining 19, with S/G ratio ≤ 4.00 and ≥0.25, can be essentially considered as equally distributed (Fig. 5, Supplementary File S5). To verify if this distribution had a statistical support we examined the DE data obtained by edgeR for the pairwise comparison S-G during our initial analysis involving all four samples. The 9 miRNAs found over-represented in the S sample were supported by reliable mean CPM (range 3344–161759) and their differential distribution had very good statistical support (FDR values range 0.0057-3.61E-16); a similar situation was found for the 11 miRNAs over-represented in salivary glands (Supplementary File S5). These observations indicate that An. coluzzii saliva does not simply mirror salivary gland miRNA content and that a selected subset of miRNAs are preferentially conveyed to saliva.
Figure 5.
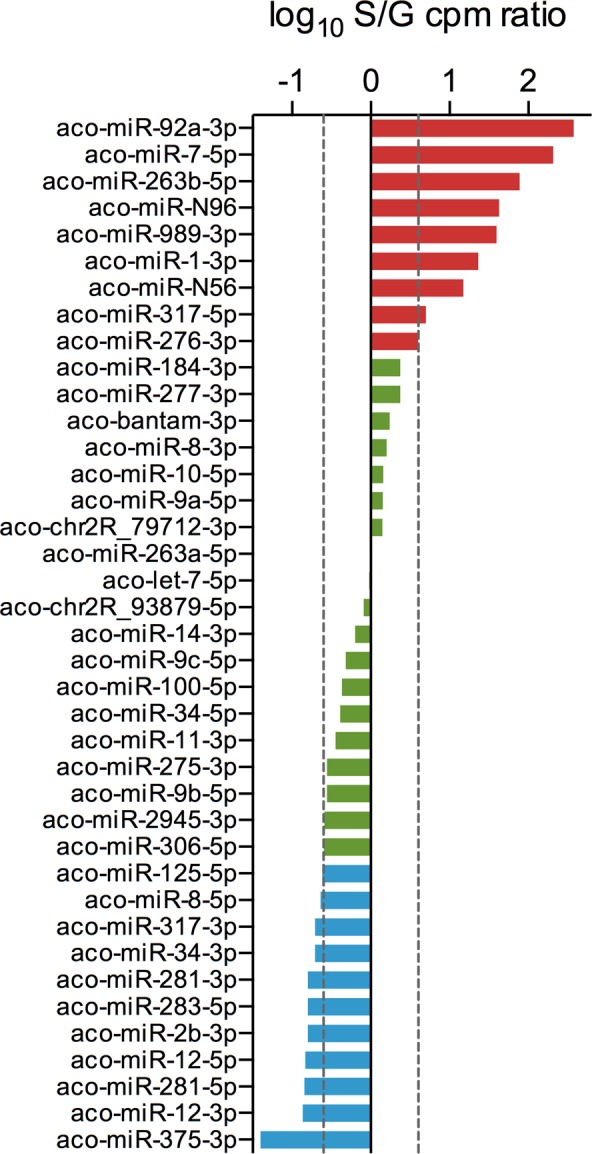
Asymmetric distribution of miRNAs in saliva and salivary glands. The mean CPM of the 30 most abundant miRNAs in saliva and salivary glands of An. coluzzii were compared and the S/G ratio calculated. For these 39 miRNAs the log10 of the S/G ratio is reported. miRNAs with S/G ratio > 4.0 or <0.25 are reported in red and light blue, respectively. In green are shown miRNAs whose S/G ratio was ≥0.25 or ≤4.0. Dashed lines mark the limits of 4-fold overexpression in saliva and salivary glands.
Target prediction
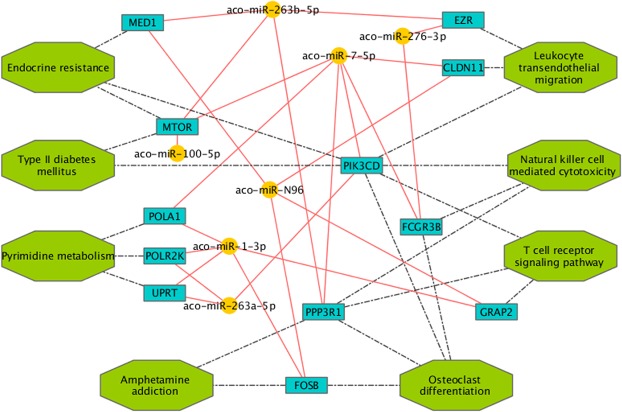
MicroRNAs regulate gene expression by binding the 3′-UTR of mRNA targets31,33,35. Since mosquitoes inject saliva into their hosts during blood feeding, and considering that miRNAs in anopheline saliva may have evolved to play some role in vector-host interactions, we were wondering about potential targets of the most abundant miRNAs found in saliva. Based on the observation that the seed region (nucleotides 2–8 of the miRNAs) plays a crucial role in recognition and binding to mRNAs, as well as on a few other features as free energy or site accessibility, several bioinformatics tools employing different algorithms have been developed for target prediction72,73. However, the output of these software packages typically consists of a large number of potential targets, which makes the identification of genuine targets a difficult task. Moreover, predictions usually require some type of downstream validation, which can be challenging, especially in complex biological contexts as the interface between the mosquito and its vertebrate host. Nevertheless, the use of more than one bioinformatics tool and the selection of transcripts targeted by multiple miRNAs may help identify relevant pathways. Therefore, we employed three different bioinformatics tools (TargetSpy, miRanda and PITA) to predict human mRNA targets of the 8 most abundant miRNAs in saliva; only transcripts expressed in human skin according to transcriptomic (FPKM > 1.0)74 and proteomic75 studies were considered for the prediction. As a control the analysis was also performed employing 8 male and 8 female miRNAs not found in saliva or salivary glands. In order to avoid common biases, such as the different average 3′UTR length of genes belonging to different functional categories76, the pathway enrichment analysis of predicted miRNA targets was restricted to those mRNAs targeted by at least two salivary miRNAs and by none of the control miRNAs. KEGG pathway enrichment analysis, performed using the WebGestalt tool77, yielded some moderately enriched (p-value < 0.05, FDR > 0.05) but potentially meaningful pathways as T cell receptor signalling (hsa04660), leukocyte transendothelial migration (hsa04670) or natural killer cell mediated cytotoxicity (hsa04650) (Fig. 6). Among the predicted targets were: MTOR (Mammalian target of rapamycin) and PIK3CD (Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta), which code for proteins playing central roles in important signalling cascades and regulating the function of diverse immune cells, including mast cells, neutrophils, T cells and B cells78,79; FCGR3B (Fc fragment of IgG receptor IIIb) that is expressed on neutrophils and whose product binds IgG and plays an active role in calcium mobilization and neutrophil degranulation80; PPP3R1 (Protein phosphatase 3 regulatory subunit B, alpha) also known as Calcineurin B that is involved in activation of transcription factors of the NFAT (Nuclear factor of activated T cells) family, which are key regulators of T-cell activation81; GRAP2 (GRB2-related adaptor protein 2), encoding a member of the Grb2 family of adaptor proteins implicated in the activation of lymphocyte-specific signalling pathways82; EZR (Ezrin), which encodes a protein linking plasma membrane to cytoskeleton and shown to play relevant roles in lymphocyte activation and migration83; FOSB that encodes a subunit of the transcription factor AP-1, which binds to promoters of early response and inflammatory genes. Overall, target prediction analysis suggest that the most abundant miRNAs in the saliva of An. coluzzii have the potential to target human genes involved in inflammatory and immune responses.
Figure 6.
Target prediction analysis. Schematic representation of genes targeted by abundant miRNAs from An. coluzzii saliva and enriched categories. miRNAs are shown by yellow dots and targeted genes (blue boxes) indicated by solid red lines. Dotted lines connect genes to enriched KEGG pathways (green octagons).
miRNAs from Anopheles coluzzii saliva mimic human miRNAs
We also wondered if miRNAs found in An. coluzzii saliva may mimic known human miRNAs. Searching miRBase it was found that 11 out of the 20 most abundant miRNAs from An. coluzzii saliva were identical or almost identical to human miRNAs (Table 3). Interestingly, these human miRNAs have several targets among genes coding for chemokines, cytokines, chemokine receptors, mitogen-activated protein (MAP) kinases, transcription factors and other mediators crucially affecting inflammatory and immune responses by acting on the NF-kB pathway, Toll-like receptors (TLRs) signalling cascades and inflammasome activation (Supplementary Table S2). A few paradigmatic examples are shortly discussed below whereas a more extensive, though not exhaustive list, can be found in Supplementary Table S3. Hsa-miR-7-5p was shown to down-regulate the NF-kB pathway both directly, by targeting the NF-kB subunit RelA84, and indirectly, by acting on RNF183, an ubiquitin ligase promoting the degradation of the NF-kB inhibitor IkBα85. Hsa-miR-1-3p targets CCL286, an inflammatory chemokine that is a powerful chemotactic factor involved in the recruitment of monocytes, memory T cells and natural killer (NK) cells to sites of inflammation produced either by tissue injury or infection87. Hsa-miR-184 downregulates NFAT1, a key transcription factor that controls the expression of a wide array of cytokines (IFNγ, GM-CSF, IL-3, IL-4, IL-2, TNFα) and plays a crucial role in the initiation of Th1 immune response88. Hsa-miR-92a-3p has, among other targets, also CCL889, a chemokine displaying chemotactic activity for monocytes, lymphocytes, basophils and eosiniphils. Hsa-miR-200b-5p was shown to target MyD88 in macrophages, a key mediator of TLR signalling and NF-kB activation90. Finally, hsa-miR-125b-5p may act on two key transcription factors in B cell differentiation as the interferon regulatory factor 4 (IRF4) and PRDM191. These observations reinforce the target prediction analysis reported above clearly pointing to the ability of abundant miRNAs from An. coluzzii saliva to target human inflammatory and immune responses.
Table 3.
Human orthologues among the 20 most abundant saliva miRNAs.
| An. coluzzii ID | nt | H. sapiens ID | seed | mm | alignment |
|---|---|---|---|---|---|
| aco-miR-276-3p | 22 | — | — | — | — |
| aco-miR-263a-5p | 24 | — | — | — | — |
| aco-miR-7-5p | 23 | hsa-miR-7-5p | Y | 0 | 1-23/1-23 |
| aco-miR-1-3p | 22 | hsa-miR-1-3p | Y | 1 | 1–22/1–21 |
| aco-miR-100-5p | 22 | hsa-miR-100-5p | Y | 0 | 1–22/1–22 |
| aco-miR-184-3p | 21 | hsa-miR-184 | Y | 0 | 1-21/1–21 |
| aco-miR-N96 | 23 | — | — | — | — |
| aco-miR-263b-5p | 21 | — | — | — | — |
| aco-miR-8-3p | 23 | hsa-miR-141-3p | Y/N | 2 | 1–21/1–21 |
| aco-let-7-5p | 21 | hsa-let-7a-5p | Y | 1 | 1–21/1–21 |
| aco-bantam-3p | 22 | — | — | — | — |
| aco-miR-92a-3p | 22 | hsa-miR-92a-3p | Y | 1 | 1–22/1–22 |
| aco-miR-8-5p | 22 | hsa-miR-200b-5p | Y | 2 | 1–22/1–22 |
| aco-miR-34-5p | 20 | hsa-miR-34c-5p | Y | 2 | 2–20/2–20 |
| aco-miR-N56 | 23 | — | — | — | — |
| aco-miR-10-5p | 21 | hsa-miR-10a-5p | Y/N | 0 | 1–21/2–22 |
| aco-miR-14-3p | 21 | — | — | — | — |
| aco-miR-125-5p | 22 | hsa-miR-125b-5p | Y | 0 | 1–22/1–22 |
| aco-miR-306-5p | 22 | — | — | — | — |
| aco-chr2R_79712-3p | 22 | — | — | — | — |
nt, miRNA length; seed, fully (Y) or partially (Y/N) conserved seed (nt 2–8); mm, number of mismatches in the aligned region; alignment, nucleotide position of aligned An. coluzzii to H. sapiens miRNAs. Human orthologues of An. coluzzi miRNAs with fully conserved seeds are in bold.
Comparison of saliva miRNAs among different species
Saliva miRNA repertoires have been previously studied in a few BFAs, specifically the mosquitoes Ae. aegypti and Ae. albopictus and the tick I. ricinus40,41. When we compared the top 30 miRNAs from An. coluzzii saliva to the 30 most abundant miRNAs found in the saliva of Ae. aegypti, Ae. albopictus and I. ricinus we found that 17 were also present in the saliva of Aedes mosquitoes and 16 in I. ricinus saliva; more specifically, 15 miRNAs were common to the three mosquito species and 10 miRNAs to both mosquitoes and the hard tick (Table 4, Supplementary File S6). We also mined a collection of miRNA profiles from the saliva of 46 healthy human subjects39 to verify if the 11 miRNAs from An. coluzzii saliva mimicking human miRNAs (Table 3) were also present in human saliva. We found that only 3 miRNAs (hsa-miR-141-3p, hsa-let-7a-5p, hsa-miR-92a-3p) were among the top 50 miRNAs in human saliva, other 3 were completely absent (hsa-miR-1-3p, hsa-miR-184, hsa-miR-200b-5p) and the remaining were present at low abundance (rank 67 to 248; Supplementary File S6 and Table 4). Overall, these observations support the idea that the presence of miRNAs in An. coluzzii saliva, at least as far as the most abundant miRNAs are concerned, it is most likely not serendipitous and that they may play some functional role.
Table 4.
Conservation of the top 30 miRNAs from An. coluzzi saliva.
| An. coluzzii ID | H. sapiens a | Ae. aegypti b | Ae. albopictus b | I. ricinus c | B. malayi d | H. polygyrus d |
|---|---|---|---|---|---|---|
| aco-miR-276-3p | ✔ | ✔ | ✔ | |||
| aco-miR-263a-5p | ✔ | ✔ | ✔ | ✔ | ||
| aco-miR-7-5p | ✔ | |||||
| aco-miR-1-3p | ✔ | ✔ | ||||
| aco-miR-100-5p | ✔ | ✔ | ✔ | ✔ | ✔ | |
| aco-miR-184-3p | ✔ | ✔ | ||||
| aco-miR-N96 | ||||||
| aco-miR-263b-5p | ||||||
| aco-miR-8-3p | ✔ | ✔ | ✔ | ✔ | ||
| aco-let-7-5p | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| aco-bantam-3p | ✔ | ✔ | ✔ | ✔ | ||
| aco-miR-92a-3p | ✔ | ✔ | ✔ | |||
| aco-miR-8-5p | ✔ | ✔ | ✔ | |||
| aco-miR-34-5p | ✔ | ✔ | ✔ | ✔ | ||
| aco-miR-N56 | ||||||
| aco-miR-10-5p | ✔ | ✔ | ✔ | |||
| aco-miR-14-3p | ✔ | ✔ | ||||
| aco-miR-125-5p | ✔ | ✔ | ✔ | ✔ | ||
| aco-miR-306-5p | ✔ | |||||
| aco-chr2R_79712-3p | ||||||
| aco-miR-281-3p | ✔ | ✔ | ||||
| aco-miR-281-5p | ✔ | ✔ | ||||
| aco-miR-317-3p | ✔ | ✔ | ✔ | |||
| aco-miR-9a-5p | ✔ | ✔ | ✔ | |||
| aco-chr3R_160384-3p | ✔ | |||||
| aco-miR-277-3p | ✔ | ✔ | ||||
| aco-miR-275-3p | ✔ | |||||
| aco-miR-989-3p | ✔ | |||||
| aco-miR-317-5p | ||||||
| aco-chr2R_93879-5p | ✔ | ✔ | ✔ | |||
| total | 3 | 17 | 17 | 16 | 9 | 7 |
| human | 3 | 8 | 8 | 9 | 7 | 5 |
The presence among the top 50 in human saliva, the top 30 in the saliva of Ae. aegypti, Ae. albopictus and I. ricinus and the top 30 exosomal miRNAs from the parasitic nematodes Brugia malayi and Heligmosomoides polygyrus are reported. The total number of conserved miRNAs and the number of those homologous to human miRNAs are shown at the bottom. An. coluzzi miRNAs mimicking human miRNAs are in bold. Extended data are reported in Supplemental File S6. 1. Yeri A. et al. 2017 Sci Rep 7:44061; 2. Maharaj P.D. et al. 2015 PLoS Negl Trop Dis 9:e0003386; 3. Hackenberg M. et al.40 RNA 23(8):1259–1269; 4. Zamanian M. et al. 2015 PLoS Negl Trop Dis 9:e0004069; 5. Buck A.H. et al. 2014 Nat Commun 5:5488.
Interestingly, the parasitic nematodes Brugia malayi and Heligmosomoides polygyrus (which infect humans and mice, respectively) were shown to secrete exosomal vesicles enriched in miRNAs identical/homologous to host miRNAs with known immunomodulatory roles. These exosomes could be internalized by target cells in vitro and H. polygyrus-derived exosomes could suppress innate immunity in vivo when administered to mice51,52, suggesting they may play a role in host manipulation. Therefore, we also compared the 30 most abundant exosomal miRNAs from these nematodes to the top 30 from An. coluzzi saliva and found 9 and 7 miRNAs in common with B. malayi and H. polygyrus, respectively (Table 4, Supplementary File S6).
Discussion
Transcriptomic, proteomic and genomic studies performed in the last decades shed light on the complexity and functions of salivary protein repertoires of hematophagous arthropods. Saliva of BFAs carries hundreds of proteins whose anti-haemostatic, anti-inflammatory and immunomodulatory activities have a highly adaptive value in hematophagy providing blood feeders with the ability to manipulate host responses at the feeding site4,6,23. However, despite the considerable progress, the understanding of the complex interactions taking place at the vector-host interface is still limited since we completely ignore the function of a rather large fraction of BFA salivary proteins and we just started getting deeper insights into the role of saliva in transmission of vector-borne pathogens27,30,92–96. Moreover, the finding of miRNAs in the saliva of Aedes mosquitoes41 and Ixodes ticks40 suggests that saliva of BFA may have a more complex composition than initially predicted.
To get insights into miRNA composition in the saliva of anopheline mosquitoes we performed a small RNA-Seq study on adult female salivary glands and saliva of the African malaria vector An. coluzzii. The number of reads mapping to mature miRNAs for the S sample was limited, most likely because of the difficulty of getting large amounts of saliva and to the paucity of small RNA content: indeed, comparable number of reads mapping to Aedes albopictus (12,075) and Aedes aegypti (298,283) miRNAs were obtained in the only study presently available on miRNAs from mosquito saliva41. While the most abundant miRNAs in the S sample were supported by a reliable number of counts, the least abundant had sometime less than 50 reads mapped (Supplementary File S2, worksheet aco_S_77). For this reason, to avoid introducing any bias, we used the S sample just to assemble a saliva catalogue of An. coluzzii miRNAs and restricted differential expression analysis to the G, F and M samples. Overall, evidence for the expression of 214 mature miRNAs was obtained, with 36 being putative novel miRNAs of An. coluzzii and An. gambiae. Almost all of these 36 novel miRNAs appeared conserved among members of the An. gambiae species complex, with a few also present in other anopheline and culicine mosquitoes and occasionally in other blood feeders. Using PCR amplification we provided evidence of expression for a subset of these bona fide novel miRNAs and found very good correlation between RNAseq and real time PCR data (Supplementary Fig. S4). These observations suggest that most of these miRNAs are real and mainly expressed at low levels in members of the An. gambiae species complex.
Differential expression analysis identified 38 miRNAs enriched in female salivary glands as compared to adult females: these may regulate endogenous genes involved in salivary gland physiology and/or blood feeding. Ten of these G-enriched miRNAs were also abundant in saliva (among the top 30) and, therefore, are likely to be injected into the vertebrate skin during blood feeding with the potential to target host genes at the biting site. In addition, 68 miRNAs appeared sex-biased, with 50 enriched in females and 18 in males, and they are expected to be implicated in sexual dimorphism. A list of miRNAs differentially abundant in the samples analysed in this study is provided in Supplementary Table S4 and may represent a useful starting point for further investigations.
One of the main goals of our experimental design was to enquire whether salivary glands and saliva from Anopheles mosquitoes have an identical or somewhat different miRNA composition. Indeed, an enrichment for selected miRNAs in saliva, as compared to salivary glands, has been previously reported in the hard tick Ixodes ricinus40; however, it was unknown if this was the case also for mosquitoes since the Aedes study of Maharaj and collaborators (2015) was focused on the effect of chikungunya virus infection and did not include a parallel analysis of salivary glands. The An. coluzzii saliva miRNA catalogue comprised 77 mature miRNAs, a number which is comparable to what previously found in Ae. albopictus (67 miRNAs) and Ae. aegypti (103 miRNAs)41. Although, as mentioned above, the least represented miRNAs had small number of counts, the 30 most abundant (top 30) were found in all five S replicates (29 cases out of 30) and appeared well supported by number of reads (range 52–4291) and CPM (range 3097–161,759) (Supplementary File S2). Moreover, since the five S replicates were processed in two batches and exhibited an overall lower correlation (see heatmap, Supplementary Fig. 1), we also compared the lists of the top 30 miRNAs from the two saliva sets and found a 77% overlap. When the relative abundance of the top 30 miRNAs in salivary glands and saliva of An. coluzzii was compared, we found groups of miRNAs overrepresented (FC > 4) in S or in G and others roughly equally distributed (Fig. 5), which indicated that specific miRNAs may be preferentially directed toward the secretory pathway or retained in salivary glands. It should be pointed out that asymmetric distribution of miRNAs between source cells and their exosomes has been previously reported and that multiple mechanisms appear to underlie their sorting into exosomes43, including recognition of exomotifs by RNA Binding Proteins97 or non-templated nucleotide addition (NTA) at the 3′-end of the miRNAs98. More specifically, 3′-end uridylated miRNA isoforms appeared overrepresented in exosomes and 3′-end adenylated in their mother cells98. We could not find evidence of enrichment of previously described exomotifs97 in An. coluzzii miRNAs abundant in saliva. However, when we determined the fraction of U and A non-templated addition to the 3′-end of miRNAs from saliva and non-saliva samples, a higher proportion of uridylation in saliva miRNAs was found (Supplementary Fig. S6). A similar situation was reported for saliva miRNAs from the tick I. ricinus and interpreted as an indirect evidence of exosomal origin of saliva miRNAs40, which may be the case for anopheline mosquitoes as well.
Comparing the top 30 saliva miRNAs from An. coluzzii, Ae. aegypti, Ae. albopictus and I. ricinus, it was found that 15 miRNAs were evolutionary conserved between Anopheles and Aedes mosquitoes and 10 miRNAs were common to both mosquitoes and the hard tick (Table 4, Supplementary File S6). Moreover, since 11 An. coluzzii saliva miRNAs were essentially identical to human endogenous miRNAs, we also wondered if they were abundant in human saliva. However, when a relatively large collection of human saliva miRNA profiles39 was searched, only 3 of these miRNAs were included among the top 50 miRNAs in human saliva (Supplementary File S6, Table 4). Furthermore, the 30 most abundant saliva miRNAs from An. coluzzii were also compared to exosomal miRNAs from the parasitic nematodes B. malayi and H. polygyrus. These miRNAs, many of which mimic host miRNAs, were previously suggested to play roles in manipulation of host innate immunity51,52. Interestingly, An. coluzzi saliva shared 9 miRNAs with B. malayi and 7 miRNAs with H. polygyrus (Table 4, Supplementary File S6).
The asymmetric distribution and the conservation of sets of miRNAs in the saliva of blood feeding arthropods (and parasitic nematodes) suggest a possible functional role for saliva miRNAs and, obviously, this leads to the question of potential host targets. Setting strict criteria to minimize false positives, and selecting only targets predicted by multiple tools, we could identify some potentially meaningful candidate target genes involved in human immune and inflammatory responses (Fig. 6). Moreover, the presence of human homologues among the most abundant miRNAs in An. coluzzi saliva came as an unexpected help for target analysis. Indeed, a quite large body of literature reporting experimentally validated human miRNA targets is available, and these 11 human miRNAs were found to target several genes and pathway involved in human inflammatory and immune responses (Supplementary Tables S2–S3). We believe that these observations point to the fascinating hypothesis that miRNAs in mosquito saliva, perhaps enclosed within exosomes, may target vertebrate host cells involved in immune and inflammatory responses.
The question whether miRNAs from body fluids play biological functions is still debated44,45; it has been calculated that miRNAs expressed below ~100 copies per cell have little regulatory capacity99 and that the concentration of extracellular miRNAs may be too low to exert in vivo effect100. However, there are several indications that exosomal miRNAs can be transferred in vitro to recipient cells where they can affect gene expression44,101 and the majority of miRNAs from human saliva was shown to be concentrated in exosomes102. Moreover, circulating exosomal miRNAs from adipose tissue have been recently shown to regulate gene expression in the liver46, reinvigorating the hypothesis of an in vivo functional role of exosomal miRNAs. According to this scenario, BFAs may manipulate host responses not only using salivary proteins but also taking advantage of salivary miRNAs. Haemostatic, inflammatory and immune responses of vertebrates are known to be both complex and redundant, and, to efficiently deal with their hosts, BFAs evolved a cocktail of salivary proteins of similar redundancy and complexity4,23. However, while salivary proteins may evoke in vertebrate hosts inactivating antibody responses that may affect blood feeding efficiency, miRNAs are not immunogenic and, therefore, would provide hematophagous arthropods with a precious additional tool. The evolutionary advantage of such a combined strategy seems obvious for ticks, which can stay attached to their hosts for days, or for parasitic nematodes, that establish long-term interaction with the host. On the contrary, the benefit does not appear equally evident for blood feeders as mosquitoes or sand flies that take their blood meals in a timescale of minutes, not compatible with the time of action of miRNAs. Nevertheless, even if not providing an immediate reward to the individual, the modulation of the host antibody response to salivary proteins (which could be achieved for example by targeting antigen presenting cells) may represent a longer-term evolutionary advantage for the species. Providing further evidence that miRNAs from saliva of BFAs play a physiological role in host manipulation (and perhaps in pathogen transmission) will require further experimentation that may be challenging considering the intricacy of the vector-host-pathogen relationships. Anyhow, we believe that our study contributes to a better understanding of function and complexity of blood feeding arthropod saliva and opens perspectives for novel investigations.
Methods
Mosquito rearing and sample collection
An. coluzzii mosquitoes (Xag, 2 R+, 2 L+, 3 R+, 3 L+; colony originally collected in Cameroon), formerly known as An. gambiae M molecular form103, were reared under standard insectary conditions (28 ± 1 °C, 60–70% humidity, 14:10 hours light:dark photoperiod). Adults 2–6 days post-emergence (dpe) were used for all the experiments reported here. Salivary glands were dissected in Phosphate Buffered Saline (PBS), collected in RNAlater (Sigma-Aldrich, R0901), kept overnight at 4 °C and then stored at −20 °C until needed. Adult males and females were immersed in RNAlater and stored as above. Saliva was collected from adult females as previously described104,105 with some modifications. Briefly, mosquitoes were deprived of wings and legs, immobilized on double-stick tape on a microscope slide, and their proboscis was inserted into a 10 µl pipet tip filled with 2–3 µl of RNAlater/PBS (50% v/v). Pilocarpine (1% w/v in PBS) was applied to the thorax and mosquitoes were left salivating for 10–15 minutes; afterwards the RNAlater/PBS solution containing saliva was ejected into eppendorf tubes containing 30 µl of RNAlater. Saliva samples (batches of saliva from 20 to 70 mosquitoes) were stored at −20 °C until used for RNA extraction. All samples were collected in low-binding tubes (Sigma-Aldrich, Z666505).
RNA extraction, library preparation and RNA-sequencing
In a pilot experiment, performed on duplicates, small RNA was extracted from saliva collected from 97 and 98 mosquitoes, respectively, using the miRNeasy Serum/Plasma kit (QIAGEN). In a second experiment 80 salivary glands, 5 adult females, 5 adult males and saliva collected from 126–128 mosquitoes (all in triplicates) were used for small RNA extraction according to the miRNeasy Micro kit (salivary glands, adult males and females) and the miRNeasy Serum/Plasma kit (saliva) protocols (QIAGEN). miRNA validation was performed using as template the small RNA fraction (<200 nt) extracted from adult An. coluzzii females using the miRNeasy Micro kit. Concentration and purity of small RNA were evaluated determining the absorbance at 260 and 280 nanometers by a BioTek SynergyHT (Take3 Module). RNA quality control and libraries preparation were performed by the EMBL Genomic Core Facility (EMBL, Heidelberg, DE). RNA quality and integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Small RNA libraries were prepared using the TruSeq Small RNA Sample preparation kit (Illumina). Fifty base pair, single end sequencing was performed on an Illumina HiSeq2000 platform. An Illumina MiSeq platform (read length 75, single end sequencing) was used for the pilot saliva duplicated samples.
Reads Mapping
Raw reads were first quality control checked by FastQC106 and then trimmed using cutadapt 1.9.1107 to remove 3′ adapters and discard reads shorter than 14 nucleotides. Processed reads from each sample were mapped to the An. gambiae AgamP4 genome assembly (version v2.00, downloaded from VectorBase108,109) using Bowtie110 (-n 0 -l 18 -e 80). Indeed, although the An. coluzzii genome has been sequenced (Mali-NIH strain, AcolM1 assembly), the reference An. gambiae PEST genome was preferred for several reasons. First, An. gambiae and An. coluzzii only recently have been classified as different species103, being formerly considered as incipient species and known as An. gambiae S and M molecular forms, respectively. Second, genome assemblies and annotations are very different for quality and accuracy: AcolM1 consists of 10,521 scaffolds with an N50 of 4,437 Kb, whereas AgamP4 includes 8 scaffolds with an N50 of 49,364 Kb and with the assembly mapped to chromosomes18. Finally, the reference An. gambiae PEST genome is actually a chimera of S and M molecular forms111. Anyhow, we also attempted using AcolM1 but, as expected, the number of reads mapping to the genome was 10% to 26% lower (depending from the sample), which further supported our choice. Reads aligned to AgamP4 were then mapped (-n 0 -l 18 -e 80–norc) to a collection of An. gambiae rRNA sequences obtained from VectorBase by the BioMart tool. After subtraction of ribosomal RNAs the remaining reads were mapped (-n 0 -l 18 -a–best–strata -e 80–norc) to a list of miRNA precursors and other non coding RNAs from An. gambiae (including tRNAs, snoRNAs, snRNAs, 7SL, 7SK and RNase P) and finally to An. gambiae transcripts and repeats downloaded from VectorBase (-n 0 -l 18 -a–best–strata -e 80). The list of miRNA precursors included a total of 273 hairpins (Supplemental File 1): among these 175 were previously known An. gambiae hairpins (66 retrieved from miRBase release 21, 59 from53, 41 from54, 9 from55) and the remaining 98 were predicted (see below). The collection of mature miRNAs consisted of 438 miRNAs (5p + 3p, Supplemental File 2): 339 previously known An. gambiae miRNAs (131 from miRBase, 118 from53, 81 from54 and 9 found by55) and 99 additional predicted miRNAs (see below).
Prediction of novel miRNAs by miRDeep* and MapMi
Reads from all samples which did not map to An. gambiae rRNAs or known small RNAs were combined and used to predict novel miRNAs by employing the miRDeep* software69. A cutoff score of 0 was used to select the most reliable novel miRNA predictions. We also used MapMi70 to map miRNAs from other species (arthropods, human and viruses retrieved from miRBase release 21) to the An. gambiae genome (threshold 25, max mature mismatch 3). This way we compiled a set of loci potentially orthologous to miRNAs validated in other species. miRNAs found by MapMi whose genomic coordinates overlapped those of known or miRDeep*-predicted miRNAs were identified through the BEDTools software112 and discarded. Secondary structure predictions and minimal free energy calculations were performed using RNAfold113.
miRNA validation by real-time PCR amplification
Validation of a subset of the putative novel miRNAs identified in this study was performed by the Stem-loop Reverse-Transcription Polymerase Chain Reaction (slRT-PCR) technique71 using as template small RNA from adult An. coluzzii females. First-strand cDNA was generated in a 20 µL reaction volume from 1 µg of small RNA using the SuperScript II Reverse Transcriptase (Invitrogen) according to manufacturer’s instruction and specific stem-loop primers (0.1 µM). Nine miRNAs were selected for validation according to their CPM values and choosing three miRNAs for each of the tree following abundance categories: (i) high (CPM ≥ 100; aco-miR-N96, aco-miR-N56, aco-miR-N951; range 103–489), (ii) medium (20 ≤ CPM < 100; aco-miR-N966, aco-miR-N149, aco-miR-N629; range 23–73) and (iii) low (CPM < 20; aco-miR-N1044, aco-miR-N645, aco-miR-N135; range 7–17). Real time PCR amplifications were performed in a final volume of 20 µl including 2X PowerUP SYBR Green Master Mix (Applied Biosystem), specific forward and universal reverse primers (1 µM each) and 2 µl of the specific first strand cDNA reaction. Amplification was as follows: initial holding stage of 2 min at 50 °C and 2 min at 95 °C followed by 40 cycles (30 sec. 95 °C, 1 min 60 °C). Melting curves were obtained for each miRNA to verify for the absence of unspecific amplification products with detection steps every 0.3 °C. All RT-qPCR reactions were performed in biological and technical triplicates. A list including the specific stem-loop and reverse primers, as well as the universal primers is provided in Supplementary Table S5.
Conservation of novel miRNAs
Putative orthologues of novel miRNAs were searched using the BLAST tool at the VectorBase web site109. The genomes of several anopheline, a few culicine mosquitoes (Aedes aegypti, Aedes albopictus and Culex quinquefasciatus), the sand flies Phlebotomus papatasi and Lutzomyia longipalpis, the tsetse fly Glossina morsitans, the stable fly Stomoxys calcitrans, the bugs Rhodnius prolixus and Cimex lectularius, the human body louse Pediculus humanus, the tick Ixodes scapularis and of the non blood feeding Diptera Drosophila melanogaster and Musca domestica were searched. A miRNA was considered as putatively conserved in a given species if the BLASTn search yielded (i) ≥70% identity over ≥70% of the length of miRNA precursors and/or (ii) ≥90% identity over the entire length and fully conserved seed sequence for mature miRNAs.
Quantification and differential expression of miRNAs
Read counts for each An. gambiae small RNA were computed from SAM files using a Python custom script. Reads with multiple highest score mappings were discarded. Expression values were calculated as count per millions (CPM) and used for sample clustering. Reads mapping to precursor miRNAs were assigned to mature miRNAs based on their mapping position; an overhang of maximum 3 nucleotides for each side of the mature form was tolerated. Differential expression analysis of mature miRNAs with 1 CPM in at least three samples was performed using glmFIT and glmLRT functions provided by the edgeR software package114,115. Fold change (FC) and false discovery rates (FDR) were calculated to provide statistical validation (Supplementary File S4).
3′-end non-templated U and A addition
To identify reads representing 3′-end non-templated additions of uridine and adenine residues, miRNAs we first searched for tags carrying at least one substitution following the 18 nt long seed region. Thereafter, for reads presenting these mismatches, the subsequence beginning at the first substitution site was evaluated for the presence of only Ts or As.
Target analysis
Host genes putatively targeted by An. gambiae miRNAs were searched using the miRNAconsTarget program from sRNAtoolbox116, which employs the prediction software TargetSpy, miRanda and PITA117–119. The 8 most abundant miRNAs found in An. coluzzii saliva (aco-miR-276-3p, aco-miR-263a-5p, aco-miR-7-5p, aco-miR-1-3p, aco-miR-100-5p, aco-miR-184-3p, aco-miR-N96, aco-miR-263b-5p) were used as query. Eight male (aco-chrX_329630-5p, aco-miR-2c-5p, aco-miR-10367-5p, aco-miR-219-5p, aco-miR-10375a-3p, aco-chr3L_119935-5p, aco-miR-10372a-5p, aco-miR-10370-3p) and 8 female (aco-miR-989-5p, aco-chr3R_150582-5p, aco-miR-10362-5p, aco-miR-N420, aco-chr3L_130625-5p, aco-miR-N148, aco-chr2L_42099-5p, aco-miR-10359-5p) miRNAs not found in saliva or salivary glands were used as a control set (Supplementary File S2). Only miRNA-mRNA interactions predicted by all three programs were taken into consideration. Prediction was done using 3′UTRs (>30 nt) of transcripts expressed in human skin74,75, which were downloaded from Ensembl120 using the BioMart tool121. The corresponding genes were provided as background for the KEGG pathway enrichment analysis that was performed using the WebGestalt tool77.
Supplementary information
Acknowledgements
BA research was supported by the Italian Ministry of Education, University and Research (PRIN 2015JXC3JF) and by Sapienza University of Rome (C26A14AKKH).
Author Contributions
B.A. conceived and designed the study. B.A., C.F. and F.L. performed experiments. V.B. constructed libraries and made RNA-seq. F.S., M.D.L. and F.L. provided reagents. A.C. and B.A. performed the bioinformatic analysis. B.A., A.C., C.F. and R.A.C. analysed data. B.A. wrote the first manuscript draft. All authors contributed to writing the final manuscript, gave the final approval and agreed to be responsible for all aspects of this work.
Data Availabilty
Small RNA-Seq data have been submitted to the NCBI GEO repository with accession number GSE:120658 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE120658) and will be publicly released upon manuscript acceptance. Other data generated during this study have been included as Supplementary Information.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-39880-1.
References
- 1.WHO. World Malaria Report, http://apps.who.int/iris/bitstream/10665/259492/1/9789241565523-eng.pdf (2017).
- 2.Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen JF. Structure and mechanism in salivary proteins from blood-feeding arthropods. Toxicon. 2010;56:1120–1129. doi: 10.1016/j.toxicon.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcà B, Ribeiro JMC. Saliva of hematophagous insects: a multifaceted toolkit. Curr Opin Insect Sci. 2018;29:102–109. doi: 10.1016/j.cois.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci (Landmark Ed) 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narasimhan, S., Schleicher, T. R. & Fikrig, E. In Arthropod vector: controller of disease transmission Vol. 2 (eds Wikel, S. K., Aksoy, S. & Dimopoulos, G.) Ch. 14, 249–300 (Elsevier/Academic Press and imprint of Elsevier, 2017).
- 7.Arcà B, et al. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Arcà B, et al. An updated catalogue of salivary gland transcripts in the adult female mosquito. Anopheles gambiae. J Exp Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- 9.Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol. 2007;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009;10:57. doi: 10.1186/1471-2164-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribeiro JM, et al. An annotated catalogue of salivary gland transcripts in the adult female mosquito. Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro JM, Martin-Martin I, Arca B, Calvo E. A Deep Insight into the Sialome of Male and Female Aedes aegypti Mosquitoes. PLoS One. 2016;11:e0151400. doi: 10.1371/journal.pone.0151400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro JMC, Martin-Martin I, Moreira FR, Bernard KA, Calvo E. A deep insight into the male and female sialotranscriptome of adult Culex tarsalis mosquitoes. Insect Biochem Mol Biol. 2018;95:1–9. doi: 10.1016/j.ibmb.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–732. doi: 10.1016/S0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 16.Arcà B, Lombardo F, Struchiner CJ, Ribeiro JM. Anopheline salivary protein genes and gene families: an evolutionary overview after the whole genome sequence of sixteen Anopheles species. BMC Genomics. 2017;18:153. doi: 10.1186/s12864-017-3579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dritsou V, et al. A draft genome sequence of an invasive mosquito: an Italian Aedes albopictus. Pathog Glob Health. 2015;109:207–220. doi: 10.1179/2047773215Y.0000000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neafsey DE, et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choumet V, et al. The salivary glands and saliva of Anopheles gambiae as an essential step in the Plasmodium life cycle: a global proteomic study. Proteomics. 2007;7:3384–3394. doi: 10.1002/pmic.200700334. [DOI] [PubMed] [Google Scholar]
- 20.Doucoure S, et al. First screening of Aedes albopictus immunogenic salivary proteins. Insect molecular biology. 2013;22:411–423. doi: 10.1111/imb.12032. [DOI] [PubMed] [Google Scholar]
- 21.Kalume DE, et al. A proteomic analysis of salivary glands of female Anopheles gambiae mosquito. Proteomics. 2005;5:3765–3777. doi: 10.1002/pmic.200401210. [DOI] [PubMed] [Google Scholar]
- 22.Wasinpiyamongkol L, et al. Blood-feeding and immunogenic Aedes aegypti saliva proteins. Proteomics. 2010;10:1906–1916. doi: 10.1002/pmic.200900626. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro JMC, Arcà B. From Sialomes to the Sialoverse: An Insight into Salivary Potion of Blood-Feeding Insects. Advances in Insect Physiology. 2009;37:59–118. doi: 10.1016/S0065-2806(09)37002-2. [DOI] [Google Scholar]
- 24.Briant L, Despres P, Choumet V, Misse D. Role of skin immune cells on the host susceptibility to mosquito-borne viruses. Virology. 2014;464–465:26–32. doi: 10.1016/j.virol.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Donovan MJ, et al. Uninfected mosquito bites confer protection against infection with malaria parasites. Infection and immunity. 2007;75:2523–2530. doi: 10.1128/IAI.01928-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontaine A, et al. Implication of haematophagous arthropod salivary proteins in host-vector interactions. Parasit Vectors. 2011;4:187. doi: 10.1186/1756-3305-4-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin L, et al. Salivary factor LTRIN from Aedes aegypti facilitates the transmission of Zika virus by interfering with the lymphotoxin-beta receptor. Nat Immunol. 2018;19:342–353. doi: 10.1038/s41590-018-0063-9. [DOI] [PubMed] [Google Scholar]
- 28.Manning JE, Morens DM, Kamhawi S, Valenzuela JG, Memoli M. Mosquito saliva: the hope for a universal arbovirus vaccine? J Infect Dis. 2018 doi: 10.1093/infdis/jiy179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider BS, Higgs S. The enhancement of arbovirus transmission and disease by mosquito saliva is associated with modulation of the host immune response. Trans R Soc Trop Med Hyg. 2008;102:400–408. doi: 10.1016/j.trstmh.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogt MB, et al. Mosquito saliva alone has profound effects on the human immune system. PLoS Negl Trop Dis. 2018;12:e0006439. doi: 10.1371/journal.pntd.0006439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 33.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 34.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 35.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582–3600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahn JH, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber JA, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeri A, et al. Total Extracellular Small RNA Profiles from Plasma, Saliva, and Urine of Healthy Subjects. Sci Rep. 2017;7:44061. doi: 10.1038/srep44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackenberg M, Langenberger D, Schwarz A, Erhart J, Kotsyfakis M. In silico target network analysis of de novo-discovered, tick saliva-specific microRNAs reveals important combinatorial effects in their interference with vertebrate host physiology. RNA. 2017;23:1259–1269. doi: 10.1261/rna.061168.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maharaj PD, Widen SG, Huang J, Wood TG, Thangamani S. Discovery of mosquito saliva microRNAs during CHIKV infection. PLoS Negl Trop Dis. 2015;9:e0003386. doi: 10.1371/journal.pntd.0003386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 43.Gebert, L. F. R. & MacRae, I. J. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol, 10.1038/s41580-018-0045-7 (2018). [DOI] [PMC free article] [PubMed]
- 44.Iftikhar H, Carney GE. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing: Potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. Bioessays. 2016;38:367–378. doi: 10.1002/bies.201500130. [DOI] [PubMed] [Google Scholar]
- 45.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: A Collision of Two Paradigms. Trends Biochem Sci. 2016;41:883–892. doi: 10.1016/j.tibs.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Thomou T, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 48.Vyas N, Dhawan J. Exosomes: mobile platforms for targeted and synergistic signaling across cell boundaries. Cell Mol Life Sci. 2017;74:1567–1576. doi: 10.1007/s00018-016-2413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemcke, H. & David, R. Potential mechanisms of miRNA mobility. Traffic, 10.1111/tra.12606 (2018). [DOI] [PubMed]
- 50.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8:e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buck AH, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zamanian M, et al. Release of Small RNA-containing Exosome-like Vesicles from the Human Filarial Parasite Brugia malayi. PLoS Negl Trop Dis. 2015;9:e0004069. doi: 10.1371/journal.pntd.0004069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biryukova I, Ye T, Levashina E. Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics. 2014;15:557. doi: 10.1186/1471-2164-15-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castellano L, et al. The germline of the malaria mosquito produces abundant miRNAs, endo-siRNAs, piRNAs and 29-nt small RNAs. BMC Genomics. 2015;16:100. doi: 10.1186/s12864-015-1257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu X, Dimopoulos G, Zhu J. Association of microRNAs with Argonaute proteins in the malaria mosquito Anopheles gambiae after blood ingestion. Sci Rep. 2017;7:6493. doi: 10.1038/s41598-017-07013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carissimo G, Pain A, Belda E, Vernick KD. Highly focused transcriptional response of Anopheles coluzzii to O’nyong nyong arbovirus during the primary midgut infection. BMC Genomics. 2018;19:526. doi: 10.1186/s12864-018-4918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lampe L, Levashina EA. MicroRNA Tissue Atlas of the Malaria Mosquito Anopheles gambiae. G3 (Bethesda) 2018;8:185–193. doi: 10.1534/g3.117.300170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu W, Criscione F, Liang S, Tu Z. MicroRNAs of two medically important mosquito species: Aedes aegypti and Anopheles stephensi. Insect molecular biology. 2015;24:240–252. doi: 10.1111/imb.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain S, et al. Blood feeding and Plasmodium infection alters the miRNome of Anopheles stephensi. PLoS One. 2014;9:e98402. doi: 10.1371/journal.pone.0098402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jain S, Rana V, Tridibes A, Sunil S, Bhatnagar RK. Dynamic expression of miRNAs across immature and adult stages of the malaria mosquito Anopheles stephensi. Parasit Vectors. 2015;8:179. doi: 10.1186/s13071-015-0772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allam M, et al. Identification and characterization of microRNAs expressed in the African malaria vector Anopheles funestus life stages using high throughput sequencing. Malar J. 2016;15:542. doi: 10.1186/s12936-016-1591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu W, et al. Comparative expression profile of microRNAs in Anopheles anthropophagus midgut after blood-feeding and Plasmodium infection. Parasit Vectors. 2017;10:86. doi: 10.1186/s13071-017-2027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng X, Wu J, Zhou S, Wang J, Hu W. Characterization and potential role of microRNA in the Chinese dominant malaria mosquito Anopheles sinensis (Diptera: Culicidae) throughout four different life stages. Cell Biosci. 2018;8:29. doi: 10.1186/s13578-018-0227-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng X, Zhou X, Zhou S, Wang J, Hu W. Analysis of microRNA profile of Anopheles sinensis by deep sequencing and bioinformatic approaches. Parasit Vectors. 2018;11:172. doi: 10.1186/s13071-018-2734-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dritsou V, et al. Non-coding RNA gene families in the genomes of anopheline mosquitoes. BMC Genomics. 2014;15:1038. doi: 10.1186/1471-2164-15-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giraldo-Calderon GI, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.An J, Lai J, Lehman ML, Nelson CC. MiRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013;41:727–737. doi: 10.1093/nar/gks1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guerra-Assuncao JA, Enright AJ. MapMi: automated mapping of microRNA loci. BMC Bioinformatics. 2010;11:133. doi: 10.1186/1471-2105-11-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kramer, M. F. Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol Chapter 15, Unit15 10, 10.1002/0471142727.mb1510s95 (2011). [DOI] [PMC free article] [PubMed]
- 72.Peterson SM, et al. Common features of microRNA target prediction tools. Front Genet. 2014;5:23. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts JT, Borchert GM. Computational Prediction of MicroRNA Target Genes, Target Prediction Databases, and Web Resources. Methods Mol Biol. 2017;1617:109–122. doi: 10.1007/978-1-4939-7046-9_8. [DOI] [PubMed] [Google Scholar]
- 74.Ahn RS, et al. Transcriptional landscape of epithelial and immune cell populations revealed through FACS-seq of healthy human skin. Sci Rep. 2017;7:1343. doi: 10.1038/s41598-017-01468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 76.Bleazard T, Lamb JA, Griffiths-Jones S. Bias in microRNA functional enrichment analysis. Bioinformatics. 2015;31:1592–1598. doi: 10.1093/bioinformatics/btv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lucas CL, Chandra A, Nejentsev S, Condliffe AM, Okkenhaug K. PI3Kdelta and primary immunodeficiencies. Nat Rev Immunol. 2016;16:702–714. doi: 10.1038/nri.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu Rev Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, et al. Crystal structure of the extracellular domain of a human Fc gamma RIII. Immunity. 2000;13:387–395. doi: 10.1016/S1074-7613(00)00038-8. [DOI] [PubMed] [Google Scholar]
- 81.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 82.Jang IK, Zhang J, Gu H. Grb2, a simple adapter with complex roles in lymphocyte development, function, and signaling. Immunol Rev. 2009;232:150–159. doi: 10.1111/j.1600-065X.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 83.Parameswaran N, Gupta N. Re-defining ERM function in lymphocyte activation and migration. Immunol Rev. 2013;256:63–79. doi: 10.1111/imr.12104. [DOI] [PubMed] [Google Scholar]
- 84.Giles KM, et al. microRNA-7-5p inhibits melanoma cell proliferation and metastasis by suppressing RelA/NF-kappaB. Oncotarget. 2016;7:31663–31680. doi: 10.18632/oncotarget.9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu Q, et al. E3 Ubiquitin ligase RNF183 Is a Novel Regulator in Inflammatory Bowel Disease. J Crohns Colitis. 2016;10:713–725. doi: 10.1093/ecco-jcc/jjw023. [DOI] [PubMed] [Google Scholar]
- 86.Wang W, et al. MiR-1-3p inhibits the proliferation and invasion of bladder cancer cells by suppressing CCL2 expression. Tumour Biol. 2017;39:1010428317698383. doi: 10.1177/1010428317698383. [DOI] [PubMed] [Google Scholar]
- 87.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weitzel RP, et al. microRNA 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood. 2009;113:6648–6657. doi: 10.1182/blood-2008-09-181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Poole E, et al. Latency-associated viral interleukin-10 (IL-10) encoded by human cytomegalovirus modulates cellular IL-10 and CCL8 Secretion during latent infection through changes in the cellular microRNA hsa-miR-92a. J Virol. 2014;88:13947–13955. doi: 10.1128/JVI.02424-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-kappaB activation. Innate Immun. 2012;18:846–855. doi: 10.1177/1753425912443903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Malumbres R, et al. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Coutinho-Abreu IV, Guimaraes-Costa AB, Valenzuela JG. Impact of Insect Salivary Proteins in Blood Feeding, Host Immunity, Disease, and in the Development of Biomarkers for Vector Exposure. Curr Opin Insect Sci. 2015;10:98–103. doi: 10.1016/j.cois.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Castro, W., Oliveira, F., Coutinho-Abreu, I. V., Kamhawi, S. & Valenzuela, J. G. In Arthropod vector: controller of disease transmission Vol. 2 (eds S. K. Wikel, S. Aksoy, & G. Dimopoulos) Ch. 5, 65–89 (Elsevier/Academic Press, and imprint of Elsevier, 2017).
- 94.Dragovic SM, et al. Immunization with AgTRIO, a Protein in Anopheles Saliva, Contributes to Protection against Plasmodium Infection in Mice. Cell Host Microbe. 2018;23:523–535 e525. doi: 10.1016/j.chom.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leitner WW, Costero-Saint Denis A, Wali T. Immunological consequences of arthropod vector-derived salivary factors. Eur J Immunol. 2011;41:3396–3400. doi: 10.1002/eji.201190075. [DOI] [PubMed] [Google Scholar]
- 96.Schleicher TR, et al. A mosquito salivary gland protein partially inhibits Plasmodium sporozoite cell traversal and transmission. Nat Commun. 2018;9:2908. doi: 10.1038/s41467-018-05374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koppers-Lalic D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 99.Mullokandov G, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coetzee M, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. doi: 10.11646/zootaxa.3619.3.2. [DOI] [PubMed] [Google Scholar]
- 104.Anderson SL, Richards SL, Smartt CT. A simple method for determining arbovirus transmission in mosquitoes. J Am Mosq Control Assoc. 2010;26:108–111. doi: 10.2987/09-5935.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossignol PA, Spielman A. Fluid Transport Across the Ducts of the Salivary Glands of a Mosquito. J. Insect Physiol. 1982;28:579–583. doi: 10.1016/0022-1910(82)90054-3. [DOI] [Google Scholar]
- 106.Andrews, S. FastQC: a quality control tool for high throughput sequence data. Available online at, http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
- 107.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 108.Megy K, et al. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40:D729–734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.VectorBase. Bioinformatics Resource for Invertebrate Vectors of Human Pathogens. Available online at, https://www.vectorbase.org/.
- 110.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 112.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rueda A, et al. sRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res. 2015;43:W467–473. doi: 10.1093/nar/gkv555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 119.Sturm M, Hackenberg M, Langenberger D, Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics. 2010;11:292. doi: 10.1186/1471-2105-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kinsella RJ, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA-Seq data have been submitted to the NCBI GEO repository with accession number GSE:120658 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE120658) and will be publicly released upon manuscript acceptance. Other data generated during this study have been included as Supplementary Information.