Abstract
Keratoconus (KC) is an ectatic corneal disease characterized by progressive thinning and irregular astigmatism, and a leading indication for corneal transplantation. KC-associated changes have been demonstrated for the entire cornea, but the pathological thinning and mechanical weakening is usually localized. We performed quantitative proteomics using Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectrometry (SWATH-MS) to analyze epithelial and stromal changes between the topographically-abnormal cone and topographically-normal non-cone regions of advanced KC corneas, compared to age-matched normal corneas. Expression of 20 epithelial and 14 stromal proteins was significantly altered (≥2 or ≤0.5-fold) between cone and non-cone in all 4 KC samples. Ingenuity pathway analysis illustrated developmental and metabolic disorders for the altered epithelial proteome with mitochondrion as the significant gene ontology (GO) term. The differential stromal proteome was related to cellular assembly, tissue organization and connective tissue disorders with endoplasmic reticulum protein folding as the significant GO term. Validation of selected protein expression was performed on archived KC, non-KC and normal corneal specimens by immunohistochemistry. This is the first time to show that KC-associated proteome changes were not limited to the topographically-thinner and mechanically-weakened cone but also non-cone region with normal topography, indicating a peripheral involvement in KC development.
Introduction
Keratoconus (KC) is an asymmetric corneal ectatic disorder characterized by progressive focal thinning, that leads to myopia and irregular astigmatism with impaired visual acuity1,2. Corneal steepening and protrusion, with an eccentric thin conical apex is the typical clinical presentation, whereas central scarring is seen in many advanced cases3. The incidence of KC is estimated to range from 1:400 to 1:2,000 people worldwide4. Ethnic differences of KC incidence have also been reported5–7. It affects both genders at puberty to early mid-life, hence it significantly impacts young, working individuals and poses a considerable socioeconomic burden to the society3. About 20% of KC patients require corneal transplantation and it is one of the leading indications for corneal grafting in the US and worldwide8,9.
KC is classically defined as a non-inflammatory corneal degeneration due to the lack of cellular infiltration and neovascularization10. However, increasing evidence has revised this theory and the etiology of KC is still not completely understood1. The etiology of KC is multifactorial, including genetic, biomechanical and environmental11. The detection of a positive family history in approximately 10% of KC cases and a high correlation among monozygotic twins have suggested a genetic etiology12. Linkage and genome-wide association studies have identified possible loci and gene variants, yet they remain to be validated in larger cohorts1,13–16. External factors, such as contact lens wear, eye rubbing and ultraviolet (UV) light exposure, potentially induce corneal microtrauma and trigger the production of inflammatory mediators17. Subjects with a history of ocular allergy and atopy are also at high risk of developing KC18,19. In addition, epigenetic factors could influence the complex etiology of KC, in particular the association between genome and the environment20. MicroRNA screening on impression cytology samples has shown several down-regulated microRNAs in KC epithelia21. Mutations of miR-184 have been reported to be associated with familial severe KC22,23.
KC pathophysiology is largely defined into: (i) altered stromal composition, e.g. altered collagen and proteoglycan content causing a reduced stromal mass, lamellar slippage and reduced interweaving of collagen24,25; (ii) enzymatic imbalance causing stromal degradation26; (iii) expression of inflammation-related mediators to regulate protease cascades (e.g. tissue plasminogen activator and metalloproteinases, MMPs) leading to stromal matrix changes27–31 and (iv) oxidative stress-induced protein denaturation causing damage to DNA and mitochondrial functions, inducing apoptosis and stromal degradation32–34.
Tear film studies using proteomics and bioinformatics have shown altered proteins belonging to families of proteinases, inflammatory cytokines, cell adhesion molecules, glycoproteins and transporters in samples from KC patients, compared to controls29,30,35–39. The findings of elevated interleukin-6, tumor necrosis factor-α and matrix metalloproteinase-9 (MMP-9) in KC tear samples have highlighted the cytokine imbalance and inflammatory mediators on the ocular surface28,40,41. Studies on aqueous humor protein profiles in KC patients have implicated altered biological processes, including the regulation of proteolysis and response to hypoxia and oxidative stress42. A recent report on saliva proteomics has identified significant hormonal metabolite changes (including IL16, myoinositol and 1-methyl-histidine) associated with pro-inflammatory processes in KC versus healthy control subjects43. However, changes in tear film, aqueous humor and saliva content may not necessarily reflect the intracorneal processes, in particular the structural thinning. Recent studies on isolated KC epithelia and stroma have illustrated altered expression of cytokeratins and cytoskeleton, matrix components and regulatory proteins, suggesting that various degenerative pathways in association with inflammation, changes of innate immune functions, oxidative stress, abnormal mitochondrial functions, and cell death occur in KC25,44–47.
KC severity is related to the degree of topographic deformation (thinning and protrusion) in a localized cone region, of which the apex is often inferiorly decentered from the pupil axis48. Outside this ectatic zone, the non-cone region is topographically normal49. Whether the pathological cellular changes of the corneal epithelium and stroma are only limited to the areas of topographical abnormality and central thinning, or are widespread to the entire cornea is unknown.
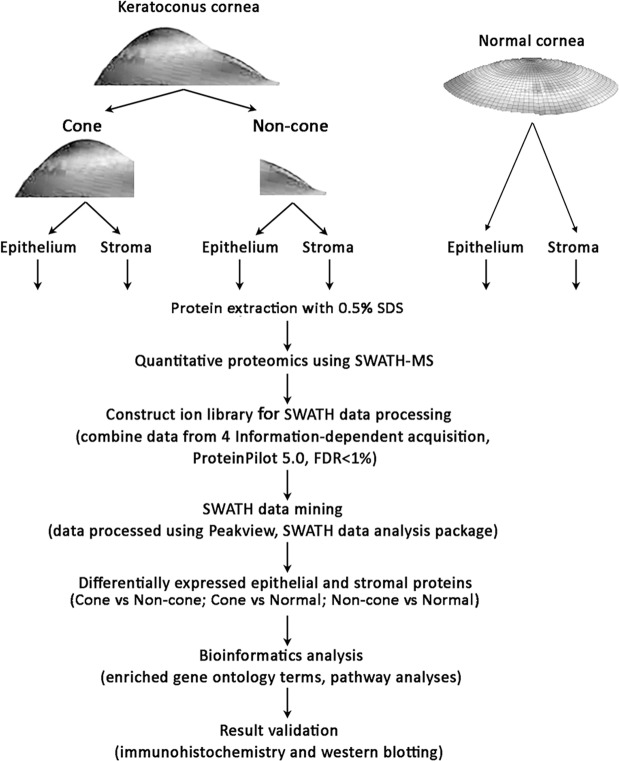
We conducted a comprehensive screening of protein expression in the corneal epithelia and stroma of the respective cone and non-cone regions from four KC patients using a robust high-performance, high-sensitivity, label-free liquid chromatography with tandem mass spectrometry (LC-MS/MS) linked to SWATH (Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra) (SWATH-MS)50,51. In this pilot study, with paired cone versus non-cone comparison and with age-matched normal controls, we identified candidate epithelial and stromal proteins that were differentially expressed in tissues. We examined the enriched gene ontology (GO) terms and putative signalling pathway changes using web-based DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources and Ingenuity Pathway Analysis (IPA, Qiagen). Figure 1 shows the schematic summary of this study. Selected proteins were validated on additional archived KC samples, and compared to corneal scar samples (non-KC cases) and normal corneal tissues, by immunohistochemistry and western blotting.
Figure 1.
Quantitative proteomic workflow of keratoconus cornea (cone and non-cone regions) with separated corneal epithelium and stroma versus normal corneal samples using a SWATH-MS approach.
Results
Clinical analysis of KC patients
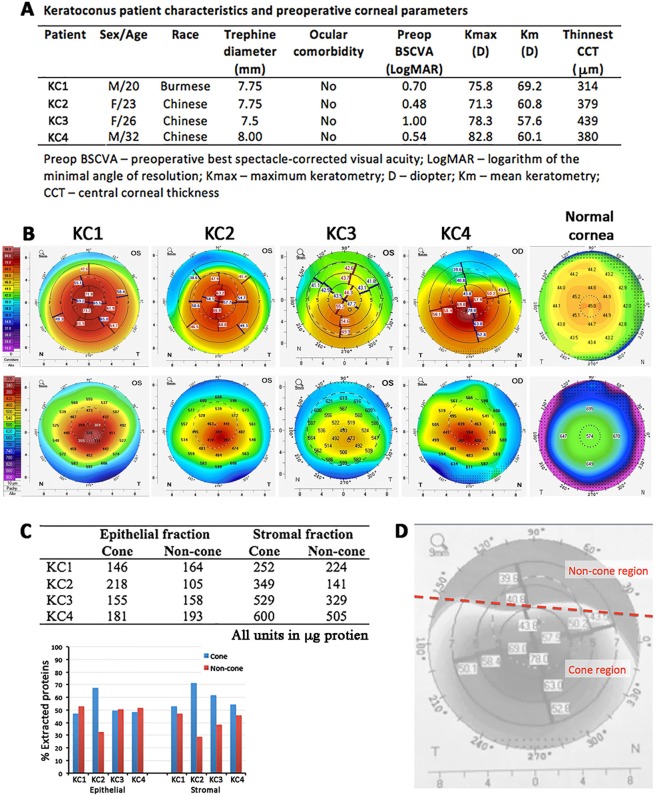
The clinical characteristics of 4 patients with advanced KC are summarized in Fig. 2A. The mean pre-operative best spectacle-corrected visual acuity (BSCVA) was 0.68 ± 0.23 (range 1.0 to 0.48) LogMAR, the mean maximum curvature power (Kmax) was 77.1 ± 4.8 (patient range 71.3 to 82.8 diopters, D, versus normal 42.0 to 46.0 D), the mean overall keratometric values (Km) was 61.9 ± 5.0 D (patient range 57.6 to 69.2 D versus normal 42.0 to 45.0 D) and the mean thinnest central corneal thickness (CCT) was 378 ± 51 μm (patient range 314 to 439 μm versus normal 524 ± 25 μm). The pre-operative topographic evaluation of KC corneas by Pentacam-Scheimpflug imaging (Pentacam, Oculus Inc., Lynnwood, WA, USA) displayed corneal thickness and anterior curvature maps to indicate the position of the thinnest point (cone apex) compared to normal cornea (Fig. 2B).
Figure 2.
Patient information and cornea samples. (A) Patient demographic information and preoperative corneal parameters. (B) Pentacam images of keratoconus corneas displaying anterior curvature (measured in diopters D, top line) and corneal thickness maps (measured in µm, bottom line) to indicate the position of cone areas, compared to normal corneal topography. The numbers from 0 to 8 on the x and y axis as well as the 3, 5 and 7 rings in the top row indicate the diameter in mm from the corneal center/apex. (C) Extracted protein quantities of epithelial and stromal fractions from KC cone and non-cone samples (patient 1 to 4).(D) A schematic diagram showing the separation of cone and non-cone regions according to the pre-operative topographic data.
KC cone and non-cone epithelial and stromal protein samples
SDS-extracted protein fractions of cone epithelium, cone stroma, non-cone epithelium and non-cone stroma from 4 KC corneas and normal epithelium and normal stroma from 2 age-matched control corneas were quantified. Without pooling samples, this preserved the unique protein features of individual KC cornea. Except for KC2 samples, the other KC pairs (KC1, 3, and 4) had similar extracted protein levels from cone and non-cone tissues (48.3 ± 1.2% for cone epithelial and 56.3 ± 4.7% for cone stromal) (Fig. 2C). KC2 had ~2-fold more epithelial and stromal proteins extracted from cone than in non-cone region (Supplemental Fig. S1).
Proteins identified and quantified in KC corneal tissues by SWATH-MS
The experimental scheme is depicted in Fig. 1. A constant of 100 μg protein per sample were loaded to SWATH-MS analysis. A spectral library contained 2980 distinct proteins (1997 epithelial and 983 stromal) identified with >95% confidence and global false discovery rate (FDR) <1%. Among them, 1626 epithelial and 799 stromal proteins were quantifiable (Supplemental Data File S1). There were 587 proteins expressed in both epithelial and stromal samples (Supplemental Fig. S2). The strong concordance of technical replicates was verified by using the standard yeast digest sample (Promega, MA, USA) (R2 = 0.998, from 2 individual runs, Supplemental Fig. S3).
Epithelial proteomes
KC cone versus normal corneal epithelial tissue
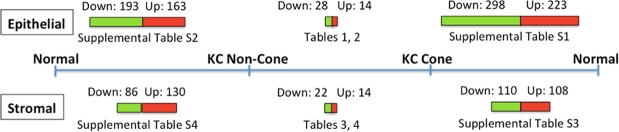
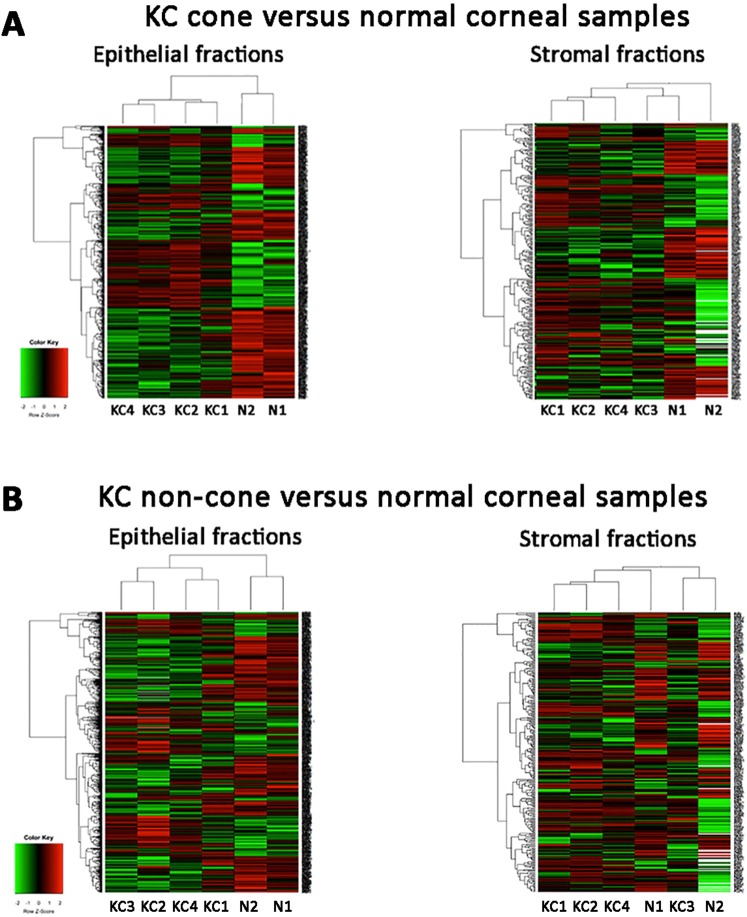
A list of 223 proteins were significantly upregulated (p < 0.05) and 298 significantly downregulated in all 4 KC cone samples compared to normal corneal epithelia (Fig. 3; Supplemental Data File S1 and data of mean fold changes in Supplemental Data File S2A). Similar to other studies comparing entire KC and normal corneas, our data of cone versus normal showed increased epithelial proteins including vimentin, S100A4, KRT12 and 14, decorin, IL18 (interleukin-18); and reduced proteins included transketolase, pyruvate transferase, LOX (lysyl oxidase), ZO-1 and SOD (superoxide dismutase). Heat map analysis showed distinct clustering between KC cone and normal epithelial protein profiles (Fig. 4A). Using DAVID function annotation tools, the enriched GO terms predicted for these altered proteins were significantly associated to cell-cell adhesion (p < 0.001, Benjamini test; enrichment score EnS:20.1), mitochondrial electron transport (p = 0.013; EnS:4.3), nuclear-transcribed mRNA catabolic process, nonsense-mediated decay (p < 0.001; EnS:3.9), cell redox homeostasis (p < 0.001; EnS:3.3), mRNA splicing via spliceosome (p = 0.03; EnS:3.1), gluconeogenesis (p = 0.006; EnS:2.9), and negative regulation of endopeptidase activity (p = 0.03; EnS:2.2) (Supplemental Table S1). The significant KEGG pathways were oxidative phosphorylation (p = 0.002; EnS:5.3), Parkinson’s disease (p = 0.002; EnS:5.3), glycoxylate and dicarboxylate metabolism (p = 0.015; EnS:2.1) (Supplemental Table S1). Similar biological events were replicated using IPA method, including EIF2 signalling (p < 0.001), mitochondrial dysfunction (p < 0.001), oxidative phosphorylation (p < 0.001), caveolar-mediated endocytosis signalling (p < 0.001) and tight junction (p < 0.001). In the physiological and functional category, the affected pathways were (1) protein synthesis, gene expression, RNA post-transcriptional modification; (2) developmental disorders, hereditary disorders and metabolic diseases; (3) RNA molecular transport and trafficking; (4) dermatological disease and conditions and (5) cancer, organismal injury and abnormalities.
Figure 3.
A bar chart summarizing the epithelial and stromal protein changes of KC non-cone versus normal, KC cone versus normal and KC cone versus non-cone samples. The bar lengths are in proportion to number of proteins with significant changes (p < 0.05); red-colored bars represent up-regulation and green-colored bars represent down-regulation. The tables showing the pathway results for each comparison are stated. Note: both normal are the same samples.
Figure 4.
Heat maps of an unsupervised hierarchical clustering of proteins with fold changes (>2 and <0.5). The colors in the map displayed the relative expression values. Green indicated the lowest expression, black for the intermediate expression, and red for the highest expression. The numerical values gave the actual values on a log2 scale, which were associated with each color. The color scale bar is shown at the lower left corner. (A) KC cone epithelium and stroma were compared to normal corneal samples. (B) KC non-cone epithelium and stroma were compared to normal corneal samples.
KC non-cone versus normal corneal epithelial tissue
A total of 163 proteins were significantly upregulated (p < 0.05) and 193 significantly downregulated in all 4 KC non-cone samples compared to normal corneal epithelia (Fig. 3; Supplemental Data File S2B). Heat map showed distinct dendrographic profiles between non-cone versus normal epithelium (Fig. 4B). Using DAVID bioinformatics, the significant enriched GO terms were identified to be cell-cell adhesion (p < 0.001, Benjamini test; enrichment score EnS:17.5), mitochondrial electron transport (p = 0.03; EnS:5.3), gluconeogenesis (p = 0.03; EnS:3.7), translation (p = 0.03; EnS:3.2), response to reactive oxygen species (p < 0.001; EnS:2.7) and actin cytoskeleton (p = 0.046; EnS:2.1) (Supplemental Table S2). The significant KEGG pathways were oxidative phosphorylation (p < 0.001; EnS:5.3), Parkinson’s disease (p < 0.001; EnS:5.3), glycolysis/gluconeogenesis (p < 0.001; EnS:3.7), cysteine and methionine metabolism (p = 0.03; EnS:3.1) and pyruvate metabolism (p = 0.03; EnS:3.1) (Supplemental Table S2). IPA replicated similar biological events, which were mitochondrial dysfunction (p < 0.001), phagosome maturation (p < 0.001), oxidative phosphorylation (p < 0.001), EIF2 signalling (p < 0.001) and glycolysis (p < 0.001). The affected physiological and functional categories included (1) cell morphology, cellular assembly, function and organization; (2) RNA post-transcriptional modification, cell death and survival (3) lipid metabolism; (4) cardiovascular disease, system development and (5) developmental, hereditary and metabolic disorders.
Differential expression of epithelial proteins in KC cone versus non-cone
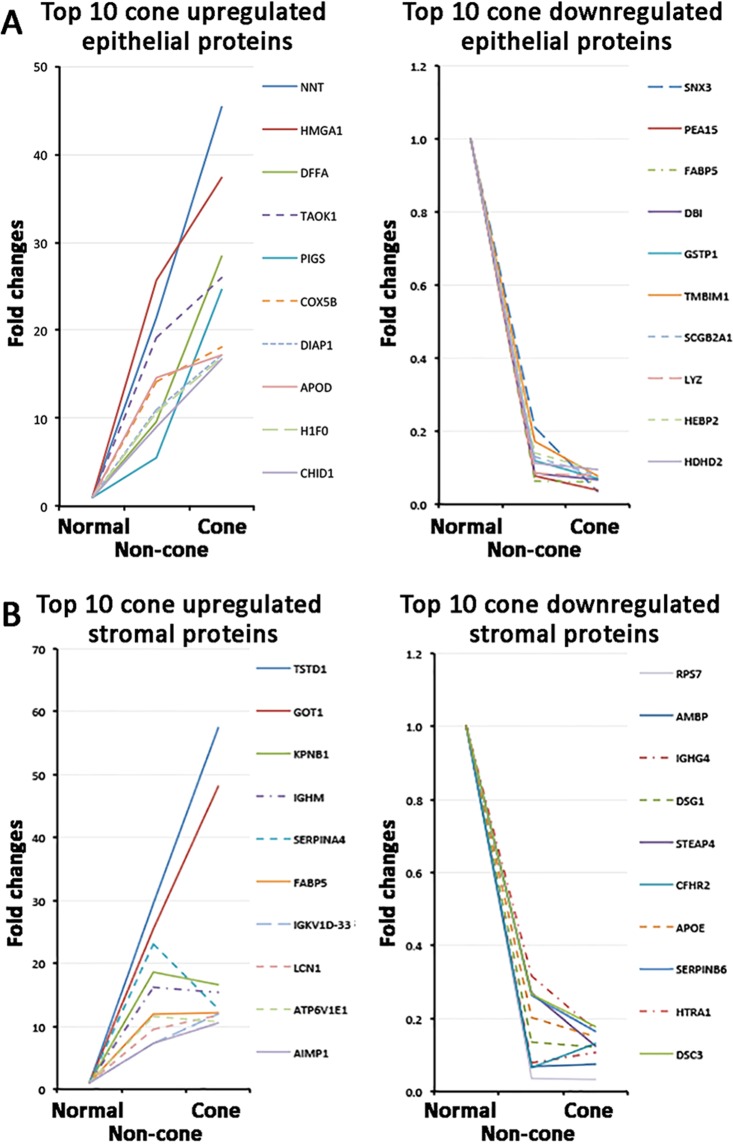
Quantitative protein analysis in separated KC cone and non-cone tissues allowed the spectral examination of selected proteins along the pathogenic development from non-cone to cone stage. In cone epithelial proteome, we selected the top 10 significantly up-regulated proteins compared to normal corneal epithelium, and examined their relative expression in non-cone epithelia. Figure 4A showed these proteins possessed similar trend of elevated expression from normal, through non-cone, to the cone stage. Among them, NNT, DFFA and PIGS showed more expeditious increase (>2 folds) in cone versus non-cone than in non-cone versus normal samples. In contrast, all cone downregulated epithelial proteins were detected low in non-cone samples (Fig. 5B). In view of the differentially regulated expression, a detailed comparison of KC cone versus non-cone epithelial proteomes was conducted. Consistently found in all 4 KC pairs, there were 14 upregulated (>2 folds) and 28 downregulated proteins (<0.5 fold) (Table 1; Fig. 3). Among them, 5 were significantly upregulated (NDUFB11, GAA, DFFA, RPL28, NNT) and 15 significantly downregulated in cone epithelia (VMA5A, IGFBP6, PPP2R5D, SNX3, EIF3G, FH, NPC2, STX4, GLS, PFDN4, ARFIP1, PAFAH1B1, TOMM22, EIF2S2, ADA10) (p < 0.05). These proteins were submitted to the function annotation tool of DAVID bioinformatics, and only one significant enriched GO term was recognized: GO:0005739-mitochondrion (p = 0.048, Benjamini test; enrichment score EnS: 2.4) (Table 2). In addition, IPA identified one biochemical interactive network: developmental disorders, hereditary and metabolic diseases that could associate with the altered protein expression between KC cone and non-cone epithelia (Fig. 6A).
Figure 5.
Spectral changes of top 10 differentially regulated cone epithelial (A) and stromal proteins (B) from normal to non-cone to cone stages.
Table 1.
Differentially expressed KC epithelial protein from pairwise cone versus non-cone comparison.
| UniPro Accession No. | Protein Symbol | Protein name | Mean fold changes | SD | CV | p | |
|---|---|---|---|---|---|---|---|
| ( A ) Upregulated epithelial proteins | |||||||
| 1 | Q9NX14 | NDUFB11 | NADH:Ubiquinone oxidoreductase subunit B11 | 24.26 | 17.06 | 1.50 | 0.015* |
| 2 | P62854 | RS26 | Ribosomal protein S26 | 20.34 | 28.34 | 1.61 | 0.382 |
| 3 | Q9NR31 | SAR1A | Secretion associated Ras related GTPase 1A | 18.73 | 15.85 | 1.07 | 0.340 |
| 4 | P26885 | FKBP2 | FK506 binding protein 2 | 12.35 | 12.28 | 1.21 | 0.506 |
| 5 | P07477 | PRSS1 | Protease, serine 1 | 11.4 | 20.13 | 1.95 | 0.349 |
| 6 | Q6IAA8 | LTOR1 | Late endosomal/lysosomal adaptor, MAPK And MTOR activator 1 | 9.04 | 14.59 | 1.72 | 0.121 |
| 7 | Q9Y3E1 | HDGR3 | Hepatoma-derived growth factor, related protein 3 | 8.57 | 13.00 | 1.70 | 0.247 |
| 8 | Q9UNP9 | PPIE | Peptidylprolyl isomerase E | 7.69 | 6.32 | 1.02 | 0.146 |
| 9 | P10253 | GAA | Glucosidase α, acid | 6.67 | 4.76 | 1.09 | 0.022* |
| 10 | Q96S52 | PIGS | Phosphatidylinositol glycan anchor biosynthesis class S | 4.37 | 3.23 | 0.90 | 0.127 |
| 11 | Q13162 | PRDX4 | Peroxiredoxin 4 | 4.32 | 3.83 | 1.03 | 0.154 |
| 12 | O00273 | DFFA | DNA fragmentation factor subunit α | 3.09 | 1.68 | 0.58 | 0.034* |
| 13 | P46779 | RPL28 | Ribosomal protein L28 | 3.08 | 1.47 | 0.55 | 0.036* |
| 14 | Q13423 | NNT | Nicotinamide nucleotide transhydrogenase | 2.57 | 1.22 | 0.51 | 0.037* |
| ( B ) Downregulated epithelial proteins | |||||||
| 1 | O00534 | VMA5A | Von Willebrand factor A domain containing 5A | 0.2 | 0.02 | 1.00 | 0.020* |
| 2 | O60493 | SNX3 | Sorting nexin 3 | 0.24 | 0.12 | 0.64 | 0.007* |
| 3 | P07954 | FH | Fumarate hydratase | 0.24 | 0.11 | 0.42 | 0.002* |
| 4 | Q14738 | PPP2R5D | Protein phosphatase 2 regulatory subunit B′delta | 0.24 | 0.37 | 1.14 | 0.050* |
| 5 | Q5T4S7 | UBR4 | Ubiquitin protein ligase E3 component N-Recognin 4 | 0.25 | 0.18 | 1.87 | 0.304 |
| 6 | P24592 | IGFBP6 | Insulin-like growth factor binding protein 6 | 0.28 | 0.03 | 1.08 | 0.033* |
| 7 | O75821 | EIF3G | Eukaryotic translation initiation factor 3G | 0.3 | 0.2 | 0.73 | 0.013* |
| 8 | O94925 | GLS | Glutaminase | 0.31 | 0.13 | 0.55 | 0.019* |
| 9 | O00217 | NDUS8 | NADH:Ubiquinone oxido reductase core subunit S8 | 0.34 | 0.13 | 0.99 | 0.091 |
| 10 | P23800 | LOX | Lysyl oxidase | 0.37 | 0.29 | 1.51 | 0.321 |
| 11 | P14324 | FPPS | Farnesyl diphosphate synthase | 0.37 | 0.24 | 0.6 | 0.153 |
| 12 | P43487 | RANG | RAN binding protein 1 | 0.37 | 0.35 | 1.11 | 0.383 |
| 13 | P61916 | NPC2 | NPC intracellular cholesterol transporter 2 | 0.38 | 0.12 | 0.94 | 0.023* |
| 14 | Q9NQP4 | PFDN4 | Prefoldin subunit 4 | 0.39 | 0.12 | 0.41 | 0.006* |
| 15 | Q12846 | STX4 | Syntaxin 4 | 0.41 | 0.24 | 0.61 | 0.027* |
| 16 | P07204 | TRBM | Thrombomodulin | 0.41 | 0.19 | 0.63 | 0.082 |
| 17 | O60936 | NOL3 | Nucleolar protein 3 | 0.43 | 0.39 | 0.75 | 0.066 |
| 18 | P53367 | ARFIP1 | ADP ribosylation factor interacting protein 1 | 0.43 | 0.17 | 0.37 | 0.015* |
| 19 | P43034 | PAFAH1B1 | Platelet activating factor acetylhydrolase 1b regulatory subunit 1 | 0.44 | 0.09 | 0.18 | 0.001* |
| 20 | O15305 | PMM2 | Phosphomannomutase 2 | 0.45 | 0.33 | 0.66 | 0.067 |
| 21 | P20042 | EIF2S2 | Eukaryotic translation initiation factor 2β | 0.45 | 0.17 | 0.35 | 0.015* |
| 22 | P41222 | PTGDS | Prostaglandin D2 synthase | 0.45 | 0.42 | 0.96 | 0.195 |
| 23 | Q96H20 | SNF8 | SNF8, ESCRT-II complex subunit | 0.46 | 0.46 | 0.84 | 0.177 |
| 24 | Q96I99 | SUCB2 | Succinate-CoA ligase GDP-forming β | 0.47 | 0.42 | 0.89 | 0.066 |
| 25 | P01033 | TIMP1 | TIMP1 metallopeptidase inhibitor 1 | 0.48 | 0.32 | 0.59 | 0.052 |
| 26 | O14672 | ADAM10 | ADAM metallo peptidase domain 10 | 0.49 | 0.06 | 0.12 | 0.000* |
| 27 | Q9NS69 | TOMM22 | Translocase of outer mitochondrial membrane 22 | 0.49 | 0.21 | 0.38 | 0.015* |
| 28 | P46782 | RS5 | Ribosomal protein S5 | 0.49 | 0.36 | 0.86 | 0.054 |
These proteins had same changes in all 4 KC sample pairs. Note: Mean fold changes from all 4 KC sample pairs; SD – standard deviation; CV: coefficient of variation; p: significance value (paired Student’s t-test; *p < 0.05 denotes statistically significance).
Table 2.
Enriched Gene Ontology terms and KEGG pathways identified for differentially expressed proteins between KC cone and non-cone epithelial samples.
| Enrichment score | Biological events | Proteins (UniPro Accession no.) | p | |
|---|---|---|---|---|
| ( A ) Enriched GO terms ( DAVID ) | ||||
| 1 | 2.36 | GO:0005739 ~ mitochondrion | P14324, Q9NS69, O94925, Q9NX14, Q96I99, P07954, Q9NQP4, O00217, O60936, Q13162, Q13423 | 0.048* |
| 2 | 1.36 | GO:0003723 ~ RNA binding | Q9UNP9, P46779, O75821, O60936, P46782, P20042 | 0.479 |
| ( B ) KEGG pathways ( DAVID ) | ||||
| 1 | 2.36 | hsa01100: Metabolic pathways | O94925, Q9NX14, P41222, P43034, O00217, P14324, P54819, P10253, Q96I99, P07954, O15305, Q13126, Q13423 | 0.321 |
The biological events were ranked using enrichment scores. *Adjusted Benjamini p < 0.05 represents statistical significance.
Figure 6.
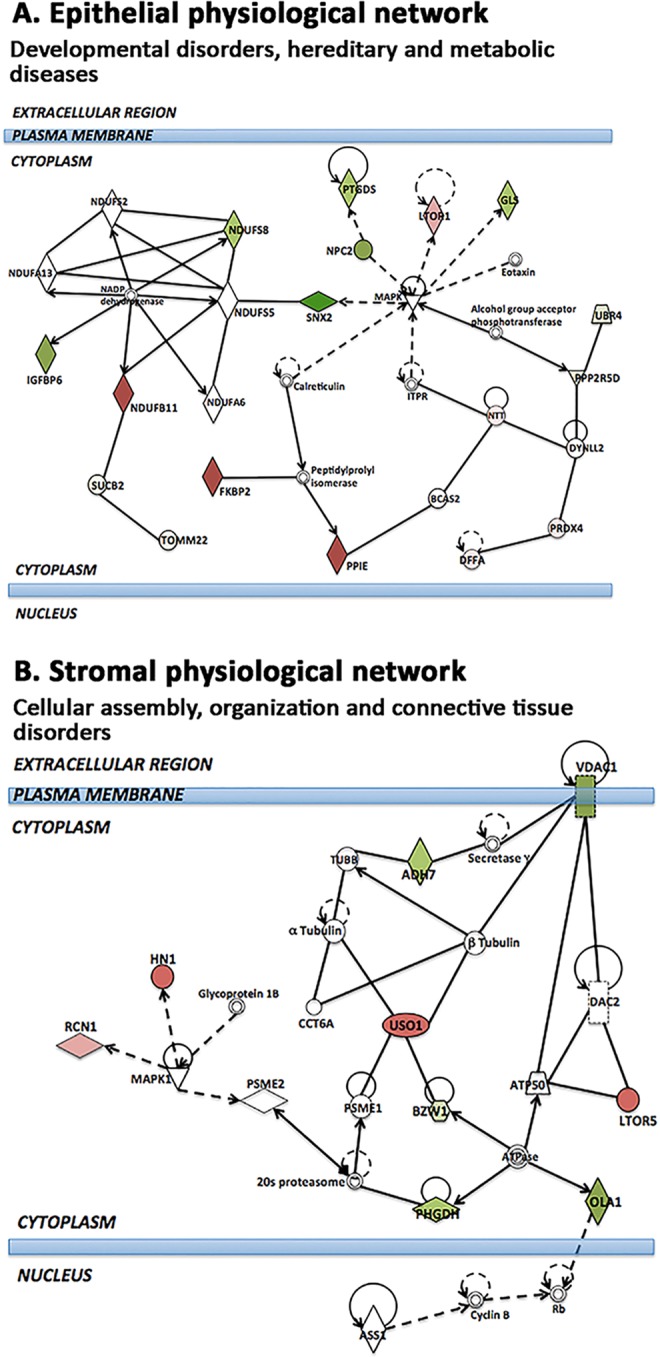
IPA interactive networks identified between KC cone and non-cone comparison. (A) Epithelial-derived interactome: developmental disorders, hereditary and metabolic diseases. This network shows the epithelial dysfunction due to MAPK signalling-associated changes in cell metabolism (reduced mitochondrial GLS and cell growth-related PPP2R5D, UBR4, IGFBP6, SNX2). The up-regulated PRDX4, NDUFB11 and FKBP2 suggest potential immune-related changes. (B) Stromal-derived interactome: cellular assembly, tissue organization and connective tissue disorders. The up-regulated microtubule protein HN1, vesicle transporter USO1 and LTOR5 as well as calcium binding RCN1, and the downregulated voltage channel VDAC1 on cell membrane and cell survival-associated ADH7, BZW1 (histone regulation), PHGDH (electron transfer in ATP synthesis) and calcium responsive OLA1 suggest perturbation of cell survival and interstitial ECM. Each network displays the genes/gene products as nodes (different shapes representing the functional classes of gene products) and the biological relationships between the nodes as lines. The length of each line reflects the amount of literature evidence supporting this node-to-node relationship. The color intensity of each node indicates the degree of upregulation (red) or downregulation (green) of the respective gene transcript. Genes in white shape are predictive to interact with the colored gene products that appear in this scheme.
Stromal proteomes
KC cone versus normal corneal stromal tissue
A list of 108 proteins were significantly upregulated (p < 0.05) and 110 significantly downregulated in all 4 KC cone samples, when compared to normal corneal stroma (Fig. 3; Supplemental Data File S2C; raw data in Supplemental Data File S1). They included reported proteins like Col1A2, MMP9 and SOD. Heat map showed distinct dendrographic clustering in cone versus normal stromal proteomes (Fig. 4A). Using DAVID, these proteins were associated to significant enriched GO terms: cell-cell adhesion (p < 0.001; EnS:10.7), extracellular region (p < 0.001, EnS:7.9), protein folding (p < 0.001, EnS:5.4), classical complement pathway (p < 0.001, EnS 5.1), Fc ε receptor signalling pathway (p < 0.001, EnS:5.1), GTP binding (p = 0.035, EnS:3), intermediate filament (p = 0.011, EnS:2.8), negative regulation of endopeptidase activity (p < 0.001, EnS:2.7) and response to ROS (p = 0.012, EnS:2.4) (Supplemental Table S3). It also revealed 2 significant KEGG pathways: hsa04610: complement and coagulation cascades (p = 0.006, EnS:2.9) and hsa00010:glycolysis/gluconeogenesis (p = 0.046, EnS:2). IPA showed biological pathways including LXR/RXR activation (p < 0.001), acute phase response signalling (p < 0.001), complement system (p < 0.001), FXR/RXR activation (p < 0.001) and glycolysis (p < 0.001). In the physiological and disease category, the KC cone stromal proteome affected cell death and survival, dermatological disease and conditions, inflammatory disease and response.
KC non-cone versus normal corneal stromal tissue
We detected 130 proteins significantly upregulated (p < 0.05) and 86 significantly downregulated in all 4 KC non-cone proteomes compared to that of normal corneal stroma (Fig. 3; Supplemental Data File S2D). There was no clear clustering between non-cone and normal stromal proteomes as revealed by heat map analysis (Fig. 4B). The function annotation of DAVID showed these proteins were related to significant enriched GO terms, namely cell-cell adhesion (p < 0.001, EnS:9.3), classical complement activation pathway (p < 0.001, EnS:6.5), Fc γ-receptor signalling pathway involved in phagocytosis (p < 0.001, EnS:6.5), immune response (p = 0.01, EnS:6.5), extracellular region (p < 0.001, EnS:5.6), oxidoreductase activity (p < 0.001, EnS:3.4), gluconeogenesis (p = 0.016, EnS:3.4), immunoglobulin receptor binding (p = 0.001, EnS:3.1), unfolded protein binding (p = 0.007, EnS:2.9), protease binding (p = 0.021, EnS:2.8) and collagen catabolic process (p = 0.003, EnS:2.4) (Supplemental Table S4). Two significant KEGG pathways were noted - hsa00010: glycolysis/gluconeogenesis (p = 0.007; EnS:2.5) and hsa04512:ECM-receptor interaction (p = 0.023; ErS:2.4) (Supplemental Table S4). Similar biological events were revealed using IPA and the top 5 canonical pathways were LXR/RXR activation, FXR/RXR activation, acute phase response signalling, complement system and glycolysis. The affected physiological and functional category were (1) cell-to-cell signalling; (2) inflammatory response and abnormalities (3) haematological disorders; (4) dermatological diseases, inflammatory disorders and response and (5) tissue infection and inflammation.
Differential expression of stromal proteins in KC cone versus non-cone
The top 10 significantly up- and down-regulated cone stromal proteins compared to normal stroma were also altered in non-cone samples. We observed 2 cone upregulated proteins (TSTD1, GOT1) had a nearly linear elevation from normal, through non-cone, to the cone stage, but the fold changes were <2 (Fig. 5B). Others had minor fluctuations between cone and non-cone samples. On the other hand, all cone downregulated stromal proteins were also detected low in non-cone samples (Fig. 5B). When KC cone and non-cone stromal proteomes were compared in pair-wise manner, there were 14 upregulated and 22 downregulated proteins in all 4 pairs of KC pairs (Table 3; Fig. 3). These included the KC reported protein, LOX, which was more suppressed in cone than non-cone stroma (0.5 ± 0.2 fold). Two proteins (SYTC, RUVBL2) were significantly upregulated and 12 were significantly downregulated (TACSTD2, VIT, STEAP4, SAR1A, PHDGH, ABCB11, SF3B3, H2AJ, MIF, NQO1, BZW1, ADH7) in cone stroma. By DAVID function annotation, the proteome changes between cone and non-cone stroma did not associate with any significant enriched GO terms (Table 4). However, one KEGG pathway was significantly noted (hsa04141: protein processing in endoplasmic reticulum, p = 0.008, Benjamini test; enrichment score EnS:1.8). IPA identified one putative biochemical network: cellular assembly, tissue organization and connective tissue disorders (Fig. 6B).
Table 3.
Differentially expressed KC stromal protein from pairwise cone versus non-cone comparison.
| UniPro Accession No. | Protein Symbol | Protein name | Mean fold changes | SD | CV | p | |
|---|---|---|---|---|---|---|---|
| ( A ) Upregulated stromal proteins | |||||||
| 1 | P26639 | SYTC | Threonyl-tRNA synthetase | 12.22 | 6.32 | 2.75 | 0.021* |
| 2 | Q9Y230 | RUVBL2 | RuvB like AAA ATPase 2 | 11.56 | 5.18 | 1.75 | 0.025* |
| 3 | P52597 | HNRPF | Heterogeneous nuclear ribonucleoprotein F | 8.72 | 8.77 | 1.01 | 0.156 |
| 4 | Q15293 | RCN1 | Reticulocalbin 1 | 8.38 | 5.82 | 0.92 | 0.185 |
| 5 | Q04828 | AK1C1 | Aldo-keto reductase family 1 member C1 | 7.79 | 5.32 | 1.63 | 0.214 |
| 6 | O43504 | LTOR5 | Late endosomal/lysosomal adaptor, MAPK And MTOR activator 5 | 6.71 | 4.22 | 1.08 | 0.130 |
| 7 | P27169 | PON1 | Paraoxonase 1 | 4.94 | 3.81 | 0.77 | 0.230 |
| 8 | P01714 | IGLV3-19 | Immunoglobulin lambda variable 3-19 | 4.83 | 7.06 | 1.46 | 0.250 |
| 9 | O43396 | TXNL1 | Thioredoxin like 1 | 4.29 | 2.85 | 0.88 | 0.292 |
| 10 | Q9UK76 | HN1 | Hematological and neurological expressed 1 | 3.75 | 3.28 | 0.99 | 0.109 |
| 11 | P01602 | IGKV1-5 | Immunoglobulin kappa variable 1–5 | 3.17 | 2.03 | 0.64 | 0.056 |
| 12 | P0CG06 | LAC3 | Immunoglobulin lambda constant 3 (Kern-Oz + Marker) | 3.02 | 2.17 | 0.79 | 0.091 |
| 13 | O60763 | USO1 | USO1 vesicle transport factor | 2.84 | 2.33 | 0.82 | 0.093 |
| 14 | P14780 | MMP9 | Matrix metalloproteinase 9 | 2.38 | 0.97 | 0.41 | 0.251 |
| ( B ) Downregulated stromal proteins | |||||||
| 1 | P09758 | TACSTD2 | Tumor-associated calcium signal transducer 2 | 0.32 | 0.14 | 0.55 | 0.006* |
| 2 | P34896 | SHMT1 | Serine hydroxymethyl transferase 1 | 0.32 | 0.31 | 0.98 | 0.088 |
| 3 | Q9NTK5 | OLA1 | Obg like ATPase 1 | 0.32 | 0.28 | 0.88 | 0.053 |
| 4 | Q9H0W9 | C11orf54 | Chromosome 11 open reading frame 54 | 0.34 | 0.32 | 0.93 | 0.092 |
| 5 | Q6UXI7 | VIT | Vitrin | 0.35 | 0.09 | 0.55 | 0.047* |
| 6 | P11166 | SLC2A1 | Solute carrier family 2 member 1 | 0.39 | 0.42 | 1.06 | 0.131 |
| 7 | O95342 | ABCB11 | ATP binding cassette subfamily B11 | 0.4 | 0.14 | 0.36 | 0.017* |
| 8 | Q9NR31 | SAR1A | Secretion associated Ras related GTPase 1A | 0.4 | 0.23 | 0.58 | 0.046* |
| 9 | P15559 | NQO1 | NAD(P)H quinone dehydrogenase 1 | 0.42 | 0.14 | 0.50 | 0.021* |
| 10 | Q9BTM1 | H2AJ | H2A histone family member J | 0.42 | 0.15 | 0.35 | 0.016* |
| 11 | Q687X5 | STEAP4 | STEAP4 metalloreductase | 0.44 | 0.12 | 0.74 | 0.041* |
| 12 | P14174 | MIF | Macrophage migration inhibitory factor | 0.44 | 0.18 | 0.41 | 0.025* |
| 13 | Q15393 | SF3B3 | Splicing Factor 3b Subunit 3 | 0.44 | 0.25 | 0.56 | 0.042* |
| 14 | P18510 | IL1RN | Interleukin 1 receptor agonist | 0.44 | 0.36 | 0.81 | 0.068 |
| 15 | P62318 | SMD3 | Small nuclear ribonucleoprotein D3 polypeptide | 0.46 | 0.37 | 0.82 | 0.171 |
| 16 | O43175 | PHGDH | Phosphoglycerate dehydrogenase | 0.47 | 0.25 | 0.70 | 0.035* |
| 17 | Q7L1Q6 | BZW1 | Basic leucine zipper and W2 domain 1 | 0.47 | 0.23 | 0.48 | 0.047* |
| 18 | Q9BTV4 | TMM43 | Transmembrane protein 43 | 0.48 | 0.35 | 0.73 | 0.091 |
| 19 | P55769 | NH2L1 | SNU13 homolog, small nuclear ribonucleoprotein (U4/U6.U5) | 0.48 | 0.34 | 0.59 | 0.086 |
| 20 | P40394 | ADH7 | Alcohol dehydrogenase 7 (Class IV), Mu Or Sigma polypeptide | 0.49 | 0.01 | 0.35 | 0.016* |
| 21 | P23800 | LOX | Lysyl oxidase | 0.49 | 0.24 | 1.88 | 0.276 |
| 22 | P21796 | VDAC1 | Voltage dependent anion channel 1 | 0.49 | 0.36 | 0.72 | 0.217 |
These proteins had same changes in all 4 KC sample pairs. Note: Mean fold changes from all 4 KC sample pairs; SD – standard deviation; CV: coefficient of variation; p: significance value (paired Student’s t-test; *p < 0.05 denotes statistical significance).
Table 4.
Enriched Gene Ontology terms and KEGG pathways identified for differentially expressed stromal proteins in KC cone and non-cone comparison.
| Enrichment score | GO terms | Proteins (UniPro Accession no.) | p | |
|---|---|---|---|---|
| ( A ) Enriched GO terms ( DAVID ) | ||||
| 1 | 2.26 | GO:0098609 ~ cell-cell adhesion | O60763, Q7L1Q6, P61026, P09758, P14618, Q9NTK5 | 0.342 |
| 2 | 1.85 | GO:0055114 ~ Oxidation-reduction | O43396, P40394, Q687X5, P15559, O43175, Q04828 | 0.785 |
| 3 | 1.73 | GO:0005739 ~ Mitochondrion | O43396, P34896, P51572, P14618, P50213, P21796 | 0.956 |
| 4 | 1.34 | GO:0005525 ~ GTP binding | Q9NR31, Q9NVJ2, P61026, Q9NTK5, P61224 | 0.454 |
| 5 | 1.2 | GO:0000398 ~ mRNA splicing, via spliceosome | P52597, P55769, P62318, Q15393 | 0.816 |
| 6 | 1.15 | GO:0038096 ~ Fc-gamma receptor signaling pathway involved in phagocytosis | O15145, P01714, P01602, P0CG06 | 0.676 |
| 7 | 1.15 | GO:0038095 ~ Fc-epsilon receptor signaling pathway | P62837, P01714, P01602, P0CG06 | 0.848 |
| 8 | 1.15 | GO:0006958 ~ Complement activation, classical pathway | P01714, P01602, P0CG06 | 0.759 |
| 9 | 1.04 | GO:0006886 ~ Intracellular protein transport | Q9NR31, O60763, Q9UNH7, P51572 | 0.774 |
| ( B ) KEGG pathways ( DAVID ) | ||||
| 1 | 1.77 | hsa04141: Protein processing in endoplasmic reticulum | Q9NR31, P62837, P14314, P39656, P04843, P51572, P05198 | 0.008* |
| 2 | 1.73 | hsa01100: Metabolic pathways | P34896, P14618, P50213, O43175 | 0.331 |
| 3 | 1.2 | hsa03040: Spliceosomes | P55769, P62318, Q15393 | 0.883 |
The pathways were ranked using enrichment scores. *Adjusted Benjamini p < 0.05 represents statistical significance.
Tissue expression of selected proteins
We selected 4 proteins for validation study using a different cohort of 4 advanced KC corneas compared to 4 non-KC samples and 2 normal corneal samples (Supplemental Table S5, representative hematoxylin-eosin pictures in Fig. 7A). These proteins were chosen based on their involvement in mitochondria (for NDUFB11), protein trafficking (for STEAP4), immune and inflammatory response (for IL1RN), and response to oxidative stress (for ADH7) (more explanation in Discussion) as well as the availability of high quality antibodies for immunohistochemistry on paraffin sections (Supplemental Table S6).
Figure 7.
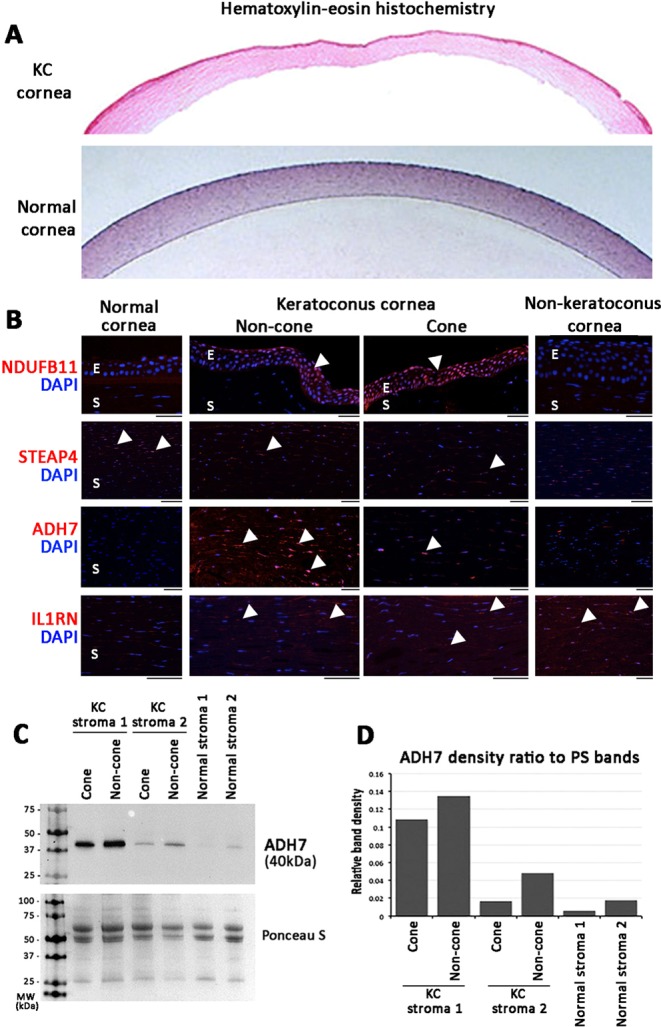
Immunohistochemistry showing the tissue expression of selected proteins in archived normal, KC and non-KC corneal specimens. (A) Hematoxylin-eosin images of KC and normal corneas. (B) Strong nuclear signal (arrowheads) of NDUFB11 was detected in KC epithelia (labeled E) but faint in non-KC and normal cornea. STEAP4 metalloproteinase was faintly stained in KC stroma (labelled S) than in normal corneal stroma. ADH7 was clearly detected in KC non-cone stroma (arrowheads), and was less in cone stroma. Negligible expression was found in normal and non-KC corneal stroma. IL1RN was mildly expressed in KC and non-KC stroma but faint in normal cornea. (C) Western blotting of ADH7 in KC and normal stromal fractions. (D) Band densitometry showed ADH7 was upregulated in KC non-cone than in cone stromal fractions. Normal corneal stromal samples had reduced expression. Full-length gel images are shown in Supplemental Fig. S4.
The KC-upregulated NDUFB11 (NADH:ubiquinone oxidoreductase subunit B11; UniPro Q9NX14) was positively stained in corneal epithelia in 3 of 4 KC specimens and was negative in normal cornea (0/2) and non-KC samples (0/4) (Fig. 7B). Similarly, ADH7 (alcohol dehydrogenase 7; P40394) was positively stained in KC samples (4/4), particularly in non-cone region and negligibly in non-KC and normal corneas (Fig. 7B). This was consistent to the proteomic data (KC cone/normal: 6.5 ± 2.6 folds; KC non-cone/normal: 11.4 ± 6.3 folds; KC cone/non-cone: 0.49 ± 0.08 fold, p = 0.016) (Table 3). Using western blotting, ADH7 was revealed at ~40 kDa position and expressed stronger in non-cone stromal samples than in cone samples (Fig. 7C). Analyzed by band densitometry, ADH7 expression was elevated in KC non-cone by 27% to 2.8 folds, when compared to the corresponding cone stroma (Fig. 7D). On the contrary, STEAP4 metalloproteinase (UniPro Q687X5) was faintly stained in KC stroma (1/4) than in normal corneal stroma (2/2) (Fig. 7B). This result matched to the stromal proteomic data that STEAP4 was suppressed in KC cone and non-cone stroma (Table 3). IL1RN (Interleukin 1 receptor agonist; P18510) was detected in KC cone and non-cone stroma as well as in non-KC samples, however the normal corneal stroma showed negligible staining (Fig. 7B).
Discussion
This study investigated the epithelial and stromal proteomes of 2 topographically divergent regions of KC cornea, i.e. cone with thin ectatic topography and non-cone with normal topography, using label-free SWATH-MS. This is the first proteome study that compared variations in cone and non-cone tissues of human KC corneas. Previous studies have examined whole KC epithelial or stromal tissue in comparison to normal corneas to obtain KC-associated expression patterns. However, little is known about the biological variations in these 2 regions and whether the pathological biological changes are restricted to the ectatically abnormal cone or extends to the rest of cornea. In this study, we reported proteins with expression changes (in the range of ≥2 folds or ≤0.5 fold) consistently found in all 4 KC samples. Our results showed that the proteome changes were not limited to the topographically thinner and mechanically weakened cone region but also in the non-cone part, indicating a peripheral involvement in the pathogenic KC development. Using DAVID functional annotation to identify enriched GO terms and IPA to depict biochemical interactive networks, the epithelial proteome change between cone and non-cone was related to cell metabolism with mitochondrial involvement, whereas the stromal alterations were linked to cellular assembly and tissue organization. Using immunohistochemistry and western blotting, four selected proteins with reported activities on mitochondria, oxidative stress, immune and inflammatory responses were validated to follow the same trend of changes as seen in our proteomic study.
KC pathogenesis has been known to be complex and a variety of cellular and extracellular processes are involved in the corneal ectatic changes and focal thinning1,11. In the past, KC was defined as a non-inflammatory corneal degenerative disorder due to the lack of cellular infiltration and neovascularization10. However, increasing evidence has contradicted this theory and implicated local inflammatory and immune responses as well as oxidative stress as the potential triggering factors29,30,35,41. This is consistent with our data, which showed the involvement of immune pathways in both cone and non-cone stroma, such as Fc γ- and ε-receptor signalling as well as complement cascade. In addition, the enzymatic imbalance, including the upregulation of MMPs and reduced LOX activity, affects collagen polymerization and its crosslinking with elastin fibers52,53. The disorientation of stromal collagen pattern, in addition to the metabolic changes, altered cell-cell interactions and cell death, would reduce the biomechanical strength of stroma and its stability. Measurement of stress-strain curves showed a reduced elastic modulus in advanced KC corneas54. This biomechanical weakening will drive the topographical changes of focal distortion and thinning55.
The topographic deformation is localized to the cone region. Using Brillouin frequency shift microscopy, the cone region showed mechanically weakening with a lower elastic modulus, while the peripheral non-cone had similar modulus to the normal corneas56. However, this biomechanical data did not show if the biological changes are restricted to the cone or extended to the rest of cornea. Our data has located target protein changes in cone stroma, including the reduction of Col1A1 and 2 (both ~0.3 fold as control), Col6A1 to 3, DSC3, FMOD, BGN, TSP4 and TIMPs, and increase of serpin family members (A4, A5, F2) and MMPs, which were indicative of stromal matrix dysregulation (Supplemental Data File S2C). This would affect collagen fibril orientation and interweaving and increased fibril degradation resulting in loss of stromal matrix stabilization and stromal mass57. In contrast, the expression of Col1A1 and 2 (both ~0.5 fold as control), BGN and TIMP2 were less reduced in non-cone stroma. This might cause insignificant stromal alterations and thinning in non-cone region55,56.
Our data also showed the upregulation of various immune-related regulators and mediators in cone stroma (such as IGKV1D-33, 2D-28, IL18, C2, C4A, C9, CFD, CFH, TF, TOLIP) and this predicted the classical and alternative complement cascades, which could activate collagenases and metalloproteinases29,31 and trigger the prostaglandin pathway (PTGDS and PTGS2) in inflammatory responses. Cell metabolic dysregulation in cone stroma was suggested by the altered expression of ribosomal proteins (RPS7, RPL11, RPS21, RLAO), IGFBP, ASPH, YWHAZ, histone family (HIST2H2BE, H3F3A) and interleukin 1 receptor antagonist IL1RN (Supplemental Data File S2C). Our validation study using archived KC samples showed that IL1RN was detected in KC cone and non-cone stroma but negligible in normal stroma by immunohistochemistry (Fig. 7B). This IL1 cytokine family member regulates the activities of IL1α and β and modulates IL1-related immune and inflammatory responses. Earlier report by Nowak et al. had shown a substitution c.214 + 242C > T of IL1RN in an Ecuadorian KC family and proposed the altered interaction between IL1 and its cell surface receptor in KC etiology58. Moreover, downregulation of retinol binding protein RBP3, integrin ITGA6 and lamin LMNB would suppress cell growth and cell-cell interactions among stromal keratocytes resulting in reduced stromal homeostasis.
This differential protein expression was further inspected by the spectral changes for the top 10 cone upregulated stromal proteins in non-cone region (Fig. 5B). We observed 2 types of changes. Type 1 with linear elevation from normal via non-cone to cone stage was found for TSTD1 and GOT1, which were the top upregulated proteins in cone stroma. TSTD1 (thiosulfate sulfurtransferase like domain containing 1) is a mitochondrial complex I enzyme for ATP production under hypoxia condition59 and relates to prostaglandin metabolism in oxidative stress-mediated cell death60. GOT1 is also a mitochondrial enzyme for ATP production via tricarboxylic acid cycle. Using L-cysteine as substrate, it regulates the level of mercaptopyruvate in producing hydrogen sulfide, which acts as a synaptic modulator61 and a substrate of sulfurtransferase TSTD1. Whether this relates to the cell metabolic changes and loss of corneal sensitivity in KC cornea is yet to be illustrated. The remaining proteins belonged to Type 2, which had elevation from normal to non-cone but stable or decrease in cone stage. SERPINA4 (serine proteinase inhibitor 4, kallistatin), an inflammation and oxidative stress mediator, has been found to suppress oxidative stress-associated NAD(P)H oxidase expression62. IGHM, IGKV1D-33 and LCN1 are related to inflammatory responses; ATP6V1E1 and FABP5 for mitochondrial ATP production; and AIMP1, KPNB1 for cellular metabolism.
Proteins that were altered in non-cone stromal proteome (compared to normal) were involved in similar biological pathways as in cone stroma, such as cell-cell adhesion, complement cascade, immune responses and oxidoreductase activity (Supplemental Table S4). This illustrated that considerable biological events have occurred in non-cone stroma even though it is topographically similar to normal. Since many stromal proteins are structural and their turnover is slow63, the pathological changes may remain undetectable until additional triggering factors are combined. This could be related to protein folding, Fc receptor activation, ROS responses and collagen degradation, which were identified for cone stromal proteome changes (Supplemental Table S3). Activation of Fc γ and ε-receptors could trigger the release of potent inflammatory mediators, which stimulate proteases30,64,65. Increased cell death due to oxidative stress-related protein misfolding and mistrafficking as well as collagen degradation would lead to the stromal matrix degeneration as a major KC process. This was validated when we compared KC cone and non-cone stroma in a pairwise manner. IPA identified the cellular assembly and tissue organization as the altered functional pathway. The altered protein expression in all 4 KC samples underscored cytoskeletal regulation, such as the increased microtubule protein HN1, vesicle transporter USO1 and LTOR5 as well as calcium binding RCN1, and the downregulated voltage channel VDAC1 on cell membrane and cell survival-associated STEAP4 (mitochondrial), ADH7 (defence against oxidative stress), BZW1 (histone H4 regulation), PHGDH (electron transfer in ATP synthesis) and calcium responsive OLA1 (Supplemental Table S7). The results were validated using immunohistochemistry and western blotting. Stronger ADH7 expression was found in KC non-cone than in cone stroma. Besides its anti-oxidation activity, ADH7 participates in retinoic acid (RA) synthesis and retinoid X receptor (RXR) signalling66 that could regulate stromal cell survival and matrix homeostasis. Loss of RA synthesis has been shown to reduce corneal stromal thickness and increase apoptosis, resulting in corneal thinning67. Additionally, the suppressed STEAP4 metalloproteinase could indicate mitochondrial damage and oxidative stress, affecting stromal keratocyte function and viability68. We also demonstrated that while some proteins followed linear expression change from normal over non-cone to cone stage, others did not. Hence, in addition to the theory of stromal degeneration in association to inflammation, immune deregulation, oxidative stress and cell death in KC development25,44–47, our data provided new insights showing that stromal cell metabolic changes, viability and collagen degradation would contribute to the focal degeneration and topographic deformation in the cone stromal region.
Though many researchers have proposed that abnormal stromal metabolism is the primary site of corneal dysfunction in KC, various histopathological studies have highlighted the epithelial abnormalities and postulated that insults to the epithelium cause a release of proteolytic enzymes (such as MMP9, IL6 and LOX) to degrade the stromal collagen and matrix components1,53,69,70. In vivo confocal imaging studies further described that the corneal epithelium was affected in correlation to the KC severity71–73. Altered gene expression related to Wnt, Hedgehog and Notch signalling in the entire KC epithelium was also demonstrated by a recent transcriptomic study47. However the initial changes of epithelium in KC progression and in the 2 topographically different regions remain unclear. Our proteome analysis had revealed altered protein expression in KC non-cone epithelium and was related to cell-cell adhesion, mitochondrial electron transport, ATP synthesis, ROS responses and cytoskeleton. Hence, considerable biological changes have occurred in the non-cone epithelium. Additional changes of RNA metabolism in the cone epithelium are linked to the altered protein synthesis, cellular stress and cell death. Cell stress-related expression of cytokines and mediators could activate proteolytic enzymes to degrade collagen matrix and weaken the cornea. Abnormal autophagy and apoptosis as a consequence of oxidative stress has been reported in KC epithelia74, leading to the reduction of epithelial cell number and thinner apical epithelium. When comparing between KC cone and non-cone epithelial proteomes, the changes were associated to cell metabolism as the enriched GO term and mitochondria as the KEGG pathway (Table 2). This was supported by IPA physiological network, which showed developmental disorders and metabolic diseases. In addition, the altered epithelial proteome changes encoded cell growth and immune response, including LTOR1 in MAPK signalling; cell survival-related IGFBP6, PPP2R5D, SUCB2 (ATP synthesis); mitochondrial NDUFS8, NDUFB11, GLS and TOMM22; PPIE, SNX2, FKBP2 in protein folding and vesicle trafficking; UBR4 in protein degradation; and immune-related PRDX4, NPC2 (Supplemental Table S7). We also validated the altered expression of NDUFB11 in archived KC epithelia compared to non-KC and normal corneal epithelia. This mitochondrial inner membrane protein belongs to multisubunit NADH:ubiquinone oxidoreductase (complex 1) transferring electrons from NADH to ubiquinone of the mitochondrial respiratory chain. This altered expression between KC cone and non-cone epithelia was supported by previous finding of nonsense mutation in NDUFB11 in association to microphthalmia (with symptoms of microcornea and corneal topographic changes) with linear skin defects75.
In this study, the KC cone and non-cone tissues were separated by manual dissection according to the pre-operative topographic data. Intra-operatively, the cone and non-cone regions were carefully demarcated by marking on the epithelium using a surgical skin marker. This warranted a precise separation of cone and non-cone tissues under dissecting microscope. Subsequent bright-field imaging and tissue dimension showed similar differences as in the Pentacam pictures. As the cone apices were decentered inferiorly from the pupil axis, the non-cone regions were obtained from the superior portion and varied between 1/3 to 1/2 of the overall resected corneal tissue, depending on the cone location. Likewise, the inferior region, where cone was located, varied between 1/2 to 2/3 of the resected corneal tissue. These dimension ratio variations were replicated by the extracted stromal protein quantities, of which the cone fractions were often greater than non-cone fractions (Fig. 2C). In order to attain a reproducible separation of cone region from the excised cornea, topography-guided laser-assisted dissection could be designed instead of manual cutting. However, the issues of tissue volume changes (stromal edema) under ex vivo storage and thermal effect on protein degradation could be the major concerns.
Even though this pilot study appeared to be limited by the sample size, we were able to identify protein groups with consistent expression changes (≥2 folds or ≤0.5 fold) in all sample pairs, indicating their significant roles in KC progression. As anticipated, a small number of proteins (20 epithelial and 14 stromal) were significantly altered between cone and non-cone samples and subsequent IPA illustrated only single network affected in either region. Future study with larger sample size would improve the precision and assure these proteins to be involved in the biological changes in KC pathogenesis and to derive the potential mechanisms of action. In addition, while KC corneas were separated into cone and non-cone regions, the normal cornea was taken as a whole. Whether the central and peripheral corneal regions possess different protein composition and expression profiles, as well as whether this plausible difference influences the pathogenic development of KC cone and non-cone regions await to be confirmed.
Our study has demonstrated that the non-cone region undergoes biological and biochemical changes, even though it is topographically normal. Hence, the altered pathophysiology is not limited to the topographically thinner and mechanically weakened cone apex but involves the non-cone region as well. The variation in protein expression has provided us with more information to understand the disease mechanism and progression of KC. Future work focusing on these early biochemical changes may elucidate new biomarkers for early KC in topographically normal eyes.
Methods
Human corneal samples
Four KC Asian patients with mean age 25.3 ± 5.1 years (range, 20–32) and gender 1:1 (male:female) (Fig. 2A) were recruited at the Singapore National Eye Centre. The study was performed in accordance with the Declaration of Helsinki and approved by SingHealth Centralized Institutional Review Board (CIRB 2016/2141). Written informed consents were obtained from all participants. Disease diagnosis was made by a corneal specialist (JSM). This was based on the findings on corneal topographies and clinical signs assessed with slit lamp biomicroscopy, and manifest refraction. Normal corneal epithelia were collected from two Chinese patients (aged 26 and 34) with normal corneal topographies undergoing photorefractive keratectomies for refractive error corrections. Normal corneal stroma was obtained from research grade corneal tissues (donor age: 31 and 37 years) deemed unsuitable for transplantation (clear corneas and corneal endothelial cell count >2200/mm2) and collected by The Lions Eye Institute for Transplant and Research Inc. (Tampa, FL, USA), following institutional review board approval. Consent was taken at the time of retrieval by the next of kin for use in research. Donor corneas were procured within 24 h after death, preserved in Optisol-GS storage medium (Bausch-Lomb, Rochester, NY, USA) and transported on ice to the laboratory. For the validation of candidate protein expression, an additional cohort of archived pathological KC samples (n = 4), corneal deep scar samples (n = 4) and normal cornea samples (n = 2) (Supplemental Table S5), were obtained from the Pathology Division, Singapore General Hospital, Singapore under valid CIRB approval. They were processed for paraffin embedding and sectioning for immunohistochemistry.
Corneal tissue processing and protein preparation
KC corneas were immediately placed in ice-cold phosphate buffer saline (PBS, Invitrogen, Carlsbad, CA, USA) and delivered on ice to the laboratory. Cone and non-cone regions were separated under dissecting microscopy, with reference to the pre-operative topographic data (Fig. 2A,B) and the demarcation of cone and non-cone tissue marked intra-operatively on the corneal epithelium by a surgical skin marker (Medline, IL). Cone and non-cone regions were separated under dissecting microscopy, according to the pre-operative topographic data (Fig. 2D). Epithelia of both cone and non-cone tissues were carefully scraped and collected, leaving the corneal stroma intact. All samples were processed for protein extraction as described previously25. In brief, tissue fragments were placed in 0.5% sodium dodecylsulfate (SDS, Sigma, St Louis, MO, USA) in PBS (150 μl), homogenized by a 2 ml Biomasher (USA Scientific, Ocala, FL, USA) and high-density sonication for 30 sec on ice. After centrifugation (15,000 × g, 10 min, 4 °C), the supernatant was collected as KC cone epithelial, cone stromal, non-cone epithelial, and non-cone stromal fractions, together with normal corneal epithelial and normal corneal stromal fractions. The protein profile and quality was assessed by 4–20% gradient SDS-polyacrylamide gel electrophoresis (SDS-PAGE, BioRad, Herculus, CA, USA), followed by Coomassie brilliant blue R-250 (Sigma) visualization (Supplemental Fig. S1) and imaged with ChemiDoc system (BioRad).
Quantitative proteomics (SWATH-MS analysis)
Protein samples (at constant 100 μg protein loads, to eliminate quantitative changes due to the corneal volume variation in the thinned cone region compared to the relatively thicker non-cone region) were trypsinized and peptides were processed for LC-MS/MS analysis and ion library data were collected by Information dependent acquisition (IDA) analysis. IDA data from four runs were combined and processed using ProteinPilot 5.0 (AB Sciex) with database search using uniprot_sprot_Sept16. Protein identification with False Discovery Rate (FDR) <1% and other parameter settings were similar as previously reported76. SWATH data were processed using SWATH quantitation function in Peakview 2.2. Protein quantification was calculated using 5–6 transitions per peptide and 1–10 peptides (99% peptide confidence level). The individual protein quantity was normalized to the total ion current. All procedure details were referred to Supplemental Information.
MS data analysis
The ratios of signal intensity of test samples and internal control were used to perform clustering analysis. The one with fluorescence intensity higher than 800 after subtracting the background was considered as an expressed signal. Clustering was done using Cluster 3 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv) (clustering type: hierarchical clustering, Distance metric: Pearson correlation).
Differential protein expression, pathway and statistical analyses
SWATH data derived epithelial and stromal proteome lists from KC cone, non-cone and normal corneal samples. The ratios were calculated from the respective comparisons (cone versus normal; non-cone versus normal and pairwise comparison between cone and non-cone samples). We selected stringent cut-off values of ≥2 folds (upregulation) and ≤0.5 fold (downregulation) between samples51,77. Smaller fold changes (>0.5 fold to <2 folds) could be due to technical variations, hence the proteins may be non-representative of real changes, unable to activate certain pathways in situ, and unable to cause pathological effects, in particular the structural and biomechanical alterations in this KC study. The geometric means and standard deviation (SD) were obtained and T-test based on the log transformation of ratios was used to calculate P values. To ensure reliable data collection, we employed stringent filtering that only proteins having consistent change in all 4 KC cone and non-cone sample pairs were analyzed. Proteins with statistically significant changes (p < 0.05) without the concordant changes in all 4 KC pairs were excluded.
Gene Ontology (GO) term analysis to identify enriched biological themes was done by inputting database to web-based DAVID Bioinformatics Resources v6.8 (NIAID, NIH, USA). Adjusted Benjamini P values were calculated by Modified Fisher Exact test with the smaller P values representing higher chance of enrichment.
Ingenuity Pathway Analysis (IPA, Qiagen, Hilden, Germany) to identify the cellular distribution of proteins and association of proteins with disease, molecular and cellular functions, physiological system development and canonical pathways.
Selected protein validation
Immunohistochemistry
The deparaffinized sections were rehydrated and heated in sodium citrate buffer for antigen retrieval. The samples were permeabilized, blocked with 0.15% saponin (Sigma), 2% bovine serum albumin (Sigma) and 5% normal goat serum (Gibco), then incubated with primary antibodies (Supplemental Table S6) followed by fluorescein-conjugated IgG secondary antibody (Jackson ImmunoRes, West Grove, PA, USA) and 4,6-diamidino-2-phenylindole nuclear staining and fluorescent signal was visualized under fluorescence microscopy (Zeiss, Oberkochen, Germany).
Western blotting and band densitometry
Corneal stromal lysates were denatured in 8 M urea/2% SDS buffer with 1% glycerol, resolved by 4–20% SDS-PAGE (BioRad) and transferred onto nitrocellulose membrane (BioRad). After blocking with 5% nonfat milk (Sigma), the nitrocellulose membrane was incubated with primary antibodies (Supplemental Table S6), followed by appropriate horseradish peroxidase-conjugated Ig secondary antibodies (Jackson ImmunoRes). Staining signals were revealed by enhanced chemiluminescence (ThermoFisher, Waltham, MA, USA) and captured using ChemiDoc XRS gel imaging system (BioRad) as grey-scale high-resolution images. Band densitometry was performed by Quantity One 1D analysis (BioRad) and target protein expression was calculated by band density subtracted with background, then normalized to the intensity of Ponceau S (Sigma) stained total protein profile.
Supplementary information
Acknowledgements
We thank Lion Eye Institute for Transplant and Research (Tampa, FL, USA) for their assistance with procurement of research grade donor corneas for normal stromal samples. We also thank Ocular Proteomics Platform and Experimental Microscopy Platform of Singapore Eye Research Institute for proteomics and fluorescence microscopy studies; and Division of Anatomical Pathology and Cytology, Pathology, Singapore General Hospital for the archived pathological samples. We thank Mr Abdelsattar Farrag, SNEC, for organizing patient information. Funding support from Singapore National Research Foundation under its Translational and Clinical Research (TCR) program (NMRC/TCR/008-SERI/2013), National Medical Research Council, Ministry of Health, Singapore.
Author Contributions
G.Y., M.F. and J.M. conceived and designed experiments; M.A., W.K., A.C. and J.M. sample provision; G.Y., M.F., H.O. and Y.L. performed research and analyzed data; L.Z., L.D. proteomic data collection, processing, biostatistics; G.Y., M.F., L.Z. and J.M. wrote the paper. All authors reviewed and approved the manuscript.
Data Availability
All data are included in the text, supplemental information and Supplemental Data Files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Gary Hin-Fai Yam and Matthias Fuest contributed equally.
Contributor Information
Gary Hin-Fai Yam, Email: gary.yam@gmail.com.
Jodhbir S. Mehta, Email: jodmehta@gmail.com
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39182-6.
References
- 1.Davidson AE, Hayes S, Hardcastle AJ, Tuft SJ. The pathogenesis of keratoconus. Eye. 2014;28:189–195. doi: 10.1038/eye.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sahebjada S, et al. Assessment of anterior segment parameters of keratoconus eyes in an Australian population. Optom Vis Sci. 2014;91:803–809. doi: 10.1097/OPX.0000000000000295. [DOI] [PubMed] [Google Scholar]
- 3.Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998;42:297–319. doi: 10.1016/S0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 4.Sharif, R., Bak-Nielsen, S., Hjortdal, J. & Karamichos, D. Pathogenesis of Keratoconus: The intriguing therapeutic potential of Prolactin-inducible protein. Prog Retin Eye Res (2018). [DOI] [PMC free article] [PubMed]
- 5.Jonas JB, Nangia V, Matin A, Kulkarni M, Bhojwani K. Prevalence and associations of keratoconus in rural maharashtra in central India: the central India eye and medical study. Am J Ophthalmol. 2009;148:760–765. doi: 10.1016/j.ajo.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Millodot M, Shneor E, Albou S, Atlani E, Gordon-Shaag A. Prevalence and associated factors of keratoconus in Jerusalem: a cross-sectional study. Ophthalmic Epidemiol. 2011;18:91–97. doi: 10.3109/09286586.2011.560747. [DOI] [PubMed] [Google Scholar]
- 7.Gokhale NS. Epidemiology of keratoconus. Indian J Ophthalmol. 2013;61:382–383. doi: 10.4103/0301-4738.116054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero-Jimenez M, Santodomingo-Rubido J, Wolffsohn JS. Keratoconus: a review. Cont Lens Anterior Eye. 2010;33:157–166. doi: 10.1016/j.clae.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Gain P, et al. Global survey of corneal transplantation and eye banking. JAMA Ophthalmol. 2016;134:167–173. doi: 10.1001/jamaophthalmol.2015.4776. [DOI] [PubMed] [Google Scholar]
- 10.Krachmer JH, Feder RS, Belin MW. Keratoconus and related noninflammatory corneal thinning disorders. Surv Ophthalmol. 1984;28:293–322. doi: 10.1016/0039-6257(84)90094-8. [DOI] [PubMed] [Google Scholar]
- 11.Khaled ML, et al. Molecular and histopathological changes associated with keratoconus. BioMed Res Int. 2017;2017:7803029. doi: 10.1155/2017/7803029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowak DM, Gajecka M. The genetics of keratoconus. Middle East Afr J Ophthalmol. 2011;18:2–6. doi: 10.4103/0974-9233.75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bisceglia L, et al. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive loci. Invest Ophthalmol Vis Sci. 2009;50:1081–1086. doi: 10.1167/iovs.08-2382. [DOI] [PubMed] [Google Scholar]
- 14.Sahebjada S, et al. Evaluating the association between keratoconus and the corneal thickness genes in an independent Australian population. Invest Ophthalmol Vis Sci. 2013;54:8224–8228. doi: 10.1167/iovs.13-12982. [DOI] [PubMed] [Google Scholar]
- 15.Sahebjada S, et al. Association of the hepatocyte growth factor gene with keratoconus in an Australian population. PloS One. 2014;9:e84067. doi: 10.1371/journal.pone.0084067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong SS, et al. Genetic associations for keratoconus: a systematic review and meta-analysis. Sci Rep. 2017;7:4620. doi: 10.1038/s41598-017-04393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galvis V, Tello A, Barrera R, Nino CA. Inflammation in keratoconus. Cornea. 2015;34:e22–23. doi: 10.1097/ICO.0000000000000499. [DOI] [PubMed] [Google Scholar]
- 18.Kemp EG, Lewis CJ. Measurement of total and specific IgE levels in the management of a family exhibiting a high incidence of keratoconus. Acta Ophthalmol. 1984;62:524–529. doi: 10.1111/j.1755-3768.1984.tb03963.x. [DOI] [PubMed] [Google Scholar]
- 19.Bawazeer AM, Hodge WG, Lorimer B. Atopy and keratoconus: a multivariate analysis. Br J Ophthalmol. 2000;84:834–836. doi: 10.1136/bjo.84.8.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMonnies CW. Epigenetic mechanisms might help explain environmental contributions to the pathogenesis of keratoconus. Eye Contact Lens. 2014;40:371–375. doi: 10.1097/ICL.0000000000000078. [DOI] [PubMed] [Google Scholar]
- 21.Wang YM, et al. Histological and microRNA signatures of corneal epithelium in keratoconus. J Refract Surg. 2018;34:201–211. doi: 10.3928/1081597X-20171215-02. [DOI] [PubMed] [Google Scholar]
- 22.Hughes AE, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89:628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner J, et al. Mutational analysis of miR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci. 2013;54:5266–5272. doi: 10.1167/iovs.13-12035. [DOI] [PubMed] [Google Scholar]
- 24.Akhtar S, et al. Ultrastructural analysis of collagen fibrils and proteoglycans in keratoconus. Acta Ophthalmol. 2008;86:764–772. doi: 10.1111/j.1755-3768.2007.01142.x. [DOI] [PubMed] [Google Scholar]
- 25.Chaerkady R, et al. The keratoconus corneal proteome: loss of epithelial integrity and stromal degeneration. J Proteomics. 2013;87:122–131. doi: 10.1016/j.jprot.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackiewicz Z, et al. Collagenolytic proteinases in keratoconus. Cornea. 2006;25:603–610. doi: 10.1097/01.ico.0000208820.32614.00. [DOI] [PubMed] [Google Scholar]
- 27.Lema I, et al. Inflammatory response to contact lenses in patients with keratoconus compared with myopic subjects. Cornea. 2008;27:758–763. doi: 10.1097/ICO.0b013e31816a3591. [DOI] [PubMed] [Google Scholar]
- 28.Lema I, Sobrino T, Duran JA, Brea D, Diez-Feijoo E. Subclinical keratoconus and inflammatory molecules from tears. Br J Ophthalmol. 2009;93:820–824. doi: 10.1136/bjo.2008.144253. [DOI] [PubMed] [Google Scholar]
- 29.Jun AS, et al. Subnormal cytokine profile in the tear fluid of keratoconus patients. PloS One. 2011;6:e16437. doi: 10.1371/journal.pone.0016437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balasubramanian SA, Mohan S, Pye DC, Willcox MD. Proteases, proteolysis and inflammatory molecules in the tears of people with keratoconus. Acta Ophthalmol. 2012;90:e303–309. doi: 10.1111/j.1755-3768.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 31.Ionescu C, et al. Inflammatory biomarkers profile as microenvironmental expression in keratoconus. Dis Markers. 2016;2016:1243819. doi: 10.1155/2016/1243819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chwa M, et al. Increased stress-induced generation of reactive oxygen species and apoptosis in human keratoconus fibroblasts. Invest Ophthalmol Vis Sci. 2006;47:1902–1910. doi: 10.1167/iovs.05-0828. [DOI] [PubMed] [Google Scholar]
- 33.Arnal E, Peris-Martinez C, Menezo JL, Johnsen-Soriano S, Romero FJ. Oxidative stress in keratoconus? Invest Ophthalmol Vis Sci. 2011;52:8592–8597. doi: 10.1167/iovs.11-7732. [DOI] [PubMed] [Google Scholar]
- 34.Wojcik KA, Kaminska A, Blasiak J, Szaflik J, Szaflik JP. Oxidative stress in the pathogenesis of keratoconus and Fuchs endothelial corneal dystrophy. Int J Mol Sci. 2013;14:19294–19308. doi: 10.3390/ijms140919294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lema I, Brea D, Rodriguez-Gonzalez R, Diez-Feijoo E, Sobrino T. Proteomic analysis of the tear film in patients with keratoconus. Mol Vis. 2010;16:2055–2061. [PMC free article] [PubMed] [Google Scholar]
- 36.Pannebaker C, Chandler HL, Nichols JJ. Tear proteomics in keratoconus. Mol Vis. 2010;16:1949–1957. [PMC free article] [PubMed] [Google Scholar]
- 37.Acera A, et al. Changes in tear protein profile in keratoconus disease. Eye. 2011;25:1225–1233. doi: 10.1038/eye.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh A, Zhou L, Ghosh A, Shetty R, Beuerman R. Proteomic and gene expression patterns of keratoconus. Indian J Ophthalmol. 2013;61:389–391. doi: 10.4103/0301-4738.116056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karamichos D, et al. Tear metabolite changes in keratoconus. Exp Eye Res. 2015;132:1–8. doi: 10.1016/j.exer.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lema I, Duran JA. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology. 2005;112:654–659. doi: 10.1016/j.ophtha.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 41.Shetty R, et al. Elevated expression of matrix metalloproteinase-9 and inflammatory cytokines in keratoconus patients is inhibited by cyclosporine A. Invest Ophthalmol Vis Sci. 2015;56:738–750. doi: 10.1167/iovs.14-14831. [DOI] [PubMed] [Google Scholar]
- 42.Soria J, et al. Label-free LC-MS/MS quantitative analysis of aqueous humor from keratoconic and normal eyes. Mol Vis. 2015;21:451–460. [PMC free article] [PubMed] [Google Scholar]
- 43.McKay TB, et al. Endocrine and metabolic pathways linked to keratoconus: implications for the role of hormones in the stromal microenvironment. Sci Rep. 2016;6:25534. doi: 10.1038/srep25534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen K, et al. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006;82:201–209. doi: 10.1016/j.exer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava OP, Chandrasekaran D, Pfister RR. Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas. Mol Vis. 2006;12:1615–1625. [PubMed] [Google Scholar]
- 46.Joseph R, Srivastava OP, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011;92:282–298. doi: 10.1016/j.exer.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 47.You J, et al. RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Res. 2018;8:389. doi: 10.1038/s41598-017-18480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan JK, Belin MW, Borgstrom M. Assessing progression of keratoconus: novel tomographic determinants. Eye Vis. 2016;3:6. doi: 10.1186/s40662-016-0038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caroline, P. J. & Norman, C. W. Corneal Topography in the Diagnosis and Management of Keratoconus. In: Corneal Topography (Schanzlin, D. J. & Robin, J. B. eds), Springer, New York, NY. pp 75–93 (1992).
- 50.Sidoli S, et al. Sequential Window Acquisition of all Theoretical Mass Spectra (SWATH) analysis for characterization and quantification of histone post-translational modifications. Mol Cell Proteomics. 2015;14:2420–2428. doi: 10.1074/mcp.O114.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Y, et al. Quantitative proteomics by SWATH-MS reveals sophisticated metabolic reprogramming in hepatocellular carcinoma tissues. Sci Rep. 2017;7:45913. doi: 10.1038/srep45913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kothapalli CR, Gacchina CE, Ramamurthi A. Utility of hyaluronan oligomers and transforming growth factor-beta1 factors for elastic matrix regeneration by aneurysmal rat aortic smooth muscle cells. Tissue Eng Part A. 2009;15:3247–3260. doi: 10.1089/ten.tea.2008.0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pahuja N, et al. Differential molecular expression of extracellular matrix and inflammatory genes at the corneal cone apex drives focal weakening in keratoconus. Invest Ophthalmol Vis Sci. 2016;57:5372–5382. doi: 10.1167/iovs.16-19677. [DOI] [PubMed] [Google Scholar]
- 54.Edmund C. Corneal elasticity and ocular rigidity in normal and keratoconic eyes. Acta Ophthalmol. 1988;66:134–140. doi: 10.1111/j.1755-3768.1988.tb04000.x. [DOI] [PubMed] [Google Scholar]
- 55.Maguire LJ, Bourne WM. Corneal topography of early keratoconus. Am J Ophthalmol. 1989;108:107–112. doi: 10.1016/0002-9394(89)90001-9. [DOI] [PubMed] [Google Scholar]
- 56.Scarcelli G, Besner S, Pineda R, Yun SH. Biomechanical characterization of keratoconus corneas ex vivo with Brillouin microscopy. Invest Ophthalmol Vis Sci. 2014;55:4490–4495. doi: 10.1167/iovs.14-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morishige N, et al. Second-harmonic imaging microscopy of normal human and keratoconus cornea. Invest Ophthalmol Vis Sci. 2007;48:1087–1094. doi: 10.1167/iovs.06-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowak DM, et al. Substitution at IL1RN and deletion at SLC4A11 segregating with phenotype in familial keratoconus. Invest Ophthalmol Vis Sci. 2013;54:2207–2215. doi: 10.1167/iovs.13-11592. [DOI] [PubMed] [Google Scholar]
- 59.Fu M, et al. Hydrogen sulfide (H2S) metabolism in mitochondria and its regulatory role in energy production. Proc Nat Acad Sci USA. 2012;109:2943–2948. doi: 10.1073/pnas.1115634109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang EY, et al. Inhibition of prostaglandin reductase 2, a putative oncogene overexpressed in human pancreatic adenocarcinoma, induces oxidative stress-mediated cell death involving xCT and CTH gene expressions through 15-keto-PGE2. Plos One. 2016;11:e0147390. doi: 10.1371/journal.pone.0147390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Mato M, et al. Human recombinant glutamate oxaloacetate transaminase 1 (GOT1) supplemented with oxaloacetate induces a protective effect after cerebral ischemia. Cell Death Dis. 2014;5:e992. doi: 10.1038/cddis.2013.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chao J, Bledsoe G, Chao L. Protective role of kallistatin in vascular and organ injury. Hypertension. 2016;68:533–541. doi: 10.1161/HYPERTENSIONAHA.116.07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meek KM, Knupp C. Corneal structure and transparency. Prog Retin Eye Res. 2015;49:1–16. doi: 10.1016/j.preteyeres.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaki NM, Tirelli N. Gateways for the intracellular access of nanocarriers: a review of receptor-mediated endocytosis mechanisms and of strategies in receptor targeting. Expert Opin Drug Deliv. 2010;7:895–913. doi: 10.1517/17425247.2010.501792. [DOI] [PubMed] [Google Scholar]
- 65.Piperi C, Papavassiliou AG. Molecular mechanisms regulating matrix metalloproteinases. Curr Top Med Chem. 2012;12:1095–1112. doi: 10.2174/1568026611208011095. [DOI] [PubMed] [Google Scholar]
- 66.Duester G. Alcohol dehydrogenase as a critical mediator of retinoic acid synthesis from vitamin A in the mouse embryo. J Nutr. 1998;128:459S–462S. doi: 10.1093/jn/128.2.459S. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, Dolle P, Ghyselinck NB, Duester G. Endogenous retinoic acid signaling is required for maintenance and regeneration of cornea. Exp Eye Res. 2017;154:190–195. doi: 10.1016/j.exer.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xue X, et al. Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc Nat Acad Sci USA. 2017;114:E9608–E9617. doi: 10.1073/pnas.1712946114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson SE, Hong JW. Bowman’s layer structure and function: critical or dispensable to corneal function? A hypothesis. Cornea. 2000;19:417–420. doi: 10.1097/00003226-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 70.Reinstein DZ, Gobbe M, Archer TJ, Silverman RH, Coleman DJ. Epithelial, stromal and total corneal thickness in kertoconus: 3D display with artermis very-high frequency digital ultrasound. J Refract Surg. 2010;26:259–271. doi: 10.3928/1081597X-20100218-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ucakhan OO, Kanpolat A, Ylmaz N, Ozkan M. In vivo confocal microscopy findings in keratoconus. Eye Contact Lens. 2006;32:183–191. doi: 10.1097/01.icl.0000189038.74139.4a. [DOI] [PubMed] [Google Scholar]
- 72.Mocan MC, Yilmaz PT, Irkec M, Orhan M. In vivo confocal microscopy for the evaluation of corneal microstructure in keratoconus. Curr Eye Res. 2008;33:933–939. doi: 10.1080/02713680802439219. [DOI] [PubMed] [Google Scholar]
- 73.Bitirgen G, Ozkagnici A, Bozkurt B, Malik RA. In vivo corneal confocal microscopic analysis in patients with keratoconus. Int J Ophthalmol. 2015;8:534–539. doi: 10.3980/j.issn.2222-3959.2015.03.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shetty R, et al. Oxidative stress induces dysregulated autophagy in corneal epithelium of keratoconus patients. PloS One. 2017;12:e0184628. doi: 10.1371/journal.pone.0184628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Rahden VA, et al. Mutations in NDUFB11, encoding a complex I component of the mitochondrial respiratory chain, cause microphthalmia with linear skin defects syndrome. Am J Hum Genet. 2015;96:640–650. doi: 10.1016/j.ajhg.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen L, et al. Global metabonomic and proteomic analysis of human conjunctival epithelial cells in response to hyperosmotic stress. J Proteome Res. 2015;14:3982–3995. doi: 10.1021/acs.jproteome.5b00443. [DOI] [PubMed] [Google Scholar]
- 77.Yenihayat, F. et al. Comparative proteome analysis of the tear samples in patients with low-grade keratoconus. Int Ophthalmol, 10.1007/s10792-017-0672-6 (2017). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the text, supplemental information and Supplemental Data Files.