Abstract
While the majority of the natural carotenoid pigments are based on 40-carbon (C40) skeleton, some carotenoids from bacteria have larger C50 skeleton, biosynthesized by attaching two isoprene units (C5) to both sides of the C40 carotenoid pigment lycopene. Subsequent cyclization reactions result in the production of C50 carotenoids with diverse and unique skeletal structures. To produce even larger nonnatural novel carotenoids with C50 + C5 + C5 = C60 skeletons, we systematically coexpressed natural C50 carotenoid biosynthetic enzymes (lycopene C5-elongases and C50-cyclases) from various bacterial sources together with the laboratory-engineered nonnatural C50-lycopene pathway in Escherichia coli. Among the tested enzymes, the elongases and cyclases from Micrococcus luteus exhibited significant activity toward C50-lycopene, and yielded the novel carotenoids C60-flavuxanthin and C60-sarcinaxanthin. Moreover, coexpression of M. luteus elongase with Corynebacterium cyclase resulted in the production of C60-sarcinaxanthin, C60-sarprenoxanthin, and C60-decaprenoxanthin.
Introduction
Carotenoids are a class of natural pigments covering yellow, orange and red colors. More than 750 carotenoids have been identified in various plants, fungi, and microorganisms1, and a wide range of essential biological functions have been described, with light-harvesting, photoprotection, antioxidatant, and pro-vitamin activities, and roles in the control of membrane fluidity2. In recent years, carotenoids have been increasingly considered in applications as therapeutic or bioelectronic materials3,4. Small changes in carotenoid structures have been associated with large differences in material properties5 and biological activities3,6. Hence, the discovery of the structurally novel carotenoids is eagerly awaited.
Most carotenoids have C40 backbones, although a few microbial carotenoids have C30 backbones. The structural diversity of carotenoids is mainly derived from differences in desaturation levels of the C40 backbone, types of end groups, and functional patterns of cyclization, oxidation, and esterification. Further structural diversity of carotenoids can be achieved by liberating carotenoids from their C40 backbones. For example, carotenoid cleavage enzymes can be used to produce a range of carotenoids with different-backbone sizes (C14, C20, or C28 carbons)7. These include precursors of bixin (C24+1), crocetin (C20), retinals (C20), and other hormonal compounds such as strigolactones (C13) and abscisic acid (C15), as reviewed previously3.
Another source of structure and size diversity comes from the attachment of additional isoprene units (C5) to C40 carotenoids. Since C45 and C50 carotenoids were first reported in the 1960s8,9, many of these carotenoids have been identified in prokaryotes, including gram-positive bacteria such as Micrococcus10, Corynebacterium11, Dietzia12, and extreme halophilic archaea such as Halobacterium or Haloarcula13. Although the natural functions of these carotenoids are not fully understood, contributions to cell membrane fluidity and permeability have been suggested13. In addition, the attachment of C5 units reportedly confers remarkable antioxidant activities to carotenoids14, and the resulting compounds have been considered for their potential as therapeutic agents13–15.
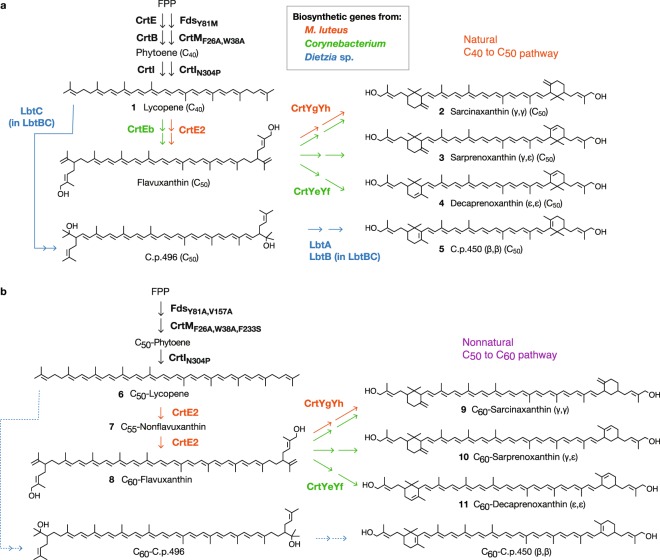
To date, four natural biosynthetic pathways for C50 carotenoids has been reported10–12,16. All known C50 carotenoids are derived from lycopene, a four-step enzymatic desaturation product of phytoene, which is produced by the condensation of two molecules of geranylgeranyl diphosphate (C20). One pathway from extremely halophillic archaea Haloarcula japonica yields acyclic C50 carotenoid bacterioruberin16 as the final product, whereas three others produce cyclic C50 carotenoids (Fig. 1a). In the Micrococcus luteus carotenoid biosynthetic pathway, lycopene elongase (CrtE2) attaches two additional isoprene units (DMAPP) to the 2,2′-position of lycopene to produce the acyclic C50 carotenoid flavuxanthin, and subsequent γ-cyclization by cyclase (CrtYgYh) produces sarcinaxanthin (γ,γ-ring)10. C50 carotenoid synthesis in Corynebacterium also proceeds via the flavuxanthin intermediate, but the expressed cyclase (CrtYeYf) is less specific and yields a mixture of decaprenoxanthin10,11 (ε,ε-ring), sarprenoxanthin10 (ε,γ-ring) and sarcinaxanthin, when expressed in E. coli. In contrast, the bacterium Dietzia, reportedly produces C50 carotenoids with β,β-rings (C.p.450)12 via a different acyclic intermediate (Fig. 1a).
Figure 1.
C5-elongation and cyclization pathways for natural (C50) and nonnatural (C60) carotenoids. (a) Natural C40-to-C50 carotenoid pathway. Using lycopene (C40) as a substrate, the lycopene elongases CrtEb from Corynebacterium and CrtE2 from M. luteus attaches two isoprene units to C40. The resulting flavuxanthin (C50) is then cyclized by the γ- and/or ε-cyclases CrtYe/Yf (Corynebacterium) or CrtYg/Yh (M. luteus) to produce sarcinaxanthin, sarprenoxanthin, or decaprenoxanthin (all C50). LbtABC genes from Dietzia sp. CQ4 produces the β,β-cyclic C50 carotenoid C.p.450 via the independent intermediate C.p.496. (b) Nonnatural C50-to-C60 carotenoid pathway construction. Combined expression of Corynebacterium and M. luteus elongases and cyclases resulted in the conversion of laboratory-generated C50-lycopene to C60 carotenoids with γ and/or ε-cyclic ends. We could not obtain C60 counterparts of β-end C50 carotenoids (indicated in arrows with dashed lines).
In a previous study, we constructed a nonnatural C50 backbone carotenoid pathway in which two geranylfarnesyl diphosphates (C25PP) are condensed to produce C50-phytoene (Fig. 1b). This compound is subsequently subjected to a six-step desaturation reaction resulting in the synthesis of C50-lycopene. Using a metabolic filtering approach17, we extended this C50-lycopene pathway to specifically produce the nonnatural C50 backbone carotenoids C50-β-carotene, C50-zeaxanthin, C50-canthaxanthin, and C50-astaxanthin. In this study, we systematically expressed elongase and cyclase in engineered E. coli strains harboring enzymes of the C50-lycopene synthetic pathway and established pathways for the synthesis of the novel carotenoids C60-flavuxanthin, C60-sarcinaxanthin, C60-sarprenoxanthin, and C60-decaprenoxanthin (Fig. 1b).
Results
Activities of elongase and cyclase in the natural C40 pathway
We selected Corynebacterium glutamicum, M. luteus, and Dietzia sp. CQ4 as sources of lycopene elongases and C50 cyclases, because the C50 carotenoid pathways from these organisms have been previously reconstructed in E. coli10–12. The carotenoid cluster (shown in Supplementary Fig. 1) of C. glutamicum11 comprises crtEb (lycopene elongase), crtYeYf (heterodimeric C50 ε/γ-cyclase), and lycopene-producing genes (crtEBI). The expression of this gene cluster in E. coli produces sarprenoxanthin, decaprenoxanthin, and sarcinaxanthin via the acyclic intermediate flavuxanthin10,11 (Fig. 1a). The corresponding gene cluster in M. luteus10 encodes crtE2 (lycopene elongase) and crtYgYh (C50 γ-cyclase), and coexpression of these genes with crtEBI results in specific accumulation of sarcinaxanthin10. Uniquely, the carotenogenic gene cluster of Dietzia sp. CQ412 encodes a gene for elongase (lbtC) that is fused in frame with a single subunit of cyclase (lbtB), which forms a heterodimer with another unit of cyclase (lbtA). Expression of lbtB with an lbtA gene product results in the production of a functional C50 β-cyclase12.
Although different in organization, all of the elongase and cyclase genes are clustered in small <1.7-kb regions (Supplementary Fig. 1), allowing one-step PCR amplification from genomic DNA. We cloned these DNA fragments into a plasmid that encodes Pantoea ananatis phytoene desaturase variant17 crtIN304P, a variant that can desaturate both (C40-) phytoene and C50-phytoene (see Fig. 2a for plasmid construct). In addition to these three operons (for C. glutamicum, M. luteus and Dietzia sp.), we cloned the elongase and cyclase genes from Corynebacterium efficiens. The carotenoid gene cluster of this organism has exactly the same organization as that of C. glutamicum, and in our homology analyses, DNA sequence identity of the two clusters (from crtE to crtEb, see Supplementary Fig. 1) was 64.3%. Yet, the functions of the C. efficiens carotenoid pathway and its products remain uncharacterized.
Figure 2.
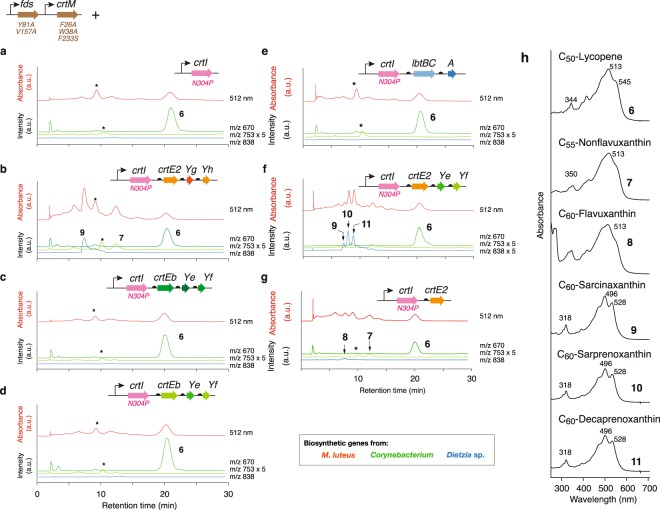
Lycopene elongases and C50 cyclases function in the natural C40-to-C50 pathway. HPLC chromatograms of carotenoid extracts from E. coli cells harboring plasmids with genome-derived natural gene cluster (a) or RBS-redesigned artificial operons (b), together with lycopene biosynthetic genes. Peak numbers correspond with those indicated in c and Fig. 1. (c) Absorbance spectra of indicated compounds.
To characterize the functions of elongase and cyclase in the context of natural (C40) pathway (Fig. 1a) in E. coli, we used an engineered E. coli strain that constitutively expresses fdsY81M and crtMF26A,W38A genes (crtEB equivalent17) and produces phytoene (C40). We introduced plasmids encoding phytoene desaturase (a crtI variant), elongases and cyclases into this strain, and after culturing for 48 h, we extracted carotenoids using acetone and analyzed these using high performance liquid chromatography (HPLC) (Fig. 2a). These analyses showed that cells expressing C. glutamicum crtYeYfEb produced a mixture of (C50-) sarcinaxanthin (2), (C50-) sarprenoxanthin (3), and (C50-) decaprenoxanthin (4), with significant quantities of their substrate (C40-) lycopene (1). Cells expressing C. efficiens crtYeYfEb indistinguishable composition of carotenoids as those from the C. glutamicum gene cluster. Cells expressing M. luteus crtE2YgYh produced a single peak of (C50-) sarcinaxanthin (2), whereas expression of Dietzia sp. CQ4 lbtABC resulted in the specific production of C.p.450 (C50). Despite the differences in the source of lycopene biosynthetic genes, promoters, RBS sequences, E. coli strain or culture condition, product distribution of the pathway we constructed was very similar to those from previous reports10.
To determine whether these enzymes can metabolize C50-lycopene, we coexpressed plasmids encoding the elongase and cyclase enzymes in another E. coli strain that constitutively expresses fdsY81A,V157A and crtMF26A,W38A,F233S and produces C50-phytoene17. In this strain, the phytoene desaturase mutant (CrtIN304P) can desaturate C50-phytoene to produce C50-lycopene selectively17, providing a sole substrate for the elongase and cyclases in this stain. However, we observed no novel peaks but C50-lycopene in carotenoid fractions from this strain.
Ribosome binding site (RBS)-optimized cyclase/elongase results in higher production of cyclic C50 carotenoids
In the previous section, we observed (C40-) lycopene accumulated in cells expressing elongase and cyclases (Fig. 2a), suggesting the presence of inefficiencies of this pathway, and room for improvement. The present four carotenoid operon constructs share similar operon organization, in which open reading frames (ORF) for cyclases and elongases are partially overlapping. Specifically, the start codon of M. luteus crtYg is 28 base pairs upstream of the stop codon of the crtE2 gene, and the start codon of crtYh overlaps with the stop codon of the crtYg gene. In C. glutamicum and C. efficiens, crtYe and crtYf overlap by 4 bases, and in Dietzia, lbtA and lbtBC genes also overlap by 4 bases, as is commonly observed in various gene clusters containing carotenoid operons10. We investigated translation initiation rates using the RBS calculator18–20 and found RBS scores as low as 0.01 (Table 1), suggesting very low translation initiation rates and likely formation of stable secondary mRNA structures. In the native operons with overlaps in translational stop and start of ORFs, the translational re-initiation is likely ensuring efficient translation in vivo.
Table 1.
Ribosome binding site strengths of original and designed operons.
| Organism | Gene | RBS strength in v1 plasmid | Designed RBS strength in v2 plasmid |
|---|---|---|---|
| M. luteus | crtE2 | 812 | 4296 |
| crtYg | 0.01 | 1525 | |
| crtYh | 140 | 4700 | |
| Dietzia sp. CQ4 | lbtA | 968 | 4106 |
| lbtBC | 15 | 4700 | |
| C. glutamicum | crtYe | 4493 | 3891 |
| crtYf | 4342 | 5886 | |
| crtEb | 389 | 5066 | |
| C. efficiens | crtYe | 776 | 1113 |
| crtYf | 33 | 2663 | |
| crtEb | 93 | 1990 |
To improve expression levels of elongases and cyclases in E. coli, we redesigned our artificial operons (Fig. 2b) by (1) separating the ORFs to yield tandem and distinct non-overlapping reading frames, and (2) to provide stronger RBSs, designed using RBS calculator to have RBS strengths between 1,000 - 5,000 (Table 1). These RBS strengths range were chosen since it was previously confirmed optimal for expressing various carotenoid biosynthetic genes in the plasmid backbone we use in this study17,21 (i.e. p15A, pUC). When expressed with (C40-) lycopene pathways, all of these new constructs with engineered RBSs led to improved production of natural C50 carotenoids and significant reductions in unconverted lycopene contents (Fig. 2b).
Production of nonnatural cyclic C60 carotenoids
Following the RBS engineering of elongases and cyclases, we cotransformed the new constructs into the strains that selectively synthesize C50-phytoene (Fig. 3). From the cells expressing M. luteus crtE2YgYh, we observed novel peaks 7 and 9 in HPLC chromatograms, and these were eluted earlier than that of C50-lycopene (6) (Fig. 3a,b). Peak 9 exhibited absorbance maxima at 496 nm; shorter than that for C50-lycopene (9, 512 nm). The m/z value (837) of peak 9 matched that of C60-sarcinaxanthin (C60H84O2, Fig. 1), and the carotenoid from peak 9 had absorption maxima at 460, 494, and 527 nm (Fig. 3h). Furthermore, analyses using positive ion electrospray ionization time-of-flight mass spectrometry (ESI TOF MS) at m/z 837.6542 MH+ indicated a molecular formula for this carotenoid of C60H84O2 (C60H85O2 calcd. for 837.6550) (Supplementary Fig. 2). The structure of this carotenoid was determined as C60-sarciniaxanthin using nuclear magnetic resonance (1H-NMR) with correlation spectroscopy (COSY) and rotating frame overhause effect spectroscopy (ROESY) (Table 2). In addition, we identified peak 7 as the C60 pathway intermediate C55-nonflavuxanthin based on an absorbance maximum of 513 nm and a m/z value of 753 (C55H76O).
Figure 3.
Lycopene elongases and C50 cyclases function in the C50-to-C60 pathway. (a–g) HPLC chromatogram of carotenoid extracts from E. coli cells expressing genes for C50-phytoene production with indicated genes. The indicated peak numbers correspond with those in Fig. 1. Peaks labelled with asterisks correspond to unidentified non-carotenoid compounds. (h) Absorbance spectra of the indicated peaks.
Table 2.
1H NMR data for C60-sarcinaxanthin and C60-flavuxanthin in CDCl3.
| Position | C60-Sarcinaxanthin (9) | Position | C60-Flavuxanthin (8) | ||||
|---|---|---|---|---|---|---|---|
| d | Mult. | J (Hz) | d | Mult. | J (Hz) | ||
| H-2 (2′) | 1.28 | m | H-2 (2′) | 2.08 | m | ||
| H-3 (3′) | 1.18 | m | H2-3 (3′) | 1.56 | m | ||
| 1.71 | m | ||||||
| H-4 (4′) | 2.05 | m | H2-4 (4′) | 2.00 | m | ||
| 2.35 | m | ||||||
| H-6 (6′) | 2.48 | d | 10 | H-6 (6′) | 5.93 | d | 11 |
| H-7 (7′) | 5.83 | d | 15.5, 10 | H-7 (7′) | 6.48 | dd | 15, 11 |
| H-8 (8′) | 6.12 | d | 15.5 | H-8 (8′) | 6.24 | d | 15 |
| H-10 (10′) | 6.12 | d | 12 | H-10 (10′) | 6.18 | d | 12 |
| H-11 (11′) | 6.62 | dd | 15, 12 | H-11 (11′) | 6.63 | dd | 15, 12 |
| H-12 (12′) | 6.34 | d | 15 | H-12 (12′) | 6.36 | d | 15 |
| H-14 (14′) | 6.23 | d | 11 | H-14 (14′) | 6.23 | d | 11 |
| H-15 (15′) | 6.64 | dd | 15, 11 | H-15 (15′) | 6.64 | dd | 15, 11 |
| H-16 (16′) | 6.38 | d | 15 | H-16 (16′) | 6.38 | d | 15 |
| H-18 (18′) | 6.27 | br. d | 10 | H-18 (18′) | 6.27 | br. d | 10 |
| H-19 (19′) | 6.64 | m | H-19 (19′) | 6.64 | m | ||
| H3-20 (20′) | 0.96 | s | H-20 (20′) | 4.70 | br. S | ||
| H-20 (20′) | 4.78 | br. S | |||||
| H3-21 (21′) | 0.73 | s | H3-21 (21′) | 1.63 | s | ||
| H-22 (22′) | 4.53 | s | H3-22 (22′) | 1.80 | s | ||
| H-22 (22′) | 4.76 | s | |||||
| H3-23 (23′) | 1.98/1.99 | s | H3-23 (23′) | 1.97 | s | ||
| H3-24 (24′) | 1.98/1.99 | s | H3-24 (24′) | 1.98/1.99 | s | ||
| H3-25 (25′) | 1.98/1.97 | s | H3-25 (25′) | 1.98/1.97 | s | ||
| H-26 (26′) | 1.72 | m | |||||
| H-26 (26′) | 2.24 | dd | 14, 5.5 | H2-26 (26′) | 2.10 | m | |
| H-27 (27′) | 5.43 | m | H-27 (27′) | 5.36 | t | 5 | |
| H3-29 (29′) | 1.67 | s | H3-29 (29′) | 1.67 | s | ||
| H2-30 (30′) | 4.03 | s | H2-30 (30′) | 4.00 | s | ||
See Supplementary Fig. S4 for the numbering of carotenoid structure.
The crtE2 gene expression (without crtYgYh) from M. luteus resulted in a new peak 8 with an m/z value of 838 and an absorbance maximum of 513 nm (Fig. 3g). The carotenoid from peak 8 showed absorption maxima at 460, 494, and 527 nm (Fig. 3h). Positive ion ESI TOF MS at m/z 857.6107 M + Na+ revealed the molecular formula C60H82O2 (C60H82O2Na calcd. for 857.6212) (Supplementary Fig. 3) for this carotenoid, and structural analyses using 1H-NMR with COSY and ROESY revealed the carotenoid C60-flavuxanthin (Table 2).
We did not observe any novel carotenoid peaks in experiments using elongase and cyclase genes from C. glutamicum, C. efficiens, or Dietzia sp. CQ4 (Fig. 3c–e). Hence, the lycopene elongases (CrtEb in Corynebacterium and LbtC in Dietzia sp. CQ4) at least fail to convert C50-lycopene in E. coli. However, the activities of the cyclases (CrtYeYf from Corynebacterium and LbtAB from Dietzia sp. CQ4) were still unknown, since the first elongase step failed to provide the substrate (C60-flavuxanthin or C60-C.p.496) for these cyclase enzymes.
Because Corynebacterium and M. luteus pathways share the intermediate flavuxanthin, we generated a chimera operon containing M. luteus crtE2 and C. efficiens crtYeYf. Following expression in cells, we detected three carotenoid peaks (Fig. 3f) (9, 10, and 11) with identical absorption spectra (Fig. 3h), indicating the presence of the same chromophore. Although we could not determine the NMR spectroscopy of the compound from the associated chromatographic peaks, their absorption spectra, m/z values (838) and retention times strongly indicate that peaks 9, 10, and 11 were C60-sarcinaxanthin (as shown in Fig. 3b), C60-sarprenoxanthin, and C60-decaprenoxanthin, respectively. These results also indicate that cyclases (crtYeYf) from Corynebacterium are functional in the C50-to-C60 pathway.
Discussion
In this study, we demonstrated that carotenoid elongation and cyclization enzymes from natural C50 carotenoid pathways metabolize C50-lycopene to novel nonnatural C60 carotenoids. These carotenoids (C60-flavuxanthin, C60-sarcinaxanthin, C60-sarprenoxanthin, and C60-decaprenoxanthin) are larger than any known natural carotenoids, and their absorbance spectra are red-shifted by as much as 58 nm compared with the natural counterpart (440 nm vs. 496 nm). With rare γ-ring structures, these C60 carotenoids provide a unique set of accessible carotenoid structures with as yet unknown functions. Natural C50 carotenoids were previously discovered in thicker membranes of halophillic bacteria, which survive in extreme hypersaline and low-temperature environments13,14,16. The present nonnatural C60 carotenoids are interesting candidates for functional characterization in extremophiles. These future studies may lend understanding to the roles of long-chain carotenoids in nature.
Through the coexpression of natural carotenoid enzymes, we have increased the number of laboratory-generated C50 carotenoid pathways. Many carotenoid enzymes from C40 and C30 pathways exhibit significant substrate promiscuity and accept a wide range of substrates with recognizable locally specific22 structures. Consequently, these enzymes are active in nonnatural pathway contexts22–25 without any mutations. However, there is increasing reports on carotenoid modifying enzymes that exhibit unexpected selectivity against non-cognate but very similar substrates. For example, lycopene ε-cyclases from plants (LCYe) cyclize only one end of the acyclic substrate lycopene and leave the other end un-cyclized26. Hence, ε-cyclases likely possess mechanisms for avoiding cyclization of non-cognate substrates. Similarly, the β-carotene 15,15′ cleavage enzyme BCMO1 only accepts β-carotene (β,β-end) as a substrate and does not act on ε-carotene (ε,ε-end). However, after removing the ε-end via 9′,10′ cleavage by BCMO2, BCMO1 precisely cleaves the 15,15′ bond and liberates retinal27.
While the elongases from Corynebacterium and Dietzia sp. CQ4 exhibited a significant activity towards natural substrate (C40-lycopene), they did not show any activity towards C50-lycopene. Considering that enzymes from both strains have significant activity in the C40-to-C50 context, it is unlikely that their inability to act on C50-lycopene reflects a lack of activity in E. coli. The more likely alternative is that some of these enzymes have higher substrate/size specificity and resist C50-lycopene as a substrate. Kim et al. showed that the Corynebacterium elongase CrtEb is functional in another non-cognate context and acts on the ψ-end of the C30 carotenoid 4,4′-diaponeurosporene25. Given this unpredictability of promiscuous functions toward non-cognate substrates, studies of several accessible gene candidates are required to identify genes that perform intended nonnatural tasks. Also, it is interesting how or whether Dietzia elongase and cyclase and Corynebacterium elongase, which were found non-functional in C50-to-C60 context in the present work, can acquire these new activities by mutations.
Methods
Strains and reagents
E. coli XL10-Gold cells were used for cloning, and XL1-Blue cells were used for carotenoid production. All enzymes were purchased from New England Biolabs. Lennox-LB Broth Base was purchased from Life Technologies, BactoTM Yeast Extract, and BactoTM Tryptone were purchased from BD Biosciences, and all other chemicals and reagents were obtained from Nacalai Tesque (Kyoto, Japan). The antibiotics carbenicillin and chloramphenicol were used at 30 and 50 µg/mL, respectively.
Plasmid construction
pUCara-crtIN304P, a plasmid encoding a crtIN304P gene downstream of an arabionse inducible araBAD promoter, was derived from a previous study17. The plasmids pUCara-crtIN304P-crtYeYfEbCg, pUCara-crtIN304P-crtYeYfEbCe, pUCara-crtIN304P-crtE2YgYhMl, and pUCara-crtIN304P-lbtABCCQ4 were constructed by amplifying genes for elongase and cyclase from various genomes using the primers listed in Supplementary Table 1 and cloning these into the ApaI/SpeI restriction site of pUCara-crtIN304P. The plasmids pAC-fdsY81A,V157A-crtMF26A,W38A,F233S and pAC-fdsY81M-crtMF26A,W38A were derived from the previous study17. The fds and crtM variant genes on these plasmids were expressed constitutively under lac promoter. Supplementary Table 2 shows the DNA sequence of the designed RBS construct. These sequences were concatenated and were then inserted into the ApaI/SpeI restriction site of pUCara-crtIN304P without spacer sequences.
Culture conditions
Single colonies were inoculated into 2 ml of LB media with antibiotics in culture tubes and were shaken at 37 °C for 16 h. Overnight cultures of 2 mL were diluted 100-fold into 40 mL of fresh Terrific Broth media in 200 mL flasks and were then shaken at 200 rpm in an incubator at 30 °C. After 8 h, 0.2% (w/v) arabinose inducer was added and cells were cultured for an additional 40 h.
Product extraction and purification
Cell cultures were centrifuged at 3,270 × g for 15 min at 4 °C. Cell pellets were washed with 10 ml of 0.9% (w/v) NaClaq and were then repelleted by centrifugation. Products were extracted by vigorously vortexing for 5 min in 10 ml of acetone containing 30-mg/L butylated hydroxytoluene. One-mL aliquots of hexane and 35-ml aliquots of 1% (w/v) NaClaq were then added, and the samples were centrifuged at 3,250 × g for 15 min. After collecting the product-containing hexane phase, the solvent was evaporated in a vacuum concentrator. Extracts were finally dissolved in 15–50-μl aliquots of tetrahydrofuran:methanol (6:4) for further analysis.
HPLC–MS analysis for compound identification and quantification
Aliquots (2–15 μL) of final extracts were analyzed using Shimadzu Prominence HPLC system equipped with LCMS-2020 MS spectrometer, a photodiode array (PDA) detector with Waters Spherisorb ODS2 Analytical Columns (4.6 × 250 mm, 5 µm, PSS831915). Mobile phases comprised acetonitrile/tetrahydrofuran/methanol (58:7:35) at a flow rate of 2 mL/min (Fig. 2a), acetonitrile/tetrahydrofuran/methanol (58:4:38) at a flow rate of 1 mL/min (Fig. 2b), and acetonitrile/tetrahydrofuran/methanol (30:7:63) at a flow rate of 1.5 mL/min (Fig. 3). Elutes were detected using PDA (200–700 nm) and APCI-MS. Mass scans were performed from m/z 10 to m/z 1000 with a 300 °C interface temperature, a 300 °C DL, a ± 4500-V interface voltage, and a neutral DL/Qarray with N2 as the nebulizing gas.
ESI TOF MS spectra were acquired using a Waters Xevo G2S Q TOF mass spectrometer (Waters Corporation, Milford, CT, USA) equipped with an Acquity UPLC system with scanning from m/z 100 to 1,500 with a capillary voltage of 3.2 kV, a cone voltage of 40 eV, and a source temperature of 120 °C. Nitrogen was used as the nebulizing gas at a flow rate of 30 L/h. MS/MS spectra were measured using a quadrupole-TOF MS/MS instrument with argon as a collision gas at a collision energy of 30 V. 1H NMR (500 MHz) including COSY and ROESY spectra were generated using a Varian UNITY INOVA 500 spectrometer in CDCl3 with tetramethylsilane as an internal standard.
Supplementary information
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology [JSPS KAKENHI Grants 15H04189, 15K14228, and 16H06450], the Hamaguchi Foundation for the Advancement of Biochemistry, the Futaba Electronics Memorial Foundation, and the Shorai Foundation for Science and Technology. L.L. is supported by a JSPS fellowship for young scientists [JSPS KAKENHI Grant 15J07486].
Author Contributions
S.W., L.L., M.F. and D.U. conceived and designed the study. S.W. and L.L. performed experiments. M.F., L.L. and D.U. analyzed the results. T.M. performed NMR and MS/MS analysis. S.K.N., K.S. and D.U. supervised the study. M.F., L.L. and D.U. wrote the paper.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ling Li and Maiko Furubayashi contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39289-w.
References
- 1.Mercadante, A., Egeland, E., Britton, G., Liaaen-Jensen, S. & Pfander, H. Carotenoids handbook. (eds Britton, G., Liaaen-Jensen, S., Pfander, H.) (Birkhäuser Basel, 2004).
- 2.Britton, G. Functions of intact carotenoids in Carotenoids (eds Britton G., Liaaen-Jensen S., Pfander H.) 189–212 (Birkhäuser Basel, 2008).
- 3.Alvarez R, Vaz B, Gronemeyer H, de Lera AR. Functions, therapeutic applications, and synthesis of retinoids and carotenoids. Chem. Rev. 2014;114:1–125. doi: 10.1021/cr400126u. [DOI] [PubMed] [Google Scholar]
- 4.Irimia-Vladu M, Sariciftci NS, Bauer S. Exotic materials for bio-organic electronics. J. of Mater. Chem. 2011;21:1350–1361. doi: 10.1039/c0jm02444a. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto, H., Uragami, C., Yukihira, N., Gardiner, A. T. & Cogdell, R. J. Understanding/unravelling carotenoid excited singlet states. J. R. Soc. Interface15, 10.1098/rsif.2018.0026 (2018). [DOI] [PMC free article] [PubMed]
- 6.Albrecht M, Takaichi S, Steiger S, Wang ZY, Sandmann G. Novel hydroxycarotenoids with improved antioxidative properties produced by gene combination in Escherichia coli. Nat. Biotechnol. 2000;18:843–846. doi: 10.1038/78443. [DOI] [PubMed] [Google Scholar]
- 7.Walter MH, Strack D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011;28:663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 8.Liaaen-Jensen S, Hertzberg S, Weeks OB, Schwieter U. Bacterial carotenoids XXVII. C50-carotenoids. 3. Structure determination of dehydrogenans-P439. Acta Chem. Scand. [A]. 1968;22:1171–1186. doi: 10.3891/acta.chem.scand.22-1171. [DOI] [PubMed] [Google Scholar]
- 9.Weeks OB, Andrewes AG, Brown BO, Weedon BC. Occurrence of C40 and C45 carotenoids in the C50 carotenoid system of Flavobacterium dehydrogenans. Nature. 1969;224:879–882. doi: 10.1038/224879a0. [DOI] [PubMed] [Google Scholar]
- 10.Netzer R, et al. Biosynthetic pathway for gamma-cyclic sarcinaxanthin in Micrococcus luteus: heterologous expression and evidence for diverse and multiple catalytic functions of C50 carotenoid cyclases. J. Bacteriol. 2010;192:5688–5699. doi: 10.1128/JB.00724-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krubasik P, Kobayashi M, Sandmann G. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur. J. Biochem. 2001;268:3702–3708. doi: 10.1046/j.1432-1327.2001.02275.x. [DOI] [PubMed] [Google Scholar]
- 12.Tao L, Yao H, Cheng Q. Genes from a Dietzia sp. for synthesis of C40 and C50 beta-cyclic carotenoids. Gene. 2007;386:90–97. doi: 10.1016/j.gene.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Heider SA, Peters-Wendisch P, Wendisch VF, Beekwilder J, Brautaset T. Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl. Microbiol. Biotechnol. 2014;98:4355–4368. doi: 10.1007/s00253-014-5693-8. [DOI] [PubMed] [Google Scholar]
- 14.Abbes, M. et al. Biological properties of carotenoids extracted from Halobacterium halobium isolated from a Tunisian solar saltern. BMC ComplementaryAltern. Med. 13, 255, 10.1186/1472-6882-13-255 (2013). [DOI] [PMC free article] [PubMed]
- 15.Mandelli F, Miranda VS, Rodrigues E, Mercadante AZ. Identification of carotenoids with high antioxidant capacity produced by extremophile microorganisms. World J. Microbiol. Biotechnol. 2012;28:1781–1790. doi: 10.1007/s11274-011-0993-y. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, et al. Complete Biosynthetic Pathway of the C50 Carotenoid Bacterioruberin from Lycopene in the Extremely Halophilic Archaeon Haloarcula japonica. J. Bacteriol. 2015;197:1614–1623. doi: 10.1128/JB.02523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furubayashi M, et al. A highly selective biosynthetic pathway to non-natural C50 carotenoids assembled from moderately selective enzymes. Nat. Commun. 2015;6:7534. doi: 10.1038/ncomms8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian T, Salis HM. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res. 2015;43:7137–7151. doi: 10.1093/nar/gkv635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furubayashi, M. et al. A High-Throughput Colorimetric Screening Assay for Terpene Synthase Activity Based on Substrate Consumption. PLoS ONE9, e93317, 10.1371/journal.pone.0093317 (2014). [DOI] [PMC free article] [PubMed]
- 22.Umeno D, Tobias AV, Arnold FH. Diversifying carotenoid biosynthetic pathways by directed evolution. Microbiol. Mol. Biol. Rev. 2005;69:51. doi: 10.1128/MMBR.69.1.51-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee PC, Momen AZ, Mijts BN, Schmidt-Dannert C. Biosynthesis of structurally novel carotenoids in Escherichia coli. Chem. Biol. 2003;10:453–462. doi: 10.1016/S1074-5521(03)00103-0. [DOI] [PubMed] [Google Scholar]
- 24.Mijts BN, Lee PC, Schmidt-Dannert C. Identification of a carotenoid oxygenase synthesizing acyclic xanthophylls: Combinatorial biosynthesis and directed evolution. Chem. Biol. 2005;12:453–460. doi: 10.1016/j.chembiol.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S. H., Kim, M. S., Lee, B. Y. & Lee, P. C. Generation of structurally novel short carotenoids and study of their biological activity. Sci. Rep.6, 21987, 10.1038/Srep21987 (2016). [DOI] [PMC free article] [PubMed]
- 26.Cunningham FX, Jr., Gantt E. One ring or two? Determination of ring number in carotenoids by lycopene epsilon-cyclases. Proc. Natl. Acad. Sci. USA. 2001;98:2905–2910. doi: 10.1073/pnas.051618398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison EH, Quadro L. Apocarotenoids: Emerging Roles in Mammals. Annu. Rev. Nutr. 2018;38:153–172. doi: 10.1146/annurev-nutr-082117-051841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.