Abstract
Copper-only superoxide dismutases (SODs) represent a new class of SOD enzymes that are exclusively extracellular and unique to fungi and oomycetes. These SODs are essential for virulence of fungal pathogens in pulmonary and disseminated infections, and we show here an additional role for copper-only SODs in promoting survival of fungal biofilms. The opportunistic fungal pathogen Candida albicans expresses three copper-only SODs, and deletion of one of them, SOD5, eradicated candidal biofilms on venous catheters in a rodent model. Fungal copper-only SODs harbor an irregular active site that, unlike their Cu,Zn-SOD counterparts, contains a copper co-factor unusually open to solvent and lacks zinc for stabilizing copper binding, making fungal copper-only SODs highly vulnerable to metal chelators. We found that unlike mammalian Cu,Zn-SOD1, C. albicans SOD5 indeed rapidly loses its copper to metal chelators such as EDTA, and binding constants for Cu(II) predict that copper-only SOD5 has a much lower affinity for copper than does Cu,Zn-SOD1. We screened compounds with a variety of indications and identified several metal-binding compounds, including the ionophore pyrithione zinc (PZ), that effectively inhibit C. albicans SOD5 but not mammalian Cu,Zn-SOD1. We observed that PZ both acts as an ionophore that promotes uptake of toxic metals and inhibits copper-only SODs. The pros and cons of a vulnerable active site for copper-only SODs and the possible exploitation of this vulnerability in antifungal drug design are discussed.
Keywords: copper, yeast, superoxide dismutase (SOD), superoxide ion, fungi, Candida, Cu-only SOD, metalloenzyme, mycocide, virulence, fungi, virulence
Introduction
Superoxide dismutases (SODs)3 are metalloenzymes that catalyze the disproportionation of the free radical superoxide anion to hydrogen peroxide and oxygen. Widely distributed from bacteria to humans, SODs play diverse roles in oxidative stress protection and in signaling involving reactive oxygen species (ROS). The eukaryotic intracellular Cu,Zn superoxide dismutase (SOD1) has been particularly well-characterized with regard to catalytic activity, structure, and function since its initial discovery 5 decades ago (1). In addition to intracellular SOD1, animals express an extracellular Cu,Zn-SOD (EC-SOD) to disproportionate extracellular sources of superoxide. EC-SOD is highly similar to intracellular SOD1 with regard to metal binding properties, sequence, and structure (2, 3).
Eukaryotic Cu,Zn-SODs have a distinctive Greek-key, β-barrel core structure and active site containing the catalytic copper ion and neighboring zinc that stabilizes the protein and aids in catalysis (4–7). In addition, Cu,Zn-SODs have a charged loop VII also known as the electrostatic loop (ESL) believed to guide superoxide anion to the active site (8–11). Structurally, the ESL helps shield the copper ion from solvent and stabilizes copper and zinc binding to the enzyme (12–14). Eukaryotic Cu,Zn-SODs have an exceptionally high affinity for copper, and features that contribute to this tight binding include the ESL and the zinc atom (13, 15–18). Such a high affinity for copper is necessary in the intracellular environment, where free copper is virtually unavailable (19), and allows copper to move along gradients of increasing affinity to pass from the copper chaperone CCS to the SOD1 enzyme (17, 20). EC-SOD acquires copper intracellularly and relies upon the copper chaperone ATOX1 and copper-transporting ATPases to deliver copper to the Golgi, where a direct handoff of copper from the ATPase to cuproenzymes has been proposed (21–24). Copper affinity gradients may also drive the copper loading of EC-SOD as has been shown for SOD1 (17).
We recently discovered a new class of eukaryotic copper-containing SODs that are exclusively extracellular and differ from the Cu,Zn family in structure and mechanism of copper acquisition (25). These copper-only SODs, which lack zinc-binding residues, are unique to the fungal kingdom and oomycete species (26). Structural analyses of copper-only SOD5 from the opportunistic fungal pathogen Candida albicans show that this enzyme lacks both zinc and sequences of the ESL, resulting in an open access copper site, compared with the buried copper atom of Cu,Zn-SODs (25). Unlike mammalian EC-SOD, copper-only SOD5 does not rely on copper loading through the secretory pathway, but rather is delivered to the cell surface as an apoprotein that acquires copper extracellularly (25). Currently, no data are available regarding the biophysical properties of copper binding to copper-only SODs. As this enzyme does not acquire copper inside the cell, where copper availability is extremely low, do copper-only SODs exhibit the same high affinity binding for copper as eukaryotic Cu,Zn-SODs? Additionally, fungal copper-only SODs lack both ESL residues and zinc that stabilize copper binding to Cu,Zn-SODs (25). Thus, it is possible that copper-only SODs have a decreased affinity for copper compared with their bimetallic counterparts, but this has never been tested.
Understanding the biochemical and biophysical properties of copper-only SODs is important in that members of this family have been shown to be vital for fungal pathogenesis (27–29). As extracellular enzymes, they represent the first line of defense against the oxidative attack of host immune cells (29–33). Copper-only SODs additionally participate in signaling pathways involving ROS (34), which may also promote fungal survival in the host. Copper-only SODs are important virulence factors in two pulmonary fungal pathogens. A deletion in the single copper-only SOD of Histoplasma capsulatum (SOD3) results in a dramatic loss of virulence in a mouse model of pulmonary infection (29). The fungal pathogen Paracoccidioides brasiliensis also encodes a single copper-only SOD3 that is required for fungal virulence and protection against host oxidative stress (28). In the case of the opportunistic fungal pathogen C. albicans, where much of the biochemical research on copper-only SODs has been completed, there are three extracellular copper-only SODs, SOD4–6. The organism can colonize a wide range of tissues, and blood stream infections are potentially fatal (35). Thus far, the requirement for copper-only SODs in C. albicans pathogenesis has been studied using a tail vein injection model for disseminated candidiasis, and loss of just one of the three SODs (SOD5) resulted in attenuation of virulence (27). However, in the clinic, one of the most common sources of systemic candidiasis are contaminated medical devices, such as catheters (36–40). No in vivo models have yet tested the importance of C. albicans copper-only SODs in these clinically relevant models for Candida.
Here, we show that deletion of just a single copper-only SOD (SOD5) of C. albicans is sufficient to totally inhibit fungal survival in a rat catheter model, underscoring the importance of these SODs in pathogenesis. Using C. albicans SOD5 as a model, we describe a possible Achilles heel of the enzyme. Specifically, we observe that SOD5 binds copper with a stability constant that is predicted to be substantially lower than that for mammalian Cu,Zn-SOD1. Screens for SOD5 inhibitors reveal an array of metal-binding chelators that selectively inhibit the copper-only SOD over a mammalian Cu,Zn-SOD1, consistent with the vulnerability of copper-only SODs to loss of their essential copper co-factor.
Results
Copper-only SOD5 promotes fungal survival in the host
In humans, life-threatening Candida bloodstream infections are often seeded by fungus-colonized implanted devices such as intravenous catheters, which account for between 25 and 40% of hospital-acquired candidemia infections (36–40). We therefore sought to expand the analysis of copper-only SODs in virulence to include studies of catheter infiltrations with C. albicans. Using a rat intravenous catheter model described previously (41), we compared the ability of WT C. albicans and an isogenic sod5Δ/Δ strain to form biofilms on the implanted device. As seen in Fig. 1A, the WT SC5314 clinical isolate formed robust biofilms consisting of fungal and host cells and extracellular matrix as described previously (41). By comparison, the sod5Δ/Δ null yeast exhibited no visible biofilms on this implanted device, with no detectable growth at ×1000 magnification (Fig. 1B). This sod5Δ/Δ defect in vivo is not due to an inherent defect in biofilm formation, as sod5Δ/Δ mutants were comparable with WT in their ability to assemble biofilms in vitro (Fig. 1C). The in vivo defect is likely due to clearance of fungal cells by the host immune response. Neutrophils represent a primary host defense against Candida biofilms on medical devices (41, 42), and planktonic sod5Δ/Δ cells are reportedly hypersensitive to killing by neutrophils (43). We show here that sod5Δ/Δ strains in biofilms are also hypersensitive to killing by neutrophils (Fig. 1D), providing a possible explanation for the in vivo biofilm defect. Overall, these studies show that elimination of just one of the three copper-only SOD proteins in C. albicans is sufficient to prevent fungal colonization of catheters, a critical source of potentially fatal fungal infections. This finding corroborates the notion that copper-only SODs are important mediators of fungal infections (27–29, 44).
Figure 1.
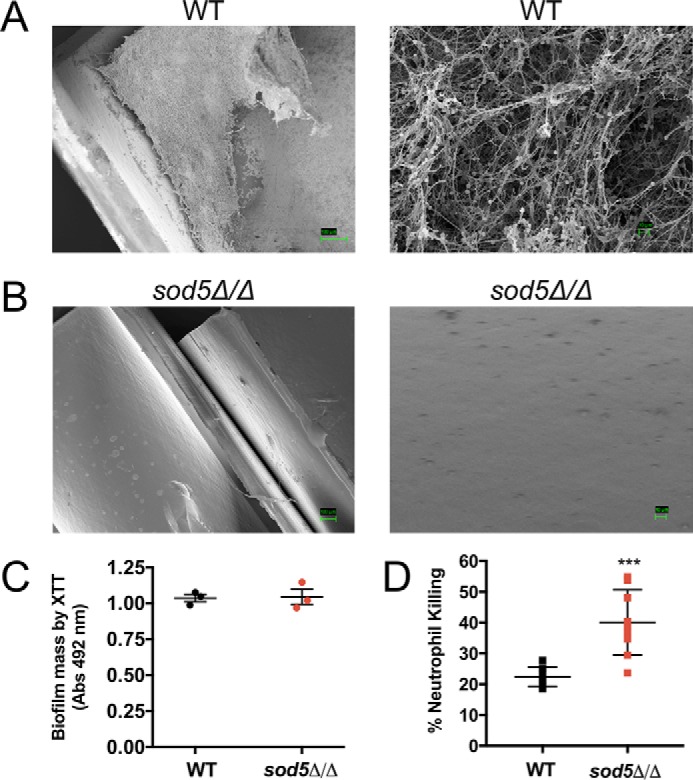
C. albicans copper-only SOD5 is essential for fungal survival in a rat intravenous catheter biofilm model. A and B, SC5314 (WT) or the isogenic sod5Δ/Δ strain were tested for biofilm formation in a rat intravenous catheter model as described under “Experimental Procedures.” Shown are scanning EM images taken at 250× and 1000× (WT) and at 150× and 1000× (sod5Δ/Δ) magnification. Results are representative of two independent experiments. C, biofilms from WT and the sod5Δ/Δ mutant were grown in vitro for 24 h, and biofilm mass was assessed by XTT reduction. Representative data are shown from the average of three samples from two experimental trials. Error bars, S.D. D, 24 h in vitro biofilms from WT and sod5Δ/Δ were tested for neutrophil killing as described under “Experimental procedures.” Results represent the average of nine samples over three experimental trials. Error bars, S.D. An unpaired t test was used to determine statistical significance. ***, p < 0.0009.
The labile active site of copper-only SOD5
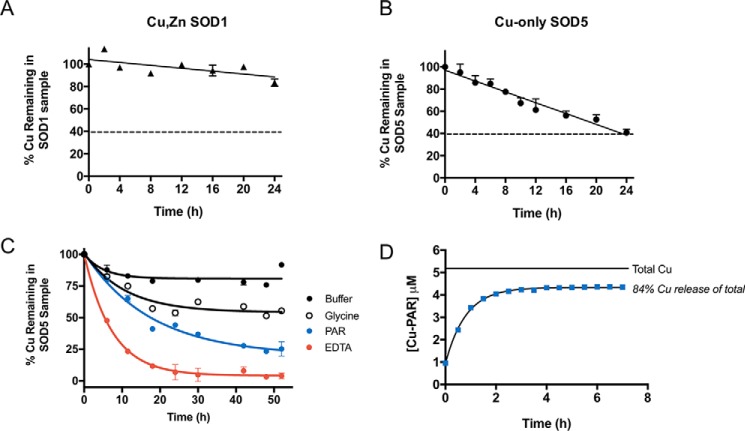
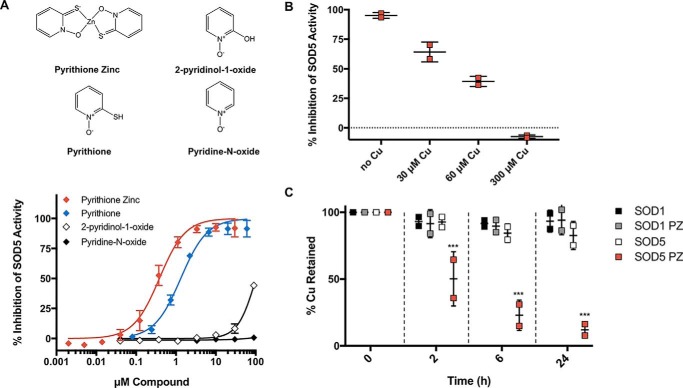
Based on the importance of copper-only SODs in fungal pathogenesis, we searched for possible vulnerabilities of these enzymes that may be exploited in the design of enzyme inhibitors. Given the more open active site of C. albicans copper-only SOD5 and the absence of zinc, we tested whether its copper co-factor was more labile than that of mammalian Cu,Zn-SOD1 and therefore more vulnerable to chelating agents. The dimeric mammalian Cu,Zn-SODs are known to be highly resistant to chelation by an excess of EDTA (15, 16, 18, 45). Consistent with these previous studies, bovine Cu,Zn-SOD1 retains 90% of its copper co-factor following 24 h of dialysis against a ∼275-fold molar excess of EDTA (Fig. 2A). By comparison, we noted that copper-only SOD5 loses copper to chelation much more rapidly than Cu,Zn-SOD1 (Fig. 2B) with a half-life of copper loss estimated to be ∼7.5 h. These findings suggested that the affinity of copper-only SOD5 for copper may be less than that of Cu,Zn-SOD1.
Figure 2.
Analysis of copper binding to copper-only SOD5. A–C, Cu,Zn-SOD1 (A) or copper-only SOD5 (B and C) were subjected to dialysis against the designated chelators at pH 8.0. Following the indicated time points, copper levels in the SOD-containing samples were measured by AAS and plotted as a percentage of total copper associated with the SOD prior to dialysis. A and B, dialysis against 1 mm EDTA in a Tris buffer was carried out at 4 °C using a system in which the dialysate can capture a maximum of 60% of the total copper subject to dialysis, represented by a dotted line (see “Experimental procedures”). C, dialysis against KPO4 buffer alone or buffer containing 1 mm glycine, 0.25 mm PAR, or 1 mm EDTA at 25 °C using a system in which the dialysate can capture a maximum of 97% of the total copper subject to dialysis (see “Experimental procedures”). Results represent averages of duplicate copper measurements and are representative of two experimental trials. Measurements obtained with glycine and PAR were used to estimate KD values for SOD5 binding to copper as defined in Table 1 and described under “Experimental procedures.” D, copper-only SOD5 was incubated with PAR for the indicated times as described under “Experimental procedures,” and PAR binding to copper was determined by absorbance at 500 nm. Total copper, the amount of copper associated with SOD5 prior to dialysis. 84% of this copper reacts with PAR at equilibrium as indicated. Values represent averages of triplicate samples and are representative of three independent experiments. KD estimations were calculated as described under “Experimental procedures” and defined in Table 1.
Mammalian Cu,Zn-SOD1 appears to bind copper with extraordinary affinity, and the enzyme is resistant to copper loss in the presence of chelators such as EDTA (16) and 2-pyridinecarboxylate (15), even after multiple days or weeks of dialysis, making it challenging to estimate copper-binding stability constants for native SOD1. Zinc-free bovine SOD1 has been shown to lose copper more quickly than the holo-Cu,Zn enzyme (16), and log K (stability constant) values of ∼15–16 have been estimated for zinc-free SOD1 (Table 1) (15). Additionally, treatment with reducing agents or denaturants has enabled measurements of copper binding to human Cu,Zn-SOD1 with reported log K of ∼15.6 (Cu(I)) and ∼17.2 (Cu(II)), respectively (17, 18). Such perturbations used to measure copper binding to Cu,Zn-SOD1 are not required for copper-only SOD5, where 100% of Cu(II) removal is achieved in less than 24 h of incubation in the presence of EDTA (Fig. 2B). Through equilibrium dialysis against glycine (Cu(II) log β′2 = 11.8) or 4-(2-pyridylazo)resorcinol (PAR) (Cu(II) log K′a1 = 12.1), we estimated the log K of Cu(II) binding to SOD5 to be 15.7 or 14.5, respectively, at pH 8.0 (Table 1). We further confirmed these values using a separate, previously described method for KD determination based on PAR–Cu complex formation, which resulted in a log K of Cu(II) binding to SOD5 = 15.5. It is noteworthy that these estimates obtained with copper-only SOD5 approximate that of zinc-deficient Cu,Zn-SOD1 (Table 1), consistent with the reported role of zinc in stabilizing copper binding to the enzymes (15, 16). Overall, our results indicate that fungal copper-only SOD5 binds copper with a stability constant that is predicted to be substantially lower than that for native mammalian Cu,Zn-SOD1.
Table 1.
Copper binding to Cu,Zn-SOD1 versus copper-only SOD5
| Protein | Chelator | Log K |
|---|---|---|
| Human Cu,Zn-SOD1 | PAR | 17.2a |
| Bovine Cu,Zn-SOD1 | 2-Pyridinecarboxylate | 15.6; 16.1b |
| C. albicans copper-only SOD5 | Glycine, PARD, PARC | 15.7, 14.5, 15.5c |
Metal chelators as chemical inhibitors of copper-only SOD5
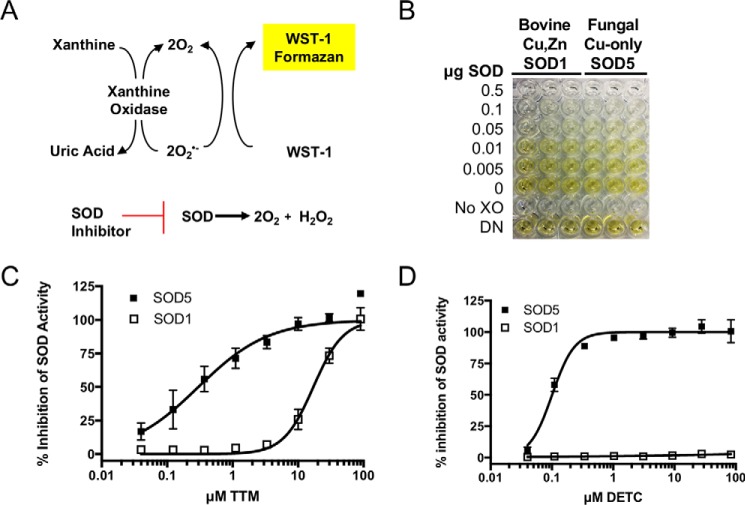
Our studies with Cu(II) binding to copper-only SOD5 suggested that this enzyme with its unusual open active site might be hypersensitive to inhibition by metal chelators or other selective small molecules. To this end, we optimized a high-throughput assay for SOD activity based on the water-soluble tetrazolium salt WST-1 (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfo-phenyl)-2H-tetrazolium, monosodium salt) (46, 47). In this system, superoxide is generated by xanthine oxidase and is detected by its reduction of the WST-1 tetrazolium salt to WST-1 formazan, a brightly yellow-colored compound visualized by absorbance at 450 nm (Fig. 3, A and B). Superoxide reduction of WST-1 is inhibited in the presence of SOD, providing a robust and stable assay for SOD activity with a Z′ = 0.9 (see “Experimental procedures”), equally effective for bovine Cu,Zn-SOD1 and fungal copper-only SOD5 (Fig. 3B). To validate this assay for revealing SOD inhibitors, we examined a known inhibitor of Cu,Zn-SOD1, ammonium tetrathiomolybdate (TTM), that works by chelating the copper co-factor of Cu,Zn-SOD1 (48) and has a very high affinity for Cu(I) (49). TTM showed dose-dependent inhibition of both Cu,Zn-SOD1 and copper-only SOD5, with more potent activity against SOD5 (Fig. 3C). The IC50 of TTM for bovine Cu,Zn-SOD1 in our assay was ∼17 μm, whereas fungal copper-only SOD5 required 50-fold less TTM for comparable inhibition. We also tested the Cu(II) and Cu(I) chelator diethyldithiocarbamate (DETC), reported as an inhibitor of both intracellular Cu,Zn-SOD1 (50, 51) and extracellular Cu,Zn EC-SOD from mammals (52). We observed that copper-only SOD5 is extremely sensitive to chemical inhibition by DETC, with an IC50 of ∼100 nm (Fig. 2D), whereas Cu,Zn-SOD1 examined in parallel showed no inhibition even at 100 μm DETC. The copper-only SOD5 is indeed hypersensitive to select copper chelators.
Figure 3.
A sensitive assay for SOD inhibitors reveals an enhanced sensitivity of copper-only SOD5 to TTM and DETC copper chelators. A, schematic showing the basis for the SOD assay whereby superoxide produced by xanthine oxidase reduces WST-1 to formazan, a reaction blocked by SOD. Inhibitors of SOD allow WST-1 formazan formation in a dose-dependent manner. B, application of the WST-1 reduction assay for SOD activity in a 96-well format shows equal reactivity of bovine Cu,Zn-SOD1 and fungal copper-only SOD5. Controls are no xanthine oxidase (No XO) and denatured enzyme, heated for 1 h at 90 °C (DN). C and D, ammonium tetrathiomolybdate (TTM, C) and diethyldithiocarbamate (DETC, D) were tested for dose-dependent inhibition against Cu,Zn-SOD1 and copper-only SOD5 in the WST-1 reduction assay. The chelators were added at t = 0 to the SOD activity assay containing 1.5 μg/ml SOD (90 nm SOD monomers), and the reaction proceeded for 45 min at 37 °C as described under “Experimental procedures.” Results are the averages of duplicate samples from three independent experiments. Error bars, S.E.
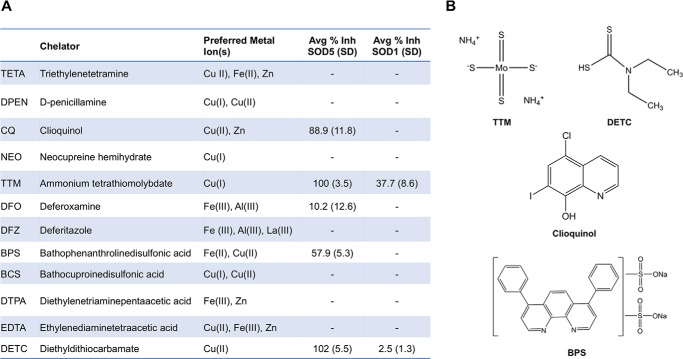
We next tested a small panel of known metal chelators with varying metal preferences and donor ligands (Fig. 4A) (49, 53–59). Aside from TTM and DETC, we identified two additional chelators with potent and selective inhibition of copper-only SOD5 over Cu,Zn-SOD1, including clioquinol (CQ) a well-documented Cu(II) and zinc chelator (55, 60–63), and bathophenanthroline disulfonic acid (BPS), a well-characterized Fe(II) chelator (64) that can also bind Cu(II) (59). The structures of these chelators are shown in Fig. 4B, and their efficacy in inhibiting SOD5 is shown in Fig. 4A. It is noteworthy that not all reported copper chelators showed preferential inhibition of copper-only SOD5. Several strong Cu(II) chelators including triethylenetetramine (TETA) and diethylenetriaminepentaacetic acid (DTPA) showed no inhibition against SOD5. CQ appears to coordinate Cu(II) with moderate affinity (55), and BPS has only been characterized as an Fe(II) chelator but is predicted to form a bis complex with Cu(II) (59). Physical interactions between the chemical compound and the SOD5 protein are likely to play key roles in dictating inhibitor efficacy as well as chelator metal affinities, as has been reported for chemical inhibitors of Cu,Zn-SOD1 (65).
Figure 4.
Targeted screen of metal chelators as inhibitors of copper-only SOD5. A, list of known metal chelators tested for reactivity with Cu,Zn-SOD1 and copper-only SOD5. Preferred Metal Ion(s), metal selectivity as previously published: TETA (53, 54), DPEN (54), CQ (54, 55), NEO (54, 56), TTM (49), DFO (57), DFZ (58), BPS (59), BCS (54, 59), DTPA (59), EDTA (54), and DETC (54). Avg % Inh, average percent inhibition of copper-only SOD5 and Cu,Zn-SOD1 with 10 μm compound as described in the legend to Fig. 3. Values are averages of two independent experimental trials with S.D. –, no detectable inhibition. B, structure of chelators showing selective inhibition of copper-only SOD5.
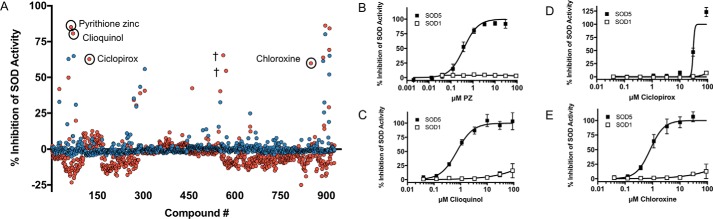
With a Z′ value of 0.9 and excellent reproducibility, we were confident that the WST assay could be applied to screen compounds on a larger scale for selective SOD5 inhibitors. We randomly screened ∼900 compounds from a library of drugs approved by the Food and Drug Administration, the Johns Hopkins Clinical Compound Library (66, 67). No inhibitors were identified that showed a preference for Cu,Zn-SOD1 over copper-only SOD5. However, a number of compounds showed >50% inhibition of copper-only SOD5 with virtually no inhibition of Cu,Zn-SOD1 at 10 μm drug (Fig. 5A). Notably, the Cu(II) chelator CQ identified in our targeted approach (Fig. 4) was also identified as a SOD5-specific inhibitor in the random library screen, validating our high-throughput platform. In addition to CQ, pyrithione zinc (PZ), ciclopirox (CP), and chloroxine (CX) were identified as selective inhibitors against copper-only SOD5 versus Cu,Zn-SOD1 (Fig. 5, A–E). These compounds, together with chelators from our targeted approach, were further validated and analyzed to determine IC50 values (Table 2). Interestingly, all of the top inhibitors of copper-only SOD5 detected in our random screen are predicted to bind metals. CX is in the same 8-hydroxyquinoline family as CQ (68). 8-Hydroxyquinoline derivatives are known to form a complex with Cu(II), providing a potential mechanism for activity against SOD5 (69). CP is a known antifungal that is a member of the hydroxamic acid class of inhibitors, whereby hydroximate can chelate enzyme-bound metal co-factors (70, 71). PZ not only binds zinc but also copper (72–74). PZ was the most effective compound revealed in our random screen with nanomolar inhibition against 90 nm copper-only SOD5 (Fig. 5A). PZ is a well-known antifungal used in anti-dandruff shampoo (75–77), and the antifungal mechanism has been attributed to its ability to act as a metal ionophore, bringing toxic levels of copper into the cell (73, 78). Our findings here indicate that PZ may have another target in fungi, namely the extracellular copper-only SODs. We therefore chose to focus on PZ for further study.
Figure 5.
High-throughput screen of a Food and Drug Administration–approved drug library for copper-only SOD5 inhibitors. A, approximately 900 compounds were tested at 10 μm for inhibition of Cu,Zn-SOD1 (blue) or copper-only SOD5 (red) using the high-throughput WST-1 assay of Fig. 3. The x axis numbers refer to an assigned number for each compound in the library. SOD5-selective inhibitors are labeled and circled. †, two compounds (magnesium carbonate and dimenhydrinate) appeared to have selective inhibition against SOD5 in the initial screen, but inhibition was not reproducible in follow-up studies. Results are the average of duplicate samples. B–E, SOD5-selective inhibitors were tested for dose-dependent inhibition, and IC50 values were determined for each compound using the WST-1 assay. Results are the average of nine samples over three experimental trials.
Table 2.
Average IC50 values for top compounds with selective inhibition against copper-only SOD5 versus Cu,Zn-SOD1
| Compound | Average IC50 ± S.E.a |
|
|---|---|---|
| Copper-only SOD5 | Cu,Zn-SOD1 | |
| Diethyldithiocarbamic acid | 100 ± 6 nm | NDb |
| Ammonium tetrathiomolybdate | 300 ± 69 nm | 17 ± 1.4 μm |
| Pyrithione Zinc | 400 ± 44 nm | ND |
| Clioquinol | 723 ± 128 nm | ND |
| Chloroxine | 816 ± 150 nm | ND |
| Bathophenanthroline sulfonic acid | 30 ± 1.6 μm | ND |
| Ciclopirox | 30 ± 18 μm | ND |
a Results are averages of nine samples over three experimental trials.
b ND, not defined; IC50 for Cu,Zn-SOD was not defined due to insufficient or absent inhibition of enzyme activity over the dose range of compounds tested (0.04–90 μm).
Characterization of pyrithione zinc as a selective copper-only SOD5 inhibitor
PZ is formed as a result of pyrithione (PT) coordination with Zn(II) (79), and PT is known to bind other metals, including Cu(II) (72–74). Is this metal-binding property important for PZ inhibition of SOD5? As PT forms a metal coordination complex based on its sulfur and oxygen donor ligands, we tested a series of PT derivatives (Fig. 6A, top) with substitutions at the thiol group for inhibition of copper-only SOD5. As seen in Fig. 6A (bottom), only PT and PZ showed clear dose-dependent inhibition; 2-pyridinol-1-oxide and pyridine-N-oxide with substitutions at the thiol group exhibited poor or undetectable inhibition of copper-only SOD5. Thus, metal binding capacity involving the thiol groups is likely important for the inhibition observed, but not the zinc atom itself. Because the free thiols of PT are easily oxidized in air (72), our subsequent studies focused primarily on PZ.
Figure 6.
Removal of copper from SOD5 by pyrithione derivatives. A (top), structures of PT derivatives as described under “Results.” Bottom, inhibition of SOD5 activity by the indicated compounds was tested as in Fig. 3. Results are the averages of nine samples over three experimental trials. Error bars, S.D. B, effects of copper on PZ inhibition of SOD5 were examined by preincubating the indicated amounts of CuSO4 with 3 μm PZ for 30 min prior to assaying SOD5 activity as in Fig. 3. Results show the mean values from two experimental trials. Wide bar, mean; error bars, S.D. C, the ability of PZ to remove copper from SOD was tested by dialyzing Cu,Zn-SOD1 and copper-only SOD5 against 100 μm PZ (gray and red squares, respectively) or against buffer without PZ (black and white squares, respectively) for the indicated times, as discussed under “Experimental procedures.” The SOD-containing samples were analyzed for copper content using AAS. Results are from two independent experiments. Error bars, S.D. PZ-treated SOD was compared with untreated SOD, and an unpaired t test was used to determine statistical significance. ***, p < 0.0001.
How does PZ inhibit SOD5? As one possibility, PZ (and PT) might act to bind the essential copper co-factor. Consistent with this notion, we observe that a 100-fold molar excess of copper over PZ was able to completely reverse the inhibition of copper-only SOD5 by PZ (Fig. 6B). To directly test the ability of PZ to remove the copper co-factor, we dialyzed copper-only SOD5 and Cu,Zn-SOD1 against PZ or buffer control and monitored copper loss from the SOD. We find that SOD5 lost over 50% of its copper to PZ following only 2 h dialysis, whereas Cu,Zn-SOD1 was refractory to copper loss in the presence of PZ (Fig. 6C). Together, our results support a copper-dependent mechanism for PZ inhibition of copper-only SOD5.
Pyrithione zinc effects on secreted SOD5 and on C. albicans cultures
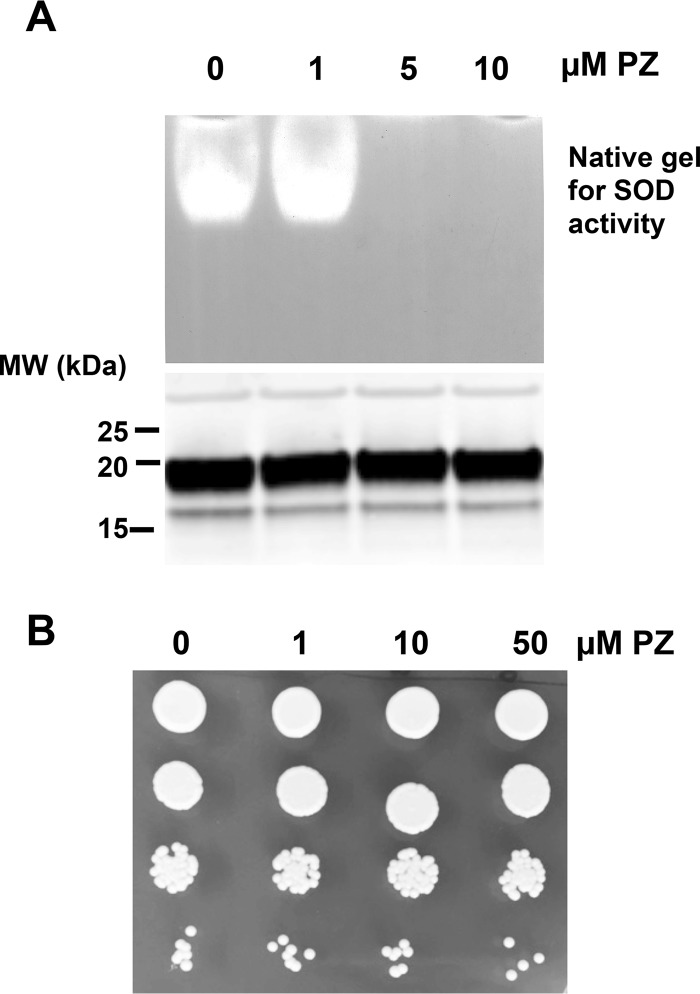
The aforementioned biochemical studies have all utilized recombinant SOD5 expressed in Escherichia coli and reconstituted in vitro with Cu(II). We therefore sought to test PZ activity against the glycosylated enzyme produced in the native host C. albicans. We used a strain of C. albicans that expresses a secreted SOD5 that is glycosylated but is missing the glycosylphosphatidylinositol anchor for attachment to the cell wall (25). Our previous studies have shown that SOD5 is secreted in an apo form and is activated by extracellular copper (25). In the experiment of Fig. 7A, SOD5 that accumulated in the extracellular medium was activated upon the addition of 5 μm copper; protein levels were monitored by immunoblotting, and enzyme activity was monitored by a native gel assay for SOD. This glycosylated enzyme is indeed susceptible to PZ, as activity was abolished following a 1-h treatment with 5 μm PZ (Fig. 7A).
Figure 7.
PZ effects in cultures of C. albicans secreting SOD5. C. albicans secreting SOD5 were cultured to mid-log phase as described under “Experimental procedures.” The extracellular growth medium (A) or cells (B) were treated with the indicated concentrations of PZ or DMSO control (0 PZ) for 1 h prior to analysis SOD5 (A) or cell viability (B). A, the growth medium (no cells) was supplemented with 5 μm CuSO4 for 10 min to activate SOD5 prior to PZ treatment. Medium was analyzed for SOD5 activity by a native gel assay (top), and SOD5 protein levels were analyzed by immunoblotting (bottom) as described under “Experimental procedures.” MW, molecular weight markers (kDa). B, cells were washed with sterile water, and 7000, 700, 70, and 7 cells were spotted on enriched YPD medium. Cells were photographed after 2 days.
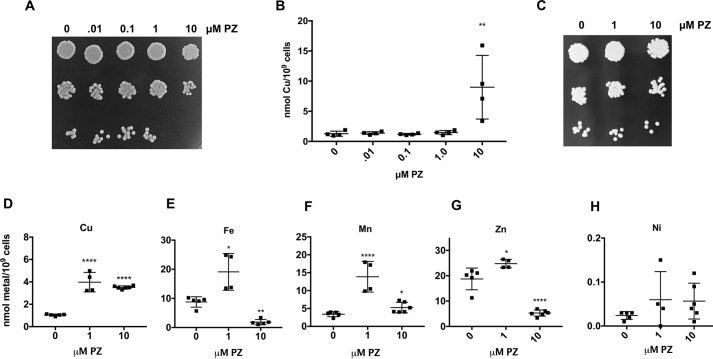
PZ is an established antifungal, and the mechanism has been ascribed to its ability to act as a metal ionophore, carrying toxic metals into the cell (73, 78). In studies that have been conducted with Malassezia globosa, Malassezia restricai, and the baker's yeast Saccharomyces cerevisiae, PZ toxicity has been associated with hyperaccumulation of metals, including copper (78) and zinc (80). The effect of PZ on growth and cellular metals in C. albicans has not been reported previously. We observed that within a 1-h incubation of PZ (the same time frame whereby PZ inhibits SOD5), there is no effect on cell viability (Fig. 7B). However, following prolonged (20-h growth) exposures to PZ, we found that 10 μm PZ was associated with >85% killing of the cells as defined by cfu (Fig. 8A). Furthermore, the toxicity seen with prolonged exposure to PZ was associated with an increase in intracellular copper (Fig. 8B). To examine effects of sublethal PZ treatment on C. albicans, we treated log-phase cells with 1 or 10 μm PZ for 6 h and assessed cell viability and intracellular metal levels. Although 6-h PZ treatment was not associated with loss in cell viability (Fig. 8C), it did result in significantly increased levels of intracellular copper (Fig. 8D). Iron, manganese, and zinc levels also rose at 1 μm PZ but less so at the higher dose, and if anything, intracellular iron and zinc decreased at 10 μm PZ (Fig. 8, E–G). PZ can therefore impact intracellular levels of multiple metals in C. albicans.
Figure 8.
Effects of PZ on C. albicans growth and intracellular metals. A–D, C. albicans strain SC5314 was either seeded at an A600 of 0.2 and grown for 20 h (A and B) or seeded at an A600 of 0.5 and grown for 6 h (C–H) in SC medium with varying concentrations of PZ or DMSO control. Cells were plated for cfu as in Fig. 7 (A and C) or analyzed for total cellular copper or the indicated metals (B and D–H) as described under “Experimental procedures.” A, 10 μm PZ incubated with cells for 20 h results in killing of 85% of C. albicans. B and D–H, results are from 4–6 samples from two or three independent experiments. Wide bar, mean; error bars, S.D. A one-way analysis of variance test for multiple comparisons was used to determine statistical significance of changes in PZ-treated samples compared with untreated controls. *, p < 0.05; **, p < 0.01; ****, p < .0001.
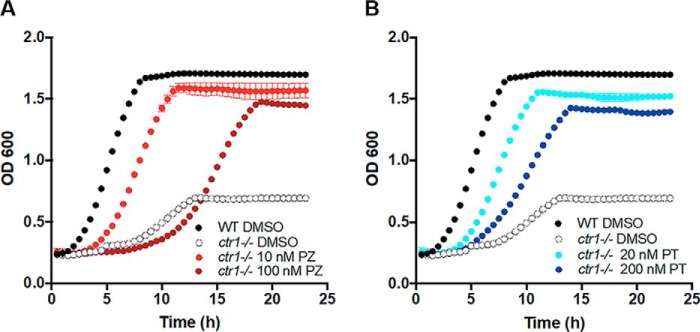
Ionophores, particularly those that bind metals, can have variable effects, depending on dose. For example, the copper ionophore elesclomol is an effective anticancer agent that causes oxidative damage to cancer cells by delivering toxic concentrations of copper to the mitochondria (81). More recently, however, low levels of elesclomol were seen to rescue copper deficiency in mammalian cells (82). We therefore tested whether PZ can also have both toxic and beneficial properties by addressing whether, at very low doses, it can rescue copper deficiency in C. albicans. For these purposes, we used a ctr1Δ/Δ mutant defective in copper uptake that grows poorly due to profound copper deficiency (83). Here, we demonstrate that as little as 10 nm PZ can rescue growth of the ctr1Δ/Δ strain, close to WT levels (Fig. 9A). The presence of zinc is not important for this rescue, and PT on its own can rescue the ctr1 deficiency (Fig. 9B). Therefore, as is the case with other metallophores, PZ and PT can either result in toxicity through metal overload or be beneficial to the cell by ameliorating metal starvation. Our studies implicate a third property of such agents: enzyme inactivation through metal chelation.
Figure 9.
Supplementation of copper as a nutrient by PZ or PT. SC5314 or the isogenic ctr1Δ/Δ strain were grown overnight in a 24-well plate at 30 °C with shaking in SC starting at an OD of 0.05 in the presence of the indicated concentration of PZ (A) or PT (B) or of DMSO as a vehicle control. Growth was monitored by absorbance at 600 nm at 30-min intervals over 24 h. Results are the average of three replicates and are representative of two independent experiments.
Discussion
Fungal infections are becoming increasingly prevalent across the globe, accompanied by an rise in drug-resistant pathogens and increased mortality due in part to emerging resistant strains of Candida and Cryptococcus species among others (84). This public health vulnerability warrants an urgent search for novel antifungal drugs and drug targets to combat the diverse array of current and emerging fungal pathogens. Here, we describe a potential new drug target worthy of consideration: the extracellular copper-only superoxide dismutases that are important for fungal pathogenesis, unique to the fungal kingdom, and selectively targetable by small molecules, leaving their highly conserved mammalian Cu,Zn-SOD counterparts unperturbed.
The importance of copper-only SODs in virulence was underscored in these studies by demonstrating a role for C. albicans SOD5 in promoting fungal survival in a rat model for catheter biofilms. C. albicans encodes three extracellular copper-only SODs, and loss of just one (SOD5) was sufficient to eliminate biofilm formation in this animal model. This deficiency is likely due to increased susceptibility to fungal killing by neutrophils through ROS, consistent with in vitro studies (32) (Fig. 1D), although this may not represent the whole story. In addition to dealing with host-derived superoxide, SOD5 partners with the recently identified fungal NADPH oxidase enzyme Fre8 to convert fungus-derived superoxide to H2O2 for downstream signaling and morphogenesis (34). Mutants in fre8Δ similarly exhibit a deficiency in biofilm formation in the rat catheter model (34), although not to the extent seen here with sod5Δ/Δ null. The potent virulence defects of sod5Δ/Δ cells in this model likely reflect a combined loss in fungal defense against host ROS and fungal morphogenesis defects related to Fre8.
We provide evidence that the affinity for Cu(II) is substantially lower in the case of C. albicans copper-only SOD5 compared with mammalian Cu,Zn-SOD1. Features that contribute to the high-affinity copper binding of eukaryotic Cu,Zn-SODs include the extended ESL (13) and zinc co-factor (15, 16), attributes missing in copper-only SOD5 (25). Contributions from zinc may be sufficient to explain the apparent disparity in binding affinities, as the stability constants we estimated for copper-only SOD5 approximate that of zinc-deficient SOD1 (15). In any case, the differential Cu(II)-binding properties may represent adaptations to the metal ion environments of these SODs. Cu,Zn-SOD1 resides in an intracellular milieu that is virtually devoid of bioavailable copper (19), and thermodynamic gradients help drive copper from high-affinity sites on GSH and the copper chaperone CCS to the ultimate high-affinity copper site in SOD1 (17, 20). Even the extracellular Cu,Zn-SOD3 of animals is believed to acquire copper inside the cells through a pathway requiring copper chaperones (21, 22). By contrast, extracellular copper-only SOD5 is charged with copper outside the cell from extracellular copper sources that are likely to be far more bioavailable (25). To date, there have been no fungal copper-only SODs predicted to be inside the cell (26, 44); their copper-binding properties appear compatible with an extracellular, but not intracellular, existence.
Interestingly, Cu,Zn-SODs of bacteria are also all extracellular or periplasmic, and these prokaryotic SODs appear to bind copper with less stability than mammalian Cu,Zn-SOD1, as they are more sensitive to inactivation by EDTA (86). Bacterial Cu,Zn-SODs can also be activated with copper from extracellular sources (87), except perhaps under copper starvation conditions (88). Thus, a lower copper-binding stability for extracellular SODs over intracellular SODs may be a unifying principal across microbes. In the case of copper-only SOD5, this may have the added advantage of preventing mis-metallation by noncopper ions (e.g. zinc). Cu,Zn-SOD1 can readily bind zinc as well as copper in the copper site (7), but copper-only SOD5 cannot stably bind zinc (25). Such metal selectively should prove beneficial to a SOD that acquires its metal from an extracellular environment of mixed metal pools.
We posit that the unique copper binding properties of fungal copper-only SODs (relatively lower affinity and open access site) makes them ideal candidates for chemical inhibition by metal-binding compounds. Indeed, through targeted and random screens, we have identified a number of metal-binding compounds that selectively inhibit copper-only SOD5 over mammalian Cu,Zn-SOD1. Interestingly, the metal-binding selectively of chelators did not perfectly correlate with their ability to inhibit SOD5, with some well-known copper chelators like d-penicillamine (DPEN), EDTA, and neocuproine hemihydrate (NEO) unable to inhibit SOD5, whereas BPS, a well-characterized Fe(II) chelator, was able to selectively inhibit SOD5. Additionally, serum contains a number of copper-binding proteins (ceruloplasmin, albumin, and transcuprein) (89), yet SOD5 maintains activity in the presence of serum, as defined by its ability to guard against the ROS attack of host immune cells (27, 31, 43) (Fig. 1D). Thus, not all copper-binding agents will inhibit SOD5 activity, and molecule structure and the ability to dock with the open active site of SOD5 must play important roles in determining efficacy as inhibitors. Based on the importance of copper-only SODs in pathogenesis, it will be worthwhile to fine-tune metal-binding compounds to maximize their efficacy as copper-only SOD inhibitors. Such inhibitors may augment the host defense involving ROS and represent potential new classes of antifungals.
The role of copper in infectious disease is multifaceted, as the metal is not only an essential nutrient but potentially toxic. The dual essential and toxic aspects of copper at the host–pathogen interface have been well-documented (90), and there is re-emerging interest in harnessing copper's toxicity in the development of antimicrobials, particularly in the use of copper-complexing compounds and copper ionophores (69, 91). Consistent with other studies (73, 78, 80), we show that the metallophore PZ can cause C. albicans toxicity through overload of copper and other metals. However, PZ can also remove copper from extracellular SOD5, representing another potential means of attacking the fungal pathogen. Interestingly, at very low doses that are not inhibitory to fungal growth or SOD5, PZ can actually be beneficial to the fungus and can rescue copper deficiency by delivering copper as a nutrient. These data are similar to what has been shown for Elesclomal, an anticancer drug that acts through copper toxicity at high doses but can also be used at low levels to rescue copper deficiency by selectively bringing copper into the mitochondria (81, 82). Caution should be taken in designing metallophores as anti-microbials, and both the potentially toxic and beneficial properties must be considered.
Experimental procedures
C. albicans strains and growth conditions
C. albicans strains used in this study include strain KC2 (ura3Δ::imm434/ura3Δ::imm434) engineered to secrete SOD5 (26), the clinical isolate SC5314, and mutant derivatives of SC5314, including the sod5Δ/Δ strain CA-JG201 (34) and the ctr1Δ/Δ mutant CA-E002. The latter was constructed using the SAT1-flipper cassette method (92) as follows. Deletion of a single CTR1 allele was accomplished with plasmid pECCTRL, whereby CTR1 residues −724 to −373 and +911 to +1232 were inserted, respectively, into the SacI and NotI and the XhoI and KpnI sites of pSFS2 (92). The cassette was mobilized by KpnI and SacI digestion and used to transform SC5314 by electroporation; the resultant ctr1Δ/+ mutant strain CA-EC001 was verified by PCR. Deletion of the second CTR1 allele employed a similar strategy using the pSFS2-based plasmid pECCTRS, harboring CTR1 sequences −373 to −3 and +609 to +910 and transformation of CA-EC001, creating the ctr1Δ/ctr1Δ strain CA-EC002.
Cultures of C. albicans cells were typically maintained at 30 °C in YPD, a yeast extract, peptone-based medium with 2% (w/v) glucose. For experiments involving the KC2 SOD5-expressing strain, cells were grown in a synthetic complete (SC) medium with 0.67% yeast nitrogen base and 2% dextrose (w/v) lacking cysteine and methionine (SC−Cys−Met) to induce expression of SOD5 under MET3 as described (25). Experiments involving SC5314 and the ctr1Δ/Δ strain were conducted with cells grown in SC medium. To assess cell viability, cells were harvested from liquid cultures by centrifugation, washed in sterile water, and spotted onto YPD plates in serial dilutions. Plates were incubated for 48 h at 30 °C prior to calculating cfu. To assess cell growth, SC5314 and ctr1Δ/Δ cells were started at an OD of 0.5 in 1 ml in a 24-well plate with or without drug and incubated at 30 °C with continuous shaking in a microplate reader (Biotek Eon) which measured the A600 every 30 min for 24 h.
In vivo and in vitro fungal biofilms and neutrophil-killing assays
A rat intravenous catheter biofilm model was used as described previously in which a catheter was surgically implanted in the jugular vein (41). 24 h later, C. albicans strains SC5314 and CA-JG201 were instilled in the catheter lumen at 106 cells/ml and flushed after 6 h. Following 24 h of biofilm growth, catheters were removed and fixed in 4% formaldehyde, 1% glutaraldehyde in PBS overnight. Catheters were then washed with PBS, treated with 1% osmium tetroxide, followed by another wash in PBS. Catheter samples were then dehydrated with ethanol washes finalized with a critical point drying step and mounted on aluminum stubs. After sputter-coating with platinum, a scanning electron microscope (LEO 1530) was used to image samples at 3 kV.
In vitro biofilms were grown as described previously (93). Briefly, C. albicans strains SC5314 and CA-JG201 were resuspended in RPMI-MOPS buffer at 1.5 × 106 cells/ml, and 200 μl was added to each well of a 96-well plate. Cells were incubated for 24 h at 37 °C with 5% CO2. Biofilms were either subjected to cell viability tests using an 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay as described previously (93) or washed in Dulbecco's PBS and used in neutrophil killing assays as follows. Human neutrophils were obtained from blood derived from volunteer donors with written informed consent through a protocol approved by the University of Wisconsin Internal Review Board. Primary human neutrophils were purified using the MACSxpress Neutrophil Isolation and MACSxpress Erythrocyte Depletion kits (Miltenyi Biotec Inc., Auburn, CA) as described previously (94). Neutrophils were added at 1.5 × 106 cells/well to C. albicans biofilms, which represented an effector/target ratio of 1:2. Cells were incubated for 4 h at 37 °C with 5% CO2 in RPMI 1640 (without phenol red) supplemented with 2% heat-inactivated fetal bovine serum and glutamine (0.3 mg/ml). An XTT-based metabolic assay was used to estimate C. albicans viability (94). 90 μl of 9:1 XTT working solution (0.75 mg/ml XTT in Dulbecco's PBS with 2% glucose/phenazine methosulfate, 0.32 mg/ml in double-distilled H2O) was added to each well. Following a 25-min incubation, samples were transferred to a Falcon 96-well U-bottom plate and centrifuged at 1200 × g for 3 min to pellet cells. Supernatants (110 μl) were transferred to a 96-well flat bottom plate, and absorption was read at 492 nm. A neutrophil-only control was used and subtracted for their contribution to the XTT values. Values were compared with wells without neutrophils to determine percent killing.
Biochemical analysis of recombinant C. albicans SOD5 from E. coli
Recombinant C. albicans SOD5 was expressed in E. coli, using expression plasmids encoding residues 27–181 with an N-terminal His10 tag and a tobacco etch virus protease cleavage site as described previously (25). The protein was purified and loaded with copper according to published procedures (25), and aliquots were stored in 25 mm acetate, pH 5.5, at −80 °C at concentrations of 5 mg/ml. Bovine Cu,Zn-SOD1 (Sigma catalog no. S8409, lot SLBN4913V) was obtained as an ammonium sulfate suspension. Prior to experimental analyses, 500-μl aliquots of SOD5 or 500 μl of Cu,Zn-SOD1 were dialyzed against 1 liter of appropriate reaction buffers (see below) at 4 °C overnight with one buffer replacement, using a 10,000 molecular weight cut-off dialysis device.
To compare the Cu(II) binding stability of Cu,Zn-SOD1 versus copper-only SOD5 in the presence of EDTA (Fig. 2, A and B), a rapid equilibrium dialysis plate with inserts (Thermo Fisher Scientific, 90006) was used. Enzymes were diluted in dialysis buffer (10 mm Tris, pH 8.0) to a final concentration of 0.17 mg/ml (SOD5) and 0.13 mg/ml (SOD1). At time 0, 400 μl of SOD1 or SOD5 with 1 mm EDTA was added to the sample chamber, and 600 μl of 10 mm Tris, 1 mm EDTA, pH 8.0, was added to the dialysis chamber. The plate was shaken at 4 °C for 24 h under aerobic conditions on an HSI Red Rotor shaker at maximum speed, and 25 μl of samples were collected from both the enzyme solution and the dialysate at designated time points up to 24 h. Samples were analyzed for copper content by atomic absorption spectroscopy (AAS) using an AAnalyst600 graphite furnace atomic absorption spectrometer (PerkinElmer Life Sciences).
For KD and binding constant estimations through equilibrium dialysis (Fig. 2C and Table 1), 10 μm SOD5 in 25 mm KPO4 buffer, pH 8.0, was dialyzed against buffer alone or against buffer containing 1 mm glycine, 0.25 mm PAR, or 1 mm EDTA in a minidialysis device (Thermo Fisher Scientific, 88401). Samples were shaken at 25 °C under aerobic conditions on an HSI Red Rotor shaker at speed 5, and 10-μl samples from the enzyme chamber were collected at the designated time points up to 52 h. Copper levels were determined using AAS as described above. Equilibrium was reached for the glycine sample by 30 and 52 h for PAR and EDTA. Initial and final concentrations of SOD5-associated copper and PAR or glycine associated copper were used to determine the KD of SOD5 and Cu(II) using a previously described approach based on competition for the same metal ion with two ligands (95). In our case, the competing ligands are glycine or PAR with different known Cu(II)-binding affinities (95). The two-ligand competition method relies on the final equilibrium concentrations of metal-bound protein (MP), free ligand (L'), metal-bound ligand (ML or ML2) and free protein (P), as listed in Reactions 1 and 2.
The apparent rate constants for Reactions 1 and 2, K′a and β′2, can be calculated using a pH-dependent α coefficient (αH-L) and the known affinity constant for each chelator and Cu(II) in Equation 1 or 2 (95).
| (Eq. 1) |
| (Eq. 2) |
The KD of SOD5 and Cu(II) can be determined using Equation 3 for the ligand (PAR) that has only one reported binding constant for Cu(II) and Equation 4 for ligands that form 1:1 or 1:2 complexes (glycine) as described previously (95). Note that for bidentate ligands, if K′a1[L′] ≥ 100 ≥ 1 and [ML2] ≥ 100[ML] ≫ [M], then free metal [M] and [ML] can be ignored in the final calculation for KD(SOD5–Cu) (95), and these conditions were met for our experiments.
| (Eq. 3) |
| (Eq. 4) |
The apparent binding constants, equilibrium concentrations of each species (Cu(II)-bound ligand, Cu(II)-bound protein, free ligand, and free protein), and final calculated values for the KD of SOD5 and Cu(II) at pH 8.0 and 25 °C using glycine and PAR are listed in Table S1. We estimated the log K for SOD5 and Cu(II) under these conditions to be 14.5 and 15.7 based on average values obtained with PAR and glycine, respectively, from two independent experiments.
As an independent method for estimating KD for SOD5 and Cu(II) (Fig. 2D and Table 1), we monitored copper release from SOD5 by PAR–Cu(II) complex formation through absorbance at 500 nm as described (18). 10 μl of 65 μm Cu(II)-SOD5 in 100 mm KPO4 buffer, pH 7.4, was added to 130 μl of the same buffer containing 100 μm PAR. These SOD-containing samples were plated in triplicate in a 96-well plate that also contained known amounts of CuSO4 standards (as determined by AAS) and 100 μm PAR to formulate a standard curve. Transparent adhesive film was added to prevent evaporation; the plate was incubated with shaking at 37 °C for 12 h, and the absorbance at 500 nm was measured at 30-min intervals in a microplate reader (Biotek Eon). The standard curve was used to determine the amount of Cu(II) bound to PAR (i.e. Cu(II) released from SOD5) at each time point until equilibrium was achieved. Using equilibrium concentrations of SOD5–Cu(II) and PAR–Cu(II), we used the previously calculated KD(PAR–Cu) value (2.6 × 10−15 m (18)) in Equation 5 to determine the average KD(SOD5–Cu) to be 3.6 × 10−16 m. The average log K of SOD5 binding to Cu(II) from three independent experiments under these conditions was determined to be 15.5.
| (Eq. 5) |
WST-1 assay for SOD activity and compound library screening
Copper-only C. albicans SOD5 and bovine Cu,Zn-SOD1 enzymatic activity was monitored using a WST-1–based reaction (46, 47). WST-1 (Dojindo) was stored as stable 30 mm aliquots in water at 4 °C. Just prior to assaying, recombinant SOD5 and SOD1 were diluted in 50 mm KPO4 buffer to 0.06 mg/ml stocks. Drugs and other compounds were dissolved in DMSO or water and diluted in DMSO. The Johns Hopkins Clinical Compound Library version 1.3 was provided in 96-well plate format, and master drug plates were made in DMSO at 400 μm. Drugs or DMSO alone were added to the assay plate, followed by 300 ng of SOD1 or SOD5 enzyme and reaction buffer (50 mm KPO4, pH 7.8, 0.1 mm xanthine, 0.2 mm EDTA, 0.3 mm WST-1, and 0.16 milliunits of xanthine oxidase) to a final volume of 200 μl. Plates were incubated at 37 °C for 45 min, and then each well was measured for absorbance at 450 nm. Three controls in triplicate were included on each plate, including a no drug/no DMSO control, DMSO vehicle alone, and no SOD enzyme. Percent inhibition of SOD activity was determined for all compounds by subtracting the A450 of the DMSO control from sample A450 and dividing by the no SOD control.
Inhibition values were plotted using Prism version 7 (GraphPad Software, Inc.), and nonlinear regression analysis was used to calculate an IC50 value for each compound. The quality of our assay for high-throughput screening was evaluated using the calculations for a Z′ factor using control data from five independent experiments (85). Z′ factors between 0.5 and 1.0 indicate a robust assay (85).
Biochemical analysis of SOD5 and metals in cultures of C. albicans
For analysis of SOD5 secreted from C. albicans, the KC2 strain expressing secreted SOD5 (25) (lacking the glycosylphosphatidylinositol anchor) was grown to late log phase (OD = 12). Cells were removed by centrifugation, and the extracellular medium was incubated with 5 μm CuSO4 for 10 min to activate secreted SOD5 (25), followed by a 1-h incubation at 30 °C with PZ or DMSO control. The medium was then concentrated (25), and SOD5 activity was analyzed by native gel electrophoresis and nitro blue tetrazolium staining as described previously using growth medium from the equivalent of ∼50 A600 units of cells (25). To determine SOD5 protein levels, the concentrated medium was treated with 500 units of endoglycosidase H (New England Biolabs, P0702S) at 37 °C overnight to deglycosylate SOD5, and the equivalent of 5 A600 units of cells was subjected to denaturing gel electrophoresis on a 4–12% BisTris gel, followed by immunoblotting using anti-SOD5 antibody (1:5000) and a secondary anti-rabbit antibody (1:10,000, Alexa Fluor 680) (26). SOD5 immunoreactivity was visualized using an Odyssey imaging system (LI-COR Biosciences).
To monitor metals in cells, 10 A600 units of cells were pelleted and washed twice with 10 mm Tris, 1 mm EDTA, pH 8.0, followed by two washes in deionized water. Pellets were then digested in 500 μl of 20% nitric acid at 100 °C overnight. Cell debris was removed by centrifugation at 20,000 × g for 5 min, and the supernatant was diluted in 14.5 ml of water to a final concentration of 2% nitric acid. Samples were analyzed for copper content by AAS as described above and for total metal content using inductively coupled plasma MS semiquantitative mode (Agilent 7700, University of Maryland, School of Pharmacy, Mass Spectrometry Center). Metal content was normalized to cell number.
Statistical and curve-fitting analyses
All statistical and curve-fitting analyses were completed using Prism version 7 (GraphPad Software). Individual tests and statistical significance are noted in each figure.
Ethics statement
Experiments involving the rat catheter model were approved by the University of Wisconsin (protocol DA0031, MV1947) Institutional Animal Care and Use Committees following guidelines under the Animal Welfare Act, the Institute of Laboratory Animal Resources Guide for the Care and Use of Laboratory Animals, and Public Health Service Policy. Studies involving human peripheral blood neutrophils were approved by the University of Wisconsin-Madison Internal Review Board under protocol 2013 1758. Blood was obtained from volunteer donors with written informed consent through a protocol approved by the University of Wisconsin Internal Review Board and compliant with the principles of the Declaration of Helsinki.
Author contributions
N. G. R., R. L. P., J. E. N., and V. C. C. conceptualization; N. G. R., R. L. P., D. A., and V. C. C. formal analysis; N. G. R. and D. A. validation; N. G. R., E. M. C., H. S., and J. E. N. investigation; N. G. R., E. M. C., R. L. P., and J. E. N. methodology; N. G. R. writing-original draft; N. G. R., E. M. C., R. L. P., D. A., J. E. N., and V. C. C. writing-review and editing; D. A., J. E. N., and V. C. C. supervision; V. C. C. project administration.
Supplementary Material
Acknowledgments
We thank Julie Gleason and Joseph Beckman for helpful discussions regarding fungal biofilms and copper binding experiments. We also thank David Sullivan for helpful discussions and provision of the clinical compound drug library.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1GM50016 and RO1 AI119949 (to V. C. C.), R01 AI073289 (to D. R. A.), F31 DK111114 (to E. M. C.), and F32 GM112320 (to R. L. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
- SOD
- superoxide dismutase
- EC-SOD
- extracellular Cu,Zn-SOD
- PZ
- pyrithione zinc
- ESL
- electrostatic loop
- ROS
- reactive oxygen species
- WST-1
- 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt
- PAR
- 4-(2-pyridylazo)resorcinol
- TETA
- triethylenetetramine
- DPEN
- d-penicillamine
- DTPA
- diethylenetriaminepentaacetic acid
- TTM
- tetrathiomolybdate
- DETC
- diethyldithiocarbamate
- BPS
- bathophenanthroline disulfonic acid
- CQ
- clioquinol
- CP
- ciclopirox
- CX
- chloroxine
- PT
- pyrithione
- SC
- synthetic complete
- XTT
- 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
- AAS
- atomic absorption spectroscopy
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. McCord J. M., and Fridovich I. (1969) Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244, 6049–6055 [PubMed] [Google Scholar]
- 2. Antonyuk S. V., Strange R. W., Marklund S. L., and Hasnain S. S. (2009) The structure of human extracellular copper-zinc superoxide dismutase at 1.7 Å resolution: insights into heparin and collagen binding. J. Mol. Biol. 388, 310–326 10.1016/j.jmb.2009.03.026 [DOI] [PubMed] [Google Scholar]
- 3. Hjalmarsson K., Marklund S. L., Engström A., and Edlund T. (1987) Isolation and sequence of complementary DNA encoding human extracellular superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 84, 6340–6344 10.1073/pnas.84.18.6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellerby L. M., Cabelli D. E., Graden J. A., and Valentine J. S. (1996) Copper-zinc superoxide dismutase: why not pH-dependent? J. Am. Chem. Soc. 118, 6556–6561 10.1021/ja953845x [DOI] [Google Scholar]
- 5. Potter S. Z., Zhu H., Shaw B. F., Rodriguez J. A., Doucette P. A., Sohn S. H., Durazo A., Faull K. F., Gralla E. B., Nersissian A. M., and Valentine J. S. (2007) Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J. Am. Chem. Soc. 129, 4575–4583 10.1021/ja066690+ [DOI] [PubMed] [Google Scholar]
- 6. Tainer J. A., Getzoff E. D., Beem K. M., Richardson J. S., and Richardson D. C. (1982) Determination and analysis of the 2 Å-structure of copper, zinc superoxide dismutase. J. Mol. Biol. 160, 181–217 10.1016/0022-2836(82)90174-7 [DOI] [PubMed] [Google Scholar]
- 7. Valentine J. S., Pantoliano M. W., McDonnell P. J., Burger A. R., and Lippard S. J. (1979) pH-dependent migration of copper(II) to the vacant zinc-binding site of zinc-free bovine erythrocyte superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 76, 4245–4249 10.1073/pnas.76.9.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fisher C. L., Cabelli D. E., Hallewell R. A., Beroza P., Lo T. P., Getzoff E. D., and Tainer J. A. (1997) Computational, pulse-radiolytic, and structural investigations of lysine-136 and its role in the electrostatic triad of human Cu,Zn superoxide dismutase. Proteins 29, 103–112 [DOI] [PubMed] [Google Scholar]
- 9. Fisher C. L., Cabelli D. E., Tainer J. A., Hallewell R. A., and Getzoff E. D. (1994) The role of arginine 143 in the electrostatics and mechanism of Cu,Zn superoxide dismutase: computational and experimental evaluation by mutational analysis. Proteins 19, 24–34 10.1002/prot.340190105 [DOI] [PubMed] [Google Scholar]
- 10. Getzoff E. D., Cabelli D. E., Fisher C. L., Parge H. E., Viezzoli M. S., Banci L., and Hallewell R. A. (1992) Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature 358, 347–351 10.1038/358347a0 [DOI] [PubMed] [Google Scholar]
- 11. Getzoff E. D., Tainer J. A., Weiner P. K., Kollman P. A., Richardson J. S., and Richardson D. C. (1983) Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature 306, 287–290 10.1038/306287a0 [DOI] [PubMed] [Google Scholar]
- 12. Banci L., Bertini I., Cabelli D. E., Hallewell R. A., Tung J. W., and Viezzoli M. S. (1991) A characterization of copper/zinc superoxide dismutase mutants at position 124. Zinc-deficient proteins. Eur. J. Biochem. 196, 123–128 10.1111/j.1432-1033.1991.tb15794.x [DOI] [PubMed] [Google Scholar]
- 13. Hayward L. J., Rodriguez J. A., Kim J. W., Tiwari A., Goto J. J., Cabelli D. E., Valentine J. S., and Brown R. H. Jr. (2002) Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 277, 15923–15931 10.1074/jbc.M112087200 [DOI] [PubMed] [Google Scholar]
- 14. Seetharaman S. V., Winkler D. D., Taylor A. B., Cao X., Whitson L. J., Doucette P. A., Valentine J. S., Schirf V., Demeler B., Carroll M. C., Culotta V. C., and Hart P. J. (2010) Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R. Biochemistry 49, 5714–5725 10.1021/bi100314n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirose J., Ohhira T., Hirata H., and Kidani Y. (1982) The pH dependence of apparent binding constants between apo-superoxide dismutase and cupric ions. Arch. Biochem. Biophys. 218, 179–186 10.1016/0003-9861(82)90334-4 [DOI] [PubMed] [Google Scholar]
- 16. Pantoliano M. W., Valentine J. S., Burger A. R., and Lippard S. J. (1982) A pH-dependent superoxide dismutase activity for zinc-free bovine erythrocuprein. Reexamination of the role of zinc in the holoprotein. J. Inorg. Biochem. 17, 325–341 10.1016/S0162-0134(00)80093-8 [DOI] [PubMed] [Google Scholar]
- 17. Banci L., Bertini I., Ciofi-Baffoni S., Kozyreva T., Zovo K., and Palumaa P. (2010) Affinity gradients drive copper to cellular destinations. Nature 465, 645–648 10.1038/nature09018 [DOI] [PubMed] [Google Scholar]
- 18. Crow J. P., Sampson J. B., Zhuang Y., Thompson J. A., and Beckman J. S. (1997) Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J. Neurochem. 69, 1936–1944 [DOI] [PubMed] [Google Scholar]
- 19. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., and O'Halloran T. V. (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284, 805–808 10.1126/science.284.5415.805 [DOI] [PubMed] [Google Scholar]
- 20. Morgan M. T., Nguyen L. A. H., Hancock H. L., and Fahrni C. J. (2017) Glutathione limits aquacopper(I) to sub-femtomolar concentrations through cooperative assembly of a tetranuclear cluster. J. Biol. Chem. 292, 21558–21567 10.1074/jbc.M117.817452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeney V., Itoh S., Wendt M., Gradek Q., Ushio-Fukai M., Harrison D. G., and Fukai T. (2005) Role of antioxidant-1 in extracellular superoxide dismutase function and expression. Circ. Res. 96, 723–729 10.1161/01.RES.0000162001.57896.66 [DOI] [PubMed] [Google Scholar]
- 22. Qin Z., Itoh S., Jeney V., Ushio-Fukai M., and Fukai T. (2006) Essential role for the Menkes ATPase in activation of extracellular superoxide dismutase: implication for vascular oxidative stress. FASEB J. 20, 334–336 10.1096/fj.05-4564fje [DOI] [PubMed] [Google Scholar]
- 23. Barry A. N., Otoikhian A., Bhatt S., Shinde U., Tsivkovskii R., Blackburn N. J., and Lutsenko S. (2011) The lumenal loop Met672-Pro707 of copper-transporting ATPase ATP7A binds metals and facilitates copper release from the intramembrane sites. J. Biol. Chem. 286, 26585–26594 10.1074/jbc.M111.229039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Köhn B., Ponnandai Shanmugavel K., Wu M., Kovermann M., and Wittung-Stafshede P. (2018) A luminal loop of Wilson disease protein binds copper and is required for protein activity. Biophys. J. 115, 1007–1018 10.1016/j.bpj.2018.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gleason J. E., Galaleldeen A., Peterson R. L., Taylor A. B., Holloway S. P., Waninger-Saroni J., Cormack B. P., Cabelli D. E., Hart P. J., and Culotta V. C. (2014) Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. U.S.A. 111, 5866–5871 10.1073/pnas.1400137111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peterson R. L., Galaleldeen A., Villarreal J., Taylor A. B., Cabelli D. E., Hart P. J., and Culotta V. C. (2016) The phylogeny and active site design of eukaryotic copper-only superoxide dismutases. J. Biol. Chem. 291, 20911–20923 10.1074/jbc.M116.748251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martchenko M., Alarco A. M., Harcus D., and Whiteway M. (2004) Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15, 456–467 10.1091/mbc.e03-03-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamayo D., Muñoz J. F., Lopez Á., Urán M., Herrera J., Borges C. L., Restrepo Á., Soares C. M., Taborda C. P., Almeida A. J., McEwen J. G., and Hernández O. (2016) Identification and analysis of the role of superoxide dismutases isoforms in the pathogenesis of Paracoccidioides spp. PLoS Negl. Trop. Dis. 10, e0004481 10.1371/journal.pntd.0004481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Youseff B. H., Holbrook E. D., Smolnycki K. A., and Rappleye C. A. (2012) Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 8, e1002713 10.1371/journal.ppat.1002713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Broxton C. N., and Culotta V. C. (2016) SOD Enzymes and microbial pathogens: surviving the oxidative storm of infection. PLoS Pathog. 12, e1005295 10.1371/journal.ppat.1005295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frohner I. E., Bourgeois C., Yatsyk K., Majer O., and Kuchler K. (2009) Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 71, 240–252 10.1111/j.1365-2958.2008.06528.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miramón P., Dunker C., Windecker H., Bohovych I. M., Brown A. J., Kurzai O., and Hube B. (2012) Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One 7, e52850 10.1371/journal.pone.0052850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schatzman S. S., and Culotta V. C. (2018) Chemical warfare at the microorganismal level: a closer look at the superoxide dismutase enzymes of pathogens. ACS Infect. Dis. 4, 893–903 10.1021/acsinfecdis.8b00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rossi D. C. P., Gleason J. E., Sanchez H., Schatzman S. S., Culbertson E. M., Johnson C. J., McNees C. A., Coelho C., Nett J. E., Andes D. R., Cormack B. P., and Culotta V. C. (2017) Candida albicans FRE8 encodes a member of the NADPH oxidase family that produces a burst of ROS during fungal morphogenesis. PLoS Pathog. 13, e1006763 10.1371/journal.ppat.1006763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim J., and Sudbery P. (2011) Candida albicans, a major human fungal pathogen. J. Microbiol. 49, 171–177 10.1007/s12275-011-1064-7 [DOI] [PubMed] [Google Scholar]
- 36. Percival S. L., Suleman L., Vuotto C., and Donelli G. (2015) Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 64, 323–334 10.1099/jmm.0.000032 [DOI] [PubMed] [Google Scholar]
- 37. Duggan S., Leonhardt I., Hünniger K., and Kurzai O. (2015) Host response to Candida albicans bloodstream infection and sepsis. Virulence 6, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Komshian S. V., Uwaydah A. K., Sobel J. D., and Crane L. R. (1989) Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev. Infect. Dis. 11, 379–390 10.1093/clinids/11.3.379 [DOI] [PubMed] [Google Scholar]
- 39. Miranda L. N., van der Heijden I. M., Costa S. F., Sousa A. P., Sienra R. A., Gobara S., Santos C. R., Lobo R. D., Pessoa V. P. Jr., and Levin A. S. (2009) Candida colonisation as a source for candidaemia. J. Hosp. Infect. 72, 9–16 10.1016/j.jhin.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 40. Richet H., Roux P., Des Champs C., Esnault Y., Andremont A., and French Candidemia Study Group (2002) Candidemia in French hospitals: incidence rates and characteristics. Clin. Microbiol. Infect. 8, 405–412 10.1046/j.1469-0691.2002.00446.x [DOI] [PubMed] [Google Scholar]
- 41. Nett J. E., and DRA. (2015) Fungal biofilms: in vivo models for discovery of anti-biofilm drugs. Microbiol. Spectr. 3, 10.1128/microbiolspec.MB-0008-2014 10.1128/microbiolspec.MB-0008-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nett J. E., Zarnowski R., Cabezas-Olcoz J., Brooks E. G., Bernhardt J., Marchillo K., Mosher D. F., and Andes D. R. (2015) Host contributions to construction of three device-associated Candida albicans biofilms. Infect. Immun. 83, 4630–4638 10.1128/IAI.00931-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fradin C., De Groot P., MacCallum D., Schaller M., Klis F., Odds F. C., and Hube B. (2005) Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol. Microbiol. 56, 397–415 10.1111/j.1365-2958.2005.04557.x [DOI] [PubMed] [Google Scholar]
- 44. Robinett N. G., Peterson R. L., and Culotta V. C. (2018) Eukaryotic copper-only superoxide dismutases (SODs): a new class of SOD enzymes and SOD-like protein domains. J. Biol. Chem. 293, 4636–4643 10.1074/jbc.TM117.000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Battistoni A., Folcarelli S., Cervoni L., Polizio F., Desideri A., Giartosio A., and Rotilio G. (1998) Role of the dimeric structure in Cu,Zn superoxide dismutase: pH-dependent, reversible denaturation of the monomeric enzyme from Escherichia coli. J. Biol. Chem. 273, 5655–5661 10.1074/jbc.273.10.5655 [DOI] [PubMed] [Google Scholar]
- 46. Peskin A. V., and Winterbourn C. C. (2017) Assay of superoxide dismutase activity in a plate assay using WST-1. Free Radic. Biol. Med. 103, 188–191 10.1016/j.freeradbiomed.2016.12.033 [DOI] [PubMed] [Google Scholar]
- 47. Peskin A. V., and Winterbourn C. C. (2000) A microtiter plate assay for superoxide dismutase using a water-soluble tetrazolium salt (WST-1). Clin. Chim. Acta 293, 157–166 10.1016/S0009-8981(99)00246-6 [DOI] [PubMed] [Google Scholar]
- 48. Juarez J. C., Betancourt O. Jr., Pirie-Shepherd S. R., Guan X., Price M. L., Shaw D. E., Mazar A. P., and Doñate F. (2006) Copper binding by tetrathiomolybdate attenuates angiogenesis and tumor cell proliferation through the inhibition of superoxide dismutase 1. Clin. Cancer Res. 12, 4974–4982 10.1158/1078-0432.CCR-06-0171 [DOI] [PubMed] [Google Scholar]
- 49. Smirnova J., Kabin E., Järving I., Bragina O., Tõugu V., Plitz T., and Palumaa P. (2018) Copper(I)-binding properties of de-coppering drugs for the treatment of Wilson disease: α-lipoic acid as a potential anti-copper agent. Sci. Rep. 8, 1463 10.1038/s41598-018-19873-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cocco D., Calabrese L., Rigo A., Argese E., and Rotilio G. (1981) Re-examination of the reaction of diethyldithiocarbamate with the copper of superoxide dismutase. J. Biol. Chem. 256, 8983–8986 [PubMed] [Google Scholar]
- 51. Misra H. P. (1979) Reaction of copper-zinc superoxide dismutase with diethyldithiocarbamate. J. Biol. Chem. 254, 11623–11628 [PubMed] [Google Scholar]
- 52. Marklund S. L. (1984) Properties of extracellular superoxide dismutase from human lung. Biochem. J. 220, 269–272 10.1042/bj2200269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kodama H., Murata Y., Iitsuka T., and Abe T. (1997) Metabolism of administered triethylene tetramine dihydrochloride in humans. Life Sci. 61, 899–907 10.1016/S0024-3205(97)00592-4 [DOI] [PubMed] [Google Scholar]
- 54. Ding X., Xie H., and Kang Y. J. (2011) The significance of copper chelators in clinical and experimental application. J. Nutr. Biochem. 22, 301–310 10.1016/j.jnutbio.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 55. Ferrada E., Arancibia V., Loeb B., Norambuena E., Olea-Azar C., and Huidobro-Toro J. P. (2007) Stoichiometry and conditional stability constants of Cu(II) or Zn(II) clioquinol complexes: implications for Alzheimer's and Huntington's disease therapy. Neurotoxicology 28, 445–449 10.1016/j.neuro.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 56. Gordge M. P., Meyer D. J., Hothersall J., Neild G. H., Payne N. N., and Noronha-Dutra A. (1995) Copper chelation-induced reduction of the biological activity of S-nitrosothiols. Br. J. Pharmacol. 114, 1083–1089 10.1111/j.1476-5381.1995.tb13317.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kiss T., and Farkas E. (1998) Metal-binding ability of desferrioxamine B. J. Inclus. Phenom. Mol. 32, 385–403 10.1023/A:1008046330815 [DOI] [Google Scholar]
- 58. Hider R. C., Kong X., Abbate V., Harland R., Conlon K., and Luker T. (2015) Deferitazole, a new orally active iron chelator. Dalton Trans. 44, 5197–5204 10.1039/C5DT00063G [DOI] [PubMed] [Google Scholar]
- 59. Haas K. L., and Franz K. J. (2009) Application of metal coordination chemistry to explore and manipulate cell biology. Chem. Rev. 109, 4921–4960 10.1021/cr900134a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bush A. I. (2002) Metal complexing agents as therapies for Alzheimer's disease. Neurobiol. Aging 23, 1031–1038 10.1016/S0197-4580(02)00120-3 [DOI] [PubMed] [Google Scholar]
- 61. Li C., Wang J., and Zhou B. (2010) The metal chelating and chaperoning effects of clioquinol: insights from yeast studies. J. Alzheimers Dis. 21, 1249–1262 10.3233/JAD-2010-100024 [DOI] [PubMed] [Google Scholar]
- 62. Pushie M. J., Nienaber K. H., Summers K. L., Cotelesage J. J., Ponomarenko O., Nichol H. K., Pickering I. J., and George G. N. (2014) The solution structure of the copper clioquinol complex. J. Inorg. Biochem. 133, 50–56 10.1016/j.jinorgbio.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 63. You Z., Ran X., Dai Y., and Ran Y. (2018) Clioquinol, an alternative antimicrobial agent against common pathogenic microbe. J. Mycol. Med. 28, 492–501 10.1016/j.mycmed.2018.03.007 [DOI] [PubMed] [Google Scholar]
- 64. Cowart R. E., Singleton F. L., and Hind J. S. (1993) A comparison of bathophenanthrolinedisulfonic acid and ferrozine as chelators of iron(II) in reduction reactions. Anal. Biochem. 211, 151–155 10.1006/abio.1993.1246 [DOI] [PubMed] [Google Scholar]
- 65. Dong X., Zhang Z., Zhao J., Lei J., Chen Y., Li X., Chen H., Tian J., Zhang D., Liu C., and Liu C. (2016) The rational design of specific SOD1 inhibitors via copper coordination and their application in ROS signaling research. Chem. Sci. 7, 6251–6262 10.1039/C6SC01272H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chong C. R., Chen X., Shi L., Liu J. O., Sullivan D. J. Jr. (2006) A clinical drug library screen identifies astemizole as an antimalarial agent. Nat. Chem. Biol. 2, 415–416 10.1038/nchembio806 [DOI] [PubMed] [Google Scholar]
- 67. Sanders N. G., Sullivan D. J., Mlambo G., Dimopoulos G., and Tripathi A. K. (2014) Gametocytocidal screen identifies novel chemical classes with Plasmodium falciparum transmission blocking activity. PLoS One 9, e105817 10.1371/journal.pone.0105817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lind S. E., Park J. S., and Drexler J. W. (2009) Pyrithione and 8-hydroxyquinolines transport lead across erythrocyte membranes. Transl. Res. 154, 153–159 10.1016/j.trsl.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 69. Dalecki A. G., Crawford C. L., and Wolschendorf F. (2017) Copper and antibiotics: discovery, modes of action, and opportunities for medicinal applications. Adv. Microb. Physiol. 70, 193–260 10.1016/bs.ampbs.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 70. Lee R. E., Liu T. T., Barker K. S., Lee R. E., and Rogers P. D. (2005) Genome-wide expression profiling of the response to ciclopirox olamine in Candida albicans. J. Antimicrob. Chemother. 55, 655–662 10.1093/jac/dki105 [DOI] [PubMed] [Google Scholar]
- 71. Subissi A., Monti D., Togni G., and Mailland F. (2010) Ciclopirox: recent nonclinical and clinical data relevant to its use as a topical antimycotic agent. Drugs 70, 2133–2152 10.2165/11538110-000000000-00000 [DOI] [PubMed] [Google Scholar]
- 72. Doose C. A., Szaleniec M., Behrend P., Müller A., and Jastorff B. (2004) Chromatographic behavior of pyrithiones. J. Chromatogr. A 1052, 103–110 10.1016/j.chroma.2004.08.028 [DOI] [PubMed] [Google Scholar]
- 73. Reeder N. L., Xu J., Youngquist R. S., Schwartz J. R., Rust R. C., and Saunders C. W. (2011) The antifungal mechanism of action of zinc pyrithione. Br. J. Dermatol. 165, 9–12 10.1111/j.1365-2133.2011.10571.x [DOI] [PubMed] [Google Scholar]
- 74. Helsel M. E., White E. J., Razvi S. Z., Alies B., and Franz K. J. (2017) Chemical and functional properties of metal chelators that mobilize copper to elicit fungal killing of Cryptococcus neoformans. Metallomics 9, 69–81 10.1039/C6MT00172F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Marks R., Pearse A. D., and Walker A. P. (1985) The effects of a shampoo containing zinc pyrithione on the control of dandruff. Br. J. Dermatol. 112, 415–422 10.1111/j.1365-2133.1985.tb02314.x [DOI] [PubMed] [Google Scholar]
- 76. Naldi L., and Diphoorn J. (2015) Seborrhoeic dermatitis of the scalp. BMJ Clin. Evid. 2015, 1713 [PMC free article] [PubMed] [Google Scholar]
- 77. Veien N. K., Pilgaard C. E., and Gade M. (1980) Seborrhoeic dermatitis of the scalp treated with a tar/zinc pyrithione shampoo. Clin. Exp. Dermatol. 5, 53–56 10.1111/j.1365-2230.1980.tb01665.x [DOI] [PubMed] [Google Scholar]
- 78. Reeder N. L., Kaplan J., Xu J., Youngquist R. S., Wallace J., Hu P., Juhlin K. D., Schwartz J. R., Grant R. A., Fieno A., Nemeth S., Reichling T., Tiesman J. P., Mills T., Steinke M., et al. (2011) Zinc pyrithione inhibits yeast growth through copper influx and inactivation of iron-sulfur proteins. Antimicrob. Agents Chemother. 55, 5753–5760 10.1128/AAC.00724-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Barnett B. L., Kretschmar H. C., and Hartman F. A. (1977) Structural characterization of bis(N-oxopyridine-2-thionato)zinc(III). Inorg. Chem. 16, 1834–1838 10.1021/ic50174a002 [DOI] [Google Scholar]
- 80. Park M., Cho Y. J., Lee Y. W., and Jung W. H. (2018) Understanding the mechanism of action of the anti-dandruff agent zinc pyrithione against Malassezia restricta. Sci. Rep. 8, 12086 10.1038/s41598-018-30588-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nagai M., Vo N. H., Shin Ogawa L., Chimmanamada D., Inoue T., Chu J., Beaudette-Zlatanova B. C., Lu R., Blackman R. K., Barsoum J., Koya K., and Wada Y. (2012) The oncology drug elesclomol selectively transports copper to the mitochondria to induce oxidative stress in cancer cells. Free Radic. Biol. Med. 52, 2142–2150 10.1016/j.freeradbiomed.2012.03.017 [DOI] [PubMed] [Google Scholar]
- 82. Soma S., Latimer A. J., Chun H., Vicary A. C., Timbalia S. A., Boulet A., Rahn J. J., Chan S. S. L., Leary S. C., Kim B. E., Gitlin J. D., and Gohil V. M. (2018) Elesclomol restores mitochondrial function in genetic models of copper deficiency. Proc. Natl. Acad. Sci. U.S.A. 115, 8161–8166 10.1073/pnas.1806296115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mackie J., Szabo E. K., Urgast D. S., Ballou E. R., Childers D. S., MacCallum D. M., Feldmann J., and Brown A. J. (2016) Host-imposed copper poisoning impacts fungal micronutrient acquisition during systemic Candida albicans infections. PLoS One 11, e0158683 10.1371/journal.pone.0158683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Geddes-McAlister J., and Shapiro R. S. (2019) New pathogens, new tricks: emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann. N.Y. Acad. Sci. 1435, 57–78 10.1111/nyas.13739 [DOI] [PubMed] [Google Scholar]
- 85. Zhang J. H., Chung T. D. Y., and Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- 86. Gabbianelli R., D'Orazio M., Pacello F., O'Neill P., Nicolini L., Rotilio G., and Battistoni A. (2004) Distinctive functional features in prokaryotic and eukaryotic Cu,Zn superoxide dismutases. Biol. Chem. 385, 749–754 [DOI] [PubMed] [Google Scholar]
- 87. Fenlon L. A., and Slauch J. M. (2017) Cytoplasmic copper detoxification in Salmonella can contribute to SodC metalation but is dispensable during systemic infection. J. Bacteriol. 199, e00437–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Osman D., Patterson C. J., Bailey K., Fisher K., Robinson N. J., Rigby S. E., and Cavet J. S. (2013) The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 87, 466–477 10.1111/mmi.12107 [DOI] [PubMed] [Google Scholar]
- 89. Linder M. C. (2016) Ceruloplasmin and other copper binding components of blood plasma and their functions: an update. Metallomics 8, 887–905 10.1039/C6MT00103C [DOI] [PubMed] [Google Scholar]
- 90. García-Santamarina S., and Thiele D. J. (2015) Copper at the fungal pathogen-host axis. J. Biol. Chem. 290, 18945–18953 10.1074/jbc.R115.649129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Djoko K. Y., Goytia M. M., Donnelly P. S., Schembri M. A., Shafer W. M., and McEwan A. G. (2015) Copper(II)-bis(thiosemicarbazonato) complexes as antibacterial agents: insights into their mode of action and potential as therapeutics. Antimicrob. Agents Chemother. 59, 6444–6453 10.1128/AAC.01289-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Jiménez-Lopez C., Collette J. R., Brothers K. M., Shepardson K. M., Cramer R. A., Wheeler R. T., and Lorenz M. C. (2013) Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot. Cell 12, 91–100 10.1128/EC.00290-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nett J. E., Cain M. T., Crawford K., and Andes D. R. (2011) Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J. Clin. Microbiol. 49, 1426–1433 10.1128/JCM.02273-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Johnson C. J., Cabezas-Olcoz J., Kernien J. F., Wang S. X., Beebe D. J., Huttenlocher A., Ansari H., and Nett J. E. (2016) The extracellular matrix of Candida albicans biofilms impairs formation of neutrophil extracellular traps. PLoS Pathog. 12, e1005884 10.1371/journal.ppat.1005884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Xiao Z., and Wedd A. G. (2010) The challenges of determining metal-protein affinities. Nat. Prod. Rep. 27, 768–789 10.1039/b906690j [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.