Abstract
Spontaneous formation of isoaspartates (isoDs) often causes protein damage. l-Isoaspartate O-methyltransferase (PIMT) repairs isoD residues by catalyzing the formation of an unstable l-isoaspartyl methyl ester that spontaneously converts to an l-aspartyl residue. PIMTs are widely distributed in all three domains of life and have been studied most intensively in connection with their role in protein repair and aging in plants and animals. Studies of bacterial PIMTs have been limited to Escherichia coli, which has one PIMT. The α-proteobacterium Rhodopseudomonas palustris has three annotated PIMT genes, one of which (rpa2580) has been found to be important for cellular longevity in a growth-arrested state. However, the biochemical activities of these three R. palustris PIMTs are unknown. Here, we expressed and characterized all three annotated PIMT proteins, finding that two of them, RPA0376 and RPA2838, had PIMT activity, whereas RPA2580 did not. RPA0376 and RPA2838 single- and double-deletion mutants did not differ in longevity from WT R. palustris and did not exhibit elevated levels of isoD residues in aged cells. Comparative sequence analyses revealed that RPA2580 belongs to a separate phylogenetic group of annotated PIMT proteins present in the α-proteobacteria. Our results suggest that this group of proteins is not involved in repair of protein isoD residues. In addition, the bona fide bacterial PIMT enzymes may play a different or subtler role in bacterial physiology than previously suggested.
Keywords: bacteria, bacterial metabolism, bioinformatics, enzyme catalysis, enzyme kinetics, enzyme purification, bacterial longevity, methyltransferase, protein damage, protein repair, Rhodopseudomonas palustris
Introduction
We have been engaged in studies of how nonspore forming bacteria are able to remain viable in a growth-arrested state for long periods of time. Our model organism is the phototrophic α-proteobacterium Rhodopseudomonas palustris, which maintains viability for months after entering a state of growth arrest when incubated in light, a condition under which it can generate ATP. Among a set of genes we identified as required for R. palustris longevity is rpa2580 (1), annotated as encoding a possible protein l-isoaspartate O-methyltransferase (PIMT).2 l-Isoaspartate (isoD) is a β-amino acid that accumulates in aged proteins because of spontaneous deamidation and isomerization of aspartates and asparagines. IsoD residues introduce kinks in protein backbones, which can lead to the loss of activity (2–5). PIMT is an almost universally distributed enzyme that repairs this damage by transferring a methyl group from S-adenosylmethionine (SAM) to the side chain of an isoD residue. The formed methyl ester cyclizes to l-succinimide with an accompanying release of methanol. l-Succinimide then spontaneously converts to l-aspartate ∼25% of the time (Fig. 1) (5–7). The repairing function of PIMT and its connection to longevity have been well-documented in Xenopus laevis (8), Drosophila melanogaster (9), potato tubers (10), barley seeds (11), and other eukaryotes (12–14). However, this enzyme has received scant attention in microbes. Escherichia coli encodes a PIMT enzyme, named PCM, that repairs isoD damage in vitro (15) and enhances the survival of stationary phase E. coli subjected to secondary stresses such as oxidative stress or high pH (16, 17).
Figure 1.
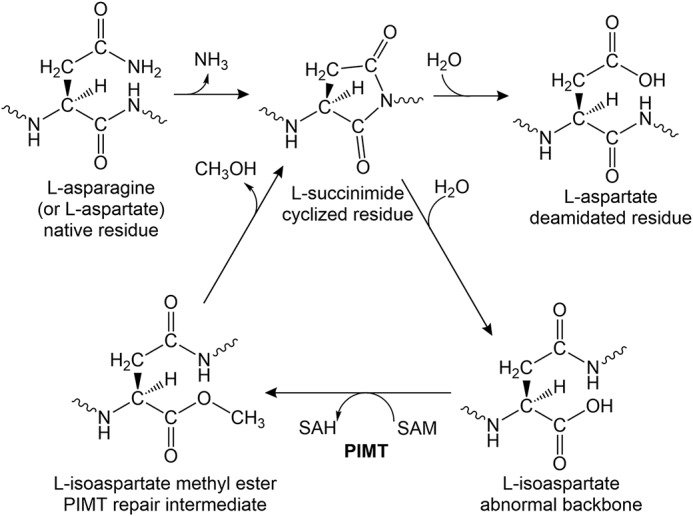
The pathway of l-isoaspartyl repair by PIMTs. l-Aspartyl and l-asparaginyl residues can spontaneously cyclize to l-succinimides. The succinimides undergo spontaneous hydrolysis resulting in a regeneration of l-aspartyl residues with a frequency of ∼25%, but this is also accompanied by the generation of abnormal l-isoaspartyl residues with a frequency of ∼75%. l-Isoaspartates introduce kinks into the backbone of proteins, which can compromise structure and activity. The repair enzyme PIMT transfers a methyl group from SAM to the side chain of l-isoaspartate, forming l-isoaspartate methyl ester. The methyl ester is unstable and spontaneously converts back to l-succinimide, which again hydrolyzes to form a combination of l-aspartyl and l-isoaspartyl residues.
It turns out that R. palustris has three genes annotated to encode PIMT enzymes (18). Because of its importance for R. palustris longevity (1), we decided to investigate the biochemical function of RPA2580, as well as its two other PIMTs. We found that two of the three proteins had PIMT activity in vitro. However, neither of the two PIMT-active proteins was important for R. palustris longevity, even though aged cells had relatively high levels of isoD damage. The longevity protein, RPA2580, did not have PIMT activity in our assays. Bioinformatics analysis also supported the notion that RPA2580 belongs to a group that has diverged from functional PIMTs.
Results
Sequence analysis of R. palustris PIMT genes
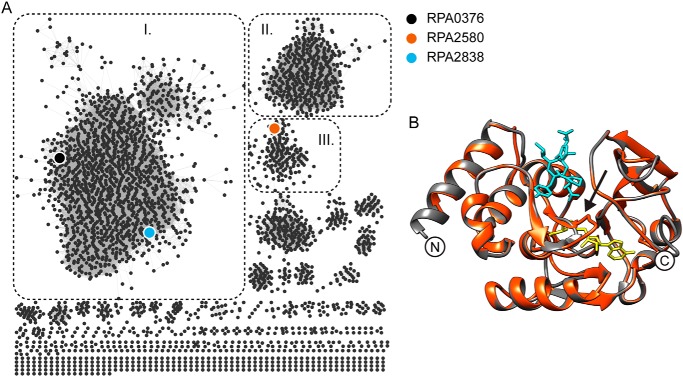
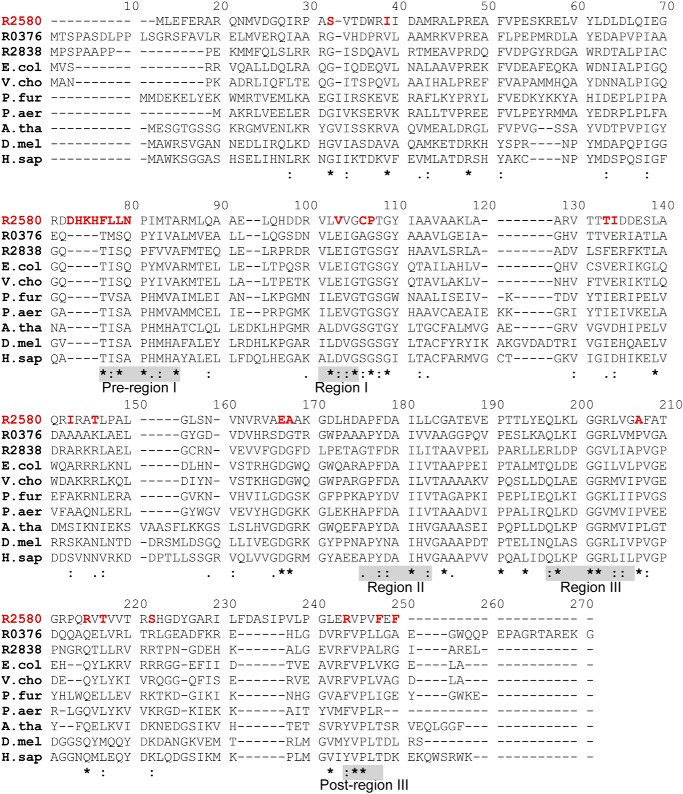
A survey of 5149 proteobacterial genomes revealed that 2669 of them have at least one PIMT gene and many have multiple PIMT sequences for a total of 4040 sequences (19). Three genes, rpa0376, rpa2580, and rpa2838, are annotated to encode PIMT proteins in R. palustris (18). To understand their divergence at the amino acid level, we analyzed the sequence similarity network of all the proteins containing a PIMT domain (PFAM family PF01135) (Fig. 2A) (20). The largest network, which includes RPA0376 and RPA2838, contains only bacterial and archaeal PIMTs (Fig. 2A, group I). The eukaryote PIMTs belong to a distinct phylogenetic grouping (Fig. 2A, group II). A separate network that includes RPA2580 has ∼700 sequences, all of which come from members of the α-proteobacteria (Fig. 2A, group III). PIMT proteins share a similar overall structure across different species (Fig. 2B). The most conserved region is near the catalytic center, which binds the methyl donor SAM and the methyl acceptor isoD-containing substrate (Fig. S1) (21–23). RPA0376 and RPA2838 share conserved residues with other PIMTs. RPA2580, however, differs at scattered locations around these conserved areas (Fig. 3). Most significantly, RPA2580 is missing part of the conserved pre-region I, which contains a serine residue (position 79 in the alignment; Fig. 3) that is critical for the catalytic activity of PIMTs (24). Instead, RPA2580 has an sequence of mostly charged residues (Fig. 3) that forms a loop in the deduced structure of RPA2580 near the active site of PIMTs, possibly shielding the SAM binding pocket (arrow in Fig. 2B). This feature is conserved in >85% of the sequences in group III (Fig. 2A). Another unique feature of group III PIMTs is their region I, which contains a proline instead of an otherwise conserved glycine residue (position 107 in the alignment; Fig. 3). Among other PIMTs, this region is highly conserved and directly interacts with SAM via hydrogen-bonding and hydrophobic interactions (25). Finally, sequences in group III end with a C-terminal FXF motif right after post-region III (position 248 in the alignment; Fig. 3). This is in contrast to other PIMTs, which usually have an extended and highly variable C-terminal tail.
Figure 2.
Sequence analysis of R. palustris PIMTs. A, sequence similarity network of all protein sequences containing the PIMT Pfam domain PF01135. Group I, which contains RPA0376 and RPA2838, includes only bacterial and archaeal PIMTs. Group II contains only eukaryotic PIMTs. Group III containing RPA2580 has ∼700 sequences, all of which come from α-proteobacteria. B, a predicted structure of RPA2580 (red) threaded onto the structure of P. furiosus PIMT (black; Protein Data Bank code 1JG3). S-Adenosyl homoserine is shown in yellow, and the peptide VYP-isoD-HA is shown in cyan. The arrow points to the loop that is present in the predicted structure of RPA2580 that does not align with the P. furiosus structure.
Figure 3.
Sequence divergence of RPA2580 from PIMT proteins. The alignment of the following 10 PIMT proteins is shown: R2580, RPA2580 (Q6N6N4); R0376, RPA0376 (Q6NCU3); R2838, RPA2838 (Q6N5Y0); E.col, PIMT in E. coli (P0A7A5); V.cho, PIMT in Vibrio cholera (Q9KUI8); P.fur, PIMT in P. furiosus (Q8TZR3); P.aer, PIMT in Pyrobaculum aerophilum (Q8ZYN0); A.tha, PIMT isoform 3 in Arabidopsis thaliana (Q64J17–3); D.mel, PIMT in D. melanogaster (Q27869); and H.sap, PIMT in Homo sapiens (P22061). Pre-region I and post-region III, which are characteristic of PIMTs, and regions I, II, and III, which are characteristic of this family of methyltransferases, are highlighted by shaded boxes (28). RPA2580 residues divergent from those in other PIMTs are highlighted with red type. Conserved PIMT amino acids that may not be conserved in RPA2580 are labeled as follows: asterisk, identical; colon, highly similar; period, similar.
RPA0376 and RPA2838 have PIMT activity in vitro, but RPA2580 does not
The genes rpa0376, rpa2580, and rpa2838 were cloned, expressed, and purified as His-tagged proteins. We then tested their abilities to repair isoD damage in vitro. Ovalbumin, which spontaneously accumulates isoDs over time, was used as the methyl accepting substrate for this assay. The E. coli PIMT, PCM, was used as a positive control (16). RPA0376 transferred methyl groups to ovalbumin at a rate of ∼6 nmol/min/mg, which is comparable with that seen by others for PIMTs (26). RPA2838 also showed significant PIMT activity in our assays but only at ∼10% the level of PCM and RPA0376. RPA2580 had no detectable PIMT activity (Fig. 4). It should be noted that the His-tagged form of RPA2580 used in enzyme assays complemented the longevity phenotype of a RPA2580 deletion mutant in trans (Fig. S2). Some PIMTs are also active in methylating d-aspartates. However, we did not attempt to measure this activity in this study.
Figure 4.
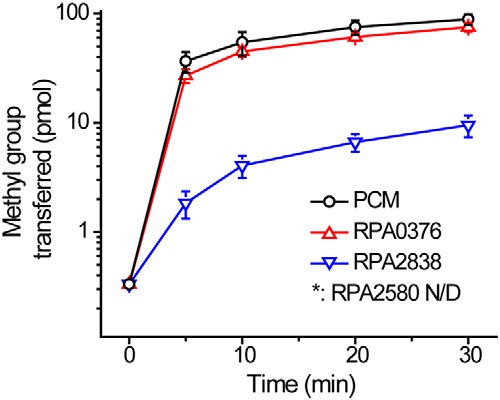
RPA0376 and RPA2838 have PIMT activity, whereas RPA2580 does not. Methyltransferase activity of R. palustris and E. coli PIMTs. Each reaction contained 0.5 μm enzyme, 10 μm [14C]SAM as the methyl donor, and 560 μm ovalbumin as the methyl acceptor. All the reactions were performed at 30 °C. The error bars represent the standard deviation of at least three replications.
Because much of the sequence divergence of group III PIMTs is located around the SAM-binding site (Figs. 2B and 3), we wondered whether RPA2580 is compromised in its ability to interact with SAM. As expected, the bona fide PIMT enzymes PCM and RPA0376 were able to bind SAM with micromolar Kd values (Fig. S4). The dissociation constant of RPA0376 with SAM was 69 ± 17 μm. This is higher than the Kd of PCM (1.5 ± 0.5 μm), but the difference might not be essential for its function in vivo, because the intracellular concentration of SAM is greater than these values, at least in E. coli (27). By contrast, RPA2580 did not bind to SAM in our assays. We tried to replace the loop in RPA2580 with the corresponding RPA0376 sequence to restore its SAM-binding capability (56EGRDDHKHFLLNPI → 56EGGQTISQPI). However, when this construct was expressed, less than 1% of protein was soluble, suggesting that this loop is an integral part of RPA2580 folding.
Recognition of isoD-containing peptides
Not all isoD residues are the same. PIMT proteins recognize isoD with vastly different efficiencies depending on the amino acid sequences surrounding the damaged residue (26). When we measured the Michaelis–Menten kinetic constants for RPA0376 and RPA2838 using four different isoD-containing peptides, we found that RPA0376 had the highest affinity for KASA-isoD-LAKY (Km = 8.0 μm) and the lowest affinity for YVS-isoD-GHG (Km = 2 mm) (Table 1). RPA2838 also had a low affinity for YVS-isoD-GHG (Km = 3 mm), but its highest affinity was for peptide VYP-isoD-HA (Km = 3.4 μm) (Table 1). For comparison, human PIMT has a high affinity for VYP-isoD-HA (Km = 0.3 μm) and an intermediate affinity for YVS-isoD-GHG (Km = 16 μm) (26). Consistent with the ovalbumin experiment, RPA2580 had no detectable activity with any of the peptides tested. With regard to Vmax calculations, it should be noted that we used 10 μm SAM in all our measurements. It is possible that this concentration was not sufficient to saturate the enzymes.
Table 1.
Michaelis–Menten kinetic constants for R. palustris PIMT proteins
| RPA0376 |
RPA2838 |
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | nmol/min/mg | μm | nmol/min/mg | |
| KASA-isoD-LAKY | 8.0 ± 0.39 | 19 ± 0.28 | 120 ± 8.9 | 0.99 ± 0.01 |
| VYP-isoD-HA | 45 ± 2.1 | 20 ± 0.32 | 3.4 ± 0.2 | 1.1 ± 0.03 |
| VYR-isoD-RR | 30 ± 1.6 | 15 ± 0.31 | 400 ± 28 | 1.1 ± 0.05 |
| YVS-isoD-GHG | 2600 ± 450 | 13 ± 1.7 | 3100 ± 540 | 0.96 ± 0.14 |
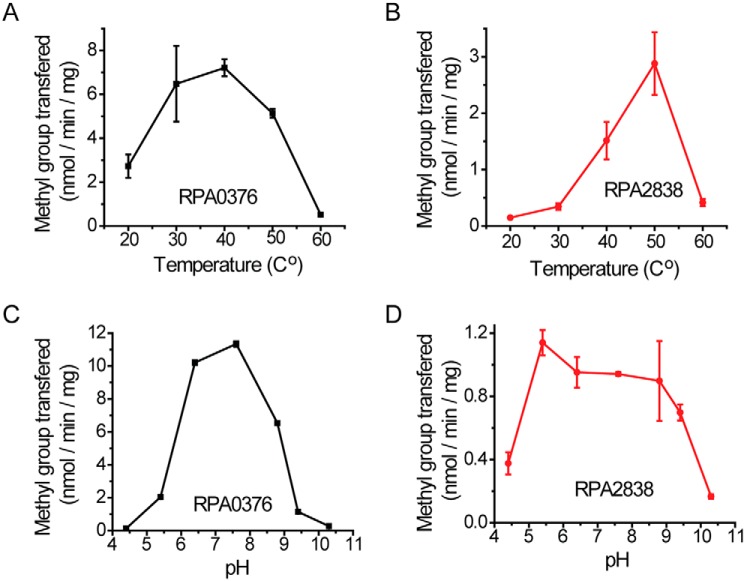
The effect of temperature and pH on R. palustris PIMT activities
PIMTs tend to function best at temperatures above 40 °C (28). We measured the temperature dependence of the isoD-repairing activity of RPA0376 and RPA2838 with VYP-isoD-HA. We found that RPA0376 had optimal activity over a wide temperature range from 30 to 50 °C. PIMT activity was ∼50% lower at 20 °C and 90% lower at a higher temperature of 60 °C (Fig. 5A). RPA2838 had a very different temperature profile. Its activity increased sharply when the temperature was increased from 30 to 50 °C. A further increase to 60 °C resulted in a loss of activity (Fig. 5B). We also looked at the activities of these enzymes at different pH levels. When the pH was lower than 6 or higher than 9, the activity of RPA0376 dropped to below 20% of its maximum value (Fig. 5C). RPA2838 has a broader pH optimum range with similar activities over a pH range from pH 5.4 to 9.4. However, its activity was low compared with that of RPA0376, regardless of the pH value (Fig. 5D).
Figure 5.
The effect of temperature and pH on R. palustris PIMT activities. A and B, the effect of temperature. C and D, the effect of pH. For all experiments, each reaction contained 0.5 μm enzyme, 10 μm [14C]SAM as the methyl donor, and 50 μm VYP-isoD-HA as the methyl acceptor. The error bars represent the standard deviation of at least three replications.
IsoD repair activity does not affect the longevity of growth-arrested R. palustris
We used RPA0376 to measure the isoD content of protein in lysates of R. palustris cells harvested on entry to stationary phase and following growth arrest. When cultures entered the stationary phase, the amount of isoD in cells was undetectable. After 30 days of growth arrest, a significant amount of isoD residues had accumulated (Table 2). However, a R. palustris strain deleted of all three annotated PIMT genes (ΔTRI-PIMT) did not accumulate more isoD residues compared with the WT strain in the same conditions (Table 2). This is contrary to what would be expected if the PIMTs were active in the repair of isoD residues. Similar results have been reported for E. coli, in which the deletion of its pcm gene did not lead to additional accumulation of isoD residues (29). Consistent with our in vitro enzymatic experiments, the same assay performed with RPA2580 did not detect any isoD repair activity with all the lysates tested.
Table 2.
The quantity of isoD residues in R. palustris cell lysates
WT, CGA009; ΔTRI-PIMT, CGA009 deleted of rpa0376, rpa2580, and rpa2838.
| Samples | Day 0 |
Day 30 |
||
|---|---|---|---|---|
| WT | ΔTRI-PIMT | WT | ΔTRI-PIMT | |
| isoD (pmol/mg protein in lysate) | <10 | <10 | 903 ± 45 | 692 ± 9 |
As shown previously (1), longevity was compromised in a rpa2580 mutant (Δrpa2580) (Fig. 6). We found that mutants with deletions in either rpa0376 or rpa2838 survived just as well as the WT after 30 days of growth arrest (Fig. 6). A double mutant with deletions in both genes also had a WT longevity phenotype (Fig. S3). The strain ΔTRI-PIMT showed compromised longevity, but only at level comparable with a Δrpa2580 mutant (Fig. S3).
Figure 6.
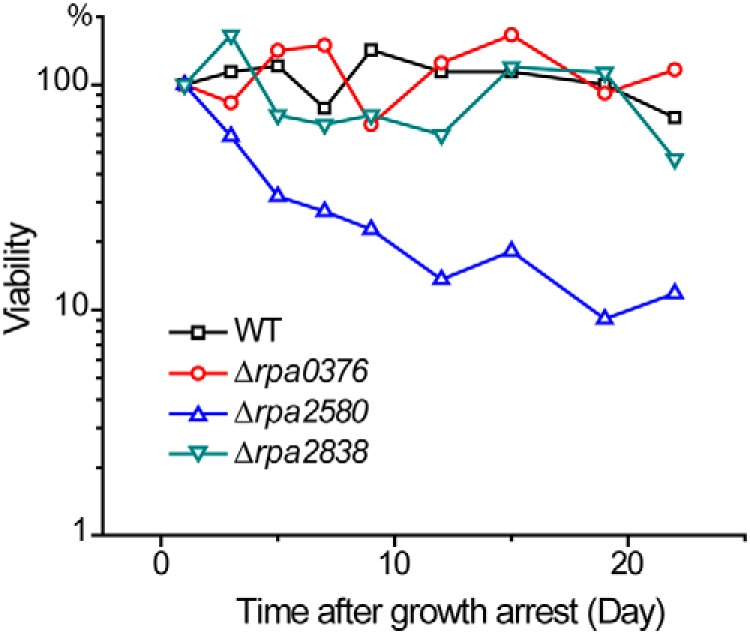
R. palustris Δrpa0376 and Δrpa2838 mutants have WT longevity. The PIMT deletion mutants were grown to stationary phase, and then their longevities were measured. The Δrpa0376 and Δrpa2838 mutants maintained ∼100% viability after 20 days of growth arrest. By comparison, the Δrpa2580 mutant showed compromised longevity. Assays performed with double or triple PIMT gene deletions have longevity phenotypes consistent with these observations (Fig. S3). At least three biological replications were performed for each strain (Fig. S5).
Discussion
R. palustris encodes three proteins annotated by several annotation algorithms as being putative protein l-isoaspartyl methyltransferases. All three are classified as members of the PFAM sequence family PF01135, but the sequence conservation for belonging to this PFAM category is somewhat less for RPA2580 than for RPA0376 and RPA2838 (Fig. 3). We determined that RPA0376 and RPA2838 are bone fide PIMTs that repair isoD residues in proteins and peptides in vitro. RPA0376 behaved similarly to the previously described E. coli PIMT, whereas RPA2838 showed significantly lower activities and a different optimal temperature and optimal pH profile. Despite the clear involvement of PIMTs in protein repair and in mitigation of aging in a variety of eukaryotes, we could not detect a similar role for PIMTs in the longevity of growth-arrested R. palustris cells. The quantity of isoD residues in aged R. palustris proteins was not increased by mutations in the two PIMT genes (Table 2), and a PIMT double mutant was not compromised in longevity (Fig. S3). In previous work, stationary phase cells of an E. coli PIMT mutant were less viable than the WT parent when subjected to secondary stresses (16, 17). However, as with R. palustris, the amount of isoD damage detected was not higher in an E. coli PIMT mutant subjected to stress conditions (17, 29). We know from RNA-seq experiments that the R. palustris PIMTs are expressed in growing and growth-arrested cells (1). It could be that proteins with isoD damage are still partially active. Considering there are multiple molecules of the same proteins in a given cell and that growth-arrested R. palustris cells are less active, the accumulation of isoDs may not be as detrimental to cells as we originally assumed. Another possibility that is more consistent with our data showing that PIMT mutants do not have increased levels of isoD is that the primary physiological role of prokaryotic PIMTs is something other than the repair of proteins damaged by spontaneous formation of isoD residues (2, 31).
Some α-proteobacterial genes annotated as PIMTs form a distinct phylogenetic group to which RPA2580 belongs (Fig. 2A). This group of proteins are characterized by an extended loop at pre-region I, mostly made up of charged residues, and a C terminus FXF motif right after post-region III (Fig. 3). RPA2580 is important for R. palustris longevity but does not have PIMT activity. A mutated E. coli PIMT enzyme, PCME104A, that was unable to repair isoD damage in vitro, increased heat shock survival better than WT PCM when both proteins were overexpressed (32). It was suggested that heat shock protective response was induced because of overexpression of a protein that was prone to misfolding. We cannot exclude that RPA2580 could be playing a similar role in R. palustris but at normal levels of expression and under conditions that do not involve heat shock. The expression level of RPA2580 in stationary phase growth-arrested cells is not higher than its levels in growing cells.
Other possibilities are that RPA2580 is active on an alternative protein substrate or that it methylates an alternative amino acid, such as aspartate or glutamate, on which the methyl group donated from SAM is known to be more stable. Although RPA2580 did not bind SAM in our experiments, it is possible that a SAM-binding site is exposed upon interaction of RPA2580 with a target substrate or a chaperone.
A final thought is that RPA2580 might sense isoD-containing proteins or peptides as a sign of cellular growth arrest. The groove in RPA2580 that interacts with an isoD residue is exposed and largely intact in our model (Fig. 2). A recent study showed that an E. coli PIMT mutant formed a larger percentage of quiescent, antibiotic-resistant, persister cells than the WT. The authors suggest that in situations where cells cannot repair isoDs, they may respond by becoming persisters, and this may protect cells from the consequences of protein damage (30). This still begs the question of how isoD protein damage might trigger persister formation. R. palustris is distinct from E. coli in its ability to remain almost 100% viable in growth arrest for weeks. We do not fully understand how R. palustris longevity relates to bacterial persistence, but on the surface, they do have a number of features in common.
Experimental procedures
Protein structure modeling and comparison
SWISS-MODEL (33) was used to predict the structure of RPA2580. The crystal structure of Pyrococcus furiosus PIMT (Protein Data Bank code 1JG1) (25) was used as the model for the prediction. To analyze the sequence conservation of PIMT structures, the sequence of RPA0376 was searched against all the assembled bacteria genomes in IMG/M (19). The top 250 hits of the search were analyzed, and sequence conservation was visualized using Chimera (34). The location of PIMT ligands was visualized by superimposing P. furiosus PIMT structures (Protein Data Bank codes 1JG1 and 1JG3) (25) on the predicted RPA0376 structure.
Sequence analysis
Multiple sequence alignments were performed using Clustal Omega (35) and edited using BioEdit. To analyze the PIMT sequence similarity networks, all protein sequences containing Pfam domain PF01135 were acquired and analyzed with Enzyme Similarity Tool (20). We limited the length of targeted sequences to 140–300 amino acids to exclude proteins with multiple domains. The resulting sequence similarity network was visualized using Cytoscape (36).
Protein expression and purification
All genes were cloned into pET30b vector (Novagen) as a construct with a His tag fused to the N terminus. The plasmids were transformed into E. coli strain TunerTM (Novagen), and cells were grown in Luria–Bertani (LB) medium until the A600 nm reached 0.4–0.8. Cultures were then induced with 1 mm of iso-propyl-β-d-thiogalactopyranoside and grown at 16 °C overnight. Cell pellets were resuspended in lysis buffer (50 mm Na2HPO4–NaH2PO4, pH 7.4, 300 mm NaCl, 10 mm imidazole, and 10% glycerol), and the mixture was disrupted by passing through a French pressure cell. The cell extracts were clarified by centrifugation at 15,000 rpm for 30 min. The proteins were purified by binding to His-Pur (ThermoFisher) resin, washed with the lysis buffer containing 30 mm imidazole, and eluted with the same buffer containing 500 mm imidazole. Imidazole was removed using Econo-Pac® 10DG (Bio-Rad) desalting columns. The purified proteins were concentrated to 2–20 mg ml−1 and suspended in storage buffer (25 mm Na2HPO4–NaH2PO4, pH 7.4, 150 mm NaCl, and 50% glycerol).
Methyltransferase assays
Methyltransferase activity was measured using a vapor diffusion method as described before (28). In general, each reaction contained the methyl-donor [14C]SAM (PerkinElmer Life Sciences), the methyl acceptor ovalbumin (Sigma), isoD-containing peptides or R. palustris lysates, and the protein to be tested. The assays were performed at 30 °C and in a buffer of 50 mm Bis-Tris-HCl, pH 6.4, unless noted otherwise. The reactions were adjusted to a final volume of 40 μl, incubated for the desired time, and stopped by adding 40 μl of stop buffer (1% SDS and 0.2 m NaOH). 60 μl of stopped reaction was immediately spotted onto a piece of prefolded filter paper. The folded filter paper was wedged in the neck of a 20-ml scintillation vial containing 5 ml of Safety Solve high–flash point mixture (Research Products International). The vial was then capped and left at room temperature for 2 h. During this time, [14C]methyl ester was hydrolyzed and released as [14C]methanol, which subsequently diffused into the counting fluor. The filter paper was then removed, and the vial contents were counted by liquid scintillation. For each experimental set, a negative control was performed without the methyl acceptor.
SAM-binding assays
All the reactions were performed in a buffer of 25 mm Na2HPO4–NaH2PO4, pH 7.4, 150 mm NaCl, and 5% glycerol. A final concentration of 0.5 μm [14C]SAM (PerkinElmer Life Sciences) was mixed with various concentrations of tested proteins. The reactions of 100 μl were incubated at 30 °C for 1 h and then applied to ZebaTM spin desalting columns (Thermo Scientific) to remove the unbound [14C]SAM. 80 μl of eluted protein was counted by liquid scintillation to measure the bound [14C]SAM. For each experimental set, a negative control was performed without the protein. Each series of protein dilution was performed three times individually, and the data were fitted to a one-to-one binding model to calculate the dissociation constant of SAM.
The effect of temperature on methyltransferase activity
The assays were carried out in a buffer of 50 mm Bis-Tris-HCl, pH 6.4. All the reaction components were pre-equilibrated at the required temperature. Each reaction contained 0.5 μm RPA0376 or RPA2838, 10 μm [14C]SAM as the methyl group donor, and 50 μm VYP-isoD-HA as the methyl group acceptor. For the reactions performed at 20–40 °C, the reaction time was 10 min. For the reactions performed at 50–60 °C, the reaction time was 5 min.
The effect of pH on methyltransferase activity
The assays were carried out at 30 °C. The following buffers were used to adjust the pH of the reactions: pH 4.4 and 5.4, citric acid and Na2HPO4; pH 6.4, Bis-Tris-HCl; pH 7.6, NaH2PO4 and Na2HPO4; pH 8.8, Tris-HCl; and pH 9.4 and 10.3, NaHCO3 and Na2CO3. Each reaction contained 0.5 μm RPA0376 or RPA2838, 10 μm [14C]SAM as the methyl group donor, and 50 μm VYP-isoD-HA as the methyl group acceptor. For RPA0376, the reaction time was 10–20 min. For RPA2838, the reaction time was 40–60 min.
PIMT assays with R. palustris cell lysates as substrate
All strains of R. palustris were grown anaerobically at 25 °C with illumination in defined mineral medium with acetate or succinate unless noted otherwise. The following steps were performed on ice or at 4 °C. Approximately 50 ml of R. palustris culture were collected by centrifugation at 4,000 rpm for 10 min. The cell pellets were resuspended to 2 mg/ml of 50 mm Na2HPO4–NaH2PO4, pH 7.4, 300 mm NaCl, and 10% glycerol. To break the cells, 0.5 ml of 0.1 mm zirconica/silica beads (BioSpec Products) were added into the mixture and vigorously shaken six times, 1 min each time with a bead beater. The mixture was clarified by centrifugation at 15,000 rpm for 10 min. The lysates were further centrifuged at 60,000 rpm for 30 min, collected, and adjusted to a protein concentration of 0.2 mg/ml. PIMT assays were performed at 37 °C for 2 h. For each experiment, a control of no enzyme added and a control of no cell lysate added were performed. The readings of these two negative controls were subtracted from the final results.
Determination of R. palustris longevity
In-frame deletions of R. palustris genes were created using suicide vector pJQ200SK as described before (37, 38). All strains and plasmids used are listed in Table S1. To determine the longevity of a strain, the culture was grown to a growth-arrested state caused by exhaustion of the carbon source, and cell longevity was monitored by counting the colony-forming units as described previously (1).
Author contributions
L. Y. data curation; L. Y. formal analysis; L. Y. investigation; L. Y. methodology; L. Y. writing-original draft; C. S. H. conceptualization; C. S. H. supervision; C. S. H. funding acquisition; C. S. H. validation; C. S. H. project administration; C. S. H. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Amy Schaefer (University of Washington) and Prof. Steven Clarke (UCLA) for valuable discussions.
This work was supported by Grant W911NF-15-1-0150 from the U.S. Army Research Office (ARO). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S5.
- PIMT
- l-isoaspartate O-methyltransferase
- isoD
- l-isoaspartate
- SAM
- S-adenosylmethionine.
References
- 1. Pechter K. B., Yin L., Oda Y., Gallagher L., Yang J., Manoil C., and Harwood C. S. (2017) Molecular basis of bacterial longevity. MBio 8, e01726–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reissner K. J., and Aswad D. W. (2003) Deamidation and isoaspartate formation in proteins: unwanted alterations or surreptitious signals? Cell. Mol. Life Sci. 60, 1281–1295 10.1007/s00018-003-2287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Galletti P., Ciardiello A., Ingrosso D., Di Donato A., and D'Alessio G. (1988) Repair of isopeptide bonds by protein carboxyl O-methyltransferase: seminal ribonuclease as a model system. Biochemistry 27, 1752–1757 10.1021/bi00405a055 [DOI] [PubMed] [Google Scholar]
- 4. Johnson B. A., Langmack E. L., and Aswad D. W. (1987) Partial repair of deamidation-damaged calmodulin by protein carboxyl methyltransferase. J. Biol. Chem. 262, 12283–12287 [PubMed] [Google Scholar]
- 5. Brennan T. V., Anderson J. W., Jia Z., Waygood E. B., and Clarke S. (1994) Repair of spontaneously deamidated HPr phosphocarrier protein catalyzed by the l-isoaspartate-(d-aspartate) O-methyltransferase. J. Biol. Chem. 269, 24586–24595 [PubMed] [Google Scholar]
- 6. Geiger T., and Clarke S. (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides: succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 262, 785–794 [PubMed] [Google Scholar]
- 7. Stephenson R. C., and Clarke S. (1989) Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 264, 6164–6170 [PubMed] [Google Scholar]
- 8. Szymanska G., Leszyk J. D., and O'Connor C. M. (1998) Carboxyl methylation of deamidated calmodulin increases its stability in Xenopus oocyte cytoplasm: implications for protein repair. J. Biol. Chem. 273, 28516–28523 10.1074/jbc.273.43.28516 [DOI] [PubMed] [Google Scholar]
- 9. Chavous D. A., Jackson F. R., and O'Connor C. M. (2001) Extension of the Drosophila lifespan by overexpression of a protein repair methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 98, 14814–14818 10.1073/pnas.251446498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar G. N., Houtz R. L., and Knowles N. R. (1999) Age-induced protein modifications and increased proteolysis in potato seed-tubers. Plant Physiol. 119, 89–100 10.1104/pp.119.1.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mudgett M. B., Lowenson J. D., and Clarke S. (1997) Protein repair l-isoaspartyl methyltransferase in plants: phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds. Plant Physiology 115, 1481–1489 10.1104/pp.115.4.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim E., Lowenson J. D., MacLaren D. C., Clarke S., and Young S. G. (1997) Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc. Natl. Acad. Sci. U.S.A. 94, 6132–6137 10.1073/pnas.94.12.6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kagan R. M., Niewmierzycka A., and Clarke S. (1997) Targeted gene disruption of the Caenorhabditis elegans l-isoaspartyl protein repair methyltransferase impairs survival of dauer stage nematodes. Arch. Biochem. Biophys. 348, 320–328 10.1006/abbi.1997.0362 [DOI] [PubMed] [Google Scholar]
- 14. Doyle H. A., Gee R. J., and Mamula M. J. (2003) A failure to repair self-proteins leads to T cell hyperproliferation and autoantibody production. J. Immunol. 171, 2840–2847 10.4049/jimmunol.171.6.2840 [DOI] [PubMed] [Google Scholar]
- 15. Fu J. C., Ding L., and Clarke S. (1991) Purification, gene cloning, and sequence analysis of an l-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J. Biol. Chem. 266, 14562–14572 [PubMed] [Google Scholar]
- 16. Visick J. E., Cai H., and Clarke S. (1998) The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J. Bacteriol. 180, 2623–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hicks W. M., Kotlajich M. V., and Visick J. E. (2005) Recovery from long-term stationary phase and stress survival in Escherichia coli requires the l-isoaspartyl protein carboxyl methyltransferase at alkaline pH. Microbiology 151, 2151–2158 10.1099/mic.0.27835-0 [DOI] [PubMed] [Google Scholar]
- 18. Larimer F. W., Chain P., Hauser L., Lamerdin J., Malfatti S., Do L., Land M. L., Pelletier D. A., Beatty J. T., Lang A. S., Tabita F. R., Gibson J. L., Hanson T. E., Bobst C., Torres J. L., et al. (2004) Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22, 55–61 10.1038/nbt923 [DOI] [PubMed] [Google Scholar]
- 19. Markowitz V. M., Chen I. M., Chu K., Szeto E., Palaniappan K., Pillay M., Ratner A., Huang J., Pagani I., Tringe S., Huntemann M., Billis K., Varghese N., Tennessen K., Mavromatis K., et al. (2014) IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 42, D568–D573 10.1093/nar/gkt919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gerlt J. A., Bouvier J. T., Davidson D. B., Imker H. J., Sadkhin B., Slater D. R., and Whalen K. L. (2015) Enzyme Function Initiative–Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 10.1016/j.bbapap.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiefer F., Arnold K., Künzli M., Bordoli L., and Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guex N., Peitsch M. C., and Schwede T. (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30, (Suppl. 1) S162–S173 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- 23. Grigoriev I. V., Nordberg H., Shabalov I., Aerts A., Cantor M., Goodstein D., Kuo A., Minovitsky S., Nikitin R., Ohm R. A., Otillar R., Poliakov A., Ratnere I., Riley R., Smirnova T., et al. (2012) The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40, D26–D32 10.1093/nar/gkr947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bennett E. J., Bjerregaard J., Knapp J. E., Chavous D. A., Friedman A. M., Royer W. E. Jr, and O'Connor C. M. (2003) Catalytic implications from the Drosophila protein l-isoaspartyl methyltransferase structure and site-directed mutagenesis. Biochemistry 42, 12844–12853 10.1021/bi034891+ [DOI] [PubMed] [Google Scholar]
- 25. Griffith S. C., Sawaya M. R., Boutz D. R., Thapar N., Katz J. E., Clarke S., and Yeates T. O. (2001) Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its l-isoaspartyl peptide substrate. J. Mol. Biol. 313, 1103–1116 10.1006/jmbi.2001.5095 [DOI] [PubMed] [Google Scholar]
- 26. Lowenson J. D., and Clarke S. (1991) Structural elements affecting the recognition of l-isoaspartyl residues by the l-isoaspartyl/d-aspartyl protein methyltransferase: implications for the repair hypothesis. J. Biol. Chem. 266, 19396–19406 [PubMed] [Google Scholar]
- 27. Halliday N. M., Hardie K. R., Williams P., Winzer K., and Barrett D. A. (2010) Quantitative liquid chromatography-tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal. Biochem. 403, 20–29 10.1016/j.ab.2010.04.021 [DOI] [PubMed] [Google Scholar]
- 28. Villa S. T., Xu Q., Downie A. B., and Clarke S. G. (2006) Arabidopsis protein repair l-isoaspartyl methyltransferases: predominant activities at lethal temperatures. Physiol. Plant 128, 581–592 10.1111/j.1399-3054.2006.00772.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Visick J. E., Ichikawa J. K., and Clarke S. (1998) Mutations in the Escherichia coli surE gene increase isoaspartyl accumulation in a strain lacking the pcm repair methyltransferase but suppress stress-survival phenotypes. FEMS Microbiol. Lett. 167, 19–25 10.1111/j.1574-6968.1998.tb13202.x [DOI] [PubMed] [Google Scholar]
- 30. VandenBerg K. E., Ahn S., and Visick J. E. (2016) (p)ppGpp-dependent persisters increase the fitness of Escherichia coli bacteria deficient in isoaspartyl protein repair. Appl. Environ. Microbiol. 82, 5444–5454 10.1128/AEM.00623-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patananan A. N., Capri J., Whitelegge J. P., and Clarke S. G. (2014) Non-repair pathways for minimizing protein isoaspartyl damage in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 289, 16936–16953 10.1074/jbc.M114.564385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kindrachuk J., Parent J., Davies G. F., Dinsmore M., Attah-Poku S., and Napper S. (2003) Overexpression of l-isoaspartate O-methyltransferase in Escherichia coli increases heat shock survival by a mechanism independent of methyltransferase activity. J. Biol. Chem. 278, 50880–50886 10.1074/jbc.M308423200 [DOI] [PubMed] [Google Scholar]
- 33. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 35. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fixen K. R., Baker A. W., Stojkovic E. A., Beatty J. T., and Harwood C. S. (2014) Apo-bacteriophytochromes modulate bacterial photosynthesis in response to low light. Proc. Natl. Acad. Sci. U.S.A. 111, E237–E244 10.1073/pnas.1322410111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Quandt J., and Hynes M. F. (1993) Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21 10.1016/0378-1119(93)90611-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.