Abstract
The DNA damage response relies on protein modifications to elicit physiological changes required for coping with genotoxic conditions. Besides canonical DNA damage checkpoint–mediated phosphorylation, DNA damage–induced sumoylation has recently been shown to promote genotoxin survival. Cross-talk between these two pathways exists in both yeast and human cells. In particular, sumoylation is required for optimal checkpoint function, but the underlying mechanisms are not well-understood. To address this question, we examined the sumoylation of the first responder to DNA lesions, the ssDNA-binding protein complex replication protein A (RPA) in budding yeast (Saccharomyces cerevisiae). We delineated the sumoylation sites of the RPA large subunit, Rfa1 on the basis of previous and new mapping data. Findings using a sumoylation-defective Rfa1 mutant suggested that Rfa1 sumoylation acts in parallel with the 9-1-1 checkpoint complex to enhance the DNA damage checkpoint response. Mechanistically, sumoylated Rfa1 fostered an interaction with a checkpoint adaptor protein, Sgs1, and contributed to checkpoint kinase activation. Our results suggest that SUMO-based modulation of a DNA damage sensor positively influences the checkpoint response.
Keywords: DNA damage; DNA damage response; small ubiquitin-like modifier (SUMO); checkpoint control; Saccharomyces cerevisiae; DNA damage checkpoint; DNA damage induced sumoylation; RPA; sumoylation, Sgs1, checkpoint kinase activation
Introduction
The DNA damage response (DDR)4 is an intricate cellular system that senses and signals the presence of DNA lesions, promotes their repair, and arrests the cell cycle to provide time for repair (1). These coordinated functions are critical for genomic stability and cellular survival under genotoxic conditions. Not surprisingly, DDR defects in humans underlie the pathology of several genome instability syndromes and can fuel tumorigenesis. DDR employs multiple types of protein modifications that can quickly and reversibly change the properties of numerous proteins to favor their functions in coping with DNA damage (2). The reversible nature of these modifications is important for turning off the DDR after DNA damage conditions are removed or adapted to. DDR dampening deficiency is also undesirable and can lead to prolonged cell cycle arrest and in extreme cases, cell death (3).
The best-studied aspect of DDR is the highly conserved DNA damage checkpoint (or DDC) pathway. When cells experience increased levels of DNA lesions, lesion processing can generate large amounts of single-stranded DNA (ssDNA). The first responder to this signal is the trimeric RPA complex (composed of Rfa1–3) that has high affinity to ssDNA (4). The RPA-coated ssDNA filament then serves as a platform to recruit checkpoint kinases and cofactors. As an example, in the budding yeast model system, RPA can recruit the apical DDC kinase Mec1 to DNA damage sites via direct binding between Rfa1 and the Mec1 cofactor Ddc2 (5). In addition, RPA helps to recruit the ring-shaped 9-1-1 complex (Rad17-Mec3-Ddc1) that can bind to the junction of ssDNA and dsDNA. 9-1-1 then promotes Mec1 activation via allosteric stimulation of its kinase activity (6, 7). In addition, 9-1-1 can facilitate the recruitment of another Mec1 activator, Dpb11, to DNA damage sites (8). Like 9-1-1, Dpb11 and another Mec1 activator, the DNA helicase-nuclease Dna2, can directly stimulate Mec1 kinase activity (7, 9). The existence of multiple DNA damage sensors and Mec1 activators can ensure a robust checkpoint response such that the lack of one factor can be compensated by the presence of the others (10). Activation of the Mec1 kinase initiates the phosphorylation of various downstream factors. Two of these are the Rad9 scaffold protein and the Sgs1 helicase. Sgs1 and Rad9 are adaptor proteins that interact with a downstream kinase Rad53 to relay the checkpoint-signaling cascade from Mec1 and enhance Rad53 phosphorylation and activation (11, 12). Activated Rad53 can diffuse away from DNA damage sites and induce additional phosphorylation events required for coping with DNA damage conditions (13).
Recent studies in yeast and human cells have shown that in parallel with the DNA damage checkpoint pathway, coordinated protein sumoylation upon DNA damage also plays critical roles in DNA repair and cellular survival of genotoxic conditions (14). Such DNA damage-induced sumoylation (or DDIS) can also be initiated by RPA-ssDNA filament. In yeast, the direct binding between the Rfa2 subunit of RPA and the Siz2 SUMO E3 recruits Siz2 to DNA break sites and enables the sumoylation of RPA itself as well as the Rad52 and Rad59 recombinational proteins (15). The combined effect of these sumoylation events can promote recombinational repair (16). Additional sumoylation events induced by DNA damage also stimulate other types of DNA repair, such as nucleotide excision repair and base excision repair (14).
How the checkpoint response and DDIS coordinate and influence each other is just beginning to be understood. On one hand, the lack of Mec1 and other checkpoint proteins in yeast or their counterparts in human cells can increase the level of DNA damage–induced sumoylation, suggesting a possible compensatory effect (16–18). On the other hand, the lack of sumoylation enzymes impairs checkpoint-mediated phosphorylation of multiple substrates, such as the Rad53 kinase in yeast, suggesting that sumoylation may stimulate the checkpoint response (17, 18). One mechanism underlying this stimulatory effect in human cells is that the sumoylation of the Ddc2 homolog, ATRIP, can enhance ATRIP association with the human Mec1 homolog ATR and with other checkpoint proteins (18). Whether additional sumoylation events also contribute to the robustness of the checkpoint is unclear (Fig. 1A). To address this question, we examined RPA because its involvement in both the checkpoint and DDIS makes it well-positioned for mediating the cross-talk between the two pathways. Among the three subunits of RPA, the sumoylation of its largest subunit, Rfa1, is the most abundant (19). We mapped the Rfa1 sumoylation sites and examined how lack of Rfa1 sumoylation influences checkpoint activity. Our data suggest that Rfa1 sumoylation enhances its association with the Sgs1 checkpoint adaptor protein and positively affects the Mec1 checkpoint response.
Figure 1.
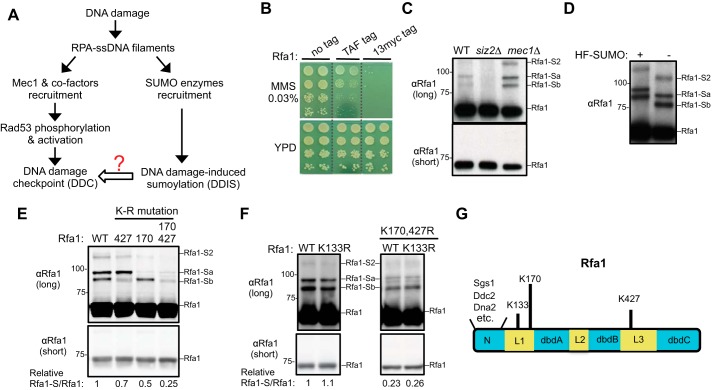
Examining the sumoylation of untagged Rfa1 and previously identified Rfa1 sumoylation sites. A, simplified schematic of the DNA damage response involving the Mec1 checkpoint and the DNA damage–induced sumoylation, with a focus on the events that require RPA-coated ssDNA. B, addition of a TAF or Myc tag on the C-terminal of Rfa1 sensitizes the cells toward DNA damaging agents. 10-fold serial dilution of cells of the indicated genotypes were spotted and growth was assessed after incubation at 30 °C for 3 days. The dashed lines indicate removal of superfluous rows. C, detection of the sumoylation of untagged Rfa1 in MMS conditions. Cells were treated with 0.02% MMS, and protein extracts from the indicated strains were examined by immunoblotting using anti-Rfa1–specific antibody (15). Rfa1-Sa and -Sb denote mono-sumoylated Rfa1 species, and Rfa1-S2 denotes di-sumoylated Rfa1 species. Molecular weight is indicated at the left of the blots. D, Rfa1 sumoylation bands exhibit expected up-shifts when a larger variant of SUMO containing His6-FLAG tag (HF-SUMO) is present. To better detect band shift, WT cells were treated with 0.3% MMS to increase Rfa1 sumoylation levels. E, mutation of lysines 170 and 427 to arginine reduces Rfa1 sumoylation. Specifically, rfa1Δ cells were supplemented with CEN-based plasmids expressing either WT or mutant Rfa1 driven by Rfa1 endogenous promoter. Cells of the indicated genotypes were treated and examined as in panel D. rfa1-K170R and rfa1-K427R reduced the signals of the Rfa1-Sa and -Sb sumoylation bands, respectively. The relative ratios of the sumoylated Rfa1 signals to those of unmodified Rfa1 are indicated below. F, mutation of lysine 133 to arginine does not reduce Rfa1 sumoylation in WT background (left) or in rfa1-K170, 427R background (right). Experiments were done and labels are indicated as in panel E. G, schematic of the budding yeast Rfa1 protein domains. Some known interactors for the Rfa1 N-terminal domain are indicated. The three lysine residues previously reported as Rfa1 sumoylation sites and representative proteins that bind to the Rfa1 N-terminal domain are also indicated.
Results
Endogenously expressed, untagged Rfa1 is modified by mono- and di-SUMO
Previous studies examining DNA damage–induced sumoylation of RPA mostly used epitope-tagged Rfa1 (16, 19). However, we found that various tags added to the Rfa1 C terminus reduced damage survival (Fig. 1B). This raised the question whether and how untagged Rfa1 was sumoylated. To address this, we examined endogenously expressed untagged Rfa1 after treating yeast cells with genotoxin methyl methanesulfonate (MMS). Total protein extract was made under denaturing conditions to minimize the loss of sumoylation during extraction and then subjected to Western blot analyses using a well-established anti-Rfa1 antibody (20, 21). As shown in Fig. 1C, whereas short exposure of the blots revealed only the unmodified Rfa1 band, long exposure revealed three additional Rfa1 bands. Based on several criteria, we concluded that these additional bands represented the Rfa1 sumoylated forms. First, removal of the Siz2 SUMO E3 eliminated these Rfa1 forms without affecting the unmodified Rfa1 (Fig. 1C). Second, a larger SUMO variant containing a His6-FLAG tag retarded gel migration of these Rfa1 forms, but not the unmodified Rfa1 (Fig. 1D). Third and as seen for the sumoylated forms of multiple other proteins, mec1Δ increased the levels of these Rfa1 forms (Fig. 1C) (17).
Analogous to most other sumoylated proteins, the relative amount of sumoylated Rfa1 was low and estimated at ∼3% of total Rfa1 (Fig. 1, C and D). As suggested previously, the low level of sumoylated forms detected could be because of the highly dynamic nature of sumoylation and desumoylation as well as possible loss of sumoylated forms during extraction (22). It is commonly found that mono-sumoylation causes ∼20 kDa up-shift of the protein on gels and di-sumoylation causes ∼40 kDa up-shift (22). In addition, the position of SUMO conjugation on a protein also influences the protein gel migration patterns (23, 24). Based on these general principles, it is likely that the two sumoylated Rfa1 bands exhibiting ∼15–20 kDa up-shift from the unmodified Rfa1 form represent mono-sumoylated species (Rfa1-Sb and Rfa1-Sa) (Fig. 1, C and D), whereas the other Rfa1 sumoylation band with a mobility shift of ∼40 kDa represents di-sumoylation species of Rfa1 (Rfa1-S2) (Fig. 1, C and D).
Rfa1 sumoylation is largely abolished by mutating four lysine residues
A previous study showed that simultaneously mutating three Rfa1 lysines (Lys-133, Lys-170, Lys-427 (K133, K170, and K427)) reduced the sumoylation levels of epitope-tagged Rfa1 proteins when a SUMO variant with all lysines mutated was used (16). Whether each of these lysines contributes to Rfa1 sumoylation in the absence of tags is unclear. To address this, we examined the effect of single lysine mutation in cells containing untagged Rfa1 and WT SUMO. We found that mutating K170 to arginine (K170R) reduced the intensity of the Rfa1-Sa band, whereas the K427R mutation reduced the intensity of the Rfa1-Sb band, suggesting that K170 and K427 are sumoylation sites responsible for the bulk of the Rfa1-Sa and Rfa1-Sb species, respectively (Fig. 1E). When these two mutations were combined (K170, 427R), total Rfa1 sumoylation levels were reduced to ∼25% of WT, and di-sumoylation band (Rfa1-S2) mostly disappeared (Fig. 1E). This result suggested that Rfa1-S2 likely represents species with both K170 and K427 modified by SUMO. In contrast, K133 mutation to arginine alone did not reduce Rfa1 sumoylation levels, nor did it impact the sumoylation levels when combined with K170, 427R (Fig. 1F). We concluded that K170 and 427, but not K133, are sumoylation sites of untagged Rfa1, and that the residual ∼25% of Rfa1 sumoylation in the rfa1-K170, K427R mutant is because of additional sumoylation sites. We note that K170 and K427 are located in the linker regions outside the DNA and protein-binding domains of Rfa1 (Fig. 1G).
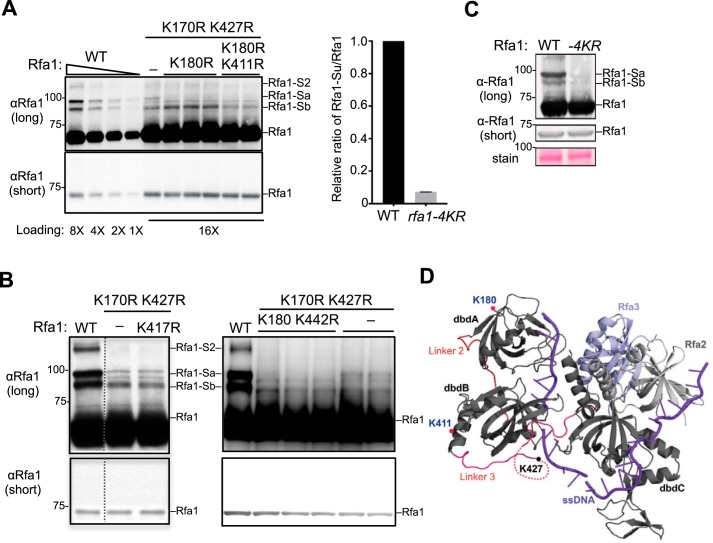
Next, we used a candidate approach to determine the additional sumoylation sites on Rfa1. As noted above, the location of sumoylation sites can differentially affect the gel migration patterns of sumoylated protein forms. In principle, sumoylation at sites close to each other likely causes similar gel retardation patterns because the peptide branch points on the sumoylated protein have similar locations. We thus reasoned that the remaining Rfa1-Sa and -Sb species seen in rfa1-K170, K427R mutants could be because of sumoylation at sites close to K170 and K427, respectively. To test this idea, we examined all the lysines located within a 20 amino acid radius from K170 (K180) or K427 (K411, K417, and K442) in a rfa1-K170, K427R background. We found that K180R specifically reduced the residual intensity of the Rfa1-Sa band, whereas K411R reduced the residual intensity of the Rfa1-Sb band (Fig. 2A). In contrast, neither K417R nor K442R further reduced sumoylation (Fig. 2B). Collectively, these analyses suggest K180 and K411 are two additional sumoylation sites on Rfa1. Indeed, replacing the RFA1 gene with a rfa1–4KR mutant wherein all four sites (K170, 180, 411, 427) were changed to arginine reduces the overall Rfa1 sumoylation by ∼90% without affecting Rfa1 protein levels (Fig. 2C). Based on the structure of RPA bound to ssDNA, K180 and K411, like K170 and K427, are not in contact with DNA; thus their mutations to arginine are unlikely to affect RPA binding of ssDNA (Fig. 2D) (25).
Figure 2.
Identification of two additional Rfa1 sumoylation sites. A, Mutating lysines 180 and 411 to arginine further reduces Rfa1 sumoylation. Adding the K180R mutation to rfa1-K170, 427R reduced the intensity of the Rfa1-Sa band. Further addition of the K411R mutation to rfa1-K170, 180, 427R (to give rfa1–4KR) reduced the intensity of the Rfa1-Sb band. The relative ratios of sumoylated Rfa1 forms to the unmodified Rfa1 form for WT and rfa1–4KR strains were quantified from two biological duplicates shown in the graph on the right. As Rfa1 sumoylation levels were low in rfa1-K170, 427R background, better visualization of this modification was achieved by using mec1Δ sml1Δ cells treated with 0.02% MMS, which are known to increase Rfa1 sumoylation. Note: For this experiment, rfa1Δ cells were supplemented with CEN-based plasmids expressing either WT or mutant Rfa1 driven by Rfa1 endogenous promoter. B, mutating lysine 417 and 442 to arginine does not affect Rfa1 sumoylation. Experiments were done as in panel A. Adding the K417R mutation to rfa1-K170, 427R had no effect on the intensity of all Rfa1 sumoylation bands. Similarly, adding the K442R mutation to rfa1-K170, 180, 427R did not further affect Rfa1 sumoylation. The data presented here are from the same immunoblot and dashed line indicates removal of superfluous lanes. C, rfa1–4KR as the only copy of Rfa1 in its endogenous locus greatly reduced Rfa1 sumoylation when cells were treated with 0.2% MMS for 2 h. rfa1–4KR did not affect Rfa1 protein levels as shown in the short exposure of the immunoblots. D, crystal structure of Ustilago maydis RPA in complex with ssDNA (purple) (25). Rfa1 is represented by its dbd A, B, and C. The locations of three of the four sumoylation sites are indicated. The K170 residue of Rfa1 was not included in the crystal structure.
rfa1–4KR jointly with a ddc1 mutant rescue the persistent checkpoint and damage sensitivity of srs2Δ cells
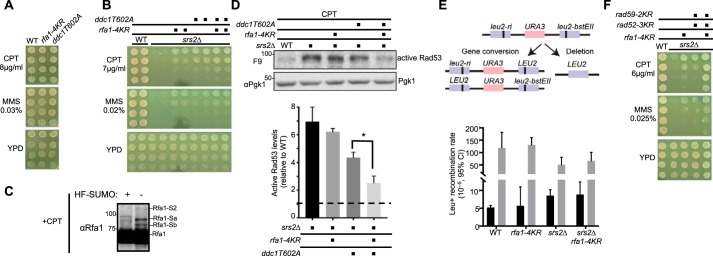
As described above, reducing Siz 1/2 SUMO E3–mediated sumoylation diminishes the Mec1 checkpoint in genotoxin conditions (17). We asked whether Rfa1 sumoylation, which is largely mediated by Siz2, contributes to this SUMO-mediated stimulation of checkpoint (15). We found that rfa1–4KR did not sensitize cells to genotoxins such as camptothecin (CPT) or MMS, nor did it affect normal growth (Fig. 3A). As multiple redundant mechanisms are involved to generate the checkpoint response, disrupting a single mechanism often does not lead to observable DNA damage sensitivity. For example, mutating the Ddc1 phosphorylation site required for Dpb11 binding (ddc1-T602A) is known to confer WT level of damage survival (Fig. 3A) (7). In this situation, a strain background wherein persistent checkpoint causes genotoxin sensitivity provides a useful context to query checkpoint impairment. Previous studies have shown that removing the DNA helicase Srs2 leads to hyper-checkpoint and subsequent DNA damage sensitivity (26), and both defects can be rescued by deleting checkpoint genes (27, 28). Consistent with this observation, we found that ddc1-T602A also improved srs2Δ cell survival on CPT- and MMS-containing media (Fig. 3B). Significantly, rfa1–4KR had similar effects as ddc1-T602A, and their combined mutants had a stronger suppressive effect on srs2Δ cells on CPT media (Fig. 3B). We confirmed that Rfa1 was sumoylated after CPT treatment (Fig. 3C) with a similar pattern as observed after MMS treatment (Fig. 1, C and D).
Figure 3.
rfa1–4KR, in combination with ddc1-T602A, reduces persistent checkpoint and DNA damage sensitivity of srs2Δ cells. A, rfa1–4KR and ddc1-T602A do not affect cell growth or sensitivity to genotoxins. 3-fold serial dilutions of cells of the indicated genotypes were spotted on media containing the indicated concentration of genotoxins and plates were grown at 30 °C for 3 days. B, rfa1–4KR or ddc1T602A suppressed srs2Δ sensitivity to CPT and MMS. Experiments were done as in panel A. C, Rfa1 sumoylation is observed after treatment with 16 μg/ml CPT for 4 h. Experiments were done as in Fig. 1D, except that cells were treated with CPT. D, the rfa1–4KR ddc1T602A double mutant reduces the levels of active Rad53 in srs2Δ cells. G1 cells were released into the cell cycle in media containing 16 μg/ml CPT for 6 h. Protein extracts were examined by immunoblotting using antibody specifically against activated Rad53 (F9) (29). Pgk1 was used as a loading control. The signals of the active Rad53 were calculated and normalized to those of Pgk1. The mean of three biological isolates per genotype is graphed relative to WT (dashed line), with error bars representing S.E. and asterisk indicating significant difference (p < 0.05). E, rfa1–4KR does not affect intra-chromosomal recombination. Top, schematic of the intra-chromosomal recombination assay used. In brief, two different leu2 defective alleles with mutations in the 5′ or 3′ end of the gene are separated by a URA3 gene. Gene conversion or deletion events can produce a LEU2 gene that supports growth on media lacking leucine and can be scored as Leu+ colonies. Bottom, Leu+ recombination rates as measured by fluctuation analysis (n = 24) are plotted for normal growth (black) or MMS-treated (gray) conditions. Error bars represent the median with 95% confidence interval. F, mutating the three Rad52 sumoylation sites (rad52–3KR) and two Rad59 sumoylation sites (rad59–2KR) also suppresses srs2Δ sensitivity to genotoxins. Combined mutation of rfa1–4KR and these rad52 and rad59 alleles showed enhanced srs2Δ suppression.
Next, we examined the activation of the Rad53 kinase, a hallmark of the Mec1 checkpoint pathway, using an antibody specifically recognizing the active form of Rad53 (29). After cells were treated with CPT, srs2Δ cells contained higher levels of active form of Rad53 (Fig. 3D). ddc1-T602A reduced the level of this Rad53 form in srs2Δ cells by ∼40%, and rfa1-4KR further decreased it by another ∼25%, although rfa1-4KR alone did not yield a statistically significant reduction of active Rad53 levels. Taken together, these genetic and biochemical data suggest that Rfa1 sumoylation contributes toward Rad53 activation, but its effect is buffered by other redundant mechanisms.
rfa1-4KR does not affect the rate of homologous recombination in srs2Δ cells
Besides reducing checkpoint activation, the DNA damage sensitivity of srs2Δ cells can also be suppressed by removing pro-recombination factors (28). It was thought that the srs2Δ damage sensitivity is partly because of accumulation of toxic recombination intermediates, as the helicase activity of Srs2 can dismantle these structures (30). Given that RPA not only supports checkpoint activation but also participates in recombinational repair, we assessed whether rfa1-4KR could affect homologous recombination. To this end, we used an intra-chromosomal recombination assay that produces recombinants via either gene conversion or deletion (31). Similar to previous findings (32), spontaneous recombination rate increases in srs2Δ cells (Fig. 3E). However, rfa1-4KR did not influence recombination rates in srs2Δ cells or in otherwise WT cells (Fig. 3E). Similar findings were observed after cells were treated with DNA damaging agents for 2 h before assaying for recombination products (Fig. 3E). This suggests that Rfa1 sumoylation is unlikely to affect intra-chromosomal recombination.
It is known that Rad52 and Rad59 sumoylation can promote recombinational repair. Using previously established sumoylation-deficient alleles of Rad52 and Rad59 (16, 33), we found that their combined mutants also improved the survival of srs2Δ in DNA damage conditions, consistent with the notion that reducing recombination could reduce srs2Δ damage sensitivity. Interestingly, when the rad52 and rad59 sumoylation-deficient alleles were combined with rfa1-4KR, the suppressive effect on srs2Δ was greater (Fig. 3F). This additive effect is consistent with the notion that Rfa1 sumoylation affects a different aspect of biology than Rad52 and Rad59 sumoylation.
Rfa1 sumoylation promotes its association with Sgs1
We next asked how Rfa1 sumoylation could contribute to checkpoint activation. A commonly reported effect of sumoylation is to enhance protein–protein interactions, as the SUMO moiety on a modified protein can provide another interacting interface to its binding partner (34). Considering the multiple protein–protein interactions involved in the checkpoint circuitry, we considered the possibility that sumoylated Rfa1 may promote its association with another checkpoint factor(s), thus enhancing the overall efficiency of checkpoint activation.
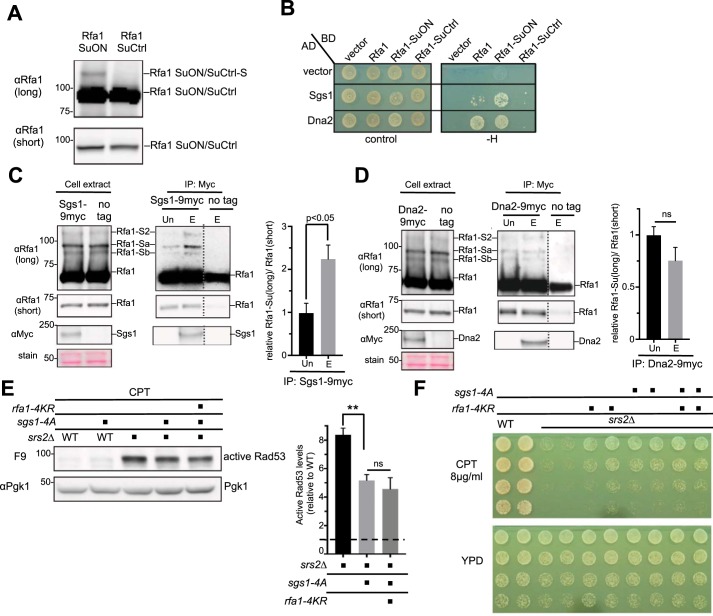
To test this idea, we queried among Rfa1 interactors involved in the Mec1 checkpoint pathway and found that the DNA helicase Sgs1 and the nuclease Dna2 were reported to also interact with SUMO (35, 36). We thus used the yeast two-hybrid assay (Y2H) to test whether Sgs1 and Dna2 can better interact with Rfa1 when it can be more abundantly sumoylated. Previous studies have shown that a SuON (for SUMO ON) tag with strong SUMO-binding affinity can increase the sumoylation of a fusion protein, likely by increasing local SUMO concentration (37, 38). A control tag, SuCtrl (for SUMO control) tag, has the same sequence as SuON except for a point mutation that abolishes SUMO binding (37, 38). We confirmed that Rfa1 sumoylation was enhanced by Rfa1-SuON compared with Rfa1-SuCtrl, such that Rfa1 sumoylation could be detected even without DNA damage treatment (Fig. 4A). In the yeast two-hybrid assay, both Sgs1 and Dna2 showed interaction with Rfa1 as expected (12, 39), although the Sgs1-Rfa1 interaction was consistently weaker than that of Dna2-Rfa1 (Fig. 4B). Moreover, Sgs1 showed better interaction with Rfa1-SuON compared with Rfa1 or Rfa1-SuCtrl, whereas Dna2 interaction with Rfa1-SuON was similar to that with Rfa1 (Fig. 4B). Next, we tested whether the interaction between Rfa1 and Sgs1 or Dna2 could be enhanced when Rfa1 sumoylation was increased in a more physiological setting. Co-immunoprecipitation analyses were performed to examine whether Sgs1 or Dna2 showed preferential interaction with the sumoylated form of Rfa1 compared with unmodified Rfa1. We observed that immunoprecipitated Sgs1 was able to co-immunoprecipitate sumoylated Rfa1, and the ratio of sumoylated to unmodified Rfa1 was significantly higher in the eluate fraction than in the unbound fraction (Fig. 4C). In contrast, Dna2 co-immunoprecipitated sumoylated Rfa1 less efficiently, and the ratio of sumoylated to unmodified Rfa1 was similar between the eluate and unbound fractions (Fig. 4D). The combination of the above data suggests that Sgs1 showed a preferential interaction with the sumoylated form of Rfa1.
Figure 4.
Rfa1 sumoylation enhances Sgs1 association, and Sgs1 checkpoint-defective mutant rescues srs2Δ defects. A, Rfa1-SuON increases Rfa1 sumoylation levels. Cells containing yeast two-hybrid plasmids used in panel B were examined under normal growth conditions. Rfa1 sumoylation was detected in cells containing Rfa1-SuON but not Rfa1-SuCtrl, although both constructs had similar unmodified protein levels. B, Sgs1 shows an enhanced interaction with Rfa1-SuON in yeast two-hybrid assay. All constructs support growth on control plates (SC-Trp-Leu), but only the ones that interact support growth on −H (SC-Trp-Leu-His) plates. C and D, Sgs1, but not Dna2, exhibits an enhanced interaction with sumoylated Rfa1. After cells were treated with 0.2% MMS for 2 h, Myc-tagged Sgs1 or Dna2 was immune-captured on myc-conjugated beads. The unbound (Un) and the bead eluate (E) fractions were examined by immunoblotting. Sgs1 and Dna2 were detected in the eluted, but not the unbound, fraction, suggesting successful immunoprecipitation and elution of these proteins. Rfa1 was present in both fractions, allowing comparison of the relative levels of sumoylated forms of Rfa1 between unbound and eluate fractions. Mean ± S.E. from three biological duplicates are plotted, with the value from unbound fractions set as 1. Controls using untagged Sgs1 and Dna2 showed a small amount of Rfa1 bound to beads because of nonspecific bead binding as reported previously (12). ns, not statistically significant. E, sgs1-4A reduces the active Rad53 levels in srs2Δ cells upon CPT treatment. Experiments and quantification were done as in Fig. 3D. Mean of three biological isolates per genotype were shown and error bars represent S.E. Double asterisks indicate a significant difference with a p value < 0.01. ns, not statistically significant. F, genetic analysis showing that a checkpoint-defective mutant of Sgs1, sgs1-4A, suppresses the DNA damage sensitivity of srs2Δ. sgs1-4A and rfa1-4KR were epistatic in their suppression of srs2Δ. Experiments were done as in Fig. 3A.
A checkpoint-defective sgs1 mutant reduces srs2Δ damage sensitivity and hyper-checkpoint
Our data so far suggest a model wherein sumoylated Rfa1 promotes interaction with Sgs1 and this may contribute to the high levels of checkpoint in srs2Δ cells. This model predicts that reducing Sgs1-mediated checkpoint functions should have similar suppressive effects as rfa1-4KR in srs2Δ cells, even though it is well-known that sgs1Δ and srs2Δ are synthetic lethal (28). To test this prediction, we used a Sgs1 phosphorylation-deficient mutant (sgs1-4A) known to impair Mec1 checkpoint activation because of its reduced interaction with Rad53 (12). We found that sgs1-4A indeed reduced active Rad53 levels in srs2Δ cells under CPT treatment and adding rfa1-4KR did not enhance this effect (Fig. 4E). Remarkably, this suppression correlated with an improvement in srs2Δ survival, and this effect of sgs1-4A was epistatic with rfa1-4KR (Fig. 4F). This result is consistent with our model that Rfa1 sumoylation positively influences the Sgs1-mediated checkpoint pathway.
Discussion
The Mec1 checkpoint and DNA damage–induced sumoylation are two important mechanisms that support the cellular response to external damages (2, 14). How they coordinate and influence each other is not well-understood. Previous studies reported SUMO-mediated enhancement of the checkpoint response in yeast and human cells (17, 18). Here we examined the effects of sumoylation of the RPA large subunit in this cross-talk. Our findings suggest that Rfa1 sumoylation positively contributes to the Mec1 checkpoint and this is mediated by fostering the RPA association with a checkpoint adaptor protein, Sgs1.
We found that two of the three previously reported sumoylation sites (K170 and K427) were indeed responsible for specific mono-sumoylation isoforms of untagged Rfa1. This result is consistent with the fact that K170 and K427, but not the third site, K133, were identified in the MS analyses as sumoylation sites (16). We further identified two additional lysines (K180 and K411) whose mutations further reduced sumoylation levels in rfa1-K170, 427R background. The lack of detection of these two sites in MS analyses is likely because they only account for a smaller portion of Rfa1 sumoylation. We showed that the rfa1–4KR mutant with all four sites mutated lost ∼90% Rfa1 sumoylation without affecting protein level or overall RPA functions, as the mutant supports WT growth and damage survival. However, in srs2Δ cells, which provide a sensitive context for detecting perturbation of checkpoint function, rfa1-4KR and a partial defective checkpoint mutant ddc1-T602A improved cell damage survival in additive manner. This finding is consistent with the result that ddc1-T602A rfa1–4KR could better reduce Rad53 hyperactivation in srs2Δ cells than ddc1-T602A alone. The additive effects in both assays suggest that rfa1-4KR and ddc1-T602A weaken checkpoint response by different mechanisms. Indeed, our further tests provided evidence that Rfa1 sumoylation fostered interaction with a checkpoint adaptor protein Sgs1, and that a sgs1 checkpoint-defective mutant (sgs1-4A) acts epistatically with rfa1-4KR in srs2Δ suppression. Taken together, our data suggest a working model that enhancing the Rfa1–Sgs1 interaction by Rfa1 sumoylation favors Rad53 activation and this effect is independent of Ddc1-based stimulation of checkpoint functions. This model expands our understanding of how RPA and Sgs1 collaborate in checkpoint activation (12).
Although we found that rfa1-4KR did not affect spontaneous or MMS-induced intra-chromosomal recombination by itself or in srs2Δ cells, it remains possible that Rfa1 sumoylation might be involved in other types of recombination processes or DNA repair contexts. Interestingly, human Rfa1 sumoylation was suggested to contribute to recombinational repair (40). We note that even though Rfa1 sumoylation is conserved, the modification sites are not the same in yeast and human proteins. The yeast Rfa1 sumoylation sites are not in contact with ssDNA as suggested by the corresponding lysine in the Ustilago maydis RPA-ssDNA structure (25). Consistently, mutating these sites supports normal growth and genotoxin resistance, unlike rfa1 DNA-binding mutants. These observations suggest that the yeast rfa1 sumoylation-defective mutant unlikely affects ssDNA binding, although biochemical tests are required to formally exclude this possibility. In contrast to the yeast Rfa1, a human RPA1 sumoylation site (K577) in its DBD-C domain is expected to contact ssDNA based on the U. maydis RPA-ssDNA structure data (25). It is thus possible that RPA sumoylation may have evolved with a more direct role in modulating RPA-ssDNA interaction in humans.
Besides supporting the DNA damage response, RPA is involved in many other DNA metabolism processes, such as various forms of DNA repair, DNA replication, and telomere maintenance, through interacting with ssDNA and with more than a dozen proteins (4). It is thus possible that RPA modifications could play a role in guiding RPA functions in other specific contexts. Our characterization of the Rfa1 sumoylation sites provides a valuable reagent for further exploration of how this modification can modulate additional genomic maintenance processes in the future.
Materials and methods
Yeast strains and genetic techniques
Standard procedures were used in cell growth and medium preparation. Strains used are listed in Table 1 and were isogenic to W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3, 112 trp1-1 rad5-535). rfa1-4KR mutant allele was generated using URA3-based pop-in-pop-out method as described previously (41) to produce marker-less allele replacement at the endogenous locus. All alleles were verified by sequencing. Yeast spotting assays were performed with standard procedures and at least two biological duplicates were used for each genotype.
Table 1.
Strains and plasmids used in this study
All strains are derivatives of W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11, 15 leu2-3, 112 trp1-1 rad5-535) (43). Only one strain is listed for each genotype, but at least two independent isolates of each genotype were used in the experiments.
| Genotype | Source | |
|---|---|---|
| Strain | ||
| T193-7c | MATa siz2Δ::KAN | This study |
| G13 | MATa mec1Δ::TRP1 sml1Δ::HIS3 | R. Rothstein |
| X3579-11d | MATa HF-Smt3::LEU2 | This study |
| T1435-14d | rfa1Δ::TRP1 pRS416-Rfa1-WT | This study |
| T1260-8-3a | rfa1Δ::TRP1 pRS416-rfa1-K427R | This study |
| T1259-9-1c | rfa1Δ::TRP1 pRS416-rfa1-K170R | This study |
| T1258-7-4c | rfa1Δ::TRP1 pRS416-rfa1-K170,427R | This study |
| T1387-t19 | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-Rfa1 | This study |
| T1327-1-5b | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K133R | This study |
| T1389-t12 | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K170,427R | This study |
| T1390-3b | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K133,170,427R | This study |
| T1264-6-5c | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K170,417,427R | This study |
| T1391-3b | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K170,180,427R | This study |
| T1344-1c | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K170,180,297,427,442R | This study |
| T1394-1b | mec1Δ::TRP1 sml1Δ::HIS3 rfa1Δ::TRP1 pRS416-rfa1-K170,180,411,427R | This study |
| Z417-17 | rfa1-K170,180,411,427R | This study |
| T436-3 | MATa Rfa1-Taf::KAN | Lab collection |
| T701-5a | MATα Rfa1-13myc::HIS3 | Lab collection |
| X7442-3a | MATa ddc1T602A::natNT2 | This study |
| X7442-4c | MATa srs2Δ::HIS3 | This study |
| X7442-3b | MATa srs2Δ::HIS3 rfa1-K170,180,411,427R | This study |
| X7442-21c | MATa srs2Δ::HIS3 ddc1T602A::natNT2 | This study |
| X7442-11a | MATa srs2Δ::HIS3 ddc1T602A::natNT2 rfa1-K170,180,411,427R | This study |
| X7684-11c | leu2-ri::URA3::leu2-bsteii | This study |
| X7684-16a | leu2-ri::URA3::leu2-bsteii rfa1-K170,180,411,427R | This study |
| X7684-11a | leu2-ri::URA3::leu2-bsteii srs2Δ::HIS3 | This study |
| X7684-16c | leu2-ri::URA3::leu2-bsteii srs2Δ::HIS3 rfa1-K170,180,411,427R | This study |
| X7258-17c | MATa srs2Δ::HIS3 rfa1-K170,180,411,427R | This study |
| X7444-8d | srs2Δ::HIS3 rad52-K43,44,253R rad59-K207,228R | This study |
| X7444-7b | srs2Δ::HIS3 rfa1-K170,180,411,427R rad52-K43,44,253R rad59-K207,228R | This study |
| X6584-1a | Sgs1-9myc::KANMX4 | Lab collection |
| T2022-11 | Dna2-9myc::hphMX6 | This study |
| X7467-19c | MATa srs2Δ::HIS3 | This study |
| X7467-19b | MATa sgs1-4A | This study |
| X7467-4d | MATa srs2Δ::HIS3 sgs1-4A | This study |
| X7467-22c | MATa srs2Δ::HIS3 rfa1-K170,180,411,427R | This study |
| X7738-5b | MATa srs2Δ::HIS3 rfa1-K170,180,411,427R sgs1-4A | This study |
| pJ69-4 | TRP1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS::GAL1-HOS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. 44 |
| Plasmid | ||
| pXZ578 | pRS416-Rfa1 | This study |
| pXZ580 | pRS416-rfa1-K170R | This study |
| pXZ581 | pRS416-rfa1-K427R | This study |
| pXZ583 | pRS416-rfa1-K170,427R | This study |
| pXZ595 | pRS416-rfa1-K133R | This study |
| pXZ594 | pRS416-rfa1-K133,170,427R | This study |
| pXZ587 | pRS416-rfa1-K170,417,427R | This study |
| pXZ612 | pRS416-rfa1-K170,180,297,427,442R | This study |
| pXZ647 | pRS416-rfa1-K170,180,427R | This study |
| pXZ623 | pRS416-rfa1-K170,180,411,427R | This study |
| p514 | pGBT9-Rfa1 | Ulrich and co-workers 45 |
| pXZ747 | pGBKT7-Rfa1-SuON | This study |
| pXZ749 | pGBKT7-Rfa1-SuCtrl | This study |
| pXZ668 | pGADT7-Sgs1 | Lab collection |
| pXZ744 | pGADT7-Dna2 | This study |
Detection and quantification of unmodified and sumoylated Rfa1
Cells were treated as indicated in the text before collection. Cells were then lysed by bead beating in the presence of 20% TCA that can both denature proteins and minimize the loss of sumoylated forms. The pellets were recovered by centrifugation and incubated with 1× Laemmli buffer at 95 °C for 5 min to recover proteins. Subsequently, proteins were separated on 3–8% Tris acetate gels (Life Technologies) followed by Western blotting with anti-Rfa1 antibody (a kind gift from S. Brill). Accurate quantification of protein bands was achieved by scanning the Western blots using a LAS-3000 luminescent image analyzer (Fujifilm) with a linear dynamic range of 104. The signal intensities of nonsaturated bands were measured using ImageJ software. For graphs, data are shown as mean ± S.E. except in Fig. 3E. Statistical differences were determined using Student's t tests.
Synchronization and detection of the active form of Rad53
Log phase cultures were arrested in G1 by treatment with 5 μg/ml α-factor for 1.5 h. G1 cells were then released into YPD media containing 100 μg/ml protease (Sigma) and 16 μg/ml CPT at 30 °C for 6 h. Then 2 × 108 cells were collected and protein extract prepared by standard TCA method as described above. Proteins were separated on gradient gels (Bio-Rad) followed by Western blotting with the F9 antibody (a kind gift from M. Foiani and D. Piccini) to detect active Rad53 levels. Pgk1 was detected by using anti-Pgk1 antibody (22C5D8, Invitrogen) and used as loading control. Quantification of Rad53 levels was done as described above.
Measurement of recombination rates
Recombination rates were measured using the leu2-ri::URA3::leu2-bsteii recombination assay (31), and the rates were calculated using fluctuation analysis based on the Lea-Coulson method of the median (42). Briefly, cells were grown in YPD to mid-log phase and the appropriate numbers of cells were then plated on SC-LEU and SC plates. For measurement of induced recombination rate, cells were grown in YPD to mid-log phase and then treated with 0.03% MMS for 2 h after which they were plated on SC-LEU and SC plates. Colonies were counted after incubation at 30 °C for 2 days. Each test was performed with 12 colonies obtained from two spore clones for each genotype and was repeated twice.
Yeast two-hybrid assay
This assay was performed as described previously (35). In brief, the Gal4 activation domain and Gal4 binding domain constructs were transformed into reporter strains (pJ69-4 a and α) and cells were grown on SC-Trp-Leu plates at 30 °C for 48 h. Protein–protein interactions were then assessed by growth of reporter strains on SC-Trp-Leu-His plates. The Rfa1-SuOn– and Rfa1-SuCtrl–containing binding-domain plasmids were constructed by fusing Rfa1 with either the SuOn tag or SuCtrl tag (38) at its C terminus.
Co-immunoprecipitation and quantification
For Fig. 4, C and D, log phase cultures were treated with 0.2% MMS for 2 h. Cells were harvested and lysed by bead beating in TMG-125 buffer. DNA was digested by incubation with Benzonase for 30 min at 4 °C. Lysates were cleared by centrifugation and incubated in TMG-125 buffer with Protein G agarose beads and anti-myc antibody for 2 h at 4 °C. After incubation, beads were washed with TMG-125 and proteins were eluted with Laemmli buffer. Proteins were separated on gradient gels followed by Western blotting with antibodies against Rfa1 (a kind gift from S. Brill) and myc-tag (9E10, Bio X Cell). Quantification and statistical analyses were done as described above for sumoylated Rfa1. Briefly, signals of sumoylated Rfa1 from the long exposure blots are divided by that of unsumoylated Rfa1 from the short exposure blots, to use signal values within a linear range of detection. This ratio was set as 1 for unbound fractions to derive the -fold change between the unbound and eluate fractions.
Author contributions
N. D., L. W., and X. Z. conceptualization; N. D., L. W., and X. Z. data curation; N. D., L. W., and X. Z. formal analysis; N. D., L. W., and X. Z. investigation; N. D. and L. W. methodology; N. D. and X. Z. writing-original draft; N. D., L. W., and X. Z. writing-review and editing; X. Z. supervision; X. Z. funding acquisition.
Acknowledgments
We are very grateful to Steve Brill for providing the anti-Rfa1 antibody and to M. Foiani and D. Piccini for providing the F9 antibody. We thank H. Klein, S. Gasser, R. Rothstein, and H. Ulrich for sharing strains and plasmids. We also thank C. Cremona for identifying impairment of Rfa1 functions by its C-terminal tagging; Chris Lima, Lumir Krejci, and Philip Zegerman for communication of unpublished data on Rfa1 sumoylation; and Jennifer Guo for help in genetic analyses.
This work was supported by NIGMS, National Institutes of Health Grant GM080670 (to X. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- DDR
- DNA damage response
- DDC
- DNA damage checkpoint
- ssDNA
- single-stranded DNA
- DDIS
- DNA damage-induced sumoylation
- MMS
- methyl methanesulfonate
- CPT
- camptothecin
- SuON
- SUMO ON
- SuCtrl
- SUMO control
- YPD
- yeast extract peptone dextrose (media)
- TMG-125 buffer
- 10 mm Tris-HCl pH 8.0, 4 mm MgCl2, 10% glycerol, 140 mm NaCl, 0.1 mm EDTA, 40 mm N-ethylmaleimide, 0.5% Tween 20 and Roche cOmplete ULTRA EDTA-free protease inhibitor.
References
- 1. Harper J. W., and Elledge S. J. (2007) The DNA damage response: Ten years after. Mol. Cell 28, 739–745 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- 2. Polo S. E., and Jackson S. P. (2011) Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 25, 409–433 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cussiol J. R., Dibitetto D., Pellicioli A., and Smolka M. B. (2016) Slx4 scaffolding in homologous recombination and checkpoint control: Lessons from yeast. Chromosoma 126, 45–58 10.1007/s00412-016-0600-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maréchal A., and Zou L. (2015) RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25, 9–23 10.1038/cr.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ball H. L., Ehrhardt M. R., Mordes D. A., Glick G. G., Chazin W. J., and Cortez D. (2007) Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 27, 3367–3377 10.1128/MCB.02238-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Majka J., Binz S. K., Wold M. S., and Burgers P. M. (2006) Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J. Biol. Chem. 281, 27855–27861 10.1074/jbc.M605176200 [DOI] [PubMed] [Google Scholar]
- 7. Navadgi-Patil V. M., and Burgers P. M. (2009) The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol. Cell 36, 743–753 10.1016/j.molcel.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Navadgi-Patil V. M., and Burgers P. M. (2008) Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 283, 35853–35859 10.1074/jbc.M807435200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar S., and Burgers P. M. (2013) Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 27, 313–321 10.1101/gad.204750.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zou L. (2013) Four pillars of the S-phase checkpoint. Genes Dev. 27, 227–233 10.1101/gad.213306.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz M. F., Duong J. K., Sun Z., Morrow J. S., Pradhan D., and Stern D. F. (2002) Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9, 1055–1065 10.1016/S1097-2765(02)00532-4 [DOI] [PubMed] [Google Scholar]
- 12. Hegnauer A. M., Hustedt N., Shimada K., Pike B. L., Vogel M., Amsler P., Rubin S. M., van Leeuwen F., Guénolé A., van Attikum H., Thomä N. H., and Gasser S. M. (2012) An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 31, 3768–3783 10.1038/emboj.2012.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrison J. C., and Haber J. E. (2006) Surviving the breakup: The DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235 10.1146/annurev.genet.40.051206.105231 [DOI] [PubMed] [Google Scholar]
- 14. Sarangi P., and Zhao X. (2015) SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem. Sci. 40, 233–242 10.1016/j.tibs.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chung I., and Zhao X. (2015) DNA break-induced sumoylation is enabled by collaboration between a SUMO ligase and the ssDNA-binding complex RPA. Genes Dev. 29, 1593–1598 10.1101/gad.265058.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Psakhye I., and Jentsch S. (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- 17. Cremona C. A., Sarangi P., Yang Y., Hang L. E., Rahman S., and Zhao X. (2012) Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the Mec1 checkpoint. Mol. Cell 45, 422–432 10.1016/j.molcel.2011.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C. S., Ouyang J., Mori E., Nguyen H. D., Maréchal A., Hallet A., Chen D. J., and Zou L. (2014) SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes Dev. 28, 1472–1484 10.1101/gad.238535.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burgess R. C., Rahman S., Lisby M., Rothstein R., and Zhao X. (2007) The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 27, 6153–6162 10.1128/MCB.00787-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ulrich H. D., and Davies A. A. (2009) In vivo detection and characterization of sumoylation targets in Saccharomyces cerevisiae. Methods Mol. Biol. 497, 81–103 10.1007/978-1-59745-566-4_6 [DOI] [PubMed] [Google Scholar]
- 21. Brill S. J., and Stillman B. (1991) Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 5, 1589–1600 10.1101/gad.5.9.1589 [DOI] [PubMed] [Google Scholar]
- 22. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- 23. Sacher M., Pfander B., Hoege C., and Jentsch S. (2006) Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8, 1284–1290 10.1038/ncb1488 [DOI] [PubMed] [Google Scholar]
- 24. Pfander B., Moldovan G. L., Sacher M., Hoege C., and Jentsch S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 10.1038/nature03665 [DOI] [PubMed] [Google Scholar]
- 25. Fan J., and Pavletich N. P. (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 26, 2337–2347 10.1101/gad.194787.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vaze M. B., Pellicioli A., Lee S. E., Ira G., Liberi G., Arbel-Eden A., Foiani M., and Haber J. E. (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10, 373–385 10.1016/S1097-2765(02)00593-2 [DOI] [PubMed] [Google Scholar]
- 27. Yeung M., and Durocher D. (2011) Srs2 enables checkpoint recovery by promoting disassembly of DNA damage foci from chromatin. DNA Repair 10, 1213–1222 10.1016/j.dnarep.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 28. Klein H. L. (2001) Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bermejo R., Doksani Y., Capra T., Katou Y. M., Tanaka H., Shirahige K., and Foiani M. (2007) Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 21, 1921–1936 10.1101/gad.432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marini V., and Krejci L. (2010) Srs2: The “Odd-Job Man” in DNA repair. DNA Repair 9, 268–275 10.1016/j.dnarep.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu H., Boone C., and Klein H. L. (2004) Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol. Cell. Biol. 24, 7082–7090 10.1128/MCB.24.16.7082-7090.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colavito S., Macris-Kiss M., Seong C., Gleeson O., Greene E. C., Klein H. L., Krejci L., and Sung P. (2009) Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 37, 6754–6764 10.1093/nar/gkp748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altmannova V., Eckert-Boulet N., Arneric M., Kolesar P., Chaloupkova R., Damborsky J., Sung P., Zhao X., Lisby M., and Krejci L. (2010) Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 38, 4708–4721 10.1093/nar/gkq195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao X. (2018) SUMO-mediated regulation of nuclear functions and signaling processes. Mol. Cell 71, 409–418 10.1016/j.molcel.2018.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bonner J. N., Choi K., Xue X., Torres N. P., Szakal B., Wei L., Wan B., Arter M., Matos J., Sung P., Brown G. W., Branzei D., and Zhao X. (2016) Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 16, 368–378 10.1016/j.celrep.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F. J., Jeram S. M., Li Z., Bahr S., Andrews B. J., Boone C., and Raught B. (2009) Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell 33, 124–135 10.1016/j.molcel.2008.12.025 [DOI] [PubMed] [Google Scholar]
- 37. Almedawar S., Colomina N., Bermúdez-López M., Pociño-Merino I., and Torres-Rosell J. (2012) A SUMO-dependent step during establishment of sister chromatid cohesion. Curr. Biol. 22, 1576–1581 10.1016/j.cub.2012.06.046 [DOI] [PubMed] [Google Scholar]
- 38. Wei L., and Zhao X. (2016) A new MCM modification cycle regulates DNA replication initiation. Nat. Struct. Mol. Biol. 23, 209–216 10.1038/nsmb.3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bae K. H., Kim H. S., Bae S. H., Kang H. Y., Brill S., and Seo Y. S. (2003) Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 31, 3006–3015 10.1093/nar/gkg422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dou H., Huang C., Singh M., Carpenter P. B., and Yeh E. T. (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39, 333–345 10.1016/j.molcel.2010.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reid R. J., Lisby M., and Rothstein R. (2002) Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350, 258–277 10.1016/S0076-6879(02)50968-X [DOI] [PubMed] [Google Scholar]
- 42. Hall B. M., Ma C. X., Liang P., and Singh K. K. (2009) Fluctuation analysis CalculatOR: A web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics 25, 1564–1565 10.1093/bioinformatics/btp253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao X., and Blobel G. (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 10.1073/pnas.0500537102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davies A. A., Huttner D., Daigaku Y., Chen S., and Ulrich H. D. (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol. Cell 29, 625–636 10.1016/j.molcel.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]