Abstract
VqmA is a highly conserved transcriptional regulator of the quorum-sensing system of Vibrio cholerae, a major human pathogen that continues to imperil human health. VqmA represses biofilm formation and plays an important role in V. cholerae pathogenicity in the human host. Although VqmA's biological function is well understood, the molecular mechanisms by which its specific ligand (and effector), 3,5-dimethylpyrazine-2-ol (DPO), controls transcription of the target gene, vqmR, remain obscure. To elucidate the molecular mechanism of DPO binding, we used structural analyses and biochemical assays to study the V. cholerae VqmA–DPO–DNA complex. These analyses revealed that VqmA contains an N-terminal homodimer domain (PAS) and a C-terminal DNA-binding domain (DBD). We observed that VqmA directly binds to a DPO molecule via a compact hydrophobic pocket, consisting of a six-stranded antiparallel β-sheet and several α-helices. We also found that the VqmA dimer interacts with the quasi-palindromic sequence of the vqmR promoter through its DBD. The results of the biochemical studies indicated that a water atom and VqmA residues Phe-67 and Lys-101 play a key role in effector recognition, which is also assisted by Tyr-36 and Phe-99. This is the first molecular level view of the VqmA dimer bound to DPO and DNA. The structure–function analyses presented here improve our understanding of the complex mechanisms in the transcriptional regulation of VqmA in Vibrio spp. and may inform the design of drugs to manage V. cholerae infections.
Keywords: quorum sensing, transcription promoter, protein-DNA interaction, protein structure, X-ray crystallography, DPO, ligand binding, Vibrio cholerae, virulence, VqmA
Introduction
Quorum sensing (QS)3 is a kind of cell–cell communication process used by many bacteria, and it helps bacteria to monitor their population density and synchronize their behavior in response to changes in their environment (1). The production and detection of signal molecules, called autoinducers, support the operation of QS. QS signal molecules are detected by proper regulators, which serve as signal receptors. Once the autoinducer concentration reaches a specific threshold, the autoinducer–receptor complex binds to palindromes and regulates QS-dependent processes, such as bioluminescence, secretion of virulence factors, and biofilm formation (2). Relying on the QS system, bacteria synchronize their behaviors on a population-wide scale and function like multicellular organisms (3).
The Gram-negative bacterium Vibrio cholerae is a major human pathogen that continues to imperil human health. The two confirmed autoinducers are CAI-1 ((S)-3-hyroxytridecan-4-one), as detected by the histidine sensor kinase receptors CqsS, and AI-2 (4,5-dihydroxy-2,3-pentanedione), as detected by LuxPQ in V. cholerae (4, 5). Recently, a novel signal–receptor pair involved in regulating biofilm formation was detected in V. cholerae (6). This new QS system depends on the transcription factor VqmA, which is activated by DPO (3,5-dimethylpyrazin-2-ol), an extracellular small molecule that accumulates at a high cell density (6–8). The new autoinducer DPO is produced from threonine and alanine and indirectly regulates biofilm formation by binding to VqmA (6). VqmA, in complex with DPO, activates expression of the vqmR gene, encoding VqmR sRNA, which represses the genes required for V. cholerae biofilm formation (6).
VqmA is a member of the LuxR family proteins, which are thought to share a ligand-binding region and DNA-binding domain (9). Similar to many other LuxR family members, VqmA possesses effector-binding sites in the N-terminal PAS domain and conservative DNA-binding domains in the C-terminal (6, 8). However, compared with other LuxR family proteins, which exhibit relative conservative in the DNA-binding domain (10), VqmA shows very weak homology of the conserved ligand-binding domains. The latter likely reflects the large ligand diversity of LuxR proteins, which exhibit broad ligand diversity (11). In addition, homologs of the VqmA receptor are limited to the Vibrio genus and one vibriophage VP882 (6, 12), and this limitation also implies that VqmA may be a Vibrio-specific drug target. Furthermore, the newly discovered VqmA encoded by the VP882 can also be activated by DPO and initiates the phage lysis process, which can also manage V. cholerae infections (12).
In this study, the crystal structure of the V. cholerae VqmA–DPO–DNA complex was determined at 2.51 Å resolution using X-ray diffraction. Our results revealed the ligand-binding mechanism of VqmA and verified the conservation of residues that contribute to DPO binding in VqmA by mutational studies. This information will help to design highly specific small molecules for the VqmA receptor. In addition, the crystal structure results further confirmed the specific recognition region of the VqmA protein in the promoter of the vqmR gene and will improve our understanding of the complex mechanisms in the transcriptional regulation of VqmA in Vibrio genus.
Results
The preparation of VqmA–DPO–DNA complex
Because we had tried our best to crystallize and failed to obtain apo-VqmA crystals and VqmA-binding DPO complex crystals, we speculated that the conformation of VqmA was unstable, and it may be necessary to stabilize their DNA-binding domain with the help of target dsDNA. Therefore, we have tried to screen the target dsDNA sequences of VqmA.
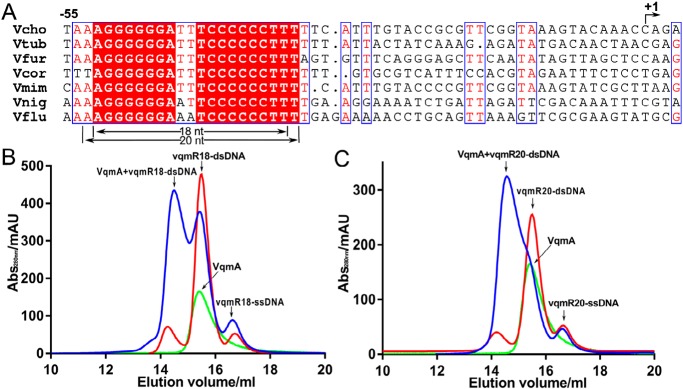
Based on the experimental results from a previous study (8), we roughly determined the target dsDNA sequence of VqmA within the vqmR promoter region, according to the characteristics of the palindrome sequence that transcription factors often bind (Fig. 1A). To determine the target dsDNA sequence of VqmA more accurately, we selected a variety of DNA sequences of different lengths from the known sequence regions above (Table S2). The binding ability of VqmA to each sequence was tested by gel filtration. It found that when the target DNA sequence containing a special quasi-palindrome sequence and exceeded 18 bp, VqmA could bind to the dsDNA effectively (Fig. 1B). Therefore, we chose the 18-bp quasi-palindrome DNA sequence (vqmR18) as our target dsDNA for crystallization.
Figure 1.
To determine the target dsDNA of VqmA. A, sequence alignment of vqmR genes. The sequences, including the promoter regions, of vqmR genes from different Vibrio species were aligned. The conserved region indicate the VqmA recognition sequence. Vcho, V. cholerae; Vtub, Vibrio tubiashii; Vfur, Vibrio furnissii; Vcor, Vibrio coralliilyticus; Vmim, Vibrio mimicus; Vnig, Vibrio nigripulchritudo; Vflu, Vibrio fluvialis. B, gel-filtration chromatography of VqmA and vqmR18 DNA showing that when VqmA was incubated with vqmR18 DNA, the peak position of its mixture (blue) is earlier than that of single vqmR18 DNA (red) and single VqmA (green). C, gel-filtration chromatography of VqmA and vqmR20 DNA showing that VqmA was incubated with vqmR20 DNA, the peak position of its mixture (blue) is earlier than that of single vqmR20 DNA (red) and single VqmA (green).
Overall structure of the VqmA–DPO–DNA complex
To explore the interaction between VqmA and its target dsDNA (5′-AGGGGGGATTTCCCCCCT-3′, from the vqmR promoter), we solved the VqmA–DPO–DNA complex crystal structure. The crystal structure of the VqmA complex was initially determined by selenium single-wavelength anomalous diffraction at a 2.51 Å resolution. The SeMet derivative crystal belonged to the I212121 space group. Unfortunately, in this structure, the noncrystallographic symmetry of the VqmA dimer was superimposed on the crystallographic symmetry, which meant that two VqmA molecules and the dsDNA of the VqmA–DPO–DNA complex belonged to two different asymmetric units, but the dsDNA in the two asymmetric units was not symmetric. Therefore, although the electron densities of the protein and DPO were clear, the electronic density of dsDNA could not be completed theoretically. A similar phenomenon was reported in CED-4 apoptosome (13). To overcome this problem, we re-screened the crystallization conditions and found a new crystal form. The new crystal form belonged to the P212121 space group. By comparing the unit-cell constants of these crystal forms, there are slight differences in the b and c axes (Table S1), which may lead to the shift of the symmetric axes. In the crystal form of the P212121 space group, there was a VqmA–DPO–DNA complex in one asymmetric unit. The dsDNA duplex formed a complex with a VqmA dimer, which bound DPO in its PAS domains. VqmA was homodimeric, the same as the other HTH superfamily members (14–17).
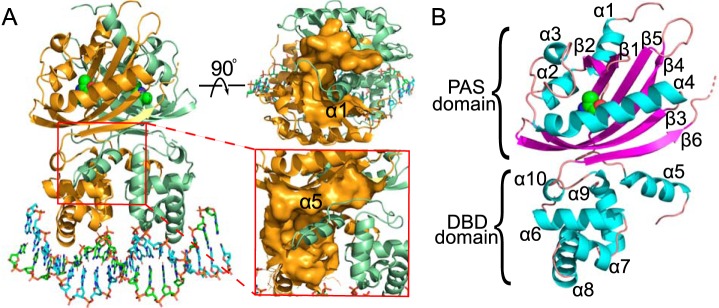
To facilitate observation of the VqmA monomer structure, one VqmA monomer was extracted from the structure of the VqmA complex. In the monomer, the VqmA structure was separated into two domains, the N-terminal PAS domain and C-terminal DNA-binding domains (PAS and DBD, respectively) (Fig. 2B). The PAS domain consisted of five α-helices and six antiparallel β-strands in the order of α1-β1-β2-α2-α3-α4-β3-β4-β5-β6. The PAS domain was connected to the DBD via one α-helix (α5) and a flexible loop. The DBD had the canonical HTH domain structure, with four α-helices (α6, α7, α8, and α9). On the whole, there were two regions that contributed to the interaction of homodimer. One region, located at the top of the PAS domain, acted by binding α1. Another region, located at the PAS-DBD interface, stabilized the interaction of homodimer and linked the DNA-binding domain with the PAS domain by binding α5 (Fig. 2A).
Figure 2.
Structure of the VqmA–DPO–DNA complex. A, structural overview of VqmA complexed with dsDNA and DPO. The two monomers are shown as cartoons colored in orange and pale green. DPO molecules bound to each monomer are shown as spheres. The top view of the structure shows the location of α1 in the upper right corner. The enlarged view of the structure shows the location of α5 in the bottom right corner. Secondary structure elements (α-helices and β-strands) are labeled in black. B, the structure of the VqmA monomer, α-helices and β-strands are shown as cartoons colored in cyan and purple, respectively.
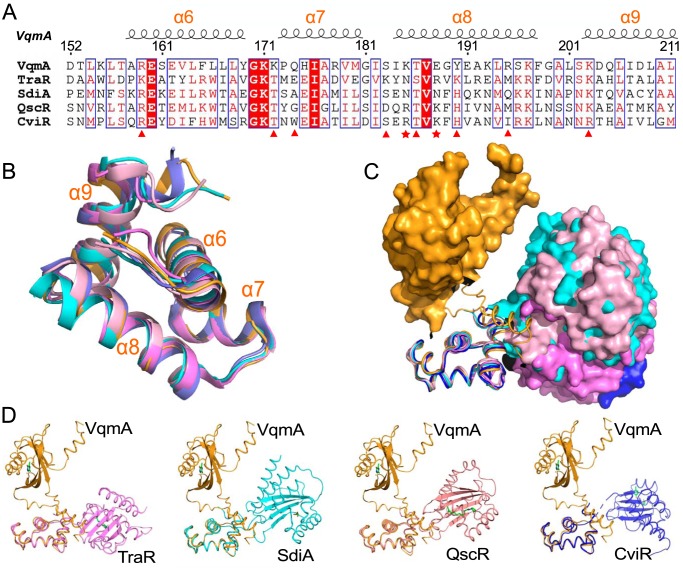
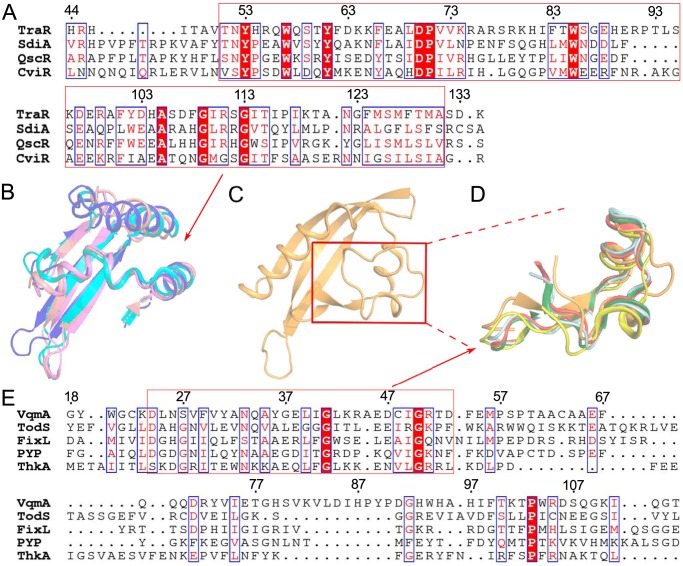
To gain insight into the particularities of the VqmA structure, two domains were compared with the corresponding structures of other family members. The DBD region was relatively conserved. We compared the DBD region of VqmA with several proteins with known structures of the LuxR family in sequence and three-dimensional conformation (17–21). The results showed that although VqmA and other LuxR family proteins were not very conservative in sequence, but they were consistent in three-dimensional conformation (Fig. 3). The more interesting result is that the PAS domain of VqmA was quite different from that of other characterized LuxR proteins (17–20), but similar to that of some signal proteins of other non-LuxR family proteins (Fig. 4 and Fig. S1) (22–25). However, although the overall folds of the PAS region were similar, the precise positions of various structural elements differed substantially, especially in the regions controlling ligand specificity (Fig. S1). Therefore, the key to our study is to explore the specific binding residues and thus determine the molecular mechanism.
Figure 3.
Homologous comparison of DBD of LuxR family proteins with known structures. A, structure-based multiple sequence alignment of DBD domains (VqmA, TraR (PDB ID 1L3L), SdiA (PDB ID 4LGW), QscR (PDB ID 6CC0), and CviR (PDB ID 3QP5)). The residues directly involved in interacting with DNA backbone and providing specific recognition are marked with red triangles and red pentagons, respectively. B, structure comparison of the DBD of VqmA (orange) and other domains (TraR (PDB ID 1L3L), violet; SdiA (PDB ID 4LGW), cyan; QscR (PDB ID 6CC0), pink; CviR (PDB ID 3QP5), blue) via superimposition. C, structural superimpositions of VqmA (orange) with TraR (violet), SdiA (cyan), QscR (pink), and CviR (blue). The C-terminal DBDs presented by the backbone traces were used for the structural overlap, and the N-terminal PAS domains are presented as surface-filling models. D, structural superimpositions of VqmA (orange) with TraR (violet), SdiA (cyan), QscR (pink), CviR (blue), respectively. Ligands are represented as green stick models.
Figure 4.
Homologous comparison of PAS domains with known structures. A, structure-based multiple sequence alignment of PAS domains of LuxR family proteins (TraR (PDB ID 1L3L), SdiA (PDB ID 4LGW), QscR (PDB ID 6CC0), and CviR (PDB ID 3QP5)). B, structure comparison of the conserved sequence in PAS domains of LuxR family proteins (TraR (PDB ID 1L3L), violet; SdiA (PDB ID 4LGW), cyan; QscR (PDB ID 6CC0), pink; CviR (PDB ID 3QP5), blue) via superimposition. C, the part of the PAS domain of VqmA having the same orientation as the structure comparison of B. D, structure comparison of the conserved sequence in PAS domains of non-LuxR family proteins (VqmA, orange; TodS (PDB ID 5HWW), yellow; FixL (PDB ID 1DP6), red; PYP (PDB ID 1F98), silver; ThkA (PDB ID 3A0S), dark green). E, structure-based multiple sequence alignment of PAS domains of non-LuxR family proteins (VqmA, TodS (PDB ID 5HWW), FixL (PDB ID 1DP6), PYP (PDB ID 1F98), ThkA (PDB ID 3A0S)).
Interaction of VqmA with DPO
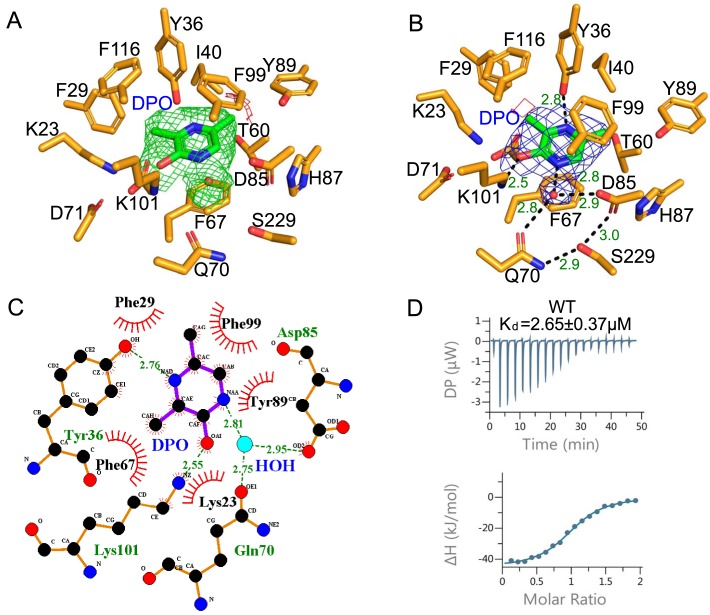
VqmA promotes the transcription of vqmR after binding a DPO molecule (6). The binding affinity of VqmA and DPO was analyzed using isothermal titration calorimetry (ITC) to understand the first step in DPO transcriptional activation of the vqmR gene by VqmA. The VqmA–DPO interaction was exothermic and exhibited an intermediate binding affinity (Kd, 2.65 μm) (Fig. 5D). The interaction was predominantly enthalpy driven (ΔH, −46.3 kJ/mol; −TΔS, 14.4 kJ/mol), indicating that the effector-binding energy of VqmA was mainly provided by hydrophilic interactions rather than hydrophobic interactions. The effector recognition mode of the VqmA structure bound to DPO and dsDNA was determined using co-crystallization. There were two VqmA molecules in one asymmetric unit. In the VqmA–DPO–DNA complex structure, a VqmA molecule bound one DPO in a hydrophobic pocket between α4 and the β-sheet (Fig. 2B). Therefore, one VqmA dimer simultaneously bound two DPO molecules. Furthermore, according to the electron density of crystal structure, DPO interacted with the β4-strand and α2-helix of the PAS domain, and the polar hydroxyl of DPO was mainly fixed by Lys-101 residues. The two N atoms of DPO were fixed by the Tyr-36 residue and one water molecule. The water molecule mediated the contacts of the N atom of DPO with two key residues (Gln-70 and Asp-85) and acted as a bolt to control the binding or unbinding of DPO. In addition, the apolar six-membered ring portion of DPO was sandwiched by two layers of VqmA residues (Phe-99 from the β4-strand; Phe-67 from the α4-helix) (Fig. 5B). Phe-67 was located below the six-membered ring of DPO and was 3.7 Å away, and the benzene ring of Phe-67 and DPO were close to parallel. Phe-99 was ∼4.5 Å above the six-membered ring of DPO, and the dihedral angle of the Phe-99 benzene ring and DPO was ∼30 degrees. Furthermore, there were two hydrogen bonds formed by Ser-229 with Gln-70 and Asp-85, and they might act as secondary barriers to resist external interference (Fig. 5B).
Figure 5.
DPO recognition by VqmA. A, the omit difference map of VqmA without DPO with σ level 3.0 colored green and σ level −3.0 colored red. The DPO added later is shown in green. B, the VqmA–DPO interaction observed in the PAS domain. VqmA is shown as orange sticks. The DPO molecule bound to VqmA is depicted as green sticks. A water molecule is shown as a red sphere. DPO and a water molecule were merged with the electron density (2Fo − Fc map contoured with σ level 1.0 colored with blue, Fo − Fc map contoured with σ level 3.0 colored with red and green). C, two-dimensional diagram of the interactions of VqmA with the DPO. D, the ITC profile for the interaction of WT VqmA with DPO. The ITC profiles of VqmA's mutants are shown in Fig. S2.
To verify the specific interactions observed in the VqmA–DPO–DNA complex structure, mutations were introduced into the DPO-binding pocket of VqmA. As expected, the single mutants F67A, Q70A, and K101L did not exhibit any detectable binding affinity to DPO in the ITC experiment (Table S3). The Y36F, F99A, and S229A mutants markedly decreased the binding affinity. To further understand the importance of Phe-67 and Phe-99, the single alanine mutations F67A and F99A were changed to single isoleucine mutations F67I and F99I, respectively. This change led to some interesting results: F67I regained a weak DPO binding capability, but the DPO binding capability of F99I was strongly enhanced and similar to that of WT VqmA (Fig. S2), which implied that Phe-99 mainly provides the steric hindrance needed for DPO binding and that Phe-67 provides the interaction force via a π-π stacking conformation. These results indicate that the water atom, Phe-67, and Lys-101 play a key role in effector recognition, which is also assisted by Tyr-36 and Phe-99 (Fig. 5).
The effect of DPO on VqmA
The effect of DPO on VqmA was demonstrated in the following three experiments.
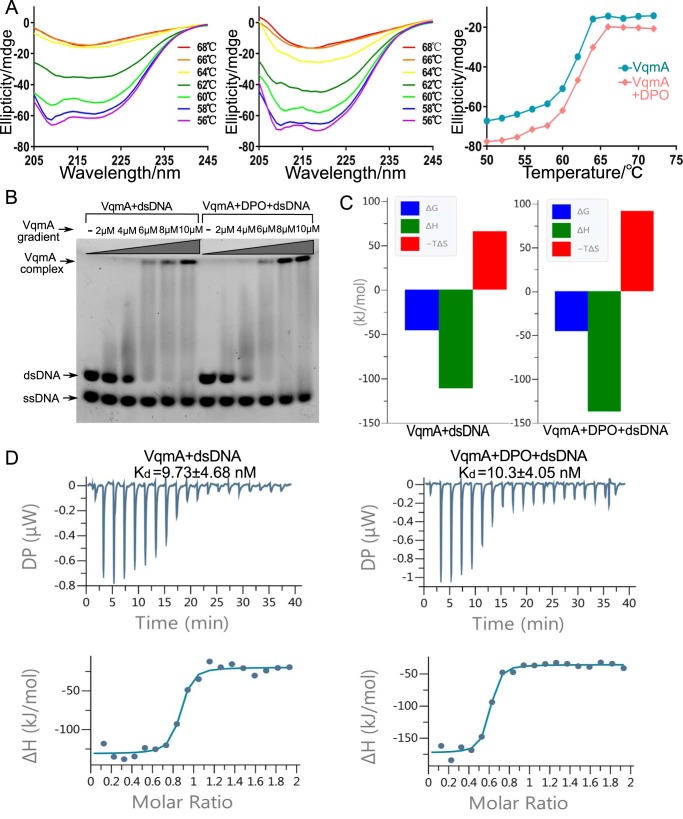
First, the thermostability of VqmA in the absence or presence of DPO was measured with a CD spectrometer. The temperature-dependent CD experiment was analyzed at 180–260 nm from 20 to 90 °C at intervals of 2 °C to determine melting temperature (Tm). In the absence of DPO, the melting temperature of VqmA was 62 °C, and its thermostability was almost unchanged when DPO was present (Fig. 6A).
Figure 6.
The effect of DPO on VqmA. A, CD spectra of VqmA with no DPO (left) and VqmA with 5 μm DPO (middle). The change trend of the CD spectra results due to the changing temperature at 216 nm (right). B, EMSA for VqmA with no DPO (left) and VqmA with 10 μm DPO (right). C, thermodynamic parameters (ΔG in blue, ΔH in green, −TΔS in red in kJ/mol) determined by ITC for the strong binding of DNA with apo-VqmA and DPO-binding VqmA. D, ITC profiles for the interactions of DNA with apo-VqmA and DPO-binding VqmA.
Second, the electrophoretic mobility shift assay (EMSA) experiments on VqmA in the presence or absence of DPO demonstrated no significant differences in DNA binding capacity (Fig. 6B).
Third, the ITC experiments on VqmA in the presence or absence of DPO showed approximate Kd values in DNA binding capacity (Fig. 6D). However, compared with the absence of DPO, the value of ΔH and −TΔS increased significantly with DPO binding (Fig. 6C, Table S4). It indicated that the entropy-driven and enthalpy-driven effects of VqmA binding to DNA were more obvious in the presence of DPO, where more obvious conformational changes may occur. The DNA binding and protein stability data suggested that although DPO has no obvious effect on improving the stability and binding ability of protein, to a certain extent, it promotes the more obvious conformation change of protein binding to DNA.
Interaction of VqmA with operator DNA
As a transcription factor, V. cholerae VqmA recognizes quasi-palindromic sequences (Fig. 1A) within the vqmR promoter region (8). We further probed the interaction of the VqmA protein with recognition dsDNA using biophysical methods. In an EMSA, the addition of the VqmA protein altered the mobility of the operator dsDNA (Fig. 6B). The ITC assay using operator DNA titrated against VqmA revealed the relatively high dsDNA-binding affinity of VqmA, with a dissociation constant (Kd) of 9.73 nm (Fig. 6D).
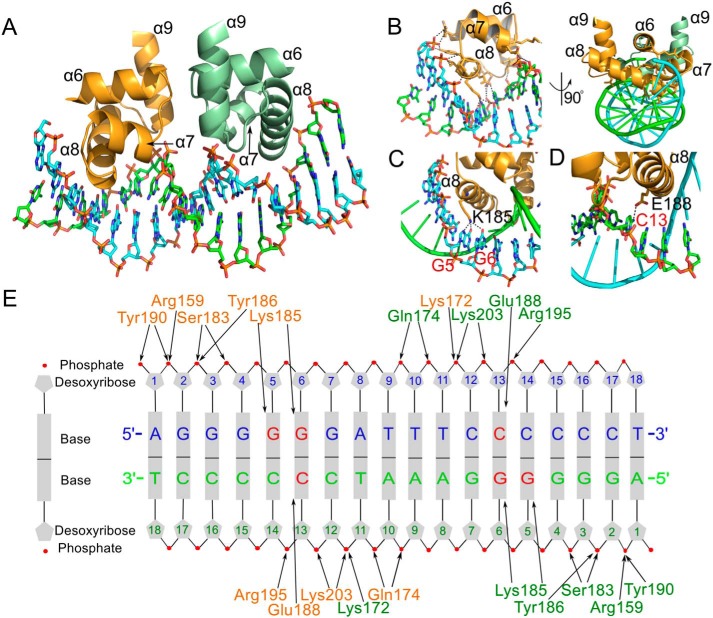
The molecular interaction of VqmA with operator DNA was visualized in the crystal structure of VqmA in complex with an 18-bp dsDNA containing the VqmA recognition sequence (Fig. 7). In the structure of the complex, the DBD of the VqmA dimer are located on the vertical axis of the dsDNA. The VqmA dimer symmetrically recognizes the quasi-palindromic sequence of the dsDNA using the two DBDs, with a 1:1 stoichiometry of VqmA dimer to dsDNA. The VqmA monomers simultaneously recognize the major groove of dsDNA, forming a major groove interaction site (Fig. 7, A and B). The major groove interaction site is primarily generated by inserting the α8 helix into the major groove of dsDNA (Fig. 7B). In the major groove interaction site, eight VqmA residues (Lys-172, Gln-174, Ser-183, Lys-185, Tyr-186, Glu-188, Tyr-190, and Arg-195) from the α7 and α8 helices make direct contacts with A1, G2, G3, G4, G5, G6, T10, T11, C12, C13, and C14. The side chain of Lys-185 forms hydrogen bonds with the G5 and G6 bases, and the side chain of Glu-188 forms hydrogen bonds with the C13 base. These direct contacts between the VqmA residues and DNA bases are buttressed by DNA backbone interactions located on both sidewalls of the major groove (Fig. 7E). To provide a suitable base recognition environment, Arg-159, Lys-172, Gln-174, Ser-183, Tyr-186, Tyr-190, Arg-195, and Lys-203 of VqmA make contacts with the DNA backbone at A1, G2, G3, G4, T10, T11, C12, C13, and C14.
Figure 7.
VqmA–dsDNA interaction observed in the VqmA complex structure. A, overall VqmA–dsDNA structure. The DBD of VqmA dimer are depicted as ribbons (orange and pale green), and dsDNA is shown as a ribbon (light blue and light green). B, detailed view of the residues of α8 in the major groove interaction site. C, detailed view of the Lys-185 residue interaction with G5 and G6. D, detailed view of the Glu-188 residue interaction with C13. E, a schematic diagram of the VqmA–dsDNA interaction. VqmA residues in the each monomer are shown in orange and green. DNA bases that VqmA makes contacts with are labeled in red.
DNA base specificity of VqmA
In the structure of the VqmA complex, each VqmA monomer makes direct contacts with three nucleotide bases at G5, G6, and C13 (Fig. 7E). The base specificity of VqmA observed in the VqmA complex structure coincides with the experimental results of Papenfort and co-workers (7, 8). When the conserved DNA bases (G4, G5, and G6) were all changed, VqmA binding was eliminated (8). These data show that VqmA is a direct transcriptional activator of the vqmR promoter and that interaction with a conserved region in the vqmR promoter is required for binding. Thus, our structural study, combined with previous mutational analysis, demonstrates that the VqmA dimer symmetrically recognizes vqmR promoter dsDNA (G5-G6-NNNNNN-C13-C14) containing two palindromic repeats of 2-bp dsDNA that are separated by a 6-bp spacer.
Discussion
The QS of V. cholerae contains many complex regulatory networks and play key roles in its pathogenicity (26–31). However, the clear molecular mechanisms of numerous key regulators remain obscure. As shown by previous studies, VqmA is a repressor of biofilm formation (6, 7, 26); yet, its structural mechanism remains unknown. Here, we determined the crystal structure of the VqmA–DPO–DNA complex. The combined biochemical results of this study and the crystal structure of VqmA should provide a better understanding of the regulatory mechanisms of VqmA.
In summary, the structural studies of VqmA performed in this study provide insights into DPO regulation of transcription factors. In a broader sense, the discovery of DPO as a VqmA ligand breaks new ground in the understanding of LuxR-type proteins (6). Its novel ligand and compact ligand-binding cavity suggest that VqmA, and potentially other LuxR family proteins, specifically detect DPO signal, allowing bacteria to make a reliable, targeted and adaptive response.
Similar to LuxR and many other members of LuxR family, the VqmA dimer possesses two ligand-binding sites in N-terminal domains and two conserved DNA-binding domains for identifying the palindromic sequence of targeted DNA in the C terminus (Fig. 2A). However, as reported, the binding site of the PAS region varies greatly with the different effector molecules (32–34).
Like most PAS domain regulators, VqmA has a binding pocket that is mainly composed of a variety of hydrophobic residues. Here, we show that Phe-67 plays a key role in binding to the six-membered ring of the DPO molecule. Compared with the PAS domains of other characterized LuxR proteins, similar to them, the PAS domain of VqmA has the typical β-sheet fold of the PAS domain, and the order is 2-1-5-4-3. However, the PAS domain of VqmA has a unique β6-strand, and is quite different from them on α-helix folds (Fig. S1). In addition, the ligand-binding sites of characterized LuxR proteins differ greatly from that of VqmA as mentioned above (17–20, 35, 36). They are located in the cavity regions between the β-sheet, α3, α5, and α7. The residues related to ligand binding are mainly located in β3, α3, α5, and α7, which is the most dissimilar region between VqmA and these characterized LuxR proteins (Fig. 4 and Fig. S1). Moreover, the PAS domain of VqmA has no conservative sequence similar to these LuxR family proteins. By contrast, it has some conservative regions compared with other PAS proteins (Fig. 4), such as PYP of Halorhodospira halophile, which plays a role as a light sensor in the photocycle by binding a chromophore (24); ThkA of Thermotoga maritima, which might be responsible for the interaction of TrrA with the ThkA DHp domain (25); FixL of Bradyrhizobium diazoefficiens, which acts as an important oxygen sensor (23, 37); and TodS of Pseudomonas putida, which is a sensor kinase that responds to various monoaromatic compounds (22). This structural information implies that the PAS of VqmA plays a role as a sensor and delivers the signal to the DBD by DPO-induced structural rearrangements, thereby regulating its DNA-binding activity.
Furthermore, the interaction of α5 with β6 and β3, to a certain extent, influences the compactness of the DBD and indirectly affects the DNA binding capacity of VqmA through the interaction of α5 with α6 (Fig. 8E). The interaction of DPO with Lys-101 and Asp-85 may stabilize the conformation of the β-sheet, thus making α5, the tail of the PAS domain, more stable in the PAS-DBD interface and stabilizing the interaction of the β-sheet of the PAS domain with β6. In addition, the interaction of DPO with Phe-67 and Gln-70 may play a key role in interaction of α4 with the β-sheet. Therefore, our structural analysis suggests that DPO enhances the VqmA–DNA interaction through an allosteric mechanism in which DPO rearranges the PAS domain of VqmA and indirectly impels the DBD into a conformation that is more compatible with DNA binding.
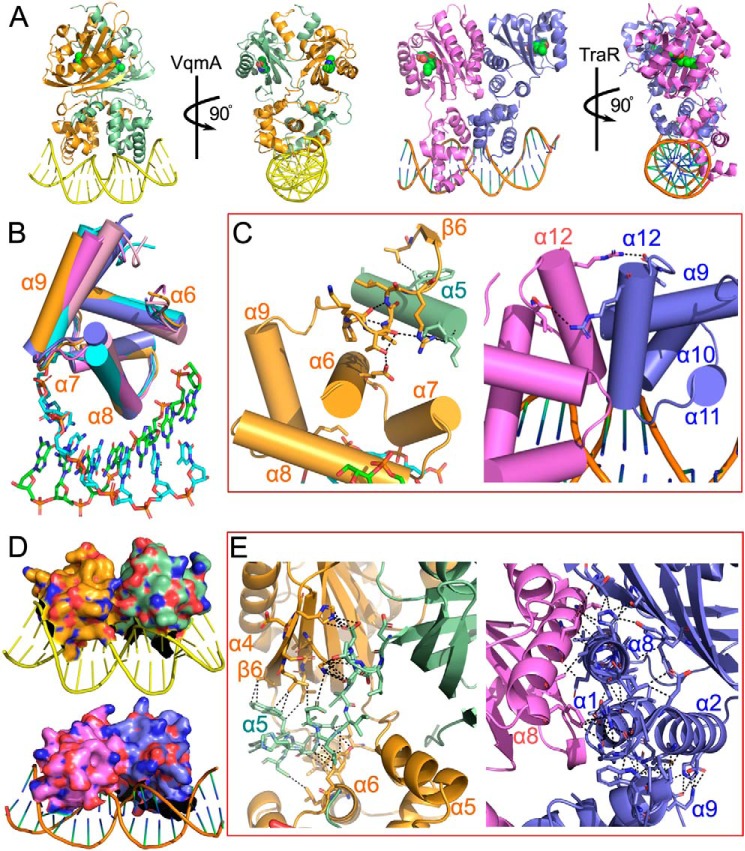
Figure 8.
Structural analysis of VqmA and TraR. A, structure comparison of the VqmA ternary complex and TraR ternary complex. B, structure comparison of the DBD of VqmA (orange) and other DBD domains of LuxR family proteins (TraR (PDB ID 1L3L), violet; SdiA (PDB ID 4LGW), cyan; QscR (PDB ID 6CC0), pink; CviR (PDB ID 3QP5), blue). C, the necessary conditions for the stability of the last α-helix conformation in the DBD of VqmA and TraR. The interactions of the α9–β6 loop (orange) with α6 (orange) and α5 (pale green) are shown in black in VqmA. The interactions of the α12 (violet) with another α12 (blue) are shown in black in TraR. D, the comparison of dimer interfaces of DBD of VqmA and TraR. E, the conduction layers between DBD and PAS domain of VqmA and TraR. The interactions of α5 (pale green) with α6 (orange) and β-sheet (orange) are shown in black in VqmA. The interactions of α1, α2, α8 (blue) with α9 (blue) and β-sheet (blue) are shown in black in TraR.
As with most HLH-superfamily proteins, VqmA shows high structural conservation with many transcription factors in its DNA-binding domain (17–20), but there are a lot of differences between the key residues associated with DNA binding (Fig. 3). As shown by sequence alignment, some of the key residues (Arg-159, Tyr-186, and Lys-203) of VqmA are conservative in the residues interacting with the DNA backbone. However, although all the residues providing specific recognition (Lys-185 and Glu-188) are charged residues like other DBD, there are obvious differences in electronegativity (Fig. 3A). These differences may be due to their different functions and the specificity of base recognition.
By comparison with other DBD of characterized LuxR proteins, all in the state of ligand binding, but only VqmA and TraR are DNA binding. The swing of the last α-helix conformation in the DBD is perhaps a good explanation (Fig. 8B). Therefore, we speculate that the conformational stability of β6 influences the stabilization of the DBD, especially the stabilization of α9 (Fig. 8C). In the structure of TraR complex (17, 35), we observed similar phenomena that the α12 of each subunit is stabilized by another α12 of the opposite subunit in the form of hydrogen bonds (Fig. 8C).
Compared with the TraR–DNA complex (17), as one of the few known DNA complexes in the LuxR family, the VqmA–DPO–DNA complex contains many similarities, but also has many differences. On the whole, both of them bind to DNA in the form of homologous dimers, and each monomer has a PAS domain and DBD, which are far apart (Fig. 8A). These two distant domains are connected by α-helices and flexible linkers, and the α-helices are charged with the task of interacting with another monomer. The difference is that the two domains of TraR have their own symmetrical axes, so the TraR dimer is asymmetrical, but the VqmA dimer is almost completely symmetrical. In addition, the connecting α-helices in TraR are still under the control of its own monomer, responsible for the stability of its own PAS domain and DBD. However, in VqmA, the connecting α-helix is more like a tango dancer's arm stretching out and embracing the waist of its partner (Fig. 8A). In the PAS domain, both have a typical β-sheet-fold. However, the binding specificity of ligands determines that their conformations around the binding pockets are quite different (Fig. S1). In the DBDs, both of them bind the major groove of DNA in the form of dimer with its third α-helices. However, the dimer interface area of the DBD of VqmA is only 175 Å2, which is obviously smaller than that of TraR (550 Å2) (Fig. 8D). This implies that the dimer conformation of DBD of VqmA is not mainly stabilized by the interaction between two DBD, but by the PAS domains or the connecting α-helices. Obviously, PAS domains have no direct control over DBD, so the connecting α-helices have become the key to stabilize DBD dimer conformation (Fig. 8A). This conjecture is also confirmed in the detailed diagram of the connecting α-helices (Fig. 8E). In the TraR–DNA complex, the connecting α-helices have some interaction with the DBD, but they can only stabilize their own DBD and cannot control their partner's DBD (Fig. 8E). In VqmA, the connecting α-helix is located between the PAS domain and DBD of its partner, and interacts closely with them, thus controlling the DBD of its partner, which has a positive contribution to the formation of DBD dimer. Therefore, the connecting α-helix in VqmA plays a more direct role in the dimerization of its DBD, so the stability of the α-helix directly affects the effectiveness of DNA binding to VqmA. In addition, according to the previous speculation of DPO binding stable the connecting α-helix conformation, it is revealed that DPO can fix the dimer structure of the DBD by stabilizing the connecting α-helix conformation, thus effectively regulating the transcription of targeted DNA.
Our structural model displays that VqmA binds with DNA effectively in the form of dimer. The results of biochemical studies suggest that DPO may result in more obvious conformational changes and presumably activates transcription. Certainly, this conjecture needs further verification. Thereafter, we will further explore the effect of DPO on the arrangements of VqmA, so as to explain more clearly the mechanism between VqmA and DNA.
On the whole, this is the first molecular-level view of the VqmA dimer bound to DPO and DNA. The structure of the VqmA–DPO–DNA complex provides the structural basis for the mechanism of transcriptional regulation by VqmA and offers a direction for the design of novel compounds to manage V. cholerae infections.
Experimental procedures
Construction of VqmA expression vectors
The full-length VqmA gene (residues 1–246) was synthesized based on GenBankTM entry NC_002506 with optimized codons. The VqmA-pET22b expression vector was constructed with C-terminal His tag using NdeI/XhoI sites. For overexpression, the VqmA-pET22b expression vector was transformed into Escherichia coli strain BL21(DE3). Y36F, F67A, F67I, Q70A, F99A, F99I, K101I, and S229A mutants were prepared using the QuikChange protocol. The sequences of these mutants were verified by DNA sequencing.
Expression and purification of VqmA protein
E. coli BL21(DE3) cells carrying the VqmA expression vector were grown in LB medium at 37 °C, and VqmA overexpression was induced by adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.4 mm when A600 reached 0.8. The cells were further grown for ∼20 h at 18 °C and harvested by centrifugation. The bacterial pellet was re-suspended by adding lysis buffer A (25 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 5% glycerol) and then lysed by passing through a microfluidizer (1,250–1,500 bar; 1 bar = 100 kilopascal) twice. The supernatant containing the soluble VqmA protein was incubated in a 5-ml nickel-nitrilotriacetic acid column (GE Healthcare) that had been equilibrated with lysis buffer A for 1 h at 4 °C. The column was washed with buffer B (25 mm Tris-HCl, pH 7.5, 200 mm NaCl, 25 mm imidazole, 5% glycerol) and buffer C (25 mm Tris-HCl, pH 7.5, 1 m NaCl, 5% glycerol). The VqmA protein was eluted with a solution of 300 mm imidazole, 25 mm Tris, pH 7.5, and 200 mm NaCl. Finally, VqmA was loaded into a Superdex 200 16/60 column (GE Healthcare), which was equilibrated with buffer D (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 5% glycerol). The resulting protein peak was concentrated to 30 mg/ml using a 30-kDa MWCO Amicon Ultra. Aliquots were snap-frozen in liquid nitrogen and stored at −80 °C until they were used for crystallization. Selenomethionine-labeled VqmA (SeMet-VqmA) protein was expressed in E. coli BL21(DE3) cells using the M9 medium protocol. SeMet–VqmA and VqmA mutations were purified using same protocol as above.
Crystallization and structure determination of VqmA
VqmA complex was crystallized at 18 °C using the co-crystallization method and hanging drop vapor diffusion method. The native VqmA complex was crystallized under the condition of 3.85% polyethylene glycol 3350 and 0.165 m K2HPO4/NaH2PO4, pH 7.5. SeMet–VqmA complex crystals were produced in a similar condition to the native crystals. VqmA was co-crystallized with DPO molecule and an 18-bp dsDNA (5′-AGGGGGGATTTCCCCCCT-3′), containing the vqmR operator sequence, in a drop comprising 1 μl of 14 mg/ml of VqmA–dsDNA complex and 0.5 μl of 3.85% polyethylene glycol 3350 and 0.165 m K2HPO4/NaH2PO4, pH 7.5. The crystals were flash-cooled at 100 K in the presence of 20% glycerol. X-ray diffraction data were collected using a Eiger X 16M on beamline BL17U1 at the Shanghai Synchrotron Radiation Facility (SSRF), Shanghai, People's Republic of China (38). The datasets were processed using the Xia2 software suite (39), and Aimless and Pointless in the CCP4 program package (39). The SeMet–VqmA complex structure was determined by single-wavelength anomalous diffraction phasing on the X-ray diffraction data collected at the peak wavelength (0.97915 Å) of selenium. The initial phases were calculated using CRANK2 (40). The solvent-flattened electron density map could be partly autotraced by Phenix (41), and the atomic model was fit with the program Coot (42). The stereochemical quality of the final model was assessed with MolProbity (43). Crystallographic statistics for the final model were show in Table S1. Figures were prepared with PyMOL (44).
Isothermal titration calorimetry
ITC experiments were performed using a MicroCal PEAQ-ITC instrument (Malvern). A total of 500 μm DPO in a syringe was titrated into the sample cell containing 50 μm VqmA protein at 25 °C with stirring at 750 rpm. A total of 0.4 and 2 μl of DPO were injected for the initial injection and the other injections, respectively. Before the ITC experiments, the VqmA protein was dialyzed against reaction buffer (25 mm Tris, pH 7.5, and 100 mm NaCl) for 12 h at 4 °C and DPO was dissolved in the reaction buffer used for dialysis. VqmA mutations used the same method as mentioned above for the WT protein. Data fitting and evaluation were performed with the PEAQ-ITC Analysis software (MicroCal) using the one-site binding model (45).
CD spectroscopy
CD spectra of WT and DPO-binding VqmA (30 μm in 10 mm NaCl, pH 7.5) were measured with a CD spectrometer (Chirascan, Applied Photophysics Ltd.) using a 0.1-cm path length quartz cuvette. Temperature-dependent CD experiments were analyzed at 180–260 nm from 20 to 90 °C at intervals of 2 °C to determine melting temperature (Tm).
Electrophoretic mobility shift assay
An EMSA was performed using the VqmA protein and operator dsDNA in the presence or absence of DPO to monitor the VqmA–dsDNA interaction and the activation effect of DPO on this interaction. For the interaction assay, the purified VqmA protein (0–10 μm) was incubated with 4 μm dsDNA (5′-AGGGGGGATTTCCCCCCT-3′) labeled with 5′-end FAM in 25 mm Tris, pH 7.5, and 100 mm NaCl. The samples were electrophoretically separated on 8–16% gradient polyacrylamide gels in Tris/boric acid/EDTA (TBE) buffer for 150 min at 120 V. For the DPO-induced activation assay, a mixture of 4 μm VqmA dimer protein and 4 μm dsDNA (5′-AGGGGGGATTTCCCCCCT-3′) was incubated with 0–10 μm DPO. The samples were electrophoretically separated on 8–16% gradient polyacrylamide gels in TBE buffer.
Gel-filtration chromatography
Gel-filtration chromatography was performed using a Superdex 200 Increase 10/300 GL column (GE Healthcare) in 25 mm Tris, pH 7.5, and 100 mm NaCl for the analysis of the interaction of VqmA with the operator DNA. To determine the appropriate length of targeted DNA, VqmA was incubated with some target DNA of different lengths, and the VqmA–DNA complex was applied to the column. The elution profiles of the samples were monitored by UV absorption at 280 nm.
Accession number
The atomic coordinates and structure factors for VqmA complex have been deposited in the Protein Data Bank with the code number 6IDE.
Author contributions
H. W., M. L., H. G., H. Z., B. L., Q. X., C. X., and F. Y. software; H. W., H. G., and H. Z. formal analysis; H. W., M. L., H. G., H. Z., B. L., Q. X., and C. X. validation; H. W., M. L., H. G., B. L., and Q. X. investigation; H. W., B. L., Q. X., and C. X. visualization; H. W., F. Y., and J. H. writing-original draft; M. L., H. Z., and C. X. methodology; F. Y. and J. H. conceptualization; F. Y. and J. H. resources; F. Y. funding acquisition; F. Y. and J. H. writing-review and editing; J. H. project administration.
Supplementary Material
Acknowledgments
We thank the staff at the Laboratory of Physical Biology, Shanghai Institute of Applied Physics, for technical help with the GBOX-Chemi-XL1.4 Imaging System. We also thank Xinmiao Lu and Zeyuan Guan for discussions.
This work was supported by National Natural Science Foundation Grants 31570740 and 81330076. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Tables S1–S4.
The atomic coordinates and structure factors (code 6IDE) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- QS
- quorum sensing
- DBD
- DNA-binding domain
- DPO
- 3,5-dimethylpyrazin-2-ol
- ITC
- isothermal titration calorimetry
- PAS
- Per-Arnt-Sim domain
- SeMet
- selenomethionine
- HTH
- helix turn helix
- EMSA
- electrophoretic mobility shift assay.
References
- 1. Bassler B. L., and Losick R. (2006) Bacterially speaking. Cell 125, 237–246 10.1016/j.cell.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 2. Fuqua W. C., Winans S. C., and Greenberg E. P. (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 10.1128/jb.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waters C. M., and Bassler B. L. (2005) Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- 4. Bassler B. L., Wright M., and Silverman M. R. (1994) Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13, 273–286 10.1111/j.1365-2958.1994.tb00422.x [DOI] [PubMed] [Google Scholar]
- 5. Miller M. B., Skorupski K., Lenz D. H., Taylor R. K., and Bassler B. L. (2002) Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314 10.1016/S0092-8674(02)00829-2 [DOI] [PubMed] [Google Scholar]
- 6. Papenfort K., Silpe J. E., Schramma K. R., Cong J.-P., Seyedsayamdost M. R., and Bassler B. L. (2017) A Vibrio cholerae autoinducer–receptor pair that controls biofilm formation. Nat. Chem. Biol. 13, 551–557 10.1038/nchembio.2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Z., Hsiao A., Joelsson A., and Zhu J. (2006) The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J. Bacteriol. 188, 2446–2453 10.1128/JB.188.7.2446-2453.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papenfort K., Förstner K. U., Cong J.-P., Sharma C. M., and Bassler B. L. (2015) Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 112, E766–E775 10.1073/pnas.1500203112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuqua C., Parsek M. R., and Greenberg E. P. (2001) Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468 10.1146/annurev.genet.35.102401.090913 [DOI] [PubMed] [Google Scholar]
- 10. Brennan R. G., and Matthews B. W. (1989) The helix-turn-helix DNA binding motif. J. Biol. Chem. 264, 1903–1906 [PubMed] [Google Scholar]
- 11. Fuqua C., Winans S. C., and Greenberg E. P. (1996) Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751 10.1146/annurev.micro.50.1.727 [DOI] [PubMed] [Google Scholar]
- 12. Silpe J. E., and Bassler B. L. (2019) A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 176, 1–13 10.1016/j.cell.2018.1010.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qi S., Pang Y., Hu Q., Liu Q., Li H., Zhou Y., He T., Liang Q., Liu Y., Yuan X., Luo G., Li H., Wang J., Yan N., and Shi Y. (2010) Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141, 446–457 10.1016/j.cell.2010.03.017 [DOI] [PubMed] [Google Scholar]
- 14. Benezera R., Davis R. L., Lockshon D., Turner T., and Weintraub H. (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Ann. N.Y. Acad. Sci. 61, 49–59 [DOI] [PubMed] [Google Scholar]
- 15. Xu C., and Rosen B. P. (1997) Dimerization is essential for DNA binding and repression by the ArsR metalloregulatory protein of Escherichia coli. J. Biol. Chem. 272, 15734–15738 10.1074/jbc.272.25.15734 [DOI] [PubMed] [Google Scholar]
- 16. Luscombe N. M., Austin S. E., Berman H. M., and Thornton J. M. (2000) An overview of the structures of protein-DNA complexes. Genome Biol. 1, reviews001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang R. G., Pappas K. M., Pappas T., Brace J. L., Miller P. C., Oulmassov T., Molyneaux J. M., Anderson J. C., Bashkin J. K., Winans S. C., and Joachimiak A. (2002) Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417, 971–974 10.1038/nature00833 [DOI] [PubMed] [Google Scholar]
- 18. Kim T., Duong T., Wu Ca. A., Choi J., Lan N., Kang S. W., Lokanath N. K., Shin D., Hwang H. Y., and Kim K. K. (2014) Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor. Acta Crystallogr. 70, 694–707 [DOI] [PubMed] [Google Scholar]
- 19. Chen G., Swem L. R., Swem D. L., Stauff D. L., O'Loughlin C. T., Jeffrey P. D., Bassler B. L., and Hughson F. M. (2011) A strategy for antagonizing quorum sensing. Mol. Cell 42, 199–209 10.1016/j.molcel.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wysoczynski-Horita C. L., Boursier M. E., Hill R., Hansen K., Blackwell H. E., and Churchill M. E. A. (2018) Mechanism of agonism and antagonism of the Pseudomonas aeruginosa quorum sensing regulator QscR with non-native ligands. Mol. Microbiol. 108, 240–257 10.1111/mmi.13930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koh S., Hwang J., Guchhait K., Lee E. G., Kim S. Y., Kim S., Lee S., Chung J. M., Jung H. S., and Lee S. J. (2016) Molecular insights into toluene sensing in the TodS/TodT signal transduction system. J. Biol. Chem. 291, 8575–8590 10.1074/jbc.M116.718841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gong W., Hao B., and Chan M. K. (2000) New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry 39, 3955–3962 10.1021/bi992346w [DOI] [PubMed] [Google Scholar]
- 24. Brudler R., Meyer T. E., Genick U. K., Devanathan S., Woo T. T., Millar D. P., Gerwert K., Cusanovich M. A., Tollin G., and Getzoff E. D. (2000) Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein. Biochemistry 39, 13478–13486 10.1021/bi0009946 [DOI] [PubMed] [Google Scholar]
- 25. Yamada S., Sugimoto H., Kobayashi M., Ohno A., Nakamura H., and Shiro Y. (2009) Structure of PAS-linked histidine kinase and the response regulator complex. Structure 17, 1333–1344 10.1016/j.str.2009.07.016 [DOI] [PubMed] [Google Scholar]
- 26. Hurley A., and Bassler B. L. (2017) Asymmetric regulation of quorum-sensing receptors drives autoinducer-specific gene expression programs in Vibrio cholerae. PLOS Genetics 13, e1006826 10.1371/journal.pgen.1006826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hammer B. K., and Bassler B. L. (2007) Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 104, 11145–11149 10.1073/pnas.0703860104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jung S. A., Chapman C. A., and Ng W.-L. (2015) Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLOS Pathogens 11, e1004837 10.1371/journal.ppat.1004837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waters C. M., Lu W., Rabinowitz J. D., and Bassler B. L. (2008) Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190, 2527–2536 10.1128/JB.01756-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Antonova E. S., and Hammer B. K. (2011) Quorum-sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae. FEMS Microbiol. Lett. 322, 68–76 10.1111/j.1574-6968.2011.02328.x [DOI] [PubMed] [Google Scholar]
- 31. Bardill J. P., Zhao X., and Hammer B. K. (2011) The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol. 80, 1381–1394 10.1111/j.1365-2958.2011.07655.x [DOI] [PubMed] [Google Scholar]
- 32. Tian H., McKnight S. L., and Russell D. W. (1997) Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11, 72–82 10.1101/gad.11.1.72 [DOI] [PubMed] [Google Scholar]
- 33. Morais Cabral J. H., Lee A., Cohen S. L., Chait B. T., Li M., and Mackinnon R. (1998) Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell 95, 649–655 10.1016/S0092-8674(00)81635-9 [DOI] [PubMed] [Google Scholar]
- 34. Kim B. S., Jang S. Y., Bang Y. J., Hwang J., Koo Y., Jang K. K., Lim D., Kim M. H., and Choi S. H. (2018) QStatin, a selective inhibitor of quorum sensing in Vibrio species. mBio 9, e02262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vannini A., Volpari C., Gargioli C., Muraglia E., Cortese R., De Francesco R., Neddermann P., and Marco S. D. (2002) The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21, 4393–4401 10.1093/emboj/cdf459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao Y., Martinez-Yamout M. A., Dickerson T. J., Brogan A. P., Wright P. E., and Dyson H. J. (2006) Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl-homoserine lactones. J. Mol. Biol. 355, 262–273 10.1016/j.jmb.2005.10.041 [DOI] [PubMed] [Google Scholar]
- 37. Gong W., Hao B., Mansy S. S., Gonzalez G., Gilles-Gonzalez M. A., and Chan M. K. (1998) Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. U.S.A. 95, 15177 10.1073/pnas.95.26.15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q.-S., Zhang K.-H., Cui Y., Wang Z.-J., Pan Q.-Y., Liu K., Sun B., Zhou H., Li M.-J., Xu Q., Xu C.-Y., Yu F., and He J.-H. (2018) Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nuclear Sci. Tech. 29, 68 10.1007/s41365-018-0398-9 [DOI] [Google Scholar]
- 39. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ness S. R., de Graaff R. A., Abrahams J. P., and Pannu N. S. (2004) Crank: new methods for automated macromolecular crystal structure solution. Structure 12, 1753–1761 10.1016/j.str.2004.07.018 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Schroedinger, LLC, New York [Google Scholar]
- 45. Wiseman T., Williston S., Brandts J. F., and Lin L.-N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 10.1016/0003-2697(89)90213-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.