Abstract
Zebrafish gata4/5/6 genes encode transcription factors that lie on the apex of the regulatory hierarchy in primitive myelopoiesis. However, little is known about the roles of microRNAs in gata4/5/6-regulated processes. Performing RNA-Seq deep sequencing analysis of the expression changes of microRNAs in gata4/5/6-knockdown embryos, we identified miR-210-5p as a regulator of zebrafish primitive myelopoiesis. Knocking down gata4/5/6 (generating gata5/6 morphants) significantly increased miR-210-5p expression, whereas gata4/5/6 overexpression greatly reduced its expression. Consistent with inhibited primitive myelopoiesis in the gata5/6 morphants, miR-210-5p overexpression repressed primitive myelopoiesis, indicated by reduced numbers of granulocytes and macrophages. Moreover, knocking out miR-210 partially rescued the defective primitive myelopoiesis in zebrafish gata4/5/6-knockdown embryos. Furthermore, we show that the restrictive role of miR-210-5p in zebrafish primitive myelopoiesis is due to impaired differentiation of hemangioblast into myeloid progenitor cells. By comparing the set of genes with reduced expression levels in the gata5/6 morphants to the predicted target genes of miR-210-5p, we found that foxj1b and slc3a2a, encoding a forkhead box transcription factor and a solute carrier family 3 protein, respectively, are two direct downstream targets of miR-210-5p that mediate its inhibitory roles in zebrafish primitive myelopoiesis. In summary, our results reveal that miR-210-5p has an important role in the genetic network controlling zebrafish primitive myelopoiesis.
Keywords: zebrafish, microRNA (miRNA), microRNA mechanism, post-transcriptional regulation, transcription factor, foxj1b, gata4/5/6, miR-210-5p, primitive myelopoiesis, slc3a2a
Introduction
Myelopoiesis is a tightly regulated developmental process in which mature myeloid cells are generated. In vertebrates, myeloid cells arise from two successive waves during development: a primitive wave producing a limited number of myeloid cells and a definitive wave giving rise to all hematopoietic lineages including myeloid cells throughout lifetime. Zebrafish, unlike mammals, have a robust primitive myeloid pathway, which makes this model animal a powerful tool to study vertebrate myelopoiesis as well as human myeloid malignancies. In zebrafish, the primitive wave arising from both anterior lateral plate mesoderm (ALPM)4 and posterior lateral plate mesoderm (PLPM), while a definitive wave occurs in the ventral wall of the dorsal aorta, which is analogous to the aorta-gonads-mesonephros region in mammals (1). Many studies have revealed the necessary role of a number of coding genes in primitive myelopoiesis, including spi1b, irf8, gfi1ab, tal1, and etv2 (2–5). However, the roles of noncoding genes in zebrafish primitive myelopoiesis are still poorly understood.
Recent discoveries have implicated that microRNAs (miRNAs), a group of noncoding post-transcriptionally regulatory RNAs (∼22 nt), are widely involved in murine hematopoiesis and zebrafish myelopoiesis (6–8). In murine myeloid progenitors, knocking out Dicer1, an essential miRNA processing RNase III enzyme, leads to abnormal myelopoiesis and depleted macrophages (9). Several miRNAs, such as miR-223, miR-146a, miR-21, and miR-196b have been intensively studied and shown to play important roles in murine and zebrafish definitive myelopoiesis (7, 10–12). These findings suggest an important role of miRNA during myelopoiesis in zebrafish, but essential players involved in primitive myelopoiesis are yet to be discovered.
Previously, others' and our works have proved that the transcriptional factors Gata4/5/6 lie on the apex of the regulatory hierarchy in zebrafish primitive myelopoiesis (13, 14). But whether there are miRNAs mediating the roles of Gata4/5/6 in the primitive myelopoiesis remains unknown. In the present work, we aim to study the miRNAs involved in the regulatory network of zebrafish primitive myelopoiesis. We searched for gata4/5/6-regulated miRNAs by comparing their expression changes of gata4/5/6-knockdown embryos to those of control embryos. Our results revealed that miR-210-5p functions downstream of gata4/5/6 and excess miR-210-5p plays an inhibitory role in zebrafish primitive myelopoiesis. In addition, the restrictive role of miR-210-5p was shown to be achieved by affecting the differentiation of hemangioblast into myeloid progenitor cells. Furthermore, we demonstrated that foxj1b and slc3a2a are two direct target genes of miRNA-210-5p to mediate its inhibitory roles in zebrafish primitive myelopoiesis.
Results
miR-210-5p works downstream of gata4, gata5, and gata6 to inhibit zebrafish primitive myelopoiesis
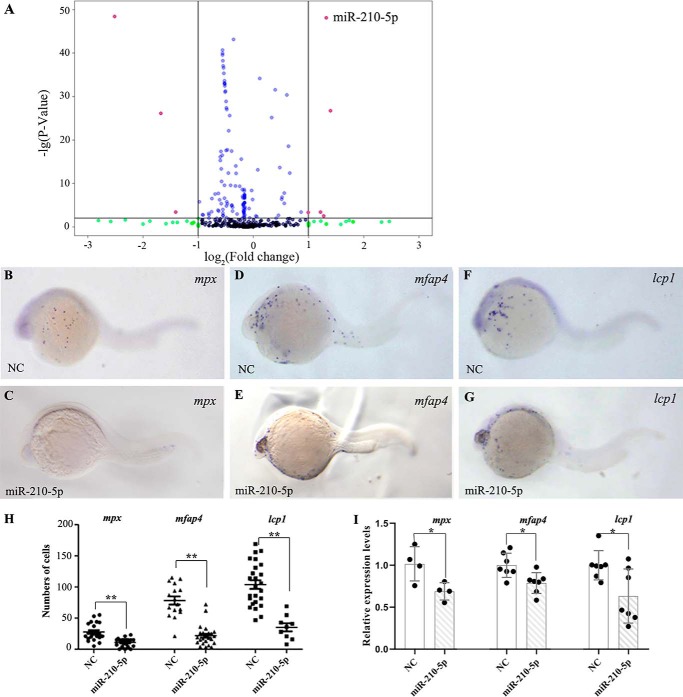
Previous works showed that gata4/5/6 lie at the hierarchical apex of the regulatory network of the hemangioblast formation and primitive myelopoiesis in zebrafish (13, 14). To explore the roles of miRNAs in zebrafish primitive myelopoiesis, we employed deep sequencing to analyze the difference of miRNA expression profiles between gata4/5/6-knockdown embryos (gata5/6 morphants) and control embryos. Because it is reported that morpholinos knocking down gata5 and gata6 also lead to the loss of gata4 expression in zebrafish embryos (15), we used a combination microinjection of gata5 and gata6 morpholinos for knocking down all the three genes. With the Illumina Solexa platform, the deep sequencing for miRNAs generated 12.3 million reads from control embryos and 13.1 million reads from gata5/6 morphants. Among them, about 7.16 and 6.83 million unique reads were annotated, respectively. We analyzed the expressions of 223 miRNAs and found the expression levels of 8 miRNAs exhibited a significant difference (|logFC| > 1; p < 0.01) in gata4/5/6-knockdown embryos compared with controls (Fig. 1A). Among them, 5 were up-regulated and 3 were down-regulated, which was confirmed by quantitative RT-PCR (qRT-PCR) (data not shown). To test the function of these miRNAs during primitive myelopoiesis, we used miRNA mimics or inhibitors, which were chemically synthesized dsRNA oligos and could be used to mimic or inhibit the function of mature endogenous miRNAs in vivo (16), to achieve overexpression or inhibition of the 8 candidate miRNAs in WT embryos. The results revealed that the microinjection of miR-210-5p mimic led to the significant (p < 0.01) inhibition of primitive myelopoiesis with reduced numbers of granulocytes (mpx+ cells), macrophages (mfap4+ cells), as well as pan-leukocyte (lcp1+ cells) as shown by whole mount in situ hybridization (Fig. 1, B–H) in the zebrafish embryos at 26 hpf. Consistently, microinjection of miR-210-5p mimic significantly (p < 0.05) reduced the expression levels of mpx, mfap4, and lcp1 in the embryos at 26 hpf as demonstrated by qRT-RCR (Fig. 1I). However, the formation of both granulocytes and macrophages were not reduced together in the embryos microinjected with mimics or inhibitors of the other 7 miRNAs including miR-129-5p, miR-722, miR-7b, miR-30a, miR-125c, miR-738, and miR-153a, respectively (Fig. S1). These results suggest that miR-210-5p plays a restrictive role in zebrafish primitive myelopoiesis, and this miRNA was chosen for further study.
Figure 1.
miR-210-5p functions downstream of gata4/5/6 to regulate zebrafish primitive myelopoiesis. A, volcano plots showing the results from deep sequencing analysis for the expression changes of miRNAs in gata4/5/6-knockdown embryos. The miRNAs with significant expression changes (logFC>1 or LogFC<−1; p < 0.01) are marked in red dots. B–G, whole mount in situ hybridized embryos at 26 hpf showing the expression patterns of marker genes for primitive myelopoiesis including mpx (B and C), mfap4 (D and E), and lcp1 (F and G) in the embryos microinjected with NC (B, D, and F) and miR-210-5p mimic (C, E, and G), respectively. Embryos were viewed laterally, and positioned head left. The images in B, D, and F were reused in Fig. S1, A–C because these results were from one ISH experiment and shared the same controls. H, scatter plot showing the numbers of mpx, mfap4, and lcp1 expression cells (shown in y axis) in the embryos microinjected with NC or miR-210-5p mimic (shown in x axis), respectively. I, qRT-PCR results showing the relative expression levels of mpx, mfap4, and lcp1 (shown in y axis) in the 26 hpf embryos microinjected with NC or miR-210-5p mimic (shown in x axis), respectively. The gene expression levels in the NC group were all normalized as 1.0. The fold-changes of gene expression levels in the miR-210-5p mimic group were shown relative to the NC group, respectively. NC, mimic negative control; miR-210-5p, miR-210-5p mimic; **, p < 0.01; *, p < 0.05.
To confirm the restrictive role of miR-210-5p in zebrafish primitive myelopoiesis, we first tested whether microinjection of the miR-210-5p mimic into the zebrafish embryos did lead to the overexpression of the mature miR-210-5p. To do this, we performed whole mount in situ hybridization to examine the expression of miR-210-5p. The results showed that miR-210-5p was widely expressed in the embryos microinjected with mimic negative control (NC) at 14 hpf when primitive myeloid progenitors start to form (Fig. S2A) and the ones at 26 hpf when myeloid cells are formed (Fig. S2B), whereas the expression of miR-210-5p was significantly increased in the whole embryos at both 14 hpf (Fig. S2C) and 26 hpf (Fig. S2D) when the embryos were microinjected with the miR-210-5p mimic. To exclude the possibility that the increased expression of miR-210-5p was due to the denaturation of the double strand RNA (dsRNA) itself (miR-210-5p mimic) into the single strand RNA that is recognized by the LNA microRNA Detection Probes against miR-210-5p during whole mount in situ hybridization, we performed an in vitro assay by incubating the dsRNA with RNase A under the same conditions (without adding yeast tRNA) as the experiment of whole mount in situ hybridization, and the results showed that the dsRNA was still present after overnight incubation at 65 °C (Fig. S2E). The results support that the findings from the whole mount in situ hybridization represent the increase of mature miRNA-210-5p in the embryos microinjected with miRNA-210-5p mimic. To further confirm the results, we performed qRT-PCR examination to detect the mature miR-210-5p, and the results showed that the microinjection of miR-210-5p mimic caused significantly (p < 0.01) increased expression of mature miR-210-5p in the embryos at both 14 and 26 hpf (Fig. S2F). Taken together, our results verified that microinjection of miR-210-5p mimic did result in the increase of mature miR-210-5p in zebrafish embryos.
We next performed knocking-down gata4/5/6 by microinjecting gata5/6 MOs (14) or overexpressing gata4/5/6 by microinjecting gata4/6 mRNAs (13) into 1-cell stage zebrafish embryos and then examined the expression changes of miR-210-5p. Compared to its expression pattern in the embryos microinjected with control MO (Fig. 2, A and B), we found that expression of miR-210-5p was significantly increased in the gata5/6 morphants (Fig. 2, C and D) but greatly reduced in the gata4/5/6-overexpressed embryos (Fig. 2, E and F) at 14 and 26 hpf, respectively. Consistently, the results from qRT-PCR confirmed the significantly (p < 0.01; p < 0.05) up-regulated expression of miR-210-5p in the gata5/6 morphants compared with control embryos at both 14 and 26 hpf (Fig. 2G). The results were consistent with the deep sequencing data and supported that miR-210-5p functions downstream of gata4/5/6 to negatively regulate zebrafish primitive myelopoiesis.
Figure 2.
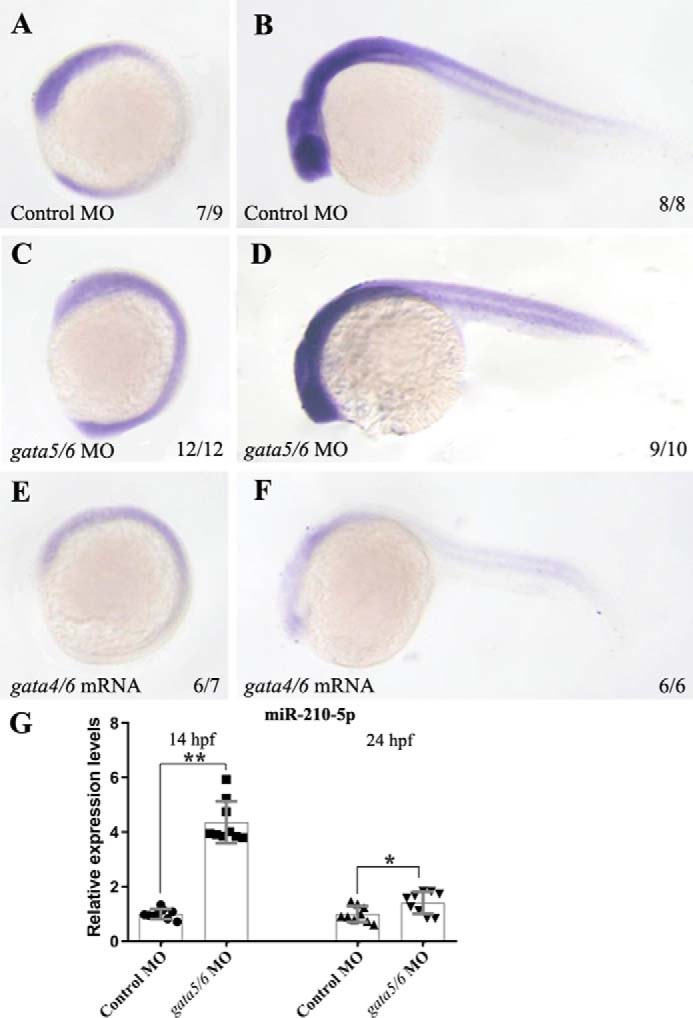
Gata4/5/6 inhibits the expression of miR-210-5p in zebrafish embryos at 14 and 26 hpf. WT embryos at 1-cell stage were microinjected with gata5/6 MOs (C and D), gata4/6 mRNAs (E and F), or control MO (A and B), respectively. When reaching 14 (A, C, and E) and 26 (B, D, and F) hpf, the microinjected embryos were fixed for in situ hybridization analysis, respectively. In the embryos microinjected with control MO, the miR-210-5p was found widely expressed at 14 hpf (A) or 26 hpf (B). The embryos microinjected with gata5/6 MOs displayed significant increased expression of miR-210-5p (C and D), whereas the ones microinjected with gata4/6 mRNAs exhibited significantly reduced expression of miR-210-5p (E and F) at 14 and 26 hpf, respectively. Embryos were positioned lateral and head left. G, results from qRT-PCR showing that the expression of mature miR-210-5p (shown in y axis) was significantly increased in gata5/6 morphants compared with the embryos microinjected with control MO (shown in y axis). The expression level of miR-210-5p in the control MO group was normalized as 1.0 and the fold-change of miR-210-5p expression level in gata5/6 morphant group was shown relative to the control MO group, respectively. **, p < 0.01; *, p < 0.05.
Knocking out miR-210 partially rescue the defective primitive myelopoiesis occurring in zebrafish gata4/5/6-knockdown embryos
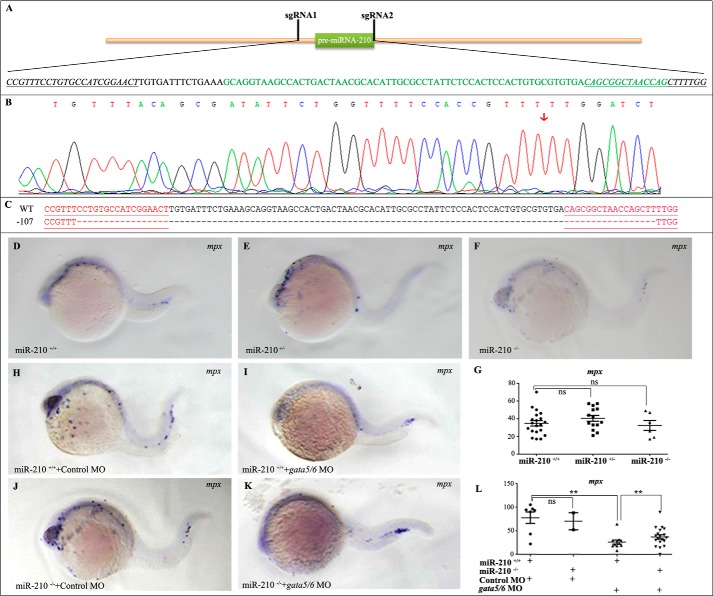
To gain further insight into the regulatory mechanisms of miR-210-5p during zebrafish primitive myelopoiesis, we generated miR-210 knockout zebrafish using CRISPR/Cas9. Microinjecting a pair of sgRNAs and Cas9 mRNA into zebrafish embryos at 1-cell stage, we obtained zebrafish mutants carrying a mutated allele of miR-210 (miR-210nju49/+). The mutated allele contains a deletion of the 107-bp genomic sequence encoding pre-miR-210 (Fig. 3, A–C). Analyses on the homozygous mutant of miR-210 (miR-210nju49/nju49) revealed that the mutant embryos developed normally with no significant differences (p > 0.05) in expressions of the granulocyte marker gene mpx compared with WT embryos at 26 hpf (Fig. 3, D–G). Furthermore, the homozygous mutant zebrafish grew normally to adult with fertility. This result indicated that lack of miR-210 has no effect on zebrafish primitive myelopoiesis although excess miR-210-5p inhibits it.
Figure 3.
Knocking out miR-210 partly rescues the primitive myelopoiesis in zebrafish embryos. A, schematic showing the recognition sites of two sgRNAs in the genomic sequence of pre-miRNA-210. sgRNA1 and sgRNA2 are designed to recognize the 5′ upstream and 3′ downstream of the miR-210 sequence (shown in green), respectively. sgRNA-recognition sites are underlined and italic. B, Sanger sequencing results showing an edit (mutated allele with a 107-bp deletion) was generated. Red arrow points to the location of deletion. C, DNA sequences showing the mutated allele (nju49) with 107 bp deletion. Sequences underlined in red show the recognition sites of two sgRNAs, respectively. D–G, knocking out miR-210 did not affect the primitive myelopoiesis in zebrafish embryos. Whole mount in situ hybridized embryos showing that there was no significant difference in the number of mpx+ cells between WT (D), heterozygous (E), and homozygous (F) embryos at 26 hpf. G, scatter plot showing the number of mpx+ cells counted from the embryos typically shown in D–F. H–K, whole mount in situ hybridized embryos showing microinjection of control MO did not affect the formation of mpx+ cells in miR-210 knockout embryos compared with their WT siblings (H and J), whereas knocking down gata4/5/6 by microinjecting gata5/6 MOs partially rescued the formation of mpx+ cells in the miR-210-5p knockout embryos (I and K). L, scatter plot showing the number of mpx+ cells counted from the embryos typically shown in H–K. Embryos are positioned laterally and head left. **, p < 0.01; ns, no significance (p > 0.05).
To confirm that excess miR-210-5p plays a restrictive role in primitive myelopoiesis by working downstream of gata4/5/6, we knocked down gata4/5/6 in miR-210 mutant embryos by microinjecting gata5/6 MOs into the fertilized eggs derived from inbreeding of miR-210nju49/+ zebrafish. Performing whole mount in situ hybridization or qRT-PCR analyses on the morphants, we found that both the number of mpx+ cells and the expression levels of mpx decreased dramatically (p < 0.01) in WT embryos microinjected with gata5/6 MOs compared with those with control MO (Fig. 3, H, I, and L), but these reduced mpx+ cells were partially (p < 0.01) rescued in miR-210−/− embryos (Fig. 3, I, K, and L), whereas the number of mpx+ cells and the expression levels of mpx had no change (p > 0.05) between miR-210+/+ and miR-210−/− embryos microinjected with control MO (Fig. 3, H, J, and L). The results demonstrate that miR-210-5p regulates primitive myelopoiesis by acting downstream of gata4/5/6, and the incomplete rescue effect suggests that other factors besides miR-210-5p are involved in mediating the roles of gata4/5/6 in regulating the formation of myeloid cells in zebrafish embryos.
miR-210-5p blocks the differentiation of hemangioblasts into myeloid progenitors
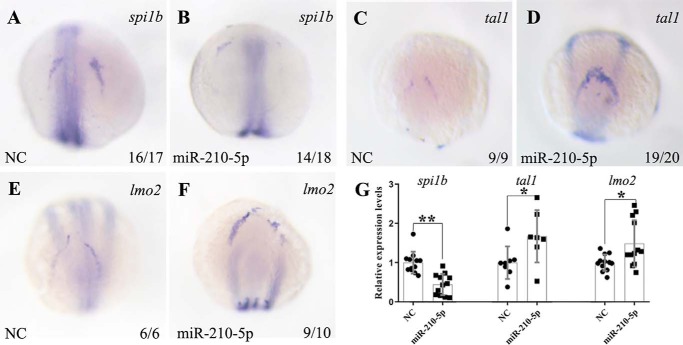
Zebrafish primitive myelopoiesis is initiated in the rostral end of ALPM to form anterior hemangioblasts marked by expressing the master regulators of tal1 and lmo2 between 3- and 5-somite stages (1, 5, 17). With development, a subset of the tal1+ precursors acquires myeloid cell fates by expressing myeloid-specific transcription factor spi1b (18, 19). Now that overexpression of miR-210-5p led to inhibition of the primitive myelopoiesis, we therefore asked whether miR-210-5p plays a restrictive role in the formation of the myeloid precursors. To answer this question, we performed whole mount in situ hybridization to examine the expressions of key transcription factors including tal1, lmo2, and spi1b at 14 hpf when the myeloid precursors are differentiated from anterior hemangioblasts. The results from whole mount in situ hybridization showed that the expressions of spi1b were significantly restricted and reduced (Fig. 4, A and B), but the expressions of hemangioblast master regulatory genes including tal1 and lmo2 were expanded and increased in the rostral end of ALPM in the miR-210-5p overexpressing embryos at 14 hpf (Fig. 4, C–F). Consistently, qRT-PCR results confirmed the significant down-regulation of spi1b (p < 0.01) and the increased expressions of tal1 and lmo2 (p < 0.05) in the miR-210-5p overexpressing embryos at 14 hpf (Fig. 4G). These results suggest that miR-210-5p plays a restrictive role in the differentiation of the hemangioblast cells into myeloid precursors in the rostral end of ALPM in the zebrafish embryos at 14 hpf.
Figure 4.
Overexpression of miR-210-5p inhibits the differentiation of anterior hemangioblast to the myeloid progenitors. A–F, whole mount in situ hybridization results showing the expressions of spi1b, tal1, and lmo2 in the 14 hpf embryos microinjected with miR-210-5p mimic or NC. The embryos are positioned dorsally and anterior top. Compared with the embryos microinjected with NC (A), overexpression of miR-210-5p inhibited the expression of spi1b (B), whereas the expressions of tal1 and lmo2 were increased in the ALPM of miR-210-5p overexpressed embryos (D and F) compared with control embryos at 14 hpf (C and E). G, qRT-PCR results showing the reduced expressions of spi1b and increased expressions of tal1 and lmo2 in the miR-210-5p overexpressed embryos at 14 hpf. The gene expression levels in NC group was normalized as 1.0 and the fold-change of gene expression level in the embryos microinjected with miR-210-5p mimic was shown relative to the NC group (shown in y axis). NC, mimic negative control; miR-210-5p, miR-210-5p mimic; *, p < 0.05; **, p < 0.01.
foxj1b and slc3a2a work as target genes of miR-210-5p to regulate the primitive myelopoiesis
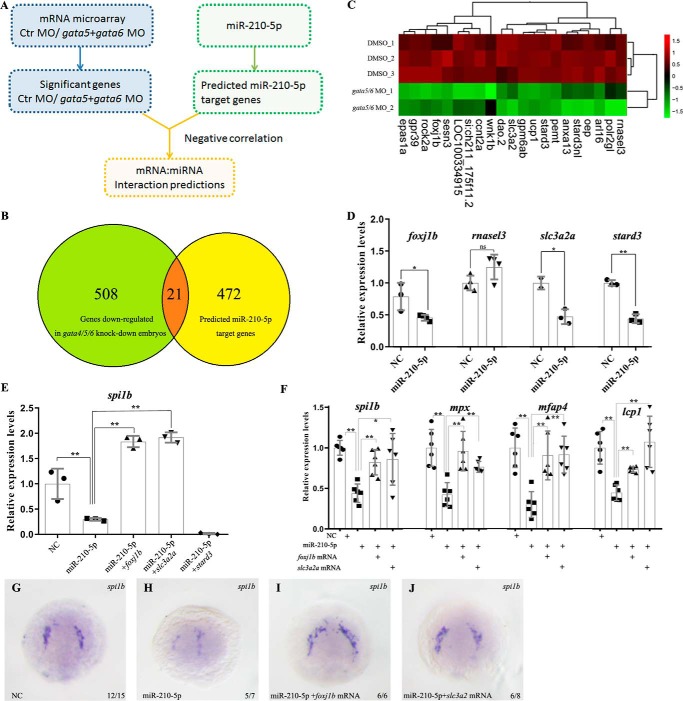
To uncover the target genes of miR-210-5p to mediate its role in inhibiting zebrafish primitive myelopoiesis, we performed microarray analysis to identify differentially expressed genes in gata4/5/6-knockdown embryos (Fig. 5A). The downstream genes of gata4/5/6 could potentially be regulated by miR-210-5p. By comparing the set of genes with reduced expression levels in gata5/6 morphants to the predicted miR-210-5p target genes, which were obtained from miRanda database (http://www.microrna.org),5 we found that 21 putative miR-210-5p downstream genes were down-regulated in gata4/5/6-knockdown embryos (Fig. 5, B and C). Among the candidate downstream targets, 4 genes including foxj1b, rnasel3, slc3a2a, and stard3, known to be involved in hematopoiesis or mesodermal development (20–23), were selected for further study. Performing qRT-RCR analyses on the embryos microinjected with the mimic of miR-210-5p, we found that the expressions of foxj1b, slc3a2a, and stard3 were significantly (p < 0.05 or p < 0.01) reduced but the expression of rnasel3 was not significantly changed (p > 0.05) in the embryos at 14 hpf (Fig. 5D). We therefore overexpressed foxj1b, slc3a2a, and stard3 (3′ UTRs were all deleted from their mRNAs) and examined whether the expression of spi1b in miR-210-5p overexpressing embryos can be rescued at 14 hpf. The qRT-PCR results showed that overexpression of foxj1b or slc3a2a, but not stard3, significantly (p < 0.01) rescued the suppressed expression of spi1b (Fig. 5E). Consistently, whole mount in situ hybridization results showed that overexpression of foxj1b or slc3a2a significantly recovered the suppressed expression of spi1b in the 14 hpf embryos overexpressed with miR-210-5p (Fig. 5, G–J). To further confirm this conclusion, we tested whether the primitive myelopoiesis at 26 hpf in miR-210-5p-overexpressed embryos can be rescued by these two genes. After co-overexpressing miR-210-5p with foxj1b or slc3a2a, we examined the expressions of spi1b, mpx, mfap4, and lcp1 in the embryos at 26 hpf. The qRT-PCR results revealed that overexpression of foxj1b or slc3a2a effectively (p < 0.05 or p < 0.01) recovered the suppressed primitive myelopoiesis at 26 hpf (Fig. 5F). Taken together, these results demonstrated that both foxj1b and slc3a2a are downstream genes of miR-210-5p involved in mediating the roles of miR-210-5p to inhibit the primitive myelopoiesis.
Figure 5.
Both foxj1b and slc3a2a work downstream of miR-210-5p to mediate its role in inhibiting zebrafish primitive myelopoiesis. A, schematic showing the workflow to screen the candidate downstream genes of miR-210-5p to mediate its role in inhibiting zebrafish primitive myelopoiesis. The candidate genes were selected by the analysis of negative correlation between the set of miR-210-5p candidate target genes and the set of genes that reduced their expressions in the gata4/5/6-knockdown embryos at 14 hpf. B, Venn map showing the candidate target genes of miR-210-5p to mediate its roles in inhibiting zebrafish primitive myelopoiesis. C, heat map showing the clustering analysis of 21 candidate genes working downstream of miR-210-5p to inhibit the primitive myelopoiesis. The horizontal lines represent different treatment groups, and the vertical columns represent the expression levels of each gene. The expression abundance of each gene is represented by the color intensity. Red represents the high expression and green represents low expression of the gene in the embryos. The upper dendritic tree represents the expression profile similarity of each gene, and the right dendritic represents the degree of similarity of the expression profile in the different treatment groups. D, qRT-PCR results showing the expression changes of candidate target genes in the miR-210-5p overexpressed embryos at 14 hpf. The expression changes of foxj1b, slc3a2a, and stard3, except rnasel3, were similar to microarray results. E, overexpressions of foxj1b and slc3a2a but not stard3 effectively rescued the expression of spi1b that was inhibited in the 14 hpf embryos overexpressed with miR-210-5p. F, overexpressions of foxj1b and slc3a2a effectively rescued the expressions of spi1b, mpx, mfap4, and lcp1 in the zebrafish embryos at 14 and 26 hpf, respectively. D–F, gene expression levels in the NC group was normalized as 1.0 and the fold-change of the gene expression level in the embryos microinjected with miR-210-5p mimic was shown relative to the NC group (shown in y axis). G–J, whole mount in situ hybridization results showing the expressions of spi1b in the 14 hpf embryos microinjected with NC, miR-210-5p mimic alone, or miR-210-5p mimic together with foxj1b or slc3a2a mRNA, respectively. Compared with the embryos microinjected with NC (G), overexpression of miR-210-5p inhibited the expression of spi1b (H), whereas overexpressions of foxj1b or slc3a2a could effectively rescue the expression of spi1b suppressed by miR-210-5p (I and J). NC, mimic negative control; miR-210-5p, miR-210-5p mimic; *, p < 0.05; **, p < 0.01; ns: no significance (p > 0.05).
To further support the conclusion, we examined the expressions of foxj1b and slc3a2a in the miR-210-5p knockout and gata4/5/6-knockdown embryos. The results from qRT-PCR showed that expressions of both foxj1b and slc3a2a were significantly (p < 0.01) up-regulated in miR-210nju49 embryos (Fig. S3, A and B). On the other hand, results from whole mount in situ hybridization revealed that the expressions of foxj1b and slc3a2a in gata4/5/6-knockdown embryos were significantly reduced (Fig. S3, C, D, G, and H). As expected, the expressions of foxj1b and slc3a2a were at least partially restored (Fig. S3, C–J) in the gata4/5/6-knockdown miR-210nju49 embryos.
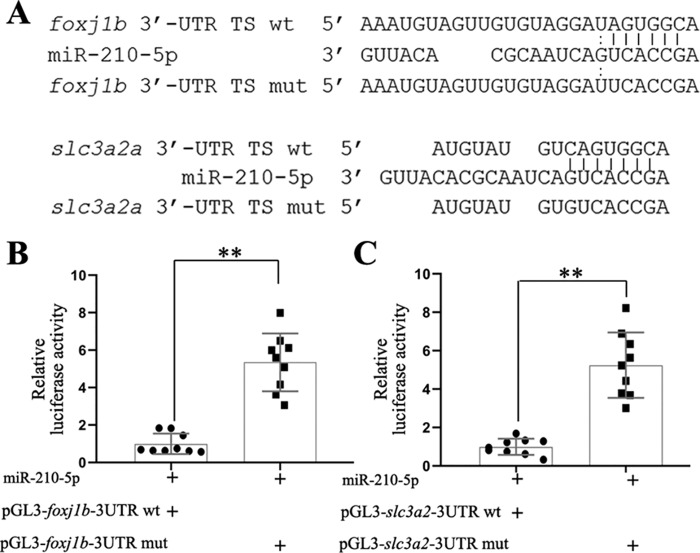
To determine whether these two genes are direct targets of miR-210-5p, we analyzed the 3′ UTRs of the two genes for putative miR-210-5p targeting sites using an online tool (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid)5 (45), which showed that there is one candidate target site of miR-210-5p in the 3′ UTRs of foxj1b or slc3a2a (Fig. 6A). To examine whether miR-210-5p regulates these genes through binding to the target sites, we performed dual-luciferase reporter assays on the expression of a firefly luciferase reporter gene fused with the WT 3′ UTR or mutated 3′ UTR of foxj1b and slc3a2a, respectively. The results showed the luciferase activities of the WT reporter of both foxj1b and slc3a2a were significantly (p < 0.01) repressed by miR-210-5p compared with the mutated reporters (Fig. 6, B and C). The results indicated that foxj1b and slc3a2a are direct target genes of miR-210-5p.
Figure 6.
Dual-luciferase reporter assay shows that foxj1b and slc3a2a are direct targets of miR-210-5p. A, the alignment of miR-210-5p target sites (in the 3′ UTR) of foxj1b and slc3a2a with miR-210-5p, and with mutated target sites, respectively. B and C, dual-luciferase reporter assay showing the relative expression levels of firefly luciferase whose coding sequence fused with WT 3′ UTR of foxj1b or slc3a2a were down-regulated when miR-210-5p was overexpressed compared to those fused with mutant ones, respectively. The expression levels of luciferase reporter in the WT 3′ UTR group was normalized as 1.0 and the fold-change of the reporter expression level in the mutated 3′ UTR group was shown relative to the WT group (shown in y axis). Three independent assays were performed with 3 repeats in each group. TS, target site; mut, mutant; miR-210-5p, miR-210-5p mimic; **, p < 0.01.
In summary, we demonstrate in this study that miR-210-5p works downstream of gata4/5/6 to inhibit zebrafish primitive myelopoiesis by repressing the differentiation of anterior hemangioblast into myeloid progenitor cells, and foxj1b and slc3a2a act as two direct target genes of miR-210-5p to mediate its role in repressing zebrafish primitive myelopoiesis.
Discussion
MicroRNAs are an abundant and conserved class of small RNAs. They play important regulatory functions on gene activity. Advances in next-generation sequencing have permitted the discovery of many miRNAs involved in myelopoiesis, but the role of miRNAs in zebrafish primitive myelopoiesis remains elusive. For example, a recent attempt to screen for miRNAs involved in spi1b (pu.1)-dependent macrophage development in zebrafish has been reported by Ghani and colleagues (7). Their results revealed a spi1b-orchestrated miRNA program during myeloid differentiation. In this study, we searched for miRNAs regulating zebrafish primitive myelopoiesis using gata4/5/6-knockdown embryos. From the deep-sequencing data, we found 8 candidates and identified one miRNA, miR-210-5p, as an inhibitory regulator in zebrafish primitive myelopoiesis (Fig. 1). Unlike the research performed by Ghani and colleagues (7), our study focused on the gata4/5/6-dependent miRNAs that are involved in myeloid differentiation. As a result, our miRNA profile differs from the spi1b-regulated miRNAs, which potentially includes miRNAs lying genetically upstream of spi1b during myeloid development, that is, miR-210-5p.
Functions of miR-210 have been related to hypoxia and tumor-suppressing. The expression level of miR-210 is higher in different cells with hypoxia responses (24–26). In agreement with this, miR-210 expression is also elevated in animals subjected to transient focal ischemia and in a variety of human cancers (27–30). Based on these results, miR-210 was considered as a potential biomarker of hypoxia, breast cancer, and acute coronary syndrome (31–33). Interestingly, it is reported that the low-level of miR-210 expression is associated with poorer outcome for pediatric acute lymphoblastic leukemia treated with chemotherapeutic drugs in clinic (34). In another study, it is reported that high levels of miR-210 associate with poor prognosis in acute myeloid leukemia patients (35). The result indicates the important role of miR-210 in acute myeloid leukemia and myeloid differentiation. However, these published studies all focused on miR-210-3p, whereas the function of miR-210-5p remains elusive although it is also conserved in different vertebrates during evolution (data not shown). In this study, we demonstrate that zebrafish miR-210-5p is one of the key regulators in the common pathway of primitive myelopoiesis, working downstream of gata4/5/6 (Fig. 2) and upstream of spi1b (Fig. 4) in the genetic hierarchy to block the differentiation of hemangioblasts into myeloid progenitors (Fig. 4).
In many cases, the biological functions of miRNAs can only be revealed in a context-specific manner (36). For example, the expression of miR-210 is intensively up-regulated in hypoxic states (37). Mice with deletion of miR-210 are viable at baseline and display no grossly abnormal phenotype. However, miR-210−/− mice are resistant to Fe-S-dependent pathophenotypes and pulmonary hypertension when exposed to hypoxia (38). Consistently, our results revealed that zebrafish carrying deleted miR-210 do not show any abnormal growth and development including myeloid development. However, in a gata4/5/6-deficient context, primitive myelopoiesis is partially recovered in miR-210−/− embryos. The results are in agreement with the experimental data that the expression of miR-210-5p is inhibited by the overexpressions of gata4/5/6 (Fig. 3) and overexpression of miR-210-5p inhibits the primitive myelopoiesis in zebrafish embryos (Fig. 1). Therefore, our data suggest that the restrictive role of miR-210-5p is not inevitable during normal development, but it can further worsen the primitive myelopoiesis if the gata4/5/6 signal is disturbed in zebrafish embryos.
miRNAs play important regulatory functions on gene activity mainly by interacting with the 3′ untranslated region (UTR) of target mRNAs. Previous study showed that VPM1 is a direct and functional downstream target gene of miR-210 in hypoxia-induced tumor cell metastasis (30). In this study, we identified that foxj1b and slc3a2a are two direct target genes of miR-210-5p in zebrafish primitive myelopoiesis by comparing the set of genes with reduced expression levels in gata5/6 morphants to that of the predicted miR-210-5p target genes from miRanda database5 (http://www.microrna.org). Obviously, this screening strategy excluded the potential target genes whose mRNA translations were blocked but stableness was not affected when miR-210-5p binds to their 3′ UTRs. Additionally, we only tested 4 of the 21 candidates of miR-210-5p target genes to examine whether their reduced mRNA levels were due to the binding of miR-210-5p to their 3′ UTRs. Therefore, our results cannot rule out the possibilities of other potential direct targets of this miRNA working downstream of gata4/5/6 to affect zebrafish primitive myelopoiesis. It is worth noting that we did not use the miR-210 knockout zebrafish as a model to screen the candidates of miR-210 target genes due to the reason that miR-210-5p knockout embryos exhibited normal primitive myelopoiesis in zebrafish.
Foxj1b is a transcription factor containing a forkhead box. It is expressed in the axial mesoderm and an area flanking the dorsal forerunner cells at 75% epiboly and in the PLPM and the precursor of the otic vesicle at the 7 somite-stage (39). Slc3a2a belongs to solute carrier family 3 (amino acid transporter heavy chain), member 2a. It is expressed in the yolk syncytial layer from 85% epiboly to 3-somite stage (40). To our knowledge, no previous research has reported that they were involved in primitive myelopoiesis in zebrafish. In this study, we found that the two genes were mainly expressed in the otic vesicle and yolk syncytial layer of embryos at 14 hpf (Fig. S3, C and G) but with no obvious expression in the ALPM where the primitive myelopoiesis occur. However, overexpression of foxj1b or slc3a2a greatly recovered the suppressed primitive myelopoiesis in embryos overexpressed with miR-210-5p (Fig. 5). Although it can be explained that the expression levels of foxj1b and slc3a2a in the ALPM might be too low to be detected by whole mount in situ hybridization, the mechanism underlying the two genes regulating the primitive myelopoiesis remains to be elucidated.
In summary, we report in this study that a gata4/5/6-regulated miRNA, miR-210-5p, is a restrictive regulator of zebrafish primitive myelopoiesis. It suppresses myeloid differentiation from hemangioblast through directly regulating the stableness of mRNAs of foxj1b and slc3a2a. Our results provide novel mechanistic insight into the role of miRNAs during zebrafish primitive myelopoiesis.
Experimental procedures
Ethics statement
Zebrafish were housed in the zebrafish room of the Model Animal Research Center, Nanjing University, in accordance with protocols approved by the IACUC of the Model Animal Research Center at Nanjing University. All methods were performed in accordance with the relevant guidelines and regulations.
Microinjection of morpholinos into zebrafish embryos
MOs were purchased from Gene Tools. The sequences of MOs are TGTTAAGATTTTTACCTATACTGGA (gata5 MO), AGCTGTTATCACCCAGGTCCATCCA (gata6 MO), and CCTCTTACCTCAGTTACAATTTATA (control MO) (13, 14). MOs were dissolved in nanopure water and microinjected into the embryos at 1-cell stages. The microinjection amount was 1 nl of solution containing 6.25 ng of gata5 MO and 1.25 ng of gata6 MO, or 7.5 ng of control MO per embryo (13).
Deep sequencing of miRNA transcripts
The embryos microinjected with control MO or gata5/6 MO at 1-cell stage were incubated for 14 h and collected for miRNA deep sequencing. The miRNA sequencing and profiling was performed by Shanghai Biotechnology Corporation. Briefly, total RNA was isolated from 80 of the 14 hpf embryos, and its quantity was examined by electrophoresis using Agilent Bioanalyzer 2100 (Agilent Technologies, USA). The miRNA was isolated using mirVana miRNA Isolation Kit (Life Technologies) from the total RNA. The 3′- and 5′-adaptors were added to the miRNA sequentially. cDNA was then reverse transcribed from the miRNA and amplified using a PCR (98 °C 30 s, 11 × (98 °C 10 s, 60 °C 30 s, 72 °C 15 s) for 72 °C for 10 min, 4 °C). The cDNA was separated by 6% Novex TBE PAGE Gel for collecting the 147–157-nt long fragments. The cDNA library was constructed and the quantity was examined using a commercial kit (QubitTM dsDNA HS, Agilent 2100). The concentration of the library was no lower than 0.5 ng/μl and the size of cDNA was 140–160 bp. The cDNA library was sequenced using Illumina Solexa following version 8 multiplexed single-read sequencing recipe. Fastx (fastx_toolkit-0.0.13.2) was used to preprocess the original reads of the sequence. The sequence of the joint and the low mass sequence including the fuzzy base N, sequence length less than 18 nt, were removed. The CLC genomics_workbench5.5 was applied to match the preprocessed sequence to the Sanger miRBase database (version 19.0). No mismatch was allowed during the process.
Microinjection of miRNA mimics or inhibitors into zebrafish embryos
miRNA mimics, inhibitors, miRNA mimics negative control, and miRNA inhibitors negative control were purchased from Shanghai Genepharma Corporation (Shanghai, China). Their sequences are listed in Table S1. Mimics and inhibitors were dissolved in RNase-free nanopure water and microinjected into the embryos at 1-cell stage. The amount of microinjection is 1 nl containing 10 μm miRNA mimic or inhibitor per embryo.
Whole mount in situ hybridization on examining the expression patterns of coding genes and noncoding genes
Whole mount in situ hybridizations on detecting the expression patterns of coding genes were performed as described previously (13). The RNA probes detecting the expressions of mpx, mfap4, lcp1, tal1, lmo2, and spi1b were prepared as described previously (13). The number of cells expressing mpx, mfap4, or lcp1 were counted manually under a dissecting microscope, and shown in a scatter plot, respectively. Whole mount in situ hybridizations on examining the expression patterns of miRNA were performed as described previously (41). The probe against miR-210 (miRCURY LNA microRNA Detection Probes) was purchased from a commercial company (Exiqon, Denmark). Photomicrographs were taken using a Olympus DP71 digital camera (Olympus, Japan) and further processed for brightness and contrast with Adobe Photoshop software.
Real-time RT-PCR assay
qRT-PCR was performed to examine the relative expression levels of the interested genes. For the coding genes in this study, we used ABI Stepone Plus (Applied Biosystems), as reported previously (13). Transcript levels of the examined coding genes were normalized to the actb1 mRNA level according to standard procedures. For miR-210-5p, we extracted total RNA from at least 30 embryos in each group using Direct-zol RNA MiniPrep (Zymol Research). qRT-PCR for the microRNAs was conducted using Mir-X miRNA First-strand synthesis and TB Green qRT-PCR Kit (Takara, Japan). The PCR was performed as follows: 95 °C for 10 s, 40× (95 °C of 5 s, 60 °C for 20 s), 95 °C for 15 s, 60 °C for 30 s, 95 °C for 15 s. The primers used are listed in Table S1.
In vitro synthesis of mRNA and microinjection of mRNAs into zebrafish embryos
mRNAs were in vitro synthesized, capped, and tailed using the method as described previously (13). The primers used for cloning the full-length coding sequences of gata4 and gata6 (13), foxj1b (GenBank accession number, NM_001008648.1), slc3a2a (GenBank accession number, NM_131601.2), and stard3 (NM_131662.1) are listed in Table S1. About 1 nl of solution containing 50 pg of gata4 and 25 pg of gata6, 200 pg of foxj1b, slc3a2a, or stard3 were microinjected into 1-cell stage embryos, respectively. The Cas9 mRNA was synthesized using the template derived from the expression plasmid of pXT7-Cas9 (42).
Generation of zebrafish miR-210 knockout line
To generate miR-210 knockout zebrafish, we first designed a pair of CRISPRs using the online tool (CRISPR). sgRNAs were transcribed using MAXIscript T7 Kit (Ambion) from the templates prepared by PCR amplification from a template plasmid pT7-gRNA with gene-specific primers sgRNA1F or sgRNA2F and a universal reverse primer (Table S1) as reported previously (42). We then microinjected 1 nl of solution containing 150 pg of Cas9 mRNA, 50 pg of sgRNA1, and 50 pg of sgRNA2 into 1-cell zebrafish embryos. The embryos were raised to adults as founders. The F1 mutant zebrafish were screened using the method as described before (43). Briefly, F1 generation zebrafish were genotyped by the PCR method using the genomic template prepared from the caudal fins clipped from zebrafish older than 6 weeks with a commercial kit (YSY, China). The primers used for amplifying the genomic sequences of the miR-210 were AATGTTCAGCTGCAGAACGG and GATCACACAACGCACCCATT. The PCR conditions were 95 °C for 2 min, 35 × (94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s), and a final extension of 5 min at 72 °C. F1 mutant zebrafish carrying the miR-210 knockout allele were selected and inbred to get miR-210 null progeny (F2).
Microarray analysis on the mRNAs isolated from gata4/5/6-knockdown embryos
The embryos microinjected with control MO or gata5/6 MOs at 1-cell stage were collected at 14 hpf for analyzing the gene expression changes affected by knocking down gata4/5/6. The microarray analysis was performed by Shanghai Biotechnology Corp. using zebrafish gene expression microarray 4*44K (Agilent). Total RNA extraction and purification, RNA amplification and labeling, hybridizations, as well as data acquisition by LC Sciences (USA) were used with strict adherence to Agilent's standardized protocols. Genes down-regulated in gata5/6 morphants (LogFC< −0.585, p < 0.05) were further analyzed using hierarchical clustering.
Dual-luciferase reporter assay
The 3′ UTR cDNAs of foxj1b and slc3a2a were first cloned into pGEM-T vector (Promega) by RT-PCR using the primer pairs of foxj1b-3utr-F and foxj1b-3utr-R, and slc3a2a-3utr-F and slc3a2a-3utr-R (Table S1), respectively. They were then recombined into pGL3-promoter Luciferase (Promega) using a One-Step Cloning Kit (Vazyme, China) with primer pairs of foxj1b-3UTR-F and foxj1b-3UTR-R, and slc3a2a-3UTR-F and slc3a2a-3UTR-R, respectively. The mutant 3′ UTR cDNAs of foxj1b and slc3a2a were first cloned into pUC57 vector. They were then recombined into pGL3-promoter luciferase (Promega) using One-Step Cloning Kit (Vazyme, China) with primer pairs of F-foxj1b-3UTR-MT and R-foxj1b-3UTR-MT, and F-slc3a2a-3UTR-MT and R-slc3a2a-3UTR-MT, respectively. 100 pg of reporter expression plasmids (pGL3-foxj1b-3UTR-wt or pGL3-foxj1b-3UTR-mut) or 50 pg of reporter expression plasmids (pGL3-slc3a2a-3UTR-wt or pGL3-slc3a2a-3UTR-mut), 1 pg of Renilla luciferase expression plasmid, and 20 pmol of miRNA mimic were microinjected together into zebrafish embryos at 1-cell stage. Dual-luciferase reporter assays were performed as described previously (44).
Statistical analysis
Data are presented as mean ± S.E. Statistical significance was determined using the unpaired two-tailed t test. A value of p < 0.05 (*) was considered statistically significant, and p < 0.01 (**) were considered statistically very significant.
Author contributions
W. J., D. L., N. L., P. H., J. Y., and Q. Z. data curation; W. J., D. L., N. L., Z. D., J. L., X. D., P. H., J. Y., and Q. Z. formal analysis; W. J., D. L., N. L., M. L., Z. D., J. L., X. D., Y. Y., and Q. Z. investigation; W. J., D. L., N. L., M. L., Z. D., J. L., X. D., Y. Y., and Q. Z. methodology; W. J., D. L., N. L., and Q. Z. writing-original draft; Q. Z. and D. L. conceptualization; Q. Z. supervision; Q. Z. and D. L. funding acquisition; Q. Z. project administration; Q. Z. writing-review and editing.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 31271569, 31471355, and 81500244 and State Key Laboratory of Genetic Engineering, Fudan University Grant SKLGE-1408. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ALPM
- anterior lateral plate mesoderm
- PLPM
- posterior lateral plate mesoderm
- miRNA
- microRNA
- NC
- negative control
- nt
- nucleotide(s)
- MO
- morpholino
- qRT
- quantitative RT
- sgRNA
- single guide RNA
- hpf
- hour post fertilization.
References
- 1. Hsia N., and Zon L. I. (2005) Transcriptional regulation of hematopoietic stem cell development in zebrafish. Exp. Hematol. 33, 1007–1014 10.1016/j.exphem.2005.06.013 [DOI] [PubMed] [Google Scholar]
- 2. Becker A. M., Michael D. G., Satpathy A. T., Sciammas R., Singh H., and Bhattacharya D. (2012) IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood 119, 2003–2012 10.1182/blood-2011-06-364976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karsunky H., Zeng H., Schmidt T., Zevnik B., Kluge R., Schmid K. W., Dhrsenü U., and Möröy T. (2002) Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30, 295–300 10.1038/ng831 [DOI] [PubMed] [Google Scholar]
- 4. McKercher S. R., Torbett B. E., Anderson K. L., Henkel G. W., Vestal D. J., Baribault H., Klemsz M., Feeney A. J., Wu G. E., Paige C. J., and Maki R. A. (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15, 5647–5658 10.1002/j.1460-2075.1996.tb00949.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sumanas S., Gomez G., Zhao Y., Park C., Choi K., and Lin S. (2008) Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood 111, 4500–4510 10.1182/blood-2007-09-110569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bissels U., Bosio A., and Wagner W. (2012) MicroRNAs are shaping the hematopoietic landscape. Haematologica 97, 160–167 10.3324/haematol.2011.051730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghani S., Riemke P., Schönheit J., Lenze D., Stumm J., Hoogenkamp M., Lagendijk A., Heinz S., Bonifer C., Bakkers J., Abdelilah-Seyfried S., Hummel M., and Rosenbauer F. (2011) Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood 118, 2275–2284 10.1182/blood-2011-02-335141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurkewich J. L., Hansen J., Klopfenstein N., Zhang H., Wood C., Boucher A., Hickman J., Muench D. E., Grimes H. L., and Dahl R. (2017) The miR-23a∼27a∼24–2 microRNA cluster buffers transcription and signaling pathways during hematopoiesis. PLoS Genet. 13, e1006887 10.1371/journal.pgen.1006887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alemdehy M. F., van Boxtel N. G., de Looper H. W., van den Berge I. J., Sanders M. A., Cupedo T., Touw I. P., and Erkeland S. J. (2012) Dicer1 deletion in myeloid-committed progenitors causes neutrophil dysplasia and blocks macrophage/dendritic cell development in mice. Blood 119, 4723–4730 10.1182/blood-2011-10-386359 [DOI] [PubMed] [Google Scholar]
- 10. Johnnidis J. B., Harris M. H., Wheeler R. T., Stehling-Sun S., Lam M. H., Kirak O., Brummelkamp T. R., Fleming M. D., and Camargo F. D. (2008) Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129 10.1038/nature06607 [DOI] [PubMed] [Google Scholar]
- 11. Fan H. B., Liu Y. J., Wang L., Du T. T., Dong M., Gao L., Meng Z. Z., Jin Y., Chen Y., Deng M., Yang H. T., Jing Q., Gu A. H., Liu T. X., and Zhou Y. (2014) miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood 124, 1320–1330 10.1182/blood-2013-12-545012 [DOI] [PubMed] [Google Scholar]
- 12. Velu C. S., Baktula A. M., and Grimes H. L. (2009) Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood 113, 4720–4728 10.1182/blood-2008-11-190215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang D., Jia W., Li J., Li K., and Zhao Q. (2012) Retinoic acid signaling plays a restrictive role in zebrafish primitive myelopoiesis. PloS One 7, e30865 10.1371/journal.pone.0030865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peterkin T., Gibson A., and Patient R. (2009) Common genetic control of haemangioblast and cardiac development in zebrafish. Development 136, 1465–1474 10.1242/dev.032748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peterkin T., Gibson A., and Patient R. (2007) Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev. Biol. 311, 623–635 10.1016/j.ydbio.2007.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Rooij E., Marshall W. S., and Olson E. N. (2008) Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ. Res. 103, 919–928 10.1161/CIRCRESAHA.108.183426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davidson A. J., and Zon L. I. (2004) The “definitive” (and “primitive”) guide to zebrafish hematopoiesis. Oncogene 23, 7233–7246 10.1038/sj.onc.1207943 [DOI] [PubMed] [Google Scholar]
- 18. Bennett C. M., Kanki J. P., Rhodes J., Liu T. X., Paw B. H., Kieran M. W., Langenau D. M., Delahaye-Brown A., Zon L. I., Fleming M. D., and Look A. T. (2001) Myelopoiesis in the zebrafish, Danio rerio. Blood 98, 643–651 10.1182/blood.V98.3.643 [DOI] [PubMed] [Google Scholar]
- 19. Lieschke G. J., Oates A. C., Paw B. H., Thompson M. A., Hall N. E., Ward A. C., Ho R. K., Zon L. I., and Layton J. E. (2002) Zebrafish SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev. Biol. 246, 274–295 10.1006/dbio.2002.0657 [DOI] [PubMed] [Google Scholar]
- 20. Gomes I., Sharma T. T., Edassery S., Fulton N., Mar B. G., and Westbrook C. A. (2002) Novel transcription factors in human CD34 antigen-positive hematopoietic cells. Blood 100, 107–119 10.1182/blood.V100.1.107 [DOI] [PubMed] [Google Scholar]
- 21. Lee T. Y., Ezelle H. J., Venkataraman T., Lapidus R. G., Scheibner K. A., and Hassel B. A. (2013) Regulation of human RNase-L by the miR-29 family reveals a novel oncogenic role in chronic myelogenous leukemia. J. Interferon Cytokine Res. 33, 34–42 10.1089/jir.2012.0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fogelstrand P., Féral C. C., Zargham R., and Ginsberg M. H. (2009) Dependence of proliferative vascular smooth muscle cells on CD98hc (4F2hc, SLC3A2). J. Exp. Med. 206, 2397–2406 10.1084/jem.20082845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borthwick F., Allen A. M., Taylor J. M., and Graham A. (2010) Overexpression of STARD3 in human monocyte/macrophages induces an anti-atherogenic lipid phenotype. Clin. Sci. 119, 265–272 10.1042/CS20100266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Camps C., Buffa F. M., Colella S., Moore J., Sotiriou C., Sheldon H., Harris A. L., Gleadle J. M., and Ragoussis J. (2008) hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 14, 1340–1348 10.1158/1078-0432.CCR-07-1755 [DOI] [PubMed] [Google Scholar]
- 25. Fasanaro P., D'Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M. C., and Martelli F. (2008) MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 283, 15878–15883 10.1074/jbc.M800731200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giannakakis A., Sandaltzopoulos R., Greshock J., Liang S., Huang J., Hasegawa K., Li C., O'Brien-Jenkins A., Katsaros D., Weber B. L., Simon C., Coukos G., and Zhang L. (2008) miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 7, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jeyaseelan K., Lim K. Y., and Armugam A. (2008) MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke 39, 959–966 10.1161/STROKEAHA.107.500736 [DOI] [PubMed] [Google Scholar]
- 28. Volinia S., Galasso M., Sana M. E., Wise T. F., Palatini J., Huebner K., and Croce C. M. (2012) Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. U.S.A. 109, 3024–3029 10.1073/pnas.1200010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Võsa U., Vooder T., Kolde R., Vilo J., Metspalu A., and Annilo T. (2013) Meta-analysis of microRNA expression in lung cancer. Int. J. Cancer 132, 2884–2893 10.1002/ijc.27981 [DOI] [PubMed] [Google Scholar]
- 30. Ying Q., Liang L., Guo W., Zha R., Tian Q., Huang S., Yao J., Ding J., Bao M., Ge C., Yao M., Li J., and He X. (2011) Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology 54, 2064–2075 10.1002/hep.24614 [DOI] [PubMed] [Google Scholar]
- 31. Ivan M., Harris A. L., Martelli F., and Kulshreshtha R. (2008) Hypoxia response and microRNAs: no longer two separate worlds. J. Cell. Mol. Med. 12, 1426–1431 10.1111/j.1582-4934.2008.00398.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Markou A., Yousef G. M., Stathopoulos E., Georgoulias V., and Lianidou E. (2014) Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin. Chem. 60, 197–205 10.1373/clinchem.2013.210542 [DOI] [PubMed] [Google Scholar]
- 33. Shalaby S. M., El-Shal A. S., Shoukry A., Khedr M. H., and Abdelraheim N. (2016) Serum miRNA-499 and miRNA-210: a potential role in early diagnosis of acute coronary syndrome. IUBMB life 68, 673–682 10.1002/iub.1529 [DOI] [PubMed] [Google Scholar]
- 34. Mei Y., Gao C., Wang K., Cui L., Li W., Zhao X., Liu F., Wu M., Deng G., Ding W., Jia H., and Li Z. (2014) Effect of microRNA-210 on prognosis and response to chemotherapeutic drugs in pediatric acute lymphoblastic leukemia. Cancer Sci. 105, 463–472 10.1111/cas.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang X., Chen L., Yan X., Li Y., Xiong Y., and Zhou X. (2015) Overexpression of miR-210 is associated with poor prognosis of acute myeloid leukemia. Med. Sci. Monit. 21, 3427–3433 10.12659/MSM.894812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leung A. K., and Sharp P. A. (2010) MicroRNA functions in stress responses. Mol. Cell 40, 205–215 10.1016/j.molcel.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cicchillitti L., Di Stefano V., Isaia E., Crimaldi L., Fasanaro P., Ambrosino V., Antonini A., Capogrossi M. C., Gaetano C., Piaggio G., and Martelli F. (2012) Hypoxia-inducible factor 1-α induces miR-210 in normoxic differentiating myoblasts. J. Biol. Chem. 287, 44761–44771 10.1074/jbc.M112.421255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White K., Lu Y., Annis S., Hale A. E., Chau B. N., Dahlman J. E., Hemann C., Opotowsky A. R., Vargas S. O., Rosas I., Perrella M. A., Osorio J. C., Haley K. J., Graham B. B., Kumar R., et al. (2015) Genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron-sulfur deficiency and pulmonary hypertension. EMBO Mol. Med. 7, 695–713 10.15252/emmm.201404511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian T., Zhao L., Zhang M., Zhao X., and Meng A. (2009) Both foxj1a and foxj1b are implicated in left-right asymmetric development in zebrafish embryos. Biochem. Biophys. Res. Commun. 380, 537–542 10.1016/j.bbrc.2009.01.111 [DOI] [PubMed] [Google Scholar]
- 40. Takesono A., Moger J., Faroq S., Cartwright E., Dawid I. B., Wilson S. W., and Kudoh T. (2012) Solute carrier family 3 member 2 (Slc3a2) controls yolk syncytial layer (YSL) formation by regulating microtubule networks in the zebrafish embryo. Proc. Natl. Acad. Sci. U.S.A. 109, 3371–3376 10.1073/pnas.1200642109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sweetman D., Rathjen T., Jefferson M., Wheeler G., Smith T. G., Wheeler G. N., Münsterberg A., and Dalmay T. (2006) FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev. Dyn. 235, 2185–2191 10.1002/dvdy.20881 10.1002/dvdy.20881 [DOI] [PubMed] [Google Scholar]
- 42. Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J. W., and Xi J. J. (2013) Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dong Z., Dong X., Jia W., Cao S., and Zhao Q. (2014) Improving the efficiency for generation of genome-edited zebrafish by labeling primordial germ cells. Int. J. Biochem. Cell Biol. 55, 329–334 [DOI] [PubMed] [Google Scholar]
- 44. Hu P., Tian M., Bao J., Xing G., Gu X., Gao X., Linney E., and Zhao Q. (2008) Retinoid regulation of the zebrafish cyp26a1 promoter. Dev. Dyn. 237, 3798–3808 10.1002/dvdy.21801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krüger J., and Rehmsmeier M. (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454 10.1093/nar/gkl243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.