Abstract
Macrophages are a heterogeneous population of immune cells that play central roles in a broad range of biological processes, including the resolution of inflammation. Although diverse macrophage subpopulations have been identified, the characterization and functional specialization of certain macrophage subsets in inflamed tissues remain unclear. Here we uncovered a key role of specific macrophage subsets in tissue repair using proteomics, bioinformatics and functional analysis. We isolated two hepatic monocyte-derived macrophage subpopulations: Ly6ChiCX3CR1lo macrophages and Ly6CloCX3CR1hi macrophages during distinct phases of acute liver injury and employed label-free proteomics approach to profile the proteome of these cells. We found that the endocytosis- and apoptotic cell clearance-related proteins were specifically enriched in Ly6CloCX3CR1hi macrophages at the resolution phase. Intriguingly, 12/15-lipoxygenase (Alox15), the most strongly up-regulated protein in Ly6CloCX3CR1hi macrophages, was identified as a specific marker for these macrophages. In co-culture systems, Ly6CloCX3CR1hi macrophages specifically induced hepatocyte proliferation. Furthermore, selective depletion of this population in CD11b-diphtheria toxin receptor mice significantly delayed liver repair. Overall, our studies shed light on the functional specialization of distinct macrophage subsets from different phases in the resolution of inflammation.
Introduction
Inflammation is an adaptive response that is triggered by infection or damage, with the aim of restoring tissue homeostasis1–3. However, inadequate or insufficient resolution of inflammation can result in tissue destruction, chronic inflammation and dysregulation of tissue repair, giving rise to fibrosis and cancer. Thus, it is not unexpected that resolution of inflammation is extremely tightly regulated4,5. Significant evidence implicates that macrophages play crucial roles in triggering resolution of inflammation through phagocytosis of cellular debris and releasing cytokines and growth factors that stimulate tissue repair and regeneration6,7.
After injury, circulating monocytes are abundantly recruited and then differentiate into macrophages as they migrate into the inflammatory sites8. Given that macrophages possess a striking functional and phenotypic plasticity, several studies have shown that there are distinct subsets of macrophages during different stages of inflammation and suggest that they may play unique and different roles6,9. Using CD11b-diphtheria toxin receptor (DTR) transgenic mice to selectively deplete macrophages at different stages in carbon tetrachloride-induced liver injury, Duffield et al. showed that macrophage depletion when liver fibrosis was advanced resulted in amelioration of fibrosis, whereas macrophage depletion during recovery phase, led to a failure of resolution with impaired matrix degradation10. During skeletal muscle repair, recruited monocyte-derived macrophages exhibited pro-inflammatory profiles and then converted to anti-inflammatory macrophages which stimulated myogenesis and fiber growth11, and disrupting the phenotypic switch of macrophages impaired healing and regeneration12. In a mouse model of spinal-cord injury and repair, distinct macrophage populations were found in the traumatized spinal cord. Ly6ChiCX3CR1lo macrophages homed to the sites of injured tissue in a CCL2 chemokine-dependent manner, while Ly6CloCX3CR1hi macrophages trafficked through a distinct path guided by VCAM-1, VLA-A, and CD7313. Although distinct macrophage subpopulations have been identified in various organ systems, the characterization and functional specialization of certain macrophage subsets in discrete microenvironment are not fully understood.
Acetaminophen (N-acetyl-p-aminophenol, APAP) overdose can cause severe liver injury and is the most common cause of death due to acute liver failure in many developed countries14. Previous studies have demonstrated that a substantial number of monocyte-derived macrophages were recruited into the inflamed liver15. Moreover, the resolution of hepatic damage was delayed in monocyte-deficient Ccr2−/− mice compared with WT mice16. Despite the essential protective effects of macrophages for liver repair, little is known about which macrophage subsets directly induce hepatocyte regeneration.
Mass Spectrometry (MS)-based proteomics is a powerful approach for in-depth characterization of the protein components of biological systems17. The proteome of in vitro polarized macrophages have been extensively studied18,19. However, relatively little is known about the proteomic characteristics of distinct primary macrophage populations in inflamed tissues. Here, we performed a systematic global proteomic comparison of two hepatic monocyte-derived macrophage subpopulations (Ly6ChiCX3CR1lo macrophages and Ly6CloCX3CR1hi macrophages) from distinct phases of acute liver injury. LC-MS/MS analysis of proteomic profiling revealed that the 72 h Ly6CloCX3CR1hi macrophages displayed upregulation of many wound healing- and endocytosis-related proteins relative to the 24 h Ly6ChiCX3CR1lo macrophages. Notably, the functional contribution of Ly6CloCX3CR1hi macrophages to liver repair and regeneration was further confirmed in in vitro macrophage-hepatocyte co-culture systems and in vivo conditional depletion of Ly6CloCX3CR1hi macrophages experiments.
Results
Experimental workflow for the differential proteomic study on distinct macrophage subpopulations
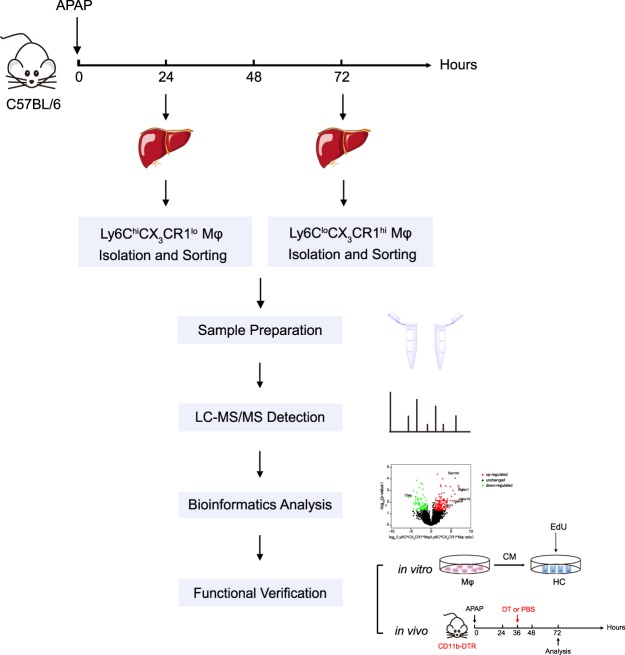
APAP-induced liver injury displays distinct injury (0–24 h) and resolution (48–72 h) phases and different monocyte-derived macrophage populations have been observed to infiltrate the inflammatory sites20. Thus, APAP-induced liver injury provides an instructive model for proteomic analysis of distinct macrophage populations. To explore the functional specialization of distinct hepatic macrophage subsets in APAP-induced liver injury, global label-free quantification (LFQ) proteomics were used. The experimental workflow was shown in Fig. 1. C57BL/6 WT mice were challenged with APAP to induce acute liver injury. Then, primary hepatic leukocytes were isolated and distinct hepatic macrophage populations (Ly6ChiCX3CR1lo macrophages and Ly6CloCX3CR1hi macrophages) were sorted by flow cytometry during the early phase and recovery phase, respectively. Then, the cells were collected and processed for proteomic profiling. Data from proteomics measurements were subjected to comprehensive bioinformatics analysis. Finally, functional validations were performed by both in vitro and in vivo experiments based on the information and clues obtained from proteomic data.
Figure 1.
Experimental workflow for the differential proteomic study on distinct macrophage subpopulations.
Characterization of distinct macrophage subsets in APAP-induced liver injury
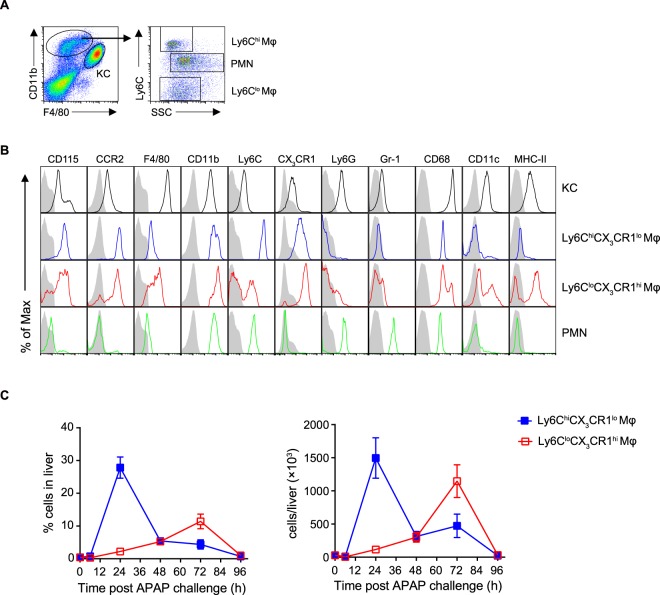
Consistent with previous reports20,21, we identified two main monocyte-derived macrophage populations infiltrating in the inflamed liver by flow cytometry: Ly6ChiCX3CR1lo and Ly6CloCX3CR1hi macrophage populations, distinguished by cell surface expression of F4/80, CD11b, Ly6C, CD115, CCR2, CX3CR1, Ly6G, Gr-1, CD68, CD11c and major histocompatibility complex class II (MHC-II) (Fig. 2A,B). Furthermore, dynamic changes in these macrophage subsets throughout the injury and recover phases of inflammation were analyzed. The number of Ly6ChiCX3CR1lo macrophages increased significantly during the early phase of inflammation, peaked at 24 h and then decreased, whereas Ly6CloCX3CR1hi macrophages became the dominant population during the resolution phase (Fig. 2C). Taken together, these results suggest that Ly6ChiCX3CR1lo macrophages and Ly6CloCX3CR1hi macrophages represented the most numerous macrophage population at the early phase and resolution phase of hepatic inflammation, respectively.
Figure 2.
Characterization of distinct macrophage subsets in APAP-induced liver injury. C57BL/6 mice were injected intraperitoneally with APAP at 400 mg/kg (body weight) to induce acute liver injury. (A) Flow cytometric analysis of hepatic leukocytes at 48 h after APAP challenge. Liver non-parenchymal cells were identified by first gating for live CD45+ leukocytes. Kupffer cells were identified as F4/80hiCD11blo. Monocyte-derived macrophages were identified as F4/80loCD11bhi. Distinct subsets of monocyte-derived macrophages and neutrophils were identified based on differential Ly6C expression and the FSC/SSC profile. (B) Flow cytometric analysis of the indicated populations as gated in (A). The shaded histograms represent the unstained controls. (C) The percentage and absolute numbers of Ly6ChiCX3CR1lo and Ly6CloCX3CR1hi monocyte-derived macrophages in the livers at each time point were determined by flow cytometry. Data shown are representative of three independent experiments (n = 3/group).
Global proteomics of distinct macrophage subsets from different phases in APAP-induced liver injury
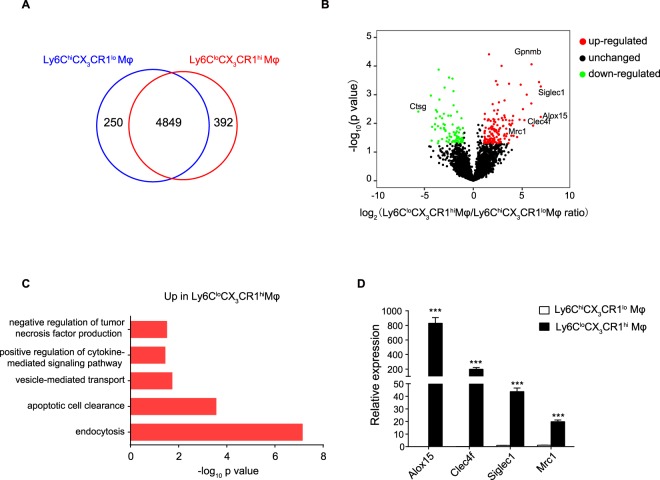
Having uncovered massive infiltration of distinct monocyte-derived macrophage subsets in injured livers during different phases of inflammation, we performed proteomic profiling of FACS-sorted 24 h Ly6ChiCX3CR1lo macrophages and 72 h Ly6CloCX3CR1hi macrophages using label-free proteomics. The proteins were identified and quantified by Maxquant software. We analyzed four independent biological replicates and identified a total of 5491 proteins for 24 h Ly6ChiCX3CR1lo macrophages and 72 h Ly6CloCX3CR1hi macrophages, of which 4849 were common between two samples (Fig. 3A). A complete list of all identified proteins can be found in Table S1. Moreover, there were 5488 (99.9%) proteins quantified with more than one unique peptide and the median value of unique peptide number was 6 (Fig. S1A). The median value of protein sequence coverage was 22.5% (Fig. S1B). Then, the differentially expressed proteins were analyzed by Perseus software using t-test methods. In total, 211 proteins were found significantly (fold change ≥2, p value ≤ 0.05) regulated between 24 h Ly6ChiCX3CR1lo macrophages and 72 h Ly6CloCX3CR1hi macrophages (Fig. 3B). From these, 137 proteins were up-regulated in 72 h Ly6CloCX3CR1hi macrophages compared with 24 h Ly6ChiCX3CR1lo macrophages (marked as red dots), whereas 74 proteins were down-regulated (marked as green dots). Figure S2A shows a heatmap of the differentially expressed genes, and a list of these genes is available in Table S2.
Figure 3.
Proteomic profiling of distinct macrophage subsets from different phases in APAP-induced liver injury. (A) Macrophage populations during distinct phases from 8–10 mice were used and pooled to acquire proteins for mass spectrometry analysis. Venn diagram of the quantified proteins in 24 h Ly6ChiCX3CR1lo macrophages and 72 h Ly6CloCX3CR1hi macrophages. (B) Volcano plot generated by differential analysis of the proteome profiles of 72 h Ly6CloCX3CR1hi macrophages compared with those of 24 h Ly6ChiCX3CR1lo macrophages. Significantly up-regulated proteins were shown as red dots and down-regulated proteins were shown as green dots. The proteins with fold change ≥2 and p < 0.05 were considered to be significant differentially expressed proteins. (C) Gene Ontology (GO) enrichment analysis of differentially expressed proteins was classified by their biological functions and arranged according to their statistical significance (-log10 p value on x axis) from DAVID. Biological processes involved in immune response were shown. (D) Validation of the indicated differentially expressed genes by qPCR, presented relative to Gapdh. Data shown are representative of three independent experiments (n = 3/group). Results represent mean ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001).
Next, Gene Ontology (GO) enrichment analysis was performed using DAVID22. Endocytosis- and apoptotic cell clearance-related proteins were identified as significant up-regulated protein function group in 72 h Ly6CloCX3CR1hi macrophages (Fig. 3C), whereas translation-, response to Gram-positive bacterium or lipopolysaccharide-, or inflammatory response-related proteins were identified as down-regulated protein function group in 72 h Ly6CloCX3CR1hi macrophages (Fig. S2B). Phagocytosis of cellular debris after tissue injury has been shown to induce an anti-inflammatory phenotype in macrophages, which is important for the resolution of inflammation6,7,23. Our proteomic analysis showed that Ly6CloCX3CR1hi macrophages displayed upregulation of several endocytosis- or wound healing-related genes (e.g., Alox15, Clec4f, Siglec1, Mrc1, etc.) (Figs 3B and S2C), indicating that Ly6CloCX3CR1hi macrophages express a gene-expression signature associated with pro-resolving or alternatively activated phenotypes. 12/15-lipoxygenase (Alox15), which is restricted expressed on alternatively activated macrophages and macrophages populations participating in the resolution of inflammation24,25, has been shown to play anti-inflammatory and tissue repair roles26,27. CLEC4F is exclusively expressed on liver resident Kupffer cells and macrophages infiltrating into the liver28. It has been reported to be involved in the phagocytosis of desialylated platelets29. Sialoadhesin (Siglec1, Cd169) was initially defined as a macrophage adhesion receptor and was then shown to play important roles in receptor-mediated internalization process30,31. Macrophage mannose receptor 1 (Mrc1, Cd206) is a highly effective endocytic receptor and is considered as a marker for alternatively activated or M2 macrophages. Since these endocytosis- or wound healing-related genes are important indicators of pro-restorative macrophages, we focused on the expression of these genes for further validation. Consistent with proteomic results, quantitative PCR (qPCR) analysis confirmed significant increase in the expression of endocytosis- or wound healing-related genes Alox15, Clec4f, Siglec1, and Mrc1 in 72 h Ly6CloCX3CR1hi macrophages compared with 24 h Ly6ChiCX3CR1lo macrophages (Fig. 3D).
Additionally, we selected a down-regulated protein associated with immune response for validation. Cathepsin G (Ctsg) has been shown to play important roles in antifungal immunity and endotoxic shock32. We performed western blot of the down-regulated protein, and found that marked decreased expression of Cathepsin G in the 72 h Ly6CloCX3CR1hi macrophages compared with the 24 h Ly6ChiCX3CR1lo macrophages (Fig. S3A,B), which was consistent with proteomic data.
Identification of Alox15 as a specific marker for Ly6CloCX3CR1hi macrophages
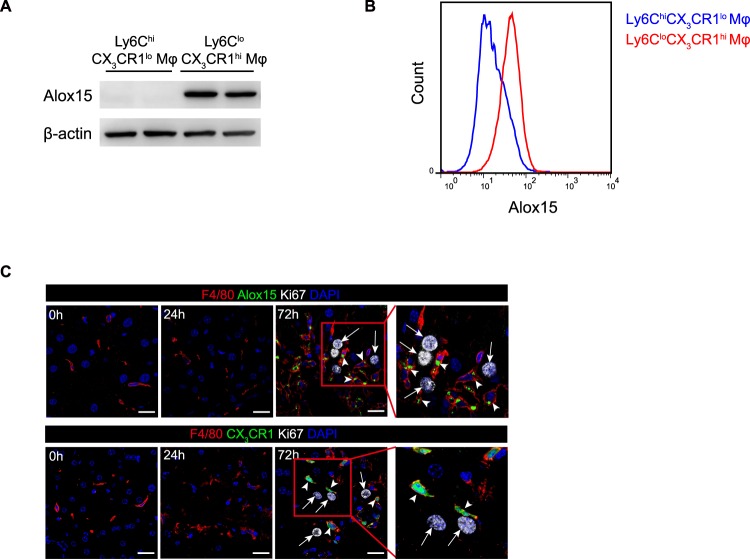
Notably, Alox15 was significantly up-regulated protein in the Ly6CloCX3CR1hi macrophages relative to the Ly6ChiCX3CR1lo macrophages, after using two-tailed t-test and Benjamini-Hochberg adjustment (Fig. 3B, Table S4). Indeed, the expression of Alox15 was restricted to Ly6CloCX3CR1hi macrophages but not Ly6ChiCX3CR1lo macrophages, revealed by LC-MS/MS. Further confirmation of the unique expression of Alox15 was achieved by qPCR, Western blotting, flow cytometry and immunofluorescence staining (Figs 4A–C, S3C). Thus, we identified Alox15 as a specific marker for Ly6CloCX3CR1hi macrophages. Importantly, using in situ fluorescence labeling of Alox15 and CX3CR1-fluorescent reporter mice, we observed that the localization of Alox15+ and CX3CR1hi macrophages was often adjacent to proliferating hepatocytes (Fig. 4C), suggesting that Ly6CloCX3CR1hi macrophages may exert positive effects on hepatocyte proliferation.
Figure 4.
Identification of Alox15 as a specific marker for Ly6CloCX3CR1hi macrophages. (A) Western blot analysis for Alox15 expression in 24 h Ly6ChiCX3CR1lo and 72 h Ly6CloCX3CR1hi macrophages. Full-length blots are shown in Fig. S3C. (B) Flow cytometric analysis for Alox15 expression in 24 h Ly6ChiCX3CR1lo and 72 h Ly6CloCX3CR1hi macrophages. Cell populations from five mice were pooled to form one group. (C) Alox15+CX3CR1hi macrophages were in close proximity to proliferating hepatocytes. White arrows indicate proliferating hepatocytes labeled with Ki67, and arrowheads indicate macrophages labeled with F4/80 and Alox15 in WT mice (upper panel) or F4/80 in Cx3cr1GFP/+ mice (lower panel). Scale bars, 20μm. Data shown are representative of three independent experiments.
Ly6CloCX3CR1hi macrophages directly accelerate hepatocyte proliferation in vitro
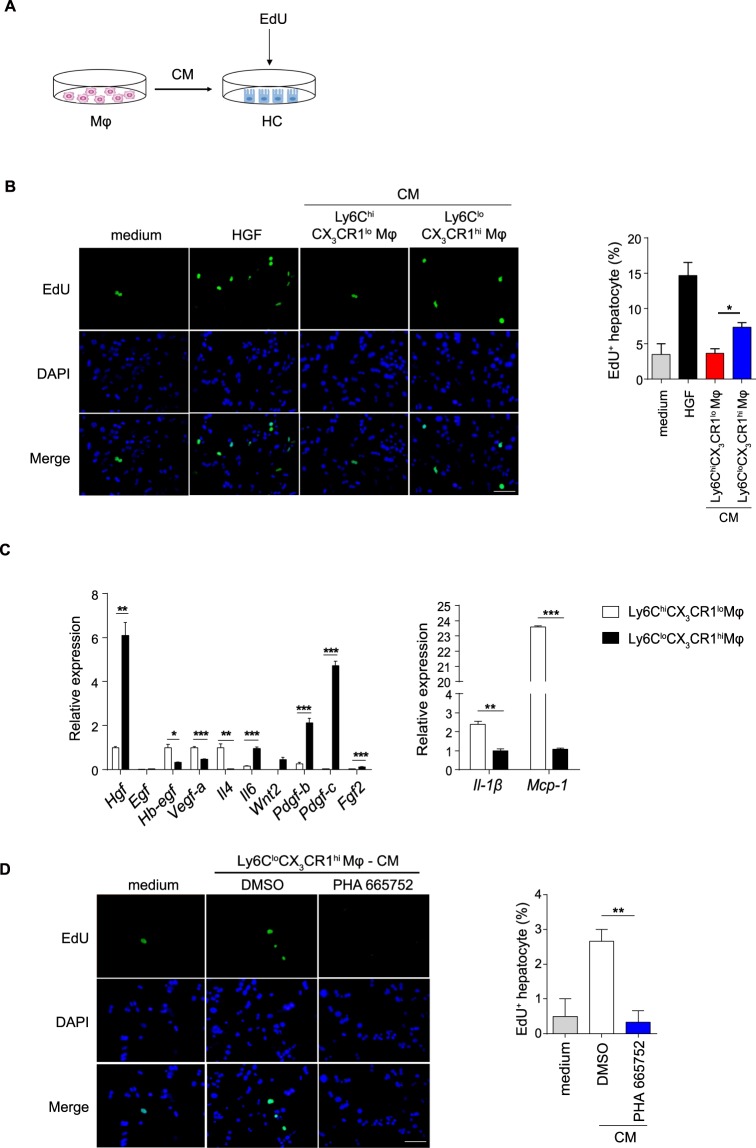
To validate the role of Ly6CloCX3CR1hi macrophages in hepatocyte proliferation, we performed a macrophage-hepatocyte co-culture system in vitro (Fig. 5A). Conditioned medium (CM) from 24 h Ly6ChiCX3CR1lo macrophages failed to support hepatocyte proliferation, whereas CM from 72 h Ly6CloCX3CR1hi macrophages led to a significant increase in the number of EdU+ hepatocytes (Fig. 5B). These results suggest that Ly6CloCX3CR1hi macrophage may mediate hepatocyte proliferation through secretory signals.
Figure 5.
Ly6CloCX3CR1hi macrophages directly accelerate hepatocyte proliferation in vitro. (A) Schematic of the experimental design. Normal hepatocytes (HCs) were co-cultured with the CM from 24 h Ly6ChiCX3CR1lo macrophages or 72 h Ly6CloCX3CR1hi macrophages. (B) Representative images of hepatocytes pulsed with EdU and the quantification of hepatocyte proliferation are shown. Scale bar, 50 μm. (C) Differential expression of the indicated genes measured by qPCR in 24 h Ly6ChiCX3CR1lo macrophages and 72 h Ly6CloCX3CR1hi macrophages, presented relative to Gapdh. (D) Hepatocytes were co-cultured with the CM from 72 h Ly6CloCX3CR1hi macrophages or supplemented with the c-Met kinase inhibitor PHA665752 (2.5 μM). Representative images of hepatocytes pulsed with EdU (left panel) and the quantification of hepatocyte proliferation (right panel) are shown. Scale bar, 50 μm. The data shown are representative of at least two independent experiments (n = 3/group). The results represent means ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001).
Having uncovered that Ly6CloCX3CR1hi macrophages specifically stimulated hepatocyte proliferation, we wanted to identify which Ly6CloCX3CR1hi macrophage-derived hepatotropic cytokines were involved in liver regeneration. We performed a candidate-based screen of established hepatocyte growth factors by qPCR33–36. qPCR analysis identified marked increased expression of Hgf, Il-6, Wnt2, Pdgf-b, Pdgf-c, Fgf2 accompanied by decreased expression of the pro-inflammatory genes Il1β and Mcp-1 in the Ly6CloCX3CR1hi macrophages compared with the Ly6ChiCX3CR1lo macrophages (Fig. 5C). Among these genes, the mRNA levels of HGF were the most significantly up-regulated (Fig. 5C). Thus, we speculated that Ly6CloCX3CR1hi macrophage-derived HGF might be involved in the pro-regeneration effect. To examine this hypothesis, we stimulated the cultured hepatocytes with conditioned medium from Ly6CloCX3CR1hi macrophages supplemented with the c-Met kinase inhibitor given that c-Met is the only known high-affinity receptor for HGF37,38. The addition of the c-Met kinase inhibitor in conditioned medium from Ly6CloCX3CR1hi macrophages decreased EdU incorporation (Fig. 5D), indicating that the proliferative actions of Ly6CloCX3CR1hi macrophages are mediated, in part, via the HGF/c-Met pathway.
Selective depletion of Ly6CloCX3CR1hi macrophages delays liver regeneration and repair in vivo
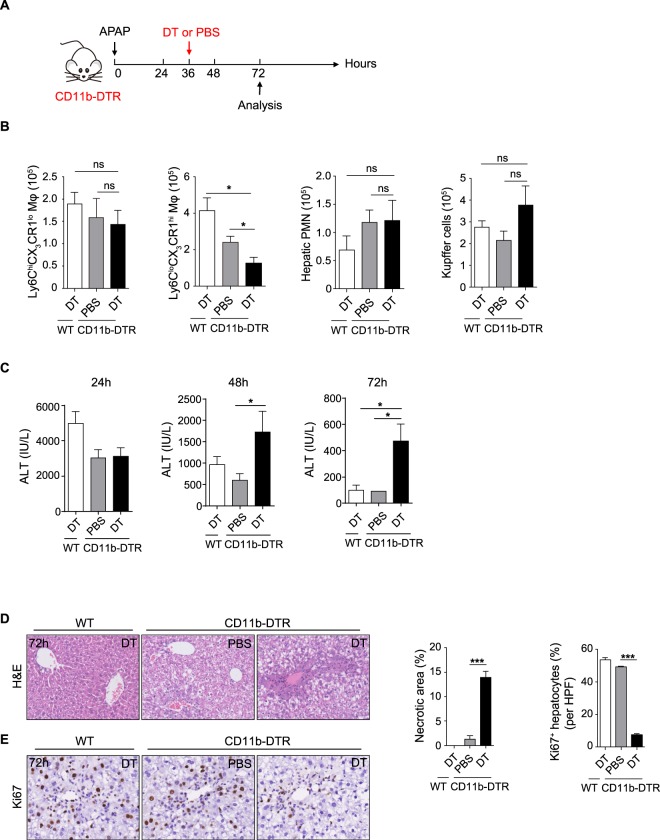
To further confirm the role of Ly6CloCX3CR1hi macrophages in liver regeneration and repair in vivo, a well-described selective macrophage depletion strategy in CD11b- DTR transgenic mice was used10. We punctually ablated Ly6CloCX3CR1hi macrophages in CD11b-DTR mice during the resolution phase of liver injury, which is when Ly6CloCX3CR1hi macrophages predominate (Fig. 6A). Using this strategy, we observed that the number of hepatic Ly6CloCX3CR1hi macrophages was specifically reduced (Fig. 6B). Consistent with previously reports10, there was no significant reduction in the number of neutrophils and Kupffer cells after DT treatment, which may be due to the insensitivity of these cells to DT. Selective ablation of Ly6CloCX3CR1hi macrophages resulted in higher serum levels of ALT, more necrotic areas and a significantly lower number of Ki67+ proliferating hepatocytes during the resolution phase compared to the control mice (Fig. 6C–E). Collectively, these results strongly suggest that Ly6CloCX3CR1hi macrophages are required for optimal liver regeneration and repair.
Figure 6.
Selective depletion of Ly6CloCX3CR1hi macrophages delays liver regeneration and repair in vivo. (A) Schematic of the experimental design. WT and CD11b-DTR mice were challenged with APAP and were given DT or PBS at 36 h after APAP injection. (B) Flow cytometric analysis of distinct hepatic monocyte-derived macrophage populations, Kupffer cells and neutrophils at 72 h. (C–E) Serum ALT levels at the indicated time points (C), histological characterization (D), and IHC staining for Ki67 per high-powered field (HPF) (E) in liver sections at 72 h were evaluated. The data shown are representative of two independent experiments (n = 3–5/group). The results represent means ± SEM (*p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
Our exploratory proteomic studies of distinct macrophage populations from different phases provide evidence for better understanding the functional specialization of distinct macrophage subsets during the resolution of inflammation. Based on the proteomic results, we speculated that, unlike 24 h Ly6ChiCX3CR1lo macrophages, 72 h Ly6CloCX3CR1hi macrophages may mediate wound healing and tissue repair. Indeed, recent studies have showed that Ly6ChiCX3CR1lo macrophages aggravated APAP hepatotoxicity and inhibition of monocyte recruitment by blocking CCL2 or CCR2/CCR5 attenuated liver damage39. Here, we demonstrated the specific role of Ly6CloCX3CR1hi macrophages in the resolution of APAP-induced liver injury. The macrophage-hepatocyte co-culture experiments suggested that Ly6CloCX3CR1hi macrophages could specifically induce hepatocyte proliferation. Furthermore, specific ablation of Ly6CloCX3CR1hi macrophages delayed liver repair. Thus, our results provide evidences of a certain macrophage subpopulation at resolution phase of inflammation that directly induce tissue repair and regeneration, with important implications for treating inflammatory conditions.
In our study, we compared two different macrophage populations at different time points to analyze their gene expression profile. Since the two macrophage subsets predominated at different stages of inflammation and it was difficult to isolate enough cells at the same time points, we did not provide a more direct comparison about the molecular phenotype of these two macrophage subsets. However, based on previous reports21,39, the comparison between macrophages at different time points may also represent a reasonable approximation. In addition, it has been reported that Ly6ChiCX3CR1lo macrophages can convert to Ly6CloCX3CR1hi macrophages during the resolution phase20,21. Thus, the comparison of 24 h Ly6ChiCX3CR1lo macrophages and 72 h Ly6CloCX3CR1hi macrophages may also show the functional changes of macrophages before and after phenotypic transition.
Our comparative proteomic analysis indicated that 72 h Ly6CloCX3CR1hi macrophages exhibited a reparative protein expression profile. Furthermore, Alox15 was the most strongly up-regulated protein in Ly6CloCX3CR1hi macrophages and has been identified as a specific marker for Ly6CloCX3CR1hi macrophages by LC-MS/MS and subsequent validations. Previous studies have shown that Alox15 was restricted expressed in certain macrophage populations including alternatively activated macrophages and macrophages participating in the resolution of inflammation24,40,41. Consistent with these findings, our proteomic data and subsequent validation suggest that Alox15 was a specific marker for Ly6CloCX3CR1hi macrophages, which exhibited a pro-resolving profile. Furthermore, several lines of evidence suggest that Alox15 plays an anti-inflammatory and tissue-repair role in various contexts. Deletion of Alox15 leads to exacerbation of inflammation and tissue damage during chronic inflammatory disorders such as arthritis42. Alox15 can also contribute to generation of pro-resolving lipid mediators such as lipoxin A4, resolving E1, and protectin D1, which are responsible for resolution of inflammation43–45. Recent studies have identified Alox15 as a central factor orchestrating the sorting of apoptotic cells (ACs) and the clearance of ACs was confined to a population of Alox15-expressing, alternatively activated resident macrophages27. Thus, it is tempting to speculate that Alox15 is a major factor mediating tissue repair in macrophages. Further studies will be required to determine the specific contribution of Alox15 in Ly6CloCX3CR1hi macrophages to wound healing process.
Additionally, in vitro co-culture system suggested that 72 h Ly6CloCX3CR1hi macrophages directly stimulated hepatocyte proliferation. Selective depletion of this population in CD11b-diphtheria toxin receptor mice further confirmed the key role of Ly6CloCX3CR1hi macrophages in liver repair and regeneration. Although we observed that the pro-regenerative effects of Ly6CloCX3CR1hi macrophages on hepatocytes relied on the HGF/c-Met pathway, we cannot rule out the possibility that additional factors other than HGF might contribute to the pro-resolving effects on macrophages.
Methods
Mice
All animal procedures were approved by the Institutional Animal Care and Utilization Committee at Beijing Institute of Lifeomics. C57BL/6 wild-type (WT) mice were purchased from Charles River in Beijing (Vital River). CD11b-DTR mice were kindly provided by Dr. Honglin Wang (Shanghai Jiao Tong University). Cx3cr1gfp/+ mice were generously provided by Dr. Zhihua Liu (Institute of Biophysics, Chinese Academy of Sciences). All mice were maintained in our specific pathogen-free facilities. Sex- and age- matched controls were used, and all experiments were performed in accordance with the Institutional Animal Care and Utilization Committee-approved protocols.
For selective macrophage depletion, CD11b-DTR mice were injected intravenously (i.v.) with 30 ng/g body weight of DT (List Biological Labs) at 36 h post APAP challenge.
APAP-induced hepatotoxicity and assays for liver injury
Mice were fasted for 16 hours and injected intraperitoneally (i.p.) with acetaminophen (Sigma Aldrich) at 400 mg/kg. Serum alanine aminotransferase (ALT) levels were evaluated by diagnostic kits. Liver specimens from APAP-challenged mice were fixed in 4% paraformaldehyde and embedded with paraffin for hematoxylin and eosin (H&E) staining and immunohistochemistry analyses. The sections were stained with H&E or with monoclonal Rabbit anti-mouse Ki67 (clone SP6, ab16667, Abcam). Necrotic areas and percentage of Ki67-positive cells were quantified by ImageJ software.
Primary hepatic cell isolation
Liver leukocytes were isolated as previously described15. After in situ two-step collagenase perfusion, the livers were homogenized and filtered through a 70 μm nylon mesh. The cell suspension was centrifuged for 5 minutes at 500 g, the cell pellet was then re-suspended in 15 ml 35% Percoll (GE Healthcare) containing 100 U/ml heparin and was centrifuged for 15 minutes at 500 g. The resulting cell pellet containing leukocytes was then lysed for erythrocytes by 3-min incubation with red blood cell lysis solution (BD Biosciences). For hepatic macrophage purification, isolated hepatic leukocytes were stained for cell-surface markers. Then, distinct hepatic macrophage subpopulations (F4/80loCD11bhiLy6G−Ly6ChiCX3CR1lo or F4/80loCD11bhiLy6G−Ly6CloCX3CR1hi) were sorted by FACSAria III (BD Biosciences).
For primary mouse hepatocytes isolation, liver perfusion and digestion were performed as described above. Following digestion, the liver was dissociated and filtered to obtain single cell suspensions. Hepatocytes were collected by centrifugation for 3 minutes at 50 g.
Flow cytometry and FACS sorting
Hepatic leukocytes were incubated with Fc blocking reagent (CD16/32, eBioscience) for 20 minutes followed by incubation with fluorescently-conjugated antibodies directed against mouse (all from eBioscience unless specified otherwise): CD45 (30-F11), F4/80 (BM8), CD11b (M1/70), Ly6C (HK1.4), CD115 (AFS98), CCR2 (475301, R&D systems), CX3CR1 (SA011F11, Biolegend), Ly6G (1A8, BD Biosciences), Gr-1(RB6-8C5) and CD11c (N418). CD68 (FA-11, AbD Serotec) was stained extracellularly and subsequently intracellularly using Fixation-and-Permeabilization buffers (eBioscience) according to the manufacturer’s protocol. Experiments were performed using the LSRFortessa cell analyzer (BD Biosciences) and the acquired data were analyzed with FlowJo software (Tree Star). For cell sorting, FACSAria III (BD Biosciences) were used and the purity of sorted cells was routinely more than 95%.
Sample preparation for LC-MS/MS analysis
FACS-sorted cells were lysed by 8 M urea and the cell debris was pelleted by centrifugation at 10000 g for 5 min. Then, the clarified lysate was transferred into a new vial. The lysate was diluted 1:8 with 50 mM NH4HCO3 and reduced by DTT (10 mM). Then it was alkylated with 50 mM IAA. The digestion was performed with trypsin (1:50) at 37 °C overnight. The resulting peptides were acidified with 0.5% formic acid, and the supernatant was collected. In each experiment, approximately 15 μg of peptides measured by nanodrop A280 method were loaded for fractionation. The amount of peptides was controlled to be equal between each replicate. The first dimension separation by reversed phase chromatography was performed using in-house C18 (3μm, 150 Å, Agela) 200 μl tip. The peptide mixtures were sequentially separated with gradient elution buffer (6, 9, 12, 15, 18, 21, 24, 30 and 35% buffer B) and then cross combined into six fractions and dried. The peptides were analyzed by Q-Exactive mass spectrometer and Orbitrap Fusion coupled to an Easy-nLC 1000 system (Thermo Fisher Scientific). Samples were dissolved in 0.1% formic acid and loaded onto a 12 cm reversed phase column (150 nm id) packed with C18 resin (1.9 μm). A binary solvent system (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid in acetonitrile) was used for peptide separation. The gradient was set as follows: 5–8% B for 8 min, 8–22% B for 50 min, 22–32% B for 12 min, 32–90% B for 1 min, and 90% B for 7 min. The constant flow rate was 300 nl/min. Data dependent acquisition mode was used to automatically pick peptides for MS2 fragmentation. When Q-Exactive was used, top 20 method was used and the dynamic exclusion duration was set 18 s. In MS1, the scan range was set 300–1400 m/z, and the resolution of MS1 was set 70,000. The AGC target was 3e6, and the maximum injection time was 60 ms. In MS2, the resolution of MS2 was set 15,000. The AGC target was 5e4, and the maximum injection time was 80 ms. When Orbitrap Fusion was used, the peptides were cross combined into three fractions. Top speed method with a cycle time of 3 s was used and the dynamic exclusion duration was set 18 s. In MS1, the scan range was set 300–1400 m/z, and the resolution of MS1 was set 120,000. The AGC target was 5e5, and the maximum injection time was 100 ms. In MS2, iontrap was used. The AGC target was 5e3, and the maximum injection time was 35 ms.
Data analysis of proteomic raw files
Raw MS data were processed using Maxquant (version 1.5.2.8) software46. In this software, the identified peptides were divided into two cases, unique peptides which are unique to the specific proteins and razor peptides which are found in more than one protein. As previously reported47, MaxQuant resolves this issue by collapsing all proteins that cannot be distinguished based on the identified peptides into protein groups. We used the default setting of the software that razor peptides contribute only to the quantification of the protein with the larger number of identifications. ‘Label free quantification’ was processed with Maxquant software. It was based on MS1 quant, and all the peak isotopes were used for MS1 quant. Normalization was performed using a solution named MaxLFQ based on peptide ion intensity. After summing up intensities with normalization factors as free variables, the software determined their quantities via a global optimization procedure based on achieving the least overall proteome variation. It was done purely from the data obtained and without the addition of external quantification standards or reliance on a fixed set of “housekeeping” proteins. A final protein level quantification value was generated by summing all identified peptide intensities. Detailed algorithm and protocols referred to previous reports47,48. The Mus musculus protein database (2018-05-29) was downloaded from Uniprot and only 16,978 canonical sequences which were annotated as “reviewed” were used for database searching. Trypsin was selected as the proteolytic enzyme. Two missed cleavages was allowed. Cysteine carbamidomethylation was set as the fixed modification. N-terminal acetylation, methionine oxidation were set as the variable modifications. The false discovery rate was set ≤1% at spectra level and protein level. Label-free quantitation was performed. The database search results were then processed with Perseus (1.5.8.5) software49. After removing the reversed and contaminating proteins (such as BSA), the LFQ values were log2 transformed and used for comparison between different experiment conditions. T-test was performed to obtain the differential expressed proteins with Perseus (1.5.8.5) software.
Western blot analysis
Macrophage protein extracts were prepared according to standard protocols. Cell lysates were separated by 8% or 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). The following antibodies were used: Cathepsin G (ab197354, Abcam), Alox15 (sc-32940, Santa Cruz) and β-actin (A5441, Sigma Aldrich).
Quantitative RT-PCR
Total RNA was extracted using RNeay Mini Kit (Qiagen) according to the manufacturer’s protocol and cDNA was made using Reverse Transcription kit (Promega). The cDNA was used for quantitative qPCR analysis on an iCycler iQ5 Real-Time PCR detection system (BioRad). The expression of target gene was normalized to the expression of the housekeeping gene, Gapdh. Relative gene expression was calculated using the standard 2−ΔΔCt method. A full list of the primer sequences is available in Table S3.
Immunofluorescence
Mouse liver tissues were fixed in 4% paraformaldehyde and then embedded in OCT (Sakura). 5 μm Frozen sections were prepared using a Cryotome FSE cryostat (Thermo-Fisher Scientific). The tissue sections were incubated in the blocking buffer (5% donkey serum, 0.3% Triton-X 100 in PBS) at room temperature for 1 hr followed by the staining with primary antibodies. The following primary antibodies were used: Rat anti-mouse F4/80 (clone CI:A3-1, ab6640, Abcam), Rabbit anti-mouse Ki67 (clone SP6, ab16667, Abcam), goat anti-mouse Alox15 antibody (clone H-235, sc-32940, Santa Cruz). Then slides were washed and incubated for 1 h with the following secondary antibodies: donkey anti-rat Alexa Fluor 488, donkey anti-goat Alexa Fluor 594 and donkey anti-rabbit Alexa Fluor 647 (Jackson ImmunoResearch Laboratories). Sections were counterstained with 4′6-diamidino-2-phenylindole dihydrochloride (DAPI) before being mounted. All immunofluorescence staining was performed in the dark. Imaging was performed using a Zeiss LSM 880 and images were processed using Zeiss ZEN software.
Determination of hepatocyte proliferation by EdU labeling
Primary mouse hepatocytes were isolated from WT mice. Ly6ChiCX3CR1lo macrophages and Ly6CloCX3CR1hi macrophages were isolated at the indicated time points. Conditioned medium (CM) was collected from 2 × 105 Ly6ChiCX3CR1lo or Ly6CloCX3CR1hi macrophages, filtered through a 0.22 μm filter, and added to 1 × 104 hepatocytes. Hepatocytes were treated with macrophage-derived CM or HGF (50 ng/ml, Peprotech) for 12 h, followed by EdU (5-ethynyl-2′-deoxyuridine, 20 μM) pulsing for an additional 36 h. In another series of experiments, isolated hepatocytes were treated with Ly6CloCX3CR1hi macrophage-derived CM supplemented with or without the c-Met kinase inhibitor PHA665752 (2.5 μM, R&D systems). Hepatocytes undergoing DNA synthesis were visualized using the EdU Imaging Kit (Life Technologies). Imaging was performed using Olympus IX71 inverted fluorescence microscopes and EdU-positive cells were quantified by ImageJ software.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5 software. Data are presented as mean ± SEM. Statistical significance was determined by unpaired, two-tailed, Student’s t test; p < 0.05 was considered statistically significant. Group allocation and outcome assessment was performed in a blinded manner. No exclusion criteria were applied, and all samples were included in data analysis.
Supplementary information
Acknowledgements
This work was supported by National Key R&D Program of China (2018YFA0507500), Chinese State Key Program in Basic Research (2014CBA02000, 2013CB910802), Chinese National Natural Science Foundation Projects (81500481, 81400615) and Beijing Science Program for the Top Young (2015000021223TD04). We would like to thank Dr. Honglin Wang (Shanghai Jiao Tong University) for the kind gift of CD11b-DTR mice, and Dr. Zhihua Liu (Institute of Biophysics, Chinese Academy of Sciences) for the kind gift of Cx3cr1gfp/+ mice. We thank Proteomics Facility, Flow Cytometry Facility, Animal Facility and Imaging Facility of National Center for Protein Sciences. Beijing (NCPSB) (Ms. Ping Wu) for their assistance.
Author Contributions
W.Y. performed the experiments, analyzed the data and wrote the manuscript. X.Z. performed mass spectrometry and analyzed the proteomic data. Y.T. performed the Immunofluorescence experiments. Y.W. provided technical or material support. L.T. and F.H. conceived and supervised the study.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011958.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fuchu He, Email: hefc@nic.bmi.ac.cn.
Li Tang, Email: tangli08@aliyun.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39007-6.
References
- 1.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529:307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugimoto MA, Sousa LP, Pinho V, Perretti M, Teixeira MM. Resolution of Inflammation: What Controls Its Onset? Frontiers in immunology. 2016;7:160. doi: 10.3389/fimmu.2016.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 4.Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nature immunology. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- 5.Buckley CD, Gilroy DW, Serhan CN, Stockinger B, Tak PP. The resolution of inflammation. Nature reviews. Immunology. 2013;13:59–66. doi: 10.1038/nri3362. [DOI] [PubMed] [Google Scholar]
- 6.Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44:450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefater JA, III., Ren S, Lang RA, Duffield JS. Metchnikoff’s policemen: macrophages in development, homeostasis and regeneration. Trends in molecular medicine. 2011;17:743–752. doi: 10.1016/j.molmed.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nature reviews. Immunology. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 9.Aurora AB, Olson EN. Immune modulation of stem cells and regeneration. Cell stem cell. 2014;15:14–25. doi: 10.1016/j.stem.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffield JS, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. Journal of Clinical Investigation. 2005;115:56–65. doi: 10.1172/jci200522675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into anti-inflammatory macrophages to support myogenesis. The Journal of experimental medicine. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mounier R, et al. AMPKalpha1 regulates macrophage skewing at the time of resolution of inflammation during skeletal muscle regeneration. Cell metabolism. 2013;18:251–264. doi: 10.1016/j.cmet.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Shechter R, et al. Recruitment of beneficial M2 macrophages to injured spinal cord is orchestrated by remote brain choroid plexus. Immunity. 2013;38:555–569. doi: 10.1016/j.immuni.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernal W, Auzinger G, Dhawan A, Wendon J. Acute liver failure. Lancet (London, England) 2010;376:190–201. doi: 10.1016/s0140-6736(10)60274-7. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z-X, Govindarajan S, Kaplowitz N. Innate immune system plays a critical role in determining the progression and severity of acetaminophen hepatotoxicity. Gastroenterology. 2004;127:1760–1774. doi: 10.1053/j.gastro.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 16.Holt MP, Cheng L, Ju C. Identification and characterization of infiltrating macrophages in acetaminopheninduced liver injury. Journal of leukocyte biology. 2008;84:1410–1421. doi: 10.1189/jlb.0308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cravatt BF, Simon GM, Yates JR., III. The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 18.Brown J, Wallet MA, Krastins B, Sarracino D, Goodenow MM. Proteome bioprofiles distinguish between M1 priming and activation states in human macrophages. Journal of leukocyte biology. 2010;87:655–662. doi: 10.1189/jlb.0809570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C, et al. Proteomic Identification of Interferon-Induced Proteins with Tetratricopeptide Repeats as Markers of M1 Macrophage Polarization. Journal of proteome research. 2018;17:1485–1499. doi: 10.1021/acs.jproteome.7b00828. [DOI] [PubMed] [Google Scholar]
- 20.Zigmond E, et al. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. Journal of immunology. 2014;193:344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 21.Ramachandran P, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3186–3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis G, Jr., et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 23.Brancato SK, Albina JE. Wound macrophages as key regulators of repair: origin, phenotype, and function. The American journal of pathology. 2011;178:19–25. doi: 10.1016/j.ajpath.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stables MJ, et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood. 2011;118:e192–208. doi: 10.1182/blood-2011-04-345330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Gronert K, et al. A role for the mouse 12/15-lipoxygenase pathway in promoting epithelial wound healing and host defense. The Journal of biological chemistry. 2005;280:15267–15278. doi: 10.1074/jbc.M410638200. [DOI] [PubMed] [Google Scholar]
- 27.Uderhardt S, et al. 12/15-lipoxygenase orchestrates the clearance of apoptotic cells and maintains immunologic tolerance. Immunity. 2012;36:834–846. doi: 10.1016/j.immuni.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Yang CY, et al. CLEC4F is an inducible C-type lectin in F4/80-positive cells and is involved in alphagalactosylceramide presentation in liver. Plos one. 2013;8:e65070. doi: 10.1371/journal.pone.0065070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, et al. Sialylation on O-glycans protects platelets from clearance by liver Kupffer cells. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:8360–8365. doi: 10.1073/pnas.1707662114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nature reviews. Immunology. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delputte PL, et al. Porcine sialoadhesin (CD169/Siglec-1) is an endocytic receptor that allows targeted delivery of toxins and antigens to macrophages. Plos one. 2011;6:e16827. doi: 10.1371/journal.pone.0016827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tkalcevic J, et al. Impaired immunity and enhanced resistance to endotoxin in the absence of neutrophil elastase and cathepsin G. Immunity. 2000;12:201–210. doi: 10.1016/S1074-7613(00)80173-9. [DOI] [PubMed] [Google Scholar]
- 33.Michalopoulos GK, DeFrances MC. Liver regeneration. Science (New York, N.Y.) 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 34.Taub R. Liver regeneration: from myth to mechanism. Nature reviews. Molecular cell biology. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 35.Ding BS, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J, et al. Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science (New York, N.Y.) 2014;343:416–419. doi: 10.1126/science.1244880. [DOI] [PubMed] [Google Scholar]
- 37.Christensen JG, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer research. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 38.Huh CG, et al. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossanen JC, et al. Chemokine (C-C motif) receptor 2-positive monocytes aggravate the early phase of acetaminophen-induced acute liver injury. Hepatology. 2016;64:1667–1682. doi: 10.1002/hep.28682. [DOI] [PubMed] [Google Scholar]
- 40.Heydeck D, et al. Interleukin-4 and -13 induce upregulation of the murine macrophage 12/15-lipoxygenase activity: evidence for the involvement of transcription factor STAT6. Blood. 1998;92:2503–2510. [PubMed] [Google Scholar]
- 41.Schif-Zuck S, et al. Saturated-efferocytosis generates pro-resolving CD11b low macrophages: modulation by resolvins and glucocorticoids. European journal of immunology. 2011;41:366–379. doi: 10.1002/eji.201040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kronke G, et al. 12/15-lipoxygenase counteracts inflammation and tissue damage in arthritis. Journal of immunology. 2009;183:3383–3389. doi: 10.4049/jimmunol.0900327. [DOI] [PubMed] [Google Scholar]
- 43.Bannenberg GL, Aliberti J, Hong S, Sher A, Serhan C. Exogenous pathogen and plant 15-lipoxygenase initiate endogenous lipoxin A4 biosynthesis. The Journal of experimental medicine. 2004;199:515–523. doi: 10.1084/jem.20031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brys L, et al. Reactive Oxygen Species and 12/15-Lipoxygenase Contribute to the Antiproliferative Capacity of Alternatively Activated Myeloid Cells Elicited during Helminth Infection. The Journal of Immunology. 2005;174:6095–6104. doi: 10.4049/jimmunol.174.10.6095. [DOI] [PubMed] [Google Scholar]
- 45.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature protocols. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 48.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics: MCP. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat Methods, 10.1038/nmeth.3901 (2016). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD011958.