Abstract
Multiple regulatory mechanisms including post-translational modifications (PTMs) confer complexity to the simpler genomes and proteomes of Mycobacterium tuberculosis (Mtb). PTMs such as glycosylation play a significant role in Mtb adaptive processes. The glycoproteomic patterns of clinical isolates of the Mycobacterium tuberculosis complex (MTBC) representing the lineages 3, 4, 5 and 7 were characterized by mass spectrometry. A total of 2944 glycosylation events were discovered in 1325 proteins. This data set represents the highest number of glycosylated proteins identified in Mtb to date. O-glycosylation constituted 83% of the events identified, while 17% of the sites were N-glycosylated. This is the first report on N-linked protein glycosylation in Mtb and in Gram-positive bacteria. Collectively, the bulk of Mtb glycoproteins are involved in cell envelope biosynthesis, fatty acid and lipid metabolism, two-component systems, and pathogen-host interaction that are either surface exposed or located in the cell wall. Quantitative glycoproteomic analysis revealed that 101 sites on 67 proteins involved in Mtb fitness and survival were differentially glycosylated between the four lineages, among which 64% were cell envelope and membrane proteins. The differential glycosylation pattern may contribute to phenotypic variabilities across Mtb lineages. The study identified several clinically important membrane-associated glycolipoproteins that are relevant for diagnostics as well as for drug and vaccine discovery.
Introduction
Tuberculosis (TB) is a major threat to public health, causing more than three deaths per minute globally. The causative agent is Mycobacterium tuberculosis (Mtb), and the TB crisis is exacerbated by the emergence of multi-drug-resistant (MDR) and extensively drug-resistant (XDR) Mtb strains. This situation highlights the urgent need for a comprehensive understanding of virulence and pathogenicity determinants of the M. tuberculosis complex (MTBC) to pave the way for the development of alternative TB control and prevention. Post-translational modifications (PTMs) including protein glycosylation are crucial in this regard. The unique Mtb glycoconjugates in the cell envelope are the predominant basis for host–pathogen interactions, antigenicity, and virulence determination and constitute one of the major components causing antimicrobial resistance (AMR) in Mtb1–6. Glycosylation in Mtb has mainly been detected in surface-exposed proteins and in some other membrane proteins7. The unique structure, antigenicity and essentiality of Mtb cell envelope glycoconjugates for mycobacterial growth provide opportunities for the development of novel drugs, vaccines, diagnostics and biomarkers1.
Mtb glycoproteins play a critical role in a number of biological activities including cell adhesion and invasion, protein stability, localization, and maintenance of protein conformation8–12. Other functions influenced by glycosylation are cellular signaling, AMR, immunomodulation, intracellular bacterial survival, biofilm formation, protein complex formation, antigenicity, pathogenicity and virulence8–12. Recently, it has been shown that protein glycosylation was associated with low cell envelope permeability and AMR in the multi-resistant Mycobacterium abscessus13. Furthermore, glycosylation protects proteolytically-sensitive cleavage sites, thereby maintaining the membrane-association of the protein by its lipid anchor, and may also be linked to protein export10,14.
The Mtb cell envelope is composed of an inner plasma membrane, a cell wall core with an outer mycomembrane, and an outermost layer, known as the capsule composed of polysaccharides, lipids and proteins1. The cell wall core is composed of peptidoglycan (PG) covalently linked via phosphoryl-N-acetylglucosaminosylrhamnosyl to arabinogalactan (AG), which in turn is esterified to α-alkyl, β-hydroxy long-chain mycolic acids, forming the mycolyl arabinogalactan-peptidoglycan (mAGP) complex1,15. This complex is essential for bacterial viability and is the basis of susceptibility and resistance to many anti-TB drugs including ethambutol (EMB) and ethionamide (ETH)12,16. Mannose-capped lipoarabinomannan (LAM), one of the key Mtb virulence factors, is a surface-exposed lipoglycan anchored to the inner and outer membranes via a mannosyl phosphate inositol17. The soluble components of the cell envelope include free lipids, proteins, LAM, and phosphatidylinositol mannosides (PIMs), which are signaling effector molecules in bacterial pathogenesis and disease processes15. The nature and amounts of the mycomembrane and capsular material vary among Mtb isolates and is likely to impact significantly on the pathogen phenotype and outcome of the pathogen-host interaction9,18.
PG is a polymer of alternating N-acylated muramic acid (MurNac) and N-acetylglucosamine (GlcNac) residues linked in a β (1 → 4) configuration with cross-linked peptides of varying composition attached to the muramyl moieties19,20. PG glycosyltransferases use the lipid-linked donor precursor for the synthesis of oligo-β-(1 → 4)-[GlcNAc-β-(1 → 4)-MurNAc(peptide)] glycan strands21. In contrast to most other bacteria, muramic acid moieties are N-glycolylated (oxidized) in mycobacteria15. In Mtb, MurNGly, MurNAc and Mur residues are present in the precursor pool and in the PG20.
The membrane-bound glycosyltransferases or oligosaccharyltransferases (OSTs) catalyze the transfer of the monosaccharide moiety of an activated nucleotide-sugar substrate from lipid carriers to acceptor substrates, such as monosaccharides, oligosaccharides, proteins, lipids, small organic molecules, and DNA, and hence produce a wide variety of biomolecules8,22. Glycosidases are enzymes involved in both the degradation of glycans and the removal of monosaccharides to form intermediates that are acted upon by glycosyltransferases for the biosynthesis of glycans22.
Campylobacter jejuni and Neisseria meningitidis have well-characterized bacterial N-linked and O-linked glycosylation systems, respectively23. In N-linked protein glycosylation, an oligosaccharide is transferred by N-OST from a lipid donor to asparagines (N) located within the well-recognized consensus sequence D/E-Y-N-X-S/T and N-X-S/T (Y≠P, X≠P) of proteins24,25. Bacterial O-OSTs are responsible for the reversible attachment of glycans to hydroxyl groups of serine (S), threonine (T) and tyrosine (Y) residues, with no apparent sequence specificity26,27.
Glycoproteomics is likely to identify Mtb virulence factors because glycoproteins on the bacterial cell envelope are used by mycobacteria to enable their entry into the primary human host cell, the macrophage28. It has been proposed that Mtb interacts with mannose receptors (MRs) on host cells via mannosylated proteins to enter the macrophages29. Despite the vital importance of these proteins in Mtb pathogenesis, our current knowledge of Mtb glycoproteins is still limited, and only a few secreted and cell wall-associated glycoproteins have been described to date8,28,30,31. Previous studies have used laboratory strains as model systems to study glycosylation in Mtb. However, only a few sub-groups within the genetically conserved MTBC appear to cause extensive outbreaks with different clinical presentation and AMR32–35. In this study, we employed qualitative and quantitative mass spectrometry and bioinformatics to explore the glycoproteomic patterns of clinical isolates from four lineages of the MTBC, lineages 3, 4, 5 and 7, to investigate the role of protein glycosylation in Mtb adaptation, survival and AMR.
Our study reveals the presence of a number of glycoproteins that play roles in MTBC virulence and pathogenesis. These include proteins involved in pathogen-host interaction, transport and biosynthesis of MTBC cell envelope components, and drug efflux pumps, which are attractive pharmacological targets. Furthermore, we found quantitative differences in glycosylation patterns among the different lineages of MTBC that may potentially contribute to explaining their phenotypic characteristics.
Results
Abundance of both O- and N-glycosylation profile among members of the MTBC
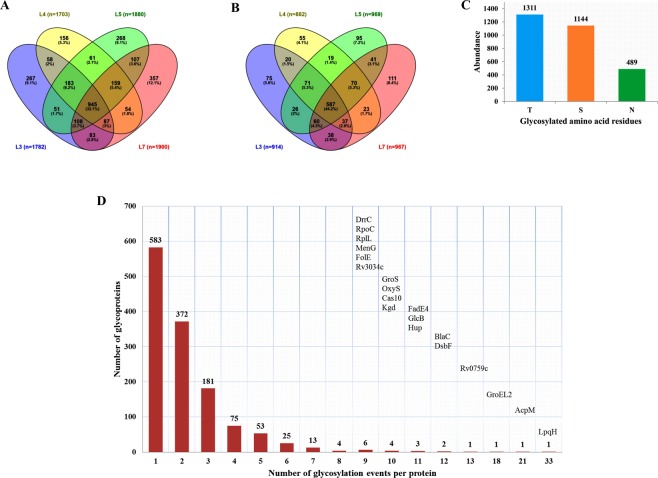
After filtering the data for potential contaminants and hits to the reverse database, the peptide intensities were log2-transformed. For protein identification, the data was further filtered using localization probability of 0.7, PEP of 0.05 and having valid values in at least one sample resulted in 2944 class-I glycosylation events derived from 1325 unique proteins in MTBC strains representing lineages 3, 4, 5 and 7. The term “glycosylation event” is used to avoid confusion when a single glycosylation site is glycosylated by more than one type of glycan residues. O-glycosylation constituted 2455 (83%) of the events identified (1311 events at T and 1144 events at S residues) and the remaining 489 sites (17%) were glycosylated at N residues (Fig. 1A,C, Supplementary Table S1). Comparative glycoproteomic analysis among the four MTBC lineages revealed that 945 (32.1%) of the total glycosylation events identified were shared amongst the four lineages (Fig. 1A). Comparison at the level of unique glycoproteins revealed that 44.2% of the glycoproteins were shared among the four lineages, irrespective of the glycosylation site and glycan residues (Fig. 1B).
Figure 1.
Abundance of glycosylation and glycoproteins in MTBC. Venn diagram showing the number of glycosylation events identified among the four lineages (N = 2944) (A), and the number of glycoproteins identified among the four lineages (N = 1325) (B), the number of N- and O-glycosylation events identified (N = 2944) (C), and number of glycosylation events identified per individual glycoprotein (D).
Among the 57 most common naturally occurring glycan residues, deoxyhexoses (DeoxyHex) was the most frequently identified glycan residue in our search, followed by Hept, pent, Hex, HexN, HexNac/GlcNac, MurNGly and MurNac (Supplementary Table S3). We also identified sugar molecules attached to both ADP and UDP in comparable proportions, which may contribute to an activated nucleotide-sugar substrates for OST (Supplementary Table S3).
The glycosylation events identified per protein ranged from 1 to 33 (Fig. 1D, Supplementary Table S2). The lipoprotein LpqH was found to be the most highly glycosylated protein with no less than 33 events, hosting 24 events on T and 9 events on S residues (Fig. 1D). Notably, all glycan residues found on LpqH were composed of solely hexoses, while a cocktail of glycan residues were observed in other proteins harboring many glycosylation events (Supplementary Table S1). Among the 33 glycosylation events detected on LpqH, 20 events were common across all four lineages. Other highly glycosylated proteins included AcpM, GroeL2, BlaC, DsbF, FadE4 and HupB (Fig. 1D, Supplementary Table S2). Several hypothetical proteins were also glycosylated.
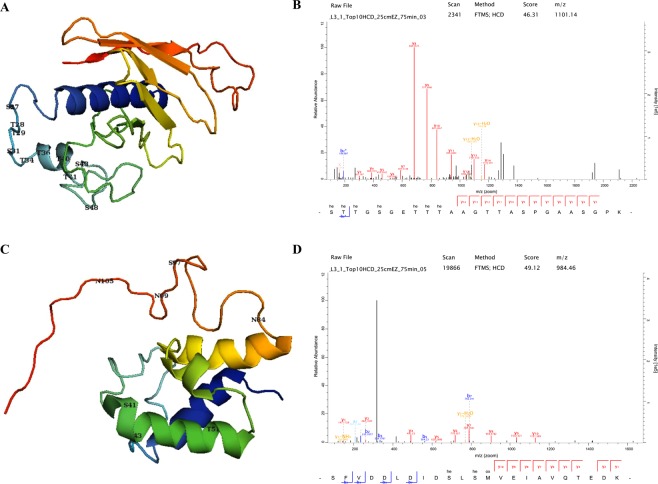
LpqH and AcpM have glycosylation sites concentrated in the interacting domains
The glycosylation sites in LpqH clustered in the N-terminus, densely located between residues 27–48 of the 159 amino acid protein (Fig. 2A). The amino acid residues from 41–60 are known to be involved in the binding of LpqH with the host MR. The four sites, T40, T41, S43 and Ser48 are located in this binding domain region of LpqH. In AcpM, three glycosylation sites in the helix S41, S43 and T51 are found in the carrier protein (CP) domain profile (Fig. 2C).
Figure 2.
The 3D models, acetylation sites and representative spectra of LpqH (A,B) and AcpM (C,D). The glycosylation sites in LpqH are clustered in the N-terminus between residues 27–48 of the 159 amino acid protein (A). The four sites, T40, T41, S43 and S48 were located in this binding domain of LpqH. In AcpM, three sites in the helix, S41, S43 and T51, were found in the carrier protein (CP) domain profile (C). (he = hexose).
No apparent amino acid sequence specificity for Mtb glycosyltransferases
Comparing the 31-mer unique sequences of all peptides containing a glycosylation site by WebLogo yielded a “consensus” sequence atlas (Fig. 3). The distribution of the amino acids flanking the modified site showed a relatively high propensity for R, L, A, V, P and G residues (Fig. 3).
Figure 3.
Glycosylation motif analysis. The N- and O-glycosylation motif generated from the high confidence identification indicates a higher likelihood of basic R, hydrophobic P, A, V and L, interspersed with Polar G; and some hydrophilic S and T around the O-glycosylation site. In addition, the glycosylation motifs seem to cluster predominantly at the N-terminus as indicated by the black boxes on the left. The height of each amino acid indicates its relative frequency at that specific position.
The glycolipoproteins and glycoproteins identified are involved in diverse biological functions
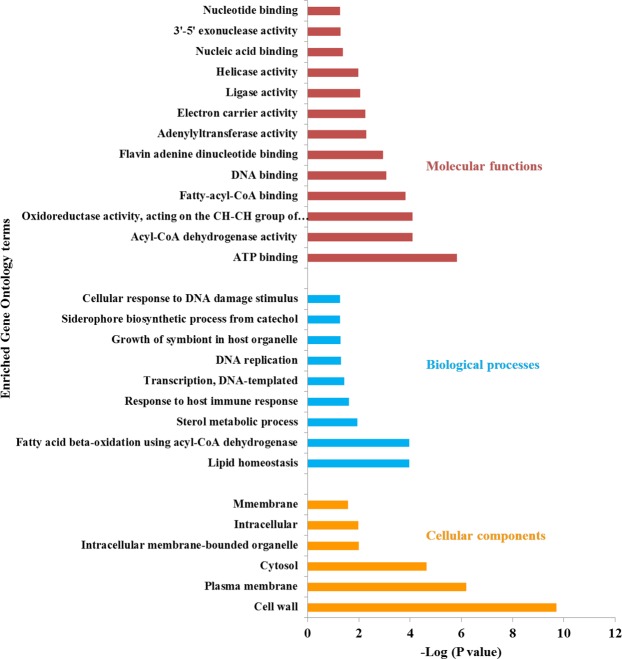
Based on the Gene Ontology (GO) analysis, fatty acid metabolism and lipid homeostasis, growth of symbiont in the host cell and responses to the host immune system were highly enriched biological processes. The cell wall and the plasma membrane were the two highly enriched cellular components of the glycoproteins identified. The molecular functions include ATP binding, oxidoreductase activity, acyl-CoA dehydrogenase activity, fatty-acyl-CoA binding, helicase activity, DNA binding, electron carrier activity and ligase activity (Fig. 4). The roles of the Mtb glycoproteins identified are summarized in Table 1. The non-glycosylated complement, however, encompassed proteins with functions and localization primarily in the cytoplasm. Furthermore, the GO analysis of uniquely identified glycoproteins provided strain-specific enrichment of biological processes, molecular functions (Supplementary Table S6).
Figure 4.
Gene Ontology analysis of Mtb glycoproteins. The gene ontology analysis showed that the majority of the glycoproteins identified were localized in the cell wall and plasma membrane while lipid homeostasis, fatty acid metabolism, and response to the host immune system were among the enriched biological processes.
Table 1.
Virulence-associated membrane-bound glycoproteins, proteins involved in regulation, antimicrobial resistance (AMR) and chaperone proteins identified in MTBC.
| Function | Process | Glycoproteins involved |
|---|---|---|
| Cell envelope synthesis | Mycolic acid synthesis | AcpM, MmaA1, MmaA2, MmaA3, PcaA, Pks13, FbpB and FbpC |
| PDIM synthesis and transport | PpsA, PpsC, PpsD, PpsE, PapA1, FadD26, FadD28, LppX, DrrC and MmpL7 | |
| PG synthesis | PBPs) PbpA (Rv0016c), PbpB (Rv2163c), PonA1 (Rv0050) and LdtA (Rv0116c), MurA, MurE, MurF, LprQ, FtsW, MviN, GlmS, GlmM, DacB1 and Wag31 | |
| Capsule biosynthesis | GlgM (Rv1212c), GlgB (Rv1326c), GlgE (Rv1327c), TreZ (Rv1562c) and MalQ (Rv1781c) | |
| AG | AftD (Rv0236c), DprE1, EmbC and EmbR | |
| Lipoglycans (LM, LAM and PI) | PimB (Rv2188c), EmbC, Rv1459c and Rv2181 | |
| Membrane transport proteins | Sec | SecA1, SecY, SecD, SecE2, SecF |
| Tat | TatB | |
| MmpL lipid transporters | MmpL1, MmpL3, MmpL4, MmpL5, MmpL6, MmpL8, MmpL9, MmpL10, MmpL11, MmpL12 and MmpL13b | |
| ATP-binding cassette (ABC) | DrrC, DppA, DppC, DppD, FecB, UgpC, UgpE, ProZ, CydD, MalQ, Rv2326c, Rv2041c, Rv1680, Rv3197, Rv0987, Rv1281c, Rv3092c, Rv1747, Rv1273c, Rv1739c, Rv2564, Rv0073 | |
| Type-VII secretion | EccA1, EccB1, EccCb1, EccA2, EccB2, EccC2, EccD3, EccB3, EccC4, EccB4, EccA5, EccC5, | |
| Others | CpnT, NanT, IrtA, IrtB, ArsC | |
| MCE family proteins | Mce2A, Mce2D, Mce1E/LprK, Mce3R, Mce1A, Mce4C, Mce1C, Mce3C, Mce2R, Mce1R, Mce1B, Mce2F, Mce2B, Mce3D and Apa* | |
| Regulatory proteins | DevS-DosT/DosR, PhoR, WhiB3, WhiB4, WhiB5, WhiB7, TcrA, PrrA/PrrB, MtrA/MtrB, KdpD, KdpC, MoxR3, NarL, EmbR, PdtaR, GlnB, Mce1R, KstR, BlaR, BlaI and OxyS, Rv1353c, Rv0890c, Rv3095, Rv0494, Rv0043c, RamB, Rv0081, Rv0339c | |
| Chaperones | GroS, DnaK, GroeL1, GroeL2, ClpB, ClpX, Hsp | |
| Role in AMR | BlaC, KatG, RpoC, KasA, AhpD, FadE24, AcpM, IniB, IniC, EthA, OpcA, Wag31, RpoB, EmbR, EmbC, FabG1, RpsL, Mdh, Ndh, Alr, MtrAB, Rv2994, Rv0194, LprG, GyrA and GyrB | |
| Potential drug targets | Mur enzymes, DrrC, PknD, MmpL3, GlgB, GlgE, Hpt, PbpA, PbpB, PonA1 and LdtA |
Glycosylated proteins are involved in 14 specific metabolic pathways
Through protein-protein interaction (PPI) network analysis, 14 highly interconnected clusters were identified (Fig. 5). Most of the interacting glycoproteins identified were part of common pathways involved in fatty acid and lipid metabolism, protein synthesis, pathogen-host interaction, PG, AG, mycolic acid and capsule biosynthesis, stress responses, two-component systems (TCS), energy metabolism, and DNA replication repair and recombination (3R) (Fig. 5).
Figure 5.
Protein-protein interaction networks of identified glycoproteins generated by Cytoscape. Networks are involved in lipid Metabolism & cell wall synthesis (A,D,E,H,I,J), protein synthesis (B), host-pathogen interaction (C), chaperone proteins (G), regulators (F,K), phthiocerol dimycocerosate (PDIM) synthesis (L), (sugar) transporters (M) and DNA replication, repair and recombination (N).
Glycolipoproteins involved in pathogen-host interaction
Our GO analysis a showed that most of the glycoproteins identified were localized in the cell wall and plasma membrane while lipid homeostasis, fatty acid metabolism, and response to the host immune system were among the biological processes enriched (Fig. 4). Lipoproteins were amongst the highly glycosylated Mtb proteins identified in this study, and are known to be involved in colonization, invasion, evasion of host defence and immunomodulation, cell envelope biogenesis, transport across membrane, nutrient acquisition, adhesion, cell invasion and initiation of inflammatory processes (Table 2)13,14,36–38. These glycoproteins include the LpqH, the MCE-family proteins, Apa, Heparin-binding hemagglutinin (HbhA) and LprG.
Table 2.
Representative biological activities elicited by glycolipoproteins identified from MTBC.
| Role or function | Glycolipoprotein(s) |
|---|---|
| Antigenicity | LpqH, LprG, LppX |
| Adhesion and cell invasion | LpqH, LprG, LprK, LprN, LppA, LpqG, LppX, MCE |
| Required for growth | LpqH, LprK, SugA, LpqY, LppY, LpqB |
| Signal transduction | LprF, LprA, LprG, LppR, LppX, LpqB |
| Role in AMR | LprG, BlaC |
| Cell wall metabolism | PbpB, PbpA, PonA1, LprQ, LprK, LppW, LppX, LpqY, LpqB |
| ACB transport system | UgpE, UgpC, Rv2041c, LpqY, MalQ, DppA, FecB |
| Degradation | LpqP, LpqI, Rv2672, LpqL |
| Other enzymes and metabolic activities | GgtB, Rv0526, DsbF, LppZ, LpqD, SodB, Rv0526 |
| Unknown function | Rv3693, Rv0679c, LppG, LpqU, LpqJ, LppO |
Glycosylation of proteins involved in MTBC cell envelope biogenesis
Membrane-associated proteins involved in lipid and fatty acid metabolism, cell envelope biosynthesis, pathogen-host interaction, transport, transcriptional regulation, and chaperone functions were also glycosylated (Fig. 5). After LpqH, the meromycolate extension acyl carrier protein AcpM was the second highly glycosylated protein identified with 21 glycosylation events. AcpM is involved in mycolic acid biosynthesis, a major component of the Mtb cell wall. Other glycoproteins involved in mycolic acid synthesis include methoxy mycolic acid synthases (MmaA1, MmaA2, MmaA3), mycolic acid synthase PcaA, polyketide synthases Pks13, enzymes involved in the synthesis of the Mtb cell wall components (PpsA, PpsC, PpsD, PpsE, PapA1, Rv2951c, FadD26, FadD28, LppX, DrrC and MmpL7), beta-ketoacyl-ACP synthases (KasA, KasB), mycolyltransferases (FbpB and FbpC), mycolic acid biosynthesis a protein FabG1, penicillin-binding glycoproteins (PBPs) (PbpA, PbpB, PonA1 and LdtA), proteins involved in PG biosynthesis (MurA, MurE, MurF, LprQ, FtsW, MviN, GlmS, GlmM, DacB1 and Wag31), AftD, EmbC, enzymes involved in the biosynthesis of alpha-D-glucan (GlgM, GlgB, GlgE, TreZ and MalQ), enzymes involved in biosynthesis of lipoglycans (PimB, Rv2181, MgtA, Ppm1 and Rv1459c).
Other clinically important glycoproteins identified
Clinically important glycoproteins include BlaC, chaperone proteins, TCS proteins, ESX secretion system proteins and other transporter proteins. Mtb BlaC was glycosylated at 12 sites, while the chaperones GroEL2 and GroS were found to have 18 and 10 glycosylation sites, respectively.
Glycosylation of cytoplasmic proteins in MTBC
Cytoplasmic proteins involved in translation and DNA metabolism were glycosylated (Fig. 5A,B). The cytochrome P450 proteins were also glycosylated.
MTBC strains exhibit lineage-specific glycoproteomic profiles
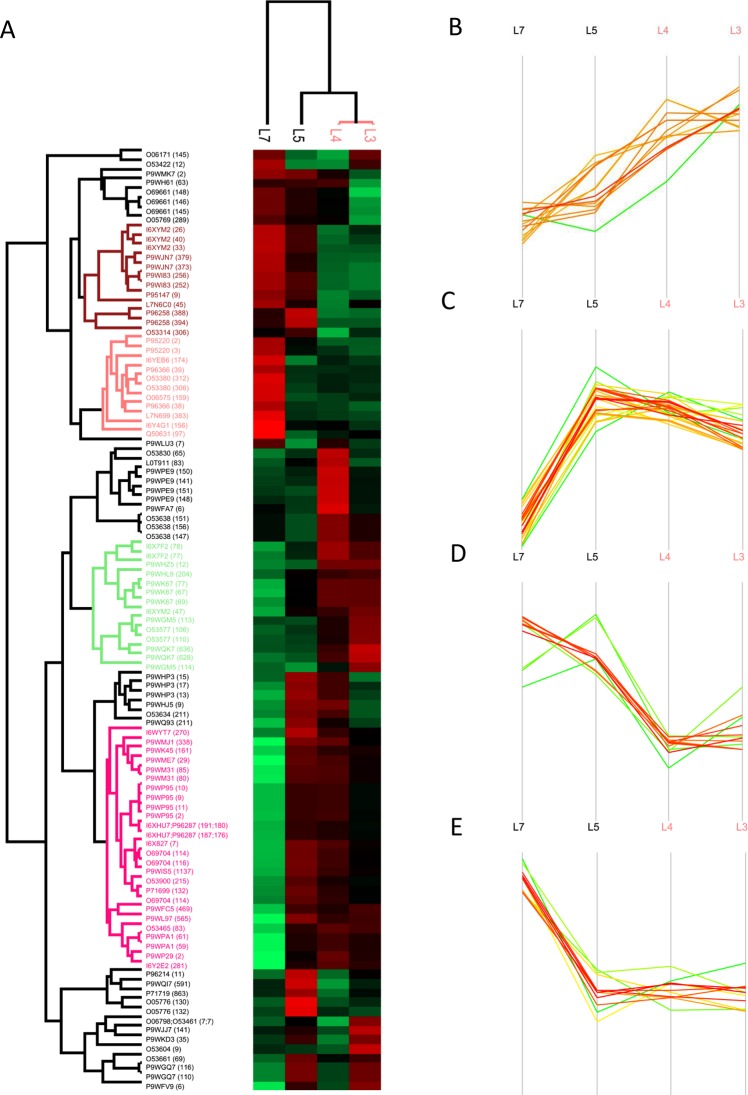
The GO analyses of exclusively identified glycoproteins provided strain-specific enrichment of biological processes and molecular functions (Supplementary Table S6). Among the 2944 glycosylation events detected, 1010 had valid values in at least six valid LFQ intensity values from the total of 12 biological replicates (50%) and were thus subjected to further quantitative analysis. The missing values were imputed from the normal distribution and the log2-transformed data was normalized to Z-scores for further statistical testing. Multiple sample test analysis at a P < 0.05 level of significance revealed that 101 sites on 67 proteins were differentially glycosylated (differential abundance of a peptide/protein glycosylated at a specific site) between the four MTBC lineages studied (Fig. 6A, Supplementary Tables S4 and S5). Notably, most of the differentially glycosylated proteins (43/67: 64%) were located in the cell wall and cell membrane or possess a membrane component. These proteins belonged to different functional categories including lipid metabolism, cell wall and cell processes, virulence, detoxification and adaptation, and hypothetical proteins (Supplementary Table S5). Clinically important differentially glycosylated proteins include the lipoarabinomannan carrier protein LprG, chaperone proteins GroEL1, class A β-lactamases BlaC, mammalian cell entry (Mce)-family protein Mce2D, peroxidase BpoB, penicillin-binding glycoprotein PbpB, and a number of proteins involved in fatty acid and lipid metabolism (Supplementary Table S5).
Figure 6.
Profile of the 101 differentially glycosylated proteins in MTBC. Hierarchical clustering of differentially glycosylated proteins (A), proteins hyper-glycosylated in lineage 3 and lineage 4 strains (B), proteins hyper-glycosylated in lineage 3, lineage 4 and lineage 5 strains (C), proteins hyper-glycosylated in lineage 5 and lineage 7 strains (D), proteins hyper-glycosylated in lineage 7 strain (E).
In the hierarchical clustering of the differentially glycosylated proteins, the modern lineages (lineage 3 and lineage 4 strains) clustered together, separated from the ancient lineages (lineage 5 and lineage 7 strains) (Fig. 6A). Four separate clusters of proteins with a particular glycosylation profile were generated. The first cluster included 14 hyper-glycosylated proteins (having a significantly higher number of glycosylation events at a specific position in a protein than the average) in lineage 3 and lineage 4 strains, encompassing LppW (S67, N69, T77), UvrA (T628, N636), PurK (S204), PPE42 (S12), FadE35 (S106, S110), LdtA (T147, S151, S156), DsbF (T47), NarL (S113, T114) and Rv3483c (T77, S78) (Fig. 6B). Cluster two included 26 glycoproteins such as Mce2D (S270), DagK (S2), LprG (N161), UvrC (T469) and VapC45 (T83) that were hyper-glycosylated in lineage 3, lineage 4 and lineage 5 strains (Fig. 6C). Membrane proteins DsbF (S26, T33, T40) and Rv0412c (388, 394), Mrp (S373, T379), ATPase MoxR3 (S306) and PknA (T252, N256) were hyper-glycosylated in ancient lineages (lineage 5 and lineage 7 strains) (Fig. 6D). Cluster four included 11 glycoproteins, such as FadD9 (T97), DacB1 (T306, T312), FadD34 (T383), short chain type dehydrogenase/reductase Rv2766c (174) and peroxidase BpoB (N159), that were hyper-glycosylated in lineage 7 strains (Fig. 6E). Virulence factors such as PbpB (S83), GroeL1 (T141, T148, S150, S151), VapC10 (S6), VapB11 (S7) and the transcriptional regulatory protein Rv0818 (S65) were hyper-glycosylated in lineage 4 strains. The alanine rich protein Rv3863 (S11), the transmembrane ATP-binding protein ABC transporter protein Rv2326c and a polyketide synthase involved in sidrophore biosynthesis MbtD (T591) were hyper-glycosylated in lineage 5 strains (Supplementary Table S4).
Discussion
Our analysis identified with high confidence a total of 2944 glycosylation events on 1325 Mtb unique proteins. To our knowledge, the discovery of such a large number of glycosylation sites in these four clinical strains from different MTBC lineages is unprecedented. About 83.4% of the glycosylation events were localized on S and T residues, indicating a possible interplay with phosphorylation. It has been reported that different glycosylation events may occur on the same S and T residues of the protein or competitively at adjacent or residues in close proximity, and hence potentially allow control of cellular signaling27. The study provides the first evidence on N-linked protein glycosylation in Mtb and Gram-positive bacteria. Protein glycosylation occurred at numerous sites on surface-exposed proteins with no apparent amino acid sequence specificity (Fig. 3)7. As previously reported, there is a relatively high propensity for R, A, P, L, G, V, S and T flanking the modified sites in a significant portion of the glycosylation sites mapped11,27,30 (Fig. 3). However, a number of suggested signature motifs were identified in nearly 17% of the events and R was enriched between the −8 and +8 positions in contrast to the D/E-Y-N-X-S/T (Y, X #P) motif proposed for N-glycosylation24. This difference might partly be due to the diversity of the glycan residues analyzed and the high degree of specificity for both their donor and acceptor substrates in the glycosyltransferases39. Comparative analysis revealed that only 32.1% of the glycosylation events and 44.2% of the glycoproteins were shared among the four lineages. The higher versatility at the level of PTMs may indicate the power of PTMs in explaining the phenotypic variability among MTBC than the proteomic studies.
DeoxyHex was the most frequently identified glycan residue in our search, followed by Hept, pent, Hex, HexN, HexNac/GlcNac, MurNGly and MurNac (Supplementary Table S3). In bacteria, 6-deoxy-hexoses, like fucose and rhamnose, are important components of cell surface glycans40. The pentose sugars arabinose and galactose are components of the heteropolysaccharide, AG, which serves to connect PG with the outer mycolic acid layer19. Bacterial heptosyltransferases are reported to be involved in O-glycosylation of autotransporters using ADP-heptose41. The presence of frequently occurring glycan residues attached to lipoproteins, extracellular polysaccharides (EPSs) and glycoproteins might alter the structure and function of these biomolecules in particular and bacterial physiology in general40. We identified both ADP- and UDP linked to different sugar molecules to form an activated nucleotide-sugar substrates for OST (Supplementary Tables S1 and S3). Most publications reported that only UDP-linked sugars were the substrates for OST8,22, while other reports showed that a particular OST, heptosyltransferase, used ADP-heptose as an activated nucleotide-sugar substrate41.
The outermost layer of the Mtb cell envelope is a major determinant of virulence and pathogenicity, and is mainly composed of proteins, polysaccharides and small amount of lipids12,42. It acts as a permeability barrier of the cell envelope, promoting the phagocytosis of Mtb43, maintaining cell integrity, regulating phagosome maturation44 and playing diverse roles in the pathogen-host interactions42,43. The gene ontology analysis showed that the majority of the glycoproteins identified were localized in the cell wall and plasma membrane while lipid homeostasis, fatty acid metabolism, and response to the host immune response were among the biological processes enriched. Besides, the PPI network analysis showed that most of these cell-envelope associated glycoproteins are involved in pathogen-host interaction and fatty acid/lipid metabolism. These cell envelope-associated glycoproteins have been shown to have a vital role in Mtb virulence and pathogenesis (reviewed in12).
Lipoproteins were amongst the highly glycosylated Mtb proteins identified in this study. Lipoproteins are a functionally diverse class of membrane-bound proteins involved in colonization, invasion, evasion of host defence and immunomodulation, cell envelope biogenesis, transport across the membrane, nutrient acquisition, adhesion, cell invasion and initiation of inflammatory processes (Table 2)13,14,36. The lipoprotein LpqH was the most densely glycosylated lipoprotein detected, with 33 N-terminally clustered O-glycosylation events, where all glycan residues were hexoses (Supplementary Table S1). Notably, these sites were densely located between residues 27–48 within the 159 amino acid protein. Some of the glycosylation sites have previously been reported as part of the MR binding domain of LpqH, as shown in the 3D model (Fig. 2)30,45. Three of the sites, T41, S43 and S48, were part of a mature protein fragment (residues 41–60) that was reported to prevent uptake of Mtb by macrophage-like U937 cells46. Altering the glycosylated Ser residues in LpqH have been shown to affect binding affinity and exposure to proteolytic cleavage10. LpqH, an immunodominant TLR2 agonist, is crucial for Mtb growth and multiplication in IFN-γ-activated macrophages as well as in IFN-γ-deficient mice47. Mannosylated LpqH is the major adhesin for the macrophage MR and DC-SIGN, and the mannose residue serves as an adhesin for binding to the host MR29.
Other groups of identified glycolipoproteins involved in pathogen-host interaction are the MCE-family of proteins36. These glycoproteins have an active role in disease development and in-host virulence12. A total of 14 glycosylation events were identified on proteins expressed from the four Mtb mce operons (mce1, mce2, mce3 and mce4). The invasion-/adhesin-like MCE family glycolipoproteins encoded by mces are located at the cell surface of Mtb and possibly involved in entry and survival inside macrophages48.
A number of other clinically important glycoproteins were identified. The cell surface glycoprotein Apa binds to DC-SIGN and surfactant protein, facilitates colonization and invasion of host cells49. Changes in the glycosylation pattern of Apa lead to a reduced stimulatory T-lymphocyte response, exhibiting the biological role of the glycan moiety50. Glycosylation is also required for proper localization of superoxide dismutases (SodB)51. The immunogenic glycoproteins MPT64 and Apa are virulence factors involved in Mtb infection of human cells and is a promising candidate for a subunit-based anti-TB vaccine12,52. Heparin-binding hemagglutinin (HbhA) glycoprotein mediates adherence to epithelial cells and is required for extrapulmonary dissemination of Mtb53. The lipoprotein LprG is another glycolipoprotein that blocks host cell phagosome-lysosome fusion, and is required for full Mtb virulence54.
Glycoproteins associated with drug efflux pumps, drug-hydrolyzing enzymes, or capable of altering Mtb cell wall permeability mediates the development of AMR (reviewed in12). These include proteins like the mycobacterial membrane protein large (MmpL) proteins, daunorubicin-dim-transport integral membrane protein ABC transporter (DrrC), class a beta-lactamase (BlaC) and LprG (Table 1). DrrC, Rv0194, Rv2994, Rv1273c and a number of MmpL glycoproteins are efflux pumps for anti-TB drugs, contributing to AMR55,56. In addition to a role in drug resistance, MmpLs are involved in the export of cell wall associated lipids and siderophores, and are attractive pharmacological targets57,58. BlaC hydrolyzes nitrocefin and other β-lactams, thereby increasing Mtb resistance towards different classes of β-lactam antibiotics4. LprG controls cell wall permeability and efflux of drugs, and therefore plays a role in Mtb susceptibility to first-line anti-TB drugs5.
The study identified a number of membrane-associated glycoproteins involved in cell envelope biosynthesis and drug efflux pumps, which are potential Mtb drug targets (Table 1, Fig. 5). AcpM was second most densely glycosylated protein involved in mycolic acid biosynthesis, one of the major components of the Mtb cell wall. Glycosylation sites Ser41, Ser43 and Thr51 were detected within the AcpM CP domain profile. Importantly, one of the glycosylation sites identified (Ser41) is the binding site for 4′-phosphopantetheine, an activator of AcpM59. Other glycoproteins involved in mycolic acid synthesis include MmaA1, MmaA2, MmaA3 and PcaA, Pks13, KasA, KasB and FabG1. Bacilli lacking all mycolic acid methyltransferases are viable but highly attenuated and hyperinflammatory in mice60. Pks13 catalyzes the last condensation step of mycolic acid biosynthesis and is essential for the mycobacterial survival61. Glycoproteins FbpB (Ag85B) and FbpC (Ag85c) also possess a mycolyltransferase activity62. These glycoproteins help to maintain the Mtb cell wall integrity by catalyzing the transfer of mycolic acids to cell wall AG, and through the synthesis of the virulence factor cord factor (trehalose 6,6′-dimycolate, TDM)62. Furthermore, FbpB and FbpC are T- and B-cell antigens and may have an application in sero-diagnostics63.
Penicillin-binding glycoproteins (PBPs) PbpA, PbpB, PonA1 and LdtA are transpeptidases involved in the synthesis of cross-linked PG that is part of the cell wall biogenesis64. Other essential glycoproteins involved in PG biosynthesis include MurA, MurE, MurF, LprQ, FtsW, MviN, GlmS, GlmM, DacB1 and Wag3165. Glycoproteins involved in PG biosynthesis, such as Mur enzymes and PBPs, are potential antibiotic targets65. Alpha-(1 → 3)-arabinofuranosyltransferase (AftD) is involved in the biosynthesis of the AG region of the mAGP complex, an essential component of the mycobacterial cell wall19. EmbC is involved in the polymerization of arabinose into the arabinan of the mycobacterial cell wall AG and is linked to resistance to EMB66.
Polyketide synthases (PpsA, PpsC, PpsD, PpsE), PapA1, Rv2951c, FadD26 and FadD28 are multifunctional enzymes involved in the synthesis of the Mtb cell wall component, PDIM and other lipids67, while the glycolipoproteins LppX, DrrC and MmpL7 are required for the translocation and localization of PDIM in the cell wall68. PDIM comprise of a number of virulence-enhancing lipids that act as defensive, offensive, or adaptive effectors of virulence69. Inactivation of mycobacterial pps and drr operons has been linked to defects in PDIM synthesis and secretion, respectively70. PknD, a regulator of MmpL7, has been proposed to be a potential anti-TB drug target71.
Glycosyltransferases such as GlgM, GlgB, GlgE, TreZ and MalQ are enzymes involved in the biosynthesis of alpha-D-glucan, a constituent of Mtb capsular polysaccharides with D-arabino-D-mannan (AM) and D-mannan1,42. These enzymes are required for Mtb virulence72. GlgE-mediated 1,4 α-glucan synthesis has been implicated in in vitro lysosomal stress and can potentially be exploited for killing intracellular Mtb73. The Thr10 glycosylation site in GlgE has been shown to be a regulatory kinase substrate and a validated anti-TB drug target74. GlgB is a potential target for inhibitors75. Glycosylated mannosyltransferases PimB and Rv2181 are involved in the biosynthesis of lipoglycans LM, LAM and phosphatidylinositol (PI)76. Mannosyltransferases MgtA, Ppm1 and Rv1459c are involved in the synthesis of immunomodulatory LM and LAM via alpha-(1 → 6)-mannopyranosyltransferase activity77. A number of glycosylated fatty acyl-AMP ligases that have been shown to play a role in cell wall biosynthesis, production of complex lipids and growth78 were identified. As discussed above, glycosylation is involved in regulating the activity of different enzymes. In this study, identification of glycosylated glycosyltransferases (with rare abundance) may play a role in regulating its function as an enzyme79. There are reports on auto-glycosylation mediated activation of glycosyltransferases in eukaryotes80–82.
Other clinically important glycoproteins identified include chaperone and TCS proteins. The differential expression of chaperone glycoproteins, such as GroeL2 and GroS, in response to heat shock have previously been reported12. TCS regulate various aspects of mycobacterial physiology, including virulence, dormancy, persistence, and drug resistance83. The glycoprotein PhoPR regulates multiple virulence-associated processes in Mtb, including the biosynthesis of polyketide-derived lipids and acyltrehaloses. The inactivation of acyltrehaloses attenuates Mtb sufficiently to make it a possible live vaccine candidate12,16. The DosR/WhiB3 regulon is associated with hypoxia and redox adaptation, while WhiB3/PhoP is involved in cell wall lipid biosynthesis84. The DevS/DosR regulon is required for full Mtb virulence and is involved in regulating stress, dormancy and hypoxia85.
Twenty glycosylation events on proteins belonging to the specialized ESX secretion system components, including the crucial T-cell antigen ESAT-6, were detected. The ESX secretion system is essential for full Mtb virulence (ESX-1) and physiological processes (ESX-3)86. Five proteins involved in the general secretion (Sec) pathway and a twin-arginine translocation pathways (TatB) were also found to be glycosylated. These specific proteins are essential for bulk export of proteins in Mtb86. CpnT, the first autotransporter-like protein to be identified in Mtb, was glycosylated at a domain that is required for the membrane localization of this protein87. Our former study showed that glycoproteins including LpqH, AcpM, GroEL1, GroEL2, DnaK, Pks13, KatG, LprK, SecA1 and a number of proteins involved in lipid metabolism and protein synthesis were highly acetylated in Mtb88, which might indicate the interaction among different PTMs in fine-tuning specific cellular processes.
A recent report demonstrated a mechanism for co-regulation of Mtb cell wall synthesis and ribosome maturation (protein synthesis), and hence glycosylation of proteins involved in these two processes (Fig. 5A,B) may have a regulatory role89. Evidence for glycosylation of DNA-binding proteins (Dps) has been observed in Salmonella enterica in response to starvation and/or oxidative stress90. This is the first report on glycosylation of those cytosolic proteins. Glycosylation of cytochrome P450 has been demonstrated in eukaryotes (CYP2W1)91 and in viral cytochrome P450 (YP_143162)92. Glycosylation in this regard may enable the proper localization of cytochrome P45014,93. Cytochrome P450 plays a role in steroid metabolism, drug deactivation, fatty acid metabolism, xenobiotic detoxification and catabolism of exogenous compounds as a source of energy94. Fatty acid metabolism is a major source of carbon and energy in Mtb95.
The PTMs identified by in vitro culture may only reflect the mycobacterial phenotype in the absence of stress, which may not completely if at all overlap patterns during infection. So, further mapping the exclusive presence and/or differential abundance of Mtb glycoproteins naturally or during exposure to environmental stress or infection may contribute to elucidate the selective advantages and survival strategies adopted by a specific pathogen. This information is fundamental for any drug or vaccine discovery process12. More than 64% of the differentially glycosylated proteins were found to be cell envelope-associated proteins. These glycoproteins are reported to be involved in Mtb virulence and pathogenesis (reviewed in12). The hierarchical clustering of the differentially glycosylated proteins coincided with the phylogeny among the MTBC, where the modern lineages (lineage 3 and lineage 4 strains) clustered together, separated from the ancient lineages (lineage 5 and lineage 7 strains) (Fig. 6A)96.
Clinically important proteins such as FadE35, LppW, LdtA, PurK, PPE42 and UvrA were hyper-glycosylated in lineage 3 and lineage 4 strains compared to the ancient lineages64. PurK has been identified to be a high-confidence drug target97. The antigen PPE42 is known to elicit humoral immune response against Mtb98. Glycoproteins including LprG, Mce2D, DagK, UvrC and VapC45 were hyper-glycosylated in all lineages except lineage 7 strains. LprG plays a role in transport and localization of the TLR2 agonists, LAM, PIM, LM and triacylglycerides to the cell surface, maintaining cell envelope integrity, and inhibition of phagosome-lysosome fusion, thereby enhancing Mtb survival inside macrophages5,54. The DagK is involved in the biosynthesis of Mtb virulence factors PI and PIMs99. The membrane proteins DsbF and Rv0412c, iron-sulfur cluster carrier protein Mrp, ATPase MoxR3 and PknA were hyper-glycosylated in lineage 5 and lineage 7 strains. A number of proteins involved in lipid metabolism such as FadD9, FadD34, and PG synthesis like DacB1, and oxidoreductases BpoB and Rv2766c, were hyper-glycosylated in lineage 7 strains. Four glycosylation sites on GroeL1, a chaperone involved in mycolic acid biosynthesis during biofilm formation100, were uniformly hyper-glycosylated in lineage 4 strains. Penicillin-binding membrane protein PbpB, another hyper-glycosylated lineage 4 strains, is an essential enzyme involved in peptidoglycan biosynthesis and has been predicted to be an important drug target101. These proteins are essential virulence factors used by Mtb for cell wall biosynthesis, stress response, immunomodulation, efficient host cell invasion, survival, growth and other physiological processes102. The relative abundance of these essential glycoproteins across the different lineages of Mtb might lead to a specific phenotype with better adaptability to the host.
Identification of glycoproteins and their function contributes to a better understanding of the pathogenesis and survival strategies adopted by Mtb. This knowledge is fundamental for diagnostic, drug or vaccine discovery process. Many anti-TB drugs target the biosynthesis of PG, MA and AG, drug efflux pumps and other virulence factors used by Mtb to efficiently invade and multiply inside the host12. Our study has identified a large number of membrane-associated glycolipoproteins involved in Mtb pathogenesis. We present a comprehensive glycoproteome map of Mtb and show that there are significant quantitative differences across the various Mtb lineages that may directly influence phenotype. Further purification and detailed functional studies addressing selected uncharacterized glycoproteins may reveal the physiological role of protein glycosylation in defining the phenotype of a bacillus. These findings expand the current understanding of the nature and diversity of Mtb glycoproteins, open a new avenue of research for identification of potential drug targets, and create opportunities to engineer glycoproteins for their clinical applications103.
Methods
Mtb strains and growth conditions
Four clinical strains representing four different Mtb lineages, lineage 3 (CAS-DELHI), lineage 4 (FSP471.1), lineage 5 (M. africanum) and lineage 7 (Aethiops vetus104) strains, were cultured on Middlebrook 7H10 agar plates for 32 days. The details of culturing, sample handling and inactivation were performed as previously described in Yimer et al.105.
Proteomic analyses
-
(i)
Preparation of cell lysates. The Mtb cell pellets were mechanically disrupted by bead beating with a MagNa Lyser (Roche, US) as described by Yimer et al.105.
-
(ii)
In-gel trypsin digestion. Gel-fractionated protein samples (100 µg) from Mtb cells grown to late exponential phase were stained using a Colloidal Blue Staining kit (Invitrogen, CA) and each gel-lane was divided into six fractions. Each fraction was subjected to in-gel reduction, alkylation, and tryptic digestion as previously described106. Proteins were reduced using 10 mM DTT, alkylated with 55 mM iodoacetamide and digested with sequence grade trypsin (Promega, 1:100; w/w) overnight at 37 °C in 50 mM NH4HCO3. The in-gel digested protein samples were extracted using 50% and 100% acetonitrile (ACN), dried by SpeedVac concentrator (Eppendorf, concentrator 5301) and re-suspended using 0.05% trifluoroacetic acid (TFA). The extracted protein samples were purified using C18 stage tips by stacking three discs from Empore and transferred to auto-sampler nano LC vials for LC-MS/MS analysis as.
-
(iii)
Nano-LC-MS/MS analysis. Peptide characterization and quantitation were performed by nano LC-MS/MS using a Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer interfaced with an EASY1000-nano-electrospray ion source (Thermo-Fisher Scientific, Biberach, Germany). The LC gradient was from 2% to 90% solvent B (0.1% FA in 97% CAN) in 50 µm × 15 cm analytical columns (PepMap RSLC, C18, 2 µm, 100 Å, Thermo Scientific) for 75 min analysis at a flow rate of 0.3 μl/min. The mass spectrometer was operated in data-dependent acquisition mode with automatic switching between MS and MS/MS scans.
The full MS scans were acquired at 70K resolution with automatic gain control (AGC) target of 1 × 106 ions between m/z = 300 to 1800 and were surveyed for a maximum injection time of 200 milliseconds (ms). Higher energy collision dissociation (HCD) was used for peptide fragmentation at normalized collision energy set to 28. The MS/MS scans were performed using a data-dependent top10 method at a resolution of 17.5K with an AGC of 5 × 104 ions at maximum injection time of 100 ms and isolation window of 2.0 m/z units. An underfill ratio of 10% and dynamic exclusion duration of 30 s was applied. For each Mtb lineage, three biological replicates were analyzed with each biological replicates fractionated into six gel bands, resulting in a total of 72 analytical runs (four lineages * three biological replicates * six fractions).
-
(iv)
Protein and PTM identification. The Maxquant software (version 1.5.7.4) was employed for protein and glycosylation site identification from the raw MS data107. The raw mass spectral data were searched against the Uniprot Mtb protein database containing 3993 protein sequences concatenated to reverse decoy database and protein sequences for common contaminants. Trypsin [KR].[^P] was specified as a cleavage enzyme with up to two missed cleavages. The “re-quantify” and “match between runs” options were utilized with a retention time alignment window of three min. Carbamidomethylation of cysteine residues was specified as a fixed modification and acetylation on protein N-terminal, conversion of N-terminal glutamine and glutamic acid to pyroglutamic acid, and oxidation of methionine were set as the variable modifications.
For the PTM analysis, a number of glycan residues were configured in the MaxQuant search at three different amino acid residues, N, S and T (Supplementary Table S3) and were set to variable modification. Both unique and razor peptides were used for the quantification of PTM abundance. Peptides with a minimum length of seven amino acids and detected in at least one or more of the replicates were considered for identification. For protein identification, a minimum of two peptides, of which at least one was unique, was required per protein group. All other parameters in MaxQuant were set to default values.
-
(v)
Bioinformatics analysis.
Statistical analysis
Statistical significance was determined with multiple-sample test at a P < 0.05 level of significance using Perseus software (version 1.6.0.7). All modified peptide spectra were validated by applying stringent site localization probability of >0.70 and PEP of <0.05 prior to further analysis. PTM sites with a minimum of one valid value from the total samples were considered for PTM site identification. Modified peptides with valid values in at least 50% of the samples were considered for label-free relative quantification analysis.
Analysis of N- and O-glycosylation motifs
A sequence logo generators WebLogo (http://weblogo.berkeley.edu/logo.cgi) was used to identify the enriched amino acid motifs flanking the glycosylated sites. The sequence windows from the identification table were used to generate the sequence motifs for the three modified amino acids S, T and N.
Gene Ontology analysis of glycosylated proteins
The biological processes, cellular component and molecular function for the identified glycoproteins were analyzed using DAVID Bioinformatics Resources 6.7. The proteins were classified by GO annotation based on three terms; molecular function (MF), biological process (BP) and cellular component (CC).
Protein-protein interaction network analysis
Protein-protein interaction (PPI) networks were generated via STRING database version 10 with a high confidence threshold of 0.7 and imported into Cytoscape software (version 3.5.0) to produce the final interaction networks. Highly interconnected clusters were identified using MCODE and ClusterOne plug-in toolkits.
Supplementary information
Acknowledgements
We thank the patients for consenting to participate in the study and selected health care facilities in the Amhara Region, Ethiopia, for facilitating the study. We are grateful to the Armauer Hansen Research Institute (AHRI), Addis Ababa, Ethiopia for facilitating the transfer of lineages 3, 4 and 7 strains to Oslo University Hospital. Funding was received from the Research Council of Norway (RCN) FRIMEDBIO project # 204747, NORBRAIN #197467 and GLOBVAC #234506 project to T.T. and #192468 to C.H.H., and from the Norwegian South-Eastern Health Authority project #2013080 to S.A.Y., G.N. and T.T.
Author Contributions
A.G.B. and T.T. conceived the study and study design. S.A.Y. and E.D.Z. performed specimen handling and cultivation. S.A.Y. collected the Mtb lineage 3, 4 and 7 strains. S.K. and A.G.B. performed the MS sample preparation. A.G.B. performed bioinformatics and statistical analyses. T.R. performed MS analysis. A.G.B. and T.T. evaluated and interpreted the data and drafted the paper. All authors edited and approved the final manuscript.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009676.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Solomon Abebe Yimer and Shewit Kalayou contributed equally.
Contributor Information
Alemayehu Godana Birhanu, Email: alexbiology97@yahoo.com.
Tone Tønjum, Email: tone.tonjum@medisin.uio.no.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39654-9.
References
- 1.Angala SK, Belardinelli JM, Huc-Claustre E, Wheat WH, Jackson M. The cell envelope glycoconjugates of Mycobacterium tuberculosis. Critical reviews in biochemistry and molecular biology. 2014;49:361–399. doi: 10.3109/10409238.2014.925420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS microbiology letters. 1994;123:11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, Cassidy C, Sacchettini JC. Crystal structure and activity studies of the Mycobacterium tuberculosis β-lactamase reveal its critical role in resistance to β-lactam antibiotics. Antimicrobial agents and chemotherapy. 2006;50:2762–2771. doi: 10.1128/AAC.00320-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nampoothiri K, et al. Molecular cloning, overexpression and biochemical characterization of hypothetical β‐lactamases of Mycobacterium tuberculosis H37Rv. Journal of applied microbiology. 2008;105:59–67. doi: 10.1111/j.1365-2672.2007.03721.x. [DOI] [PubMed] [Google Scholar]
- 5.Bianco MV, et al. Role of P27–P55 operon from Mycobacterium tuberculosis in the resistance to toxic compounds. BMC infectious diseases. 2011;11:195. doi: 10.1186/1471-2334-11-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bigi F, et al. The gene encoding P27 lipoprotein and a putative antibiotic-resistance gene form an operon in Mycobacterium tuberculosis and Mycobacterium bovis. Microbiology. 2000;146:1011–1018. doi: 10.1099/00221287-146-4-1011. [DOI] [PubMed] [Google Scholar]
- 7.Daubenspeck JM, Jordan DS, Simmons W, Renfrow MB, Dybvig K. General N-and O-Linked Glycosylation of Lipoproteins in Mycoplasmas and Role of Exogenous Oligosaccharide. PLOS ONE. 2015;10:e0143362. doi: 10.1371/journal.pone.0143362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calder B, Soares NC, de Kock E, Blackburn JM. Mycobacterial proteomics: analysis of expressed proteomes and post-translational modifications to identify candidate virulence factors. Expert Rev Proteomics. 2015;12:21–35. doi: 10.1586/14789450.2015.1007046. [DOI] [PubMed] [Google Scholar]
- 9.Torrelles JB, Schlesinger LS. Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis (Edinburgh, Scotland) 2010;90:84–93. doi: 10.1016/j.tube.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann J, O’Gaora P, Gallagher A, Thole J, Young D. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. The EMBO journal. 1996;15:3547. doi: 10.1002/j.1460-2075.1996.tb00724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann JL, Delahay R, Gallagher A, Robertson B, Young D. Analysis of post‐translational modification of mycobacterial proteins using a cassette expression system. Febs Letters. 2000;473:358–362. doi: 10.1016/S0014-5793(00)01553-2. [DOI] [PubMed] [Google Scholar]
- 12.Sonawane A, Mohanty S, Jagannathan L, Bekolay A, Banerjee S. Role of glycans and glycoproteins in disease development by Mycobacterium tuberculosis. Critical reviews in microbiology. 2012;38:250–266. doi: 10.3109/1040841X.2011.653550. [DOI] [PubMed] [Google Scholar]
- 13.Becker K, et al. Lipoprotein Glycosylation by Protein-O-Mannosyltransferase (MAB_1122c) Contributes to Low Cell Envelope Permeability and Antibiotic Resistance of Mycobacterium abscessus. Frontiers in microbiology. 2017;8:2123. doi: 10.3389/fmicb.2017.02123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacs-Simon A, Titball R, Michell SL. Lipoproteins of bacterial pathogens. Infection and immunity. 2011;79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis. 2003;83:91–97. doi: 10.1016/S1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 16.Jackson M, McNeil MR, Brennan PJ. Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol. 2013;8:855–875. doi: 10.2217/fmb.13.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaur RL, et al. LprG-mediated surface expression of lipoarabinomannan is essential for virulence of Mycobacterium tuberculosis. PLoS pathogens. 2014;10:e1004376. doi: 10.1371/journal.ppat.1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daffe M, Etienne G. The capsule of Mycobacterium tuberculosis and its implications for pathogenicity. Tubercle and lung disease: the official journal of the International Union against Tuberculosis and Lung Disease. 1999;79:153–169. doi: 10.1054/tuld.1998.0200. [DOI] [PubMed] [Google Scholar]
- 19.Alderwick LJ, Harrison J, Lloyd GS, Birch HL. The Mycobacterial cell wall—peptidoglycan and Arabinogalactan. Cold Spring Harbor perspectives in medicine. 2015;5:a021113. doi: 10.1101/cshperspect.a021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahapatra S, et al. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. Journal of bacteriology. 2005;187:2747–2757. doi: 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moran, A. P. Microbial glycobiology: structures, relevance and applications (Elsevier, 2009).
- 22.Rini, J. M. & Esko, J. D. Glycosyltransferases and glycan-processing enzymes (2017).
- 23.Tan FY, Tang CM, Exley RM. Sugar coating: bacterial protein glycosylation and host–microbe interactions. Trends in biochemical sciences. 2015;40:342–350. doi: 10.1016/j.tibs.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Kowarik M, et al. Definition of the bacterial N-glycosylation site consensus sequence. Embo j. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nita-Lazar M, Wacker M, Schegg B, Amber S, Aebi M. The NXS/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology. 2004;15:361–367. doi: 10.1093/glycob/cwi019. [DOI] [PubMed] [Google Scholar]
- 26.Zarschler K, et al. Protein tyrosine O-glycosylation—a rather unexplored prokaryotic glycosylation system. Glycobiology. 2010;20:787–798. doi: 10.1093/glycob/cwq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gloster TM, Vocadlo DJ. Mechanism, structure, and inhibition of O-GlcNAc processing enzymes. Current signal transduction therapy. 2010;5:74–91. doi: 10.2174/157436210790226537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham RL, Hess S. Mass spectrometry in the elucidation of the glycoproteome of bacterial pathogens. Current Proteomics. 2010;7:57–81. doi: 10.2174/157016410790979662. [DOI] [Google Scholar]
- 29.Diaz-Silvestre H, et al. The 19-kDa antigen of Mycobacterium tuberculosis is a major adhesin that binds the mannose receptor of THP-1 monocytic cells and promotes phagocytosis of mycobacteria. Microbial pathogenesis. 2005;39:97–107. doi: 10.1016/j.micpath.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Smith GT, Sweredoski MJ, Hess S. O-linked glycosylation sites profiling in Mycobacterium tuberculosis culture filtrate proteins. Journal of proteomics. 2014;97:296–306. doi: 10.1016/j.jprot.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Zamorano M, et al. Mycobacterium tuberculosis glycoproteomics based on ConA-lectin affinity capture of mannosylated proteins. J Proteome Res. 2009;8:721–733. doi: 10.1021/pr800756a. [DOI] [PubMed] [Google Scholar]
- 32.Peters JS, et al. Identification of Quantitative Proteomic Differences between Mycobacterium tuberculosis Lineages with AlteredVirulence. Frontiers in Microbiology. 2016;7:813. doi: 10.3389/finicb.2016.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dormans J, et al. Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clinical & Experimental Immunology. 2004;137:460–468. doi: 10.1111/j.1365-2249.2004.02551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manabe YC, et al. Different strains of Mycobacterium tuberculosis cause various spectrums of disease in the rabbit model of tuberculosis. Infection and immunity. 2003;71:6004–6011. doi: 10.1128/IAI.71.10.6004-6011.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicol MP, Wilkinson RJ. The clinical consequences of strain diversity in Mycobacterium tuberculosis. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:955–965. doi: 10.1016/j.trstmh.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS microbiology reviews. 2004;28:645–659. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Rezwan M, Grau T, Tschumi A, Sander P. Lipoprotein synthesis in mycobacteria. Microbiology. 2007;153:652–658. doi: 10.1099/mic.0.2006/000216-0. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol Microbiol. 2010;76:348–364. doi: 10.1111/j.1365-2958.2010.07110.x. [DOI] [PubMed] [Google Scholar]
- 39.Rini, J. M. & Esko, J. D. In Essentials of Glycobiology (eds rd et al.) 65–75 (Cold Spring Harbor Laboratory Press Copyright 2015–2017 by The Consortium of Glycobiology Editors, La Jolla, California. All rights reserved., 2015).
- 40.Mäki M, Renkonen R. Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology. 2003;14:1R–15R. doi: 10.1093/glycob/cwh040. [DOI] [PubMed] [Google Scholar]
- 41.Lu Q, et al. An iron-containing dodecameric heptosyltransferase family modifies bacterial autotransporters in pathogenesis. Cell Host Microbe. 2014;16:351–363. doi: 10.1016/j.chom.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Sambou T, et al. Capsular glucan and intracellular glycogen of Mycobacterium tuberculosis: biosynthesis and impact on the persistence in mice. Molecular microbiology. 2008;70:762–774. doi: 10.1111/j.1365-2958.2008.06445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torrelles JB, Azad AK, Schlesinger LS. Fine discrimination in the recognition of individual species of phosphatidyl-myo-inositol mannosides from Mycobacterium tuberculosis by C-type lectin pattern recognition receptors. The Journal of immunology. 2006;177:1805–1816. doi: 10.4049/jimmunol.177.3.1805. [DOI] [PubMed] [Google Scholar]
- 44.Fenton, M. J., Riley, L. W. & Schlesinger, L. S. In Tuberculosis and the tubercle bacillus 405–426 (American Society of Microbiology, 2005).
- 45.Parra J, et al. Scrutiny of Mycobacterium tuberculosis 19 kDa antigen proteoforms provides new insights in the lipoglycoprotein biogenesis paradigm. Sci Rep. 2017;7:43682. doi: 10.1038/srep43682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ocampo M, Curtidor H, Vanegas M, Patarroyo MA, Patarroyo ME. Specific interaction between Mycobacterium tuberculosis lipoprotein-derived peptides and target cells inhibits mycobacterial entry in vitro. Chemical biology & drug design. 2014;84:626–641. doi: 10.1111/cbdd.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henao-Tamayo M, et al. A mutant of Mycobacterium tuberculosis lacking the 19-kDa lipoprotein Rv3763 is highly attenuated in vivo but retains potent vaccinogenic properties. Vaccine. 2007;25:7153–7159. doi: 10.1016/j.vaccine.2007.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmad S, El‐Shazly S, Mustafa A, Al‐Attiyah R. The six mammalian cell entry proteins (Mce3A–F) encoded by the mce3 operon are expressed during in vitro growth of Mycobacterium tuberculosis. Scandinavian journal of immunology. 2005;62:16–24. doi: 10.1111/j.1365-3083.2005.01639.x. [DOI] [PubMed] [Google Scholar]
- 49.Ragas A, Roussel L, Puzo G, Rivière M. The Mycobacterium tuberculosis cell-surface glycoprotein apa as a potential adhesin to colonize target cells via the innate immune system pulmonary C-type lectin surfactant protein A. Journal of biological chemistry. 2007;282:5133–5142. doi: 10.1074/jbc.M610183200. [DOI] [PubMed] [Google Scholar]
- 50.Satchidanandam V, et al. The glycosylated Rv1860 protein of Mycobacterium tuberculosis inhibits dendritic cell mediated TH1 and TH17 polarization of T cells and abrogates protective immunity conferred by BCG. PLoS Pathog. 2014;10:e1004176. doi: 10.1371/journal.ppat.1004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ota F, Kizuka Y, Kitazume S, Adachi T, Taniguchi N. N‐Glycosylation is essential for the secretion of extracellular superoxide dismutase. FEBS letters. 2016;590:3357–3367. doi: 10.1002/1873-3468.12378. [DOI] [PubMed] [Google Scholar]
- 52.Rodríguez D, et al. Peptides from the Mycobacterium tuberculosis Rv1980c protein involved in human cell infection: insights into new synthetic subunit vaccine candidates. Biological chemistry. 2010;391:207–217. doi: 10.1515/bc.2010.019. [DOI] [PubMed] [Google Scholar]
- 53.Pethe K, et al. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature. 2001;412:190–194. doi: 10.1038/35084083. [DOI] [PubMed] [Google Scholar]
- 54.Becker K, Sander P. Mycobacterium tuberculosis lipoproteins in virulence and immunity–fighting with a double‐edged sword. FEBS letters. 2016;590:3800–3819. doi: 10.1002/1873-3468.12273. [DOI] [PubMed] [Google Scholar]
- 55.da Silva PEA, Von Groll A, Martin A, Palomino JC. Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunology & Medical Microbiology. 2011;63:1–9. doi: 10.1111/j.1574-695X.2011.00831.x. [DOI] [PubMed] [Google Scholar]
- 56.Gupta AK, et al. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microbial drug resistance. 2010;16:21–28. doi: 10.1089/mdr.2009.0054. [DOI] [PubMed] [Google Scholar]
- 57.Chalut C. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis (Edinburgh, Scotland) 2016;100:32–45. doi: 10.1016/j.tube.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 58.Viljoen A, et al. The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Molecular microbiology. 2017;104:889–904. doi: 10.1111/mmi.13675. [DOI] [PubMed] [Google Scholar]
- 59.Zimhony O, et al. AcpM, the Meromycolate Extension Acyl Carrier Protein of Mycobacterium tuberculosis, Is Activated by the 4′-Phosphopantetheinyl Transferase PptT, a Potential Target of the Multistep Mycolic Acid Biosynthesis. Biochemistry. 2015;54:2360–2371. doi: 10.1021/bi501444e. [DOI] [PubMed] [Google Scholar]
- 60.Barkan D, Hedhli D, Yan H-G, Huygen K, Glickman MS. Mycobacterium tuberculosis lacking all mycolic acid cyclopropanation is viable but highly attenuated and hyperinflammatory in mice. Infection and immunity. 2012;80:1958–1968. doi: 10.1128/IAI.00021-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gavalda S, et al. The Polyketide Synthase Pks13 Catalyzes a Novel Mechanism of Lipid Transfer in Mycobacteria. Chemistry & Biology. 2014;21:1660–1669. doi: 10.1016/j.chembiol.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 62.Puech V, et al. Evidence for a partial redundancy of the fibronectin‐binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Molecular microbiology. 2002;44:1109–1122. doi: 10.1046/j.1365-2958.2002.02953.x. [DOI] [PubMed] [Google Scholar]
- 63.Steingart KR, et al. Performance of purified antigens for serodiagnosis of pulmonary tuberculosis: a meta-analysis. Clinical and vaccine immunology. 2009;16:260–276. doi: 10.1128/CVI.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kieser KJ, et al. Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proceedings of the National Academy of Sciences. 2015;112:13087–13092. doi: 10.1073/pnas.1514135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annual review of biochemistry. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 66.Goude R, Amin A, Chatterjee D, Parish T. The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrobial agents and chemotherapy. 2009;53:4138–4146. doi: 10.1128/AAC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bisson GP, et al. Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis. Journal of bacteriology. 2012;194:6441–6452. doi: 10.1128/JB.01013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goude, R. & Parish, T. The genetics of cell wall biosynthesis in Mycobacterium tuberculosis (2008). [DOI] [PubMed]
- 69.Wei J, et al. Genome-wide transcription analyses in Mycobacterium tuberculosis treated with lupulone. Brazilian Journal of Microbiology. 2014;45:333–342. doi: 10.1590/S1517-83822014005000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waddell S, et al. Inactivation of polyketide synthase and related genes results in the loss of complex lipids in Mycobacterium tuberculosis H37Rv. Letters in applied microbiology. 2005;40:201–206. doi: 10.1111/j.1472-765X.2005.01659.x. [DOI] [PubMed] [Google Scholar]
- 71.Pérez J, et al. Mycobacterium tuberculosis transporter MmpL7 is a potential substrate for kinase PknD. Biochemical and biophysical research communications. 2006;348:6–12. doi: 10.1016/j.bbrc.2006.06.164. [DOI] [PubMed] [Google Scholar]
- 72.Koliwer-Brandl H, et al. Metabolic Network for the Biosynthesis of Intra- and Extracellular alpha-Glucans Required for Virulence of Mycobacterium tuberculosis. PLoS Pathog. 2016;12:e1005768. doi: 10.1371/journal.ppat.1005768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin W, et al. Transcriptional Profiling of Mycobacterium tuberculosis Exposed to In Vitro Lysosomal Stress. Infection and immunity. 2016;84:2505–2523. doi: 10.1128/IAI.00072-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leiba J, et al. Mycobacterium tuberculosis maltosyltransferase GlgE, a genetically validated antituberculosis target, is negatively regulated by Ser/Thr phosphorylation. Journal of Biological Chemistry. 2013;288:16546–16556. doi: 10.1074/jbc.M112.398503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dkhar HK, et al. Discovery of Mycobacterium tuberculosis α-1, 4-glucan branching enzyme (GlgB) inhibitors by structure-and ligand-based virtual screening. Journal of Biological Chemistry. 2015;290:76–89. doi: 10.1074/jbc.M114.589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torrelles JB, et al. Inactivation of Mycobacterium tuberculosis mannosyltransferase pimB reduces the cell wall lipoarabinomannan and lipomannan content and increases the rate of bacterial-induced human macrophage cell death. Glycobiology. 2009;19:743–755. doi: 10.1093/glycob/cwp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mishra AK, et al. Identification of a novel α (1 → 6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan‐and lipoarabinomannan‐deficient mutant. Molecular microbiology. 2008;68:1595–1613. doi: 10.1111/j.1365-2958.2008.06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Portevin D, et al. The Acyl-AMP Ligase FadD32 and AccD4-containing Acyl-CoA Carboxylase Are Required for the Synthesis of Mycolic Acids and Essential for Mycobacterial Growth Identification of The Carboxylation Product and Determination of The Acyl-Coa Carboxylase Components. Journal of Biological Chemistry. 2005;280:8862–8874. doi: 10.1074/jbc.M408578200. [DOI] [PubMed] [Google Scholar]
- 79.Varki A et al. editors. Essentials of Glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; Chapter 17, Glycosyltransferases. Available from, https://www.ncbi.nlm.nih.gov/books/NBK20718/ (1999).
- 80.El-Battari A, et al. Different glycosyltransferases are differentially processed for secretion, dimerization, and autoglycosylation. Glycobiology. 2003;13:941–953. doi: 10.1093/glycob/cwg117. [DOI] [PubMed] [Google Scholar]
- 81.Muhlenhoff M, Eckhardt M, Bethe A, Frosch M, Gerardy-Schahn R. Autocatalytic polysialylation of polysialyltransferase-1. Embo j. 1996;15:6943–6950. doi: 10.1002/j.1460-2075.1996.tb01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 83.Zhou P, Long Q, Zhou Y, Wang H, Xie J. Mycobacterium tuberculosis two-component systems and implications in novel vaccines and drugs. Critical reviews in eukaryotic gene expression. 2012;22:37–52. doi: 10.1615/CritRevEukarGeneExpr.v22.i1.30. [DOI] [PubMed] [Google Scholar]
- 84.Domenech, P. et al. Unique Regulation of the DosR Regulon in the Beijing Lineage of Mycobacterium tuberculosis. J Bacteriol199, 10.1128/jb.00696-16 (2017). [DOI] [PMC free article] [PubMed]
- 85.Converse PJ, et al. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infection and immunity. 2009;77:1230–1237. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feltcher ME, Sullivan JT, Braunstein M. Protein export systems of Mycobacterium tuberculosis: novel targets for drug development? Future microbiology. 2010;5:1581–1597. doi: 10.2217/fmb.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Danilchanka O, et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc Natl Acad Sci USA. 2014;111:6750–6755. doi: 10.1073/pnas.1400136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Birhanu, A. et al. Nε-and O-Acetylation in Mycobacterium tuberculosis Lineage 7 and Lineage 4 strains: Proteins Involved in Bioenergetics, Virulence and Antimicrobial Resistance are Acetylated. Journal of proteome research (2017). [DOI] [PMC free article] [PubMed]
- 89.Schwenk, S., Moores, A., Nobeli, I., McHugh, T. D. & Arnvig, K. B. Cell-wall synthesis and ribosome maturation are co-regulated by an RNA switch in Mycobacterium tuberculosis. bioRxiv, 232314 (2017). [DOI] [PMC free article] [PubMed]
- 90.Hanna ES, et al. Evidence for glycosylation on a DNA-binding protein of Salmonella enterica. Microbial cell factories. 2007;6:11. doi: 10.1186/1475-2859-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gomez A, et al. Colorectal cancer-specific cytochrome P450 2W1: intracellular localization, glycosylation, and catalytic activity. Molecular pharmacology. 2010;78:1004–1011. doi: 10.1124/mol.110.067652. [DOI] [PubMed] [Google Scholar]
- 92.Lamb DC, et al. The first virally encoded cytochrome p450. Journal of virology. 2009;83:8266–8269. doi: 10.1128/JVI.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bartels K-M, et al. Glycosylation is required for outer membrane localization of the lectin LecB in Pseudomonas aeruginosa. Journal of bacteriology. 2011;193:1107–1113. doi: 10.1128/JB.01507-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure. 1995;3:41–62. doi: 10.1016/S0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 95.McKinney JD, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 96.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nature genetics. 2013;45:1176. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raman K, Yeturu K, Chandra N. targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC systems biology. 2008;2:109. doi: 10.1186/1752-0509-2-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chakhaiyar P, et al. Regions of high antigenicity within the hypothetical PPE major polymorphic tandem repeat open-reading frame, Rv2608, show a differential humoral response and a low T cell response in various categories of patients with tuberculosis. J Infect Dis. 2004;190:1237–1244. doi: 10.1086/423938. [DOI] [PubMed] [Google Scholar]
- 99.Owens RM, et al. M. tuberculosis Rv2252 encodes a diacylglycerol kinase involved in the biosynthesis of phosphatidylinositol mannosides (PIMs) Mol Microbiol. 2006;60:1152–1163. doi: 10.1111/j.1365-2958.2006.05174.x. [DOI] [PubMed] [Google Scholar]
- 100.Ojha A, et al. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 101.Amir A, et al. Mycobacterium tuberculosis H37Rv: In Silico Drug Targets Identification by Metabolic PathwaysAnalysis. International journal of evolutionary biology. 2014;2014:284170. doi: 10.1155/2014/284170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forrellad MA, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Facciuolo A, Mutharia LM. Mycobacterial glycoproteins: a novel subset of vaccine candidates. Frontiers in cellular and infection microbiology. 2014;4:133. doi: 10.3389/fcimb.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nebenzahl-Guimaraes H, et al. Genomic characterization of Mycobacterium tuberculosis lineage 7 and a proposed name: ‘Aethiops vetus’. Microb Genom. 2016;2:e000063. doi: 10.1099/mgen.0.000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yimer SA, et al. Comparative Proteomic Analysis of Mycobacterium tuberculosis Lineage 7 and Lineage 4 Strains Reveals Differentially Abundant Proteins Linked to Slow Growth and Virulence. Front Microbiol. 2017;8:795. doi: 10.3389/fmicb.2017.00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 107.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009676.