Abstract
A series of oxadiazole (7a-l) and hydroxypyrazoline derivatives (8a-l) incorporating thiazole were synthesized and characterized by spectral analysis (1H-NMR, 13C-NMR, Mass, and FT-IR). The synthesized compounds were screened for their in vitro cytotoxicity against MDA-MB231 and HT-29 human cell lines. Conjugates 7d, 7e, 7f, 7i, 7l, 8a, 8b, 8i and 8l exhibited significant antiproliferative activity on both MDA-MB231 and HT-29 cell lines. Flow cytometric analysis reveals that, 7i arrests both cells lines at Go/G1 phase whereas 8i induced G0/G1 arrest only in the HT-29 cells. Furthermore, Computational interaction studies of 7i and 8i exhibited its capacity of being a plausible CDK2 and BCL-2 inhibitor respectively. In addition, DNA binding of the synthesized compounds and DNA docking of 7i and 8i demonstrated the ability to interact with DNA. Compounds 7i and 8i causes' remarkable growth inhibition of MDA-MB231 and HT-29 cells but compound 8i was considerably effective against HT-29 cells. Overall these compounds can be practiced for further drug development.
Keyword: Organic chemistry
1. Introduction
As per WHO, the burden of cancer will increase to 23.6 million new cases each year by 2030 [1]. Breast cancer is the most frequently diagnosed cancer and the leading cause of death in females, whereas colorectal cancer is the third most commonly diagnosed cancer in males and females [2]. The treatments for these cancers still remain a challenging task as chemotherapy is often ineffective, because of the intrinsic drug resistance to these tumours [3]. There is evidence to indicate that colorectal cancer cells are self-sufficient in growth signals, which escapes from apoptosis [4] Therefore, it is imperative to develop more effective drugs. Apoptosis is a morphologically and biochemically driven process, while impaired apoptosis and defects in the regulation of the cell cycle are hallmarks that contribute to cancer growth and aggressiveness [5]. With progressing knowledge of oncogenesis and apoptosis, comes an appreciation of the role played by cell-cycle regulation, in malignant transformation. Modulation of the cell cycle also contributes to chemotherapy resistance. The cyclin-dependent kinases1 (CDK1), cyclin-dependent kinases2 (CDK2) and DNA, the essential engines of the cell cycle, are therefore rational therapeutic targets. Over the last several years, a new class of anticancer therapy has been developed and extensively tested against these targets.
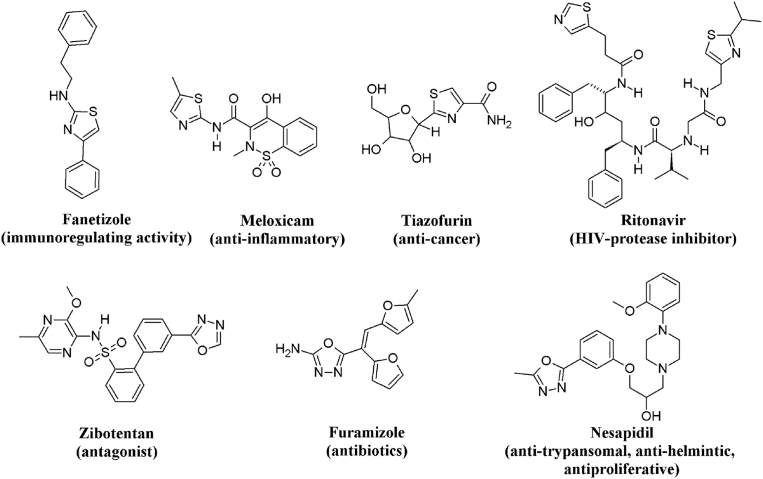
Modern studies have shown a significant fascinate in thiazole derivatives, due to far-reaching biological activities, such as anticonvulsant activity [6], anti-HIV [7], antidiabetic [8], anti-alzheimer [9], antimalarial [10], antimicrobial [11], anti-inflammatory [12], antiproliferative against MiaPaCa-2 cell line [13], antiproliferative against gastric carcinoma cells [14], antiproliferative against diffuse malignant peritoneal mesothelioma cell lines, a very aggressive form of cancer [15] and CDK1 inhibitory activity of thiazol [16]. Most of the pharmaceutical drugs such as Fanetizole, Meloxicam [17], Tiazofurin [18] and Ritonavir [19] (Fig. 1) contains thiazole rings.
Fig. 1.
Commercially available drugs containing thiazole and 1,3,4- oxadiazole.
On the other hand, there are a bunch of reports on 1,3,4-oxadiazoles exhibiting various pharmacological activities, such as anti-diabetic [20], antihypertension [21], analgesic [22], antiviral [23], anticonvulsant [24], antifungal [25] antibacterial [26], anticancer [27], anti-glycation [28], anti-inflammatory [29], antimicrobial [30], and ulcerogenic [31]. Compounds containing oxadiazole units such as Nesapidil, Furamizole, and Zibotentan [32] (Fig. 1) are currently used in clinical medicines. Additionally, pyrazolines incorporated with a variety of functional groups or substituents are found in many important biologically active compounds and significant research on this species has been carried out. They exhibit a wide range of biological activities such as antidepressant, anti-inflammatory, antimicrobial, and anticancer effects etc. In accordance with literature, pyrazoline derivatives are not useful in treatment of various cancer types, including lung, breast, colon, rectum, brain, stomach, liver, bladder, pancreas, bone, mouth, esophagus, cervix and prostate cancers, and also some of them act as cancer chemopreventive agents [33, 34, 35, 36, 37, 38]. Numerous analysis shown, pyrazoline derivatives were reported as epidermal growth factor receptor tyrosine kinase (EGFR-TK) inhibitors [39], COX-2/B-Raf inhibitors [40], aurora kinase inhibitors [41], tubulin assembling inhibitors [42], telomerase inhibitors [43]. Provoked by above-mentioned observations and in continuation of our search for potent and less toxic antiproliferative agents, we have aspired to introduce some hybrids by combining thiazole-oxadiazole and thiazole-hydroxypyrazoline pharmacophores in a molecular framework.
Dinesh et al. [44] published anticancer activity of hydroxypyrazoline against MCF-7, MDA-MB-cancer cell lines. Samir Bondock et al. [45] reported 1,3,4-oxadiazole with antitumor activities against HepG2, WI-38, VERO, MCF-7 cancer cell lines, N.C. Desai et al. [11] analyzed 1,3,4-oxadiazole clubbed thiazole with anticancer activity against HeLa cell lines and M.F. Hassan et al. [46] disclosed DNA binding studies of 1,3,4-oxadiazole and 1,3,4-thiadiazole derivatives.
Based on literature survey, the interaction of thiazole-hydroxypyrazoline derivatives in a molecular framework has so far not been studied whereas there exists a report on some of the thiazole-oxadiazole derivatives as antitubercular agent [47] but not reported as antiproliferative agents. This flurry encouraged us on synthesis, purification and characterization of thiazole-oxadiazoles and thiazole-hydroxypyrazoline class of compounds, for relatively safe alternatives to ameliorate the clinical consequences of the breast and colorectal cancers.
2. Results and discussion
2.1. Chemistry
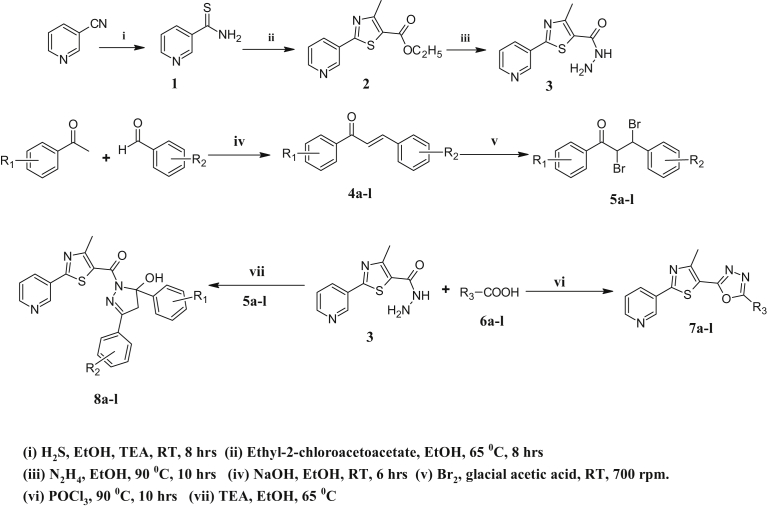
The multistep reaction sequence for the synthesis of the objective compounds 7a-l and 8a-l (Table 1) are framed in Fig. 2. 3-Cyanopyridine was taken as a starting material which on treatment with H2S gas in the presence of triethylamine in absolute alcohol, gave pyridine-3-carbothioamide 1, which on further reaction with ethyl-2-chloroacetoacetate yielded ethyl-4-methyl-2-(pyridin-3-yl)-1,3-thiazole-5-carboxylate 2. Compound 2 was further refluxed with hydrazine hydrate to give an intermediate 5-methyl-2-(pyridin-3-yl)-1,3-thiazole-4-carbohydrazide 3. Among the title compounds 7a-l were obtained by refluxing intermediate 3 with different substituted acids 6a-l, in presence of POCl3 which was confirmed by characteristic FT-IR, 1H-NMR 13C-NMR and LC-MS spectroscopic techniques. The absorption bands in the range 653, 1060 and 1568 cm−1 corresponds to (C-S), (C-O-C) and (C=N) respectively. The 1H-NMR spectrum of 7h showed eleven different types of protons at δ 2.46 (s, 3H, -CH3),7.42 (t, 1H(H3), J = 6 Hz), 7.55 (d, 2H(2H5), J = 5.6 Hz), 8.02 (dd, 1H(H4), J = 6 Hz and J = 1.2 Hz), 8.57–8.64 (3H, (8.58 (dd, 1H(H2), J = 6 Hz and J = 0.8 Hz), 8.64 (d, 2H(2H6), J = 6 Hz), 8.88 (d, 1H(H1), J = 1.2 Hz).
Table 1.
Derivatives of 7a-l and 8a-l.
| Code | R1 | R2 | R3 |
|---|---|---|---|
| 7a | - | - | furfuryl |
| 7b | - | - | 6-bromonapthyl |
| 7c | - | - | 4-CH3-Ph |
| 7d | - | - | 4-NH2-Ph |
| 7e | - | - | 2-Cl,4-NO2-Ph |
| 7f | - | - | 2-Chloropyridyl |
| 7g | - | - | 4-Cl-Ph |
| 7h | - | - | 4-Pyridyl |
| 7i | - | - | 3,4-OCH3-Ph |
| 7j | - | - | 3,4-Cl-Ph |
| 7k | - | - | -CH2-Ph |
| 7l | - | - | -CH2C(Cl)3 |
| 8a | 4-CH3 | 3-Cl | - |
| 8b | 4-CH3 | 3-NO2 | - |
| 8c | 4-CH3 | 4-CH3 | - |
| 8d | 2,4-Cl | 3,4-OCH3 | - |
| 8e | 4-CH3 | 4-F | - |
| 8f | 4-OCH3 | 4-F | - |
| 8g | 4-F | 4-F | - |
| 8h | 4-F | 4-OCH3 | - |
| 8i | 4-F | 3-Cl | - |
| 8j | 4-Cl | 4-F | - |
| 8k | 4-OCH3 | 4-Cl | - |
| 8l | 4-OCH3 | 4-Br | - |
Fig. 2.
The synthetic route to compounds 7a-l and 8a-l.
13C-NMR spectra exhibited different types of carbons, thereby confirming the structure of 7h. The LC-MS spectrum of 7h revealed the presence of molecular ion peak at 322 (M+1), which was in agreement with the molecular weight of the respective compound. Scaffolds 8a-l were obtained by condensation of chalcone-dibromides 5a-l (which were obtained from chalcones 4a-l by well-known Claisen-Schmidt reaction) with 3 in presence of catalytic amount of triethylamine using ethanol as solvent. Target scaffolds 8a-l were confirmed by characteristic FT-IR, NMR and LC-MS spectroscopic technique. Peaks at 3359, 1631, 1602 and 698 corresponds to (O-H), (C=O), (C=N) and (C-S) respectively. The 1H-NMR spectra of 8g showed that hydroxyl proton resonated as a singlet at δ 5.2. The methylene protons of hydroxypyrazoline ring appeared as two doublets at δ 3.7 and 3.4 with a germinal coupling constant (J = 18.4 Hz) indicating the magnetic non-equivalence of the two protons of the CH2 group adjacent to a chiral centre. A sharp singlet at δ 2.80 and 5.28 is assigned for methyl and OH protons. Other aromatic protons of 8g resonated as complex multiples at δ 7.06 to 7.78. Moreover, 13C-NMR spectra of 8g confirmed the presence of pyrazoline ring in which singlet at δ 29.3 and 94.6 are due to the sp [3] carbon of C-4 and C-5 respectively. Singlet at 19.0 and 165.6 are due to methyl and carbonyl carbon respectively. whereas, other aromatic carbon appeared in the expected region. Molecular ion peak at 477.61 (M+1), which was in agreement with the molecular weight of 8g confirmed the structure.
3. Biological activity
3.1. In vitro cytotoxicity
As per the IC50 data (Table 2), nine derivatives, 7d, 7e, 7f, 7i, 7l, 8a, 8b, 8i, and 8l, have shown significant inhibition on both MDA-MB231and HT 29 cancer cell lines. Considering the IC50 values for 7a-l and 8a-l, we tried to link a correlation between the cytotoxicity and molecular structure, by looking at the position and nature of the functional groups on the thiazole-oxadiazole and thiazole-pyrazoline derivatives. The presence of dimethoxy (3,4-dimethoxybenzyl), trichloromethyl, nitro combined with chloro (2-chloro-4-nitrobenzyl), 3-chloropyridyl and amine (4-aminobenzyl) in position 5 on the oxadiazole ring, correspond to compounds 7i, 7l, 7e, 7f, and 7d, respectively which exhibited highest antiproliferative activity. On the other hand, the presence of methyl (7c), chloro (7g), dichloro (7j), phenyl (7k), pyridine (7h), furan (7a) and bromo (7b), decreased the antiproliferative efficiency. The viability of MDA-MB231and HT 29 cell lines decreases with an increase in the concentration of the thiazole-oxadiazole derivatives 7a-l. The presence of 3-chlorobenzyl,4′-methylbenzyl (8a), 3-nitrobenzyl,4-methylbenzyl (8b), 3-chlorobenzyl,4′-fluorobenzyl (8i), and 4-bromobenzyl,4′-methoxybenzyl (8l) on the hydroxypyrazoline ring, exhibited highest antiproliferative activity on both the cell lines. However, the combination of 3,4-dimethoxybenzyl,2,4′-dichlorobenzyl (8d), 4-fluorobenzyl,4′-methoxybenzyl (8f), 4-fluorobenzyl,4′-fluorobenzyl (8g) and 4-chlorobenzyl,4′-methoxybenzyl (8k) were effective towards HT-29. On the other hand combination of 4-methylbenzyl,4′-methylbenzyl (8c), 4-fluorobenzyl,4′-methylbenzyl (8e), 4-methoxybenzyl,4′-fluorobenzyl (8h) and 4-fluorobenzyl,4′-chlorobenzyl (8j) decreased the antiproliferative efficiency. The viability of MDA-MB231and HT 29 cell lines decreases with an increase in the concentration of the hydroxypyrazoline derivatives 8a-l. Due to the significant antiproliferative activity of 7i on both MDA-MB231 (IC50: 10.2 ± 0.02 μM) and HT 29 (IC50: 25.91 ± 1.12 μM) cell lines and 8i MDA-MB231 (IC50: 29.50 ± 1.26 μM) and HT 29 (IC50: 20.32 ± 1.23 μM) was studied further.
Table 2.
Cytotoxicity IC50 (μM) of 7a-l and 8a-l.
| Code | MDA-MB231 | HT 29 |
|---|---|---|
| 7a | 141.53 ± 1.89 | 33.26 ± 2.85 |
| 7b | 48.88 ± 0.13 | 52.34 ± 2.85 |
| 7c | 36.35 ± 1.25 | 42.35 ± 1.12 |
| 7d | 35.67 ± 0.13 | 32.04 ± 0.89 |
| 7e | 19.88 ± 0.06 | 36.31 ± 1.23 |
| 7f | 30.05 ± 0.12 | 29.49 ± 2.16 |
| 7g | 46.31 ± 0.05 | 175.19 ± 5.64 |
| 7h | 306.99 ± 2.56 | 105.91 ± 4.45 |
| 7i | 10.2 ± 0.02 | 25.91 ± 1.12 |
| 7j | 33.87 ± 0.89 | 202.5 ± 5.64 |
| 7k | 133.69 ± 4.56 | 215.38 ± 6.89 |
| 7l | 16.89 ± 0.89 | 56.98 ± 0.86 |
| 8a | 41.90 ± 2.35 | 39.07 ± 1.12 |
| 8b | 52.05 ± 1.23 | 36.29 ± 1.12 |
| 8c | 73.33 ± 2.54 | 307.35 ± 0.12 |
| 8d | 38.92 ± 1.12 | 343.36 ± 1.23 |
| 8e | 123.66 ± 2.35 | 700.60 ± 1.16 |
| 8f | 39.10 ± 3.89 | 856.70 ± 8.53 |
| 8g | 42.45 ± 1.25 | 946.20 ± 12.13 |
| 8h | 385.11 ± 3.58 | 395.91 ± 2.56 |
| 8i | 29.50 ± 1.26 | 20.32 ± 1.23 |
| 8j | 118.88 ± 3.32 | 590.40 ± 2.56 |
| 8k | 28.01 ± 1.23 | 231.42 ± 4.12 |
| 8l | 24.78 ± 2.25 | 26.64 ± 1.16 |
3.2. Flow cytometry assay
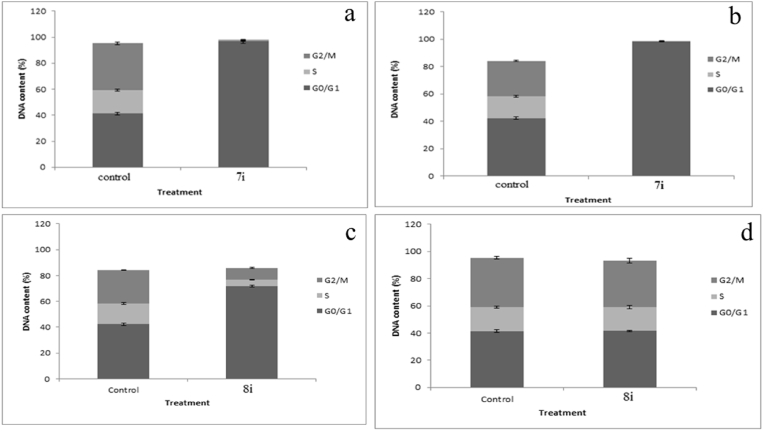
To investigate the effect of compound 7i and 8i on the progression of cell cycle, MDA-MB231 cells and HT-29 cells were treated with its IC50 concentrations, 10.2 and 25.91 μM respectively for 7i similarly 20.32 and 29.50 μM for 8i. Cell cycle distribution was analyzed after appropriate gating of cell populations in FL-2-Area vs FL-2-Width plot of PI fluorescence. The compound 7i was able to induce G0/G1 arrest in treated MDA-MB231 cells, 48 hrs after the treatment. The percentage of G0/G1 cells increased significantly from 41.33% in control (untreated) to 96.73% in cells after treatment with the test compound. These results suggest that the compound 7i brings about changes in the first phase of the cell cycle and mitosis. The compound also induced G0/G1 arrest in the HT-29 cells, thereby indicating its antiproliferative action on colorectal cancer. Analogously, 8i did not induce cell cycle arrest in treated MDA-MB231 cells, 48 h after the treatment. No difference was incurred in the DNA content in different phases of cell cycle (G0/G1, S, and G2/M) compared to control, indicating that the compound did not induce cytotoxicity via cell cycle arrest in MDA-MB231 cells. But the compound 8i induced G0/G1 arrest in the HT-29 cells. The percentage of G0/G1 phase increased from 42.36 to 71.1 % compared to that of control cells. There was a significant reduction in the cellular DNA content of S and G2/M phases compared to that of control cells. These results indicate the antiproliferative action of 8i on the colorectal cancer cells via cell cycle arrest (Fig. 3).
Fig. 3.
Flow cytometric analysis in (a) MDA-MB231 after treatment with conjugate, 7i, (b) HT 29 after treatment with conjugate, 7i, (c) HT 29 after treatment with conjugate, 8i, (d) MDA-MB231 after treatment with conjugate, 8i.
3.3. Molecular docking studies
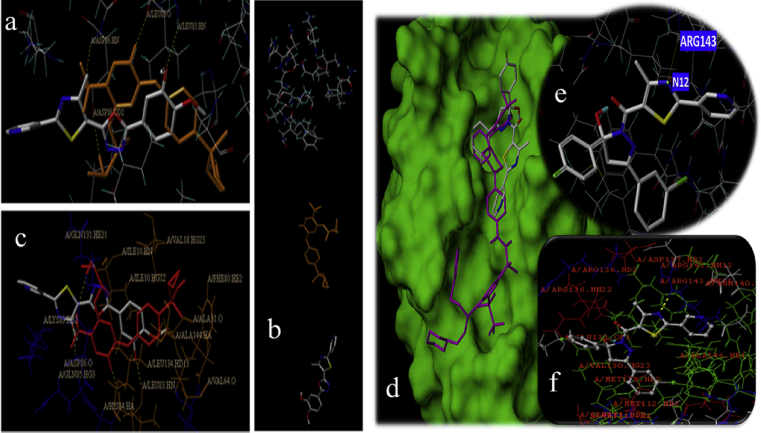
As per flowcytometric results CDK-2 (5iev) and BCL-2 proteins were selected for molecular docking of compounds 7i and 8i respectively. Compounds 7i (chemscore 3.90) and 8i (Chemscore 3.70) showed good interaction within the binding site of CDK-2 protein and BCL-2. A stable hydrogen bonding was observed with the active site amino acid LYS89 and N4 of 7i. This interaction is similar to the Roniciclib (Chemscore 5.01) interactions with ASP86 NH of CDK-2 protein. Likewise with the active site amino acid ARG124 and N4 of 8i. Which is similar to the Navitoclax (Chemscore 2.71) interactions with the BCL-2 protein. The presence of lead 7i and 8i in proximity to an active pocket of a target site CDK-2 and BCL-2 proteins respectively shows a better non-H bonding interaction efficacy. Hydrophilic sulphur present in the ligand 7i interacts with the ILeu10 amino acid. Similarly, hydrophilic GLN141, HIS84 interacts with O17, O28, and N12 group of the ligand. On the other hand, Sulphur, fluorine, and chlorine present in ligand 8i interact with the ALA146, PHE101 and valine130 amino acids respectively. Thus, GLU138 interacts with N17 and O14 groups of the ligand. From this result, it is inferred that 7i and 8i molecules are a promising cancer inhibitor of synthetic origin (Fig. 4).
Fig. 4.
(a) Binding of Compounds 7i and Roniciclib (brown) with protein target site, (b) Ligand 7i and Roniciclib binding orientation, (c) non-H Hydrogen bond interactions between 7i with binding sites of amino acids, (d) Binding of compound 8i and navitoclax (Pink) with protein target site, (e) Shows compound 8i hydrogen bonding with binding site amino acid, (f) Non-Hydrogen bond interactions between 8i with binding site amino acids.
3.4. DNA binding studies
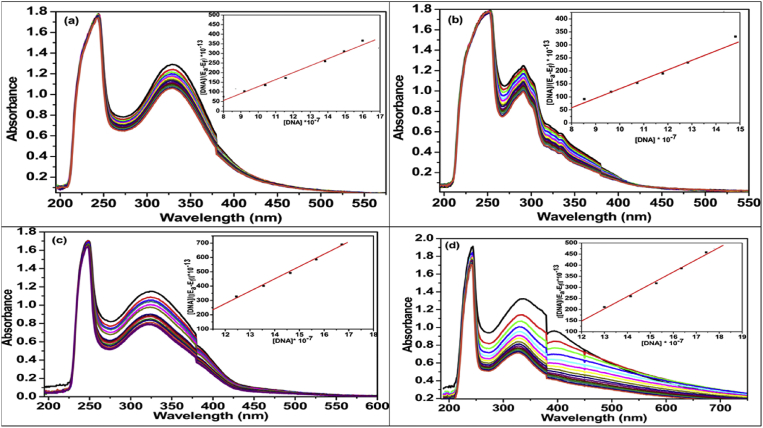
The Interactions of prepared compounds 7a-l and 8a-l with CT-DNA was monitored by absorption titrations using UV-visible spectrophotometer in 290–340 nm range. In the presence of increasing amount of DNA, the spectra of 7a-l and 8a-l (except 7e, 7j and 8b, because of forming precipitation in the buffer) compounds, showed a decrease in the intensity of the band, but the bands were shifted to either lower wavelength or higher wavelength region. The change in absorbance values with increasing amounts of CT-DNA was used to calculate the binding constant of 7a-l and 8a-l. Due to the strong stacking interaction between an aromatic chromophore and the base pairs of DNA, the binding constants concluded that 7a-l and 8a-l interacted with CT-DNA through intercalation mode, which is well supported by the available literature for similar kind of compounds [48, 49, 50, 51, 52]. It is also indicated that the compound form adducts with DNA through intercalation and was stabilized by hydrophobic and hydrogen bond interactions [53, 54, 55, 56]. Hence, the compounds revealed a stronger binding affinity towards DNA double helix (Fig. 5, Table 3).
Fig. 5.
Absorption spectra of (a) 7a, (b) 7b, (c) 8a, and (d) 8c in the absence [Top curve in each] and presence [subsequent curve] of increasing concentration of CT-DNA.
Table 3.
Wavelength shifts, % hypochromism (H%) and binding constants of 7a-l and 8a-l using calf thymus DNA.
| Compound | Free | Bound | Δƛmax (nm)a | H%b | Kb106 M−1c |
|---|---|---|---|---|---|
| 7a | 329 | 328 | 1 | 15.33 | 5.83 |
| 7b | 291 | 291 | 0 | 14.91 | 4.34 |
| 7c | 301 | 300 | 1 | 33.17 | 7.74 |
| 7d | 311 | 313 | 2 | 18.93 | 4.01 |
| 7e | - | - | - | - | - |
| 7f | 328 | 326 | 2 | 11.13 | 7.26 |
| 7g | 318 | 311 | 7 | 8.14 | 7.53 |
| 7h | 334 | 332 | 2 | 11.06 | 3.29 |
| 7i | 340 | 332 | 8 | 44.03 | 13.8 |
| 7j | - | - | - | - | - |
| 7k | 323 | 325 | 2 | 7.18 | 4.65 |
| 7l | 323 | 320 | 3 | 11.43 | 3.34 |
| 8a | 326 | 320 | 6 | 30.61 | 3.70 |
| 8b | - | - | - | - | - |
| 8c | 337 | 325 | 12 | 46.12 | 3.71 |
| 8d | 305 | 312 | 7 | 52.52 | 5.85 |
| 8e | 320 | 313 | 7 | 38.09 | 3.28 |
| 8f | 307 | 303 | 4 | 34.07 | 2.79 |
| 8g | 308 | 311 | 3 | 30.07 | 3.53 |
| 8h | 300 | 297 | 3 | 49.54 | 3.93 |
| 8i | 329 | 316 | 13 | 31.11 | 4.06 |
| 8j | 300 | 314 | 14 | 35.60 | 3.39 |
| 8k | 313 | 319 | 6 | 17.74 | 2.52 |
| 8l | 315 | 325 | 10 | 59.58 | 4.27 |
Δƛmax= (Bound – Free).
H% = [(Af-Ab)/Af)] 100, Where Af and Ab represent the absorbance of free and bound compounds
Kb = Intrinsic binding constant.
3.5. DNA docking studies
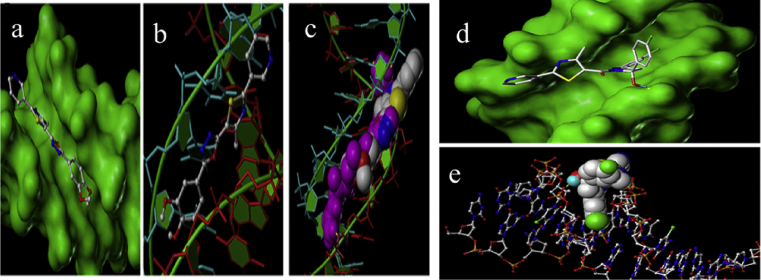
The compound 7i and 8i have been docked with a double-helical B-DNA of sequence: C-G-C-G-A-A-T-T-C-G-C-G (Fig. 6). The results obtained from experiments proposed that the synthesized compound interacted with DNA groves and intercalation. The compounds 7i and 8i fits snugly within the minor groove in the A-A-T-T center. The mode of intercalation of this complex between the DNA base pairs is primarily due to effective stacking forces between the aromatic nucleus and DNA bases [57]. Molecular docking simulation was used to gain an insight of preferential docked location and orientation of the complexes within the DNA groove. It is confirmed that the proposed structure 7i and 8i should be capable of binding to DNA base sequences.
Fig. 6.
(a) Represents the docked complex of the 7i complex with DNA. (b) Binding orientation of compound 7i in the grove of DNA. (c) The DNA binding with netropsin (CPK structure) is shown docked into DNA (stick structure), (d) Binding orientation of compound 8i in the minor grove of DNA. (e)The DNA binding compound 8i (CPK structure) is shown docked into DNA (stick structure). The H bond is shown as (yellow) line.
4. Conclusion
A series of oxadiazole derivatives 7a-l and hydroxypyrazoline derivatives 8a-l incorporating thiazole were synthesized with agreeable yield and their structures were confirmed by spectroscopic techniques. The in vitro cytotoxicity was evaluated for 7a-l and 8a-l on two human cell lines, MDA-MB231 and HT-29. Among the series, compounds 7i and 8i exhibited the most potent antiproliferative activity on both MDA-MB231 and HT-29 cell lines. It was also observed that 7i showed cell cycle arrest in Go/G1 phase on both cell lines. Whereas, 8i revealed the cell cycle arrest in Go/G1 phase on the colorectal cancer cells. Further, 7i (Chemscore 3.90) and 8i (Chemscore 3.70) showed good interaction within the binding site of the CDK-2 protein (5iev) and BCL-2 respectively. In addition, 7a-l and 8a-l (except 7e, 7j and 8b due to the formation of precipitation in the buffer) interacted with CT-DNA via intercalation mode. Furthermore, DNA docking indicated that 7i and 8i interacted with DNA groves and intercalation. Overall, these compounds act as multi-targeting agents, as they can interact with protein target cdk (7i) and BCL-2 (8i) as well as DNA, which are the essential engines for the cell cycle. Thus, these compounds can be further studied as antiproliferative lead structures, to lay the foundation for developing anticancer drugs.
5. Experimental
5.1. Materials and methods
All the reagents for the present study were purchased from commercial suppliers of Sigma-Aldrich, Spectrochem India and Himedia. Melting points were determined in an open capillary tube and were uncorrected. Thin layer chromatography (Merck silica gel 60 F254 coated aluminium plates) confirmed the purity of the products. Synthesised compounds were characterized by 1H-NMR, 13C-NMR, FT-IR, LC-MS and elemental analysis. FT-IR spectrum was recorded on Shimadzu-FTIR Infrared spectrometer (γmax in cm−1). 1H-NMR (400 MHz) and 13C-NMR spectrum, was recorded on a Bruker Advance II 400 spectrometer, with5mm PABBO BB-1H Tubes, using DMSO/CDCl3 as a solvent, using TMS as internal standard (Chemical shift in δ ppm). Elemental analysis was carried out by using VARIOEL-III (Elemental analyze system GmBH). LC-MS was obtained by using Agilent 1200 series LC and MicromasszQ spectrometer. DNA binding studies were carried out using ELICO SL 159 UV-VIS Spectrophotometer.
5.2. Synthesis of pyridine-3-carbothioamide (1)
Pyridine-3-carbothioamide [58] 1 was synthesized by passing H2S to a stirring solution (750 rpm) of 3-cyanopyridine in presence of trimethylamine (TEA) at RT for 8 hrs. The progress of the reaction was monitored by TLC. The precipitate formed in the reaction medium was filtered and recrystallized from ethanol.
5.3. Synthesis of ethyl 4-methyl-2-(pyridin-3-yl)-1,3-thiazole-5-carboxylate (2)
Mixture of Pyridine-3-carbothioamide 1 (2 g, 0.0144 mol) and ethyl-2-chloroacetoacetate (2.04 g, 0.0144 mol) was refluxed at 65 °C for 8 hrs. The progress and completion of the reaction was confirmed by TLC. The reaction mixture was cooled and poured into ice cold water, the precipitate formed was filtered and recrystallized from ethanol.
5.4. Ethyl 4-methyl-2-(pyridin-3-yl)-1,3-thiazole-5-carboxylate (2)
Yield 89%; m.p.162–165 °C; IR (cm−1): 694 (C-S), 1085 (C-O-C), 1579 (C=N), 1720 (C=O), 2932 (C-H), 3044 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 1.3 (t, 3H, -CH3, J = 4.8 Hz), 2.49 (s, 3H, -CH3), 4.39 (q, 2H, -CH2-, J = 9.6 Hz and J = 4.4 Hz), 7.47 (t, 1H(H3), J = 6.4 Hz), 8.07 (dt, 1H(H4), J = 6 Hz and J = 0.8 Hz), 8.64 (dd, 1H(H2), J = 6 Hz and J = 1.2 Hz), 8.98 (d, 1H(H1), J = 0.8 Hz); 13C-NMR (100 MHz, DMSO, δ ppm): 14.5 (-CH3), 17.6 (-CH3), 61.8 (-CH2-), 122.5, 124.7, 128.59, 134.4, 147.6, 152.3, 160.8, 161.6, 166.4. LC-MS, (m/z): 249.08 (M+1), Anal. Calcd. for C12H12N2O2S (248.30): C, 58.05; H, 4.87; N, 11.28. Found: C, 58.11; H, 4.82; N, 11.16. Figs. S1–S4.
5.5. Synthesis of 4-methyl-2-(pyridin-3-yl)-1,3-thiazole-5-carbohydrazide (3)
Compound 2 (2 g, 0.01 mol) was treated with hydrazine hydrate (1.6 g, 0.05 mol) and refluxed at 90 °C for 10 hrs, formation of product was confirmed by TLC, which was obtained by cooling the reaction mixture overnight and recrystallized from ethanol.
5.6. 4-Methyl-2-(pyridin-3-yl)-1,3-thiazole-5-carbohydrazide (3)
Yield 93%; m.p.148–151 °C; IR (cm−1): 671 (C-S), 1589 (NH2 Scissoring), 1620 (C=O), 2848 (C-H), 2916 (Ar C-H), 3352, 3332 (-NH2); 1H-NMR (400 MHz, DMSO, δ ppm): 2.49 (s, 3H, -CH3), 4.60 (d, 2H, -NH2, J = 4.8 Hz), 7.58 (dd, 1H(H3), J = 7.6 Hz and J = 4.8 Hz), 8.31 (d, 1H(H4), J = 8 Hz), 8.71 (t, 1H, J = 4.8 Hz, NH), 9.12 (d, 1H(H2), J = 1.6 Hz), 9.65 (s, 1H(H1)); 13C-NMR (100 MHz, DMSO, δ ppm): 17.4, 124.8, 126.1, 128.9, 134.2, 147.4, 151.8, 155.3, 161.3, 163.4. LC-MS, m/z: 235.04 (M+1), Anal. Calcd. For C10H10N4OS (234.27): C, 51.27; H, 4.30; N, 23.91. Found: C, 51.16; H, 4.32; N, 23.88. Figs. S5–S8.
5.7. Procedure for the synthesis of chalcone (4a-l)
4a-l are prepared by Claisen Schmidt condensation between substituted acetophenones and substituted aldehydes in presence of NaOH as base at RT. The product formed was confirmed by TLC and poured into ice cold water, the precipitate formed was filtered and recrystallized from ethanol.
5.8. Procedure for the synthesis of chalcone dibromides (5a-l)
To a solution of chalcones 4a-l (0.01 mol) in glacial acetic acid (50 mL), bromine (0.01 mol) in glacial acetic acid (25 mL) was added slowly with vigorous stirring (700 rpm) at RT for 12 hrs. The reaction mixture was poured into ice cold water the precipitate formed was filtered and recrystallized from ethanol.
5.9. Procedure for the synthesis of hydroxypyrazolines derivatives (8a-l)
To a mixture of chalcone dibromides 5a-l (0.1 mol) in absolute ethanol (7.5 mL) 4-methyl-2-(pyridin-3-yl)-1,3-thiazole-5-carbohydrazide 3 (0.1 mol) and triethylamine (1 mL) were added and the reaction mixture was heated under reflux for 10 hrs on a water bath. The contents were cooled and poured into ice cold water. The resulting hydroxypyrazolines derivatives 8a-l were collected by filtration and recrystallized from ethanol.
5.10. General procedure for the synthesis of (7a-l)
To a mixture of substituted aromatic and aliphatic acids 6a-l, (0.01 mol) in POCl3 (15 mL) 5-methyl-2-(pyridin-3-yl)-1,3-thiazole-4-carbohydrazide 3 (0.01 mol) was added. The reaction mixture was heated under reflux at 90 °C for 10 hrs on an oil bath. The contents were cooled, neutralized by sodium bicarbonate and poured into crushed ice. The resulting substituted thiazole-oxadiazoles 7a-l, were collected by filtration and recrystallized from ethanol.
5.10.1. 3-{5-[5-(furan-2-yl)-1,3,4-oxadiazol-2-yl]-4-methyl-1,3-thiazol-2-yl}pyridine (7a)
Yield 78%; mp 180–182 °C; IR (cm−1): 649 (C-S), 1041 (C-O-C), 1460 (C=C), 1525 (C=N), 2953 (C-H), 3037 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.46 (s, 3H, -CH3), 6.50 (t, 1H, O-CH = CH-CH = C, J = 6.4 Hz), 6.95 (dd, 1H, O-CH = CH-CH = C, J = 6 Hz), 7.40 (t, 1H(H3), J = 6.8 Hz), 7.78 (dd, 1H, O-CH = CH-CH = C, J = 6 Hz and 1.2 Hz), 8.07 (dt, 1H(H4), J = 6.8 Hz and J = 1.2 Hz), 8.64 (dd, 1H(H2), J = 6.2 Hz and J = 0.8 Hz), 8.98 (d, 1H(H1), J = 2.8 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.3 (-CH3), 113.9, 116.7, 120.6, 123.3, 127.8, 133.0, 139.6, 147.0, 148.2, 149.8, 152.3, 157.4, 161.2, 168.3; LC-MS, (m/z): 311 (M+1); Anal. Calcd. For C15H10N4O2S (310.41): C, 58.05; H, 3.25; N, 18.05. Found: C, 58.07; H, 3.27; N, 18.10.
5.10.2. 3-{5-[5-(6-bromonaphthalen-1-yl)-1,3,4-oxadiazol-2-yl]-4-methyl-1,3-thiazol-2-yl}pyridine (7b)
Yield 65%; m.p. 205–207 °C; IR (cm−1): 658 (C-S), 1042 (C-O-C), 1463 (C=C), 1566 (C=N), 2931 (C-H), 3041 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.50 (s, 3H, -CH3), 7.44–8.99 (m, 10H); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.35 (-CH3), 120.3, 120.5, 120.6, 123.3, 124.9, 126.7, 127.3, 127.8, 128.4, 129.4, 129.8, 130.1, 132.9, 133.0, 148.9, 149.8, 152.3, 161.2, 165.2, 168.3; LC-MS, (m/z): 449.99 (M+), 451.99 (M+2); Anal. Calcd. For C21H13BrN4OS (449.32): C, 56.13; H, 2.92; N, 12.47. Found: C, 56.10; H, 2.94; N, 12.49.
5.10.3. 3-{4-methyl-5-[5-(4-methylphenyl)-1,3,4-oxadiazol-2-yl]-1,3-thiazol-2-yl}pyridine (7c)
Yield 73%; m.p. 210–212 °C; IR (cm−1): 696 (C-S), 1016 (C-O-C), 1492 (C=C), 1577 (C=N), 2960 (C-H), 3039 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.34 (s, 3H, -CH3), 2.47 (s, 3H, -CH3), 7.29 (d, 2H(2H6), J = 6 Hz), 7.42 (t, 1H(H3), J = 6 Hz), 7.52 (d, 2H(2H5), J = 5.6 Hz), 8.02 (t, 1H(H4), J = 6 Hz), 8.59 (dd, 1H(H2), J = 6 Hz and J = 5.6 Hz), 8.8 (d, 1H(H1), J = 1.2 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.57(-CH3), 21.69(-CH3), 115.4, 120.6, 123.93, 126.9, 128.8, 129.9, 133.8, 142.6, 147.9, 151.6, 157.1, 159.2, 164.4, 165.4; LC-MS, (m/z): 334 (M+); Anal. Calcd. For C18H14N4OS (334.39): C, 64.65; H, 4.22; N, 16.75. Found: C, 64.62; H, 4.25; N, 16.72. Figs. S13–S16.
5.10.4. 4-{5-[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,3,4-oxadiazol-2-yl}aniline (7d)
Yield 70%; m.p. 236–238 °C; IR (cm−1): 673 (C-S), 1037 (C-O-C), 1471 (C=C), 1573 (C=N), 2941 (C-H), 3045 (Ar C-H), 3286, 3221 (-NH2); δ 1H-NMR (400 MHz, CDCl3, δ ppm): 2.47 (s, 3H, -CH3), 4.16 (s, 2H, -NH2), 6.73 (d, 2H(2H6), J = 6 Hz), 7.36–7.44 (7.38 (d, 2H(2H5), J = 6 Hz), 7.44 (t, 1H(H3), J = 6.8 Hz), 8.03–8.05 (dt, 1H(H4), J = 6.1 Hz, J = 1.2 Hz), 8.61 (dd, 1H(H2), J = 6.4 Hz and J = 1.2 Hz), 8.90 (d, 1H(H1), J = 1.2 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 16.6 (-CH3), 109.5, 114.5, 118.0, 127.6(d), 134.9(d), 137.0, 140.2, 149.2, 151.3, 156.2, 161.3, 165.8; LC-MS, (m/z): 336 (M+1); Anal. Calcd. For C17H13N5OS (335.38): C, 60.88; H, 3.91; N, 20.88. Found: C, 60.85; H, 3.93; N, 20.85.
5.10.5. 3-{5-[5-(2-chloro-4-nitrophenyl)-1,3,4-oxadiazol-2-yl]-4-methyl-1,3-thiazol-2-yl}pyridine (7e)
Yield 86%; m.p. 203–205 °C; IR (cm−1): 3032 (Ar C-H), 2958 (C-H), 1575 (C=N), 1516 & 1346 (N-O), 696 (C-S); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.46 (s, 3H, -CH3), 7.42 (t, 1H(H3), J = 6 Hz), 7.8 (d, 1H(H7), J = 6 Hz), 8.02 (dt, 1H(H4), J = 6 Hz and J = 2.4 Hz), 8.19 (dd, 1H(H6), J = 5.6 Hz and J = 1.8 Hz), 8.37 (d, 1H(H5), J = 1.2 Hz), 8.59 (dd, 1H(H2), J = 6 Hz and J = 1.2 Hz), 8.88 (d, 1H(H1), J = 1.2 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.7(-CH3), 114.6, 122.0, 123.6, 126.6, 128.0, 132.0, 133.9, 134.1, 147.9, 149.6, 151.9, 158.3, 160.8, 161.2, 166.5; LC-MS, m/z: 399.99 (M+), 402.99 (M+2); Anal. Calcd. For C17H10ClN5O3S (399.81): C, 51.07; H, 2.52; N, 17.57. Found: C, 51.91; H, 2.56; N, 17.14. Figs. S17–S20.
5.10.6. 3-chloro-4-{5-[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,3,4-oxadiazol-2-yl}pyridine (7f)
Yield 90%; m.p.199–201 °C; IR (cm−1): 696 (C-S), 1076 (C-O-C), 1346 (N-O, symmetric), 1516 (N-O, asymmetric), 1575 (C=N), 2958 (C-H), 3032 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.49 (s, 3H, -CH3), 7.39 (dd, 1H(H3), J = 6.4 Hz and J = 1.2 Hz), 7.71 (d, 1H(H7), J = 6 Hz), 8.09 (dt, 1H(H4), J = 6.2 Hz, J = 1.2 Hz), 8.54 (dd, 1H(H2), J = 6.6 Hz and J = 1.2 Hz), 8.63 (s, 1H(H6)), 8.73 (s, 1H(H5), 8.84 (d, 1H(H1), J = 1.2 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.8(-CH3), 120.6, 120.7, 123.3, 126.7, 127.8, 128.0, 133.0, 147.7, 148.9, 149.1, 149.8, 152.3, 161.2, 164.8, 168.3. LC-MS, (m/z): 355.81 (M+), 357.92 (M+2); Anal. Calcd. For C16H10ClN5OS (355.80): C, 54.01; H, 2.83; N, 19.68. Found: C, 54.04; H, 2.80; N, 19.65.
5.10.7. 3-{5-[5-(4-chlorophenyl)-1,3,4-oxadiazol-2-yl]-4-methyl-1,3-thiazol-2-yl}pyridine (7g)
Yield 85%; m.p.121–123 °C; IR (cm−1): 667 (C-S), 1064 (C-O-C), 1488 (C=C), 1543 (C=N), 2945 (C-H), 3031 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.41 (s, 3H, -CH3), 7.39–7.44 (dd, 3H(H3, 2H6), J = 6.2 Hz, J = 12.4 Hz), 7.51 (d, 2H(2H5), J = 6 Hz), 8.16 (dt, 1H(H4), J = 6.4 Hz, J = 1.2 Hz), 8.63 (dd, 1H(H2), J = 6.1 Hz), 8.91 (d, 1H(H1), J = 0.8 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.5(-CH3), 120.6, 122.8, 123.3, 127.8, 128.8, 128.8, 129.2, 129.2, 133.0, 135.7, 148.9, 149.8, 152.3, 161.2, 163.7, 168.3; LC-MS, (m/z): 354.8 (M+), 356.3 (M+2); Anal. Calcd. For C17H11ClN4OS (354.81): C, 57.55; H, 3.12; N, 15.79. Found: C, 57.52; H, 3.09; N, 15.76.
5.10.8. 3-{4-methyl-5-[5-(pyridin-4-yl)-1,3,4-oxadiazol-2-yl]-1,3-thiazol-2-yl}pyridine (7h)
Yield 79%; m.p.116–118 °C; IR (cm−1): 653 (C-S), 1060 (C-O-C), 1469 (C=C), 1568 (C=N), 2949 (C-H), 3035 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.46 (s, 3H, -CH3),7.42 (t, 1H(H3), J = 8 Hz), 7.55 (d, 2H(2H5), J = 5.6 Hz), 8.02 (dd, 1H(H4), J = 6 Hz and J = 1.2 Hz), 8.57–8.64 (3H, (8.58 (dd, 1H(H2), J = 6 Hz and J = 0.8 Hz), 8.64 (d, 2H(2H6), J = 6 Hz), 8.88 (d, 1H(H1), J = 1.2 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.7(-CH3), 114.6, 120.2, 123.9, 130.5, 133.9, 147.9, 151.0, 151.8, 158.2, 160.5, 162.4, 166.2; LC-MS, (m/z): 322 (M+1); Anal. Calcd. For C16H11N5OS (321.35): C, 59.80; H, 3.45; N, 21.79. Found: C, 59.76; H, 3.41; N, 21.82. Figs. S9–S12.
5.10.9. 3-{5-[5-(3,4-dimethoxyphenyl)-1,3,4-oxadiazol-2-yl]-4-methyl-1,3-thiazol-2-yl}pyridine (7i)
Yield 83%; m.p. 126–128 °C; IR (cm−1): 634 (C-S), 1139 (C-O-C), 1465 (C=C), 1510 (C=N), 2939 (C-H), 3047 (Ar C-H); 1H-NMR (400 MHz, DMSO, δ ppm): 2.47 (s, 3H, -CH3), 3.80 (s, 3H, -OCH3), 3.83 (s, 3H, -OCH3), 7.00 (d, 1H(H7), J = 6 Hz), 7.19 (d, 1H(H5), J = 1.2 Hz), 7.20–7.22 (dd, 1H(H6), J = 6 Hz, J = 1.2 Hz), 7.42 (t, 1H(H3), J = 6 Hz), 8.00–8.02 (dt, 1H(H4), J = 6 Hz, J = 1.2 Hz), 8.57–8.59 (dd, 1H(H2), J = 1.2 Hz), 8.87 (d, 1H(H1), J = 0.8 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.5(-CH3), 56.0(-OCH3), 56.1(-OCH3), 109.5, 111.2, 115.4, 115.9, 120.5, 133.7, 147.8, 149.5, 151.6, 152.3, 157.0, 159.1, 164.2, 165.3; LC-MS, (m/z): 381.07 (M+1); Anal. Calcd. For C19H16N4O3S (380.42): C, 59.99; H, 4.24; N, 14.73. Found: C, 59.96; H, 4.21; N, 14.76. Figs. S21–S24.
5.10.10. 3-{5-[5-(3,4-dichlorophenyl)-1,3,4-oxadiazol-2-yl]-4-methyl-1,3-thiazol-2-yl}pyridine (7j)
Yield 90%; m.p. 218–220 °C; IR (cm−1): 676 (C-S), 1091 (C-O-C), 1490 (C=C), 1549 (C=N), 2947 (C-H), 3034 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.52 (s, 3H, -CH3), 7.48 (m, 4H(H3, H5, H6, H7)), 8.08 (dt, 2H(H4), J = 6 Hz and J = 1.2 Hz), 8.67 (dd, 1H(H2), J = 6.2 Hz and J = 1.2 Hz), 8.93 (d, 1H(H1), J = 1.6 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.85, 120.6, 123.3, 125.5, 127.8, 128.7, 128.8, 129.9, 133.0, 133.2, 133.6, 148.9, 149.8, 152.3, 161.2, 163.7, 168.3; LC-MS, (m/z): 389.30 (M+), 391.23 (M+2), 393.41 (M+4); Anal. Calcd. For C17H10Cl2N4OS (389.25): C, 52.45; H, 2.59; N, 14.39. Found: C, 52.47; H, 2.56; N, 14.41.
5.10.11. 3-[5-(5-benzyl-1,3,4-oxadiazol-2-yl)-4-methyl-1,3-thiazol-2-yl]pyridine (7k)
Yield 71%; m.p. 190–192 °C; IR (cm−1): 690 (C-S), 1009 (C-O-C), 1485 (C=C), 1569 (C=N), 2957 (C-H), 3029 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.43 (s, 3H, -CH3), 3.93 (s, 2H, -CH2-), 7.15–7.42 (m, 5H(2H5, 2H6, H7)), 7.39 (t, 1H(H3), J = 8 Hz), 7.97 (dt, 1H(H4), J = 6.1 Hz and J = 1.2 Hz), 8.48 (dd, 1H(H2), J = 6.4 Hz and J = 0.8 Hz), 8.86 (d, 1H(H1), J = 0.8 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.2, 33.9, 120.6, 123.3, 127.8, 128.6, 128.7, 128.7, 128.9, 133.0, 134.0, 148.9, 149.8, 152.3, 161.2, 165.6, 168.3; LC-MS, (m/z): 335.51 (M+1); Anal. Calcd. For C18H14N4OS (334.39): C, 64.65; H, 4.22; N, 16.75. Found: C, 64.68; H, 4.24; N, 16.79.
5.10.12. 3-{4-methyl-5-[5-(trichloromethyl)-1,3,4-oxadiazol-2-yl]-1,3-thiazol-2-yl}pyridine (7l)
Yield 80%; m.p.169–171 °C; IR (cm−1): 677 (C-S), 1103 (C-O-C), 1483 (C=C), 1565 (C=N), 2961 (C-H), 3050 (Ar C-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.56 (s, 3H, -CH3), 7.46 (t, 1H(H3), J = 7.8 Hz), 8.07 (dt, 1H(H4), J = 6.6 Hz and J = 1.2 Hz), 8.64 (dd, 1H(H2), J = 6.2 Hz and J = 1.2 Hz), 8.89 (d, 1H(H1), J = 0.8 Hz); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.61, 92.85, 120.60, 123.32, 127.82, 133.01, 148.94, 149.87, 152.31, 161.26, 162.34, 168.30; LC-MS, (m/z): 361.81 (M+), 363.27 (M+2), 365.43 (M+4), 368.18 (M+6); Anal. Calcd. For C12H7Cl3N4OS (361.63): C, 39.85; H, 1.95; N, 15.49. Found: C, 39.88; H, 1.97; N, 15.46.
5.10.13. 3-(4-methylphenyl)-5-(3-chlorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8a)
Yield 80%; m.p.106–108 °C; IR (cm−1): 682 (C-S), 1593 (C=N), 1631 (C=O), 2845 (Ar C-H), 2918 (C-H), 3152 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.30 (s, 3H, -CH3), 2.74 (s, 3H, -CH3), 3.46 (d, 1H, -CH2-, J = 18.4 Hz), 3.76 (d, 1H, -CH2-, J = 18.4 Hz), 5.25 (d, 1H, O-H, J = 16 Hz), 7.24–9.16 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.9(-CH3), 20.0(-CH3), 49.1, 94.1, 119.9, 122.8, 123.8, 125.9, 127.4, 128.0, 128.3, 128.6, 129.2, 129.7, 131.6(d), 132.9, 134.0, 137.2, 139.0, 141.9, 146.9, 150.4, 150.9, 160.0, 162.4, 166.7; LC-MS, (m/z): 487.9 (M+), 490.4 (M+1); Anal. Calcd. For C12H7Cl3N4OS (488.98): C, 63.86; H, 4.33; N, 11.46. Found: C, 63.80; H, 4.29; N, 11.42. Figs. S25–S27.
5.10.14. 3-(4-methyl)-5-(3-nitrophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8b)
Yield 84%; m.p.108–110 °C; IR (cm−1): 698 (C-S), 1527 (N=O), 1602 (C=N), 1654 (C=O), 2842 (C-H), 2916 (Ar C-H), 3163 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.34 (s, 3H, -CH3), 2.79 (s, 3H, -CH3), 3.54 (d, 1H, -CH2-, J = 18.4 Hz), 3.86 (d, 1H, -CH2-, J = 18.4 Hz), 5.25 (d, 1H, O-H, J = 16 Hz), 7.19–9.29 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 16.6(-CH3), 21.1(-CH3), 47.4, 92.6, 122.9, 124.8, 125.3, 124.3, 127.2, 128.9, 129.4, 129.4, 132.0, 137.2, 134.5, 134.9, 135.3, 137.0, 139.0, 144.1, 147.1, 148.3, 149.2, 150.4, 151.35; LC-MS, (m/z): 499.35 (M+); Anal. Calcd. For C12H7Cl3N4OS (499.54): C, 62.51; H, 4.24; N, 14.02. Found: C, 62.59; H, 4.25; N, 14.07. Figs. S28–S30.
5.10.15. 3-(4-methylphenyl)-5-(4-methylphenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8c)
Yield 82%; m.p. 88–90 °C; IR (cm−1): 676 (C-S), 1592 (C=N), 1641 (C=O), 2847 (C-H), 2912 (Ar C-H), 3157 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.36 (s, 3H, -CH3), 2.68 (d, 6H, 2CH3), 2.80 (s, 3H, -CH3), 3.48 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 3.79 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 5.26 (s, 1H, O-H), 7.26–9.33 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 18.98(-CH3), 19.65(-CH3), 19.7(-CH3), 50.1, 92.0, 121.0(d), 127.1(d), 128.9, 129.4, 129.6, 133.9, 138.2, 140.2, 143.1, 147.9, 151.3, 152.2, 160.9(d), 163.0, 163.3(d), 165.5, 167.6, 167.6; LC-MS, (m/z): 468.81 (M+); Anal. Calcd. For C12H7Cl3N4OS (468.57): C, 69.21; H, 5.16; N, 11.96. Found: C, 69.15; H, 5.21; N, 11.88.
5.10.16. 3-(2,4-dichlorophenyl)-5-(3,4-dimethoxyphenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8d)
Yield 79%; m.p.122–124 °C; IR (cm−1): 698 (C-S), 1224 (C-O-C), 1605 (C=N), 1627 (C=O), 2851 (C-H), 2928 (Ar C-H), 3206 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.38 (s, 3H, -CH3), 3.56 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 3.82 (s, 3H, -OCH3), 3.84 (s, 3H, -OCH3), 3.88 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 5.14 (s, 1H, O-H), 7.45–9.45 (m, 10H); 13C-NMR (100 MHz, CDCl3, δ ppm): 17.2(-CH3), 49.8, 51.0, 51.2, 91.1, 116.3, 121.0, 126.2, 127.1, 128.4, 128.1, 128.9, 128.9, 129.4, 129.6, 132.9, 138.5, 139.4, 141.1, 149.9, 152.2, 153.2, 161.9, 163.1, 164.2, 165.3, 166.3, 168.6, 169.6; LC-MS, (m/z): 569.66 (M+), 571.62 (M+1), 573.60 (M+4); Anal. Calcd. For C12H7Cl3N4OS (569.46): C, 56.95; H, 3.89; N, 9.84. Found: C, 56.93; H, 3.88; N, 9.80.
5.10.17. 3-(4-methylphenyl)-5-(3-fluorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8e)
Yield 94%; m.p.124–126 °C; IR (cm−1): 698 (C-S), 1605 (C=N), 1623 (C=O), 2848 (C-H), 2917 (Ar C-H), 3306 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.39 (s, 3H, -CH3), 2.81 (s, 3H, -CH3), 3.59 (d, 1H, -CH2-, J = 18.4 Hz), 3.91 (d, 1H, -CH2-, J = 18.4 Hz), 5.23 (s, 1H, O-H), 7.26–9.31 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 18.6(-CH3), 20.6, 49.0, 92.3, 116.4(d), 122.0, 127.0, 128.35, 128.7(d), 129.0, 129.4, 134.0, 138.1, 140.4, 143.0, 148.0, 151.5, 152.4, 161.0, 163.0, 163.4, 165.6, 167.6, 167.8; LC-MS, (m/z): 473.61 (M+); Anal. Calcd. For C12H7Cl3N4OS (472.53): C, 66.09; H, 4.48; N, 11.86. Found: C, 66.14; H, 4.49; N, 11.77.
5.10.18. 3-(4-methoxyphenyl)-5-(4-fluorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8f)
Yield 81%; m.p. 160–162 °C; IR (cm−1): 698 (C-S), 1226 (C-O-C), 1605 (C=N), 1621 (C=O), 2845 (C-H), 2921 (Ar C-H), 3296 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.77 (s, 3H, -CH3), 3.54 (d, 1H, -CH2-, J = 18.4 Hz), 3.81 (s, 3H, -OCH3), 3.86 (d, 1H, -CH2-, J = 18.4 Hz), 5.14 (s, 1H, O-H), 7.21–9.43 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 18.9(-CH3), 47.9, 50.0, 50.3, 95.0, 116.3(d), 121.0(d), 127.1(d), 128.4, 128.9(d), 128.9, 129.4, 129.6, 133.9, 138.2, 140.2, 143.15, 147.9, 151.3, 152.2, 160.9(d), 163.0, 163.3(d), 165.5, 167.6, 167.6; LC-MS, (m/z): 488.70 (M+); Anal. Calcd. For C12H7Cl3N4OS (488.53): C, 63.92; H, 4.33; N, 11.47. Found: C, 63.84; H, 4.30; N, 11.45. Figs. S31–S33.
5.10.19. 3-(4-fluorophenyl)-5-(4-fluorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8g)
Yield 90%; m.p. 163–165 °C; IR (cm−1): 698 (C-S), 1602 (C=N), 1631 (C=O), 2958 (C-H), 3053 (Ar C-H), 3359 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.80 (s, 3H, -CH3), 3.43 (d, 1H, -CH2-, J = 18.4 Hz), 3.79 (d, 1H, -CH2-, J = 18.4 Hz), 5.28 (s, 1H, O-H), 7.06–9.22 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 19.0(-CH3), 48.1(-CH2-), 94.6, 115.7, 116.4, 120.8, 123.9, 125.9(d), 126.9(d), 129.0, 133.9, 139.0, 148.0, 151.5, 152.2, 161.0(d), 161.4, 163.0(d), 163.5, 165.6, 167.8; LC-MS, (m/z): 477.61 (M+1); Anal. Calcd. For C12H7Cl3N4OS (476.49): C, 63.02; H, 3.81; N, 11.76. Found: C, 63.05; H, 3.80; N, 11.77. Figs. S34–S37.
5.10.20. 3-(4-fluorolphenyl)-5-(4-methoxyphenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8h)
Yield 71%; m.p. 74–76 °C; IR (cm−1): 693 (C-S), 1221 (C-O-C), 1602 (C=N), 1625 (C=O), 2844 (C-H), 2913 (Ar C-H), 3302 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.68 (s, 3H, -CH3), 3.52 (dd, 1H, -CH2-, J = 18.4 Hz), 3.77 (s, 3H, -OCH3), 3.83 (dd, 1H, -CH2-, J = 18.4 Hz), 5.11 (s, 1H, O-H), 7.19–9.40 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 18.9(-CH3), 49.9, 50.0, 94.9, 116.29(d), 127.0, 128.9(d), 128.8, 129.5, 133.8, 138.2, 140.2, 143.0, 147.8, 151.2, 152.2, 160.8(d), 163.0, 163.3(d), 165.6, 167.7, 167.5; LC-MS, (m/z): 488.70 (M+); Anal. Calcd. For C12H7Cl3N4OS (488.53): C, 63.92; H, 4.33; N, 11.47. Found: C, 63.84; H, 4.30; N, 11.45.
5.10.21. 3-(4-fluorophenyl)-5-(3-chlorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8i)
Yield 64%; m.p. 161–163 °C; IR (cm−1): 694 (C-S), 1599 (C=N), 1631 (C=O), 2938 (C-H), 3044 (Ar C-H), 3359 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.31 (s, 3H, -CH3), 3.55 (d, 1H, -CH2-, J = 18.4 Hz), 3.83 (d, 1H, -CH2-, J = 18.4 Hz), 5.16 (s, 1H, O-H), 7.31–9.28 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 19.4(-CH3), 49.3, 92.4, 117.7, 118.4, 121.7, 124.7, 126.9(d), 127.9(d), 130.1, 134.9, 140.2, 149.3, 152.5, 153.8, 162.3(d), 162.9, 164.1(d), 164.4, 165.8, 167.5; LC-MS, (m/z): 493.44 (M+), 495.56 (M+1); Anal. Calcd. For C12H7Cl3N4OS (492.95): C, 60.91; H, 3.68; N, 11.37. Found: C, 60.92; H, 3.67; N, 11.22.
5.10.22. 3-(4-chlorophenyl)-5-(4-fluorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8j)
Yield 92%; m.p. 154–156 °C; IR (cm−1): 695 (C-S), 1600 (C=N), 1629 (C=O), 2936 (C-H), 3045 (Ar C-H), 3357 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.75 (s, 3H, -CH3), 3.51 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 3.76 (dd, 1H, -CH2-, J = 18.4 Hz, and J = 6.8 Hz), 5.23 (s, 1H, O-H), 7.25–9.21 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 19.4(-CH3), 49.4, 92.4, 117.8, 118.4, 121.76, 124.7, 126.8(d), 127.8(d), 130.0, 134.9, 140.5, 149.3, 152.5, 153.8, 162.4(d), 162.8, 164.22(d), 164.3, 165.8, 167.5; LC-MS, (m/z): 493.52 (M+), 495.47 (M+1); Anal. Calcd. For C12H7Cl3N4OS (492.95): C, 60.91; H, 3.68; N, 11.37. Found: C, 60.92; H, 3.67; N, 11.22.
5.10.23. 3-(4-methoxyphenyl)-5-(4-chlorophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8k)
Yield 77%; m.p. 85–87 °C; IR (cm−1): 697 (C-S), 1225 (C-O-C), 1602 (C=N), 1624 (C=O), 2842 (C-H), 2915 (Ar C-H), 3298 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.80 (s, 3H, -CH3), 3.55 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 3.79 (s, 3H, -OCH3), 3.86 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 5.14 (s, 1H, O-H), 7.20–9.31 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 18.6(-CH3), 50.0, 50.2, 94.9, 116.2(d), 121.1(d), 127.2(d), 128.4, 128.9(d), 128.9, 129.4, 129.7, 133.9, 138.2, 140.2, 143.1, 147.9, 151.3, 152.3, 160.9(d), 163.2, 163.4(d), 165.4, 167.5, 167.6; LC-MS, (m/z): 505.81 (M+), 507.78 (M+2); Anal. Calcd. For C12H7Cl3N4OS (504.98): C, 61.84; H, 4.19; N, 11.09. Found: C, 61.81; H, 4.18; N, 11.07.
5.10.24. 3-(4-methoxyphenyl)-5-(4-bromophenyl)-5-hydroxy-4,5-dihydro-1H-pyrazol-1-yl)[4-methyl-2-(pyridin-3-yl)-1,3-thiazol-5-yl]methanone (8l)
Yield 89%; m.p. 143–145 °C; IR (cm−1): 692 (C-S), 1223 (C-O-C), 1601 (C=N), 1620 (C=O), 2842 (C-H), 2914 (Ar C-H), 3301 (O-H); 1H-NMR (400 MHz, CDCl3, δ ppm): 2.71 (s, 3H, -CH3), 3.52 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 3.77 (s, 3H, -OCH3), 3.84 (dd, 1H, -CH2-, J = 18.4 Hz and J = 6.8 Hz), 5.11 (s, 1H, O-H), 7.18–9.38 (m, 12H); 13C-NMR (100 MHz, CDCl3, δ ppm): 18.6 (-CH3), 50.0, 50.2, 94.9, 116.2(d), 121.0(d), 127.2(d), 128.4, 128.8(d), 128.9, 129.4, 129.6, 133.8, 138.2, 140.2, 143.1, 147.9, 151.3, 152.2, 160.8(d), 163.0, 163.3(d), 165.4, 167.5, 167.6; LC-MS, (m/z): 549.61 (M+), 551.73 (M+2); Anal. Calcd. For C12H7Cl3N4OS (549.43): C, 56.84; H, 3.85; N, 10.20. Found: C, 56.88; H, 3.83; N, 10.22.
5.11. MTT assay
MDA-MB231 (triple negative breast cancer) and HT-29 (Colorectal cancer) cells, were procured form National Centre for Cell Sciences, Pune. They were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% foetal bovine serum and 1% antibiotic-antimycotic solution. The cells were maintained at 37 °C and 5% CO2 levels. They were used for the experiments after three consecutive passages.Cytotoxicity of 1,3,4-Oxadiazole derivatives was assessed by MTT assay (Mosmann, 1983). Briefly, MDA-MB231 and HT-29 cells were seeded onto 96 well microtiter plates at a density of 5,000 cells/well and incubated overnight in the humidified atmosphere. The test compounds were added at concentrations of 6.25, 12.5, 25, 50 and 100 μM. 48 hrs after drug addition, 100 μL of MTT solution (1 mg/mL) was added to the wells, 4 hrs following which the formazan crystals formed were, solubilized in DMSO. Absorbance was recorded at 570 nm and percentage cytotoxicity was calculated in comparison with control.
5.12. Flow cytometry
To concentrate the impact of compounds 7i and 8i on different phase of MDA-MB231 and HT 29 cell cycle, Flow cytometric studies were carried out. Cells were seeded at a density of 0.5 million cells in 60 mm dishes. 24 hrs after seeding, they were treated with IC50 concentration of the drug 7i and 8i, 48 hrs following drug addition, cells were harvested by trypsinization, washed with ice-cold PBS and fixed with 70% chilled ethanol. Fixed cells were stored at -20 °C for 48 hrs. Cells in ethanol were centrifuged at 3000 rpm for 20 minutes, ethanol was removed and washed with PBS. Cells were incubated with 200 μL of 50 μg/mL solution of RNase A at 37 °C for 5 hrs and added with 1 μL Propidium iodide. Cell cycle analysis was done using analytical flow cytometer (Guava EasyCyte, Merck Millipore).
5.13. Molecular docking studies
The crystal structures from PDB 5iev of Cyclin-dependent kinase 2 with Roniciclib (for 7i) and 4LVT from PDB of B-cell lymphoma 2 (BCL-2) with Navitoclax (for 8i) was used for the study. The protein was prepared by removing all water molecules and adding all hydrogen atoms. The ligands 7i and 8i was docked into the active sites using the molecular docking software SYBYL ver 7.3 (Tripos, L.P.) Surflex-Dock (BioPharmics LLC.) with the default parameters. The proprietary software is licensed to Manipal Institute of Technology, Manipal University, India. Surflex-Dock is a program for calculating the docking modes of small molecules into protein-binding sites. In this study, we have used ChemScore, a scoring function that is derived from regression against ligand-receptor binding free energies. In the docking process, the active site was defined. For each ligand, 20 conformations were generated (40 × 20 = 800 conformations) and then docked into M1 mAChR.
5.14. DNA binding studies
Electronic absorption spectroscopy, is one of the most common techniques for the investigation of the binding mode of small molecules to DNA [59]. All the experiments involving the binding of prepared compounds with CT-DNA, were carried out in double distilled water. A solution of CT-DNA in 50 mM NaCl/5 mM Tris–HCl (pH 7.2) buffer, gave a UV absorbance at 260 and 280 nm and was found to be 1.8–1.9, indicating that the DNA was sufficiently free of protein [55]. A concentrated stock solution of DNA, was prepared in 5 mM Tris–HCl/50 mM NaCl in water at pH 7.0 and the concentration of CT-DNA was determined per nucleotide by taking the absorption coefficient (6,600 dm3 mol-1 cm−1) at 260 nm. The compounds (except 5e and 5j, because of forming precipitation in buffer) were dissolved in DMF solution for all the experiments. Absorption titration experiments, were performed with a fixed concentration of the compound (10–25 μM), and increasing concentration of DNA (0–350 μM). After equilibrium was reached (ca. 5 min), the spectra were recorded against an analogous blank solution containing the same concentration of DNA. To enable quantitative comparison of the DNA binding affinities, the intrinsic binding constant (Kb) of the complexes for binding with CT-DNA were obtained by the following equation [46].
Where [DNA] is the concentration of DNA in base pair, and corresponds to the molar extinction coefficients of apparent, bound and free metal complexes respectively. A plot of versus [DNA], gave a slope and an intercept equal to ; Kb is the ratio of slope to the intercept.
5.15. DNA docking
The crystal structure 6BNA [56] from PDB with netropsin was used for the study. The DNA structure was prepared by removing the bound netropsin molecule and removing all water molecules to avoid potential interference with the docking. The binding site was defined using an atom in the center of the DNA molecule and was large enough, that it encompassed netropsin binding site.
Declarations
Author contribution statement
Rangappa Santosh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Gundibasappa K. Nagaraja, Mukunthan K. Selvam, Ashwini Prabhu: Performed the experiments.
Punchappady D. Rekha: Conceived and designed the experiments.
Panchangam M. Krishna: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors are thankful to Department of Chemistry, Mangalore University for providing laboratory facilities for performing the experiments.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.https://publications.cancerresearchuk.org/downloads/product/cs_report_world.pdf. (Accessed 12 April 2017).
- 2.Jemal A., Bray F., Melissa M., Ferlay J., Ward E., Forman D. Global cancer statistics. Cancer. J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Chan S.K., Griffith O.L., Tai I.T., Jones S.J. Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol. Biomark. Prev. 2008;17:543–552. doi: 10.1158/1055-9965.EPI-07-2615. [DOI] [PubMed] [Google Scholar]
- 4.Hardebol A.H.S., Carvalho B., de M.W., Postma C., Diemen P.M.D., Mongera S. Identification of key genes for carcinogenic pathways associated with colorectal adenoma-to-carcinoma progression. Tumour Biol. 2010;31:89–96. doi: 10.1007/s13277-009-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Siddiqui N., Ahsan W. Synthesis, anticonvulsant and toxicity screening of thiazolyl–thiadiazole derivatives. Med. Chem. Res. 2011;20:261–268. [Google Scholar]
- 7.Rawal R.K., Tripathi R., Katti S.B., Pannecouque C., Clercq E.D. Design and synthesis of 2-(2,6-dibromophenyl)-3-heteroaryl-1,3-thiazolidin-4-ones as anti-HIV agents. Eur. J. Med. Chem. 2008;43:2800–2806. doi: 10.1016/j.ejmech.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 8.Lino T., Tsukahara D., Kamata K., Sasaki K., Ohyama S., Hosaka H., Hasegawa T., Chiba M., Nagata Y., Eiki J., Nishimura T. Discovery of potent and orally active 3-alkoxy-5-phenoxy-N-thiazolyl benzamides as novel allosteric glucokinase activators. Bioorg. Med. Chem. 2009;17:2733–2743. doi: 10.1016/j.bmc.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 9.Shiradkar M.R., Akula K.C., Dasari V., Baru V., Chiningiri B., Gandhi S., Kaur R. Clubbed thiazoles by MAOS: a novel approach to cyclin-dependent kinase 5/p25 inhibitors as a potential treatment for Alzheimer's disease. Bioorg. Med. Chem. 2007;15:2601–2610. doi: 10.1016/j.bmc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Makam P., Thakur P.K., Kannan T. In vitro and in silico antimalarial activity of 2-(2-hydrazinyl)thiazole derivatives. Eur. J. Pharm. Sci. 2014;52:138–145. doi: 10.1016/j.ejps.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Desai N.C., Bhatt N., Somani H., Trivedi A. Synthesis, antimicrobial and cytotoxic activities of some novel thiazole clubbed 1,3,4-oxadiazoles. Eur. J. Med. Chem. 2013;67:54–59. doi: 10.1016/j.ejmech.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helal M.H.M., Salem M.A., El-Gaby M.S.A., Aljahdali M. Synthesis and biological evaluation of some novel thiazole compounds as potential anti-inflammatory agents. Eur. J. Med. Chem. 2013;65:517–526. doi: 10.1016/j.ejmech.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Diana P., Carbone A., Barraja P., Montalbano A., Parrino B., Lopergolo A., Pennati M., Zaffaroni N., Cirrincione G. Synthesis and antitumor activity of 3-(2-Phenyl-1,3-thiazol-4-yl)-1H-indoles and 3-(2-Phenyl-1,3-thiazol-4-yl)-1H-7-azaindoles. ChemMedChem. 2011;6:1300–1309. doi: 10.1002/cmdc.201100078. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B.L., Song L.X., Li Y.F., Li Y.L., Guo Y.Z., Zhang E., Liu H.M. Synthesis and biological evaluation of dehydroepiandrosterone-fused thiazole, imidazo[2,1-b]thiazole, pyridine steroidal analogues. Steroids. 2014;80:92–101. doi: 10.1016/j.steroids.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Carbone A., Pennati M., Parrino B., Lopergolo A., Barraja P., Montalbano A., Spano V., Sbarra S., Doldi V., De Cesare M., Cirrincione G., Diana P., Zaffaroni N. Novel 1H-Pyrrolo[2,3-b]pyridine derivative nortopsentin analogues: synthesis and antitumor activity in peritoneal mesothelioma experimental models. J. Med. Chem. 2013;56:7060–7072. doi: 10.1021/jm400842x. [DOI] [PubMed] [Google Scholar]
- 16.Parrino B., Attanzio A., Spanò V., Cascioferro S., Montalbano A., Barraja P., Tesoriere L., Diana P., Cirrincione G., Carbone A. Synthesis, antitumor activity and CDK1 inhibiton of new thiazole nortopsentin analogues. Eur. J. Med. Chem. 2017;138:371–383. doi: 10.1016/j.ejmech.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 17.Lednicer D., Mitscher L.A., George G.I. Vol. 4. Wiley; New York, USA: 1990. pp. 95–97. (Organic Chemistry of Drug Synthesis). [Google Scholar]
- 18.Jayarama N.H., Pillwein K., Craig R.N., Hoffman R., Webe G. Selective sensitivity to tiazofurin of human leukemic cells. Biochem. Pharmacol. 1986;35:2029–2032. doi: 10.1016/0006-2952(86)90737-9. [DOI] [PubMed] [Google Scholar]
- 19.Izawa K., Onishi T. Industrial syntheses of the central core molecules of HIV protease inhibitors. Chem. Rev. 2006;106:2811–2827. doi: 10.1021/cr050997u. [DOI] [PubMed] [Google Scholar]
- 20.Basu S., Prasad U.V., Barawkar D.A., De S., Palle V.P., Menon S., Patel M., Thorat S., Singh U.P., Sarma K.D., Waman Y., Niranjan S., Pathade V., Gaur A., Reddy S., Ansari S. Discovery of novel and potent heterocyclic carboxylic acid derivatives as protein tyrosine phosphatase 1B inhibitors. Bioorg. Med. Chem. Lett. 2012;22:2843–2849. doi: 10.1016/j.bmcl.2012.02.070. [DOI] [PubMed] [Google Scholar]
- 21.Bankar G.R., Nandakumar K., Nayak P.G., Thakur A., Chamallamudi M.R., Nampurath G.K. Vasorelaxant effect in rat aortic rings through calcium channel blockage: a preliminary in vitro assessment of a 1,3,4-oxadiazole derivative. Chem. Biol. Interact. 2009;181:377–382. doi: 10.1016/j.cbi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 22.Gilani S.J., Khan S.A., Siddiqui N. Synthesis and pharmacological evaluation of condensed heterocyclic 6-substituted 1,2,4-triazolo-[3,4-b]-1,3,4-thiadiazole and 1,3,4-oxadiazole derivatives of isoniazid. Bioorg. Med. Chem. Lett. 2010;20:4762–4765. doi: 10.1016/j.bmcl.2010.06.125. [DOI] [PubMed] [Google Scholar]
- 23.Hajimahdi Z., Zarghi A., Zabihollahi R., Aghasadeghi M.R. Synthesis, biological evaluation, and molecular modeling studies of new 1,3,4-oxadiazole- and 1,3,4-thiadiazole-substituted 4-oxo-4H-pyrido[1,2-a]pyrimidines as anti-HIV-1 agents. Med. Chem. Res. 2013;22:2467–2475. [Google Scholar]
- 24.Zarghi A., Tabatabai S.A., Faizi M., Ahadian A., Navabi P., Zanganeh V., Shafiee A. Synthesis and anticonvulsant activity of new 2-substituted-5-(2-benzyloxyphenyl)-1,3,4-oxadiazoles. Bioorg. Med. Chem. Lett. 2005;15:1863–1865. doi: 10.1016/j.bmcl.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Liu J., Zhang H., Yang X., Liu Z. Stereoselective synthesis and fungicidal activities of (E)-α-(methoxyimino)-benzeneacetate derivatives containing 1,3,4-oxadiazole ring. Bioorg. Med. Chem. Lett. 2006;16:2278–2282. doi: 10.1016/j.bmcl.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 26.Madhu M.S., Nagarjuna U., Padmavathi V., Padmaja A., Reddy N.V., Vijaya T. Synthesis and antimicrobial activity of pyrimidinyl 1,3,4-oxadiazoles, 1,3,4-thiadiazoles and 1,2,4-triazoles. Eur. J. Med. Chem. 2018;145:1–10. doi: 10.1016/j.ejmech.2017.12.067. [DOI] [PubMed] [Google Scholar]
- 27.Yadav M.R., Shirude S.T., Putnambekar D.S., Patel P.J., Prajapti H.B., Parmar A., Balaraman R., Giridhar R. Studies in 3,4-diaryl-1,2,5-oxadiazoles and their N-oxides: search for better COX-2 inhibitors. Acta Pharm. 2007;57(1):13–30. doi: 10.2478/v10007-007-0002-z. [DOI] [PubMed] [Google Scholar]
- 28.Taha M., Ismail N.H., Jamil W., Imran S., Rahim F., Kashif S.M., Zulkefeli M. Synthesis of 2-(2-methoxyphenyl)-5-phenyl-1,3,4-oxadiazole derivatives and evaluation of their antiglycation potential. Med. Chem. Res. 2016;25(2):225–234. [Google Scholar]
- 29.Palaska E., Sahin G., Kelicen P., Durlu N.T., Altinok G. Synthesis and anti-inflammatory activity of 1-acylthiosemicarbazides, 1,3,4-oxadiazoles, 1,3,4-thiadiazoles and 1,2,4-triazole-3-thiones. Farmaco. 2002;57:101–107. doi: 10.1016/s0014-827x(01)01176-4. [DOI] [PubMed] [Google Scholar]
- 30.Solanki N.G., Thakor M.K. World J. Pharm. Pharmaceut. Sci. 2017;6:1181–1188. [Google Scholar]
- 31.Abd-Ellah H.S., Abedl-Aziz M., Shoman M.E., Beshr E.A.M., Kaoud T.S., Ahmed A.S.F.F. New 1,3,4-oxadiazole/oxime hybrids: design, synthesis, anti-inflammatory, COX inhibitory activities and ulcerogenic liability. Bioorg. Chem. 2017;74:15–29. doi: 10.1016/j.bioorg.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Musmade D.S., Pattan S.R., Yalgatti M.S. Int. J. Pharm. Chem. 2015;5:251. [Google Scholar]
- 33.Kumar S., Bawa S., Drabu S., Kumar R., Gupta H. Anti-infect. Recent pat. Drug Discov. 2009;4:154–163. doi: 10.2174/157489109789318569. PMID: 19545230. [DOI] [PubMed] [Google Scholar]
- 34.Marella A., Ali R., Alam T., Saha R., Tanwar O., Akhter M., Shaquiquzzaman M., Alam M.M. Pyrazolines: a biological review. Med. Chem. 2013;13:921–931. doi: 10.2174/1389557511313060012. [DOI] [PubMed] [Google Scholar]
- 35.Insuasty B., Montoya A., Becerra D., Quiroga J., Abonia R., Robledo S., DaríoVélez I., Upegui Y., Nogueras M., Cobo J. Synthesis of novel analogs of 2-pyrazoline obtained from [(7-chloroquinolin-4-yl)amino]chalcones and hydrazine as potential antitumor and antimalarial agents. Eur. J. Med. Chem. 2013;67:252–262. doi: 10.1016/j.ejmech.2013.06.049. [DOI] [PubMed] [Google Scholar]
- 36.Shin S.Y., Yoon H., Hwang D., Ahn S., Kim D., Koh W., Lee D., Lim Y.H. Benzochalcones bearing pyrazoline moieties show anti-colorectal cancer activities and selective inhibitory effects on aurora kinases. Bioorg. Med. Chem. 2013;21:7018–7024. doi: 10.1016/j.bmc.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Kucukoglu K., Oral F., Aydin T., Yamali C., Algul O., Sakagami H., Gulcin I., Supuran C.T., Gul H.I. Synthesis, cytotoxicity and carbonic anhydrase inhibitory activities of new pyrazolines. J. Enzym. Inhib. Med. Chem. 2016;31:20–24. doi: 10.1080/14756366.2016.1217852. [DOI] [PubMed] [Google Scholar]
- 38.Wang H., Zheng J., Xu W., Chen C., Wei D., Ni W., Pan Y. A new series of cytotoxic pyrazoline derivatives as potential anticancer agents that induce cell cycle arrest and apoptosis. Molecules. 2017;22:1635. doi: 10.3390/molecules22101635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv P.C., Li D.D., Li Q.S., Lu X., Xiao Z.P., Zhu H.L. Synthesis, molecular docking and evaluation of thiazolyl-pyrazoline derivatives as EGFR TK inhibitors and potential anticancer agents. Bioorg. Med. Chem. Lett. 2011;21:5374–5377. doi: 10.1016/j.bmcl.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Yu M., Yang H., Wu K., Ji Y., Ju L., Lu X. Novel pyrazoline derivatives as bi-inhibitor of COX-2 and B-Raf in treating cervical carcinoma. Bioorg. Med. Chem. 2014;22:4109–4118. doi: 10.1016/j.bmc.2014.05.059. [DOI] [PubMed] [Google Scholar]
- 41.Shin S.Y., Yoon H., Hwang D., Ahn S., Kim D.W., Koh D., Lee Y.H., Lim Y. Benzochalcones bearing pyrazoline moieties show anti-colorectal cancer activities and selective inhibitory effects on aurora kinases. Bioorg. Med. Chem. 2013;21:7018–7024. doi: 10.1016/j.bmc.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Qin Y.J., Li Y.J., Jiang A.Q., Yang M.R., Zhu Q.Z., Dong H., Zhu H.L. Design, synthesis and biological evaluation of novel pyrazoline-containing derivatives as potential tubulin assembling inhibitors. Eur. J. Med. Chem. 2015;94:447–457. doi: 10.1016/j.ejmech.2015.02.058. [DOI] [PubMed] [Google Scholar]
- 43.Amin K.M., Abou-Seri S.M., Awadallah F.M., Eissa A.A.M., Hassan G.S., Abdulla M.M. Synthesis and anticancer activity of some 8-substituted-7-methoxy-2H-chromen-2-one derivatives toward hepatocellular carcinoma HepG2 cells. Eur. J. Med. Chem. 2015;90:221–231. doi: 10.1016/j.ejmech.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 44.Dinesha Viveka S., Priya B.K., Pai K.S.R., Naveen S., Lokanath N.K., Nagaraja G.K. Synthesis and pharmacological evaluation of some new fluorine containing hydroxypyrazolines as potential anticancer and antioxidant agents. Eur. J. Med. Chem. 2015;104:25–32. doi: 10.1016/j.ejmech.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 45.Bondock S., Adel S., Etman H.A., Badria F.A. Synthesis and antitumor evaluation of some new 1,3,4-oxadiazole-based heterocycles. Eur. J. Med. Chem. 2012;48:192–199. doi: 10.1016/j.ejmech.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 46.Hassan M.F., Rauf A. Spectrochim. Acta Mol. Biomol. Spectrosc. 2016;153:510–516. doi: 10.1016/j.saa.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Sambhaji T.D., Amarsinh R.D., Manisha R.B., Vijay M.K., Laxman U.N., Dhiman S., Ramrao A.M. Synthesis and antitubercular activity of new 1,3,4-oxadiazoles bearing pyridyl and thiazolyl scaffolds. Bioorg. Med. Chem. Lett. 2016;26:3646–3651. doi: 10.1016/j.bmcl.2016.05.093. [DOI] [PubMed] [Google Scholar]
- 48.Pratviel G., Bernadou J., Meunier B. DNA and RNA cleavage by metal complexes. Adv. Inorg. Chem. 1998;45:251–312. [Google Scholar]
- 49.Ali I., Waseem A.W., Saleem K., Hsei M.F. Design and synthesis of thalidomide based dithiocarbamate Cu(II), Ni(II) and Ru(III) complexes as anticancer agents. Polyhedron. 2013;56:134–143. [Google Scholar]
- 50.Santosh R., Selvam M.K., Saptami U.K., Nagaraja G.K., Madan K. Design, synthesis, DNA binding, and docking studies of thiazoles and thiazole-containing triazoles as antibacterials. Chemistry. 2018;3:3892–3898. [Google Scholar]
- 51.Santosh R., Selvam M.K., Saptami U.K., Nagaraja G.K. Synthesis, characterization, antibacterial and antioxidant studies of some heterocyclic compounds from triazole-linked chalcone derivatives. Chemistry. 2018;3:6338–6343. [Google Scholar]
- 52.Santosh R., Paul P., Selvam M.K., Raril C., Krishna P.M., Manjunatha J.G., Nagaraja G.K. One-pot synthesis of pyrimido[4,5-d]pyrimidine derivatives and investigation of their antibacterial, antioxidant, DNA-binding and voltammetric characteristics. Chemistry. 2019;4:990–996. [Google Scholar]
- 53.Ali I., Lone M.N., Hsieh M.F. N-substituted (substituted-5-benzylidine) thiazolidine-2,4-diones: crystal structure, In Silico, DNA binding and anticancer studies. Biointerface Res. Appl. Chem. 2016;6:1356–1379. [Google Scholar]
- 54.Barton J.K., Danishefsky A.T., Goldberg J.M. Tris(phenanthroline)ruthenium(II): stereoselectivity in binding to DNA. J. Am. Chem. Soc. 1984;106:2172–2176. [Google Scholar]
- 55.Wolfe A., Chimer G.H., Meechan T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry. 1987;26:6392–6396. doi: 10.1021/bi00394a013. [DOI] [PubMed] [Google Scholar]
- 56.Kopka M.L., Yoon C., Goodsell D., Pjura P. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J. Mol. Biol. 1985;183:553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- 57.Raman N., Sobha S., Thamaraichelvan A. A novel bioactive tyramine derived Schiff base and its transition metal complexes as selective DNA binding agents. Spectrochim. Acta Mol. Biomol. Spectrosc. 2011;78:888–898. doi: 10.1016/j.saa.2010.12.056. [DOI] [PubMed] [Google Scholar]
- 58.Form G.R., Raper E.S., Downie T.C. The crystal and molecular structure of 3-thioamidopyridine. Acta Crystallogr. Sec. Struct. Crystallogr. Cryst. Chem. 1973;29:776–782. [Google Scholar]
- 59.Pasternack R.F., Gibbs E.J., Villafrancas J. Interactions of porphyrins with nucleic acids. Biochemistry. 1983;22:2406–2414. doi: 10.1021/bi00279a016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.