Abstract
Biosensors, advanced microscopy, and single-cell transcriptomics are advancing plant synthetic biology.
Our knowledge of plant biology has reached the point where we can begin to rationally engineer plant form and function to meet our needs. From a bioengineer’s or synthetic biologist’s point of view, the goal of studying developmental biology is to generate a predictive model that specifies the molecular circuitry required to move a cell from one state to another. This model could then serve as a guide for harvesting the most useful parts and logic to enable the engineering of novel states and multicell behaviors. Among the most critical parts to understand from this perspective are the signaling molecules that enable intra- and intercellular communication. Several biosensors have been developed in recent years to detect plant-specific signals and secondary messengers. Many other general biosensors have been successfully implemented in plant systems. These biosensors, in combination with single cell “omics” techniques and predictive statistical frameworks, are providing the type of high resolution, quantitative descriptions of cell state that will ultimately make it possible to decode and re-engineer traits associated with higher yields and stress tolerance.
Being a plant developmental biologist today can feel like a lot like being a cryptographer piecing together fragmented messages with only a partial knowledge of the cipher. Biological signaling is rife with redundancy, feedback, and feedforward motifs acting to dampen or amplify each signal, and modulate outputs depending on position and cell identity. To crack the code of these complex genetic signal processors, it is important to be able to measure, as well as manipulate, both signals and responses. Recent advances in synthetic biology have provided a means to access such tools. Sensitive, genetically encoded reporters (biosensors), in combination with emerging single-cell transcriptomics approaches, are providing increasingly detailed molecular descriptions of cells undergoing developmental transitions (Moreno-Risueno et al., 2015; Efroni et al., 2016; Ristova et al., 2016; Cao et al., 2017). However, in many cases we are still unable to measure key signaling molecules directly with fine spatiotemporal resolution.
Several excellent reviews have been published recently that describe the application of biosensors to plant systems (Goold et al., 2018; Hilleary et al., 2018; Walia et al., 2018). Here, we review this state of the art in measuring plant signaling, using principles and tools borrowed from and inspired by engineering, as well as efforts to use this knowledge to enable rapid, rational re-engineering of plant development. We have arranged this review as an engineering cycle in which we will cover “designing” biosensors; “building” biosensors, including technologies to facilitate the use of biosensors in plants; “testing” biosensors; “modeling” signaling and development, including our perspective on integrating biosensors, systems approaches, and optimal experimental design to generate minimal predictive models of plant development; and finally “learning” about plant development, including our outlook on how biosensors paired with synthetic and systems biology approaches will advance knowledge in the future.
DESIGN
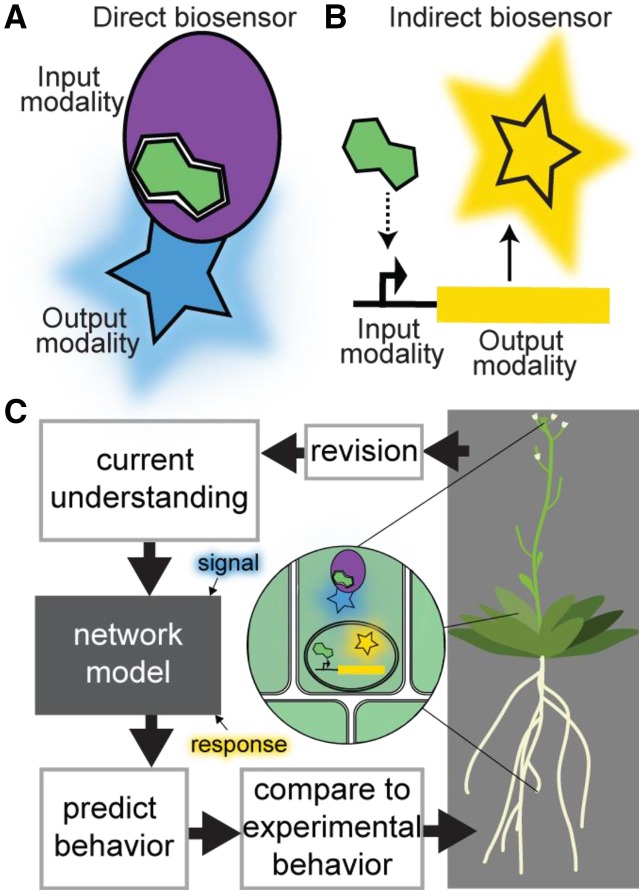
The design of any genetically encoded biosensor involves connecting an input modality, which interacts in some way with the species to be measured, to an output modality, which provides some quantifiable product (Fig. 1). These modalities may be DNA, RNA, and/or proteins. The species to be measured (analyte) may be any molecule or complex of molecules. Input modalities may be promoters that respond to the analyte, naturally occurring proteins domains or engineered novel proteins or nucleic acids, which bind (or otherwise respond) to the analyte. Each input modality offers different advantages and drawbacks. As opposed to direct biosensors, which bind to and report the concentration of the desired species, indirect biosensors have input modalities that are natural or engineered responsive promoters or protein domains, such as degrons, which require additional cellular machinery to respond to the analyte (Brunoud et al., 2012; Larrieu et al., 2015). Often referred to as “reporters,” indirect biosensors report on the status of the signaling network required to activate the responsive element. Although this complex output can be misinterpreted, indirect biosensors have facilitated numerous discoveries, particularly when paired with systems biology approaches (such as transcriptomic and other genome-scale analyses) to decipher network status (Moreno-Risueno et al., 2010; de Luis Balaguer et al., 2017; Wu et al., 2018). Such advances will be discussed further in the “Test” section.
Figure 1.
Biosensors link detection of an analyte (such as a signaling molecule) by an input modality to a quantifiable change in an output modality. A, Schematic of a direct biosensor exemplified by a signaling molecule (green) binding protein as the input modality (purple oval) with a fluorescent protein output modality (blue star). This biosensor directly measures the “signal,” i.e. concentration of the signaling molecule. B, Schematic of an indirect biosensor exemplified by a signaling molecule responsive promoter of unknown mechanism (dotted arrow) driving expression of a fluorescent protein output modality (yellow star). This biosensor provides a measure of the response of this signaling pathway. C, Using biosensors to measure both the signal and response of a developmental signaling network along with plant phenotype leads to iterative improvement of the developmental network model and our understanding of plant development. Improved understanding of auxin signaling dynamics—realized by multiple biosensors and means of functional quantification—has facilitated rational tuning of plant architecture (Guseman et al., 2015; Je et al., 2016; Wright et al., 2017; Khakhar et al., 2018; Shibata et al., 2018). Newly developed biosensors (Liao et al., 2015; Rizza et al., 2017; Wu et al., 2018), paired with functional and phenotypic quantification of development, will help crack the code underlying developmental signaling and allow rational breeding and engineering of next-generation crops.
Natural binding domains are often part of the signaling pathway one is trying to measure and may interfere with the native pathway components. The laws of thermodynamics dictate that a system cannot be measured without perturbation (Szilard, 1929), but ideally this perturbation will be controlled for and/or minimized. To study normal development, the presence of a biosensor must not alter normal development. Further, protein engineering may be used to render biosensors orthogonal to the native pathway (Rizza et al., 2017). Novel engineered binding proteins or DNA/RNA aptamers require significant investment but are less likely to interfere with the native signaling pathway, especially if potential off-target interactions are controlled for in the design and screening. Numerous methods for directing the evolution of binding modalities have been developed, including phage-display (Smith, 1985; Tan et al., 2016), microbial cell surface display (Charbit et al., 1986; Freudl et al., 1986; Agterberg et al., 1987; Schreuder et al., 1996; Boder and Wittrup, 1997; Daugherty, 2007; Liu, 2015), ribosome display (Mattheakis et al., 1994; Plückthun, 2012), and many in vitro display techniques (Joyce, 1989; Ellington and Szostak, 1990; Tuerk and Gold, 1990; Darmostuk et al., 2015; Tizei et al., 2016). These methods link the genotype and molecular phenotype of large libraries of binding proteins, allowing specific binders to a ligand of choice to be identified and amplified or further characterized. In all cases, expression in the desired host is not guaranteed and further optimization may need to be done, as the expression level of the biosensor combined with the affinity of the input modality for the species of interest determines the dynamic range of the sensor (i.e. the range of input concentrations over which the output of the sensor is quantifiable). Because of these challenges, a transient transformation system for screening expression constructs can expedite biosensor optimization.
Output modalities largely determine the spatiotemporal domain and resolutions of the biosensor measurements. Fluorescent, luminescent, or chromogenic proteins are typical output modalities. Pairs of fluorescent and/or luminescent proteins capable of Förster resonance energy transfer (FRET) or bioluminescence resonance energy transfer are also frequently used. FRET-based biosensors have the advantage of inherent ratiometric output, allowing the expression of the biosensor to be measured by specifically exciting the acceptor fluorophore, and exciting the donor fluorophore to measure the species of interest. Beyond the common issues of photobleaching and phototoxicity, fluorescence measurements in plants can be particularly challenging given autofluorescence and the potential for stimulation of endogenous photoreceptors (Mylle et al., 2013). Luminescence measurements avoid these problems, as they do not require incident light. Bioluminescence resonance energy transfer further allows tuning of the luminescent emission spectra, facilitating ratiometric measurements or measurement of multiple species at once. All light-based measurements are limited by the penetrance of light through tissue, and the numerous light-absorbing structures in some plant cells limit the useful spectrum. Fortunately, dramatic advances are continually being made in microscopy, photo detection, and protein engineering to allow high-resolution imaging across most scales in plants (Rousseau et al., 2015; Clark and Sozzani, 2017; Rios et al., 2017).
Connecting the input and output modalities is generally the most challenging and critical aspect of direct biosensor design, as the connection has a large effect on biosensor resolution and dynamic range. Direct genetically encoded biosensors are typically fusions of the sequences of the input and output modalities (Ostermeier, 2009). The most laborious task in direct biosensor engineering is creating a library of fusions and identifying members that undergo structural changes when exposed to the species of interest, which in turn alter their output. Fortunately, there is a wealth of literature containing numerous case studies (recently reviewed in Bolbat and Schultz, 2017; Sanford and Palmer, 2017), because of early work on engineering of direct biosensors and protein switches (Siegel and Isacoff, 1997; Doi and Yanagawa, 1999; Prehoda et al., 2000; Tucker and Fields, 2001; Dueber et al., 2003; Guntas and Ostermeier, 2004). Ideally the design space of structurally reasonable fusions is thoroughly explored using protein engineering techniques to vary insertional position, linker residues between the modalities, and possibly circular permutation of one or both modalities (Kanwar et al., 2013; Younger et al., 2018). Recently, advances in bioinformatics and decreasing costs of next-generation sequencing have facilitated prediction and experimental determination of sites of potential allosteric regulation (Nadler et al., 2016; Rivoire et al., 2016; Pincus et al., 2017). Folding and stability can be tuned and can also be exploited, either inadvertently or directly, to develop direct fusion biosensors (Tucker and Fields, 2001; Wright et al., 2011, 2014; Choi et al., 2015; Feng et al., 2015; Dagliyan et al., 2016).
Transcription factors are an interesting alternative for connecting input and output modalities of direct or indirect biosensors, by allowing recognition of the species of interest to drive expression of any of the above output domains or another genetic circuit (Feng et al., 2015; Khakhar et al., 2016, 2018; Younger et al., 2016, 2018). The amplification provided by transcription and translation may result in a wider dynamic range. Additionally, this modular connection allows the biosensor to regulate multiple outputs facilitating both measurement and reprogramming of cellular behavior (Faden et al., 2016; Khakhar et al., 2018; Lowder et al., 2018). However, this synthetic gene circuit approach also limits the spatiotemporal resolution of the sensor to the cellular scale and the turnover rate of the output modality.
Biosensors are not limited to detection of monomeric species. Biosensors consisting of short genetic circuits are reminiscent of the enhancer trap (O’Kane and Gehring, 1987) or yeast two-hybrid system (Fields and Song, 1989) and their numerous variants. Advances in microscopy have made possible the in vivo application of well-established methods of quantifying proteins, protein complexes, and protein-protein interactions (Magde et al., 1972; Lakowicz et al., 1992). These methods rely on simple translational fusions, similar to classical FRET-based or protein fragment complementation interaction assays (Pelletier et al., 1999), but utilize highly sensitive confocal microscopes, pulsed lasers, and computational methods to quantify interactions in vivo. It may also be possible to express antibody-like proteins fused to fluorescent proteins, or pairs of antibodies fused to split fluorescent proteins to detect native proteins or complexes (Carlin et al., 2016).
Fluorescence correlation spectroscopy (FCS) measures fluctuations in fluorescence intensity that correlate with the motion of the fluorescently labeled molecule(s) of interest to quantify diffusion (Clark et al., 2016; Clark and Sozzani, 2017). When two different molecules are measured simultaneously in different spectral channels, kinetic parameters of their binding can be inferred from cross-correlation in their diffusion. Another technique, fluorescence lifetime imaging microscopy (FLIM), aims to overcome these issues with overlap in the spectra of the two fluorophores as well as autofluorescence and photobleaching, which can result in poor signal-to-noise ratios in some instances. These issues associated with traditional wave laser microscopy can be abated by using a pulsed laser and by visualizing the time each fluorophore spends in its excited state after the pulse (fluorescent lifetime) instead of intensity. FLIM can be paired with FCS as well as FRET to measure protein-protein interactions (Boer et al., 2014; Long et al., 2017; Rios et al., 2017). These technologies will improve the sensitivity of existing biosensors and facilitate the development of new biosensor approaches.
BUILD
Direct biosensors are generally developed in microbial organisms and then shuttled into organisms less amenable to transformation. This translation between kingdoms and even translation of indirect biosensors between species is not always perfect. This can be due to a combination of issues with expression, folding, stability, and interference with or divergence of endogenous signaling pathways. In most plants, where targeted insertion is not yet possible, there is the additional complexity of integration site variation and frequent silencing (Jupe et al., 2018). Organisms allowing targeted insertion provide an ideal platform for biosensor development, as more direct comparisons of activity can be made between different biosensors. Targeted genetic insertion also allows reporter-tagging of native gene loci, reducing variation. Plants that readily perform homologous recombination, such as Physcomitrella patens and Marchantia polymorpha, deserve consideration for both the design and application of biosensors, as there is still much to be learned about their development that may inform work in other species (Cove et al., 2009; Ishizaki et al., 2013). To our knowledge, biosensors have yet to be paired with targeted transgene insertion technology (De Paepe et al., 2013) or “landing pads” for plants. This technology is currently low-efficiency and does not allow full specification of the insertion site but does provide more accurate comparison of independent transformants. Homology-directed repair has been demonstrated several times, but usually with low efficiency (Zhao et al., 2016; Čermák et al., 2017; Hahn et al., 2018). Insertional variation in expression can also be mitigated, at least in part, by ratiometric sensors. By expressing a nonfunctional, or constitutively active, version of the biosensor within the same transgene or cistron, expression of the transgene insertion site can be controlled for and higher fidelity achieved (Wend et al., 2013; Liao et al., 2015).
Another challenge across organisms is efficient assembly of unwieldy multigenic constructs. Fortunately, many new toolsets are available for the design and assembly of large and difficult constructs. Several software packages are available for the design and modeling of polycistronic cassettes for biosensors and other applications (Chen et al., 2012a; Hillson, 2014; Harris et al., 2017; Choi et al., 2018; Misirli et al., 2018; Shockley et al., 2018; Watanabe et al., 2018). Several new plant-specific toolkits for assembling the designed constructs have also been developed recently (Engler et al., 2014; Beyer et al., 2015; Shih et al., 2016; Zhu et al., 2017; Pollak et al., 2018).
One of the aspects of these tools that is most critical to the field of biosensor development is the ability to share and reproduce the design, parameterization, and measurement of biosensors between groups and study systems. Common standards for the description of genetic designs and models have been established (Hucka et al., 2015; Martínez-García et al., 2015; Cox et al., 2018), alongside tools for developing and parameterizing (Harris et al., 2017; Zhang et al., 2017; Choi et al., 2018; Shockley et al., 2018; Wandy et al., 2018; Watanabe et al., 2018), as well as visualizing and communicating these designs and models (Merchant et al., 2016; Cox et al., 2017; Der et al., 2017; Medley et al., 2018). Laboratory inventory management and electronic laboratory notebook systems have also been developed to provide a higher degree of organization and reproducibility in the wet lab (List et al., 2014, 2015; Barillari et al., 2016; Craig et al., 2017; Klavins, 2017). The ability of several of these tools to be operated in an integrative notebook environment, containing interleaved narrative with figures and code (possibly of several languages), allows science to be communicated seamlessly and reproducibly (Kluyver et al., 2016; Allaire et al., 2018; Medley et al., 2018). In the future, open sharing of transparent example notebooks documenting complete design-build-test-learn workflows integrating these tools will be the norm. Such examples will provide excellent training and teaching tools, reducing burden, and establishing reproducibility expectations for the field.
TEST
Biosensors have allowed plant biologists to visualize and quantify developmental signals and signaling machinery, as well as provided means to ask better questions as to how development is controlled. To realize our goal of understanding and re-engineering development, we must pair biosensors with systems biology to inform a predictive model of development. Use of systems biology approaches and mathematical modeling paired with transcriptional and translational reporters, cell-type–specific promoters, and enhancers have led to impressive breakthroughs (Vernoux et al., 2011; Bargmann et al., 2013; Efroni et al., 2016; Je et al., 2016; Sparks et al., 2016; de Luis Balaguer et al., 2017; Wendrich et al., 2017; Drapek et al., 2018; Shibata et al., 2018). For example, Shibata et al. used transcriptome and chromatin immunoprecipitation data to develop a gene regulatory network model controlling root hair growth. This model identified both a key positive and negative regulator of root hair growth that formed a feedback loop. This model allowed the authors to identify, and confirm experimentally, genetic manipulations with strong effects on root hair growth. Indirect biosensors paired with systems approaches have also revealed fascinating dynamics of developmental signaling that are still not completely understood, such as oscillations in auxin response within the root meristem, which determine the positions of future lateral roots (Moreno-Risueno et al., 2010; Xuan et al., 2015, 2016; Laskowski and Ten Tusscher, 2017). To track down the unknowns of developmental dynamics will require a better understanding of which signals indirect biosensors are integrating, as well as development of new direct biosensors, simultaneous measurement of multiple biosensors, and generation of dynamic omics datasets paired with these sensors.
Recently, highly sensitive ratiometric sensors of the auxin signaling network status were developed (Liao et al., 2015) based on improved knowledge of specificity within this network (Boer et al., 2014). These sensors helped revealed new domains of auxin accumulation that were previously predicted by models of auxin transport and production (Scarpella et al., 2006; Grieneisen et al., 2007; Robert et al., 2013). These models were parameterized using translational fusion biosensors, demonstrating the power of the application of multiple biosensors, as the simultaneous measurement of two species facilitates prediction of their dynamic relationship. We highly anticipate proposed future work combining these two high-sensitivity ratiometric sensors (Liao et al., 2015), as well as the development of a direct auxin biosensor (Vernoux and Robert, 2017).
A direct biosensor for gibberellin has recently revealed a strong correlation between gibberellin and cell elongation and helped to decipher the role of the light-responsive PHYTOCHROME INTERACTING FACTORs in regulating gibberellin levels (Rizza et al., 2017). Two indirect abscisic acid signaling biosensors have also recently been developed (Wu et al., 2018). These engineered abscisic acid-responsive promoters complement the detection range of existing direct abscisic acid biosensors (Jones et al., 2014; Waadt et al., 2014). These reporters helped to solidify existing knowledge of abscisic acid’s roles in the development of lateral roots and stomata. They also revealed differential regulation depending on the sequence of the core cis-regulatory element and cross-regulation of this promoter by stem cell maintenance transcription factors in the stem cell niche. This important finding highlights the importance and power of characterizing promoter-based reporters thoroughly. In the future, pairing direct and indirect biosensors to measure both signaling inputs and transcriptional outputs may facilitate inference of the intervening network and examination of how these networks interact with cell fate (Fig. 1).
Promoter-based indirect sensors have also been recently used to examine the dynamic relationship between auxin and cytokinin in both barley (Hordeum vulgare) and soybean (Glycine max; Fisher et al., 2018; Kirschner et al., 2018). These reporters functioned as expected in soybean; however, in barley, the auxin reporters DR5rev::GFP (Benková et al., 2003) and DR5v2 (Liao et al., 2015) were poorly expressed and not auxin-responsive (Kirschner et al., 2018). This interesting result compels further examination but may uncover unique paths of evolutionary divergence in auxin signaling components and root development. In soybean, auxin and cytokinin signaling reporters were observed simultaneously in premature root nodules (Fisher et al., 2018). This revealed stark differences in the auxin/cytokinin signaling ratio between premature vascular and parenchyma cells of developing nodules. This pilot study will, we hope, lead to better understanding of the complex roles hormones play in mediating symbioses (Gamas et al., 2017; Betsuyaku et al., 2018; Kunkel and Harper, 2018). Future work integrating multiple biosensors for different developmental signals or different elements within a signaling pathway will greatly improve our understanding of the connectivity and tunability of these signals and the developmental processes they regulate. Integrating nutrient biosensors with developmental signaling will also be crucial to our ability to engineer plants with low resource requirements (Chen et al., 2012b; Upadhyay and Verma, 2015; Okumoto and Versaw, 2017). Novel plant signaling mechanisms are also being revealed by biosensors, such as the recently uncovered Glu-triggered long-distance calcium signaling after wounding (Toyota et al., 2018).
FRET-FLIM and FCS have also helped decipher complex molecular interactions critical to development. FRET-FLIM was recently used to reveal cell-type–specific protein–protein interactions among the SHORTROOT, SCARECROW, and JACKDAW transcription factors, which regulate cell division and patterning in the root (Long et al., 2017). FCS has also been used to track diffusion and interaction of SHORTROOT and SCARECROW (Clark et al., 2016). These studies clearly show cell-type–specific variation in the composition, structure, and activity of complexes of these transcription factors. Future work employing these techniques to examine dynamics of transcription factor complexes, as well as hormone response complexes (Rios et al., 2017), throughout development will provide a mechanistic understanding of cell fate transitions.
MODEL
Measurements of signals alone are of limited use without a predictive framework for linking developmental signals and cell status to transcriptional and phenotypic outcomes. Formulating our current understanding in the framework of a mathematical model allows us to quantify the completeness of our understanding as the deviation between our model and experimental data. An accurate model and understanding also facilitates rational engineering of plant development (Guseman et al., 2015; Khakhar et al., 2018). If the goal of our collective science is to generate the simplest model that most completely predicts plant development, then we must accept that our model is, by definition, incomplete. To achieve a maximally informative yet simple model of development, we must carefully design experiments to minimize the uncertainty in both our model selection and parameterization (Smucker et al., 2018). Several groups have developed frameworks for computational design of the optimal set of experiments to identify the mathematical relationship among the signaling inputs, network status, and the developmental outcome, i.e. model selection (Busetto et al., 2013; Apri et al., 2014; Vanlier et al., 2014; Minas et al., 2017; Rougny et al., 2018). Other statistical frameworks aim to design optimal experiments for determining parameter uncertainty in the chosen model (Dehghannasiri et al., 2015; Fan et al., 2015; Imani et al., 2018; Mohsenizadeh et al., 2018). For example, Dehghannasiri et al. (2015) provides a method for prioritizing future experiments based on existing knowledge of a gene regulatory network and the desired intervention in the network, where intervention in this case is a therapy targeting a pathological network state. Systems biology approaches including similar frameworks have facilitated inference of networks and logic in plant development (Astola et al., 2014; Fisher and Sozzani, 2016; Ristova et al., 2016; de Luis Balaguer et al., 2017; Minas et al., 2017; Shibata et al., 2018; Varala et al., 2018). In addition to optimally improving our knowledge of developmental networks, connecting signaling network models with phenotypic outcome models are of particular importance to the goal of engineering plant development (Prusinkiewicz and Runions, 2012; O’Connor et al., 2014; Landrein et al., 2015; Mellor et al., 2017; Schnepf et al., 2018). One effort critical to the success of systems and synthetic biology in deciphering development will be the continued collaboration among and integration of statistical modeling, optimal experimental design, and dynamic, multivariate molecular genetics techniques.
LEARN
Synthetic biologists’ goals for understanding plant developmental biology are within reach. Mathematical models that integrate cell state data from systems approaches with dynamic signal data from biosensors will greatly support efforts to rationally engineer plant form and function. Such models facilitate prioritization and design of experiments to minimize model parameters and improve the certainty of remaining parameters. Implementing statistical tools to design optimal experiments to improve certainty in model selection and parameterization will allow new questions to be addressed efficiently in the context of existing knowledge.
Transdisciplinary approaches combining synthetic, systems, and computational biology are making it increasingly straightforward to quantify the dynamic behavior of signals we already know are important (the “known unknowns”) and find new signals and circuits (the “unknown unknowns”). This knowledge will be invaluable in guiding rapid improvements in the quality and quantity of the foods, fuels, fibers, and pharmaceuticals that can be produced by the next generation of crops.
Acknowledgments
We thank Marc Ostermeier and members of the Nemhauser lab for helpful discussions. We apologize to the authors of many substantive contributions to plant synthetic developmental biology that we have not included in this review due to space limitations.
Footnotes
This work was supported by the National Institutes of Health (R01-GM107084), the National Science Foundation (DBI-1402222), and the Howard Hughes Medical Institute.
Articles can be viewed without a subscription.
References
- Agterberg M, Adriaanse H, Tommassen J (1987) Use of outer membrane protein PhoE as a carrier for the transport of a foreign antigenic determinant to the cell surface of Escherichia coli K-12. Gene 59: 145–150 [DOI] [PubMed] [Google Scholar]
- Allaire J, Xie Y, McPherson J, Luraschi J, Ushey K, Atkins A, Wickham H, Cheng J, Chang W (2018) R Markdown: Dynamic Documents for R. http://markdown.rstudio.com
- Apri M, de Gee M, van Mourik S, Molenaar J (2014) Identifying optimal models to represent biochemical systems. PLoS One 9: e83664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astola L, Stigter H, van Dijk ADJ, van Daelen R, Molenaar J (2014) Inferring the gene network underlying the branching of tomato inflorescence. PLoS One 9: e89689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BOR, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, et al. (2013) A map of cell type-specific auxin responses. Mol Syst Biol 9: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillari C, Ottoz DSM, Fuentes-Serna JM, Ramakrishnan C, Rinn B, Rudolf F (2016) openBIS ELN-LIMS: An open-source database for academic laboratories. Bioinformatics 32: 638–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Betsuyaku S, Katou S, Takebayashi Y, Sakakibara H, Nomura N, Fukuda H (2018) Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol 59: 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer HM, Gonschorek P, Samodelov SL, Meier M, Weber W, Zurbriggen MD (2015) AQUA cloning: A versatile and simple enzyme-free cloning approach. PLoS One 10: e0137652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder ET, Wittrup KD (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 15: 553–557 [DOI] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589 [DOI] [PubMed] [Google Scholar]
- Bolbat A, Schultz C (2017) Recent developments of genetically encoded optical sensors for cell biology. Biol Cell 109: 1–23 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Busetto AG, Hauser A, Krummenacher G, Sunnåker M, Dimopoulos S, Ong CS, Stelling J, Buhmann JM (2013) Near-optimal experimental design for model selection in systems biology. Bioinformatics 29: 2625–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al. (2017) Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin KB, Cruz-Teran CA, Kumar JP, Gomes C, Rao BM (2016) Combinatorial pairwise assembly efficiently generates high affinity binders and enables a “mix-and-read” detection scheme. ACS Synth Biol 5: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29: 1196–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbit A, Boulain JC, Ryter A, Hofnung M (1986) Probing the topology of a bacterial membrane protein by genetic insertion of a foreign epitope: Expression at the cell surface. EMBO J 5: 3029–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Densmore D, Ham TS, Keasling JD, Hillson NJ (2012a) DeviceEditor visual biological CAD canvas. J Biol Eng 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-Q, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, Frommer WB (2012b) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Choi JH, Laurent AH, Hilser VJ, Ostermeier M (2015) Design of protein switches based on an ensemble model of allostery. Nat Commun 6: 6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Medley JK, König M, Stocking K, Smith L, Gu S, Sauro HM (2018) Tellurium: An extensible Python-based modeling environment for systems and synthetic biology. Biosystems 171: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Sozzani R (2017) Measuring protein movement, oligomerization state, and protein–protein interaction in Arabidopsis roots using scanning fluorescence correlation spectroscopy (scanning FCS). In Busch W, ed, In Plant Genomics: Methods and Protocols. Springer, New York, pp 251–266 [DOI] [PubMed] [Google Scholar]
- Clark NM, Hinde E, Winter CM, Fisher AP, Crosti G, Blilou I, Gratton E, Benfey PN, Sozzani R (2016) Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5: e14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ, Perroud P-F, Charron AJ, McDaniel SF, Khandelwal A, Quatrano RS (2009) The moss Physcomitrella patens: A novel model system for plant development and genomic studies. Cold Spring Harb Protoc 2009: pdb.emo115. [DOI] [PubMed] [Google Scholar]
- Cox RS III, McLaughlin JA, Grünberg R, Beal J, Wipat A, Sauro HM (2017) A visual language for protein design. ACS Synth Biol 6: 1120–1123 [DOI] [PubMed] [Google Scholar]
- Cox RS, Madsen C, McLaughlin JA, Nguyen T, Roehner N, Bartley B, Beal J, Bissell M, Choi K, Clancy K, et al. (2018) Synthetic Biology Open Language (SBOL) Version 2.2.0. J Integr Bioinform 15: 1613-4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig T, Holland R, D’Amore R, Johnson JR, McCue HV, West A, Zulkower V, Tekotte H, Cai Y, Swan D, et al. (2017) Leaf LIMS: A flexible laboratory information management system with a synthetic biology focus. ACS Synth Biol 6: 2273–2280 [DOI] [PubMed] [Google Scholar]
- Dagliyan O, Tarnawski M, Chu P-H, Shirvanyants D, Schlichting I, Dokholyan NV, Hahn KM (2016) Engineering extrinsic disorder to control protein activity in living cells. Science 354: 1441–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmostuk M, Rimpelova S, Gbelcova H, Ruml T (2015) Current approaches in SELEX: An update to aptamer selection technology. Biotechnol Adv 33: 1141–1161 [DOI] [PubMed] [Google Scholar]
- Daugherty PS. (2007) Protein engineering with bacterial display. Curr Opin Struct Biol 17: 474–480 [DOI] [PubMed] [Google Scholar]
- Dehghannasiri R, Yoon B-J, Dougherty ER (2015) Efficient experimental design for uncertainty reduction in gene regulatory networks. BMC Bioinformatics 16(Suppl 13): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luis Balaguer MA, Fisher AP, Clark NM, Fernandez-Espinosa MG, Möller BK, Weijers D, Lohmann JU, Williams C, Lorenzo O, Sozzani R (2017) Predicting gene regulatory networks by combining spatial and temporal gene expression data in Arabidopsis root stem cells. Proc Natl Acad Sci USA 114: E7632–E7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe A, De Buck S, Nolf J, Van Lerberge E, Depicker A (2013) Site-specific T-DNA integration in Arabidopsis thaliana mediated by the combined action of CRE recombinase and ϕC31 integrase. Plant J 75: 172–184 [DOI] [PubMed] [Google Scholar]
- Der BS, Glassey E, Bartley BA, Enghuus C, Goodman DB, Gordon DB, Voigt CA, Gorochowski TE (2017) DNAplotlib: Programmable visualization of genetic designs and associated data. ACS Synth Biol 6: 1115–1119 [DOI] [PubMed] [Google Scholar]
- Doi N, Yanagawa H (1999) Design of generic biosensors based on green fluorescent proteins with allosteric sites by directed evolution. FEBS Lett 453: 305–307 [DOI] [PubMed] [Google Scholar]
- Drapek C, Sparks EE, Marhavy P, Taylor I, Andersen TG, Hennacy JH, Geldner N, Benfey PN (2018) Minimum requirements for changing and maintaining endodermis cell identity in the Arabidopsis root. Nat Plants 4: 586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber JE, Yeh BJ, Chak K, Lim WA (2003) Reprogramming control of an allosteric signaling switch through modular recombination. Science 301: 1904–1908 [DOI] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip P-L, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Faden F, Ramezani T, Mielke S, Almudi I, Nairz K, Froehlich MS, Höckendorff J, Brandt W, Hoehenwarter W, Dohmen RJ, et al. (2016) Phenotypes on demand via switchable target protein degradation in multicellular organisms. Nat Commun 7: 12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Kuwahara H, Wang X, Wang S, Gao X (2015) Parameter estimation methods for gene circuit modeling from time-series mRNA data: A comparative study. Brief Bioinform 16: 987–999 [DOI] [PubMed] [Google Scholar]
- Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, Morey KJ, Rios X, Medford JI, Church GM, et al. (2015) A general strategy to construct small molecule biosensors in eukaryotes. eLife 4: e10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Fisher AP, Sozzani R (2016) Uncovering the networks involved in stem cell maintenance and asymmetric cell division in the Arabidopsis root. Curr Opin Plant Biol 29: 38–43 [DOI] [PubMed] [Google Scholar]
- Fisher J, Gaillard P, Fellbaum CR, Subramanian S, Smith S (2018) Quantitative 3D imaging of cell level auxin and cytokinin response ratios in soybean roots and nodules. Plant Cell Environ 41: 2080–2092 [DOI] [PubMed] [Google Scholar]
- Freudl R, MacIntyre S, Degen M, Henning U (1986) Cell surface exposure of the outer membrane protein OmpA of Escherichia coli K-12. J Mol Biol 188: 491–494 [DOI] [PubMed] [Google Scholar]
- Gamas P, Brault M, Jardinaud M-F, Frugier F (2017) Cytokinins in symbiotic nodulation: When, where, what for? Trends Plant Sci 22: 792–802 [DOI] [PubMed] [Google Scholar]
- Goold HD, Wright P, Hailstones D (2018) Emerging opportunities for synthetic biology in agriculture. Genes (Basel) 9: E341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Guntas G, Ostermeier M (2004) Creation of an allosteric enzyme by domain insertion. J Mol Biol 336: 263–273 [DOI] [PubMed] [Google Scholar]
- Guseman JM, Hellmuth A, Lanctot A, Feldman TP, Moss BL, Klavins E, Calderón Villalobos LI, Nemhauser JL (2015) Auxin-induced degradation dynamics set the pace for lateral root development. Development 142: 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F, Eisenhut M, Mantegazza O, Weber APM (2018) Homology-directed repair of a defective Glabrous gene in Arabidopsis with Cas9-based gene targeting. Front Plant Sci 9: 424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LA, Nobile MS, Pino JC, Lubbock ALR, Besozzi D, Mauri G, Cazzaniga P, Lopez CF (2017) GPU-powered model analysis with PySB/cupSODA. Bioinformatics 33: 3492–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleary R, Choi W-G, Kim S-H, Lim SD, Gilroy S (2018) Sense and sensibility: The use of fluorescent protein-based genetically encoded biosensors in plants. Curr Opin Plant Biol 46: 32–38 [DOI] [PubMed] [Google Scholar]
- Hillson NJ. (2014) j5 DNA Assembly Design Automation. In Valla S, Lale R, eds, DNA Cloning and Assembly Methods. Humana Press, Totowa, NJ, pp 245–269 [DOI] [PubMed] [Google Scholar]
- Hucka M, Bergmann FT, Hoops S, Keating SM, Sahle S, Schaff JC, Smith LP, Wilkinson DJ (2015) The Systems Biology Markup Language (SBML): Language specification for level 3 version 1 core. J Integr Bioinform 12: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imani M, Dehghannasiri R, Braga-Neto UM, Dougherty ER (2018) Sequential experimental design for optimal structural intervention in gene regulatory networks based on the mean objective cost of uncertainty. Cancer Inform 17: 1176935118790247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Johzuka-Hisatomi Y, Ishida S, Iida S, Kohchi T (2013) Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Sci Rep 3: 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Arevalo ED, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, et al. (2016) Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet 48: 785–791 [DOI] [PubMed] [Google Scholar]
- Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce GF. (1989) Amplification, mutation and selection of catalytic RNA. Presented at the Albany Conference on RNA, Rensselaerville, NY, 22–25 September 1988. In Belfort M, Shub DA, eds, RNA: Catalysis, Splicing, Evolution. Elsevier, Amsterdam, pp 83–87 [Google Scholar]
- Jupe F, Rivkin AC, Michael TP, Zander M, Motley ST, Sandoval JP, Slotkin KR, Chen H, Castanon R, Nery JR, et al. (2018) The complex architecture and epigenomic impact of plant T-DNA insertions. bioRxiv 282772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar M, Wright RC, Date A, Tullman J, Ostermeier M (2013) Protein switch engineering by domain insertion. In Keating AE, ed, Methods in Enzymology. Academic Press, New York, pp 369–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar A, Bolten NJ, Nemhauser J, Klavins E (2016) Cell-cell communication in yeast using auxin biosynthesis and auxin responsive CRISPR transcription factors. ACS Synth Biol 5: 279–286 [DOI] [PubMed] [Google Scholar]
- Khakhar A, Leydon AR, Lemmex AC, Klavins E, Nemhauser JL (2018) Synthetic hormone-responsive transcription factors can monitor and re-program plant development. eLife 7: e34702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner GK, Stahl Y, Imani J, von Korff M, Simon R (2018) Fluorescent reporter lines for auxin and cytokinin signalling in barley (Hordeum vulgare). PLoS One 13: e0196086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klavins E. (2017) Aquarium: Toward reproducible. Mol Biol [Google Scholar]
- Kluyver T, Ragan-Kelley B, Pérez F, Granger B, Bussonnier M, Frederic J, Kelley K, Hamrick J, Grout J, Corlay S, et al. (2016) Jupyter Notebooks—a publishing format for reproducible computational workflows. In Loizides F and Schmidt B, eds, Positioning and Power in Academic Publishing: Players, Agents and Agendas. IOS Press, Amsterdam, pp 87–90 [Google Scholar]
- Kunkel BN, Harper CP (2018) The roles of auxin during interactions between bacterial plant pathogens and their hosts. J Exp Bot 69: 245–254 [DOI] [PubMed] [Google Scholar]
- Lakowicz JR, Szmacinski H, Nowaczyk K, Berndt KW, Johnson M (1992) Fluorescence lifetime imaging. Anal Biochem 202: 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B, Refahi Y, Besnard F, Hervieux N, Mirabet V, Boudaoud A, Vernoux T, Hamant O (2015) Meristem size contributes to the robustness of phyllotaxis in Arabidopsis. J Exp Bot 66: 1317–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu A, Champion A, Legrand J, Lavenus J, Mast D, Brunoud G, Oh J, Guyomarc’h S, Pizot M, Farmer EE, et al. (2015) A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat Commun 6: 6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Ten Tusscher KH (2017) Periodic lateral root priming: What makes it tick? Plant Cell 29: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C-Y, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D (2015) Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210, 2, 210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- List M, Schmidt S, Trojnar J, Thomas J, Thomassen M, Kruse TA, Tan Q, Baumbach J, Mollenhauer J (2014) Efficient sample tracking with OpenLabFramework. Sci Rep 4: 4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List M, Franz M, Tan Q, Mollenhauer J, Baumbach J (2015) OpenLabNotes—An electronic laboratory notebook extension for OpenLabFramework. J Integr Bioinform 12: 274. [DOI] [PubMed] [Google Scholar]
- Liu B, editor (2015) Protein engineering and selection using yeast surface display. In Yeast Surface Display: Methods, Protocols, and Applications. Springer, New York. [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, Postma M, Zhou W, Goedhart J, Sánchez-Pérez M-I, Gadella TWJ, Simon R, Scheres B, et al. (2017) In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548: 97–102 [DOI] [PubMed] [Google Scholar]
- Lowder LG, Zhou J, Zhang Y, Malzahn A, Zhong Z, Hsieh T-F, Voytas DF, Zhang Y, Qi Y (2018) Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol Plant 11: 245–256 [DOI] [PubMed] [Google Scholar]
- Magde D, Elson E, Webb WW (1972) Thermodynamic fluctuations in a reacting system—Measurement by fluorescence correlation spectroscopy. Phys Rev Lett 29: 705–708 [Google Scholar]
- Martínez-García E, Aparicio T, Goñi-Moreno A, Fraile S, de Lorenzo V (2015) SEVA 2.0: An update of the Standard European Vector Architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res 43: D1183–D1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattheakis LC, Bhatt RR, Dower WJ (1994) An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci USA 91: 9022–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medley JK, Choi K, König M, Smith L, Gu S, Hellerstein J, Sealfon SC, Sauro HM (2018) Tellurium notebooks—An environment for reproducible dynamical modeling in systems biology. PLOS Comput Biol 14: e1006220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor N, Adibi M, El-Showk S, De Rybel B, King J, Mähönen AP, Weijers D, Bishopp A (2017) Theoretical approaches to understanding root vascular patterning: A consensus between recent models. J Exp Bot 68: 5–16 [DOI] [PubMed] [Google Scholar]
- Merchant N, Lyons E, Goff S, Vaughn M, Ware D, Micklos D, Antin P (2016) The iPlant Collaborative: Cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol 14: e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minas G, Jenkins DJ, Rand DA, Finkenstädt B (2017) Inferring transcriptional logic from multiple dynamic experiments. Bioinformatics 33: 3437–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misirli G, Nguyen T, McLaughlin JA, Vaidyanathan P, Jones TS, Densmore D, Myers C, Wipat A (2018) A computational workflow for the automated generation of models of genetic designs. ACS Synth Biol [DOI] [PubMed] [Google Scholar]
- Mohsenizadeh DN, Dehghannasiri R, Dougherty ER (2018) Optimal objective-based experimental design for uncertain dynamical gene networks with experimental error. IEEE/ACM Trans Comput Biol Bioinformatics 15: 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardımcı GG, Petricka JJ, Vernoux T, Blilou I, Alonso J, Winter CM, Ohler U, Scheres B, et al. (2015) Transcriptional control of tissue formation throughout root development. Science 350: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylle E, Codreanu M-C, Boruc J, Russinova E (2013) Emission spectra profiling of fluorescent proteins in living plant cells. Plant Methods 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler DC, Morgan S-A, Flamholz A, Kortright KE, Savage DF (2016) Rapid construction of metabolite biosensors using domain-insertion profiling. Nat Commun 7: 12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor DL, Runions A, Sluis A, Bragg J, Vogel JP, Prusinkiewicz P, Hake S (2014) A division in PIN-mediated auxin patterning during organ initiation in grasses. PLOS Comput Biol 10: e1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane CJ, Gehring WJ (1987) Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci USA 84: 9123–9127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto S, Versaw W (2017) Genetically encoded sensors for monitoring the transport and concentration of nitrogen-containing and phosphorus-containing molecules in plants. Curr Opin Plant Biol 39: 129–135 [DOI] [PubMed] [Google Scholar]
- Ostermeier M. (2009) Designing switchable enzymes. Curr Opin Struct Biol 19: 442–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier JN, Arndt KM, Plückthun A, Michnick SW (1999) An in vivo library-versus-library selection of optimized protein-protein interactions. Nat Biotechnol 17: 683–690 [DOI] [PubMed] [Google Scholar]
- Pincus D, Pandey J, Creixell P, Resnekov O, Reynolds KA (2017) Evolution and engineering of allosteric regulation in protein kinases. bioRxiv 189761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plückthun A. (2012) Ribosome display: A perspective. Methods Mol Biol 805: 3–28 [DOI] [PubMed] [Google Scholar]
- Pollak B, Cerda A, Delmans M, Álamos S, Moyano T, West A, Gutiérrez RA, Patron N, Federici F, Haseloff J (2018) Loop assembly: A simple and open system for recursive fabrication of DNA circuits. New Phytol 247593. [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA (2000) Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science 290: 801–806 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Runions A (2012) Computational models of plant development and form. New Phytol 193: 549–569 [DOI] [PubMed] [Google Scholar]
- Rios AF, Radoeva T, De Rybel B, Weijers D, Borst JW (2017) FRET-FLIM for visualizing and quantifying protein interactions in live plant cells. In Kleine-Vehn J and Sauer M, eds, Plant Hormones: Methods and Protocols. Springer, New York, pp 135–146 [DOI] [PubMed] [Google Scholar]
- Ristova D, Carré C, Pervent M, Medici A, Kim GJ, Scalia D, Ruffel S, Birnbaum KD, Lacombe B, Busch W, et al. (2016) Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci Signal 9: rs13. [DOI] [PubMed] [Google Scholar]
- Rivoire O, Reynolds KA, Ranganathan R (2016) Evolution-based functional decomposition of proteins. PLOS Comput Biol 12: e1004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants 3: 803–813 [DOI] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J (2013) Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512 [DOI] [PubMed] [Google Scholar]
- Rougny A, Gloaguen P, Langonné N, Reiter E, Crépieux P, Poupon A, Froidevaux C (2018) A logic-based method to build signaling networks and propose experimental plans. Sci Rep 8: 7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau D, Chéné Y, Belin E, Semaan G, Trigui G, Boudehri K, Franconi F, Chapeau-Blondeau F (2015) Multiscale imaging of plants: Current approaches and challenges. Plant Methods 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford L, Palmer A (2017) Recent advances in development of genetically encoded fluorescent sensors. Methods Enzymol 589: 1–49 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T (2006) Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf A, Leitner D, Landl M, Lobet G, Mai TH, Morandage S, Sheng C, Zörner M, Vanderborght J, Vereecken H (2018) CRootBox: A structural-functional modelling framework for root systems. Ann Bot 121: 1033–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuder MP, Deen C, Boersma WJ, Pouwels PH, Klis FM (1996) Yeast expressing hepatitis B virus surface antigen determinants on its surface: Implications for a possible oral vaccine. Vaccine 14: 383–388 [DOI] [PubMed] [Google Scholar]
- Shibata M, Breuer C, Kawamura A, Clark NM, Rymen B, Braidwood L, Morohashi K, Busch W, Benfey PN, Sozzani R, et al. (2018) GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 145: dev159707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih PM, Vuu K, Mansoori N, Ayad L, Louie KB, Bowen BP, Northen TR, Loqué D (2016) A robust gene-stacking method utilizing yeast assembly for plant synthetic biology. Nat Commun 7: 13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockley EM, Vrugt JA, Lopez CF (2018) PyDREAM: High-dimensional parameter inference for biological models in Python. Bioinformatics 34: 695–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY (1997) A genetically encoded optical probe of membrane voltage. Neuron 19: 735–741 [DOI] [PubMed] [Google Scholar]
- Smith GP. (1985) Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228: 1315–1317 [DOI] [PubMed] [Google Scholar]
- Smucker B, Krzywinski M, Altman N (2018) Optimal experimental design. Nat Methods 15: 559–560 [DOI] [PubMed] [Google Scholar]
- Sparks EE, Drapek C, Gaudinier A, Li S, Ansariola M, Shen N, Hennacy JH, Zhang J, Turco G, Petricka JJ, et al. (2016) Establishment of expression in the SHORTROOT-SCARECROW transcriptional cascade through opposing activities of both activators and repressors. Dev Cell 39: 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard L. (1929) On the reduction of entropy in a thermodynamic system during the intervention of intelligent beings. [in German] Z Phys 53: 840–856 [DOI] [PubMed] [Google Scholar]
- Tan Y, Tian T, Liu W, Zhu Z, J Yang C (2016) Advance in phage display technology for bioanalysis. Biotechnol J 11: 732–745 [DOI] [PubMed] [Google Scholar]
- Tizei PAG, Csibra E, Torres L, Pinheiro VB (2016) Selection platforms for directed evolution in synthetic biology. Biochem Soc Trans 44: 1165–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, Gilroy S (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Tucker CL, Fields S (2001) A yeast sensor of ligand binding. Nat Biotechnol 19: 1042–1046 [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Upadhyay LSB, Verma N (2015) Recent advances in phosphate biosensors. Biotechnol Lett 37: 1335–1345 [DOI] [PubMed] [Google Scholar]
- Vanlier J, Tiemann CA, Hilbers PA, van Riel NA (2014) Optimal experiment design for model selection in biochemical networks. BMC Syst Biol 8: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varala K, Marshall-Colón A, Cirrone J, Brooks MD, Pasquino AV, Léran S, Mittal S, Rock TM, Edwards MB, Kim GJ, et al. (2018) Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc Natl Acad Sci USA 115: 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Robert S (2017) Auxin 2016: A burst of auxin in the warm south of China. Development 144: 533–540 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia A, Waadt R, Jones AM (2018) Genetically encoded biosensors in plants: Pathways to discovery. Annu Rev Plant Biol 69: 497–524 [DOI] [PubMed] [Google Scholar]
- Wandy J, Niu M, Giurghita D, Daly R, Rogers S, Husmeier D (2018) ShinyKGode: An interactive application for ODE parameter inference using gradient matching. Bioinformatics 34: 2314–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe L, Nguyen T, Zhang M, Zundel Z, Zhang Z, Madsen C, Roehner N, Myers C (2018) iBioSim 3: A tool for model-based genetic circuit design. ACS Synth Biol [DOI] [PubMed] [Google Scholar]
- Wend S, Dal Bosco C, Kämpf MM, Ren F, Palme K, Weber W, Dovzhenko A, Zurbriggen MD (2013) A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci Rep 3: 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich JR, Möller BK, Li S, Saiga S, Sozzani R, Benfey PN, De Rybel B, Weijers D (2017) Framework for gradual progression of cell ontogeny in the Arabidopsis root meristem. Proc Natl Acad Sci USA 114: E8922–E8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CM, Wright RC, Eshleman JR, Ostermeier M (2011) A protein therapeutic modality founded on molecular regulation. Proc Natl Acad Sci USA 108: 16206–16211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RC, Khakhar A, Eshleman JR, Ostermeier M (2014) Advancements in the development of HIF-1α-activated protein switches for use in enzyme prodrug therapy. PLoS One 9: e114032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RC, Zahler ML, Gerben SR, Nemhauser JL (2017) Insights into the evolution and function of auxin signaling F-Box proteins in Arabidopsis thaliana through synthetic analysis of natural variants. Genetics 207: 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Duan L, Pruneda-Paz JL, Oh DH, Pound M, Kay S, Dinneny JR (2018) The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol 177: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Audenaert D, Parizot B, Möller BK, Njo MF, De Rybel B, De Rop G, Van Isterdael G, Mähönen AP, Vanneste S, et al. (2015) Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr Biol 25: 1381–1388 [DOI] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Möller BK, Skorzinski N, Njo MF, et al. (2016) Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351: 384–387 [DOI] [PubMed] [Google Scholar]
- Younger AK, Dalvie NC, Rottinghaus AG, Leonard JN (2016) Engineering modular biosensors to confer metabolite-responsive regulation of transcription. ACS Synth Biol 6: 311-325 [DOI] [PubMed] [Google Scholar]
- Younger AKD, Su PY, Shepard AJ, Udani SV, Cybulski TR, Tyo KEJ, Leonard JN (2018) Development of novel metabolite-responsive transcription factors via transposon-mediated protein fusion. Protein Eng Des Sel 31: 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, McLaughlin JA, Wipat A, Myers CJ (2017) SBOLDesigner 2: An intuitive tool for structural genetic design. ACS Synth Biol 6: 1150–1160 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li W-X, Mao L, Chen B, Xu Y, et al. (2016) An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep 6: 23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, Xie X, Shen R, Tan J, Li H, et al. (2017) Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant 10: 918–929 [DOI] [PubMed] [Google Scholar]