An optimized two-component transcription activator-like effector (TALE)–based platform for AND-gating in planta is discussed.
Abstract
Transcription activator-like effectors (TALEs) are bacterial Type-III effector proteins from phytopathogenic Xanthomonas species that act as transcription factors in plants. The modular DNA-binding domain of TALEs can be reprogrammed to target nearly any DNA sequence. Here, we designed and optimized a two-component AND-gate system for synthetic circuits in plants based on TALEs. In this system, named split-TALE (sTALE), the TALE DNA binding domain and the transcription activation domain are separated and each fused to protein interacting domains. Physical interaction of interacting domains leads to TALE-reconstitution and can be monitored by reporter gene induction. This setup was used for optimization of the sTALE scaffolds, which result in an AND-gate system with an improved signal-to-noise ratio. We also provide a toolkit of ready-to-use vectors and single modules compatible with Golden Gate cloning and MoClo syntax. In addition to its implementation in synthetic regulatory circuits, the sTALE system allows the analysis of protein-protein interactions in planta.
Orthogonal tools to regulate gene expression for gene regulatory networks with a desired outcome are a key element of synthetic biology. Although much effort has been invested in synthetic tools for gene regulatory networks in bacterial, yeast, or mammalian cells, comparatively little has been done in plants. Transcription activator-like effectors (TALEs) are derived from phytopathogenic bacteria of the Xanthomonas species and represent a promising and versatile tool for gene regulation in planta (Boch and Bonas, 2010). TALEs are bacterial type-III effector proteins that are secreted and translocated into plant cells, where they enter the nucleus and induce transcription of plant genes to promote virulence (Boch et al., 2014).
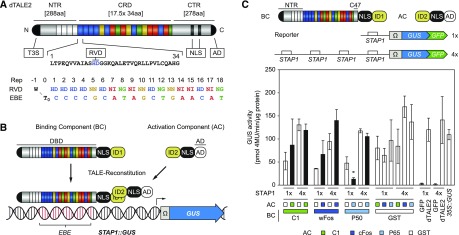
One distinct feature of this protein class is the modular DNA-binding domain, which consists of tandemly arranged, nearly identical 33 to 35 amino acid repeats (central repeat domain; Fig. 1A). Single TALE repeats mainly differ in two amino acid residues described as repeat-variable diresidue (RVD). TALEs bind DNA in a “one repeat to one base pair” manner in which the specificity is defined by the RVD of a single repeat (Boch et al., 2009; Moscou and Bogdanove, 2009). Although all possible amino acid variants were analyzed, the most abundant naturally occurring RVDs, His-Asp, Asn-Asn, Asn-Ile, and Asn-Gly, mediate specific modular binding to cytosine, guanine or adenine, adenine, and thymine, respectively (Yang et al., 2014; Juillerat et al., 2015; Miller et al., 2015). TALE-bound DNA sequences (effector binding element [EBE]) are thus defined by the number and order of the repeats (RVDs), and in turn TALEs can be designed to target almost any DNA-sequence with a corresponding sequence of repeats containing the adequate RVD-order (Geissler et al., 2011; Weber et al., 2011b; Streubel et al., 2012). One constraint is the presence of a thymine (T0) flanking the EBE at the 5′-end that is almost invariant in natural EBEs. Its importance is affected by the number of repeats and the RVD composition, especially by the RVD of the first repeat (Doyle et al., 2013; Schreiber et al., 2015).
Figure 1.
Design of the split-TALE and preliminary tests. A, Schematic representation of dTALE2 (NTR; central repeat region [CRD]; CTR; type 3 secretion signal [T3S]; RVD; NLS; AD; repeat [Rep]; EBE; invariant thymine flanking the 5` end of the RVD-defined target sequence [T0]. B, Schematic representation of the split-TALE system (BC; AC; DBD; ID; STAP). C, GUS-assay after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (1×STAP-Ω-GUS-GFP, 4×STAP-Ω-GUS-GFP) in N. benthamiana leaves. The BC contains the full-length NTR and 47 amino acids of the CTR. Color codes indicate corresponding IDs (C1, green; wFos/cFos, dark blue; P50/P65, blue; GST, white). White bars, except for the positive controls on the right side (dTALE2 and 35S::GUS), are for assays where only one component of the sTALE with an ID, either BC or AC, was expressed. In that case, the other component was fused to GST, which should not interact with the ID, and therefore served as a negative control. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. Free GFP (35S::GFP) and dTALE2 were used to monitor background activity of the STAPs (1x or 4x) and TALE-mediated transcriptional induction of the reporter, respectively. 35S::GUS serves as a positive control. Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with control AC (ID = GST; Student’s t test; *P-value ≤ 0.05).
The repeat domain is flanked by N-terminal regions (NTR) and C-terminal regions (CTR; Fig. 1A). The NTR contains signals that promote secretion and translocation, as well as essential parts of the DNA-binding domain (Gao et al., 2012; Schreiber et al., 2015; Scheibner et al., 2017). Structural analysis revealed that the repeat domain is extended into the NTR by at least 4 degenerated repeats (called Repeat-3 to 0; Deng et al., 2012; Gao et al., 2012; Mak et al., 2012). Repeat-1 contains a RVD-loop like structure with a Trp residue that coordinates T0 and facilitates compact binding between the central repeat domain and the EBE (Mak et al., 2012; Doyle et al., 2013; Meckler et al., 2013; Schreiber and Bonas, 2014; Cuculis et al., 2015). The CTR contains two to three nuclear localization signals (NLS) and an acidic activation domain (Van den Ackerveken et al., 1996; Szurek et al., 2001; Fig. 1A).
Like clustered regularly interspersed short palindromic/Cas systems, TALEs are programmable DNA binding proteins with potential for applications in synthetic biology. By replacing the naturally occurring activation domain (AD) with other executer domains, TALEs were converted into artificial sequence specific nucleases, transcription repressors, methylases, and recombinases among others (de Lange et al., 2014; Khan et al., 2017). TALEs were also converted into inducible transcription regulators. For this, researchers followed different strategies, where TALEs were kept as one peptide chain or separated into two components. One-chain approaches include monomeric receptors (Mercer et al., 2014), tobacco etch virus protease (TEVp)-site containing cyclic TALEs or cyclic transcription activator-like orthogonal repressors (Copeland et al., 2016; Lonzarić et al., 2016). Monomeric receptors undergo conformational changes upon binding to a ligand, leading to exposure of the AD and thereby to transcriptional induction (Mercer et al., 2014). TEVp-site containing cyclic TALEs and cyclic transcription activator-like orthogonal repressors are cleaved in the presence of TEVp, thereby enabling access to DNA or preventing transcriptional repression, respectively (Copeland et al., 2016; Lonzarić et al., 2016). Two-component approaches are based on the separation of the binding domain (BD) from the AD and their fusion to interacting protein domains (IDs) that interact with each other in the presence of light of a specific wavelength or a specific ligand (Li et al., 2012; Konermann et al., 2013; Hochrein et al., 2017; Lo et al., 2017; Zhao et al., 2018). These systems need to be induced by a third component (light or ligand) and therefore rather represent three-component AND-gate systems. Another described AND-gate system is based on the autocatalytic intein-mediated protein splicing, which mediates the fusion of two TALE parts on the protein level (Lienert et al., 2013). To our knowledge, none of these AND-gate systems was tested in plants.
Here, we generated and optimized a TALE-based two-component AND-gate system, which we name split-TALE (sTALE). We separated TALEs into two components (binding domain and AD) and fused them to IDs, which constitutively interact with each other. To render the sTALE system accessible to the scientific community we generated Golden Gate (GG)-based modules and vectors compatible with the syntax of the MoClo system, which is now accepted as one of the standard modular cloning systems (Engler et al., 2014; Patron et al., 2015). The sTALE system has the potential for many applications in planta including programmable signaling in synthetic circuits and analysis of protein-protein interaction.
RESULTS
Generation of sTALE Constructs
To establish a sTALE two-component system we separated the DNA-binding domain (DBD) and AD of the previously described artificial dTALE2 (Weber et al., 2011b; Brückner et al., 2015; Fig. 1A; Supplemental Fig. S1). Fusion of known IDs to the DBD and AD should allow the functional reconstitution of the TALE. According to previous studies, the TALE DBD spans almost the complete NTR and the central repeat domain (Gao et al., 2012; Mak et al., 2012; Schreiber et al., 2015). In addition, a C-terminal deletion leaving 47 amino acids after the central repeat domain (C47) was successfully used for TALE-nucleases (Mussolino et al., 2011). Based on this, we generated the TALE DBD with the complete NTR and C47 (Fig. 1A). According to previous studies, the TALE AD is located within the very last 31 amino acids of the CTR, and this part is sufficient to mediate transcriptional induction if fused to DNA binding domains (Szurek et al., 2001; Schreiber et al., 2015). Therefore, we used the last 31 amino acids of dTALE2 as the AD. In this configuration, the segment C48-C247 of the CTR, which naturally contains NLS, is missing (Van den Ackerveken et al., 1996; Fig. 1, A and B). To enable TALE-reconstitution in the nucleus we fused one ID and the SV40 NLS at the C terminus of the DBD (binding component [BC]) and the other ID with the SV40 NLS at the N terminus of the AD (activation component [AC]; Fig. 1B).
We chose three small sized IDs from non-plant organisms to avoid potential negative effects on the expression rate or interference with endogenous plant processes. These are the C1 peptide, which is the dimerization domain of repressor Lambda C1 (Bell et al., 2000), FosW and cFos–coiled coil Leu zipper domains derived from transcription factor activator protein 1 (Mason et al., 2006)– and P50-P65–IDs of the nuclear factor κB subunits P50 and P65 (Chen et al., 1998; Fig. 1A; Supplemental Fig. S2).
To monitor TALE-reconstitution we generated reporter constructs containing the GUS-GFP reporter gene fused to the omega enhancer (Ω) from the Tobacco mosaic virus (Gallie et al., 1987a) driven by one or four copies of a synthetic TALE activated promoter 1 (STAP1; Brückner et al., 2015; Fig. 1C). The rationale behind this was to allow monitoring of sTALE activity by either GUS activity or GFP fluorescence, but we focused first on GUS as a readout because of its higher sensitivity. We tested matching pairs of BC and AC together with the GUS reporter construct by Agrobacterium-mediated transient expression in Nicotiana benthamiana leaves (Fig. 1C). As a negative control we used a BC and an AC in which the ID was replaced by glutathione S-transferase (GST), a domain that should not interact with any of the other IDs.
Regardless of which ID pair was used, we observed significant background activity (Fig. 1C). The intensity of expression correlated with the number of STAPs fused to the β-glucuronidase (GUS) reporter gene (1×STAP, 4×STAP; Fig. 1C). Notably, we detected no activity if the reporter was combined with free green fluorescent protein (GFP; = 35S::GFP), confirming that the STAP1 promoter in the absence of dTALE2 does not give rise to detectable levels of expression. It is unlikely that GST interacts with every ID; therefore, we concluded that the BC alone already led to reporter induction. There was a trend in the increase of GUS expression in the presence of the matching AC, but this was not significant. Only the sTALE pair BC(P50)-AC(P65) showed an unexpected significant reduction of GUS activity if combined with the 1×STAP-GUS reporter. This effect was not detectable with 4×STAP-GUS, possibly because of the higher background activity. Given these results, we next sought to improve the scaffold of the BC to reduce its background activity.
N-terminal Truncations Reduce Background Activity of the BC
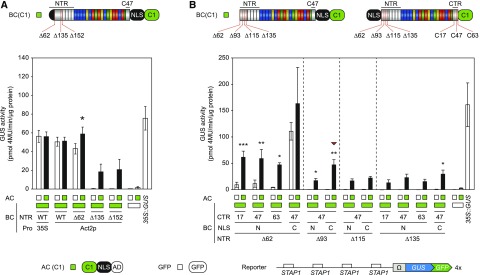
Previous studies on TALE-based hybrids applied in human and yeast cells also reported background activity of TALE-based two-component systems in the absence of inducing signals (light or ligand; Konermann et al., 2013; Hochrein et al., 2017). This problem was circumvented by the fusion of nuclear export signals to the BC, which should prevent its nuclear import in the absence of interaction with the AC. TALE-reconstitution in the cytoplasm then triggers piggyback nuclear import of the BC mediated by the NLS present in the AC (Konermann et al., 2013; Hochrein et al., 2017). We followed different strategies to reduce the background activity and focused initially on the BC alone. In the first experiments (Fig. 1), the BC was expressed under the control of the strong constitutive 35S promoter. We hypothesized that a weaker expression rate of the BC might also reduce its background activity. Furthermore, in our first design the BC contains the complete NTR, which was reported to contain the translocation signals (1-50 amino acids) and an essential part of the DBD in terms of transcriptional induction (64-288 amino acids; Szurek et al., 2002; Schreiber et al., 2015; Scheibner et al., 2017).
For further optimization of the BC we focused on the sTALE-pair BC(C1)-AC(C1) and generated a set of N- and C-terminal deletion derivatives. We used the constitutive but weaker Actin2 promoter (Act2p) for expression of the BC and scaffolds with truncated NTRs (∆N62, ∆N135, and ∆N152; Fig. 2A). To detect the transcription activity induced by single components we coexpressed either BCs or ACs with free GFP (35S::GFP). The GUS reporter construct with four copies of STAP1 was used for higher sensitivity. The expression of the BC by Act2p did not reduce the background activity (Fig. 2A). Interestingly, truncation of the NTR reduced the background activity of the BC but also led to reduced reporter induction of the reconstituted TALE. Expression of AC alone combined with free GFP induced only marginal levels of GUS activity (Fig. 2A). As shown in Figure 2A the truncation of the NTR from ∆N62 to ∆N135 completely abolished the background activity of the BC. Next, we further refined the scaffold of the BC to improve the sTALE system in terms of signal-to-noise ratio. We generated additional N-terminal truncations between ∆N63 and ∆N135 in predicted unstructured regions of the NTR (∆N93, ∆N115; Fig. 2B; Supplemental Fig. S1). Based on functional TALEN scaffolds we also shifted the NLS from the C terminus to the N terminus of the DNA binding domain and included additional C-terminal truncations (C17, C47, C63; Miller et al., 2011; Mussolino et al., 2011; Bedell et al., 2012).
Figure 2.
Optimization of the BC-scaffold. Modifications are visualized by schematic representations above the diagrams. The position of the N-terminal deletions is indicated by Δx, where x represents the position of the amino acid. The position of the C-terminal deletions is indicated by Cx, where x represents the number of amino acids after the repeat region. The diagrams show the results of GUS-assays after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (4xSTAP-Ω-GUS-GFP) in N. benthamiana leaves. The color code indicates corresponding split-TALE constructs (C1, green boxes) or free GFP (35S::GFP, white boxes). Free GFP was used as a control to monitor the background activity of the single BC and AC. 35S::GUS serves as the positive control. White bars are for assays where only one component of the sTALE, either BC or AC, was expressed. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. A, The BC was either expressed under the control of 35S or Actin2 (Act2p) promoters with no change in background activity. Truncation of the NTR up to 135 amino acids significantly reduced the background activity of the BC. Activity was significantly increased in the presence of the corresponding AC. B, Positioning of the NLS to the N terminus of the BC further reduces its background activity, which was accompanied with a loss of overall activity in the presence of the matching AC. The BC-scaffold with a deletion of 93 amino acids of the NTR, 47 amino acids of the CTR, and a C-terminal NLS possessed the best signal-to-noise ratio (indicated by a red triangle). Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with free GFP (white box; Student’s t test; *P-value ≤ 0.05, **P-value ≤ 0.01, ***P-value ≤ 0.001). Experiments were performed two times with similar results. All raw data are available in the Supplemental Data.
For comparative functional analysis of the BCs, we used the same AC (C1-NLS-AD). Comparative analysis revealed that a deletion of 93 amino acids of the NTR is sufficient to strongly reduce the background activity of the BC. The C-terminal truncations had no clear effect on the signal-to-noise ratio. Notably, fusion of the NLS to the N terminus of the DBD strongly reduces BC background activity [compare BC(NLS-∆N62-C47-C1) with BC(∆N62-C47-NLS-C1); Fig. 2B]. However, if more than 63 amino acids of the NTR are deleted, BCs with C-terminal DBD-fused NLS always showed higher activity in the presence of the matching AC. The BC(∆N93-C47-NLS-C1) gave the best signal-to-noise ratio and was used as the BC scaffold in all following experiments.
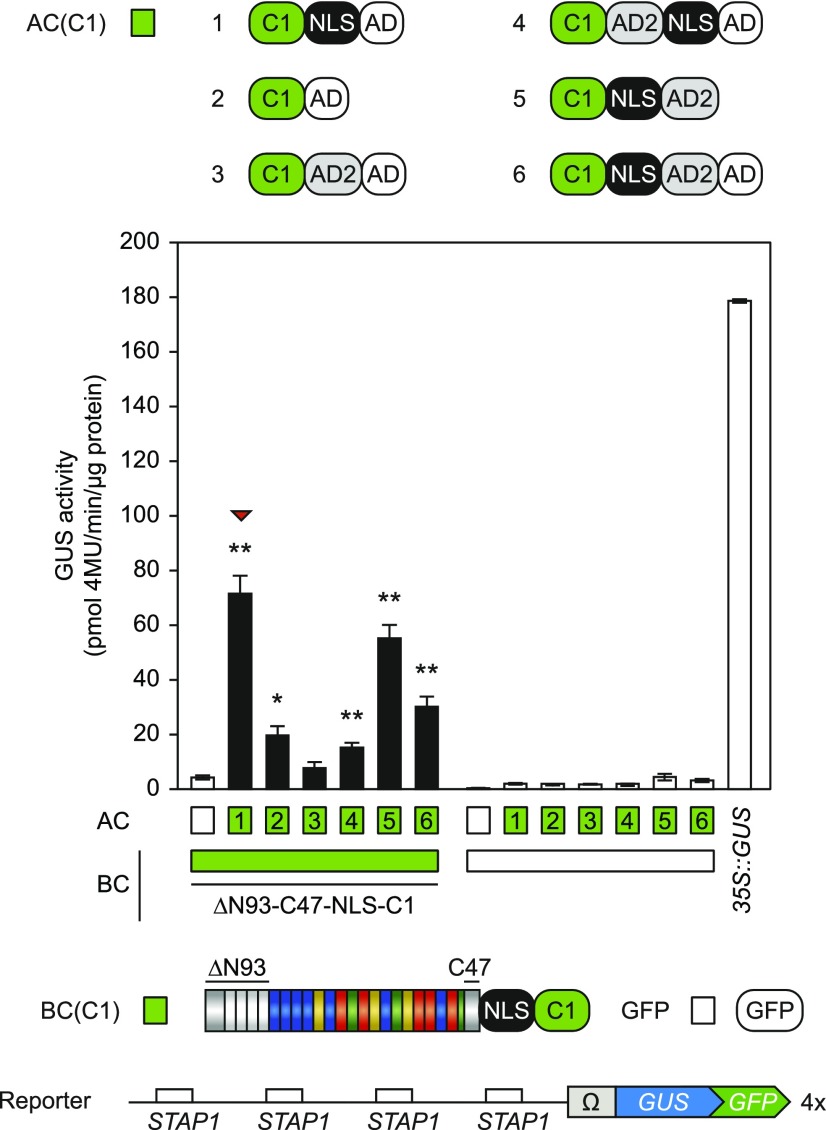
Stacking Activation Domains Does Not Improve Transactivation
We next sought to optimize the AC to improve the sTALE system further. As shown for dCas9- and TALE-based transcriptional regulators in human cells, the combination of multiple ADs had synergistic effects on transcriptional activation (Konermann et al., 2013; Chavez et al., 2016; Zhao et al., 2018). An exchange of the TALE-AD with the VP16- or GAL4-AD was shown to reduce the TALE-mediated transcriptional induction in planta, which indicates that these ADs are not optimal for plant cells (Geissler et al., 2011). The synthetic AD derived from the plant Ethylene Response Factor 2 (ERF2) was shown to be highly active in planta (Li et al., 2013). We generated new AC fusions with the AD from ERF2 (AD2) separately and in combination with the TALE-AD (AD) and tested them in the same conditions as before (Fig. 3). We also generated ACs without NLS because their small size (C1-AD, 16.6 kD; C1-AD1-AD2, 20.5 kD) should allow free diffusion into the nucleus. The AD2 exhibits similar activity compared with the TALE-AD, but a combination of both ADs within the AC did not result in higher activities. ACs without NLS are still functional although with a strongly reduced activity. All tested ACs possess low levels of background activity. The AC(C1-NLS-AD) showed the highest activity and was used as the AC scaffold in all following experiments.
Figure 3.
Optimization of the AC-scaffold. Modifications are visualized by schematic representations above the diagram (AD, TALE AD; AD2, modified AD from AtERF2). The diagram shows the GUS-assay after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (4×STAP-Ω-GUS-GFP) in N. benthamiana leaves. The color code indicates corresponding split TALE constructs (C1, green boxes) or free GFP (35S::GFP, white boxes). Free GFP was used to monitor background activity of the single BC and AC, respectively. 35S::GUS serves as the positive control. White bars are for assays where only one component of the sTALE, either BC or AC, was expressed. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. Deletion of the NLS within the AC-scaffold (2) strongly reduces split TALE activity. Stacking of AD and AD2 (6) negatively affects the activity of the AD. AC-scaffolds with AD (1) and AD2 (5) showed comparable activities, but the AC-scaffold with the TALE AD possessed the best signal-to-noise ratio (indicated by a red triangle). Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with free GFP (white box; Student’s t test; *P-value ≤ 0.05, **P-value ≤ 0.01). Experiments were performed three times with similar results. All raw data are available in the Supplemental Data.
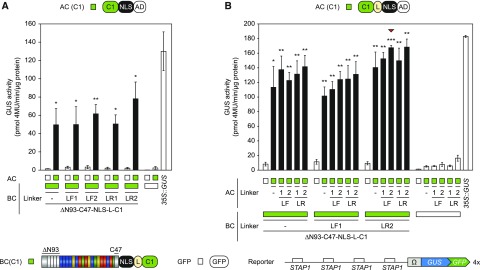
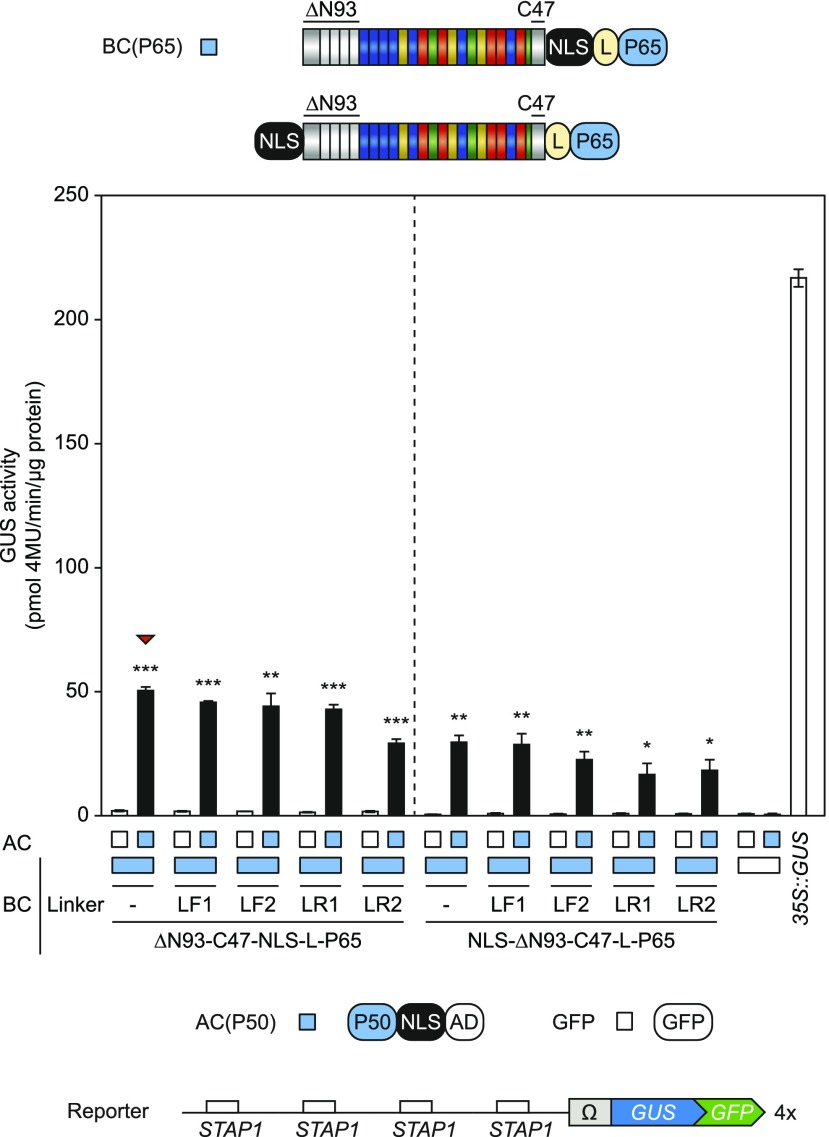
Testing a Set of Linkers to Generate the Fusions with the Activation and DNA-Binding Domains
The nature of the linker connecting the individual elements can greatly affect the functionality of protein fusions (Chen et al., 2013). To endow the sTALE system with additional options, we designed a set of linkers that allow the easy integration of interaction domains using a Golden Gate compatible cloning procedure (Fig. 4). We chose two flexible linkers (LF1, LF2) and two rigid linkers (LR1, LR2; Chen et al., 2013). We tested whether these linkers had an effect on the functionality of the optimized sTALE. Application of these linkers within the C1-based BC revealed a tendency for LR2 to improve the sTALE system (Fig. 4A). We also analyzed linker-containing C1-based ACs together with these BCs and found that ACs with LR2 showed the highest level of TALE-reconstitution (Fig. 4B). This, however, was accompanied with increased background activity of this AC alone. The combination with the best signal-to-noise ratio was BC-C1-LR2 together with AC-C1-LF2, which led to a 17.8-fold induction of 4×STAP1:GUS compared with the BC alone (Fig. 4B). Therefore, the C1-based sTALE system is suitable for synthetic AND-gate circuits in planta.
Figure 4.
Integration of linkers within the AC- and BC-scaffold. Modifications are visualized by schematic representations above and below the diagrams (Linkers: F, flexible, R, rigid). The diagrams show the results of the GUS assays after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (4×STAP-Ω-GUS-GFP) in N. benthamiana leaves. The color code indicates corresponding split-TALE constructs (C1, green boxes) or free GFP (35S::GFP, white boxes). Free GFP was used to monitor background activity of the single BC and AC, respectively. 35S::GUS serves as the positive control. White bars are for assays where only one component of the sTALE, either BC or AC, was expressed. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. A, Linker-containing BCs were combined with ACs without linkers. Flexible linkers within the BC-scaffold showed no clear effect, whereas the rigid linker 2 (LR2) led to slightly increased split TALE activity. B, Linker-containing BCs were combined with linker-containing ACs. Rigid linkers within the AC-scaffold showed a trend to positive effects on split-TALE activity. The combination of BC(LR2) and AC(LF2) possessed the best signal-to-noise ratio (indicated by a red triangle). Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with free GFP (white box; Student’s t test; *P-value ≤ 0.05, **P-value ≤ 0.01, ***P-value ≤ 0.001). Experiments were performed three times with similar results. All raw data are available in the Supplemental Data.
The Interacting Pair p65-p50 also Leads to a Functional sTALE
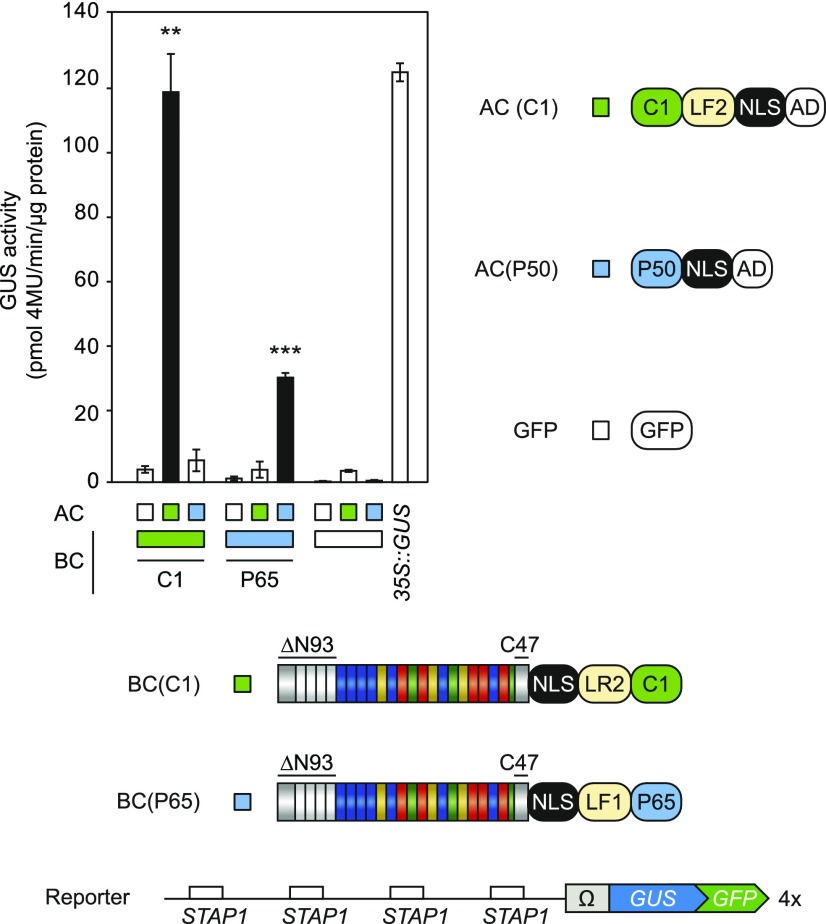
We also used these optimized sTALE scaffolds for the interaction pair P65-P50. As shown in Figure 1, the interaction between BC(P50) and AC(P65) led to transcriptional repression of the BC-mediated background activity (Fig. 1C). To avoid such negative effects, we switched the positions of P50 and P65 between the sTALE components (BC[P65] and AC[P50]; Fig. 5). As with the C1 sTALE constructs, we observed that the fusion of the NLS to the N terminus of the DBD led to reduced transactivation of GUS expression. In addition, the use of flexible linkers within the BC did not change sTALE activity significantly. However, in contrast with C1, rigid linkers within the BC seemed to have a negative effect with the P50-P65 pair (Fig. 5). This shows that different types of linkers may have different effects on the reconstitution of TALE activity depending on the interacting pairs. To confirm that TALE-reconstitution depends on the specific interaction between the corresponding IDs, we performed a cross test with all C1 and P50/P65 sTALE components (Fig. 6). A significant induction of the reporter occurs only in the presence of the matching ID. This rules out the possibility that TALE-reconstitution is driven by the direct interaction between the DBD and the AD and confirms the specificity of the interacting pairs.
Figure 5.
Optimized AC- and BC-scaffold with P65-P50 IDs. Modifications are visualized by schematic representations above the diagram (Linkers: F, flexible, R, rigid). Diagram shows GUS-assay after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (4×STAP-Ω-GUS-GFP) in N. benthamiana leaves. The color code indicates corresponding split TALE constructs (P65/P50, blue boxes) or free GFP (35S::GFP, white boxes). Free GFP was used to monitor background activity of the single BC and AC, respectively. 35S::GUS serves as the positive control. White bars are for assays where only one component of the sTALE, either BC or AC, was expressed. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. A C-terminal NLS is beneficial for sTALE activity using the ID pair P65/P50 (left). Flexible linkers showed no clear effect, whereas rigid linkers showed negative effects on split-TALE activity. The combination of BC and AC without linkers possessed the best signal-to-noise ratio (indicated by a red triangle). Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with free GFP (white box; Student’s t test; *P-value ≤ 0.05, **P-value ≤ 0.01, ***P-value ≤ 0.001). Experiments were performed two times with similar results. All raw data are available in the Supplemental Data.
Figure 6.
Cross test of C1 and P65/P50 split TALE scaffolds. The constructs used are visualized by schematic representations. The diagram shows the results of the GUS-assay after Agrobacterium-mediated transient expression of indicated split-TALE components (BC, AC) and the reporter construct (4×STAP-Ω-GUS-GFP) in N. benthamiana leaves. The color code indicates corresponding split-TALE constructs (C1, green boxes; P65/P50, blue boxes) or free GFP (35S::GFP, white boxes). Free GFP was used to monitor background activity of the single BCs and ACs, respectively. 35S::GUS serves as the positive control. White bars are for assays where only one component of the sTALE, either BC or AC, was expressed. Black bars are for assays where both functional components were expressed. The identity of the expressed component is indicated below the graph by green boxes or squares. Split-TALE activity was only measurable in the presence of the matching IDs. Error bars represent sd (sd) of three biological replicates. Asterisks indicate a significant difference in activity of the same BC tested with free GFP (white box; Student’s t test; **P-value ≤ 0.01, ***P-value ≤ 0.001). Experiments were performed two times with similar results. All raw data are available in the Supplemental Data.
Tuneable Output Strength by Different Reporter Scaffolds
As shown in Fig. 1, the number of STAPs upstream of the reporter gene affects the strength of sTALE-mediated output. To analyze the effect of STAP number we systematically tested a set of reporters that are based on either GUS, GFP, or GUS-GFP fusion and differ in the number of STAPs (Supplemental Fig. S3). We also generated GUS reporters by using different numbers of the commonly used tomato (Solanum lycopersicum) minimal Bs4 promoter fused to the EBE (EBE-Bs4min; Boch et al., 2009). As expected, sTALE activity positively correlated with the number of STAPs or EBE-Bs4min promoter fragments (Supplemental Fig. S3A). Notably, the presence of the viral translational enhancer led to strongly increased sTALE-activity using STAP-based reporters. This scaffold also outcompetes EBE-Bs4min reporters in terms of inducibility and background activity (see combination of the reporter with free GFP; Supplemental Fig. S3A).
We also qualitatively analyzed GUS activity by GUS-staining, but low GUS activity already led to strong staining, which hampers conclusions about the influence of the number of STAPs (Supplemental Fig. S3B). A fluorescent reporter would be an attractive alternative to GUS because it would allow a quick readout macroscopically using whole leaves under UV-light or on a cellular level using a fluorescence microscope. We found that the expression of GFP induced by sTALE can be robustly seen macroscopically with four STAPS and with a microscope with a minimum number of three STAPs (Supplemental Fig. S3C). In addition, a dual reporter that contains a GUS-GFP fusion showed reduced fluorescence in comparison with free GFP (Supplemental Fig. S3B).
Comparative Quantification of the sTALE-Mediated Output Strength
These observations with GFP as a reporter prompted us to evaluate more precisely the strength of the transcriptional activation by the sTALE system. During the optimization process of the sTALE system, we focused on reducing the background activity of the single components (BC and AC). Therefore, we performed the GUS experiments as end point measurements using long incubation times at 37°C (see “Material and Methods”). This does not allow a direct and quantitative comparison with other promoters (like 35S) in terms of strength.
To obtain data that better reflect the real strength of the transcriptional induction, we performed kinetics of the GUS measurements by measuring GUS activity every 10 min at 37°C (10-90 min) incubation (Supplemental Fig. S4A). With the 35S promoter, the system was already saturated after 20 min. We therefore performed assays after a short incubation time of 15 min. We compared the transcriptional activation of the GUS reporter gene by all sTALE pairs (C1-C1 and P50-P65) with that induced by a full-length TALE as well as by the Actin 2 and 35S promoters. We could confirm that the C1 sTALE-pair, with 30% of the GUS activity driven by the 35S promoter, is more active than the P50-P65 sTALE pair, with 5% of the GUS activity driven by the 35S promoter (Supplemental Fig. S4B). The GUS activity induced by the P50-P65 sTALE was comparable with that of the Actin2 promoter (Supplemental Fig. S4B).
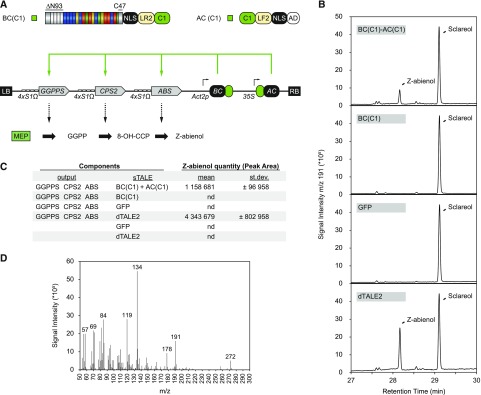
Application of the sTALE System to the Production of Z-Abienol
To show that the sTALE AND-gate system can be applied for synthetic circuits in planta, we used the optimized C1-sTALE constructs for the production of the diterpene Z-abienol. Z-abienol is a plant diterpene with a labdane backbone that can be used as starting material for the synthesis of Ambrox, a valuable ingredient for the fragrance industry with fixative properties and amber notes (Barrero et al., 1993). In tobacco (Nicotiana tabacum), the biosynthesis of abienol requires two genes encoding NtCPS2, a class II diterpene synthase that converts geranylgeranyl diphosphate (GGPP) to 8-hydroxycopalyl diphosphate (8-OH-CPP), and NtABS, a class I diterpene synthase that catalyzes the conversion of 8-OH-CPP to Z-abienol (Sallaud et al., 2012). We generated constructs containing constitutively expressed sTALE components [Act2p-BC(dN93-C47-NLS-LR2-C1)-tOCS and 35S-AC(C1-LF2-NLS-AD)-tOCS] and sTALE-inducible NtCPS2 (4×STAP1-Ω-NtCPS2-tMas), NtABS (4×STAP1-Ω-NtABS-t35S), and NtGGPPS2 (geranylgeranyl diphosphate synthase, 4xSTAP1-Ω-NtGGPPS2-tOCS) fused to four copies of STAP1 each (Fig. 7A). The production of Z-abienol requires the expression of three genes, in contrast with the previously used GUS reporter encoded by a single gene, thereby constituting a test for the capacity of the sTALE to concurrently activate the expression of several genes. The circuit was tested transiently in N. benthamiana, which does not produce Z-abienol. The three sTALE-inducible genes were combined with free GFP or full-length TALE specific for STAP1 as a negative or positive control, respectively. Leaf surface extracts were isolated 3 d after infiltration, and the presence of abienol was analyzed by gas chromatography coupled to mass spectrometry (GC-MS). We detected abienol only when we infiltrated both sTALE components (BC and AC) and in the positive control with the full-length TALE (Fig. 7B). No Z-abienol was detected in the presence of the BC only, which further confirmed the low background activity of the sTALE system.
Figure 7.
sTALE-regulated Z-abienol production. A, Schematic representation of the constructs used for Z-abienol production. BC and AC are constitutively expressed with Act2 and 35S promoters, respectively. Genes for the production of Z-abienol are fused to 4×STAP1-Ω and induced by TALE-reconstitution [geranylgeranyl diphosphate synthase (NtGGPPS2), 8-hydroxycopalyl diphoshate (8-OH-CPP) synthase (NtCPS2), Z-abienol synthase (NtABS)]. B, GC-MS chromatogram (m/z 191) of hexane leaf surface extracts after transient expression in N. benthamiana leaves. Only the portion of the chromatogram between retention times from 27 to 30 min is shown. Sclareol serves as internal standard for normalization. Z-abienol elutes at 28.20 min. Z-Abienol is produced only in the presence of the two sTALE components or the full-length dTALE2. C, Peak areas of Z-abienol normalized to sclareol. nd, Not detected. D, Mass spectrum of Z-abienol.
DISCUSSION
Development and Optimization of a Split-TALE System
We developed a TALE-based AND-gate system for applications in planta. We showed that the specific interaction of the corresponding IDs enables reconstitution of a functional TALE (Fig. 5). Turning TALEs into two-component systems was done earlier, although none of the previously developed systems were generated for plants. Most sTALE systems were light (Konermann et al., 2013; Hochrein et al., 2017; Lo et al., 2017) or ligand induced (Li et al., 2012; Lonzarić et al., 2016; Zhao et al., 2018). To our knowledge, there is only one two-component sTALE AND-gate system previously reported, which is based on intein-mediated protein splicing (Lienert et al., 2013).
Importantly, previously used TALE-based two-component systems all possess a certain level of background activity, which was circumvented by the nuclear export of the BC (Konermann et al., 2013; Hochrein et al., 2017; Zhao et al., 2018). In our initial attempts, we also encountered high levels of background activity of the BC element on its own (Figs. 1 and 2). A key step in reducing this background activity was the truncation of amino acid 62 to 92 from the N terminus (∆N93; Fig. 2B), which was also shown previously to affect the activity of wild-type TALEs (Schreiber et al., 2015). The explicit function of the NTR residues 63-92 for TALE activity is not completely understood, but it is known that residues 1-152 of the NTR also contribute to TALE DNA-binding affinity (Schreiber et al., 2015). Our NTR deletion results, however, suggest that residues 62-92 contain a cryptic transcription activation domain. Additionally, the fusion of the NLS to the truncated ΔN62 deletion of the N-terminal domain also led to reduced background activation of transcription (Fig. 2B). Biochemical analysis indicated that the DNA-binding domain is extended to residue 148 as one continuous fold (Gao et al., 2012). Residues 1-147 likely form a separate domain, but no structural information is available to date. Further investigations would be needed to show the presence of an additional cryptic AD in the NTR of TALEs.
Attempts to optimize the AC to increase transcriptional activation were less successful and could be pursued with the analysis of additional AC scaffolds. For instance, one could use multiple tandem copies of the TALE AD as recently described for the synthetic dCas9-based activator dCas9-TV, which was developed for plants (Li et al., 2017). However, as it stands our sTALE system allows gene expression levels that are around 30% of those induced by the 35S promoter for the C1 fusions and on a par with the Actin2 promoter for the P50/P65 interacting partners. These are expression levels that allow visualization of reporters with a high detection threshold, such as GFP, as well as the simultaneous induction of multiple genes as for the biosynthesis of Z-abienol (Fig. 7; Supplemental Fig. S4).
Impact of the Interacting Partners
The capacity of the C1 domain to homodimerize did not prevent the reconstitution of active transcription. In theory, homodimerization of C1 should lead to the formation of C1-AC and C1-BC homodimers, so that, assuming an equally strong interacting power of C1 in either fusions, only one-third of heterodimeric complexes should be formed between C1-AC and C1-BC. Thus, we expected a stronger transcription activation with the heterodimer P50/P65 than with C1. One possible reason for the strong activity with C1 fusions is that the C1 domain was also shown to form higher order complexes, including homotetramers and homooctamers (Senear et al., 1993; Bell et al., 2000). This could lead to the formation of multimeric complexes that contain several C1-AC bound to one C1-BC, thereby leading to increased transcriptional activation. Another possibility is that the interaction between P50 and P65 is weaker than between C1 peptides.
Applications of the sTALE System
In synthetic biology, the sTALE system can be used as an AND-gate. AND-gates allow the production of an output when, and only when, two conditions are met. In plants, these two conditions could be two spatially or temporally overlapping expression profiles conferred by two distinct promoters. Environmental signals could also be used, for example drought and wounding, and additionally combined with tissue-specific promoters. The output can be a visual reporter or more complex downstream processes, such as metabolic or signal transduction pathways. Based on previous work, it would be conceivable to develop an AND-gate system based on the GAL4 enhancer-trap system in plants (Johnson et al., 2005; Laplaze et al., 2005). However, the customizable and modular DNA-binding domain of TALEs confers much more flexibility to the sTALE system, including orthogonality as well as the possibility to transactivate endogenous genes.
One challenge for the incorporation of the sTALE system into synthetic AND-gate circuits might be the required expression rate of the single components (BC and AC), especially the expression of the AC needs to be relatively strong (with a 35S promoter) for reporter gene activation in a transient system. This could be circumvented by the introduction of a positive amplification step for the input (for higher expression of BC and AC) or the output (for signal amplification of the Reporter). This could be done, for example, by the use of a full-length TALE with a different EBE, under the control of a tissue-specific promoter that would activate the expression of the AC. Alternatively, the sTALE components could activate a full-length TALE with a different EBE, which itself would activate the output genes.
One potential application of the sTALE system is the analysis protein-protein interactions. Putative interactors can be fused to either BC or AC, and their interaction in planta can be monitored by the expression of a reporter gene, e.g. GUS or luciferase. This is analogous to the widely used yeast two-hybrid system (Fields and Song, 1989). The simple experimental setup allows analysis of protein-protein interactions within 3 d. Besides direct interactions, indirect interactions (or complex formations) can be monitored because bridging components are present in planta, which might be absent in yeast. The low background activity of our sTALE system might be an advantage over other protein-protein interaction methods like split-YFP or split-luciferase, which were reported to give rise to background signals because of their propensity to reconstitute themselves independently of the fused IDs (Horstman et al., 2014). However, a direct comparison of the sTALE and split-YFP systems would be needed to clearly demonstrate the differences of these systems. The developed sTALE scaffolds (BC and AC) can also be converted into a ligand-dependent AND-gate system by IDs, which exhibit ligand-dependent interaction such as DmrA and DmrC in the presence of rapamycin (Lonzarić et al., 2016).
A sTALE System Compatible with Golden Gate and MoClo Standards
To make the sTALE system user-friendly, we generated a set of vectors for easy cloning of putative interactors or desired IDs by GG cloning (Weber et al., 2011a, 2011b; Engler et al., 2014; Supplemental Fig. S5A). The advantage is that only one GG-module per ID is needed to fuse it either to the BC or to the AC [BsaI(AATG)-ID-(TTCG)BsaI, frame underlined]. Depending on the vector used, BCs and ACs contain flexible, rigid, or no linkers (Supplemental Fig. S5A). In addition, the BC and AC are also provided as single Level 0 GG-modules compatible with the MoClo system (Werner et al., 2012; Engler et al., 2014), which can be combined with promoters, tags, UTRs, or terminators that fulfill desired needs (Supplemental Fig. S5B). Furthermore, because of its modular nature, the TALE DNA binding domain can be modified to bind any desired DNA sequence, making it possible to induce endogenes as well as foreign genes orthogonally. We also provide a set of responsive promoters, which contain one, two, three, or four copies of STAP1 (Brückner et al., 2015; Supplemental Fig. S5B). Each STAP1 contains an EBE, which can be bound by the BC. Increasing the number of EBEs in the promoter also increases the binding events of the BC and in turn the reporter induction upon TALE-reconstitution. Based on our results, it is recommended to combine the STAPs with the translational enhancer from the Tobacco mosaic virus (Ω; Gallie et al., 1987b), because it strongly increases the expression of the reporter (Supplemental Fig. S3). The combination of STAPs with Ω also outcompetes the commonly used minimal Bs4 promoter for TALE-mediated transcriptional induction (Schornack et al., 2006; Boch et al., 2009; Supplemental Fig. S3). The strength of the output can be further fine-tuned by using STAPs of different strengths (Brückner et al., 2015).
In conclusion, the sTALE system provides a platform with potential for many applications, ranging from in planta protein-protein interaction studies to synthetic biology projects. Its compatibility with the now widely used Golden Gate system should ensure rapid adoption within the plant scientific community.
MATERIAL AND METHODS
Bacterial and Plant Growth Conditions
Escherichia coli strain DH10B [F-mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara,leu)7697 araD139 GalU galK nupG rpsL λ-] and Agrobacterium tumefaciens strain GV3101::pMP90 (Koncz and Schell, 1986) were grown in lysogeny broth/medium (LB medium [Duchefa Biochemie]: 10 g/l tryptone, 10 g/l sodium chloride, 5 g/l yeast extract) with selective antibiotics at 37°C and 28°C, respectively. Nicotiana benthamiana plants were grown in the phyto chamber (day and night temperatures of 21°C and 18°C, respectively) with 16-h light and 50% to 60% humidity.
Generation of Constructs
All constructs were generated by PCR using Thermococcus kodakaraensis Polymerase (Merck Millipore) and assembled by GG-cloning according to the MoClo system (Weber et al., 2011a; Engler et al., 2014). PCR products were purified with the Monarch PCR & DNA Cleanup kit (New England Biolabs) and cloned into corresponding vectors by GG-cloning using either BsaI (New England Biolabs) or BpiI (Thermo Fisher Scientific) depending on the level of the destination MoClo vectors (Supplemental Fig. S5) and T4 Ligase (Promega Cooperation). GG-modules coding for P50 and P65 were synthesized (Eurofins Genomics). Assembled plasmids were transformed into E. coli (DH10B) by heat shock; then transformants were plated on LB agar plates with selective antibiotics and grown overnight at 37°C. Single clones were inoculated in liquid LB media with selective antibiotics and incubated overnight at 37°C. Plasmids were isolated with the Nucleospin Plasmid preparation kit (MACHEREY-NAGEL GmbH) and analyzed by sequencing (generation of GG-modules) or control digestion (assembling of existing GG-modules). Binary vectors were transformed in Agrobacterium (GV3101::pMP90) by electroporation; then transformants were plated on LB agar plates with selective antibiotics and grown for 2 d at 28°C. Agrobacterium strains were stored as cryo-stocks (LB with selective antibiotics and 7% v/v dimethyl sulfoxide) at −80°C. Sequences of primers and of interaction domains used as well as vector maps are given in the Supplemental Tables S1 and S2 and Supplemental Figure S6, respectively.
GUS Reporter Assay
Binary vectors containing Agrobacterium strains with T-DNAs for BC, activation components (AC), free GFP (= 35S::GFP), or reporter constructs were grown on plates for 1 to 2 d at 28°C. Agrobacterium strains were resuspended in Agrobacterium infiltration media (10 mm MES, 10 mm mgCl2, 150 µM acetosyringone) and diluted to an optical density of OD600 = 0.8. Agrobacterium strains with T-DNAs for AC, BC, and reporter were mixed 1:1:1 and inoculated into 5 to 6 weeks old N. benthamiana plants with a needleless syringe. At 2 to 3 d after inoculation, two leaf discs (diameter 0.9 mm) were harvested from three different plants and analyzed by GUS assay (Kay et al., 2007). According to Kay et al., 2007, incubation of the enzymatic reaction was done for 60 to 90 min at 37°C unless stated otherwise (Supplemental Fig. S4). 35S::GFP and 35S::GUS served as negative and positive controls, respectively. Error bars represent the sd of three biological replicates. Each biological replicate was measured twice (technical replicates). The values displayed in the figures are the average of the biological replicates (each being the average of the two technical replicates). The error bars represent the sd of the biological replicates. All experiments were repeated two to three times with similar results. All raw data can be found in the Supplemental Data.
Production and Detection of Z-Abienol
Agrobacterium strains were prepared as for the GUS assays but inoculated into N. benthamiana leaves with an OD600 = 0.3. At 3 d after inoculation, 10 leaf discs (diameter 0.9 mm) were harvested, immediately transferred to 1.8 ml hexane (with 1 µM sclareol for normalization), and incubated for 2 min at room temperature using a thermo mixer at 1,400 rpm. The hexane extracts were then transferred into fresh tubes, incompletely evaporated under a flow of nitrogen, down to a volume of around 100 µL. Analysis of hexane extracts was done by GC-MS as in Sallaud et al. (2012). The peak area of sclareol in each extract was used as reference for normalization.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers J02459.1 (C1), P25799.2 (P50), Q04207.1 (P65).
SUPPLEMENTAL DATA
The following supplementary data are available.
Supplemental Figure S1. Amino acid sequence of dTALE2.
Supplemental Figure S2. Summary of the interacting domains (IDs) used in this study.
Supplemental Figure S3. Analysis of different split TALE reporters.
Supplemental Figure S4. Quantitative analysis of the optimized split TALE system.
Supplemental Figure S5. sTALE system compatible with Golden Gate and MoClo standards.
Supplemental Figure S6. Maps of plasmids used in this study.
Supplemental Data. Raw data from all N. benthamiana transient assays.
Supplemental Table S1. List of oligonucleotide primers used in this study.
Supplemental Table S2. Accession numbers and sequences of major proteins used in this study.
Acknowledgments
We thank Sylvestre Marillonnet for providing GG-based TALE modules and the MoClo system. We thank Ulla Bonas for providing lab environment and financial support in the initial phases of the project through her CRC-648 grant from the Deutsche Forschungsgemeinschaft. We also thank Wolfgang Brandt for modelling the TALE protein structure and the IPB-staff and gardeners for excellent support.
Footnotes
This work was supported by the Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research) (grant 031L0009 to A.T.).
Articles can be viewed without a subscription.
References
- Barrero AF, Alvarez-Manzaneda EJ, Altarejos J, Salido S, Ramos JM (1993) Synthesis of Ambrox(R) from (-)-Sclareol and (+)-Cis-Abienol. Tetrahedron 49: 10405–10412 [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG II, Tan W, Penheiter SG, Ma AC, Leung AY, et al. (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CE, Frescura P, Hochschild A, Lewis M (2000) Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell 101: 801–811 [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu Rev Phytopathol 48: 419–436 [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U, Lahaye T (2014) TAL effectors--pathogen strategies and plant resistance engineering. New Phytol 204: 823–832 [DOI] [PubMed] [Google Scholar]
- Brückner K, Schäfer P, Weber E, Grützner R, Marillonnet S, Tissier A (2015) A library of synthetic transcription activator-like effector-activated promoters for coordinated orthogonal gene expression in plants. Plant J 82: 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, Chari R, Ter-Ovanesyan D, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, et al. (2016) Comparison of Cas9 activators in multiple species. Nat Methods 13: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 391: 410–413 [DOI] [PubMed] [Google Scholar]
- Chen X, Zaro JL, Shen WC (2013) Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65: 1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland MF, Politz MC, Johnson CB, Markley AL, Pfleger BF (2016) A transcription activator-like effector (TALE) induction system mediated by proteolysis. Nat Chem Biol 12: 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuculis L, Abil Z, Zhao H, Schroeder CM (2015) Direct observation of TALE protein dynamics reveals a two-state search mechanism. Nat Commun 6: 7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange O, Binder A, Lahaye T (2014) From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J 78: 753–771 [DOI] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N (2012) Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335: 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Hummel AW, Demorest ZL, Starker CG, Voytas DF, Bradley P, Bogdanove AJ (2013) TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat -1. PLoS One 8: e82120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S (2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340: 245–246 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TM (1987a) The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res 15: 3257–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TM (1987b) A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res 15: 8693–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wu X, Chai J, Han Z (2012) Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res 22: 1716–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J (2011) Transcriptional activators of human genes with programmable DNA-specificity. PLoS One 6: e19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrein L, Machens F, Messerschmidt K, Mueller-Roeber B (2017) PhiReX: A programmable and red light-regulated protein expression switch for yeast. Nucleic Acids Res 45: 9193–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman A, Tonaco IA, Boutilier K, Immink RG (2014) A cautionary note on the use of split-YFP/BiFC in plant protein-protein interaction studies. Int J Mol Sci 15: 9628–9643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AA, Hibberd JM, Gay C, Essah PA, Haseloff J, Tester M, Guiderdoni E (2005) Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J 41: 779–789 [DOI] [PubMed] [Google Scholar]
- Juillerat A, Pessereau C, Dubois G, Guyot V, Maréchal A, Valton J, Daboussi F, Poirot L, Duclert A, Duchateau P (2015) Optimized tuning of TALEN specificity using non-conventional RVDs. Sci Rep 5: 8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651 [DOI] [PubMed] [Google Scholar]
- Khan Z, Khan SH, Mubarik MS, Sadia B, Ahmad A (2017) Use of TALEs and TALEN technology for genetic improvement of plants. Plant Mol Biol Report 35: 1 [Google Scholar]
- Koncz C, Schell J (1986) The promoter of Tl-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Molec Gen Genet 204: 383–396 [Google Scholar]
- Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F (2013) Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Li J, Blue R, Zeitler B, Strange TL, Pearl JR, Huizinga DH, Evans S, Gregory PD, Urnov FD, Petolino JF (2013) Activation domains for controlling plant gene expression using designed transcription factors. Plant Biotechnol J 11: 671–680 [DOI] [PubMed] [Google Scholar]
- Li Y, Moore R, Guinn M, Bleris L (2012) Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep 2: 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang D, Xiong X, Yan B, Xie W, Sheen J, Li JF (2017) A potent Cas9-derived gene activator for plant and mammalian cells. Nat Plants 3: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Torella JP, Chen JH, Norsworthy M, Richardson RR, Silver PA (2013) Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res 41: 9967–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CL, Choudhury SR, Irudayaraj J, Zhou FC (2017) Epigenetic editing of Ascl1 gene in neural stem cells by optogenetics. Sci Rep 7: 42047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonzarić J, Lebar T, Majerle A, Manček-Keber M, Jerala R (2016) Locked and proteolysis-based transcription activator-like effector (TALE) regulation. Nucleic Acids Res 44: 1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak ANS, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL (2012) The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Schmitz MA, Müller KM, Arndt KM (2006) Semirational design of Jun-Fos coiled coils with increased affinity: Universal implications for leucine zipper prediction and design. Proc Natl Acad Sci USA 103: 8989–8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsäesser J, et al. (2013) Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res 41: 4118–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF III (2014) Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth Biol 3: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Miller JC, Zhang L, Xia DF, Campo JJ, Ankoudinova IV, Guschin DY, Babiarz JE, Meng X, Hinkley SJ, Lam SC, et al. (2015) Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods 12: 465–471 [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T (2011) A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res 39: 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, et al. (2015) Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol 208: 13–19 [DOI] [PubMed] [Google Scholar]
- Sallaud C, Giacalone C, Töpfer R, Goepfert S, Bakaher N, Rösti S, Tissier A (2012) Characterization of two genes for the biosynthesis of the labdane diterpene Z-abienol in tobacco (Nicotiana tabacum) glandular trichomes. Plant J 72: 1–17 [DOI] [PubMed] [Google Scholar]
- Scheibner F, Marillonnet S, Büttner D (2017) The TAL effector AvrBs3 from Xanthomonas campestris pv. vesicatoria contains multiple export signals and can enter plant cells in the absence of the type III secretion translocon. Front Microbiol 8: 2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Meyer A, Römer P, Jordan T, Lahaye T (2006) Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol 163: 256–272 [DOI] [PubMed] [Google Scholar]
- Schreiber T, Bonas U (2014) Repeat 1 of TAL effectors affects target specificity for the base at position zero. Nucleic Acids Res 42: 7160–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T, Sorgatz A, List F, Blüher D, Thieme S, Wilmanns M, Bonas U (2015) Refined requirements for protein regions important for activity of the TALE AvrBs3. PLoS One 10: e0120214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senear DF, Laue TM, Ross JB, Waxman E, Eaton S, Rusinova E (1993) The primary self-assembly reaction of bacteriophage lambda cI repressor dimers is to octamer. Biochemistry 32: 6179–6189 [DOI] [PubMed] [Google Scholar]
- Streubel J, Blücher C, Landgraf A, Boch J (2012) TAL effector RVD specificities and efficiencies. Nat Biotechnol 30: 593–595 [DOI] [PubMed] [Google Scholar]
- Szurek B, Marois E, Bonas U, Van den Ackerveken G (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: Protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J 26: 523–534 [DOI] [PubMed] [Google Scholar]
- Szurek B, Rossier O, Hause G, Bonas U (2002) Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol Microbiol 46: 13–23 [DOI] [PubMed] [Google Scholar]
- Van den Ackerveken G, Marois E, Bonas U (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87: 1307–1316 [DOI] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S (2011a) A modular cloning system for standardized assembly of multigene constructs. PLoS One 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S (2011b) Assembly of designer TAL effectors by Golden Gate cloning. PLoS One 6: e19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Engler C, Weber E, Gruetzner R, Marillonnet S (2012) Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs 3: 38–43 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Y, Yuan P, Zhou Y, Cai C, Ren Q, Wen D, Chu C, Qi H, Wei W (2014) Complete decoding of TAL effectors for DNA recognition. Cell Res 24: 628–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang Y, Zhao Y, Ying Y, Ai R, Zhang J, Wang Y (2018) Multiple chemical inducible Tal effectors for genome editing and transcription activation. ACS Chem Biol 13: 609–617 [DOI] [PubMed] [Google Scholar]