Theoretical-experimental approaches are described for the engineering of synthetic chemical- and light-regulated (optogenetic) switches and circuits for the targeted interrogation and control of cell processes.
Abstract
Synthetic biology is an established but ever-growing interdisciplinary field of research currently revolutionizing biomedicine studies and the biotech industry. The engineering of synthetic circuitry in bacterial, yeast, and animal systems prompted considerable advances for the understanding and manipulation of genetic and metabolic networks; however, their implementation in the plant field lags behind. Here, we review theoretical-experimental approaches to the engineering of synthetic chemical- and light-regulated (optogenetic) switches for the targeted interrogation and control of cellular processes, including existing applications in the plant field. We highlight the strategies for the modular assembly of genetic parts into synthetic circuits of different complexity, ranging from Boolean logic gates and oscillatory devices up to semi- and fully synthetic open- and closed-loop molecular and cellular circuits. Finally, we explore potential applications of these approaches for the engineering of novel functionalities in plants, including understanding complex signaling networks, improving crop productivity, and the production of biopharmaceuticals.
Signaling processes are central to the organization and existence of any form of life. Exogenous and endogenous inputs are sensed and integrated by molecular networks in cells with feedback loops and Boolean logic decision making, resulting in a specific response (output). For this purpose, regulatory circuits are structured as a tightly and finely coordinated network of information with transfer and processing steps and chains, each individually fulfilling a specific task. These processes are in turn organized in time and space: within subcellular compartments (membranes, organelles, cytosol, and nuclei) and between cells and tissues. Signal mediators include proteins, nucleic acids, and small molecules (Lim, 2010). A key characteristic of biological regulatory networks is their modular architecture, in which building blocks are assembled in a combinatorial fashion. The constituent individual components perform a given distinct, particular function within the network, be it signals per se or switches (i.e. components that are able to detect an input signal and transform it into an output cue; Stein and Alexandrov, 2015).
Plants have evolved complex networks to integrate environmental, genetic (via spatial and temporal cues), developmental, and metabolic programs as well as the current physiological status. The output is a response tailored to adjust the cell welfare and function in the context of a multicellular organism (Trewavas, 2005; Sheen, 2010). These systems are constantly active, monitoring the ever-varying conditions and executing outputs following both open- and closed-loop programming principles for optimal responses. Recent advances in molecular biology, genetics, and systems biology-associated technologies have led to the identification of a huge number of signaling components, cascades, and regulatory mechanisms thereof. The field of plant signaling is growing rapidly, as is our knowledge of the complexity of these networks (Jaeger et al., 2013; Lavedrine et al., 2015). Most signaling pathways comprise many components and exhibit redundancy of function, extensive feedback control, and cross-interaction with other networks. The fine-tuning involves different types of posttranslational modifications, as exemplified by the complex mesh integrating light and hormone signaling, the circadian clock, and developmental and growth processes (Pokhilko et al., 2013; Fogelmark and Troein, 2014). In addition, there is a lack of quantitative molecular tools to interrogate and monitor the dynamics of these systems (Liu and Stewart, 2015; Samodelov and Zurbriggen, 2017). This not only hinders a comprehensive understanding of the function, regulation, and effects of signaling circuits but also the targeted manipulation of metabolic and signaling networks and, consequently, the introduction of novel functionalities into plants. In combination with modern analytical technologies, synthetic biology approaches represent the key to overcoming these limitations, and they are currently revolutionizing fundamental bacterial, yeast, and metazoan research as well as the biotechnology and biomedicine industries (Lu et al., 2009; Lienert et al., 2014).
Synthetic biology is a relatively new discipline bridging engineering with life sciences. It applies basic engineering principles for the modular, combinatorial assembly of biological parts into higher order complex signaling and metabolic structures. Key to the strategy is the implementation of mathematical modeling for the design and quantitative functional characterization of the molecular parts and for guiding the assembly, implementation, and optimization of the individual modules and networks (Ellis et al., 2009; Lim, 2010). Thus, inspired by nature, synthetic biology harnesses the modular architecture of biological systems. However, the goal is to develop novel molecular and cellular systems with desired properties and biological functionalities that are not present in nature. These properties range from gene switches and genetically encoded biosensors to fully synthetic autonomous molecular and cellular circuits and organelles as well as biohybrid smart materials and biopharmaceuticals (Brophy and Voigt, 2014; Lienert et al., 2014; Xie and Fussenegger, 2018). This field has already taken root in microbial systems as well as other higher eukaryotes. However, the generalized implementation of these approaches in the plant field lags behind.
This review is intended to serve as inspiration for plant scientists, raising interest in the field-changing potential of widely implementing synthetic biology principles. We will give an overview on the state of the technology and progress achieved with the application of synthetic biology strategies for studying, manipulating, and de novo engineering of signaling circuitry, with exemplary illustration of bacterial, yeast, and animal systems. The first implementations and future prospects in plant research will be highlighted, and the limitations and necessary technological advances for a straightforward implementation in plants will be discussed. The article is structured in three parts, following a hierarchy of molecular and realization complexity, starting off with molecular switches. Chemical-inducible devices will be introduced. In particular, the implementation of light as a trigger will be highlighted, describing the groundbreaking experimental advances enabled by optogenetics and its applications for the control of cellular processes. The concepts of orthogonality in the design of the molecular parts and the need for hand-in-hand work with theoreticians/mathematical modeling will be discussed. Further aspects include the functional combination of simple synthetic switches into molecular devices implemented in cells to perform decision-making processes, such as oscillators and molecular Boolean logic gates. Finally, we will focus on semi- or fully synthetic molecular signaling networks with open- and closed-loop control configurations and the transition into cellular devices with ad hoc functionalities for applications. For example, these systems will facilitate personalized nutrition, the production of biopharmaceuticals, and the obtainment of higher crop yields in an ecologically sustainable manner.
SYNTHETIC GENETIC SWITCHES
The rational combination of sensing and effector modules allows the wiring of inputs and outputs that are normally not functionally linked in nature, with the goal of performing novel functions. These functions range from the targeted control of a cellular process and the quantitative monitoring of a molecule to the induction of enzymatic activity or posttranslational modifications. The molecular mechanisms behind the signal integration and transfer mostly involve conformational changes. These allosteric modifications are induced by interactions between proteins, nucleic acids, and small molecules (e.g. protein/protein, small molecule/protein, and RNA/DNA; Stein and Alexandrov, 2015). Synthetic switches are engineered in a modular fashion, integrating natural and de novo-designed molecular parts. Unfortunately, switches often do not perform as expected when introduced into living systems. As in engineering, having a complete quantitative functional characterization of the modules and a supporting mathematical model contributes to straightforward and optimal implementation. A series of functional parameters of switches to be evaluated include dynamic range (ratio between maximal and basal activation), leakiness (basal activity in the absence of an inducing signal), kinetics, and reversibility of function. This is also critical when using switches as building blocks for the assembly of higher order circuits (see next section). Finally, the use of orthogonal components helps to maximize the insulation of the system, with the objective of achieving independent function and reducing unwanted effects on the endogenous networks, which are not targets of the synthetic regulation. Next, chemical- and light-inducible switches for the control of gene expression and other cellular processes will be discussed. Protein and RNA switches used for quantitative monitoring of molecules and processes (sensors) will not be discussed in this review; for a comprehensive description, see Okumoto et al. (2012) and Walia et al. (2018).
Gene Expression Control
Transcriptional Switches
The principle of autoregulation is a key architectural element in genetic or biochemical networks, shared by prokaryotic and eukaryotic cells (Freeman, 2000). Therefore, the synthesis of proteins is essentially influenced by the genetic program and cellular environment and underlies a tight regulation through gene switches. A gene switch can be considered as any natural or synthetically designed module controlling gene expression at the level of DNA, RNA, or protein (posttranslational modifications and stability; Xie and Fussenegger, 2018). Key building blocks of natural switches were first described by Jacob and Monod (1961) for the regulation of the lactose (lac) operon in Escherichia coli, which is regarded as the classic model for gene expression control. They characterized the promoter as the point of transcriptional initiation and identified controlling elements (repressors and inducers), which, upon binding with highly specific affinity to the upstream-located operator motif, quantitatively enhance or repress mRNA transcription. This binding is dependent on the presence of a metabolite that changes the conformation (allosteric regulation) of the regulator protein (Dickson et al., 1975).
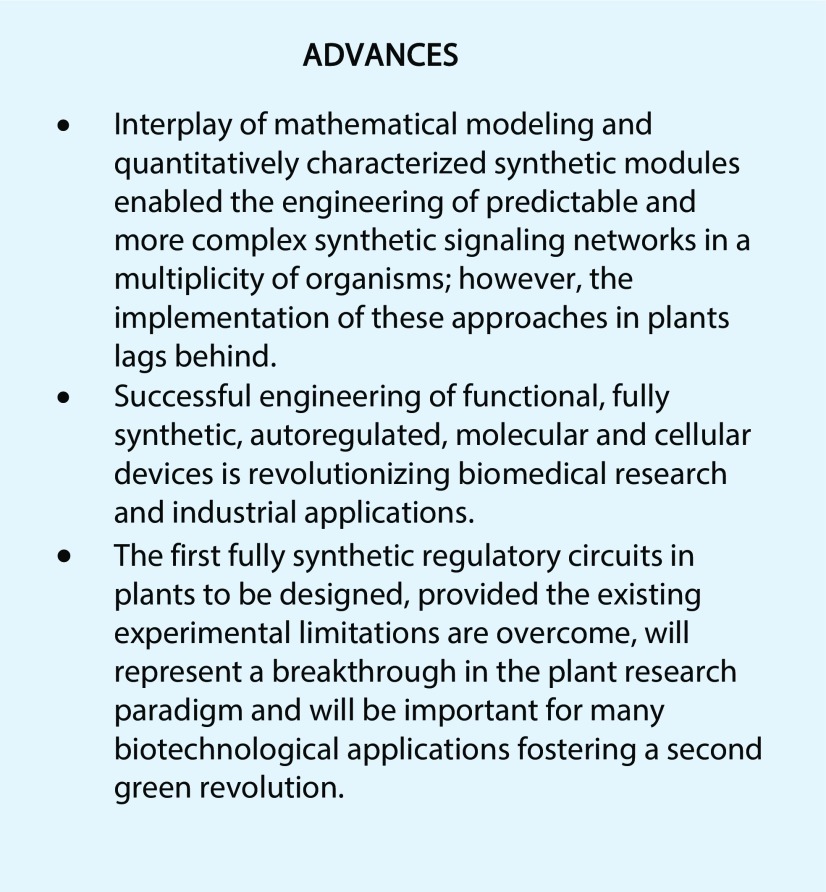
While prokaryotic gene expression circuits mostly utilize autoregulatory inhibition (negative feedback) to guarantee homeostasis, eukaryotic transcriptional regulation comprises more complex combinations of negative and positive regulators engaging in feedback loops and Boolean logic gate computing mechanisms (Savageau, 1974; Bateman, 1998; Thieffry et al., 1998; Becskei and Serrano, 2000; Freeman, 2000). A mechanistic and functional characterization of some of these simple prokaryotic regulatory elements (Beck et al., 1982; Berens et al., 1992) enabled the engineering of artificial, exogenously controlled systems of gene expression in prokaryotic and eukaryotic cells (Gardner et al., 2000; Ajo-Franklin et al., 2007). One of the first inducible gene switches is based on the tetracycline-regulated promoter of E. coli that controls the expression of the tetracycline-resistance-mediating tetA gene (Fig. 1A). In brief, a simple C-terminal fusion of the tetracycline repressor (TetR) to a transcriptional activation domain from the herpes simplex virus type 1 virion protein16 (VP16) converted the transcriptional repressor into a tetracycline-controlled transcriptional transactivator (tTA) in eukaryotic cells (Gossen and Bujard, 1992). In the absence of tetracycline, tTA binds to the cognate tet-operator region on the synthetic promoter construct, activating transcription from an adjacent minimal promoter sequence. Upon addition of tetracycline, tTA is removed from the promoter and gene expression is shut off (Fig. 1B). A reversed TetR was generated by random mutagenesis (Gossen et al., 1995), which, when fused to the VP16 domain, enables tetracycline-induced transcriptional activation (Fig. 1B). Alternatively, fusion of a transrepressor instead of a transactivator to TetR or modification of the synthetic promoter region enables other positive and negative regulation configurations (Kramer et al., 2004a). Following these simple molecular engineering principles, and modifications thereof, a vast set of chemically inducible gene switches were developed for use in yeast and animal cells sensitive to antibiotics, primary and secondary metabolites, and volatiles, among other substances (for review, see Hörner and Weber, 2012).
Figure 1.
Illustration of the natural bacterial tetracycline resistance mechanism and synthetic tetracycline-based gene expression systems. A, In the absence of tetracycline (tet), the tet repressor (TetR) is bound to its cognate tet operator (tetO) DNA-binding motif, repressing the expression of the tet resistance-mediating tetA gene. Upon increasing cellular levels of tet, tet binding induces a conformational change of TetR, leading to its dissociation from the operator sequence, and expression of tetA ensues. B, The tet-OFF system designed for use in mammalian cells is based on a synthetic switch comprising the natural TetR fused to the activating domain of VP16 of the herpes simplex virus and a synthetic promoter with a series of repeats of the tetO motif placed upstream of a minimal promoter (e.g. human cytomegalovirus minimal promoter). The system is constitutively active and is turned OFF in the presence of the antibiotic. Implementation of a reversed TetR mutant (rTetR) generates a tet-ON system: tet induces the binding of rTetR to the target sequence, which in turn induces gene expression (tet can be replaced by other antibiotics of the tetracycline family like doxycycline). Replacement of VP16 by a transrepressor such as KRAB inverts the effect of the switch (not depicted here). GOI, Gene of interest. (Adapted from Gossen et al., 1995.)
To achieve tight and predictable control over gene expression, a quantitative characterization and mathematical modeling of the regulator/promoter-switch is needed (for the implementation of mathematical modeling into synthetic circuitry approaches, refer to the detailed works of Ellis et al. [2009] and Lim [2010]). The optimization of key parameters such as strength and kinetics of expression, leakiness, etc., can be performed subsequently by reengineering the switch components. Usual approaches include the redesign of promoter regions: introduction of multiple repeats of binding sites, point mutations to alter affinity, protein engineering, and use of different transactivators/transrepressors (Ajo-Franklin et al., 2007). The incorporation of positive and/or negative feedback loop configurations (e.g. by placing the regulator under control of its own target synthetic promoter) enables a greater dynamic range of the dose-dependent response (Gossen et al., 1995; Becskei et al., 2001). Promoters can be engineered further by combining activation and repression of gene expression in a simultaneous manner, thereby facilitating a deeper insight into gene network regulation by increasing the possible regulation conditions. Studying unregulated, repressed, activated, or simultaneously repressed/activated gene expression helped develop a model for precise prediction of the behavior of genetic networks in vivo (Guido et al., 2006). Other examples include the implementation of several chemical-, hormone-, or CRISPR/Cas-inducible or repressible switches for the control of multiregulated systems, especially for pharmacological application in mammalian cells (Weber et al., 2002; Nielsen and Voigt, 2014). Broad implementation of these gene switches in cell culture and in vivo (mouse, rat, Drosophila spp., zebrafish, Caenorhabditis elegans) represented a paradigm change in the way metabolic and signaling networks can be studied and redesigned synthetically.
In plant systems, several chemically inducible switches have been developed for a temporal and quantitative regulation of expression (Table 1). For instance, these switches are triggered by IPTG (Wilde et al., 1992), antibiotics such as tetracycline (Gatz et al., 1992; Weinmann et al., 1994; Müller et al., 2014), macrolides and pristinamycin (Frey et al., 2001; Müller et al., 2014), copper (McKenzie et al., 1998), or ethanol (Caddick et al., 1998; Roslan et al., 2001). The most widely employed switch is a steroid-based system that allows precise temporal control over cellular processes in whole plants (Schena et al., 1991). Recently, gene switches comprising a Cas9-based repressor and regulatory modules of hormone signaling pathways (auxin, GA, and jasmonate) have been implemented in Arabidopsis (Arabidopsis thaliana; hormone activated Cas9-based repressor [HACRs]; Khakhar et al., 2018). The HACRs are sensitive to both exogenous hormone treatments and varying endogenous hormone levels, leading to degradation of the switch and thereby regulating target gene expression (the single guide RNA-Cas9 complex dictates the specificity). This tool can be applied to regulate hormone signaling or any other target of interest, thus allowing the manipulation of stress tolerance and yield in crop plants.
Table 1. Representative synthetic switches and regulatory circuits in plants.
AlcR, Promoter of the ALCR transcription factor; AlcA, alcohol dehydrogenase I from Aspergillus nidulans; XVE, chimeric transcription factor based on LexA-VP16-ER; OlexA, DNA-binding domain of the bacterial LexA repressor; pOp, chimeric promoter, comprising lac operators cloned upstream of a minimal cauliflower mosaic virus promoter; LhGR, transcription activator, a fusion between a high‐affinity DNA‐binding mutant of the lac repressor, lacI His-17, and the transcription‐activation domain II of GAL4 and the ligand‐binding domain of the rat glucocorticoid receptor; TraR, autoinducer-dependent transcriptional activator from Agrobacterium tumefaciens; OOHL, 3-oxooctanyl-l-homoserine lactone; lacO, lac operator; LacI, lac repressor; IPTG, isopropyl β-d-thiogalactopyranoside; ACE1, promoter of the copper-binding regulatory protein; rTetR, reversed tetracycline repressor; TetR, tetracycline repressor; tetO, tetracycline operator; GAL4, Gal-responsive transcription factor GAL4; UAS, upstream activation sequence; PiP, pristinamycin repressor protein; PIR, pristinamycin I-responsive element; E, macrolide repressor protein from E. coli; etr8, eight MphR(A) [macrolide 2′-phosphotransferase I]-binding operators; N1, 10 N1-TATA minimal promoter; NEV, three finger protein N1-ER-VP64; HACR, hormone-activated Cas9-based repressor; dCAS9, nuclease-dead Cas9; PIF6, phytochrome-interacting factor6; CarH, light-responsive transcription factor; CarO, CarH-binding site-containing operator; TIR1, TRANSPORT INHIBITOR RESPONSE1; Aux/IAA, auxin/indole-3-acetic acid protein; Trg, transmembrane signaling protein; PhoB, phosphate regulon transcriptional regulatory protein; PhoR, phosphate regulon sensor protein; VP64, four copies of the virion protein16 domain of the herpes simplex virus type 1; ABA, abscisic acid.
| Feature | System | Properties | References |
|---|---|---|---|
| Chemically inducible switches for gene expression | AlcR/AlcA | Ethanol inducible | Caddick et al. (1998) |
| Roslan et al. (2001) | |||
| Roberts et al. (2005) | |||
| XVE/OlexA | β-Estradiol inducible | Zuo et al. (2000) | |
| Curtis and Grossniklaus (2003) | |||
| Böhmdorfer et al. (2010) | |||
| pOp/LhGR | Dexamethasone inducible | Schena et al. (1991) | |
| Aoyama and Chua (1997) | |||
| TraR | OOHL inducible (quorum sensing system) | You et al. (2006) | |
| lac operator/LacI | IPTG inducible | Wilde et al. (1992) | |
| ACE1-based Cu-inducible promoter | Copper inducible | McKenzie et al. (1998) | |
| (r)TetR/tetO | Tetracycline inducible (rTet)/repressible (TetR) | Gatz et al. (1992) | |
| Weinmann et al. (1994) | |||
| Müller et al. (2014) | |||
| GAL4-UAS | Enhancer trap lines | Gardner et al. (2009) | |
| Johnson et al. (2005) | |||
| Laplaze et al. (2005) | |||
| PiP/PIR | Pristinamycin repressible | Frey et al. (2001) | |
| Müller et al. (2014) | |||
| E/etr8 | Macrolide regulated | Müller et al. (2014) | |
| 10xN1/NEV | 4-Hydroxytamoxifen inducible | Beerli et al. (2000) | |
| Cas-based gene expression | HACR | Phytohormone inducible | Khakhar et al. (2018) |
| dCAS9 | gRNA-mediated gene-specific induction | Piatek et al. (2015) | |
| Lowder et al. (2015) | |||
| Light-regulated gene expression | Phytochrome B/PIF6 | Red light induced/far-red light repressed | Müller et al. (2014) |
| Ochoa-Fernandez et al. (2016) | |||
| CarH/CarO | Green light repressed/dark induced | Chatelle et al. (2018) | |
| Synthetic riboswitch | Synthetic theophylline riboswitch in plastids | Theophylline inducible | Verhounig et al. (2010) |
| MicroRNA-based gene silencing | Artificial microRNA | Gene-specific silencing | Schwab et al. (2006) |
| Posttranslational degradation | N-terminal degradation signal (It degron) | Temperature-controlled protein degradation | Faden et al. (2016) |
| Optogenetic manipulation of endogenous signaling networks | Red light-controlled up- or down-regulation of TIR1 in combination with a ratiometric auxin sensor to monitor the manipulated signaling | Red light-controlled tuning of auxin signaling | Müller et al. (2014) |
| Synthetic ligand detection and signal relay | TgR/PhoR fusion phosphorylates PhoB-VP64 | Synthetic programmable ligand detection system | Antunes et al. (2011) |
| Synthetic ABA agonist | Cyanabactin: agonist of ABA IIIA receptors | Synthetic manipulation of transpiration and other physiological processes | Park et al. (2015) |
| Vaidya et al. (2017) |
However, chemical switches have limitations concerning defined spatiotemporal activation of the system due to abundance, administration, and diffusion of the inducer molecules as well as usual toxicity effects. Recently, light-controlled genetically encoded molecular devices have been engineered and implemented in living cells to control cellular processes, giving rise to the nascent field of optogenetics (Box 1). These devices overcome the inherent restrictions of chemically regulated switches. Light-regulated switches comprise bacterial and plant photoreceptors, such as UV-B RESISTANCE8, phototropin1/EL222/CRYPTOCHROME2, CarH, PYHTOCHROME B/A, and the bacterial phytochrome BphP1, among others (for a comprehensive list, see Kolar et al., 2018). Upon absorption of light, they undergo a conformational change leading to homo/hetero-association/dissociation (Kolar and Weber, 2017). This light-dependent protein interaction relays a signal to an output module that then fulfills a cellular function. In the last decade, a multitude of optogenetic gene switches regulated by UV-B, blue, green, red, and far-red/near infrared light have been engineered and implemented for the noninvasive control of gene expression with a precise temporal and spatial resolution in prokaryotic and eukaryotic systems (Zhang and Cui, 2015; Fig. 2).
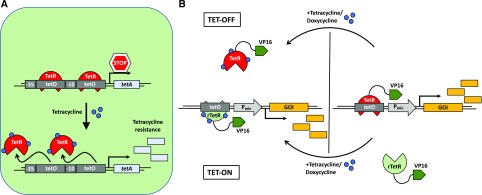
Figure 2.
Optogenetic switches. Molecular principles of light-induced signaling and optogenetic tools are illustrated. A, Natural red light-inducible signaling mediated by the plant photoreceptor phytochrome B (phyB) and optogenetic tools developed based on it. A1, The red/far-red light-perceiving photoreceptor phyB remains in its inactive Pr conformation in the dark. Upon absorption of a red light photon, the photoreceptor undergoes a conformational change, converting to its active Pfr conformation. The active form can interact with several transcription factors like the bHLH transcription factors of the PHYTOCHROME INTERACTING FACTOR (PIF) family. This interaction triggers light-signaling responses. In contrast, illumination with far-red light reconverts phyB to its inactive Pr form, abolishing the interaction with PIFs (Rockwell and Lagarias, 2006). Several optogenetic approaches make use of the red light/far-red light switchable interaction of phyB and PIFs. A2, Selective activation of intracellular signaling pathways with light. Red light illumination induces the recruitment of the cytoplasmic fusion protein consisting of a PIF, C-terminally fused to the fluorescent protein YFP and the catalytic domain of the SOS protein (SOScat), to the membrane-bound, RFP-tagged phyB. When recruited to the membrane, SOScat is capable of activating the Ras-signaling cascade and inducing nuclear transport of BFP-Erk and subsequent Erk pathway signaling. (Adapted from Toettcher et al., 2013.) A3, Construction of a phyB-PIF-based, red light-inducible split-transcription factor system. A truncated PIF6 was N-terminally fused to the tetracycline repressor (TetR), and the synthetic protein is bound to the tetracycline operator motif tetO of a synthetic reporter construct (as in Fig. 1). In the absence of light or under far-red light illumination (740 nm), there is no expression from the minimal promoter, PCMVmin. Upon illumination with red light, phyB, C-terminally fused to the VP16 transactivation domain, interacts with the PIF. The spatial proximity of the transactivator recruits the transcriptional machinery to the minimal promoter. Only in this condition is the expression of the secreted human alkaline phosphatase (SEAP) reporter gene activated. (Adapted from Müller et al., 2013a.) An adaptation of this system was engineered in Arabidopsis and tobacco (Nicotiana tabacum) cells and the moss Physcomitrella patens (Müller et al., 2014; Ochoa-Fernandez et al., 2016). A4, Reversible red light-inducible nuclear transport of phyB fusion proteins. phyB was C-terminally fused to the fluorescent protein mCherry and a nuclear export sequence (NES), while PIF3, containing an intrinsic nuclear localization sequence (NLS), was C-terminally fused to enhanced GFP (EGFP). Upon illumination with red light, the nucleocytoplasmic shuttling PIF induces nuclear transport of phyB, while far-red radiation reversed the translocation of the photoreceptor-mediated by the NES. (Adapted from Beyer et al., 2015.) B, Natural blue light-induced signal transduction mediated by the plant photoreceptor phototropin1 and the light-sensitive bacterial transcription factor EL222. A synthetic approach based on blue light-triggered conformational change of EL222 and the LOV2 domain for the dual-controlled optogenetic down-regulation of proteins in animal cells was used. B1, EL222 is a light-sensitive transcription factor from the gram-negative bacterium Erythrobacter litoralis. It contains a blue light-sensitive LOV domain and a helix-turn-helix (HTH)-DNA-binding domain. In the dark, the HTH domain is docked to the LOV core. Upon illumination with blue light, the interaction of LOV and the HTH domain is disrupted, enabling homodimerization of the protein via the HTH and subsequent binding to the C120-DNA motif (Nash et al., 2011). B2, Schematic illustration of light-induced signal transduction via the blue light plant photoreceptor phototropin1. In the dark, the kinase domain is bound to the LOV domain, inhibiting its phosphorylation activity. Under blue light, the kinase domain is released, inducing protein phosphorylation and downstream signal transduction. (Adapted from Kimura et al., 2006.) B3, The dual optogenetic system for targeted degradation and repression of expression of a protein of interest (POI) consists of a synthetic reporter module comprising the PSV40 promoter, for constitutive expression of a POI fused to the B-LID (Bonger et al., 2014) module, and the C1205-DNA-binding motif of the EL222 protein. EL222 is fused to the transrepressor KRAB. In the dark, the degron (peptide RRRG) is docked to the LOV domain of the B-LID, and KRAB-EL222 is not able to bind to the C120 motif on the reporter plasmid. In this case, the POI accumulates. Upon illumination with blue light, the degron is exposed, triggering proteasomal degradation of the POI-B-LID fusion protein. Simultaneously, KRAB-EL222 dimerizes binding to the C120 motif, repressing transcription of the POI-B-LID. (Adapted from Baaske et al., 2018.)
Contrary to most nonautotrophic organisms, the life cycle of plants requires exposure to sunlight, which might lead to nonintentional activation of the optogenetic systems. Therefore, the simple transfer of optogenetic tools developed in other organisms is challenging. While long-term experiments in dark conditions are harmful, exposure to a specific wavelength of light may interfere with the natural light-sensitive signaling and photosynthetic circuitry of the plant through their photoreceptors or light-sensitive pigments. These natural light-absorbing moieties might in turn interfere with the inducing light and the high autofluorescence of plants, posing limitations to microscopy analysis. In addition, compared with the genetic engineering of simpler organisms, generating stable transformed plants expressing the synthetic components of the switches is a lengthy process that slows down the implementation and characterization processes.
Despite these technical and experimental constraints, the first optogenetic tools have already been successfully implemented in plants (Table 1). These include a phytochrome-based red light-inducible and a CarH-based green light-regulated expression system (Müller et al., 2014; Ochoa-Fernandez et al., 2016; Chatelle et al., 2018). The former is activated by red light and inactivated by far-red light. Simple supplementation of ambient illumination in greenhouses with low intensities of far-red light keeps the system repressed. Irradiation with red light leads to quantitatively controlled activation of gene expression (Müller et al., 2014; Chatelle et al., 2018). The second strategy comprises the engineering of a green light-inducible bacterial photoreceptor, CarH. Use of green light as a stimulus minimizes the interference with endogenous plant photoreceptors, as this region of the sunlight spectrum normally does not produce physiologically active signaling responses of relevance (Chatelle et al., 2018).
Translational and Posttranslational Switches
While transcriptional gene switches currently play a major role in customized gene expression and are used for a broad range of applications, synthetic RNA-based switches constitute a complementary approach for controlling gene expression on the translational level. The most prominent components of RNA-based tools include RNA interference (RNAi; Fire et al., 1998), microRNAs (Lagos-Quintana, 2001), aptamers, and ribozymes. While RNAi, microRNAs, and ribozymes lead to cleavage or splicing of the target mRNA (Fire et al., 1998; Warashina et al., 2000; Lagos-Quintana, 2001), aptamers bind to specific targets like metal ions, small molecules, DNA, or proteins (Xiao et al., 2008). Aptamers are structured noncoding RNAs, naturally found in riboswitches that interfere with the accessibility of the ribosomes to the mRNA, affecting translational control (Breaker, 2012; Ausländer and Fussenegger, 2017). Using the in vitro selection method SELEX (for systematic evolution of ligands by exponential enrichment; Ellington and Szostak, 1990), many aptamers for new targets have been developed, such as the synthetic tetracycline-binding aptamer (Hanson et al., 2005; Xiao et al., 2008). By integrating protein-binding aptamers, the control of translational regulation via repression or alternative splicing can be achieved (Culler et al., 2010; Endo et al., 2013). In addition, fusion of the aptamer to translational repressors or enhancers permits the up- or down-regulation of the translation rate of the target protein (Pillai et al., 2004; Van Etten et al., 2012; Paek et al., 2015). Compared with transcriptional switches, translational switches can control endogenous genes without any alteration of the genomic sequence. They are relatively small in size and therefore are amenable for use in combination with transcriptional switches when the size and number of cassettes imposes an experimental limitation (Ausländer and Fussenegger, 2017).
In plants, specific RNA-based gene silencing, using artificial antisense mRNAs or microRNAs under the control of tissue-specific or inducible promoters, has been widely used for more than 20 years. However, these approaches usually suffer from off-target effects and provide limited exogenous and quantitative control and reduced efficiency (Schwab et al., 2006). Other examples for the translational control of gene expression in plants are limited to applications in plastids (Verhounig et al., 2010). Recently, Faden et al. (2016) reported a posttranscriptional switch for the in planta down-regulation of protein levels based on a temperature-controlled N-terminal degradation signal. Similar to other techniques already implemented in simpler, unicellular organisms, the transfer of the system to multicellular organisms, like plants, strongly depended on the adaptation to the corresponding physiological conditions. To test the functionality of the system for reversible protein accumulation, trichome formation was manipulated after shifting the plants from a permissive to a restrictive temperature (29°C). This led to the degradation of the protein of interest, TRANSPARENT TESTA GLABRA1, thus affecting the spatiotemporal development of trichomes (Table 1).
Switches Regulating Cellular Processes
Besides transcriptional and translational switches, a plethora of chemical- and light-regulated systems have been developed for the targeted regulation of a multiplicity of cellular processes ranging from the activation/inactivation of signaling cascades (receptors, kinases, transcription factors, etc.) and membrane trafficking to the controlled movement of organelles from one pole of the cell to the other (for review, see Hörner and Weber, 2012; Kolar and Weber, 2017). Selected examples include the utilization of optogenetic tools for (1) the control of the subcellular localization of proteins (e.g. red light; Beyer et al., 2015; Fig. 2A) and blue light-induced (Niopek et al., 2014, 2016) nuclear import and export of transcriptional effectors; and (2) the light-mediated degradation/depletion of proteins (Bonger et al., 2014; Baaske et al., 2018; Fig. 2B). A comprehensive list of approaches is reviewed elsewhere (Hörner and Weber, 2012; Kolar and Weber, 2017).
SYNTHETIC GENETIC CIRCUITS
Genetic circuits combine a series of synthetic switches into networks that can perceive a signal (exogenous or endogenous, natural or synthetic), process the information, and generate an output, normally triggering gene expression (e.g. induction of a given phenotype or change in cellular morphology) and expression of a reporter to monitor a process or activation of a metabolic pathway (Lim, 2010; Xie and Fussenegger, 2018). Simple circuits perform basic functionalities and integrate few signals. Next, we will discuss toggle switches, synthetic oscillators, and Boolean logic gates, which are built up from simple combinations of a reduced number of modules. Then we will review more complex arrays of switches integrated into cell-cell communication systems, open- and closed-loop circuit control, and synthetic cellular devices and their applicability.
Simple Circuits
Since therapeutic applications are one of the driving forces for the development of functional, robust, and complex genetic circuits, many recent technical breakthroughs have been made in mammalian cell systems. First approaches included the transfer and optimization of basic synthetic circuits, previously engineered in lower organisms. An illustrative example is a simple negative feedback circuit in yeast based on the combination of two tetracycline-inducible modules, controlling the expression of EGFP and the TetR repressor (Nevozhay et al., 2009). This loop enabled a tightly controlled, dose-dependent activation of gene expression in mammalian cells. Expression of both EGFP and TetR is regulated by the rate of influx of the inducer but subsequently restricted by the increasing level of TetR protein (Nevozhay et al., 2013).
Toggle Switches
The first combined synthetic gene switches date back to the early 2000s with the design of bistable transcriptional repression toggle switches in bacteria and mammalian cells (Ajo-Franklin et al., 2007). Here, mutual inhibition of two independent chemical- and temperature-controlled (Gardner et al., 2000) or antibiotic-inducible (Kramer et al., 2004b) promoters, each controlling the expression of the counterpart’s repressor, generates two equilibrium states of induction, switchable by the respective transient induction.
Plants also employ natural toggle switches for the control of endogenous processes, such as the CLAVATA pathway for stem cell fate. In line with this, the implementation of synthetic toggle switches in plants could open new perspectives for the development of, for instance, a programmable path of stem cell differentiation (Medford and Prasad, 2016) or trichome development. However, the intrinsic complexity of plant signaling networks restricts the straightforward transferability of already existing synthetic systems into plants. Plants integrate a wide range of biotic and abiotic external cues like light and temperature with genetic programs in an intertwined or redundant manner. This poses experimental and theoretical constraints (resources, time, lack of thorough knowledge of regulatory mechanisms, limited genetic tools, etc.). Therefore, exhaustive design and implementation phases will be needed for engineering all the synthetic circuits discussed in this article.
Oscillators
Autonomous and self-sustained oscillating gene expression patterns, like the circadian clock or the cell cycle, are crucial to sustain pulsatile cellular activities; therefore, there is much interest in understanding their regulation and function (for review, see Schibler and Sassone-Corsi, 2002; Fig. 3A). By designing and implementing synthetic oscillators, key insights on the mechanistic principles of cellular processes can be obtained, and novel functionalities could be engineered, as described below.
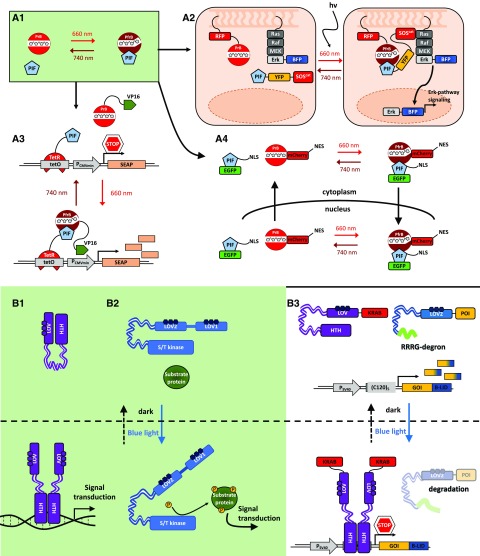
Figure 3.
Molecular principle of a natural and synthetic oscillator. A, Simplified molecular model of the circadian clock in Arabidopsis (natural oscillator). The core oscillator feedback loop consists of TOC1, CCA1, and LHY. In this core oscillator, LHY and CCA1 repress the transcription of TOC1; TOC1 in turn is a positive regulator of CCA1 and LHY. In a second loop, LHY and CCA1 are also positive regulators of three TOC1 paralogs (PRR5, PRR7, and PRR9), which in turn are negative regulators of CCA1 and LHY. In a third loop, CCA1 and LHY positively regulate GI, ELF3, ELF4, and LUX; these in turn regulate CCA1 and LHY. The circadian oscillator of Arabidopsis is illustrated here in a simplified form; for clarity, several other components involved were not included. (Adapted from McClung, 2006.) B, Scheme of a synthetic oscillator engineered by Stricker et al. (2008). This synthetic oscillator comprises positive and negative feedback loops. The araC, lacI, and yemGFP (as a readout) genes are all under the control of the hybrid synthetic promoter Plac/ara-1, comprising the activation operator site from the araBAD promoter and the repression operator site from the lacZYA promoter (Lutz and Bujard, 1997). In the presence of arabinose, the AraC protein activates the hybrid promoter and, thus, the gene expression of araC, lacI, and yemGFP, which results in two feedback loops: a positive feedback loop mediated by the produced AraC and the resulting activation of the hybrid promoter, and a negative feedback loop due to the production of the LacI protein. In the absence of IPTG, LacI negatively regulates the expression of all three genes under the control of the hybrid promoter. Both engineered feedback loops together constitute the synthetic oscillator. (Adapted from Stricker et al., 2008.)
After the discovery of the first gene regulation model (Jacob and Monod, 1961), theoreticians started developing mathematical models on genetic oscillatory networks, and ideas for synthetic circuits were proposed. The first prototypical oscillator, termed the Goodwin oscillator, utilizes a single protein that inhibits its own transcription; namely, it can be seen as a closed negative feedback loop (Goodwin, 1963, 1965). Several decades later, the advances in genetics and molecular and cell biology allowed engineers to implement this and other oscillators in living cell systems (Elowitz and Leibler, 2000; Fung et al., 2005; Stricker et al., 2008; Danino et al., 2010; Ryback et al., 2013). The first of these genetic circuits implemented in E. coli was a synthetic oscillatory network of transcriptional regulators, known as the repressilator (Elowitz and Leibler, 2000). A repressilator is defined as a subset of genes that can repress their successor in the cycle; thus, it can be seen as an extension of the one-gene Goodwin oscillator (Müller et al., 2006; Purcell et al., 2010). The Elowitz synthetic repressilator consists of a cyclic negative feedback loop composed of three repressor proteins, which are not part of any natural biological clock/oscillator, namely, LacI (E. coli), TetR (Tn10 transposon), and cI (λ phage), and their corresponding cognate promoters. However, it suffered from noisy behavior, with only 40% of the E. coli cells showing oscillations (Elowitz and Leibler, 2000). Theoretical studies revealed that by implementing a positive feedback loop, the robustness of the oscillations and the tunability of the amplitude and period could be improved (Hasty et al., 2002; Atkinson et al., 2003; Stricker et al., 2008; Purcell et al., 2010; Tomazou et al., 2018). Later, a dual-feedback oscillator developed by Stricker et al. (2008) achieved faster oscillatory periods, 99% oscillating cells, and decoupling from the cell cycle. The period was tuned by either IPTG, arabinose, or temperature (Fig. 3B). In most of these approaches, mathematical model-assisted design was essential for identifying the experimental parameters and molecular components (relative amounts thereof) used to tune the oscillations.
Autonomous, self-sustained, and tunable oscillatory behavior was also achieved in mammalian cells with an amplified negative feedback oscillatory mechanism (Tigges et al., 2009). The oscillator is based on an autoregulated sense-antisense transcription control circuit in the negative feedback loop leading to a delay in the repressive effect (Tigges et al., 2009; Purcell et al., 2010). An alternative approach applied in mammalian cells involved the combination of both natural and synthetic elements to create oscillatory behavior by manipulating amplitude, damping, and frequency in an independent fashion. For this purpose, the endogenous transcription factor p53, which is activated in response to cellular stress, and its negative regulator Mdm2 were utilized (Toettcher et al., 2010). This simple core negative feedback loop served as an example to define and modulate the dynamics of naturally occurring oscillatory systems in a controlled fashion. Considerable progress has been made recently to link different kinds of genetic circuits to functional synthetic self-regulated networks. This is necessary for integrating synthetic control into endogenous signaling networks, for instance, the Elowitz repressilator coupled to a modified quorum-sensing circuit of Vibrio fischeri and A. tumefaciens (Fernández-Niño et al., 2017).
Despite almost two decades of in vivo experiments and associated theoretical background on oscillators, there are still no oscillators implemented in plants. This represents a big experimental challenge. As discussed above, a major obstacle for the implementation of synthetic oscillatory networks in multicellular organisms like plants is the existence of a multiplicity of internal or external parameters, regulating metabolic and signaling pathways. A first attempt at this would be the engineering of hybrid oscillators, employing a similar approach to the one introduced by Toettcher et al. (2010). The introduction of synthetic orthogonal modules to achieve tight control over oscillatory parameters of an endogenous pathway minimizing cross talk could contribute to a broader understanding of oscillatory behavior in plant signaling and metabolic networks. In the future, fully synthetic systems could be implemented to bypass endogenous oscillators. A potential application of this would be the decoupling of endogenous metabolic pathways from the circadian clock to allow, for example, a prolonged bioproductive/anabolic daily phase, thereby increasing crop yield.
Boolean Logic Gates
Boolean logic gates utilize Boolean algebra to convert multiple input signals into truth values, meaning a true or false answer (1 or 0). In a simple way, cells use this mechanism for a plethora of decision-making processes (e.g. promoters integrate the information encoded in the combination of positive and negative transcriptional regulators bound at any given point in time, translating it into an output signal [gene expression]; Fig. 4). Following these principles, synthetic genetic circuits have been designed and successfully implemented in prokaryotes (Tamsir et al., 2011; Moon et al., 2012), yeast (Gander et al., 2017), and mammalian cells (Xie et al., 2011; Ausländer et al., 2012; Lebar et al., 2014) controlling various biological functions. They can integrate multiple molecular input signals following a set of algorithms and generate a response only if strictly defined conditions are met (Xie and Fussenegger, 2018). For instance, an OR gate only generates an output when either input signal A or B is present, whereas both input signals have to concur for an AND gate to be true. More complex logic gates could be built in a combinatorial fashion out of these simple ones (Xie and Fussenegger, 2018). Different transcriptional regulators were used to meet these demands, including promoters functioning as input and output (Tamsir et al., 2011; Moon et al., 2012), RNAi (Xie et al., 2011), and TALE repressor- (Gaber et al., 2014) and dCas9-based switches in bacteria (Nielsen and Voigt, 2014), yeast (Gander et al., 2017), and mammalian cells (Gao et al., 2016).
Figure 4.
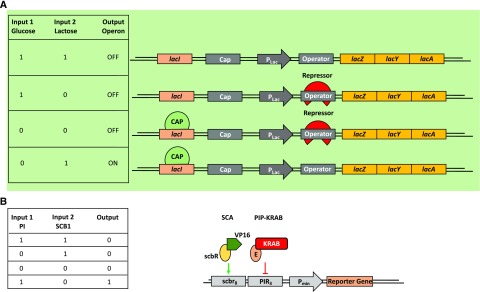
Natural and synthetically built AND NOT (NOT IF) Boolean logic gates. An AND NOT gate generates an output when only one specific single input signal is present, but not when there is no input signal, nor a second input, nor both signals. A, Truth table and scheme of the regulatory region of the Lac operon as an AND NOT (NOT IF) gate. This AND NOT gate only generates an output when lactose is the only single input available. If Glc and lactose are available in the cell, the lac operon is OFF because the catabolite activator protein, CAP, is not bound. The same is true when Glc, but no lactose, is available. In this case, the lac repressor is bound. In the case when there is neither Glc nor lactose, the lac operon is OFF because even though CAP is bound, the lac repressor prevents transcriptional initiation. Only when there is lactose, but no Glc, available is the lac operon ON. In the absence of Glc, CAP can bind, and because of the availability of lactose, the lac repressor is not bound. Both actions are necessary for transcriptional initiation of the lac operon. (Adapted from Phillips et al., 2009.) B, An example of an AND NOT (NOT IF) gate in synthetic biology. In this synthetic system, the transactivator SCA (transactivator of the streptogramin-responsive gene regulation system) and the transrepressor PIP-KRAB are constitutively expressed along with a reporter plasmid containing a chimeric SCA- and PIP-specific promoter. The absence of SCB1 [racemic 2-(1V-hydroxy-6-methylheptyl)-3-(hydroxymethyl)butanolide] enables the binding of the transactivator SCA to its corresponding promoter region. The presence of the transrepressor pristinamycin (PI) in turn prevents the binding of PIP-KRAB to its promoter. Thus, this engineered AND NOT gate generates an output only in the presence of pristinamycin and the absence of SCB1. (Adapted from Kramer et al., 2004a.)
An illustration of such a circuit using chemically controlled transcription factors was depicted in the work of Gao et al. (2016). An efficient gene activation and repression system was designed by combining plant hormone signaling components with Sp-dCas9, which enabled the manipulation of multiple gene targets in an orthogonal mammalian cell setup. To achieve this, ABA and GA phytohormone signaling components that heterodimerize in the presence of the individual hormones (PYRABACTIN RESISTANCE1-LIKE [PYL] with ABA INSENSITIVE [ABI] for ABA and GA INSENSITIVE DWARF1 [GID1] with GIBBERELLIC ACID INSENSITIVE [GAI] for GA) were fused to either a transcriptional activator (VPR) or repressor (KRAB) or to Sp-dCas9. When the corresponding hormones are added, GID1-VPR/-KRAB and GAI-Sp-dCas9 (or PYL1-VPR/-KRAB and ABI-Sp-dCas9, respectively) heterodimerize, thereby activating or repressing gene expression from a target synthetic promoter. These switches perform very well, are robust, and show almost no leakiness. Based on these characteristics, both systems were customized and combined to construct AND, OR, NAND, and NOR Boolean logic gates. A NOT IF gate was successfully built in which expression of a gene was possible only in the presence of one inducer (e.g. ABA) while it was OFF in the presence of the second one (e.g. GA; Gao et al., 2016). This approach therefore utilized phytohormone signaling components to control multiple transcriptional outputs in an orthogonal system, namely, mammalian cells. Despite its potential applicability, to our knowledge, there has not been any synthetic Boolean logic gate implemented in plants yet.
Higher Order Genetic Circuits
The characteristics of the different levels of genetic circuits are summarized in Box 2. More complex synthetic devices connecting multiple layers of signal processing, including detection of the inducer, signal transduction, and precise (nuclear) activation of the defined output, have been implemented in prokaryotic and eukaryotic cell systems. Most of these circuits partially rely on endogenous elements, utilized for a desired purpose, in combination with the integrated synthetic, orthogonal components. Here, we describe cell-cell communication systems and illustrate differential characteristics and applicability, currently in biomedicine, of open- and closed-loop circuit control configurations and prosthetic synthetic circuits (Box 2).
Cell-Cell Communication Systems
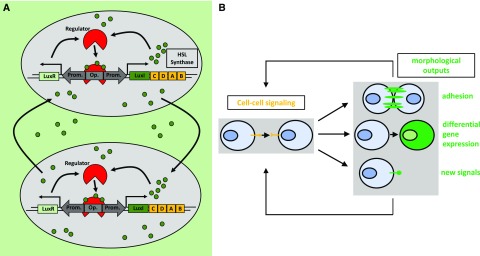
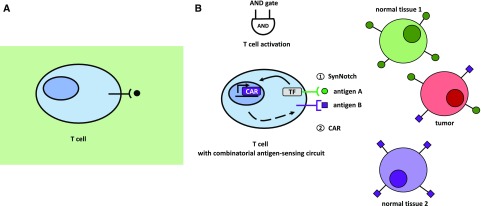
Unicellular and multicellular organisms rely on cell-cell communication mechanisms to regulate crucial life decisions (e.g. growth, development, organ identity, and metabolism/nutrition, among a wide range of processes). Bacteria, for instance, employ quorum sensing to assess the density of cells in their surroundings (Fig. 5A). Depending on the population density, genes responsible for key processes such as biofilm formation are up- or down-regulated (Fuqua et al., 1994; Abisado et al., 2018). Multicellular organisms coordinate processes such as tissue development or immune cell responses employing cell-cell communication networks (Thurley et al., 2018). Different signaling molecules are used for this purpose in unicellular and multicellular organisms, including metabolites, small RNAs, peptides, and proteins. The synthetic reconstruction or de novo engineering of these communication processes can contribute to experimental strategies to both understand these processes and develop biotechnological applications (Prindle et al., 2011). In tissue engineering approaches, tight control and manipulation of cell-cell communication is needed for the establishment of edges between different populations of cells, as achieved by Kolar et al. (2015). Targeted spatiotemporally resolved induction of cell death was engineered by using bacterial quorum sensing-regulated systems (You et al., 2004). Finally, cell adhesion through cell-cell communication was achieved by linking the synthetic notch receptor system to the expression of specific cadherin molecules and new synthetic Notch (synNotch) ligands (Toda et al., 2018; Fig. 5B). Importantly, the synNotch receptor mechanism is also utilized in potentially therapeutic engineered T-cells, which can detect given combinations of antigens (for details, see Fig. 6) instead of only one antigen (Roybal et al., 2016). These engineered combinatorial T-cells represent a breakthrough in the treatment of cancer.
Figure 5.
Cell-cell communication in bacteria and synthetic cell-cell communication networks. A, Simplified illustration of the natural homoserine lactone (HSL) quorum-sensing network in V. fischeri. Quorum sensing describes the ability of bacteria to assess the cell density of a population by sensing chemical signals that are produced by surrounding cells (Davis et al., 2015). HSLs, in the case of V. fischeri AHL, bind to the LuxR protein. LuxR then binds to its cognate operator, inducing the transcription of LuxI, which catalyzes the synthesis of AHL. AHL is able to diffuse out of the cell, accumulating in the external milieu and entering surrounding cells, thus activating the circuit in those cells. B, Engineered cell-cell communication networks in mammalian cells. Engineered cell-cell signaling via two synNotch ligand-receptor pairs was used to manipulate cell adhesion, differentiation, and the production of new cell-cell signals (Toda et al., 2018). Upon binding of the ligand to the synNotch receptor, an orthogonal transcription factor is cleaved from the cytoplasmic tail of the receptor, migrates to the nucleus, and then drives gene expression of the output proteins. These genes include fluorescent proteins as cellular markers for differentiation, several cadherins as morphological outputs, and two synNotch ligand-receptor pairs as input signals. In this way, the outputs are propagated to the next generation. (Adapted from Toda et al., 2018.)
Figure 6.
Natural and engineered combinatorial T-cells. A, Natural T-cell with its T-cell receptor, targeting only single antigens. This single-antigen recognition without any further control machinery can lead to off-target tissue damage. B, An engineered synthetic T-cell with new types of receptors specific for detecting given combinations of antigens. Upon binding of antigen A to the synNotch receptor, an orthogonal transcription factor is cleaved from the cytoplasmic tail of the receptor, which in turn activates CAR transcription. If a second antigen, antigen B, is recognized by the newly synthesized CAR receptor, the T-cell is activated. (Adapted from Roybal et al., 2016; Roybal and Lim, 2017.)
In plants, cell-cell communication also plays an important role. Key regulators such as phytohormones not only control almost every aspect of plant life, like coordinating responses between tissues and organs, but also mediate interactions with symbiotic microorganisms. An example is the phytohormone strigolactone, which can act both as an endogenous phytohormone and as an exogenous signal molecule in the rhizosphere (for review, see Morffy et al., 2016). As an exogenous signal, it recruits arbuscular mycorrhizal fungi to the root to provide the plant with nutrients (i.e. phosphate) under nutrient-limiting conditions (Akiyama et al., 2005). However, strigolactone also mediates the recognition of host roots by parasitic weeds, leading to severe yield losses (Parker, 2009). Inspired by these natural mechanisms, semi- or fully synthetic networks could be engineered to exploit novel useful symbiotic interactions under abiotic and biotic stress or to develop orthogonal signaling networks among organs. Therefore, the manipulation on command of the information flow can be used in strategies to improve crop productivity. It can also be used to abolish or reprogram detrimental or beneficial interactions between microorganisms and plants.
Open- Versus Closed-Loop Circuit Control, and Prosthetic Network Devices
Two exemplary realizations of semi-hybrid open-loop control strategies are optogenetic and radio wave-inducible devices for the in vivo regulation of blood Glc levels in mice. Both devices have been developed by integrating a synthetic input module with the native Ca2+-inducible NFAT-signaling pathway, activating the expression of genes involved in several developmental processes and immune responses (Crabtree and Olson, 2002; Crabtree and Schreiber, 2009). The optogenetic approach uses blue light to activate melanopsin and triggers a signaling cascade to ultimately induce a Ca2+ influx (Ye et al., 2011). The second circuit utilizes an engineered temperature-sensitive Ca2+ channel. This channel is bound by antibodies coated with ferrous oxide nanoparticles, which are heated with radio waves to trigger channel opening, leading to subsequent Ca2+ influx (Stanley et al., 2012).
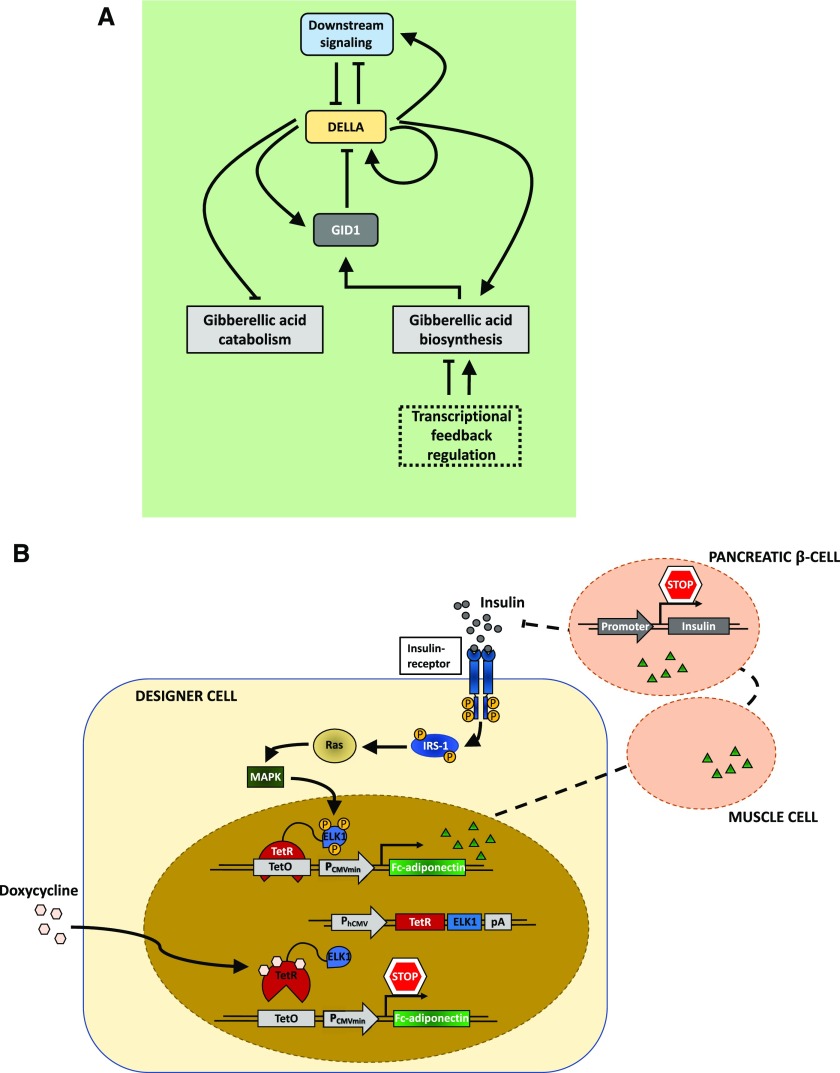
Smole et al. (2017) reported an exemplary case of a fully synthetic network that can sense an inflammatory signal in mice and produce a response to suppress this signal (Fig. 7). They engineered a synthetic device consisting of a sensor module that, upon activation by inflammation signals, triggers the expression of a transcriptional activator, GAL4-VP16. The fusion protein not only acts as an inducer of expression of anti-inflammatory proteins by the output module but also triggers the positive feedback loop of an amplifying module, leading to enhanced levels of GAL4-VP16. A fourth module constitutively expresses GAL4 lacking the transactivation domain, competing with the GAL4-VP16 for restricting the level of activation of the system, therefore acting as a thresholder device. Due to its autonomous activation by inflammatory signals, the activation of the circuit is independent of external induction. Furthermore, the system includes signal enhancement, while leakage is minimized by the thresholding module. Nevertheless, it still needs external inhibition for resetting the system to the OFF state due to the self-activating positive feedback characteristics and therefore is not strictly a closed-loop system. Ye et al. (2017) accomplished the construction of a closed-loop, prosthetic network for the self-adjusting regulation of the insulin level in vivo, consisting of an implant of encapsulated engineered HEK cells (Fig. 8). Here, perception of insulin by the cell via its native insulin receptor leads to phosphorylation of the insulin receptor substrate 1 protein, triggering a signaling cascade that induces nuclear transport of a MAPK. In the nucleus, the MAPK phosphorylates the ELK1 domain of the synthetic fusion protein TetR-ELK1, initiating the transcriptional activity of a target gene, otherwise tightly disrupted in the absence of insulin or external supplementation of doxycycline. Programming the circuit for the production of adiponectin, a therapeutic protein involved in regulating insulin homeostasis, turns the network into a closed, self-regulating loop, increasing insulin sensitivity in different tissues. The increased sensitivity subsequently leads to reduced insulin production by pancreatic β-cells. Fulfilling a function that is missing in the cellular genetic network, synthetic regulatory circuits in mammalian systems can overcome the constraints of endogenous cellular processes. This illustrates the potential of synthetic biology for developing functional therapeutic devices and tailor-made medicine. Such complexity has not been reached yet in synthetic circuitry in plants; however, the first synthetic networks have already started to be implemented in plants, as described below.
Figure 7.
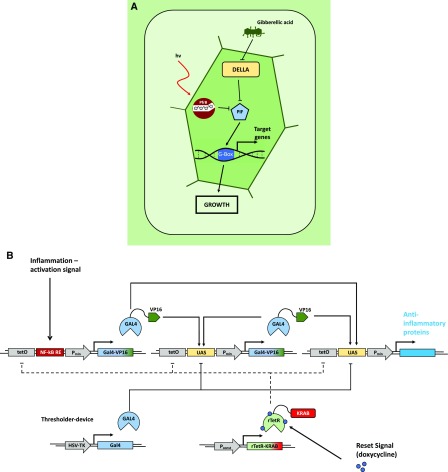
Natural and engineered open-loop regulatory circuits. A, GA3-induced degradation of DELLA proteins suppresses the repression of PHYTOCHROME INTERACING FACTORs (PIFs). The PIFs subsequently bind to G-box cis regulatory elements in the promotors of response genes, promoting growth responses. In parallel, transcription of PIFs is inhibited by the red light-induced active conformer of phytochrome B, modulating the growth promotion in response to the light conditions. (Adapted from Havko et al., 2016.) B, Schematic overview of a synthetic device for detection of inflammation signals in mammalian systems. Detection of inflammatory signals through the NF-kB-responsive element of the sensor module leads to expression of the transcriptional regulator GAL4 fused to the VP16 transactivation domain (GAL4-VP16). GAL4-VP16 subsequently binds to the UAS motif in the amplifier and effector modules, increasing the abundance of GAL4-VP16 through a self-activating positive feedback loop from the amplifier module. This triggers production of anti-inflammatory proteins via the effector module. Additionally, the system is equipped with a thresholder device, constitutively expressing GAL4 lacking the transactivation domain. GAL4 competes for binding the UAS motifs with the activating GAL4-VP16, thereby restricting the initiation of the expression of the therapeutic output. A fifth module constitutively expresses the doxycycline-inducible reversed tetracycline repressor protein (rTetR) fused to the inhibitory KRAB domain. Exogenous application of doxycycline inhibits the activation of the sensor, amplifier, and effector modules by binding to their upstream tetO motifs, thus deactivating the system. (Adapted from Smole et al., 2017.)
Figure 8.
Natural and engineered closed-loop regulatory circuits. A, Simplified model of the homeostatic regulation of GA3 metabolism and signaling in Arabidopsis. In the absence of the phytohormone GA, the regulator DELLA proteins accumulate. Through transcriptional control of GA metabolism and catabolism, DELLAs boost the level of GA and subsequently of the GA receptor GID1 proteins. Accumulation of the GID1 proteins and of GA eventually leads to GID1-mediated DELLA degradation. These feedback loops ensure GA homeostasis. (Adapted from Hedden and Thomas, 2012.) B, Schematic overview of a synthetic autoregulatory gene circuit for adjusting insulin resistance in mammalian systems. Upon binding of insulin to the insulin receptor of the designer cell, the intracellular β-subunit of the receptor is autophosphorylated. This leads to further phosphorylation of Tyr residues of the insulin receptor substrate 1 (IRS-1), among other proteins, triggering their interaction with several signaling components. Induced by this interaction, the GTPase Ras and the MAPK are activated, triggering nuclear import of the MAPK. In the nucleus, the MAPK phosphorylates the ELK1 domain of the synthetic regulator protein, consisting of the tet repressor (TetR) and the regulated activation domain of the transcription factor ELK1, expressed under the control of the constitutive human cytomegalovirus immediate early promoter (PhCMV). The hybrid transcription factor binds to the tet operator motif (tetO) in a synthetic effector device; however, the activation domain remains inactive. It gets activated and initiates the expression of the therapeutic Fc-adiponectin protein only upon MAPK-induced phosphorylation of the ELK1 domain. Subsequent secretion of Fc-adiponectin increases the sensitivity for insulin in other tissues (e.g. muscle cells), leading to a decreased insulin production of pancreatic β-cells. (Adapted from Ye et al., 2017.)
First Attempts at Genetic Circuits in Plants
Future development of complex circuitry with predictable and controllable features in plants for biotechnological applications (e.g. production of biopharmaceuticals and other fine chemicals and engineering of stress-tolerant traits and enhanced nutritional content) requires one key prerequisite: namely, to have functionally well-characterized synthetic modules and switches. However, the quantitative characterization of genetic parts in plants is a time-consuming process, and the library of available parts to be used in modular assemblies is still rather limited. Moreover, the complexity of plants as multicellular organisms still remains experimentally challenging for constructing and implementing synthetic genetic circuits with a predictable outcome and robustness. A first step toward a consistent functional and quantitative categorization of molecular switches in plants was reported by Schaumberg et al. (2016); Table 1). The authors built a simple genetic circuit in plant protoplasts, comprising two genetic transcriptional switches and a dual-luciferase output. Addition of an inducer (dexamethasone or 4-hydroxytamoxifen) activates expression of a repressor protein and a firefly luciferase, which are both under the control of the same inducible promoter but on different plasmid constructs. In this case, firefly luciferase acts as a proxy for the amount of repressor. The repressor protein, on the other hand, represses Renilla luciferase expression from a second plasmid. In this way, it is possible to obtain quantitative data on the levels of a repressor protein and correlate it with its repressing activity over a target promoter (Schaumberg et al., 2016). This approach could be expanded easily to characterize, in a standardized fashion, transcriptional regulators, promoter sequences, and higher order circuitry arising from combinations of simple modules. As a note, in a recent example following the principle of bypassing endogenous pathways (in this case, a metabolic one), South et al. (2019) engineered an alternate, synthetic glycolate metabolic route. This pathway is more efficient than the endogenous photorespiratory route, increasing photosynthetic efficiency considerably (∼40%), thereby leading to increased biomass production of tobacco plants. This example represents a milestone, fostering future similar strategies for other metabolic and signaling networks.
Optogenetically regulated systems have been implemented in plant cells (e.g. protoplasts) for the targeted control of signaling pathways. In a first approach, auxin regulatory networks were manipulated using a red light-inducible gene switch that allowed the quantitative control of the expression of the receptor of auxin, the F-box protein TIR1 (up-regulation and down-regulation upon expression of an antisense microRNA; Müller et al., 2014; Samodelov and Zurbriggen, 2017; Table 1). The effects of precisely tuning the sensitivity of the regulatory network to the hormone was monitored with a genetically encoded biosensor designed ad hoc (Wend et al., 2013). This open-loop system enabled inducible quantitative control and monitoring of a signaling network for the study of complex regulatory principles. This is performed in a simple experimental platform without the need for generating mutants (Müller et al., 2014).
Another example of an open-loop system in plants is a fully synthetic signal transduction system that could potentially be used for the programmable detection of ligands (Antunes et al., 2011). In this approach, bacterial signal transduction components were adapted to eukaryotic target sequences and consequently transferred into transgenic plants. The engineered chimeric His kinase included a bacterial receptor, Tgr, fused to the His kinase PhoR. Upon binding a redesigned periplasmic binding protein in complex with the ligand of interest, this chimeric receptor phosphorylates its cognate chimeric response regulator PhoB-VP64. The response regulator in turn activates the expression of a reporter gene. Drought, in the context of climate changes, is one of the biggest challenges to food security. One promising approach to improve plant water usage is to manipulate the ABA signaling pathway, which plays a major role in drought tolerance (Helander et al., 2016). Recent advances have been made in manipulating different aspects of ABA signaling (e.g. receptor engineering and developing an ABA agonist; Park et al., 2015; Vaidya et al., 2017; Table 1). Cyanabactin is a potent, selective agonist for one distinct ABA receptor family, namely, the subfamily of IIIA receptors. These targeted approaches help bypass pleiotropic or unwanted side effects, resulting in more specific, controllable manipulation of a given signaling network. The promising case of cyanabactin could be a model for further directed design of synthetic substances and synthetic cognate receptors.
DISCUSSION AND PERSPECTIVES
In the almost 20 years since the foundational publications of synthetic devices, synthetic biology has evolved into a mature discipline that already revolutionizes fundamental research, most noticeably biomedicine, as well as the biotechnology industry. A broad range of synthetic molecular tools, regulatory and metabolic circuitry, and even synthetic organelles and genomes have been engineered and successfully applied in bacterial, yeast, and animal systems (Brophy and Voigt, 2014). As described in this article, several synthetic biosensors and switches for the control of gene expression (including a couple of optogenetic modules), genome editing, and protein stability have already been implemented in plants (for review, see Liu et al., 2013; Braguy and Zurbriggen, 2016; Walia et al., 2018). The first approaches toward combinations of switches in plant cell systems are arising, including (1) the use of an optogenetic gene switch to control hormone signaling, coupled to a genetically encoded biosensor, as a proxy of the activity of the signaling pathway (Müller et al., 2014); and (2) a semi- and a fully synthetic transduction pathway, sensing a plant hormone or a foreign metabolite, respectively, by transducing the signal into a phenotypic response (sentinel approach; Antunes et al., 2006, 2009). However, engineering and implementation of more complex circuitry is not yet a reality in plant research. Plants are multicellular organisms with complex metabolism and highly regulated and intertwined signaling networks, integrating different environmental cues, like light and temperature, with the genetic program and metabolic status. Experimental constraints and slow generation times often make it cumbersome to implement and evaluate genetic circuits in the whole plant. Altogether, it is still challenging to build synthetic circuits with a predictable output and function.
In order to transition the plant synthetic biology field from a slow and error-prone engineering phase into a more automated, rational, and reliable discipline, a series of approaches have to be implemented. In this way, the development and introduction of advanced circuitry could be achieved, as is already the case for other organisms. In the first place, biosynthetic platforms for the rational design, construction, and quantitative characterization of a bigger number of variants of genetic parts need to be established. Toward this goal, adequate vectors and high-throughput DNA assembly methods are already in place (Patron, 2014; Vazquez-Vilar et al., 2018). However, experimental approaches to quantitatively and functionally describe synthetic modules, as well as hand-in-hand work with mathematical modelers to improve predictability and reliability, still lag behind. Finally, based on the experiences in yeast and animal cells, generalized incorporation of orthogonal components (sensing modules, signaling molecules, and output elements) in the designs will contribute to optimal functionality, including high control specificity, robustness of the networks, and a reduced cross-modulation of the endogenous pathways.
Given the creative and successful applications reported in other organisms, it is easy to imagine that engineering of synthetic circuits in plants will help solve many problems in the near future (see Outstanding Questions). One future goal is to achieve a quantitative increase in crop yield, a much-needed second Green Revolution, to satisfy the demands of the ever-growing world population (Wollenweber et al., 2005). Another goal is to improve plant stress tolerance to environmental hardships by manipulating phytohormone signaling pathways or introducing orthogonal networks, targeting key plant stress responses. First steps toward this were recently reported based on engineering the receptor for the phytohormone ABA and developing chemical agonists thereof to control the responses to drought (Park et al., 2015; Vaidya et al., 2017). A next step would be to design hybrid circuitry to overcome limitations and bypass endogenous regulation of plant signaling networks to improve the efficiency of existing cascades. Self-regulating, smart pathways that bypass endogenous regulation may be easier to design using fully synthetic circuits. These can be engineered to achieve a high target specificity and are orthogonal to the organism, reducing off-target effects. A further application of such smart plants could be the incorporation of synthetic circuitry to integrate information on environmental cues and the genetic program with long-distance synthetic signal transduction. For example, flowering time could be regulated upon computation of the nutrient availability (roots) and perception of environmental stress, thereby optimizing seed production. An alternative approach to increase productivity would be to decouple growth and development from regulatory elements, such as the circadian clock or other genetic programs, thereby achieving longer biosynthetic periods. It is evident that the possible applications of these approaches are endless and would completely reshape plant science. A long-term vision encompasses the implementation of synthetic cellular circuits, such as closed-loop prosthetic networks, which are capable of generating new functionalities, including immune system-like properties or optimized nutrient assimilation and production of high-value compounds. By virtue of the fast development and achievements in other higher eukaryotic systems, we will witness a paradigm change in experimental plant fundamental research and the development of green biotechnological applications in the near future.
Acknowledgments
We thank Leonie-Alexa Koch for fruitful discussions and comments on the article as well as reviewer 1 for the thorough review and the valuable comments and suggestions that contributed to improve the quality of the article. We apologize to our colleagues whose work could not be cited due to space constraints.
Footnotes
This work was supported by the Excellence Initiatives of the German Federal States Governments (DFG, EXC-1028-CEPLAS), a stipend from the Max-Planck-Gesellschaft (Max Planck Society), the University of Düsseldorf, and the University of Cologne.
Articles can be viewed without a subscription.
References
- Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR (2018) Bacterial quorum sensing and microbial community interactions. MBio 9: e02331-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EPS, Landgraf D, Phillips I, Silver PA (2007) Rational design of memory in eukaryotic cells. Genes Dev 21: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827 [DOI] [PubMed] [Google Scholar]
- Antunes MS, Ha SB, Tewari-Singh N, Morey KJ, Trofka AM, Kugrens P, Deyholos M, Medford JI (2006) A synthetic de-greening gene circuit provides a reporting system that is remotely detectable and has a re-set capacity. Plant Biotechnol J 4: 605–622 [DOI] [PubMed] [Google Scholar]
- Antunes MS, Morey KJ, Tewari-Singh N, Bowen TA, Smith JJ, Webb CT, Hellinga HW, Medford JI (2009) Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol 5: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes MS, Morey KJ, Smith JJ, Albrecht KD, Bowen TA, Zdunek JK, Troupe JF, Cuneo MJ, Webb CT, Hellinga HW, et al. (2011) Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE 6: e16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ (2003) Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113: 597–607 [DOI] [PubMed] [Google Scholar]
- Ausländer S, Fussenegger M (2017) Synthetic RNA-based switches for mammalian gene expression control. Curr Opin Biotechnol 48: 54–60 [DOI] [PubMed] [Google Scholar]
- Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M (2012) Programmable single-cell mammalian biocomputers. Nature 487: 123–127 [DOI] [PubMed] [Google Scholar]
- Baaske J, Gonschorek P, Engesser R, Dominguez-Monedero A, Raute K, Fischbach P, Müller K, Cachat E, Schamel WWA, Minguet S, et al. (2018) Dual-controlled optogenetic system for the rapid down-regulation of protein levels in mammalian cells. Sci Rep 8: 15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E. (1998) Autoregulation of eukaryotic transcription factors. Prog Nucleic Acid Res Mol Biol 60: 133–168 [DOI] [PubMed] [Google Scholar]
- Beck CF, Mutzel R, Barbé J, Müller W (1982) A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J Bacteriol 150: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L (2000) Engineering stability in gene networks by autoregulation. Nature 405: 590–593 [DOI] [PubMed] [Google Scholar]
- Becskei A, Séraphin B, Serrano L (2001) Positive feedback in eukaryotic gene networks: Cell differentiation by graded to binary response conversion. EMBO J 20: 2528–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Schopfer U, Dreier B, Barbas CF III (2000) Chemically regulated zinc finger transcription factors. J Biol Chem 275: 32617–32627 [DOI] [PubMed] [Google Scholar]
- Berens C, Altschmied L, Hillen W (1992) The role of the N terminus in Tet repressor for tet operator binding determined by a mutational analysis. J Biol Chem 267: 1945–1952 [PubMed] [Google Scholar]
- Beyer HM, Juillot S, Herbst K, Samodelov SL, Müller K, Schamel WW, Römer W, Schäfer E, Nagy F, Strähle U, et al. (2015) Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth Biol 4: 951–958 [DOI] [PubMed] [Google Scholar]
- Böhmdorfer G, Tramontano A, Luxa K, Bachmair A (2010) A synthetic biology approach allows inducible retrotransposition in whole plants. Syst Synth Biol 4: 133–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ (2014) General method for regulating protein stability with light. ACS Chem Biol 9: 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braguy J, Zurbriggen MD (2016) Synthetic strategies for plant signalling studies: Molecular toolbox and orthogonal platforms. Plant J 87: 118–138 [DOI] [PubMed] [Google Scholar]
- Breaker RR. (2012) Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4: a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat C, Zechner C, Khammash M (2016) Design of a synthetic integral feedback circuit: Dynamic analysis and DNA implementation. ACS Synth Biol 5: 1108–1116 [DOI] [PubMed] [Google Scholar]
- Brophy JAN, Voigt CA (2014) Principles of genetic circuit design. Nat Methods 11: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson I, Krause KP, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB (1998) An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol 16: 177–180 [DOI] [PubMed] [Google Scholar]
- Chatelle C, Ochoa-Fernandez R, Engesser R, Schneider N, Beyer HM, Jones AR, Timmer J, Zurbriggen MD, Weber W (2018) A green-light-responsive system for the control of transgene expression in mammalian and plant cells. ACS Synth Biol 7: 1349–1358 [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN (2002) NFAT signaling: Choreographing the social lives of cells. Cell (Suppl) 109: S67–S79 [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Schreiber SL (2009) SnapShot: Ca2+-calcineurin-NFAT signaling. Cell 138: 210.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler SJ, Hoff KG, Smolke CD (2010) Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330: 1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T, Mondragón-Palomino O, Tsimring L, Hasty J (2010) A synchronized quorum of genetic clocks. Nature 463: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RM, Muller RY, Haynes KA (2015) Can the natural diversity of quorum-sensing advance synthetic biology? Front Bioeng Biotechnol 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Hegemann P (2017) The form and function of channelrhodopsin. Science 357: eaan5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Abelson J, Barnes WM, Reznikoff WS (1975) Genetic regulation: The Lac control region. Science 187: 27–35 [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- Ellis T, Wang X, Collins JJ (2009) Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol 27: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338 [DOI] [PubMed] [Google Scholar]
- Endo K, Stapleton JA, Hayashi K, Saito H, Inoue T (2013) Quantitative and simultaneous translational control of distinct mammalian mRNAs. Nucleic Acids Res 41: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faden F, Ramezani T, Mielke S, Almudi I, Nairz K, Froehlich MS, Höckendorff J, Brandt W, Hoehenwarter W, Dohmen RJ, et al. (2016) Phenotypes on demand via switchable target protein degradation in multicellular organisms. Nat Commun 7: 12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LZ, Lin MZ (2015) Optical control of biological processes by light-switchable proteins. Wiley Interdiscip Rev Dev Biol 4: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Niño M, Giraldo D, Gomez-Porras JL, Dreyer I, González Barrios AF, Arevalo-Ferro C (2017) A synthetic multi-cellular network of coupled self-sustained oscillators. PLoS ONE 12: e0180155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fogelmark K, Troein C (2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLOS Comput Biol 10: e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. (2000) Feedback control of intercellular signalling in development. Nature 408: 313–319 [DOI] [PubMed] [Google Scholar]
- Frey AD, Rimann M, Bailey JE, Kallio PT, Thompson CJ, Fussenegger M (2001) Novel pristinamycin-responsive expression systems for plant cells. Biotechnol Bioeng 74: 154–163 [DOI] [PubMed] [Google Scholar]
- Fung E, Wong WW, Suen JK, Bulter T, Lee SG, Liao JC (2005) A synthetic gene-metabolic oscillator. Nature 435: 118–122 [DOI] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R, Lebar T, Majerle A, Šter B, Dobnikar A, Benčina M, Jerala R (2014) Designable DNA-binding domains enable construction of logic circuits in mammalian cells. Nat Chem Biol 10: 203–208 [DOI] [PubMed] [Google Scholar]
- Gander MW, Vrana JD, Voje WE, Carothers JM, Klavins E (2017) Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat Commun 8: 15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Xiong X, Wong S, Charles EJ, Lim WA, Qi LS (2016) Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods 13: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie JM, Poethig RS, Haseloff JP, Webb AAR (2009) GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J Exp Bot 60: 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342 [DOI] [PubMed] [Google Scholar]
- Gatz C, Frohberg C, Wendenburg R (1992) Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J 2: 397–404 [DOI] [PubMed] [Google Scholar]
- Goodwin BC. (1963) Temporal Organization in Cells: A Dynamic Theory of Cellular Control Processes. Academic Press, London [Google Scholar]
- Goodwin BC. (1965) Oscillatory behavior in enzymatic control processes. Adv Enzyme Regul 3: 425–438 [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89: 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H (1995) Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769 [DOI] [PubMed] [Google Scholar]
- Guido NJ, Wang X, Adalsteinsson D, McMillen D, Hasty J, Cantor CR, Elston TC, Collins JJ (2006) A bottom-up approach to gene regulation. Nature 439: 856–860 [DOI] [PubMed] [Google Scholar]
- Hanson S, Bauer G, Fink B, Suess B (2005) Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA 11: 503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty J, Dolnik M, Rottschäfer V, Collins JJ (2002) Synthetic gene network for entraining and amplifying cellular oscillations. Phys Rev Lett 88: 148101. [DOI] [PubMed] [Google Scholar]
- Havko NE, Major IT, Jewell JB, Attaran E, Browse J, Howe GA (2016) Control of carbon assimilation and partitioning by jasmonate: An accounting of growth-defense tradeoffs. Plants (Basel) 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Helander JDM, Vaidya AS, Cutler SR (2016) Chemical manipulation of plant water use. Bioorg Med Chem 24: 493–500 [DOI] [PubMed] [Google Scholar]
- Heng BC, Aubel D, Fussenegger M (2015) Prosthetic gene networks as an alternative to standard pharmacotherapies for metabolic disorders. Curr Opin Biotechnol 35: 37–45 [DOI] [PubMed] [Google Scholar]
- Hörner M, Weber W (2012) Molecular switches in animal cells. FEBS Lett 586: 2084–2096 [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356 [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA (2013) Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25: 820–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AAT, Hibberd JM, Gay C, Essah PA, Haseloff J, Tester M, Guiderdoni E (2005) Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J 41: 779–789 [DOI] [PubMed] [Google Scholar]
- Kaberniuk AA, Shemetov AA, Verkhusha VV (2016) A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods 13: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhar A, Leydon AR, Lemmex AC, Klavins E, Nemhauser JL (2018) Synthetic hormone-responsive transcription factors can monitor and re-program plant development. eLife 7: e34702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kagawa T, Kieber J, Araki T (2006) Phototropin and light-signaling in phototropism. Curr Opin Plant Biol 9: 503–508 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ (2004) Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci USA 101: 8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar K, Weber W (2017) Synthetic biological approaches to optogenetically control cell signaling. Curr Opin Biotechnol 47: 112–119 [DOI] [PubMed] [Google Scholar]
- Kolar K, Wischhusen HM, Müller K, Karlsson M, Weber W, Zurbriggen MD (2015) A synthetic mammalian network to compute population borders based on engineered reciprocal cell-cell communication. BMC Syst Biol 9: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar K, Knobloch C, Stork H, Žnidarič M, Weber W (2018) OptoBase: A web platform for molecular optogenetics. ACS Synth Biol 7: 1825–1828 [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F (2013) Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Fischer C, Fussenegger M (2004a) BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng 87: 478–484 [DOI] [PubMed] [Google Scholar]
- Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M (2004b) An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol 22: 867–870 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M. (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J (2005) GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56: 2433–2442 [DOI] [PubMed] [Google Scholar]
- Lavedrine C, Farcot E, Vernoux T (2015) Modeling plant development: From signals to gene networks. Curr Opin Plant Biol 27: 148–153 [DOI] [PubMed] [Google Scholar]
- Lebar T, Bezeljak U, Golob A, Jerala M, Kadunc L, Pirš B, Stražar M, Vučko D, Zupančič U, Benčina M, et al. (2014) A bistable genetic switch based on designable DNA-binding domains. Nat Commun 5: 5007. [DOI] [PubMed] [Google Scholar]
- Lienert F, Lohmueller JJ, Garg A, Silver PA (2014) Synthetic biology in mammalian cells: Next generation research tools and therapeutics. Nat Rev Mol Cell Biol 15: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA. (2010) Designing customized cell signalling circuits. Nat Rev Mol Cell Biol 11: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Stewart CN Jr (2015) Plant synthetic biology. Trends Plant Sci 20: 309–317 [DOI] [PubMed] [Google Scholar]
- Liu W, Yuan JS, Stewart CN Jr (2013) Advanced genetic tools for plant biotechnology. Nat Rev Genet 14: 781–793 [DOI] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW III, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Khalil AS, Collins JJ (2009) Next-generation synthetic gene networks. Nat Biotechnol 27: 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie MJ, Mett V, Stewart Reynolds PH, Jameson PE (1998) Controlled cytokinin production in transgenic tobacco using a copper-inducible promoter. Plant Physiol 116: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford JI, Prasad A (2016) Towards programmable plant genetic circuits. Plant J 87: 139–148 [DOI] [PubMed] [Google Scholar]
- Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA (2012) Genetic programs constructed from layered logic gates in single cells. Nature 491: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morffy N, Faure L, Nelson DC (2016) Smoke and hormone mirrors: Action and evolution of karrikin and strigolactone signaling. Trends Genet 32: 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, Gardner KH (2014) An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 10: 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Engesser R, Metzger S, Schulz S, Kämpf MM, Busacker M, Steinberg T, Tomakidi P, Ehrbar M, Nagy F, et al. (2013a) A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res 41: e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Engesser R, Schulz S, Steinberg T, Tomakidi P, Weber CC, Ulm R, Timmer J, Zurbriggen MD, Weber W (2013b) Multi-chromatic control of mammalian gene expression and signaling. Nucleic Acids Res 41: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Siegel D, Rodriguez Jahnke F, Gerrer K, Wend S, Decker EL, Reski R, Weber W, Zurbriggen MD (2014) A red light-controlled synthetic gene expression switch for plant systems. Mol Biosyst 10: 1679–1688 [DOI] [PubMed] [Google Scholar]
- Müller K, Naumann S, Weber W, Zurbriggen MD (2015) Optogenetics for gene expression in mammalian cells. Biol Chem 396: 145–152 [DOI] [PubMed] [Google Scholar]
- Müller S, Hofbauer J, Endler L, Flamm C, Widder S, Schuster P (2006) A generalized model of the repressilator. J Math Biol 53: 905–937 [DOI] [PubMed] [Google Scholar]
- Nash AI, McNulty R, Shillito ME, Swartz TE, Bogomolni RA, Luecke H, Gardner KH (2011) Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc Natl Acad Sci USA 108: 9449–9454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G (2009) Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci USA 106: 5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevozhay D, Zal T, Balázsi G (2013) Transferring a synthetic gene circuit from yeast to mammalian cells. Nat Commun 4: 1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AAK, Voigt CA (2014) Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol 10: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M (2015) CRISPR-Cas9-based photoactivatable transcription system. Chem Biol 22: 169–174 [DOI] [PubMed] [Google Scholar]
- Niopek D, Benzinger D, Roensch J, Draebing T, Wehler P, Eils R, Di Ventura B (2014) Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat Commun 5: 4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B (2016) Optogenetic control of nuclear protein export. Nat Commun 7: 10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Fernandez R, Samodelov SL, Brandl SM, Wehinger E, Müller K, Weber W, Zurbriggen MD (2016) Optogenetics in plants: Red/far-red light control of gene expression. Methods Mol Biol 1408: 125–139 [DOI] [PubMed] [Google Scholar]
- Okumoto S, Jones A, Frommer WB (2012) Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol 63: 663–706 [DOI] [PubMed] [Google Scholar]
- Paek KY, Hong KY, Ryu I, Park SM, Keum SJ, Kwon OS, Jang SK (2015) Translation initiation mediated by RNA looping. Proc Natl Acad Sci USA 112: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR (2015) Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 520: 545–548 [DOI] [PubMed] [Google Scholar]
- Parker C. (2009) Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci 65: 453–459 [DOI] [PubMed] [Google Scholar]
- Patron NJ. (2014) DNA assembly for plant biology: Techniques and tools. Curr Opin Plant Biol 19: 14–19 [DOI] [PubMed] [Google Scholar]
- Phillips R, Kondev J, Theriot J (2009) Physical Biology of the Cell. Garland Science, New York [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13: 578–589 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W (2004) Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Mas P, Millar AJ (2013) Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst Biol 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11: 198–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J (2011) A sensing array of radically coupled genetic ‘biopixels.’ Nature 481: 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell O, Savery NJ, Grierson CS, di Bernardo M (2010) A comparative analysis of synthetic genetic oscillators. J R Soc Interface 7: 1503–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GR, Garoosi GA, Koroleva O, Ito M, Laufs P, Leader DJ, Caddick MX, Doonan JH, Tomsett AB (2005) The alc-GR system: A modified alc gene switch designed for use in plant tissue culture. Plant Physiol 138: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC (2006) The structure of phytochrome: A picture is worth a thousand spectra. Plant Cell 18: 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MRH, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al. (2001) Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Roybal KT, Lim WA (2017) Synthetic immunology: Hacking immune cells to expand their therapeutic capabilities. Annu Rev Immunol 35: 229–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal KT, Williams JZ, Morsut L, Rupp LJ, Kolinko I, Choe JH, Walker WJ, McNally KA, Lim WA (2016) Engineering T cells with customized therapeutic response programs using synthetic notch receptors. Cell 167: 419–432.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryback BM, Odoni DI, van Heck RGA, van Nuland Y, Hesselman MC, Martins Dos Santos VAP, van Passel MWJ, Hugenholtz F (2013) Design and analysis of a tunable synchronized oscillator. J Biol Eng 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas F, Rojas V, Delgado V, Agosin E, Larrondo LF (2017) Optogenetic switches for light-controlled gene expression in yeast. Appl Microbiol Biotechnol 101: 2629–2640 [DOI] [PubMed] [Google Scholar]
- Samodelov SL, Zurbriggen MD (2017) Quantitatively understanding plant signaling: Novel theoretical-experimental approaches. Trends Plant Sci 22: 685–704 [DOI] [PubMed] [Google Scholar]
- Savageau MA. (1974) Comparison of classical and autogenous systems of regulation in inducible operons. Nature 252: 546–549 [DOI] [PubMed] [Google Scholar]
- Schaumberg KA, Antunes MS, Kassaw TK, Xu W, Zalewski CS, Medford JI, Prasad A (2016) Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat Methods 13: 94–100 [DOI] [PubMed] [Google Scholar]
- Schena M, Lloyd AM, Davis RW (1991) A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci USA 88: 10421–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P (2002) A web of circadian pacemakers. Cell 111: 919–922 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2010) Discover and connect cellular signaling. Plant Physiol 154: 562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smole A, Lainšček D, Bezeljak U, Horvat S, Jerala R (2017) A synthetic mammalian therapeutic gene circuit for sensing and suppressing inflammation. Mol Ther 25: 102–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR (2019) Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SA, Gagner JE, Damanpour S, Yoshida M, Dordick JS, Friedman JM (2012) Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 336: 604–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein V, Alexandrov K (2015) Synthetic protein switches: Design principles and applications. Trends Biotechnol 33: 101–110 [DOI] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J (2008) A fast, robust and tunable synthetic gene oscillator. Nature 456: 516–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamsir A, Tabor JJ, Voigt CA (2011) Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires.’ Nature 469: 212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieffry D, Huerta AM, Pérez-Rueda E, Collado-Vides J (1998) From specific gene regulation to genomic networks: A global analysis of transcriptional regulation in Escherichia coli. BioEssays 20: 433–440 [DOI] [PubMed] [Google Scholar]
- Thurley K, Wu LF, Altschuler SJ (2018) Modeling cell-to-cell communication networks using response-time distributions. Cell Syst 6: 355–367.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M (2009) A tunable synthetic mammalian oscillator. Nature 457: 309–312 [DOI] [PubMed] [Google Scholar]
- Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA (2018) Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 361: 156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Mock C, Batchelor E, Loewer A, Lahav G (2010) A synthetic-natural hybrid oscillator in human cells. Proc Natl Acad Sci USA 107: 17047–17052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA (2013) Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155: 1422–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazou M, Barahona M, Polizzi KM, Stan GB (2018) Computational re-design of synthetic genetic oscillators for independent amplitude and frequency modulation. Cell Syst 6: 508–520.e5 [DOI] [PubMed] [Google Scholar]
- Trewavas A. (2005) Green plants as intelligent organisms. Trends Plant Sci 10: 413–419 [DOI] [PubMed] [Google Scholar]
- Vaidya AS, Peterson FC, Yarmolinsky D, Merilo E, Verstraeten I, Park SY, Elzinga D, Kaundal A, Helander J, Lozano-Juste J, et al. (2017) A rationally designed agonist defines subfamily IIIA abscisic acid receptors as critical targets for manipulating transpiration. ACS Chem Biol 12: 2842–2848 [DOI] [PubMed] [Google Scholar]
- van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC (2015) Optogenetic control of organelle transport and positioning. Nature 518: 111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC (2012) Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 287: 36370–36383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Vilar M, Orzaez D, Patron N (2018) DNA assembly standards: Setting the low-level programming code for plant biotechnology. Plant Sci 273: 33–41 [DOI] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA 107: 6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia A, Waadt R, Jones AM (2018) Genetically encoded biosensors in plants: Pathways to discovery. Annu Rev Plant Biol 69: 497–524 [DOI] [PubMed] [Google Scholar]
- Warashina M, Takagi Y, Stec WJ, Taira K (2000) Differences among mechanisms of ribozyme-catalyzed reactions. Curr Opin Biotechnol 11: 354–362 [DOI] [PubMed] [Google Scholar]
- Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, Fussenegger M (2002) Macrolide-based transgene control in mammalian cells and mice. Nat Biotechnol 20: 901–907 [DOI] [PubMed] [Google Scholar]
- Weinmann P, Gossen M, Hillen W, Bujard H, Gatz C (1994) A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J 5: 559–569 [DOI] [PubMed] [Google Scholar]
- Wend S, Dal Bosco C, Kämpf MM, Ren F, Palme K, Weber W, Dovzhenko A, Zurbriggen MD (2013) A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci Rep 3: 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde RJ, Shufflebottom D, Cooke S, Jasinska I, Merryweather A, Beri R, Brammar WJ, Bevan M, Schuch W (1992) Control of gene expression in tobacco cells using a bacterial operator-repressor system. EMBO J 11: 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenweber B, Porter JR, Lübberstedt T (2005) Need for multidisciplinary research towards a second green revolution. Curr Opin Plant Biol 8: 337–341 [DOI] [PubMed] [Google Scholar]
- Xiao H, Edwards TE, Ferré-D’Amaré AR (2008) Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol 15: 1125–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Fussenegger M (2018) Designing cell function: Assembly of synthetic gene circuits for cell biology applications. Nat Rev Mol Cell Biol 19: 507–525 [DOI] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y (2011) Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333: 1307–1311 [DOI] [PubMed] [Google Scholar]
- Ye H, Baba MD, Peng R, Fussenegger M (2011) A synthetic optogentic transcription device enhances blood-glucose homeostasis in mice. Science 332: 1565–1569 [DOI] [PubMed] [Google Scholar]
- Ye H, Xie M, Xue S, Charpin-El Hamri G, Yin J, Zulewski H, Fussenegger M (2017) Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat Biomed Eng 1: 0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, Cox RS III, Weiss R, Arnold FH (2004) Programmed population control by cell-cell communication and regulated killing. Nature 428: 868–871 [DOI] [PubMed] [Google Scholar]
- You YS, Marella H, Zentella R, Zhou Y, Ulmasov T, Ho THD, Quatrano RS (2006) Use of bacterial quorum-sensing components to regulate gene expression in plants. Plant Physiol 140: 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Cui B (2015) Optogenetic control of intracellular signaling pathways. Trends Biotechnol 33: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]