RBP-L is essential for glutelin and prolamine mRNA localization in rice endosperm cells.
Abstract
The transport and targeting of glutelin and prolamine mRNAs to distinct subdomains of the cortical endoplasmic reticulum is a model for mRNA localization in plants. This process requires a number of RNA-binding proteins (RBPs) that recognize and bind to mRNA cis-localization (zipcode) elements to form messenger ribonucleoprotein complexes, which then transport the RNAs to their destination sites at the cortical endoplasmic reticulum. Here, we present evidence that the rice (Oryza sativa) RNA-binding protein, RBP-L, like its interacting RBP-P partner, specifically binds to glutelin and prolamine zipcode RNA sequences and is required for proper mRNA localization in rice endosperm cells. A transfer DNA insertion in the 3′ untranslated region resulted in reduced expression of the RBP-L gene to 10% to 25% of that in the wild-type. Reduced amounts of RBP-L caused partial mislocalization of glutelin and prolamine RNAs and conferred other general growth defects, including dwarfism, late flowering, and smaller seeds. Transcriptome analysis showed that RBP-L knockdown greatly affected the expression of prolamine family genes and several classes of transcription factors. Collectively, these results indicate that RBP-L, like RBP-P, is a key RBP involved in mRNA localization in rice endosperm cells. Moreover, distinct from RBP-P, RBP-L exhibits additional regulatory roles in development, either directly through its binding to corresponding RNAs or indirectly through its effect on transcription factors.
Intracellular mRNA localization, a process where mRNAs are transported to specific regions within the cell, is a conserved mechanism found in prokaryotes and eukaryotic organisms (Nevo-Dinur et al., 2011; Medioni et al., 2012; Blower, 2013; Weis et al., 2013; Tian and Okita, 2014). This process, which efficiently drives protein targeting, initiates in the nucleus where the cis-localization RNA sequences, also called “zipcodes,” are recognized by their corresponding trans factors, RNA-binding proteins (RBPs), forming a ribonucleoprotein complex. After nuclear export to the cytoplasm, the RNP complex undergoes extensive remodeling with the addition (e.g. myosin motor) and removal of one or more components to form a transport particle, allowing mRNA trafficking via the cytoskeleton under a translation-arrested state. Once anchored at its final destination, the RNA is partitioned in storage granules and/or processed in P bodies or translated locally, the latter process leading to the enrichment of specific proteins in localized regions of the cell. RNA localization processes, therefore, link temporal and spatial control of gene expression with protein synthesis at discrete cellular locals.
Localization of mRNAs is well studied in yeast and animal cells (Martin and Ephrussi, 2009; Nevo-Dinur et al., 2011; Medioni et al., 2012; Blower, 2013; Weis et al., 2013; Tian and Okita, 2014). In higher plants, however, few examples of RNA localization exist. Developing rice (Oryza sativa) endosperm provides one of the most optimal plant systems for the study of mRNA localization. Rice endosperm cells are large and cytoplasmic-dense—properties conducive to study at the light microscopy level. Moreover, they contain abundant RNAs that code for the major storage proteins, glutelin and prolamine, and these mRNAs are asymmetrically distributed on distinct endoplasmic reticulum (ER) subdomains of the cortical-ER complex. Prolamine mRNAs are targeted to the protein body ER (PB-ER) that delimit the prolamine intracisternal inclusion granule whereas glutelin mRNAs are localized on adjoining cisternal-ER (Choi et al., 2000; Crofts et al., 2005; Tian and Okita, 2014). The specific localization of these mRNAs enriches the newly synthesized prolamine and glutelin proteins within distinct ER subdomains, which facilitates the assembly of prolamine polypeptides into ER-bounded protein body I (PB-I) or transport and packaging of glutelin to the protein storage vacuoles (PSV). Conversely, mislocalization of these mRNAs mistargets the coded prolamine and glutelin proteins from their normal sites of deposition within the endomembrane system (Crofts et al., 2004; Washida et al., 2009a, 2009b).
A key step in regulating mRNA localization events is the cooperative interaction between mRNAs and multiple RBPs to form the core mRNP complex. We previously identified the zipcode RNA sequences located on the glutelin and prolamine mRNAs (Hamada et al., 2003; Washida et al., 2009a, 2012). Prolamine mRNA contains two zipcode RNA sequences, one located downstream of the signal peptide CDS and the other in the three-prime untranslated region (3′UTR), whereas three RNA localization elements were identified in glutelin mRNA (Hamada et al., 2003; Washida et al., 2009a). These RNA elements are required for proper targeting of those mRNAs to PB-ER and cisternal-ER, respectively, as removing one or more zipcode elements resulted in partial or complete mistargeting (Hamada et al., 2003; Washida et al., 2009b).
More than 200 RBPs potentially involved in RNA transport and localization have been identified by proteomic analysis of a cytoskeleton-enriched fraction from developing rice endosperm (Doroshenk et al., 2009, 2012). Among the identified proteins, RBP-A, I, J, K, Q, and P bind directly as multiprotein complexes to the glutelin and/or prolamine zipcode RNA sequences (Doroshenk et al., 2014; Yang et al., 2014; Chou et al., 2017; Tian et al., 2018). Mutations within RBP-P lead to a reduction in its capacity to bind to RNAs and interact with other RBPs. These deficiencies account for the mislocalization of glutelin and prolamine mRNAs transport to PB-ER and cisternal-ER (Tian et al., 2018) and infer that proper formation of a functional RNP complex is essential for correct targeting of mRNAs.
The RNA-binding protein, RBP-L, was found to be major interacting partner of RBP-P (Tian et al., 2018). Although the interaction of RBP-L was unaffected by mutations in RBP-P, it still may participate in the formation of RNP complexes that are essential for targeting of prolamine and glutelin mRNAs. However, the molecular properties of RBP-L and its role, if any, in glutelin and prolamine mRNA localization remain unclear and further studies of RBP-L are required to better understand the mechanism of mRNA localization in rice endosperm cells.
In this study, we provide direct evidence that RBP-L is required for localization of storage protein RNAs to their respective sites on the PB-ER and cisternal-ER. RBP-L binds to prolamine and glutelin RNAs in general and specifically to their zipcode sequences. RBP-L is found closely associated with the distribution of glutelin and prolamine RNAs as viewed by a combination of in situ reverse transcription (RT)-PCR and immunofluorescence microscopy. Moreover, knockdown of RBP-L gene expression by transfer DNA (T-DNA) gene insertion induces partial mistargeting of both glutelin and prolamine mRNAs, which provides direct evidence that RBP-L, like RBP-P, is required for proper localization of both glutelin and prolamine mRNAs. The T-DNA insertional rice line exhibited dwarfism, late flowering, and smaller seeds, indicating that RBP-L is also required in processes involving plant growth and development. Transcriptome analysis showed that RBP-L knockdown significantly alters the expression of prolamine family genes and several gene families of transcription factors. This regulatory pattern mediated by RBP-L is different from that observed for RBP-P (Tian et al., 2018). Overall, our results show that RBP-L, like its interacting RBP-P, is required for storage protein RNA localization and is essential for normal rice growth and development; however, RBP-L modulates molecular and cellular processes distinct from that controlled by RBP-P.
RESULTS
RBP-L Binds to Glutelin and Prolamine mRNAs In Vitro and In Vivo
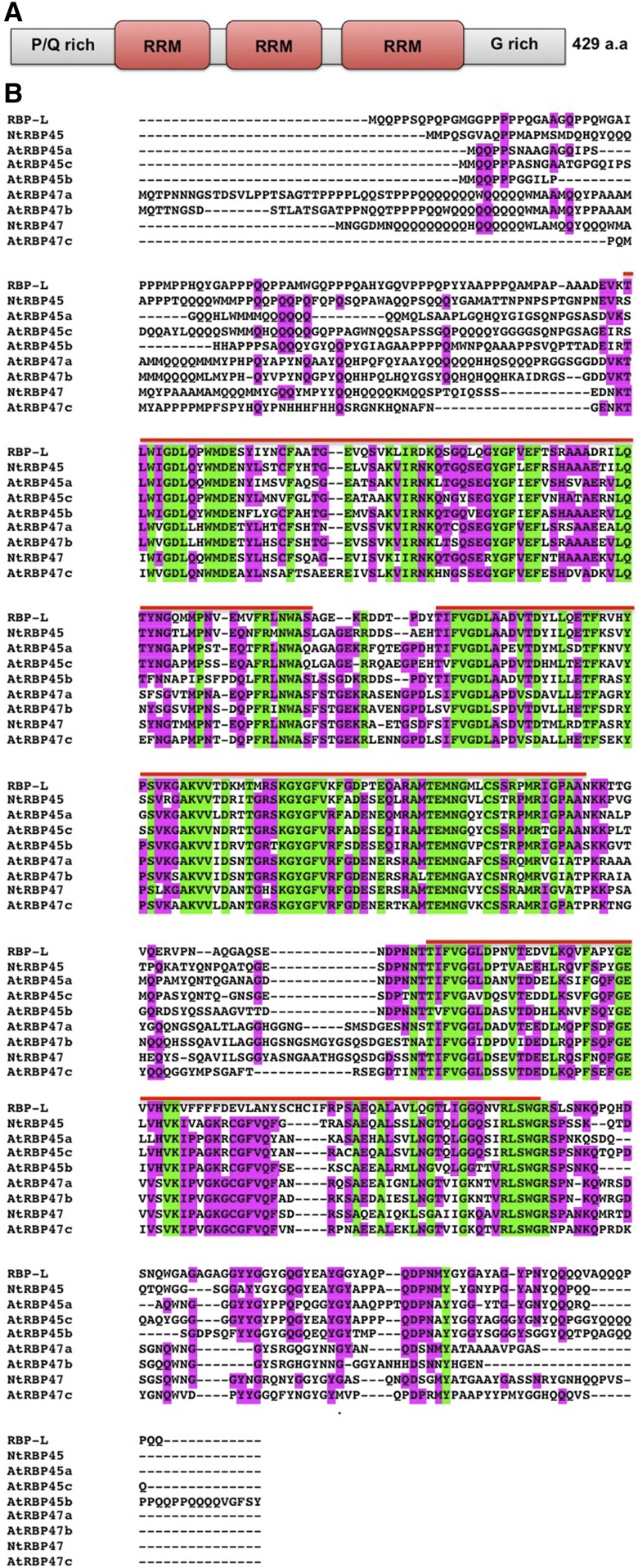
RBP-L was initially isolated from a cytoskeleton-enriched fraction (Doroshenk et al., 2009) and later identified as a member of a group of RBPs that specifically recognized the prolamine zipcode (Crofts et al., 2010). More recently, RBP-L has been identified as an interacting partner of RBP-P that specifically binds to the glutelin and prolamine zipcodes (Tian et al., 2018). RBP-L contains three RNA recognition motifs (RRMs) with flanking Prolamine-rich N-terminal and Glycine-rich C-terminal segments (Fig. 1A). It belongs to the RBP45/47 family proteins as it shares ∼50% sequence identity with the Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) RBP45s (Fig. 1B).
Figure 1.
Sequence information of RBP-L. A, Schematic structure of RBP-L. B, Protein sequence alignment of RBP-L to RBP45/47 family members from Arabidopsis and tobacco. The three RRMs are indicated by the red line above the sequences. The conserved (shared by all protein sequences) and semiconserved (shared by six out of nine protein sequences) amino acids are highlighted in green and magenta, respectively. P/Q rich, Pro- and Gln-rich domain; G-rich, Gly-rich domain; a.a., amino acids.
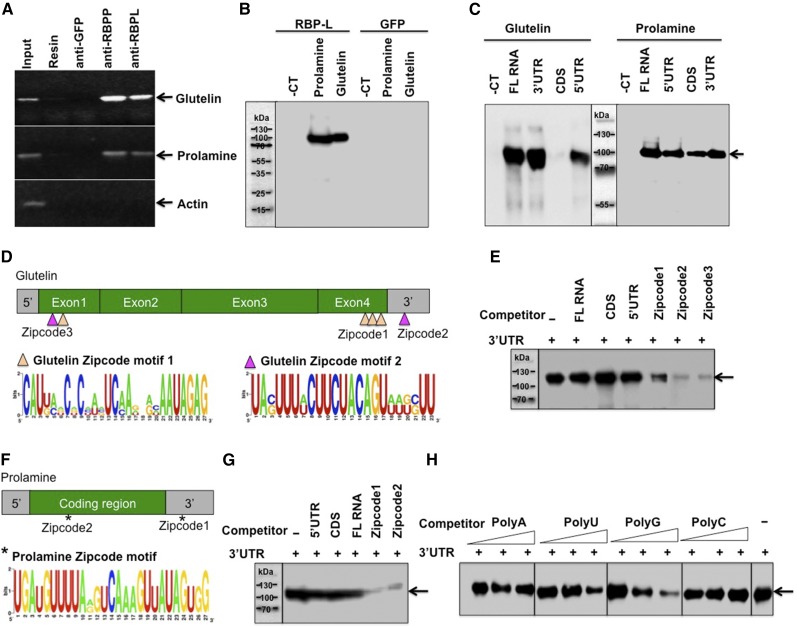
The interaction of RBP-L with the glutelin-prolamine zipcode binding RBP-P suggests that this RBP may also recognize glutelin and prolamine mRNAs as well. To directly study this capacity, we assessed whether RBP-L binds to glutelin and prolamine RNAs in vivo through RNA-immunoprecipitation (RNA-IP) and in vitro through RNA-protein UV cross-linking analyses. For RNA-IP, developing rice grains were pretreated with formaldehyde to stabilize the in vivo ribonucleoprotein complexes before homogenization to generate cell extracts. The clarified cell extracts were then incubated with protein-A agarose beads bound with immobilized RBP-L antibodies. Protein-A bound to RBP-P or green fluorescent protein (GFP) antibodies, were employed as positive and negative controls, respectively. The generated immunoprecipitants (IPs) were then heat-treated to dissociate the RNA-protein cross-links and then extracted with TRIzol to isolate the bound RNAs for subsequent complementary DNA (cDNA) synthesis and PCR using primers specific to glutelin and prolamine gene sequences. As shown in Figure 2A, both glutelin and prolamine RNAs were detected in total grain extracts and in IPs generated with anti-RBP-P but not with anti-GFP or naked protein-A resin controls. Glutelin and prolamine RNAs were also detected in the IP samples of anti-RBP-L, although the amounts of the amplified glutelin and prolamine products were lower than that captured by anti-RBP-P. Actin RNA, which was used as a PCR control, was not detectable in these IP samples, indicating that RBP-L preferentially binds to glutelin and prolamine RNAs in rice endosperm cells.
Figure 2.
RBP-L binds to glutelin and prolamine mRNAs, especially to their zipcode RNA sequences. A, RNA-IP analysis showing that RBP-L is associated with glutelin and prolamine mRNAs in vivo. The RNAs extracted from IPs generated by anti-RBP-P, anti-RBP-L, anti-GFP, and empty resin were subjected to RT-PCR to detect glutelin, prolamine, and actin gene sequences. The figure depicts an agarose gel with resolved PCR products. Anti-GFP and empty resin were used as negative controls. Input, PCR using cDNA synthesized from RNAs isolated from IPs. B, Binding activity of RBP-L to glutelin and prolamine mRNAs compared to RNA control (for detailed information, see “Materials and Methods”). The figure depicts an immunoblot where anti-DIG antibody was used to detect bound DIG-labeled RNAs (see “Materials and Methods” for detailed experimental details). GFP protein was used as negative control for the binding test. C, Binding activities of RBP-L to different region of glutelin and prolamine RNAs. Arrow indicates the location of recombinant RBP-L. D, The glutelin RNA zipcode elements, which consist of two types of motifs, zipcode motif 1 (orange triangle) and zipcode motif 2 (magenta triangle). (Top) Location and components of three glutelin zipcode elements; (bottom) consensus zipcode sequences generated by WebLogo (http://weblogo.berkeley.edu). E, Competition assays using equimolar amounts of DIG-labeled glutelin 3′UTR and the indicated unlabeled competitor RNAs. Arrows indicates the location of recombinant RBP-L. F, The prolamine zipcode elements, which consist of only a single zipcode motif (*). (Top) Location of the two zipcode elements within prolamine mRNA; (bottom) consensus sequence of prolamine zipcode. G, Competition assays using equimolar amounts of DIG-labeled prolamine 3′UTR and indicated unlabeled competitor RNAs. Arrows indicate the location of recombinant RBP-L. H, Binding specificity of RBP-L to different homoribopolymers. Unlabeled poly(U), poly(A), poly(C), and poly(G) were added at 5-, 10-, and 20-fold mole excess over DIG-labeled glutelin 3′ UTR RNA. Arrows indicates the location of recombinant RBP-L. -CT, pBlueScript KS vector sequence; 5′, 5′UTR; 3′, 3′UTR; -, no competitor.
To assess whether RBP-L can directly bind to storage protein RNAs, we performed in vitro RNA-protein UV-cross–linking assays. The purified recombinant RBP-L was incubated with digoxigenin (DIG)-labeled glutelin and prolamine RNAs, UV-cross-linked and the resulting RNA-protein complexes resolved by SDS-PAGEfollowed by immunoblot analysis using anti-DIG antibody. Under these conditions, RBP-L showed strong binding activity to full-length (FL) glutelin and prolamine RNAs (Fig. 2B), whereas no binding was seen with recombinant GFP. Further assays were performed on different regions of these RNAs. Strong positive binding affinity was detected with the 3′UTR and 5′UTR of both glutelin and prolamine RNAs. Whereas binding to the prolamine coding region of RNAs (i.e. the coding sequences, CDS) was seen, no detectable binding of RBP-L to glutelin CDS was observed (Fig. 2C).
To study the binding specificity to the various mRNA regions, we performed competition assays in which equimolar amounts of DIG-labeled 3′UTR and unlabeled competitor RNA sequences were incubated together with recombinant RBP-l to see the impact of the competitor RNAs on binding to the 3′UTR. RBP-L prefers to bind to the glutelin 3′UTR, as the FL, CDS, and 5′UTR did not noticeably compete with binding to the 3′UTR (Fig. 2E). The failure of the glutelin FL, which contains the 3′UTR, to compete against the 3′UTR indicates that the FL assumes a conformation that masks the 3′UTR sequences. Similar binding results were observed for prolamine RNAs although 3′UTR binding was slightly reduced when unlabeled prolamine FL was added as a competitor (Fig. 2G).
Further competition analysis (Fig. 2, D–G) was conducted to investigate the binding preferences between the 3′UTR and zipcode RNA sequences, the cis-elements that direct the localization of glutelin and prolamine RNA to the cisternal-ER and PB-ER, respectively. Glutelin RNA contains three zipcode RNA sequences composed of one or two sequence motifs (Fig. 2D). Zipcode 1, located at the end of exon 4, is composed of three repeats of motif 1. Zipcode 2 contains motif 2 located within the 3′UTR, whereas Zipcode 3 located at exon 1 contains both motifs 1 and 2 (Washida et al., 2009a). Binding of RBP-L to the 3′UTR of glutelin RNAs was largely reduced when unlabeled zipcode RNAs were added, indicating that RBP-L exhibited higher binding affinity to zipcode RNAs than to the glutelin 3′UTR (Fig. 2E). Unlike the weaker competitor Zipcodes 1, the Zipcodes 2, and 3 strongly competed against binding to the glutelin 3′UTR, suggesting that RBP-L prefers binding to Zipcode 2 and 3 (Fig. 2E). Both of these zipcodes contain motif 2.
Prolamine has two zipcodes (Hamada et al., 2003) containing a conserved motif located in the coding region and 3′UTR (Fig. 2F). Similar to that observed for glutelin, RBP-L showed strong binding to prolamine zipcode sequences as they strongly abolished binding to the 3′UTR (Fig. 2G). Thus, the zipcode elements are direct binding targets of RBP-L proteins. The binding to these zipcode RNAs that drives the targeting of glutelin and prolamine mRNAs to specific subdomains of the cortical-ER in developing rice endosperm cells highlights the role of RBP-L in mRNA localization.
Closer examination of the zipcode motifs suggests that RBP-L prefers binding to U-rich sequences prevalent in the two prolamine zipcodes and glutelin zipcode 2 and 3 (Fig. 2, D–G). We, therefore, tested the binding preference of RBP-L to U, A, G, or C homopolymer sequences by performing competition assays in which the recombinant proteins were incubated with DIG-labeled glutelin 3′UTR RNA in the presence of unlabeled homoribopolymers. As shown in Figure 2H, the binding activity of RBP-L to glutelin 3′UTR was substantially reduced by poly U and G, suggesting that RBP-L prefers binding to poly U and G sequences. This binding preference to homopolymer U accounts for the more selective binding of RBP-L to the U-rich zipcode motifs.
RBP-L Is Located in Both the Nucleus and Cytoplasm, Especially on the ER Membrane
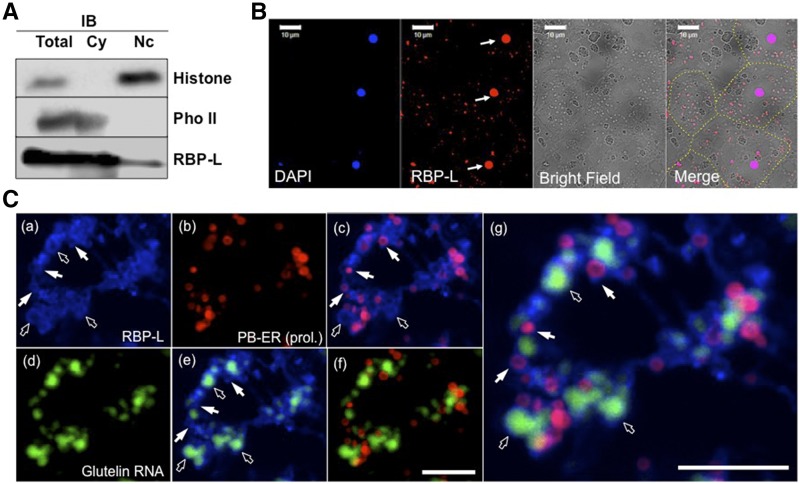
To gain further insight on the molecular function of RBP-L in seed storage protein biosynthesis, we determined the cellular location of RBP-L via subcellular fractionation of nucleus and cytoplasmic fractions isolated from developing seed extracts followed by immunoblot analysis. To determine the relative distribution of this RBP between these two subcellular compartments, the nuclear extract fraction was adjusted to the same volume as the cytoplasmic fraction. Histone H3 and phosphorylase II (Doroshenk et al., 2014) were used as marker indicators of the nucleus and cytoplasm, respectively (Fig. 3A). A specific antibody against RBP-L was generated (Supplemental Fig. S1) and used in the experiment to detect the distribution of RBP-L. As shown in Figure 3A, RBP-L was detected in both the nucleus and cytoplasm, though the amount of RBP-L in nucleus was much less than that detected in the cytoplasm.
Figure 3.
Subcellular localization of RBP-L in rice seed endosperm cells. A, The distribution of RBP-L in the nuclear and cytoplasmic subcellular fractions as revealed by immunoblot analysis. Histone H3 and starch phosphorylase II were used as marker proteins of the nuclear and cytoplasmic fractions. B, Immunofluorescence analysis to locate RBP-L (red) in rice endosperm cells using its specific antibody. The labeling was performed on 5-μm-thin sections from LR-white embedded rice grain tissue. Nuclei stained by 4′,6-diamidino-2-phenylindole (blue) are indicated by arrows and the different cells shown in the figure are distinguished by dashed yellow line. Scale bar = 10 μm. C, Triple labeling for detecting the distribution of RBP-L (blue, a), PB-ER (red, b), and glutelin RNAs (green, d). Fresh-frozen 20-μm cryosections of rice seed tissues were initially subjected to in situ RT-PCR in the presence of glutelin primers and Alexa Fluor 488-UTP (green, d) to visualize the distribution of glutelin mRNAs. The sections were then treated by immunofluorescence analysis for detection of RBP-L and then poststained with rhodamine B to visualize PB-ER (red, b), the ER membranes where prolamine mRNAs are localized. C, (e) to (g), are the merged images of (a) and (b), (a) and (d), (b) and (d), and the combined (a), (b) and (d) panels, respectively. White arrows denote colocalized RBP-Lwith rhodamine-B–labeled PB-ER membranes and open arrows indicate distribution of RBP-L protein surrounding glutelin mRNA patches. Scale bar = 20 μm. IB, immunoblot analysis; Cy, cytoplasmic; Nc, nuclear; DAPI, 4′,6-diamidino-2-phenylindole; Pho II, phosphorylase II; prol., prolamine.
We further performed indirect immuno-fluorescence labeling analysis on ultrathin sections of a LR-white embedded rice grain sample to confirm the results obtained by subcellular fractionation. In agreement with the subcellular localization studies, RBP-L was found located in 4′,6-diamidino-2-phenylindole–stained nucleus as well as particles distributed throughout the cytoplasm (Fig. 3B). These results indicate that RBP-L functions in both the nucleus and cytoplasm, locations consistent with its possessing multiple roles in RNA metabolism.
More detailed microscopic analysis was undertaken to determine the basis for the particle-like distribution of RBP-L in the cytoplasm. In particular, we explored the possible relationship of RBP-L with the cortical-ER and with the distribution of glutelin and prolamine RNAs. To maximize the simultaneous detection of RBP-L, glutelin RNAs, and PB-ER, much thicker (20-μm) frozen sections, prepared from freshly harvested, developing rice grains, were employed. After fixation with paraformaldehyde solution, the thick sections were initially subjected to in situ reverse transcription PCR (RT-PCR) in the presence of glutelin primers and fluorescently labeled Alexa488 dUTP. The grain sections were then incubated with RBP-L antibodies. After washing, the antibody-antigen reaction was labeled with AlexFluo 633-conjugated secondary antibody, followed by incubation with the lipophilic ER-staining dye, Rhodamine B, to selectively stain the PB-ER (as well as the surface of prolamine intracisternal inclusion) in the developing rice grains (Muench et al., 2000). When viewed by microscopy, RBP-L was distributed in a broad patchy pattern with some small ring-structures (Fig. 3C, a). These ring-structures coincided with Rhodamine-stained PB-ER, the ER membrane subdomain harboring prolamine mRNAs (Fig. 3C, a–c). The colocalization of RBP-L to PB-ER, the site of prolamine mRNA accumulation, is a property consistent with its role in prolamine RNA localization.
To explore the spatial relationship between RBP-L and glutelin RNAs (Fig. 3C, d and e), the distribution of RBP-L and glutelin RNAs was directly assessed. Glutelin RNAs were distributed as patches on cisternal ER adjacent to prolamine PBs (Fig. 3C, d) as previously demonstrated (Li et al., 1993; Choi et al., 2000; Hamada et al., 2003). RBP-L was located on the periphery of the glutelin mRNA patches although, in some instances, coincided with the RNA patches themselves. There were also areas where RBP-L was not associated with glutelin RNAs or PB-ER, suggesting a role independent of storage protein RNA localization. Overall, our microscopic studies show a direct relationship between the location of RBP-L and the distribution of glutelin and prolamine RNAs on the cortical-ER.
RBP-L Associates with Prolamine and Glutelin mRNAs
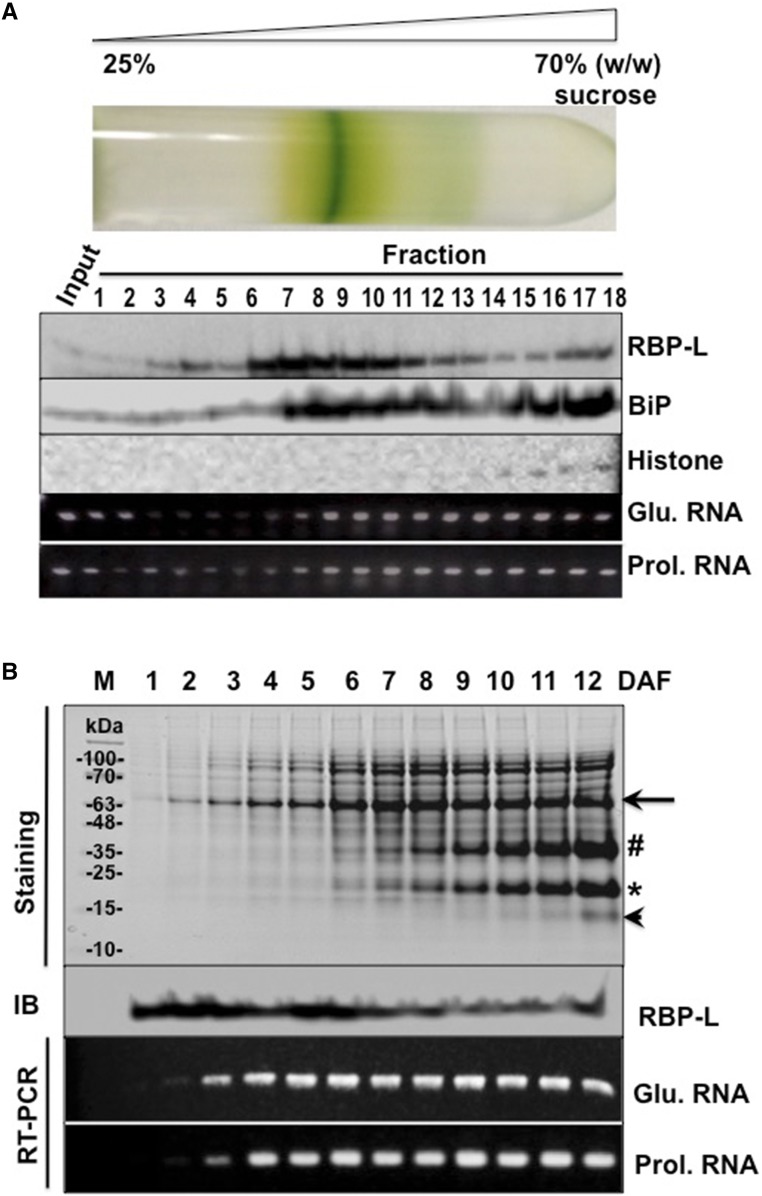
To further explore the relationship of RBP-L with prolamine and glutelin mRNAs, developing grain extracts were fractionated on 25% to 70% Suc density gradient (Fig. 4A). After centrifugation, a chlorophyll-containing membrane band contributed by chlorenchyma cells of the outer grain pericarp was noticeably present near the middle of the gradient. The relative narrow banding of this chlorophyll layer denotes the relatively good resolving power of the Suc density gradient centrifugation step.
Figure 4.
The association of RBP-L with glutelin and prolamine mRNAs in developing rice grains. A, The distribution of RBP-L, glutelin, and prolamine mRNAs, histone, and BiP as resolved by Suc density gradient centrifugation. Samples from the Suc gradient were collected from the top (25% [w/w] Suc) to bottom (70% [w/w] Suc) and subjected to SDS-PAGE, immunoblot analyses, and RT-PCR. Input, rice seed lysate sample before centrifugation. Histone H3 and BiP were used as nuclear and the ER markers, respectively. Glutelin and prolamine mRNA distribution was assessed by RT-PCR using the total RNA isolated from each fraction. B, Expression of RBP-L protein and glutelin/prolamine RNAs during rice seed development. Total proteins and RNAs extracted from developing rice seeds, collected daily from 1 to 12 DAF, were subject to SDS-PAGE (Coomassie brilliant-blue–stained gel) followed by immunoblot analysis or by RT-PCR using glutelin- and prolamine-specific primers (RT-PCR), respectively. Black arrow, Glutelin precursor; #, acidic subunit; *, basic subunit; arrowhead, prolamine polypeptide; Glu. mRNA, Glutelin RNA; Prol. mRNA, prolamine RNA; IB, immunoblot analysis.
Fractions were analyzed by immunoblotting to determine the distribution of nuclei and ER membranes using antibodies raised against Histone H3, as a marker for intact nuclei and chromatin, and the ER-resident BiP proteins. Intact nuclei and chromatin were located in the bottom four fractions, whereas BiP (ER) was distributed from fraction 7 to th]e denser regions of the Suc density gradient. By contrast, RBP-L was dispersed in a bimodal distribution in the Suc density gradient. Much of this RBP was concentrated in fractions 6–10. The amounts progressively decreased in fractions 11–14 until they slightly increased again in fractions 15–18. The presence of RBP-L in the bottom fractions of the gradient is consistent with the location of this RBP in the nucleus.
To assess the distribution of glutelin and prolamine RNAs, total RNAs were purified from each gradient fraction and subjected to RT-PCR using glutelin- and prolamine-specific primers. As shown in Figure 4A, glutelin and prolamine RNAs were dispersed throughout the gradient and, other than the initial six fractions of the gradient, paralleled the distribution pattern of ER membranes as denoted by the BiP marker protein. By contrast, the distribution of RBP-L only partially overlapped with ER membranes and storage protein RNAs. This observation is consistent with RBP-L having roles in gene expression other than transporting and locating storage protein mRNAs to the cortical ER.
To further explore the relationship between RBP-L and storage protein expression, the temporal expression of RBP-L and glutelin/prolamine RNAs during grain development were compared. Developing rice grains were collected daily to 12 days after flowering (DAF) for total protein and RNA extraction followed by immunoblotting with antibodies raised against RBP-l and by RT-PCR using glutelin- and prolamine-specific primers. Proglutelin precursor (arrow, Fig. 4B) was initially detected as early as 1 DAF followed by several days later by the appearance of processed acidic and basic glutelin subunits. The glutelin subunits steadily increasing as the grain developed. By contrast, prolamine polypeptides were expressed later with faint detection at 8 DAF and significant accumulation by 12 DAF.
RT-PCR results showed that glutelin RNAs were detected as early as 2 DAF, whereas expression of prolamine RNA was detected a day later (3 DAF). Both glutelin and prolamine RNAs maintained steady-state levels between 4 and 12 DAF. RBP-L exhibited a profile distinct from storage protein RNAs. Substantial accumulation of RBP-L was readily evident at 1 DAF and remained steady to 6 DAF before decreasing at subsequent stages of grain development. The relatively high expression of RBP-L at the beginning of grain development implicates a role for RBP-L as a general factor in the initiation and early events of grain development.
T-DNA Insertion in the 3′UTR Induces Down-Regulation of the RBP-L Gene
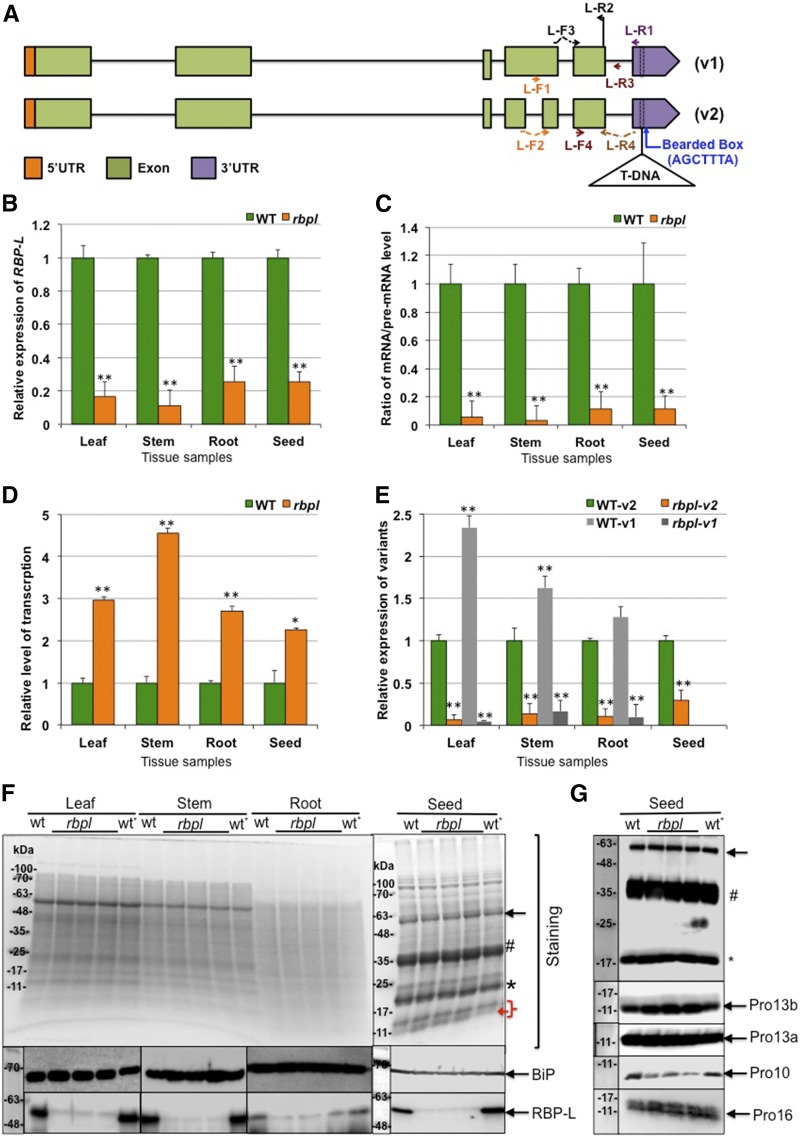
RBP-L is encoded by the rice gene with location number LOC_Os04g53440. A transgenic line with a single T-DNA insertion in the RBP-L gene, designated rbpl, was obtained from the Rice Mutant Database (http://rmd.ncpgr.cn), which contains information on over 129,000 rice japonica lines containing T-DNA insertions. The T-DNA insertion is located in the 3′UTR of the RBP-L gene, 27 basepairs downstream from the stop codon (Fig. 5A; Supplemental Fig. S2). Based on the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu), the RBP-L gene in rbpl is expressed as two splicing variants due to the absence or presence of small in-frame intron within the fourth exon (Fig. 5A). The two transcripts yield proteins of predicted molecular sizes of 50.3 kD and 46.2 kD, the latter devoid of 35 amino acids (Fig. 5A).
Figure 5.
Knockdown of RBP-L by a T-DNA insertion within its 3′UTR. A, Gene structure and T-DNA insertion site of the RBP-L gene. Two splicing variants are labeled as “v1” and “v2,” respectively. T-DNA insertion site is bordered by dashed lines in the 3′UTR. Arrows indicate the location and direction of the corresponding primers. B, Relative expression level of the RBP-L gene revealed by RT-qPCR using primer set of L-F3 and L-R2. C, The relative apparent splicing efficiency as denoted by the ratio of RBP-L mRNA/premRNA levels. RT of total RNA was performed using primer L-R1, and mRNA and premRNA levels amplified by primer sets of L-F3 + L-R2, and L-F4 + L-R3, respectively. D, Total transcription level of the RBP-L gene expression was determined by RBP-L gene transcripts (spliced and unspliced) amplified by primers L-F4 and L-R2. E, Relative expression of the two splicing variants of the RBP-L gene amplified by L-R4 with L-F1 or L-F2. All relative expression levels of RBP-L gene transcripts in rbpl line shown in (B) to (E) are normalized to that of wild type. **P value < 0.01. The expression level of the splicing variant v1 in developing seeds is not shown due to its low expression in developing rice grains. F, Expression level of RBP-L protein revealed by immunoblot using anti-RBP-L antibody. BiP levels as assessed by immunoblot analysis was used as a loading control. (Top) Coomassie brilliant-blue–stained SDS-PAGE; (center and bottom) the immunoblot results for BiP and RBP-L, respectively. The three lanes between “wt” and “wt*” present total protein samples from three individual plants of rbpl line. G, The differential expression of prolamine family proteins during grain development. Pro-10 polypeptides levels are depressed whereas Pro-13b polypeptides amounts are elevated in rbpl line. Black arrow, Glutelin precursor; #, acidic subunit; *, basic subunit; red bracket, prolamine polypeptide; red arrow, possible up-regulation of 13 kD prolamine polypeptides; wt, wild type; wt*, wild-type genotype segregated from heterozygous rbpl line (for detailed information, see “Materials and Methods”).
The relative steady-state mRNAs levels for the RBP-L gene in leaf, stem, root, and developing seed were compared between the rbpl line and wild-type. Based on quantitative PCR (qPCR) analysis using oligo(dT) as RT primer and primer pair of L-F3 and L-R2 as amplification primers (Fig. 5B), the RBP-L gene mRNA levels were depressed in the rbpl line and ranged from 11% (stem) to 25% (root and seed) of that observed in wild-type. Overall, the steady-state expression of the RBP-L gene was largely down-regulated in several tissues of the rbpl line.
We then examined how the T-DNA insertion in the 3′UTR affects the expression of the RBP-L gene at the transcriptional and posttranscriptional levels. Initially, we tested whether RNA splicing was affected. We designed a primer, L-R1, based on the 3′UTR sequence upstream of the T-DNA insertion site (Fig. 5A) for RT of RBP-L transcripts. Two pairs of specific primers (Fig. 5A) were used to amplify unspliced (premRNA, primer pair of L-F4 and L-R3) and spliced (mRNA, primer pair of L-F3 and L-R2) transcripts, enabling an estimate of splicing efficiency (mRNA/premRNA ratio). As shown in Figure 5C, splicing efficiency of RBP-L transcripts in rbpl was only 3% to 11% of that seen in wild type, suggesting that the disruption of its 3′UTR also affected processing and/or half-life of the premRNA.
We then designed specific primers (primer pair of L-F4 and L-R2) to amplify all unspliced and spliced transcripts of the RBP-L gene to assess changes at the transcriptional level. Interestingly, total levels of RBP-L RNAs were elevated 2.2-fold to over 4-fold in the rbpl line compared to that in wild type in all tissues examined (Fig. 5D). Although transcription is elevated in rbpl, levels of the final processed polyA+ containing RBP-L transcripts remain depressed.
To account for the activation of transcription of RBP-L in the rbpl line, we inspected the 3′UTR sequence of RBP-L and found a Bearded-box motif located five basepairs downstream of the T-DNA insertion site (Fig. 5A; Supplemental Fig. S2). The Bearded-box motif is a well-established 3′UTR sequence motif that mediates negative posttranscriptional regulation in Drosophila (Lai and Posakony, 1997; Leviten et al., 1997), as mutations within the Brd box motif enhanced expression of the corresponding genes (Lai and Posakony, 1997; Leviten et al., 1997). The Brd box in the 3′UTR of the RBP-L gene may also provide the same function in suppressing RBP-L expression with the loss of the Brd box enhancing premRNA expression of the RBP-L gene in rbpl. However, final expression of RBP-L at the mRNA level in the rbpl mutant remains low due to the reduction in splicing efficiency and/or mRNA turnover.
We analyzed the levels of the two splicing variants (Fig. 5E). Two pairs of primers were designed to amplify the two variants, v1 and v2. When compared to the expression of v2, we found that v1 dominated in leaf, stem, and root tissues but was absent in developing grains. The biased expression of the v2 variant in developing grains suggests that RBP-L has a more specialized role at this developmental stage.
We next assessed the expression levels of RBP-L protein in different tissues, leaf, stem, root, and seed, from wild-type and rbpl line through immunoblot analysis of total protein extracts (Fig. 5F). Although two different-sized RBP-L proteins should have been produced by the two variants, we only detect a prominent band at ∼60 kD in various tissues, a molecular size much larger than predicted for either protein variant likely due to posttranslational modification. RBP-L protein was prominently observed in wild-type tissues but only weakly detected in the rbpl line. These data are consistent with the qPCR results and confirms the down-regulation of RBP-L in the rbpl line.
Knockdown of RBP-L Affects the Expression of Storage Proteins
Glutelins are synthesized in the ER as 57-kD precursor and transported through the Golgi to the PSV where the precursors are processed into acidic (37-kD) and basic (22-kD) subunits (Takemoto et al., 2002). The prolamine gene superfamily is composed of three gene classes, which yield three kinds of polypeptides, 10-, 13-, and 16-kD prolamines based on their molecular size (Nagamine et al., 2011; Saito et al., 2012). The 13-kD prolamines (Pro-13) are further subdivided into sulfur-rich 13a and sulfur-poor 13b polypeptides, which differ by their composition of Cys residues. Pro-13 and 16-kD prolamines (Pro-16) share very high sequence similarity, whereas the 10-kD prolamines (Pro-10) share very little sequence similarity with the two other classes. Due to the high abundance of glutelins and prolamines in rice seed, accounting for 70% to 80% and 5% to 10% of the total proteins, respectively, the expression levels of glutelin and prolamine can be easily assessed by Coomassie Blue-stained SDS-PAGE (Fig. 5F, right). Whereas the accumulation of glutelins, including the 57-kD precursor and the two subunits, was not substantially altered in the rbpl line, the expression levels of Pro-13 were slightly elevated in the rbpl line. To confirm these results, we evaluated the protein expression by immunoblots using specific antibodies to each of the major prolamine classes (Fig. 5G). Consistent with the SDS-PAGE results, the expression of glutelin was unaffected in the rbpl line when compared to wild type. Whereas Pro-16 and Pro-13a levels remained the same, immunoblots revealed elevated amounts of Pro-13b in the rbpl line and decreased levels of Pro-10. These results suggest that knockdown of RBP-L expression affected the expression of specific prolamine gene classes.
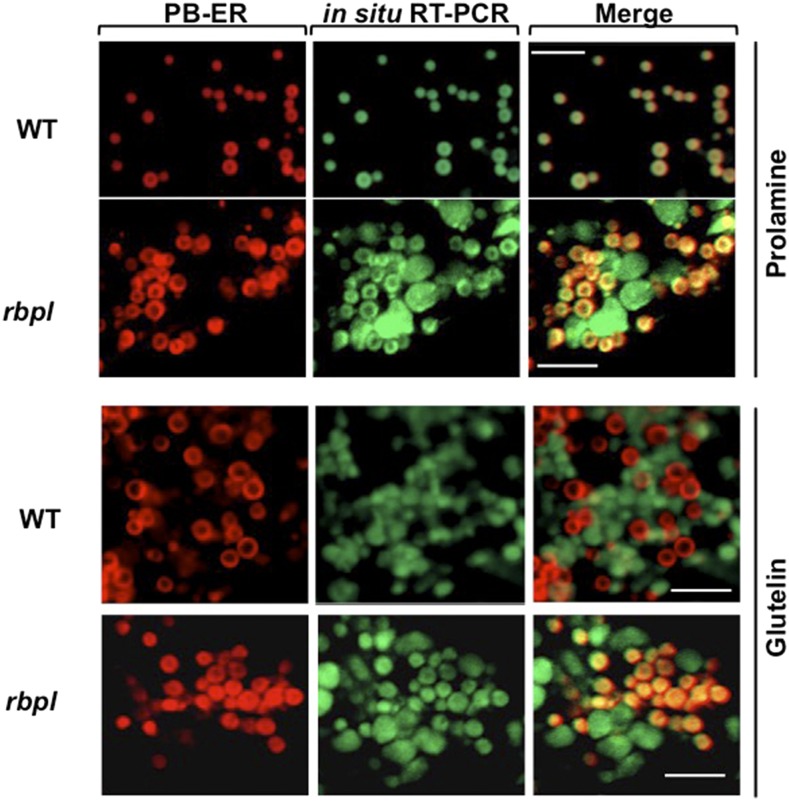
Knockdown of RBP-L Results in Mislocalization of Glutelin and Prolamine mRNAs
We performed in situ RT-PCR on rice seed to see whether the knockdown of RBP-L affected localization of glutelin and prolamine mRNAs (Fig. 6). In wild type, prolamine mRNA are transported to the PB-ER, the ER membrane selectively stained by Rhodamine B, whereas glutelin are targeted to adjacent cisternal-ER. Partial mislocalization patterns of glutelin and prolamine RNAs were observed in the rbpl line. Although prolamine RNAs were observed at its normal location on the PB-ER, it was readily detected on the cisternal-ER. Similarly, glutelin mRNAs were observed on its normal destination site on the cisternal-ER but also present on the PB-ER. The deficiency in RBP-L leads to the partial mislocalization of prolamine and glutelin mRNAs, suggesting that RBP-L is necessary and sufficient for the accurate localization of glutelin and prolamine mRNAs in developing rice endosperm cells.
Figure 6.
Knockdown of RBP-L results in the partial mislocalization of glutelin and prolamine mRNAs on the cortical-ER as revealed by in situ RT-PCR. In situ RT-PCR was performed directly on developing rice grain sections in the presence of Alexa-488-UTP (green) and specific primers to label prolamine and glutelin mRNAs. PB-ER was stained using Rhodamine B dye (red). Note that prolamine and glutelin mRNAs are localized to the PB-ER (red) and cisternal-ER, respectively, in wild type. In the rbpl line, glutelin and prolamine mRNAs are distributed on both the PB-ER and cisternal-ER. Scale bar = 5 μm. WT, wild type.
Knockdown of RBP-L Confers Dwarf and Late Flowering
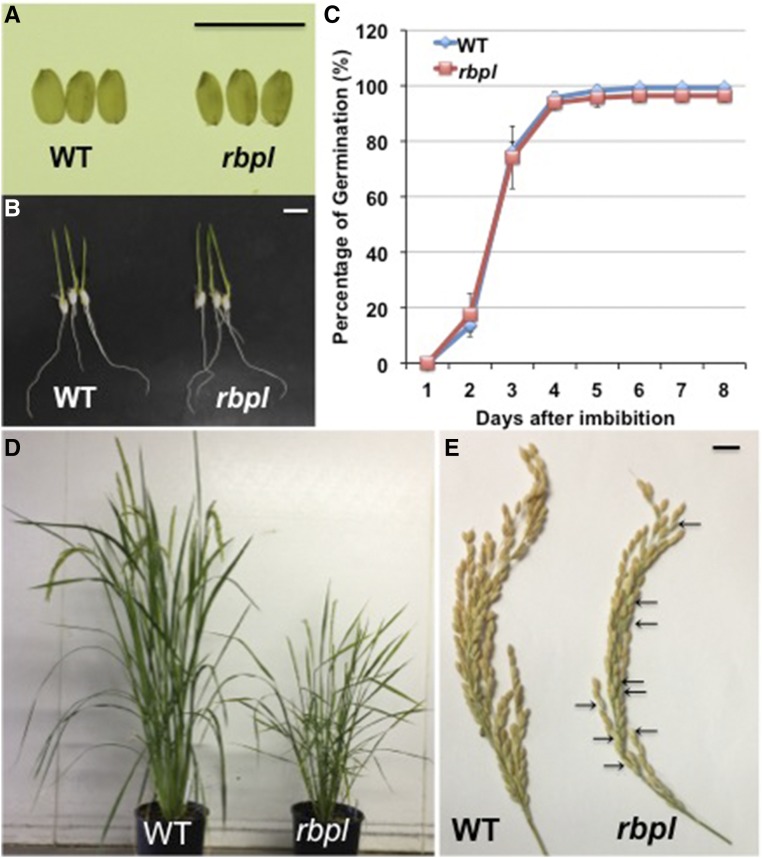
The rbpl line exhibited significant alterations in growth and development. While the rbpl line showed normal germination rates on d 5 after imbibition like wild type, subsequent growth was severely impacted as the rbpl line exhibited a semidwarf phenotype with final plant height of 47.6 cm compared to the 88.8 cm of wild type. Changes were also readily evident at the reproductive stage of rice development. The flowering time of rbpl line was delayed about a week compared to wild type. The rbpl line showed a reduced number of spikelets per panicles and significantly reduced spikelet fertility, specifically 62.8% compared to the 97.5% of wild type. Although grain size and shape produced from the homozygous rbpl line were not visibly different from that of wild type, the average seed weight was slightly lower (Fig. 7; Table 1). These phenotypes were not displayed in RBP-L wild-type plants that segregated from heterozygous rbpl (Table 1). Hence, the abnormal growth and development exhibited by the rbpl line resulted from the T-DNA disrupted RBP-L gene and not by epigenetic effects generated during the plant transformation process.
Figure 7.
Knockdown of RBP-L confers several growth defects in rbpl line in comparison to wild type. A, Seed morphology of wild-type and rbpl line. Scale bar = 1 cm. B, Germinated wild-type and rbpl seeds after 6 d of imbibition. Scale bar = 1 cm. C, Germination rate of wild-type and rbpl seeds. Seeds were considered germinated when the radicle was extended >1 cm or more. D, The rbpl line showed late flowering and dwarfism. When rbpl line plants were blooming, the grains from wild-type plants were approaching maturity. E, The rbpl lines showed lower spikelet fertility than wild type. Some empty glumes on the rbpl tassel are indicated by arrows. WT, wild type.
Table 1. Plant growth parameters of wild-type (ZH11), rbpl line and its segregated wild-type RBP-L sibling.
Each of the traits in mutants was compared to wild-type using the Student’s t test. *P value of two tailed t test < 0.05; **P value of two tailed t test < 0.01; wt*, the segregated wild-type plant from heterozygous rbpl line.
| Genotype | Mature Plant Height (cm)a | Days to Floweringa | No. of Tillers per Planta | No. of Spikelets per Panicle | Spikelet Fertility (%)a | Seed Weight (mg)b |
|---|---|---|---|---|---|---|
| WT | 88.8 ± 3.7 | 59.0 ± 0.7 | 10.2 ± 4.3 | 113.7 ± 11.0 | 97.5 ± 1.4 | 22.7 ± 1.9 |
| wt* | 89.4 ± 3.2 | 59.4 ± 1.3 | 11.2 ± 1.4 | 110.6 ± 9.7 | 97.5 ± 1.3 | 22.6 ± 2.0 |
| rbpl | 47.6 ± 3.1** | 68.8 ± 1.8** | 11.7 ± 3.7 | 99.1 ± 14.0* | 62.8 ± 9.7** | 20.9 ± 1.6** |
Data are shown as the mean ± sd (n = 10).
Data are shown as the mean ± sd (n = 100).
RBP-L Regulates the Expression of Many Genes
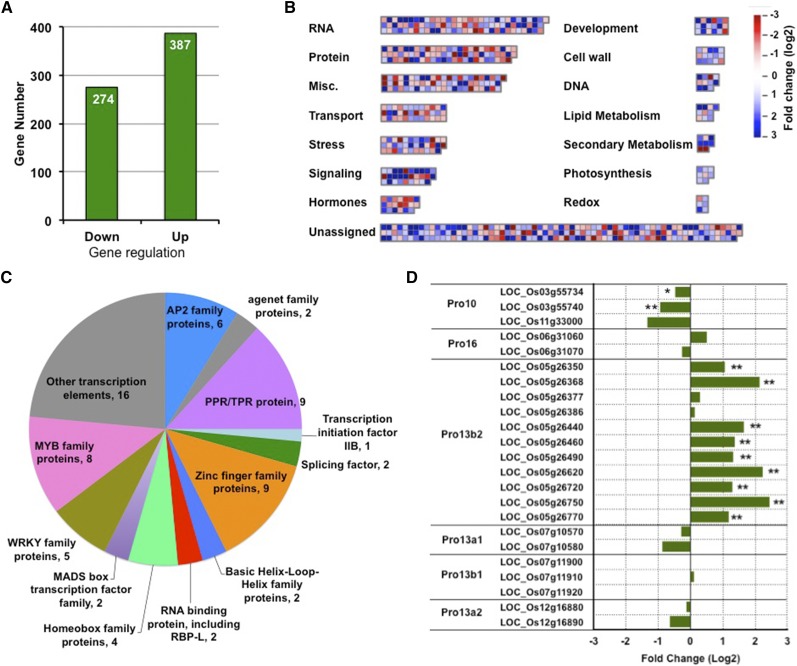
To address the underlying basis for the changes in expression of storage protein genes and other genes involved in rice development in the rbpl line, we performed high-throughput transcriptome analysis. Three biological repeat samples of total RNA were isolated from developing grains collected from three individual wild-type or three individual rbpl plants for RNA sequencing (RNA-seq). The three biological samples of wild type gave average total reads (average Reads Per Kilobase of transcript, per Million mapped reads [RPKM] > 0) of 30,223, while rbpl gave similar values of 30,151 (Supplemental Table S1), indicating that RBP-L knockdown did not result in a considerable overall change in the transcriptome. Consistent with our qPCR results, the expression of the RBP-L gene was 26% of that in wild type (Supplemental Table S2). The splicing variant1 was expressed at an almost undetectable level in grain tissue with a RPKM of 0–0.05 (Supplemental Table S2). This result confirms our earlier observation on the very low expression of variant1 in developing grains and that RBP-L variant2 is the major form regulating gene expression in developing rice grains.
To identify genes that were differentially expressed between rbpl and wild type, we used a cutoff of log2 fold change > 1 and a P value < 0.01. Based on these criteria, a total of 387 genes were up-regulated, whereas 274 genes were down-regulated in rbpl (Fig. 8A; Supplemental Table S3). We further analyzed the pathways involved by the 661 differentially expressed genes (DEGs) using MapMan software (Fig. 8B; Supplemental Table S4). A little less than a third (198) of the total were transposons, expressed proteins, and other proteins with unknown function, and were not assigned to any pathway. Among the remaining genes, the RNA metabolic pathway involving transcriptional processes was a prominent group consisting of 92 genes. These included nine pentatricopeptide repeat/tetratricopeptide repeat proteins, nine zinc finger proteins, six AP2 domain containing proteins, two Agenet-domain–containing proteins, two RRM domain RBPs (including RBP-L), two splicing factors, one transcription initiation factor, and 21 transcriptional factors from the MYB, WRKY, MADS box, basic Leu zipper, homeobox, or helix-loop-helix domain families (Fig. 8C; Supplemental Table S5). Two other highly enriched pathways consisted of miscellaneous enzyme families and protein metabolic pathways, composed of 61 and 60 genes, respectively. Other enriched processes included those involved in signaling, transport, stress, hormones, development, and cell wall, which contained 15 or more genes.
Figure 8.
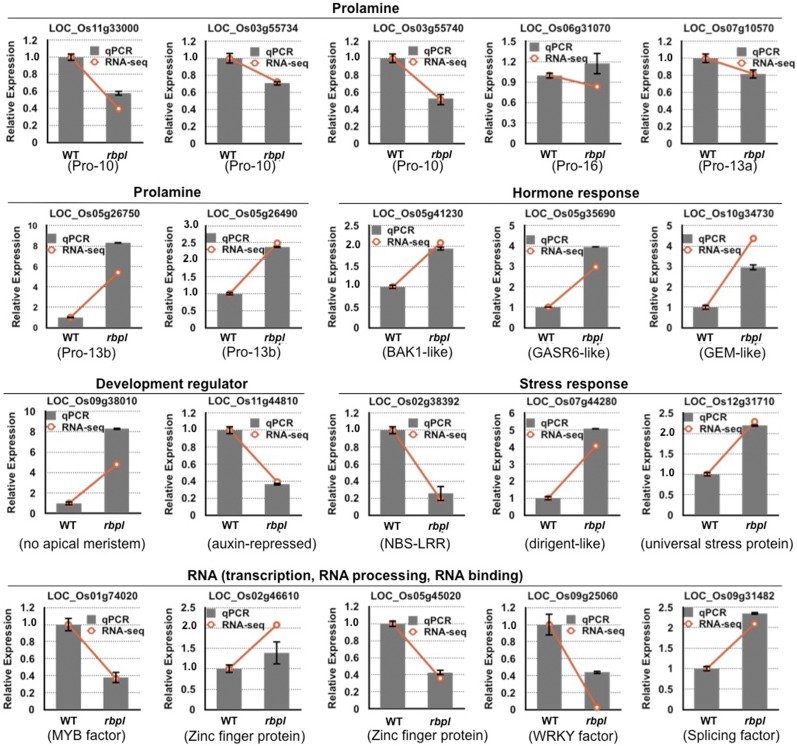
Transcriptome changes mediated by knockdown of RBP-L expression. A, Chart showing the numbers of down- and up-regulated genes (log 2 fold change > 1, P value < 0.01) in rbpl developing seeds compared to that in wild-type. B, Distribution of molecular function and biological pathways that were enriched by the 661 DEGs in the rbpl line. C, Composition of the DEGs involved in RNA metabolic pathways. D, Fold changes on gene expression of prolamine family members. Prolamine genes with relative low RPKM (<1) were not analyzed and are not shown in the chart. *P value of two-tail t test < 0.05; **P value of two-tail t test < 0.01, based on RNA-seq data. Detailed information shown in this figure can be found in Supplemental Tables S2–S6.
We then evaluated the expression of prolamine and glutelin gene families. Whereas the 15 genes from glutelin family were not significantly altered, differential expression was readily observed for members of the prolamine gene family (Fig. 8D; Supplemental Table S6). The prolamine gene family consists of 31 genes. These include three genes for Pro-10, two for Pro-16, six for Pro-13a, and 22 for Pro-13b. Based on chromosome location, the Pro-13 family was further divided into four groups, two genes for 13a1, four for 13a2, three for 13b1, and 19 for 13b2 (Fig. 8D; Supplemental Table S6). As shown in Figure 8D, many of the Pro-13b2 genes on chromosome 5 were significantly up-regulated in rbpl line. In contrast, the three Pro-10 genes were down-regulated (Fig. 8D; Supplemental Table S6), especially LOC_Os03g55734 and LOC_Os03g55740 (P values < 0.05). The up-regulation of Pro-13b and down-regulation of Pro-10 genes were in agreement with the increased protein accumulation of Pro-13 kD and decreased levels of Pro-10 kD (Fig. 5G). In general, RBP-L is a key regulator for gene expression of specific prolamine classes but not for glutelin.
qPCR analysis was further performed to verify RNA-seq results (Fig. 9). First, we tested the expression of the prolamine and glutelin genes. Due to the high sequence identity among Pro-13b genes, only two pairs of primers that showed good melting curves for qPCR were used to verify the expression of this gene class. RT-qPCR analysis indicated that the expression of the two selected Pro-13b genes were indeed up-regulated in the rbpl mutant line. We also selected three Pro-10 genes, one Pro-16 gene, and one Pro-13a gene for qPCR to verify the expression of the other prolamine genes. Consistent with RNA-seq results, the expression of the three Pro-10 genes was reduced, whereas expression of Pro-16 and Pro-13a was not significantly altered in the rbpl mutant line. Both RNA-seq and qPCR results support the view that RBP-L selectively regulates the expression of Pro-10 and Pro-13b gene subfamilies of the prolamine superfamily.
Figure 9.
Validation of DEGs in rbpl by RT-qPCR. Analysis of selected genes from prolamine family, hormone signaling pathway, development, stress response, and RNA metabolism confirmed the differentially expressed patterns determined by RNA-seq. Gray columns indicate the fold-changes based on qPCR results, whereas orange lines show the data obtained from RNA-seq. y axis, fold changes relative to that of wild-type. Error bars shown on gray column represent ses of the mean fold changes calculated from three qPCR replicates.
Several genes encoding regulatory factors of essential biological pathways were also further tested for experimental validation. These genes from four representative categories covered hormone metabolism, development regulation, stress response, and the RNA metabolic pathway. Among the hormone-related genes, LOC_Os05g41230, a putative Brassinosteroid insensitive 1-associated receptor kinase1, is an important protein that plays various roles in Brassinosteroid signaling, stress defense, and plant development (Chinchilla et al., 2009; Belkhadir and Jaillais, 2015; Liu et al., 2017). LOC_Os05g35690 encodes GASR6, which is reported to be highly expressed in rice seeds and is a key factor in regulation of Gibberellic acid biosynthesis essential for seed development (Xue et al., 2012). LOC_Os10g34730 encodes a putative GL2 expression modulator, an abscisic acid (ABA) and stress-responsive protein that functions in the ABA signaling pathway and also regulates inflorescence architecture, seed development and germination (Baron et al., 2014; Mauri et al., 2016). The expression of these hormone-responsive genes was elevated in the rbpl line. Another hormone-responsive gene, LOC_Os11g44810, which encodes an auxin-repressed protein, showed reduced expression in the rbpl line. This “auxin-repressed protein” functions in plant growth, development, and disease resistance in tobacco (Zhao et al., 2014). Another tested development-relevant gene is LOC_Os09g38010, which encodes a putative “no apical meristem” protein. No-apical-meristem proteins, which are essential for embryo formation and floral organ structure (Cheng et al., 2012), showed a 5-fold up-regulation in the rbpl line.
The RBP-L knockdown also perturbed the expression of stress-relevant genes. Three stress responsive genes, nucleotide-binding site (NBS)-Leucine-rich repeat (LRR; LOC_Os02g38392), dirigent (LOC_Os07g44280), and universal stress protein (LOC_Os12g31710) were selected for validation. NBS-LRR disease resistance proteins are characterized by their NBS and LRR domains, and respond to a variety of bacterial and fungal pathogens and insect pests (McHale et al., 2006). This gene was largely down-regulated in the rbpl line. Dirigent proteins are extracellular glycoproteins and modulate cell wall metabolism when the plants are exposed to stress (Pickel and Schaller, 2013; Paniagua et al., 2017). Universal stress protein was reported to be redox-dependent chaperones during heat and oxidative stress (Jung et al., 2015). The expression of the dirigent and universal stress protein genes were activated 4-fold and 2-fold in the rbpl mutant line, respectively.
Five genes from the RNA metabolic pathway were selected to verify the function of RBP-L in RNA regulation. Many genes affected in the rbpl mutant line are transcriptional factors (Fig. 8C), suggesting that RBP-L may function as key regulator in gene expression through its regulation or interaction with these regulatory proteins. The two tested transcription factors, a MYB transcription factor (LOC_Os01g74020) and a WRKY transcription factor (LOC_Os09g25060), showed largely reduced expression in the rbpl line confirming their regulation by RBP-L. The differential expression of three genes coding zinc finger motif-containing proteins, including splicing factor U2AF (LOC_Os09g31482) and two zinc finger transcription factors (LOC_Os02g46610 and LOC_Os05g45020) were also confirmed by qPCR analyses.
PB-I Displays an Abnormal Structure in the rbpl Line
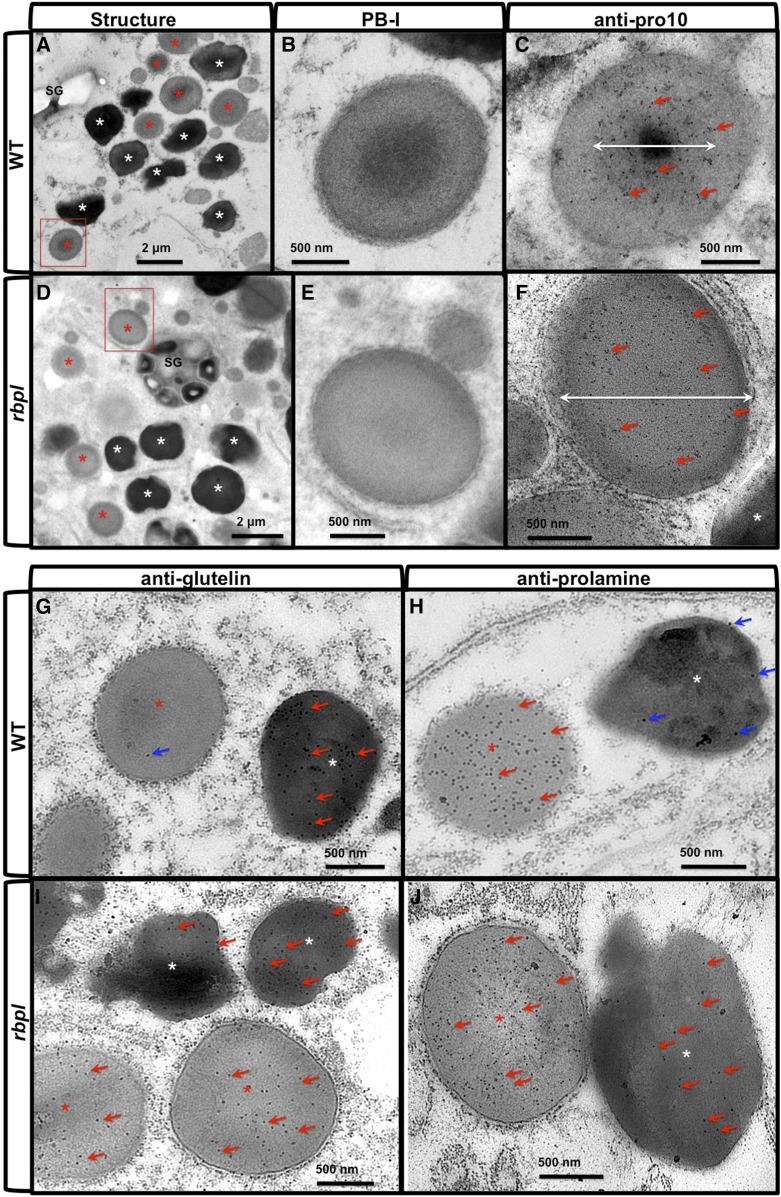
Pro-10s accumulate at the electron-dense center core in PB-I (Nagamine et al., 2011). Repression of Pro-10 expression by RNAi strategy resulted in the formation of irregular-shaped PB-I devoid of the electron-dense core (Nagamine et al., 2011). Hence, Pro-10 is required for the formation of normal PB-I where it facilitates packaging of prolamine proteins into a compact spherical structure. To determine whether the reduction of Pro-10 proteins in the rbpl line conferred structural changes within PB-I, we investigated the ultrastructure of rbpl endosperm cells via immunocytochemical transmission electron microscopy (TEM) analysis (Fig. 10, A–F). In wild-type endosperm cells, PB-I displays a spherical lamellar morphology with the usual accumulation of Pro-10 proteins in the electron-dense center core (Fig. 10, A–C). Although many PB-I in rbpl endosperm cell retain their spherical structure, there was no obvious electron-dense core at the center of PB-I. Moreover, the distribution of Pro-10 protein was not restricted to the center core region of PB-I but distributed throughout the intracisternal granule (Fig. 10, D–F). The PB-I in rbpl also displayed a more uniform electron density. These findings indicate that the reduction of Pro-10 proteins impaired the normal packaging of prolamine proteins into PB-I.
Figure 10.
PB-I structure and localization of glutelin and prolamine proteins in wild-type and rbpl line endosperm cells. A to F, Ultrastructure of PB-I in wild-type (A–C) and rbpl line (D–F) endosperm cells. B and E, Enlarged picture of the areas indicated by the red boxes in (A) and (D). C and F, Immunolabeling patterns using anti-prolamine (Pro-10) antibody and 10-nm gold particle-conjugated secondary antibody in wild type (C) and rbpl (F) PB-I, respectively. White double-headed arrows indicate the primary distribution area of Pro-10 proteins within PB-I. Scale bar = 2 μm (A and D) or 500 nm (B, C, E, and F). Red and white asterisks in (A) and (D) denote PSV and PB-I, respectively. G to J, Immunolabeling of glutelin (G, I) and prolamine (H, J) proteins using monospecific antibodies and 15-nm gold particles-conjugated secondary antibodies. G and H, wild-type. I and J, rbpl line. Red arrows denote positive gold particle labeling, and blue arrows denote slight background nonspecific labeling. Scale bar = 500 nm. WT, wild type.
As mislocalization of prolamine and glutelin mRNAs was observed in rbpl line, we studied whether the encoded proteins were also mistargeted (Fig. 10, G–J). In wild-type endosperm cells, with the exception of a couple of gold particles nonspecifically detected on PB-I, glutelin proteins are exclusively located in PSV (Fig. 10G). In rbpl endosperm cells, however, glutelin proteins were also detected in PB-I in addition to the PSV (Fig. 10I). In the case of prolamine, localization of the dominant 13-kD prolamine was determined. Similar to the analysis on glutelin, although a few nonspecific gold particle background labeling could be observed in PSV, the labeling of prolamine was exclusively restricted to PB-I in wild-type endosperm cells (Fig. 10H). However, in rbpl endosperm cells, distribution of prolamine was observed in both PB-I and PSV (Fig. 10J). These results indicate that mislocalization of glutelin and prolamine mRNAs misdirected the final deposition of their coded protein products.
DISCUSSION
In developing rice grains, proper localization of glutelin and prolamine mRNAs to distinct subdomains of the cortical ER membrane complex is necessary for the efficient packaging of their coded proteins in the PSV and PB-I, respectively. mRNA localization process is initiated by transfactors, RBPs that recognize and bind to the mRNA’s zipcode elements. Loss of function of these zipcode binding RBPs results in mislocalization of these mRNAs.
Our studies described here readily demonstrate that RBP-L is an important RBP for the transport and localization of glutelin and prolamine mRNAs. It specifically recognizes glutelin and prolamine zipcodes. RBP-L colocalizes with PB-ER, the site of prolamine RNAs and with glutelin RNA patches associated with the cisternal-ER. T-DNA insertion in the 3′UTR of RBP-L mRNA resulted in a 75% decrease in its expression in developing rice grains and led to a partial mislocalization of both glutelin and prolamine mRNAs. The mislocalization of these mRNAs also resulted in the mistargeting of the coded glutelin and prolamine proteins from their normal site of protein accumulation.
Expression of the RBP-L gene Is Regulated by its 3′UTR
The significant reduction (75% to 90%) of RBP-L gene expression in various rice tissues mediated by T-DNA insertion in the 3′UTR illustrates the importance of the 3′UTR in gene regulation. The 3′UTR plays crucial roles in gene regulation at the posttranscriptional level; these include premRNA processing, mRNA export from the nucleus to cytoplasm, mRNA stability, and translation efficiency (Mignone et al., 2002; Matoulkova et al., 2012). The underlying basis for these multiple roles is that the 3′-UTR contains cis-elements recognized by transacting factors and microRNAs (Mignone et al., 2002; Matoulkova et al., 2012). For instance, interaction of miRNA23b at the K box 1 motif located in the 3′UTR of the mouse μ-opioid receptor suppressed the translation of mouse μ-opioid receptor mRNA by inhibiting the association of mRNA with polysomes (Wu et al., 2008). Several reports on alterations in gene regulation are achieved by changes in 3′UTR sequences of plant genes. Examples include the insertion of a miniature inverted-repeat transposable element into the 3′UTR, which enhanced the transcription of the TaHSP16.9 gene in wheat (Li et al., 2014), whereas a T-DNA insertion in the 3′UTR of a flowering-relevant gene, At4g20010, severely reduced its expression in several different tissues and delayed flowering (Svensson et al., 2005).
T-DNA insertion in the 3′UTR drastically altered the expression of the RBP-L gene in rbpl rice line at the transcriptional and posttranscriptional levels. The reduction of RBP-L gene expression due to the T-DNA insertion in its 3′UTR was observed in several tissue types including leaf, stem, root, and seeds. This non-tissue–dependent regulation suggests a possible self-regulation of RBP-L by its 3′UTR. The T-DNA is inserted 27 nucleotides downstream of the TGA stop codon and, hence the mRNA is missing 251 nucleotides of its normal 3′ end including the polyadenylation signal and the negative regulator Brd-box motif (Supplemental Fig. S2). The elevated levels of total RBP-L RNAs (spliced and unspliced) suggest a possible self-regulation of RBP-L via the 3′UTR Brd-box motif, a known negative regulator. Despite the elevated levels of total (spliced and unspliced) RNA transcripts, only 3% to 11% of mature RNA transcripts was observed in the rbpl line due to lower splicing efficiency of splicing and/or RNA turnover. The 3′UTR is not known to possess any sequence or motif involved in mRNA splicing (Bilenoglu et al., 2002). One possible reason is that the truncated 3′UTR may affect the secondary structure of the RNA, which further interferes with the recognition of the splice-site by the relevant proteins. Alternatively, the loss of the normal polyadenylation signal via the T-DNA insertion in the 3′UTR may lead to a higher turnover of premRNA.
The Roles of RBP-L in Glutelin and Prolamine mRNA Localization
The reduction of RBP-L mediated the partial mislocalization of both glutelin and prolamine mRNAs (Fig. 6) suggesting that maintenance levels of RBP-L protein are critical for proper and efficient targeting of these mRNAs. This stringent condition is consistent with the requirement for two zipcodes to direct RNA localization with the presence of a single zipcode resulting in partial RNA mislocalization (Hamada et al., 2003; Washida et al., 2009a). The key regulatory performance of RBP-L in glutelin and prolamine mRNA localization is supported by its direct high-affinity binding to the zipcode RNAs that determine the final localization onto cisternal-ER and PB-ER, respectively (Fig. 2). Colocalization of RBP-L with glutelin and prolamine mRNAs on the cisternal-ER and PB-ER, respectively (Fig. 3C) further supports the involvement of RBP-L in mRNA localization.
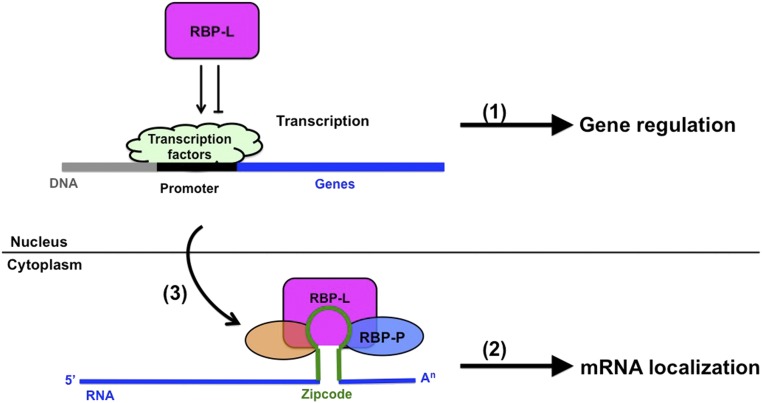
There are two underlying mechanisms on how RBP-L may modulate glutelin and prolamine mRNA localization (Fig. 11). The first mechanism consists of the direct involvement of RBP-L in glutelin and prolamine mRNA localization. RBP-L directly binds to glutelin and prolamine zipcode RNAs, which serves as an important platform to further recruit other RBPs and factors to drive mRNA transport to their destination. The process likely requires cooperation from its interacting partners, including RBP-P and RBP208, as described in our previous study (Tian et al., 2018). These RBPs may form a multiprotein complex that binds to the zipcode RNAs and precisely define the targeting of glutelin and prolamine mRNAs. The reduction in protein levels as demonstrated by the reduction of RBP-L expression in this study or their binding activity to RNAs and proteins as shown for the mutant RBP-Ps in our previous study (Tian et al., 2018), results in the mislocalization of glutelin and prolamine mRNAs.
Figure 11.
A proposed working model to show the critical roles of RBP-L in rice glutelin and prolamine mRNA localization and gene expression. (1) Through posttranscriptional control of genes that encode regulation on transcription factors, RBP-L indirectly regulates the transcription of prolamine genes, particularly Pro-10 and Pro-13b genes. This regulatory mechanism can also be extended to other genes that are regulated by RBP-L. (2) Through its direct binding affinity to glutelin and prolamine zipcode RNAs, RBP-L together with RBP-P (Tian et al., 2018) and other factors drive storage protein mRNAs to their destination on subdomains of the cortical-ER. (3) RBP-L may also regulate glutelin and prolamine mRNA localization through its regulation on the expression of other RBPs (orange oval) that are components of the mRNP complex.
In addition to its direct involvement in mRNA localization events, RBP-L may also regulate the expression of other RBPs that are involved in mRNA localization, which subsequently affects glutelin and prolamine mRNA localization in an indirect way. Based on the RNA-seq data, RBP-L is required in the gene regulation of many transcription factors and RBPs, including zinc finger proteins, pentatricopeptide proteins, and RRM-motif–containing protein (Fig. 8). The reduction of RBP-L levels result in the up- or down-regulation of these genes (Supplemental Table S5), which may also further affect the proper assembly of the mRNP complex and contribute to the mislocalization of glutelin and prolamine mRNAs.
RBP-L is an ortholog of the RBP45/47 family proteins, which assemble with nuclear poly(A)+ RNA (Lorković et al., 2000) and participate at several steps of RNA processing and maturation in the nucleus and in the export of mRNAs from the nucleus to the cytoplasm (Lorković et al., 2000; Lewandowska et al., 2004; Weber et al., 2008; Sorenson and Bailey-Serres, 2014). Given that RBP-L may also perform a similar function as reflected by its location in both the nucleus of cytoplasm, the regulation of transcription factors and RBPs by RBP-L is likely contributed to by its involvement in RNA processing, maturation, stability, and turnover of these target genes. The deficiency in RBP-L levels in the rbpl line may prevent optimal processing of premRNAs for transcription factors and RBPs in nucleus, and in turn, alter gene expression. These two mechanisms on the role of RBP-L in mRNA localization and posttranscriptional regulation are not mutually exclusive and may be coupled and further studies are required to delineate RBP-L’s function in these processes.
RBP-L Regulates the Expression of Prolamine Gene
In addition to its participation in glutelin and prolamine mRNA localization, RBP-L was found to alter the RNA levels of specific prolamine gene classes. The reduced levels of RBP-L decreased the expression of the Pro-10 genes but increased the expression of the Pro-13b genes (Fig. 8D; Supplemental Table S6). Therefore, RBP-L not only directs the localization of storage protein mRNAs to specific subdomains of the cortical-ER (Hamada et al., 2003; Crofts et al., 2004; Washida et al., 2009a) but also specifically influences the steady-state RNA levels of the Pro-10 and Pro-13b genes. This control on transcriptional levels of Pro-10 and Pro-13b is independent of RNA localization as overall expression of glutelin genes and the remaining prolamine genes classes (Pro-16 and Pro-13a) are not affected by the reduction in RBP-L.
The reduction in Pro-10 RNA levels in rbpl suggests that expression of Pro-10 is dependent on adequate levels of RBP-L. Conversely, expression of Pro-13b is inversely related to RBP-L expression. As the Pro-16 and Pro-13a prolamine gene subfamilies are not affected in rbpl, it is unlikely that RBP-L directly controls the transcription of Pro-10 and Pro-13b genes.
Mutations in the RBP-L interacting partner RBP-P also affected prolamine gene expression (Tian et al., 2018), although the regulatory pattern differed substantially from that seen for RBP-L. Unlike the differential gene expression patterns evident for Pro-10 and Pro-13b gene subfamilies in the rbpl line, RBP-P mutations affected the expression of specific prolamine genes that span four subfamilies: one Pro-10, one Pro-13a1, two Pro-13b2, and one Pro-16. Only the Pro-10 gene (LOC_Os03g55740) and one Pro-13b2 gene (LOC_05g26620) were down- and up-regulated, respectively, in both the RBP-P and RBP-L mutant lines, whereas the remaining three were not affected in the rbpl line. These observations suggest that expression of prolamine multigene family is complex where specific subfamilies and genes within a subfamily may be controlled by different regulatory mechanisms. Obviously, RBP-L has a more prominent role in governing prolamine gene expression.
Given the close homology of RBP-L to the RBP45/47 family proteins, RBP-L likely regulates gene expression indirectly by regulating other transcription factors. Transcription factors play a crucial role in regulation of gene expression (Schwechheimer et al., 1998; Lelli et al., 2012), and many transcription factors from the AP2, bZIP, MADS, and WRKY families have been reported to regulate seed development and seed storage protein gene expression (Lara et al., 2003; Arora et al., 2007; Chandler et al., 2007; Masiero et al., 2011; Zhang et al., 2011; Xue et al., 2012). In this study, the reduction of RBP-L significantly altered the expression of more than 50 transcription factors belonging to several large classes. Although the functions of the rbpl-affected transcription factors are undefined, one or more may regulate the transcription of Pro-10 and Pro-13b genes. One candidate is OsMADS78 (LOC_Os09g02830), whose transcripts showed specific expression in rice grains of 5–20 d after pollination (Arora et al., 2007), a stage where prolamine storage proteins accumulate. The expression of this gene was increased almost 3-fold in the RBP-L knockdown line, which may affect the expression of specific prolamine gene subfamilies. Clearly, the transcription factors that control the expression of the Pro-10 and Pro-13b prolamine genes require further study.
Taken together, we propose a working model on the two important roles of RBP-L in rice storage protein biosynthesis (Fig. 11). RBP-L plays an essential role in prolamine and glutelin mRNA localization in rice endosperm cells through its direct binding to the zipcode RNAs and assembly of the relevant RNP complex to transport storage protein mRNAs to their destination sites on the cortical-ER. The formation of the RNP complex also requires the zipcode binding RBP-P, which directly interacts with RBP-L in the nucleus and cytoplasm. In addition to its role in mRNA localization, RBP-L also regulates gene expression, including prolamine genes, likely through its modulation on posttranscriptional regulation of the corresponding transcription factors. This indirect gene regulation may also modulate the expression of other RBPs that are involved in the assembly of the mRNP complex for mRNA transport, which further impacts mRNA localization in rice endosperm cells.
RBP-L Plays an Important role in Rice Development
The regulatory influence mediated by the RBP-L knockdown is not limited to storage protein genes. The genes involved in RNA metabolic pathway, protein metabolism, enzymes, signaling/transport, and stress and hormone responses are enriched among the DEGs in the RBP-L knockdown line (Fig. 8; Supplemental Table S4). As RBP-L is widely expressed in various organs and tissues during the entire growth cycle of the rice plant (Supplemental Fig. S3), RBP-L has a broad function during plant growth and development. The general role of RBP-L is also reflected by the severe dwarfism, late flowering and, fewer and smaller seeds (reduced fertility) exhibited by the rbpl line. Moreover, there is indirect cytological and biochemical evidence for an extensive role for RBP-L in plant growth and development. Although RBP-L colocalizes with the location of prolamine and glutelin RNAs on the cortical-ER, it is also distributed to other areas of the cisternal-ER devoid of glutelin RNAs presumably in localizing nonstorage protein mRNAs. Moreover, the bulk of RBP-L is not strictly associated with glutelin and prolamine RNAs and ER on Suc density gradients, inferring some role other than RNA localization.
Similar to the regulation in storage protein gene expression, RBP-L may also modulate the expression of developmental-relevant genes by its direct RNA-binding properties to regulate several aspects of RNA processing pathways and/or by its indirect effects on transcription factors. Given the central role of transcription factors in controlling gene expression and, in turn, many essential aspects of plant growth and development (Magome et al., 2004; Pandey and Somssich, 2009; Dubos et al., 2010; Masiero et al., 2011; Ambawat et al., 2013; Perotti et al., 2017), the requirement of RBP-L for the proper expression of many transcription factors suggests that RBP-L plays an important role in plant development.
In addition to their roles in RNA processing, maturation, and nuclear export, RBP45/47 family proteins respond to stress stimulus of heat, salt, and hypoxia and are specific markers for stress granules (Weber et al., 2008; Yan et al., 2014; Lokdarshi et al., 2016; Chantarachot and Bailey-Serres, 2018). In Arabidopsis, RBP47 is localized to both the nucleus and cytoplasm under normal growth conditions but is redistributed and reorganized into stress granule foci in the cytoplasm upon stress treatment (Lorković et al., 2000; Lewandowska et al., 2004; Weber et al., 2008; Sorenson and Bailey-Serres, 2014). Although specific details on the role of the RBP45/47 family proteins in the stress response remains unclear, they likely facilitate stress granule assembly by recruiting cytoplasmic mRNAs (Lewandowska et al., 2004; Weber et al., 2008; Sorenson and Bailey-Serres, 2014; Lokdarshi et al., 2016). While RBP-L, a member of the RBP45/47 family, may also be involved in stress granule assembly, it is interesting to note that it influences the expression of 35 genes involved in stress response. These include heat shock proteins, defensin family proteins, NBS-LRR protein, dirigent-like protein, and universal stress protein (Fig. 8B; Supplemental Table S4).
It is noteworthy that RBP-L regulates several genes in hormone metabolism and response (Fig. 8B; Supplemental Table S4). Whereas knockdown of RBP-L expression resulted in the down-regulation of two genes coding two auxin-repressed proteins, the majority of the affected hormone-related genes were up-regulated in the rbpl mutant line. Specifically, the expression levels of one auxin-inducible protein, three auxin-responsive SAUR family members, the brassinosteroid signal transducer Brassinosteroid insensitive 1-associated receptor kinase1-like protein, the gibberellin-regulated GASA/GAST/Snakin family protein and the ABA-responsive GL2 expression modulator were up-regulated 2- to 5-fold (Supplemental Table S4). RBP-L apparently serves as negative modulator in several hormone-signaling pathways through its regulation on these key genes.
Collectively, this study demonstrates that RBP-L is an essential RBP that drives prolamine and glutelin mRNA localization in rice endosperm cells through its strong binding to their zipcode RNAs. Its proper expression is regulated by motifs located in its 3′UTR and is required for normal plant growth and development owing to its effects on many genes encoding transcription factors and other factors essential in several biological processes.
MATERIALS AND METHODS
Plant Materials and Growth Condition
Oryza sativa wild-type (ZH11) and rbpl rice lines were obtained from the Rice Mutant Database. The rbpl lines were grown at WA State University for three generations to obtain stable homozygous genotype lines. The segregated wild-type line carrying a normal RBP-l gene was collected as a control transgenic line. All wild-type and rbpl lines were grown in a walk-in growth chamber with 12-h light, 12-h dark photoperiod at 27°C. Seed germination assay was conducted as described in Tian et al. (2018). In brief, 100 sterilized rice grains were germinated in a petri dish and recorded for up to 8 d. Rice grains were regarded as germinated when the radicle reached a length of at least 1 cm.
RNA-Protein Binding Analysis
In vitro RNA-protein UV-cross–linking assay was carried out as described in Doroshenk et al. (2014). Specifically, the pBlueScript KS plasmids carrying different glutelin and prolamine RNA sequences (FL, 3′UTR, 5′UTR, and coding region sequences) and homoribopolymers of poly(A), poly(U), poly(G), or poly(C) sequence were constructed previously (Sami-Subbu et al., 2000) and used in this study. The glutelin and prolamine zipcode sequences (Hamada et al., 2003; Washida et al., 2009a, 2012) were also cloned into pBlueScript SK plasmids. Plasmid DNAs was linearized by digestion of the corresponding restriction enzymes and subjected to in vitro transcription using Ambion’s MAXIscript Kit to yield sense strand RNAs as described in Doroshenk et al. (2014). DIG-UTP was added during in vitro transcription reaction to label the transcribed RNAs. The RNAs that were transcribed from the linearized empty pBlueScript SK plasmids by digestion of the same restriction enzymes were used as negative controls.
RNA-IP with in vivo cross-linking using formaldehyde was performed as described in Doroshenk et al. (2014). Following RNA-IP, the PCR conditions were 20 cycles for glutelin and prolamine and 25 cycles for ACTIN using specific primers of each gene listed in Supplemental Table S7.
Total RNA and Protein Extraction from Rice Tissues
Total RNA extraction from rice leaves, stems, roots, and developing seeds and the subsequent RT-PCR using RBP-L gene-specific primers (shown in Supplemental Table S7) were performed as described in Doroshenk et al. (2014). Total protein from rice leaves, stems, roots, and developing seeds was obtained by extracting the tissues in buffer (0.1 m Tris-HCl, pH 8.5, 50 mm NaCl, 5% [w/v] SDS, 4 m urea, and 5% [v/v] β-mercaptoethanol) and subjected to SDS-PAGE and immunoblot analysis.
Production of Recombinant Proteins and Antibodies
FL cDNAs of RBP-L, RBP-P, BiP, and GFP were cloned into pET30a for His-tagged fusion protein expression. The purified His-tagged proteins were used to immunize New Zealand White rabbits for antibody production. Prolamine and glutelin antibodies were generated in earlier studies (Washida et al., 2009a, 2009b; Nagamine et al., 2011).
In Situ RT-PCR
In situ RT-PCR on thin sections from rice seeds was performed as described in Chou et al. (2017). The distribution of prolamine and glutelin RNAs was evaluated using specific primers for highly abundant genes, LOC_Os07g10570 and LOC_Os01g55690, respectively.
Cellular Fractionation Analysis
Cellular fractionation to isolate nuclear and cytosolic proteins were performed using the CelLytic PN Isolation/Extraction Kit (Sigma-Aldrich) as described in Doroshenk et al. (2014).
RNA-seq and RT-qPCR
Total RNA extraction from dehulled 10- to 14-d-old developing grains of wild type and the rbpl line was performed as described in Tian et al. (2018). Three biological replicates of total RNA samples from three independent plants of each rice genotype were sent to Novogene (https://en.novogene.com) for construction of RNA-seq libraries using NEBNext Ultra RNA Library Prep Kit for Illumina (https://www.neb.com) and subsequent next generation RNA-seq analysis on Illumina Sequencers. The trimming and mapping to the rice genome database (http://rice.plantbiology.msu.edu/index.shtml) were conducted using the CLC Genomics workbench (Qiagen, https://www.qiagenbioinformatics.com) as described in Tian et al. (2018). Expression of each gene based on mapping of unique reads was normalized to RPKM (Supplemental Table S1). DEGs were defined as genes with log2 fold change > 1 and P value < 0.01. A complete list of DEGs is shown in Supplemental Table S3. Pathway enrichment analysis of DEGs was performed by MapMan software (https://mapman.gabipd.org) with slight modification. Some undefined genes were manually categorized into corresponding pathway based on putative functions. Verification of RNA-seq by RT-qPCR was performed as described in Tian et al. (2018) using specific primers shown in Supplemental Table S7.
Suc Gradient Centrifugation
Two grams of dehulled developing rice seeds 10–14 d after pollination were extracted on ice in 2 mL of buffer containing 40 mm HEPES-NaOH, pH 7.2, 10 mm KCl, 3 mm MgCl2, and 0.1 mm EDTA, 100 U/mL RNase inhibitor and 0.4 m Suc and centrifuged at 1000g for 10 min at 4°C to remove starch. Two-hundred microliters of the resulting supernatant was kept as input sample and the remaining sample layered on top of a Suc density gradient consisting of equal layers of 25%, 30%, 40%, 45%, 50%, 60%, and 70% (w/w) Suc in extraction buffer and centrifuged at 175,000g for 1.5 h at 4°C. Each fraction (150 μL) of the gradient was collected and used for SDS-PAGE, immunoblot, and RT-PCR analyses.
Microscopy Analysis
Preparation of LR-white embedded TEM samples and immuno-gold labeling were prepared as described in Tian and Sun, 2011; Tian et al., 2018. The immuno-gold-labeled ultra-sections were poststained in 2% (w/v) uranyl acetate and Reynold’s lead citrate and observed on a FEI Tecnai G2 20 Twin TEM equipped with a charge-coupled device camera under a 200KV LaB6 electron source.
Immunofluorescence labeling of LR-white thin sections of seed samples was performed as described in a previous study (Doroshenk et al., 2014). For further detection of RBP-L colocalization with glutelin and prolamine mRNAs targeting sites, fresh mid-developing rice grains were cryotome-sectioned into 20-μm-thick specimens on a Leica Cryostat, subjected to in situ RT-PCR using glutelin-specific primers as mentioned above, and then fixed with 95% (v/v) ethanol for 30 s. The sample was then further labeled with anti-RBP-L antibody and AlexFluor 633-conjugated secondary antibody, and stained by Rhodamine B to label the PB-ER. The fluorescence labeling patterns were examined using a Zeiss LSM 510 Meta confocal microscope. As a negative control, endosperm sections were treated with preimmune serum (Supplemental Fig. S1) and the microscope gain adjusted to remove background labeling generated by nonspecific immunoglobulin G. These conditions were then used to observe the fluorescently labeled signals generated with RBP-L antibody.
Accession Numbers
Sequence data from this article can be found in rice genome database (http://rice.plantbiology.msu.edu/index.shtml) with their LOC number shown in the text or the GenBank/EMBL data libraries under the following NCBI accession numbers: for RBP45/47 family members from Arabidopsis and tobacco, Q9FPJ8.1 (AtRBP45a), Q9SAB3.1 (AtRBP45b), Q93W34.1 (AtRBP45c), XP_016465791.1 (NtRBP45), AtRBP47A (F4I3B3.1), AtRBP47B (Q0WW84.1), AtRBP47C (Q9SX80.1), and NtRBP47 (Q9LEB3.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Specificity test of anti-RBP-L antibody.
Supplemental Figure S2. Screening of the rbpl line and location of T-DNA insertion.
Supplemental Figure S3. Expression of RBP-L in various organs and tissues.
Supplemental Table S1. Raw data file of gene expression from wild-type and rbpl lines obtained by RNA-seq analysis.
Supplemental Table S2. Expression profile of RBP-L transcripts in wild-type and rbpl lines based on RNA-seq analysis.
Supplemental Table S3. List of DEGs in the rbpl line compared to wild type.
Supplemental Table S4. Classification of DEGs to 36 pathways (bins) by MapMan analysis.
Supplemental Table S5. List of DEGs involved in RNA metabolic pathway.
Supplemental Table S6. Expression profile of glutelin and prolamine families in wild-type and rbpl lines.
Supplemental Table S7. Sequence information of all primers used in the study.
Acknowledgments
We acknowledge support from the Franceschi Microscopy and Imaging Center at Washington State University.
Footnotes
This work is supported by the National Science Foundation (NSF grant nos. MCB-1444610 and IOS-1701061), and by the U.S. Department of Agriculture National Institute of Food and Agriculture (USDA Hatch Umbrella Project no. 0590 1015621 and Hatch grant no. 0119 WNP00119).
Articles can be viewed without a subscription.
References
- Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: An overview. Physiol Mol Biol Plants 19: 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron KN, Schroeder DF, Stasolla C (2014) GEm-Related 5 (GER5), an ABA and stress-responsive GRAM domain protein regulating seed development and inflorescence architecture. Plant Sci 223: 153–166 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y (2015) The molecular circuitry of brassinosteroid signaling. New Phytol 206: 522–540 [DOI] [PubMed] [Google Scholar]
- Bilenoglu O, Basak AN, Russell JE (2002) A 3'UTR mutation affects beta-globin expression without altering the stability of its fully processed mRNA. Br J Haematol 119: 1106–1114 [DOI] [PubMed] [Google Scholar]
- Blower MD. (2013) Molecular insights into intracellular RNA localization. Int Rev Cell Mol Biol 302: 1–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Grewe B, Werr W (2007) The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Chantarachot T, Bailey-Serres J (2018) Polysomes, stress granules, and processing bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176: 254–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Peng J, Ma J, Tang Y, Chen R, Mysore KS, Wen J (2012) NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol 195: 71–84 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B (2009) One for all: The receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Chou HL, Tian L, Kumamaru T, Hamada S, Okita TW (2017) Multifunctional RNA binding protein OsTudor-SN in storage protein mRNA transport and localization. Plant Physiol 175: 1608–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Ogawa M, Kumamaru T, Satoh H (2004) Targeting of proteins to endoplasmic reticulum-derived compartments in plants. The importance of RNA localization. Plant Physiol 136: 3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AJ, Washida H, Okita TW, Satoh M, Ogawa M, Kumamaru T, Satoh H (2005) The role of mRNA and protein sorting in seed storage protein synthesis, transport, and deposition. Biochem Cell Biol 83: 728–737 [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Crofts N, Whitelegge JP, Okita TW (2010) Isolation and identification of cytoskeleton-associated prolamine mRNA binding proteins from developing rice seeds. Planta 231: 1261–1276 [DOI] [PubMed] [Google Scholar]
- Doroshenk KA, Crofts AJ, Morris RT, Wyrick JJ, Okita TW (2009) Proteomic analysis of cytoskeleton-associated RNA binding proteins in developing rice seed. J Proteome Res 8: 4641–4653 [DOI] [PubMed] [Google Scholar]
- Doroshenk KA, Crofts AJ, Morris RT, Wyrick JJ, Okita TW (2012) RiceRBP: A resource for experimentally identified RNA binding proteins in Oryza sativa. Front Plant Sci 3: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshenk KA, Tian L, Crofts AJ, Kumamaru T, Okita TW (2014) Characterization of RNA binding protein RBP-P reveals a possible role in rice glutelin gene expression and RNA localization. Plant Mol Biol 85: 381–394 [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW (2003) Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YJ, Melencion SM, Lee ES, Park JH, Alinapon CV, Oh HT, Yun DJ, Chi YH, Lee SY (2015) Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front Plant Sci 6: 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Posakony JW (1997) The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development 124: 4847–4856 [DOI] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J (2003) Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Lelli KM, Slattery M, Mann RS (2012) Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet 46: 43–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviten MW, Lai EC, Posakony JW (1997) The Drosophila gene BEARDED encodes a novel small protein and shares 3′ UTR sequence motifs with multiple enhancer of split complex genes. Development 124: 4039–4051 [DOI] [PubMed] [Google Scholar]
- Lewandowska D, Simpson CG, Clark GP, Jennings NS, Barciszewska-Pacak M, Lin CF, Makalowski W, Brown JW, Jarmolowski A (2004) Determinants of plant U12-dependent intron splicing efficiency. Plant Cell 16: 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Z, Peng H, Liu Z (2014) A MITE insertion into the 3′-UTR regulates the transcription of TaHSP16.9 in common wheat. Crop J 2: 381–387 [Google Scholar]
- Li X, Franceschi VR, Okita TW (1993) Segregation of storage protein mRNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell 72: 869–879 [DOI] [PubMed] [Google Scholar]
- Liu J, Chen S, Chen L, Zhou Q, Wang M, Feng D, Li JF, Wang J, Wang HB, Liu B (2017) BIK1 cooperates with BAK1 to regulate constitutive immunity and cell death in Arabidopsis. J Integr Plant Biol 59: 234–239 [DOI] [PubMed] [Google Scholar]
- Lokdarshi A, Conner WC, McClintock C, Li T, Roberts DM (2016) Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 170: 1046–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Wieczorek Kirk DA, Klahre U, Hemmings-Mieszczak M, Filipowicz W (2000) RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6: 1610–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K (2004) dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 37: 720–729 [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A (2009) mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2011) The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E, Michalova E, Vojtesek B, Hrstka R (2012) The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol 9: 563–576 [DOI] [PubMed] [Google Scholar]
- Mauri N, Fernández-Marcos M, Costas C, Desvoyes B, Pichel A, Caro E, Gutierrez C (2016) GEM, a member of the GRAM domain family of proteins, is part of the ABA signaling pathway. Sci Rep 6: 22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: Adaptable guards. Genome Biol 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C, Mowry K, Besse F (2012) Principles and roles of mRNA localization in animal development. Development 139: 3263–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3: Reviews0004.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench DG, Chuong SD, Franceschi VR, Okita TW (2000) Developing prolamine protein bodies are associated with the cortical cytoskeleton in rice endosperm cells. Planta 211: 227–238 [DOI] [PubMed] [Google Scholar]
- Nagamine A, Matsusaka H, Ushijima T, Kawagoe Y, Ogawa M, Okita TW, Kumamaru T (2011) A role for the cysteine-rich 10 kDa prolamine in protein body I formation in rice. Plant Cell Physiol 52: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O (2011) Translation-independent localization of mRNA in E. coli. Science 331: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua C, Bilkova A, Jackson P, Dabravolski S, Riber W, Didi V, Houser J, Gigli-Bisceglia N, Wimmerova M, Budínská E, et al. (2017) Dirigent proteins in plants: Modulating cell wall metabolism during abiotic and biotic stress exposure. J Exp Bot 68: 3287–3301 [DOI] [PubMed] [Google Scholar]
- Perotti MF, Ribone PA, Chan RL (2017) Plant transcription factors from the homeodomain-leucine zipper family I. Role in development and stress responses. IUBMB Life 69: 280–289 [DOI] [PubMed] [Google Scholar]
- Pickel B, Schaller A (2013) Dirigent proteins: molecular characteristics and potential biotechnological applications. Appl Microbiol Biotechnol 97: 8427–8438 [DOI] [PubMed] [Google Scholar]
- Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, Kishida K, Kuroda M, Tanaka K, Morita S, Satoh S, et al. (2012) Formation mechanism of the internal structure of type I protein bodies in rice endosperm: relationship between the localization of prolamin species and the expression of individual genes. Plant J 70: 1043–1055 [DOI] [PubMed] [Google Scholar]
- Sami-Subbu R, Muench DG, Okita TW (2000) A cytoskeleton-associated RNA-binding protein binds to the untranslated regions of prolamine mRNA and to poly(A). Plant Sci 152: 115–122 [Google Scholar]
- Schwechheimer C, Zourelidou M, Bevan MW (1998) Plant transcription factor studies. Annu Rev Plant Physiol Plant Mol Biol 49: 127–150 [DOI] [PubMed] [Google Scholar]
- Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 111: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson M, Lundh D, Bergman P, Mandal A (2005) Characterisation of a T-DNA-tagged gene of Arabidopsis thaliana that regulates gibberellin metabolism and flowering time. Funct Plant Biol 32: 923–932 [DOI] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Okita TW (2014) mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr Opin Plant Biol 22: 77–85 [DOI] [PubMed] [Google Scholar]
- Tian L, Sun SSM (2011) A cost-effective ELP-intein coupling system for recombinant protein purification from plant production platform. PLoS One 6: e24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chou HL, Zhang L, Hwang SK, Starkenburg SR, Doroshenk KA, Kumamaru T, Okita TW (2018) RNA-binding protein RBP-P is required for glutelin and prolamine mRNA localization in rice endosperm cells. Plant Cell 30: 2529–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washida H, Kaneko S, Crofts N, Sugino A, Wang C, Okita TW (2009a) Identification of cis-localization elements that target glutelin RNAs to a specific subdomain of the cortical endoplasmic reticulum in rice endosperm cells. Plant Cell Physiol 50: 1710–1714 [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Kaneko S, Crofts N, Sakulsingharoj C, Kim D, Choi SB, Hamada S, Ogawa M, Wang C, et al. (2009b) Identification of cis-localization elements of the maize 10-kDa delta-zein and their use in targeting RNAs to specific cortical endoplasmic reticulum subdomains. Plant J 60: 146–155 [DOI] [PubMed] [Google Scholar]
- Washida H, Sugino A, Doroshenk KA, Satoh-Cruz M, Nagamine A, Katsube-Tanaka T, Ogawa M, Kumamaru T, Satoh H, Okita TW (2012) RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J 70: 471–479 [DOI] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Weis BL, Schleiff E, Zerges W (2013) Protein targeting to subcellular organelles via MRNA localization. Biochim Biophys Acta 1833: 260–273 [DOI] [PubMed] [Google Scholar]
- Wu Q, Law PY, Wei LN, Loh HH (2008) Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3′ untranslated region: A role for microRNA23b. FASEB J 22: 4085–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue LJ, Zhang JJ, Xue HW (2012) Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PLoS One 7: e31081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Yan Z, Wang Y, Yan X, Han Y (2014) Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Bot 65: 5933–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Crofts AJ, Crofts N, Okita TW (2014) Multiple RNA binding protein complexes interact with the rice prolamine RNA cis-localization zipcode sequences. Plant Physiol 164: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CQ, Xu Y, Lu Y, Yu HX, Gu MH, Liu QQ (2011) The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234: 541–554 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li C, Ge J, Xu M, Zhu Q, Wu T, Guo A, Xie J, Dong H (2014) Recessive mutation identifies auxin-repressed protein ARP1, which regulates growth and disease resistance in tobacco. Mol Plant Microbe Interact 27: 638–654 [DOI] [PubMed] [Google Scholar]