A biological device consisting of mammalian, fungal, and plant components drives gene expression in an oxygen-dependent manner.
Abstract
Due to the involvement of oxygen in many essential metabolic reactions, all living organisms have developed molecular systems that allow adaptive physiological and metabolic transitions depending on oxygen availability. In mammals, the expression of hypoxia-response genes is controlled by the heterodimeric Hypoxia-Inducible Factor. The activity of this transcriptional regulator is linked mainly to the oxygen-dependent hydroxylation of conserved proline residues in its α-subunit, carried out by prolyl-hydroxylases, and subsequent ubiquitination via the E3 ligase von Hippel-Lindau tumor suppressor, which targets Hypoxia-Inducible Factor-α to the proteasome. By exploiting bioengineered versions of this mammalian oxygen sensor, we designed and optimized a synthetic device that drives gene expression in an oxygen-dependent fashion in plants. Transient assays in Arabidopsis (Arabidopsis thaliana) mesophyll protoplasts indicated that a combination of the yeast Gal4/upstream activating sequence system and the mammalian oxygen sensor machinery can be used effectively to engineer a modular, oxygen-inducible transcriptional regulator. This synthetic device also was shown to be selectively controlled by oxygen in whole plants when its components were expressed stably in Arabidopsis seedlings. We envision the exploitation of our genetically encoded controllers to generate plants able to switch gene expression selectively depending on oxygen availability, thereby providing a proof of concept for the potential of synthetic biology to assist agricultural practices in environments with variable oxygen provision.
As sessile organisms, plants must adapt their developmental and metabolic programs to the surrounding environment, especially when this entails stress conditions. Such adaptation often is achieved through the perception of exogenous and endogenous stimuli via dedicated sensors, which trigger signaling cascades that, in turn, culminate in the reprogramming of gene expression and metabolism. Numerous proteins involved in the sensing of temperature (Jung et al., 2016), light quality and quantity (Possart et al., 2014), water availability (Zhu, 2002), and nutrient deficiency (Cui, 2012) have been identified in Arabidopsis (Arabidopsis thaliana) and other plant species and are currently being investigated to unveil their signaling mechanisms.
Plants successfully colonized most terrestrial niches by evolving specific adaptive traits to stress conditions, depending on the duration, intensity, and frequency at which these adverse conditions occur in different regions of our planet. However, the relatively recent transfer of crops from their area of origin and extreme climatic events cause serious limitations to productivity, since cultivated plants are exposed to environmental conditions for which they have not yet evolved any adaptations (Crane et al., 2011; Pingali, 2012; Nelson et al., 2014). To this end, breeding using simple beneficial traits may not be sufficient to rapidly develop stress-resistant crop varieties. The engineering of complex and novel traits using strategies inspired by synthetic biology, based on the fast advances in plant molecular physiology, represents a promising way to overcome such limitations. Synthetic biology strategies aim at reconstructing artificial genetic circuits, often of modular structure and composed of functionally characterized constituents, that are able to operate without interfering with the host cell functions (orthogonally; Baltes and Voytas, 2015; Liu and Stewart, 2015). The successful design of orthogonal drought responses (Yang et al., 2010), synthetic nitrogen fixation (Mus et al., 2016), or improved photosynthetic performances (Erb and Zarzycki, 2016) have shown the remarkable potential held by such approaches.
Among the adverse environmental conditions that limit yield production, flooding is one of the main causes for agricultural losses worldwide (Bailey-Serres et al., 2012). Since gas diffusion is extremely reduced in water as compared with air (Armstrong, 1980), submerged plant tissues are exposed to the concomitant decrease in oxygen availability and accumulation of CO2 and ethylene, which inhibit aerobic metabolism (Bailey-Serres and Voesenek, 2008). Considering also the reduced efficiency of photosynthesis in muddy water (Mommer and Visser, 2005), flooded or waterlogged plants suffer a general energy crisis that, if prolonged, can lead to death (Licausi and Perata, 2009). To date, crop improvement approaches aimed at enhancing resistance to transient flooding have been applied mostly to rice (Oryza sativa), where either fast elongation determinants during germination (Kretzschmar et al., 2015) or the arrest of underwater growth in adult plants (Xu et al., 2006) have been exploited. Remarkably, this last strategy relies on transcriptional regulators dedicated to the metabolic adaptation to hypoxia in plants: the Group VII Ethylene Response Factor proteins (Giuntoli and Perata, 2018).
Surprisingly, the same class of transcription factors has been shown to participate in the response to other environmental stimuli, including nitric oxide (Gibbs et al., 2014), sugar status (Loreti et al., 2018), and developmental processes (Abbas et al., 2015; Meitha et al., 2018). Therefore, efficient engineering of flood-adaptive strategies, avoiding an effect on the overall plant physiology, requires the development of a strategy to relate specific gene expression to oxygen availability in an orthogonal manner.
In order to evaluate the feasibility of this strategy, we explored possible alternative strategies of oxygen sensing developed by other organisms. In most animal cells, transcriptional adaptation to low-oxygen conditions is mediated by Hypoxia-Inducible Factor (HIF) basic helix-loop-helix regulators. This oxygen-sensing mechanism is based on the hydroxylation of specific Pro residues carried by HIF1α (Pro-402 and Pro-564; Ivan et al., 2001), catalyzed by oxygen- and oxoglutarate-dependent enzymes known as prolyl-hydroxylases (PHDs; Bruick and McKnight, 2001; Epstein et al., 2001). The hydroxylated HIFα is polyubiquitinated subsequently by a ubiquitin E3 ligase complex, which contains the substrate recognition subunit von Hippel-Lindau tumor suppressor (pVHL; Maxwell et al., 1999). Polyubiquitination leads to HIFα degradation through the 26S proteasome complex. When oxygen levels drop, HIFα is protected from proteolysis and thereby enabled to migrate into the nucleus (Depping et al., 2008). Here, heterodimerization with its cognate β-subunit (Wang et al., 1995) reconstitutes a functional transcription complex able to induce the expression of hypoxic genes.
In humans, a single HIFβ isoform and three different HIFα isoforms have been identified (Ema et al., 1997; Gu et al., 1998). Among them, HIF1α and HIF2α have been investigated extensively and are currently considered to act as the master regulators of the hypoxic response. In the case of PHD enzymes, three different isoforms have been identified (Bruick and McKnight, 2001). They differ for tissue specificity (Hirsilä et al., 2003), cellular localization (Metzen et al., 2003), and affinity for HIFα (Masson et al., 2001).
Numerous in vitro and in vivo assays have revealed the bases for the oxygen-dependent HIF-pVHL interaction. HIF1α contains two oxygen-dependent degradation domains (ODDs), called NODD or CODD depending on their proximity to the N-terminal or C-terminal end of the protein, respectively (Masson et al., 2001). The ODDs allow interaction with both pVHL (Ivan et al., 2001; Jaakkola et al., 2001) and PHDs (Bruick and McKnight, 2001; Epstein et al., 2001). In turn, pVHL activity relies on two different domains: an α-domain (spanning residues 157–171) that recruits the ubiquitination complex and a β-domain (residues 63–157) involved in the HIF-ODD interaction (Stebbins et al., 1999; Hon et al., 2002; Min et al., 2002).
Previous studies have shown that the HIF-pVHL interaction can be promoted in a heterologous yeast system, provided that PHD is present (Alcaide-German et al., 2008). In this work, we developed a transcriptional regulatory device, based on the HIF-pVHL-PHD triad, able to switch from an active to an inactive state in response to oxygen availability in plants.
RESULTS
Engineering an Oxygen-Sensing HIF-pVHL-PHD Device in Arabidopsis Protoplasts
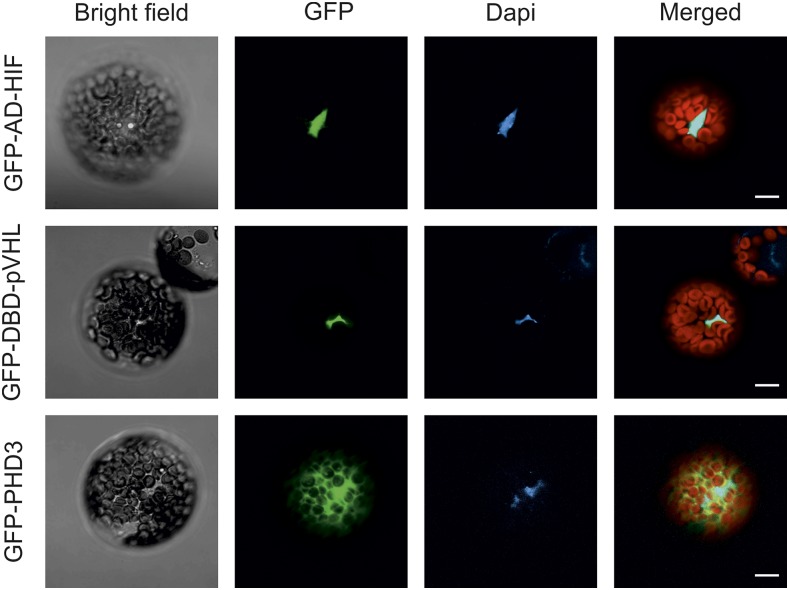
To achieve orthogonal control of gene expression in plants in an oxygen-dependent manner, we took advantage of the mammalian HIF1α and pVHL pair. We selected the essential domains required for their oxygen-driven interaction, namely a CODD and a β-domain fragment encoded by HIF1α554-576 and pVHL63-157. To reconstitute a fully functional transcriptional activator, we fused the HIF1α and pVHL sequences to the activation domain (AD) and the DNA-binding domain (DBD) of the Saccharomyces cerevisiae Gal4 transcription factor (Keegan et al., 1986) to exploit a mechanism of intermolecular complementation of Gal4 function. In this way, we generated two chimeric effector modules: AD-HIF and DBD-pVHL. Since their interaction is expected to rely on HIF1α hydroxylation at the Pro-564 residue (HIFHyp), the oxygen-sensing device had to be equipped with an additional unit, constituted by the human PHD3 enzyme (sensory module). Our choice fell on this PHD isoform due to its high activity on the CODD substrate (Appelhoff et al., 2004) and its relatively small size, which confers a homogenous distribution among cellular compartments, including the nucleus (Fig. 1). This ensured the colocalization of PHD3 with AD-HIF and DBD-pVHL, both provided with a nuclear localization sequence (Fig. 1).
Figure 1.
Subcellular localization of the components of the oxygen-sensing machinery in Arabidopsis protoplasts. AD-HIF (Gal4 AD fusion of the HIF1α CODD domain), DBD-pVHL (Gal4 DBD fusion of the pVHL β-domain), and PHD3 DNA sequences were fused in frame with an N-terminal GFP protein sequence, expressed in Arabidopsis mesophyll protoplasts, and imaged with a confocal microscope. The signal from the GFP channel is visualized in green. 4′,6-Diamidino-2-phenylidone (Dapi) staining (1 µg mL−1) marks the nuclei (blue). In the merged image, the red color is associated with chloroplast autofluorescence. Bars = 10 µm.
Therefore, we envisioned that the presence of oxygen would lead to AD-HIF hydroxylation, the association of the effector modules, and the formation of a transcriptional complex. This, in turn, would be able to induce promoters containing the Gal4-specific cis-element, called the upstream activating sequence (UAS; Guarente et al., 1982). Accordingly, we generated an output module by cloning the firefly (Photinus pyralis) luciferase reporter gene downstream of a synthetic UAS promoter (Fig. 2A). We also placed a second luciferase gene from sea pansy (Renilla reniformis) under the control of a constitutive Cauliflower mosaic virus (CaMV) 35S promoter to be used as a normalization variable proportional to the transformation efficiency. Consequently, relative luciferase activity was measured as the readout of active complex formation, arising from the HIFHyp-pVHL interaction.
Figure 2.
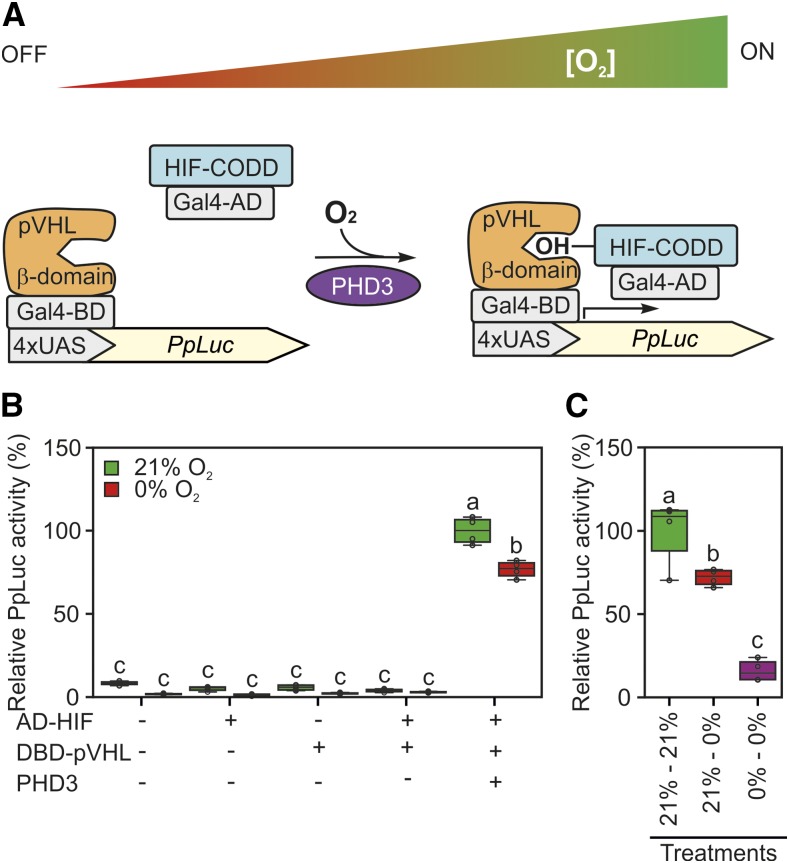
Schematization and testing of a synthetic oxygen sensor device based on the mammalian hypoxia sensing. A, Conceptual working mechanism of the device dependent on the environmental oxygen concentration. Human HIF1-α or pVHL fragments (HIF-CODD and β-domain) were fused in frame with the Gal4 AD or DBD, respectively. In the presence of oxygen, AD-HIF is hydroxylated by PHD3 and dimerizes with DBD-pVHL to induce the expression of a firefly luciferase reporter (PpLuc) controlled by a 4xUAS promoter. B, Sensor output in Arabidopsis protoplasts subjected to 18-h-long anoxia (0% oxygen) or normoxia (21% [v/v] oxygen) 12 h after transfection with plasmids bearing the modules depicted in A. Firefly luciferase activity was normalized to that of a sea pansy luciferase, coexpressed under the control of a constitutive CaMV 35S promoter. The average aerobic value produced by the full device (equipped with reporter, sensory, and effector modules) was set to 100%. C, Comparison of sensor responsiveness to an 18-h-long anoxic treatment applied either 12 h after protoplast transfection (21%–0%) or immediately after it (0%–0%). In the box plots, dots represent single data points, the black line marks the median, and the box indicates the interquartile range (IQR). Whiskers extend to data points that are lower than 1.5× IQR away from the box extremities. Different letters indicate statistically significant differences (P ≤ 0.05) calculated from two-way ANOVA followed by Tukey’s posttest.
We tested the four modules in plant cells. The constructs were expressed transiently, in different combinations, in freshly prepared protoplasts from Arabidopsis mesophyll cells. Twelve hours after the transformation, protoplasts were either subjected to anoxic treatment or maintained in normoxia. The two effector proteins, when expressed alone, did not manifest transactivation capacity, either in ambient or anoxic conditions (Fig. 2B). More strikingly, no output was produced by the combination of the effectors without concurrent PHD3 expression, revealing that plants do not possess any endogenous enzyme able to hydroxylate the Pro-564 residue borne by the HIF-CODD (Fig. 2B). Instead, we detected increased relative luminescence in aerobic protoplasts expressing the full set of sensor modules (Fig. 2B), indicating that the chosen PHD3 isoform enabled AD-HIF interaction with DBD-pVHL and subsequent promoter activation.
However, contrary to the expectations, prolonged exposure to complete anoxia exerted only a mild although significant repression of the luciferase reporter (Fig. 2B). Imposition of an anoxic atmosphere in the absence of photosynthetic activity (as a consequence of darkness) was assumed to deplete the intracellular oxygen availability below PHD affinity (Ehrismann et al., 2007). Excluding the residual catalytic activity of PHD3 under such conditions, we put forward that a slow turnover of the two interacting partners would result in the persistence of a fraction of the complex, sufficient to drive transactivation in the absence of oxygen. To evaluate this possibility, we subjected protoplasts to an immediate oxygen deprivation after transfection. In this case, we hypothesized that direct exposure of any newly formed HIF-CODD protein to an anoxic environment should prevent complex formation, thereby hindering promoter induction. Immediate exposure to the anoxic atmosphere limited the response of the output sensor considerably when compared with the level displayed after sequential aerobic and anoxic incubations (Fig. 2C). Thus, we concluded that, once formed under oxygen-replete conditions, the HIF-pVHL effector complex is characterized by a relatively long half-life, resulting in a substantial amount of transactivating complex being still present and functional during the following anaerobic treatment.
Expanding the Dynamic Range of the Oxygen Sensor
In order to increase the hypoxic responsiveness of our oxygen sensor, we decided to enhance the turnover of at least one of the three components. In principle, AD-HIF could work as the main determinant of the activation state of the device, since its hydroxylation determines whether the transcriptional complex assembles. It is then conceivable that, during the progression of the anoxic treatment, a fast recycling AD-HIF would leave only newly synthesized units, which would no longer be functional.
In the attempt to generate a fast recycling effector, we fused the N terminus of the AD-HIF chimeric protein to a known destabilizing domain, consisting of amino acid residues 21 to 30 of the KIP-RELATED PROTEIN1 (KRP1; Li et al., 2016) of Arabidopsis (Fig. 3A). This new version of the effector was tested in Arabidopsis mesophyll protoplasts. While retaining the ability to be activated specifically by oxygen in the presence of PHD3, this time the sensor indeed showed a larger reduction of promoter activity (−60% of the aerobic output) in anoxia-treated protoplasts (Fig. 3B), as compared with the first module evaluated (Fig. 2B; −25% of the aerobic output). As shown by direct comparison of the two HIF modules (Supplemental Fig. S1), the output was lower in the presence of the modified HIF than when the original effector was used. Therefore, we concluded that conjugation of the KRP1 domain modified the steady-state level of the effector complex, decreasing its stability in both aerobic and anoxic conditions. Nonetheless, more pronounced destabilization in anaerobiosis determined an actual expansion of the dynamic range of the sensor. Overall, this effect could be explained by the continuous degradation of KRP1-AD-HIF, resulting in limited persistence of the active complex, which could only accumulate under aerobic conditions when newly synthesized AD-HIF is hydroxylated by PHD3.
Figure 3.
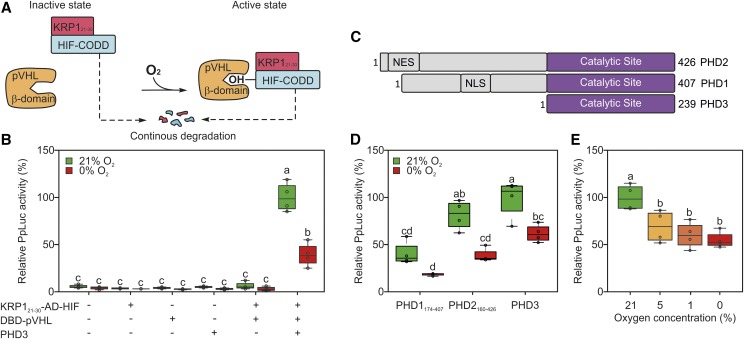
Engineering of a destabilized version of the AD-HIF protein. A, To increase HIF-CODD turnover, the AD-HIF effector module was modified with an additional destabilizing domain (KRP121-30), promoting its recycling through the 26S proteasome. For the sake of simplicity, the Gal4 domains and the reporter construct are not shown in the schematic. B, Sensor output in an Arabidopsis protoplast transiently transformed with reporter, sensory, and effector plasmids and subjected to 18 h of anoxia 12 h after transfection. C, Structures of the three known human PHD isoforms, according to the information related to the Uniprot accessions Q96KS0 (PHD1), Q9GZT9 (PHD2), and S5Q9G2 (PHD3). NES stands for nuclear export signal and NLS for nuclear localization signal. D, Sensor output in protoplasts transfected with one of the three known human PHD isoforms. Effector and reporter modules were provided equally in all transformations. E, Arabidopsis protoplasts were incubated under a range of oxygen concentrations, spanning from complete anoxia to aerobic levels. In the box plots, dots represent single data points, the black line marks the median, and the box indicates the interquartile range (IQR). Whiskers extend to data points that are lower than 1.5× IQR away from the box extremities. Different letters indicate statistically significant differences (P ≤ 0.05) among means, as assessed by two-way ANOVA (in B and D) or one-way ANOVA (in E) followed by Tukey’s posttest to correct for multiple comparisons.
Performance of the Device under Different Magnitudes of the Oxygen Stimulus and with Alternative Sensory Modules
Once we improved the mammalian-based oxygen device, we decided to evaluate its performance when the PHD3 sensory module was replaced by the other prolyl hydroxylase isoforms annotated in the human genome. The catalytic domains of PHD1 and PHD2 (Fig. 3C; Supplemental Information S1) were tested in the protoplast system. When PHD1174-407 was employed, the device produced a reduced output as compared with that associated with PHD3, due to lower aerobic reporter activity and decreased range (Fig. 3D). PHD2160-426, instead, had comparable performance to PHD3 (Fig. 2D), suggesting it as a suitable potential substitute of the latter in this kind of device.
Additionally, we explored the sensor sensitivity to distinct atmospheric oxygen levels, as we envision that the ability of the HIF-pVHL sensor to discriminate between normoxia and hypoxia would be a necessary condition for its application as a proxy for physiological hypoxia in plants. To test this aspect, we incubated protoplasts expressing the complete sensor under a range of oxygen concentrations. Significant reduction of luciferase activity was recorded when comparing aerobic protoplasts with those subjected to oxygen limitation, but no significant difference was measured among samples treated with atmospheres containing either 5% or 1% (v/v) oxygen or totally devoid of it (Fig. 3E). We concluded that the synthetic sensor had high sensitivity to relatively mild decreases in oxygen availability (represented by 5% [v/v] oxygen in our test), without being able to sense the magnitude of hypoxia further. Based on the responsiveness of the HIF-pVHL sensor to moderate drops in intracellular hypoxia, we propose it as a viable candidate to report about the occurrence of physiological hypoxia in living plant tissues.
In Silico Prediction and Testing of Mutated Variants
Despite the employment of a fast recycling HIF module, basal activity of the sensor was still observed at the end of the anaerobic incubation (Fig. 3B), indicating that a substantial amount of the transactivating complex was maintained after the removal of oxygen. Therefore, we sought to weaken the HIF-pVHL interaction to further reduce the half-life of the active complex. To do so, we adopted a rational mutagenesis approach based on available structural information. Inspection of the three-dimensional structure of a pVHL/HIF-CODD complex, provided by Min et al. (2002) and Hon et al. (2002), revealed that the association between the HIF fragment and pVHL not only involves interactions generated as a consequence of Pro-564 hydroxylation (Fig. 4A) but also occurs through other residues. In the aerobic HIFHyp-pVHL complex, HIF1⍺ Hyp-564 lies in a deep pocket, lined up by pVHL residues Trp-88, Tyr-98, Ser-111, His-115, and Trp-117, and establishes a hydrogen bond network with Ser-111, His-115, and Tyr-98 (Fig. 4A). Additionally, HIF-CODD Asp-571 and pVHL Arg-107 form a salt bridge outside of the pocket accommodating Hyp-564 (Supplemental Fig. S2A).
Figure 4.
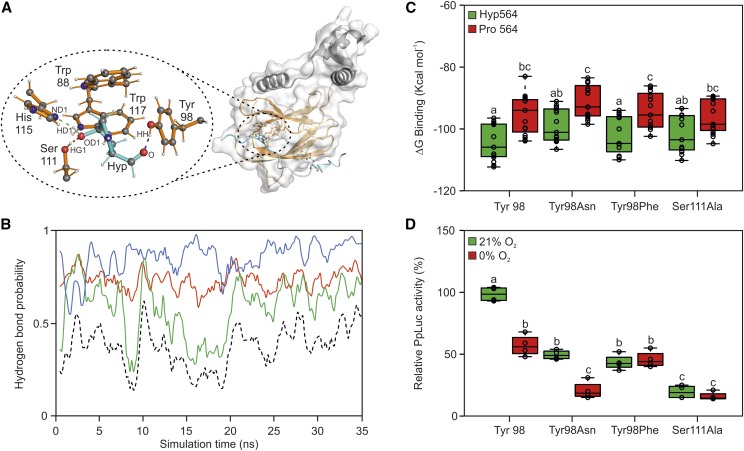
Rational mutagenesis of sensor components to increase its responsiveness to anoxia. A, Crystal structure of HIF1⍺-CODD and pVHL (Hon et al., 2002). The HIF1⍺-CODD peptide and its residues are shown in cyan, the pVHL β-domain and its residues in orange, while the α-domain is shown in gray. The oval encloses a magnified view of the hydrogen bond network established between HIF1⍺-CODD and pVHL residues. Atomic names are given according to the Protein Data Bank nomenclature. B, MD simulation of the interactions between HIFHyp and pVHL His-115, Ser-111, and Tyr-98. Hydrogen bond probability involving the donor-acceptor pairs HD1Hyp-ND1His115 are shown in green, HG1Ser111-OD1Hyp in red, and HHTyr98-OHyp in blue. The black dotted line represents the hydrogen bond probability for all three bonds to be present. C, MM-GBSA calculations of interaction stability between wild-type HIF1⍺-CODD residue 564 and pVHL residues in the case of Tyr-98Asn, Tyr-98Phe, and Ser-111Ala mutations. Data are expressed as binding free energy (ΔG binding). D, Effects of individual pVHL point mutations on luciferase transactivation in Arabidopsis protoplasts. The average aerobic output for the wild-type pVHL peptide version (Tyr-98) was set to 100%. In the box plots, dots represent single data points, the black line marks the median, and the box indicates the interquartile range (IQR). Whiskers extend to data points that are lower than 1.5× IQR away from the box extremities. Different letters indicate statistical differences (P ≤ 0.05) calculated from two-way ANOVA followed by Tukey’s posttest.
Impairment of these key interactions is expected to facilitate the dissociation of the complex. We speculated that a decreased complex stability, while being compensated by the continuous assembly of new transcriptional units under aerobic conditions, might especially constrain output production under anoxic conditions.
We applied a molecular dynamics (MD) simulation to model the distances and probability of occurrence of the pairwise interactions between HIF1⍺ Hyp-564 and the three aforementioned residues belonging to the pVHL pocket. The computational results revealed fluctuations of the hydrogen bond pattern over the simulation time and identified the hydrogen bond between the HIF564 β-carbon and pVHL Tyr-98 as the most stable interaction (Fig. 4B).
Once we described the network of noncovalent interactions centered on HIF564, we proceeded to the rational mutagenesis of the module. As a first target site, we focused on pVHL Tyr-98: the absence of other interactions, beyond the one involving HIF564, together with its side chain orientation pointing outward from the surface of the complex made its substitution unlikely to interfere with protein folding. An alternative target residue selected was Ser-111; as its side chain was oriented inward, it was exchanged with a short and neutral side chain (Ser-111Ala) to abolish the interaction.
The Tyr-98Asn, Tyr-98Phe, and Ser-111Ala variants were selected for molecular mechanics combined with generalized Born and surface area continuum solvation analysis (MM-GBSA) to predict changes in their relative binding energy with HIF564. Tyr-98Asn, a common mutation in many types of tumors (Knauth et al., 2006), had been shown before to destabilize the interaction. Substitution with Phe should provide the same structural properties as the wild-type Tyr residue, only hindering hydrogen bond formation (Supplemental Fig. S2B). In general, the MM-GBSA did not predict a reduction in complex stability for the selected variants, suggesting the potential for complex formation for any of the selected variants (Fig. 4C). Tyr-98Asn and Ser-111Ala were predicted to be the most destabilizing variants, since no statistical differences were found comparing their ∆G of binding measured in the presence of HIFHyp with that of the wild-type variant in the presence of HIFPro, representing a complete dissociated complex (Fig. 4C).
We then tested experimentally these variants in the transient protoplast system. According to the output of the sensor, all point mutations on pVHL led to weaker interaction with HIFHyp, with Ser-111Ala being the most destabilizing substitution (Fig. 3D). Tyr-98Asn and Tyr-98Phe variants were associated with a 50% loss of the aerobic output of the wild-type sensor. Remarkably, only the Tyr-98Asn preserved the oxygen regulation of the device and even showed an increased dynamic range compared with the wild type (Fig. 4D).
Due to this partial absence of correlation between MM-GBSA and experimental data, we decided to carry on with the in vivo investigation of the relevance of selected residues to the HIF-pVHL interaction. Thus, we evaluated the performance of an additional set of residues whose side chains possess mildly or dramatically different physicochemical properties. Specifically, three mutations (Tyr-98His, Tyr-98Trp, and Tyr-98Cys) were predicted to disrupt the hydrogen bond with HIF1⍺ Hyp-564, whereas another three (Tyr-98Asp, Tyr-98Glu, and Tyr-98Ile) had the potential to retain it (Supplemental Fig. S2B). We also examined the contribution of the remaining contact point between the two protein fragments as revealed by the crystal structure, the one formed by pVHL Arg-107 and HIF-CODD Asp-571. All new variants resulted in a general reduction of the aerobic interaction between the effector modules (Supplemental Fig. S2C). This was unexpected for the HIF571-pVHL107 interaction; although far apart from the pocket allocating Hyp-564, it seemed instead to be crucial for active complex stability and conformation maintenance of the pocket. Overall, the transactivation values recorded for pVHL98 variants under aerobic conditions did not appear to correlate with the predicted ability of the substituted residues to form a hydrogen bond with the β-carbon of HIF564 (Fig. 4D; Supplemental Fig. S2B). The Tyr-98Cys variant conserved a moderate interaction capacity under aerobic conditions, although, as observed for Tyr-98Phe, this interaction became independent from oxygen availability (Supplemental Fig. S2C). Strikingly, the oxygen regulation of the sensor was lost as a consequence of each individual point mutation, with the exception of Tyr-98Glu, where it was associated with lower stability of the complexes (Supplemental Fig. S2C).
Taking these results together, we could conclude that the responsiveness to oxygen of our device was ensured, beyond HIF-CODD Pro-564, by several crucial residues, likely granting the maintenance of a proper conformation of the two effectors. It can be speculated that PHD3 access to the HIF564-containing pocket was determined both by steric features of the pocket itself (relying on pVHL98, pVHL111, and HIF564) and by faraway residues (such as pVHL107 and HIF571). Regarding the performance of the synthetic sensor, the most optimized version, among those examined in the transient test system (Arabidopsis mesophyll protoplasts), turned out to be composed by a constitutively destabilized form of the HIF-CODD peptide (KRP1-AD-HIF) in combination with the wild-type form of the pVHL β-domain (DBD-pVHL).
Sensor Output Dynamics under Reversible Oxygen Inputs
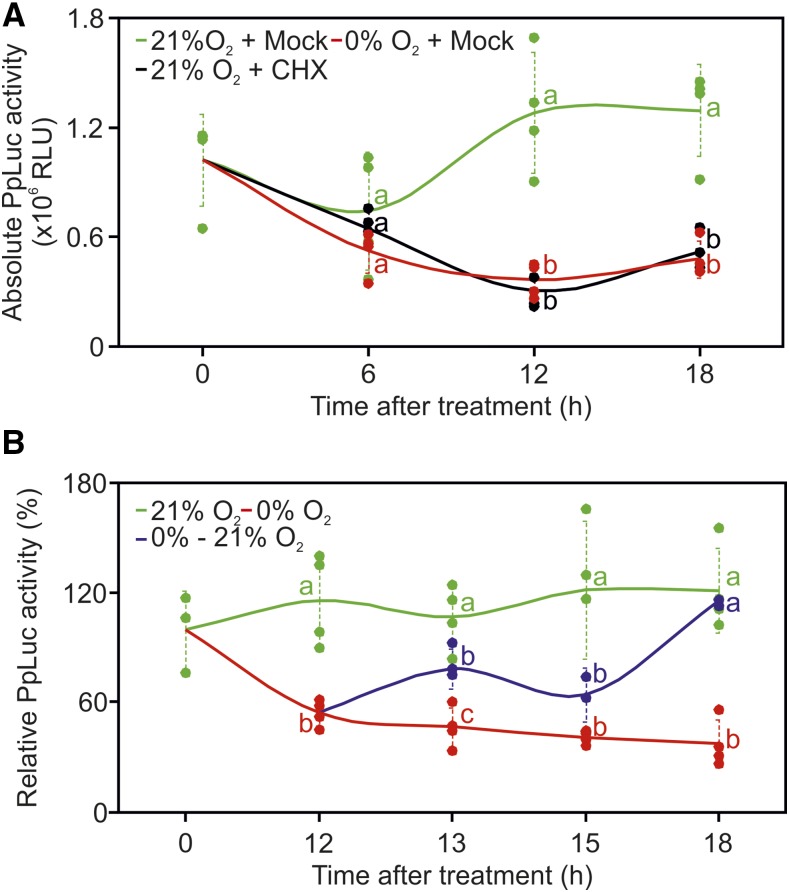
The persistence of luciferase activity observed under anaerobic conditions led us to wonder whether the high stability of the reporter protein could be masking a rapid inactivation of the device after removal of the stimulus (oxygen). Therefore, we decided to investigate the behavior of the sensor in aerobic protoplasts in which de novo reporter synthesis was blocked and compare its output with the reduction of reporter activity observed in anaerobic protoplasts (Fig. 3B). We transformed the test system with all four modules constituting the sensor. This time, to follow the dynamic behavior of the device and to assess the time of anoxia exposure required to turn it into an inactive state, we subjected protoplasts to anoxia for 6, 12, and 18 h, after a 12-h aerobic incubation to ensure the expression of all components. In parallel, we treated a subset of aerobic protoplasts with 100 μm cycloheximide (CHX) to block any de novo synthesis of the effector and reporter proteins and thereby gather information about the luciferase reporter half-life.
The sensor output remained substantially unaltered across the three aerobic time points, suggesting that the reporter protein was kept at a steady-state level during the investigated time frame (Fig. 5A). No significant difference in the output values was observed after 6 h among protoplasts treated with anoxia, normoxia, or CHX, indicating the impossibility of determining a reduction in reporter activity in such a range (Fig. 5A). In particular, the pattern displayed by CHX-treated cells revealed that full turnover of the reporter protein took place only between 12 and 18 h after the block of protein synthesis. The turnover rate measured entails that, as long as a wild-type firefly luciferase protein is used as the output, our transcriptional sensor could display responses to anoxia (i.e. output repression) with a minimum lag of 12 h. Remarkably, a perfectly overlapping trend was observed for CHX-treated and anoxic protoplasts. Anoxia, comparable to CHX application, is expected to prevent novel HIF-pVHL active complex formation; indeed, it caused a significant decrease in luminescence only after 12 h, with no further decrease at the last time point (Fig. 5A). These pieces of evidence clearly demonstrated that our oxygen-dependent device had reached a completely inactive state within 12 h from the onset of oxygen deprivation. In other words, the data suggest that the HIF-pVHL complex assembled during prior aerobic incubation had been processed within such a time frame, while the newly synthesized modules had not been able to associate together.
Figure 5.
Characterization of sensor dynamics. A, Comparison of oxygen sensor output under aerobic conditions (21% O2 + Mock), in the absence of external oxygen (0% O2 + Mock), or in the case of protein synthesis inhibition by 100 μm CHX (21% O2 + CHX) in Arabidopsis protoplasts. Absolute luciferase activity was monitored at 6, 12, and 18 h after the onset of each condition. RLU, Relative luminescence units. B, Sensor output dynamics under reversible oxygen inputs. The 12-h anoxia-treated protoplasts were transferred back to aerobic conditions for 1, 3, or 6 h (blue line), and the output was compared with that produced by cells kept under continuous anoxia (red line) or fully aerated conditions (green line) for the same time span. The luminescence output was normalized to the signal corresponding to the initial time point, set to 100%. Dotted lines show sd (n = 4), and different letters indicate statistical differences (P ≤ 0.05) between treatments at each time point, as assessed by one-way ANOVA followed by Tukey’s posttest.
After we explored the anoxia-dependent inhibition of the sensor, we moved on to evaluate its reversibility; for this purpose, anoxia-treated protoplasts were transferred back to aerobic conditions for 1, 3, or 6 h. Reoxygenation started after 12 h of anoxia, as the end point of sensor inactivation. Full restoration of control levels occurred after 6 h in air, but a response by the sensor was detected already as early as 1 h after reoxygenation (Fig. 5B). Such a fast induction dynamic was in favor of the interpretation of a quick reassembly of the preexisting components (HIFPro and pVHL) into an active complex. In conclusion, our data show that the HIF and pVHL effectors produced in the absence of oxygen cannot interact, but a HIF-pVHL complex is readily reformed as soon as the input is restored.
The HIF/pVHL Transcriptional Complex Does Not Affect Plant Endogenous Transcriptional Regulation
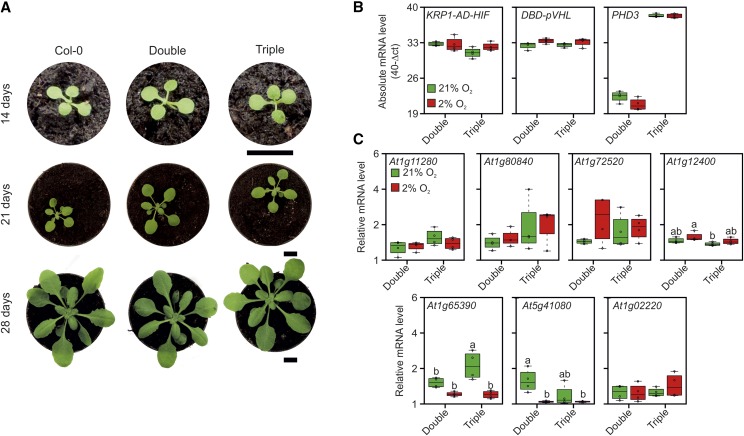
After describing it in a transient test system, we attempted to incorporate the genetically encoded synthetic oxygen sensor in Arabidopsis plants. At first, we generated independent transgenic lines expressing separate modules of the sensor and subsequently combined the modules by crossing. By this procedure, we came up with the production of two different genotypes, one bearing only the two transcriptional effectors AD-HIF and DBD-pVHL (double overexpressor) and another also encoding a PHD3 expression construct (triple overexpressor). Neither genotype exhibited phenotypic differences when compared with wild-type plants, indicating a very limited effect of the transgenes on the overall plant physiology (Fig. 6A).
Figure 6.
Molecular and phenotypic consequences of the expression of the synthetic device. A, Phenotypical analysis of 2-, 3-, and 4-week-old Arabidopsis plants overexpressing KRP1-AD-HIF and DBD-pVHL in the absence (Double) or presence (Triple) of PHD3. Bars = 1 cm. B, Absolute mRNA levels for the three transgenes (KRP1-AD-HIF, DBD-pVHL, and PHD3) assessed by quantitative PCR (qPCR) in 3-week-old plants expressing two (Double) or three (Triple) modules and subjected to aerobic (21% O2) or hypoxic (2% O2) conditions for 12 h. C, qPCR assessment of expression levels of the genes identified as induced by the microarray analysis (Table 1; Supplemental File S1) in double or triple transgenic plants under aerobic (21% O2) or hypoxic (2% O2) conditions. In the box plots, dots represent single data points, the black line marks the median, and the box indicates the interquartile range (IQR). Whiskers extend to data points that are lower than 1.5× IQR away from the box extremities. Different letters indicate statistical differences (P ≤ 0.05) calculated from two-way ANOVA followed by Tukey’s posttest.
Nonetheless, interference of the three transgenes, two of which formed a transcriptional complex, with the regular gene expression profile of the plant could not be ruled out in principle. Therefore, we looked for possible alterations at the transcriptome scale by microarray profiling of the double and triple overexpressors. By this, we aimed to detect interactions between the synthetic transcriptional complex and endogenous genomic sequences of Arabidopsis. A side consideration of the experiment was that the discovery of genomic targets could supply candidate endogenous reporters for sensor activity, overcoming the need for a fourth transgene encoding for a reporter module, such as the one used in the transient expression experiments described above.
The two genotypes exhibited a broad transcriptome overlap (Supplemental File S1), coherent with the absence of visible phenotypic differences between them. The analysis returned a small set of nine genes that were weakly up-regulated in the triple overexpressor seedlings (Table 1), with a maximum induction in the range of 2-fold. Down-regulated genes also were found (Supplemental File S1) but not taken into account, since they are unlikely to represent direct targets of an activator complex.
Table 1. Up-regulated genes in Arabidopsis plants encoding mammalian oxygen sensor components.
Fold change values (normalized expression ratios) were calculated from a microarray experiment comparing 7-d-old seedlings bearing both the effector and sensory modules (triple overexpressors) with seedlings bearing only the effector modules (double overexpressors).
| Arabidopsis Genome Initiative Code | Gene Name | Fold Change |
|---|---|---|
| At1g0220 | NAC003 | 0.97 |
| At3g30720 | QQS | 1.12 |
| At1g27730 | Zat10 | 1.14 |
| At5g41080 | GDPD2 | 1.37 |
| At1g65390 | PP2-A5 | 1.39 |
| At1g12400 | TFB5 | 1.44 |
| At1g72520 | Lox4 | 1.47 |
| At1g80840 | WRKY40 | 1.57 |
| At1g11280 | 1.91 |
To test whether the differentially expressed genes displayed any oxygen regulation, we analyzed their transcripts in rosette leaves of double and triple overexpressing plants subjected to hypoxia (2% oxygen) or maintained under normoxia as a control. We confirmed that all transgenes were expressed properly and found them unaffected by hypoxia (Fig. 6A). However, we could confirm higher expression levels in the triple overexpressors for only one of the nine selected genes, At1g65390, which codes for PHLOEM PROTEIN2 A5 (PP2-A5; Fig. 6B). Moreover, hypoxia repressed its expression in the presence of PHD3.
In summary, these data suggest that the transcriptional device based on yeast and mammalian elements is fully orthogonal to any endogenous pathways converging on transcriptional regulation, which makes it likely to operate without affecting development and growth in plants.
The UAS Promoter Activity Is Influenced by Oxygen Concentration and the Presence of PHD3 Also in Plants
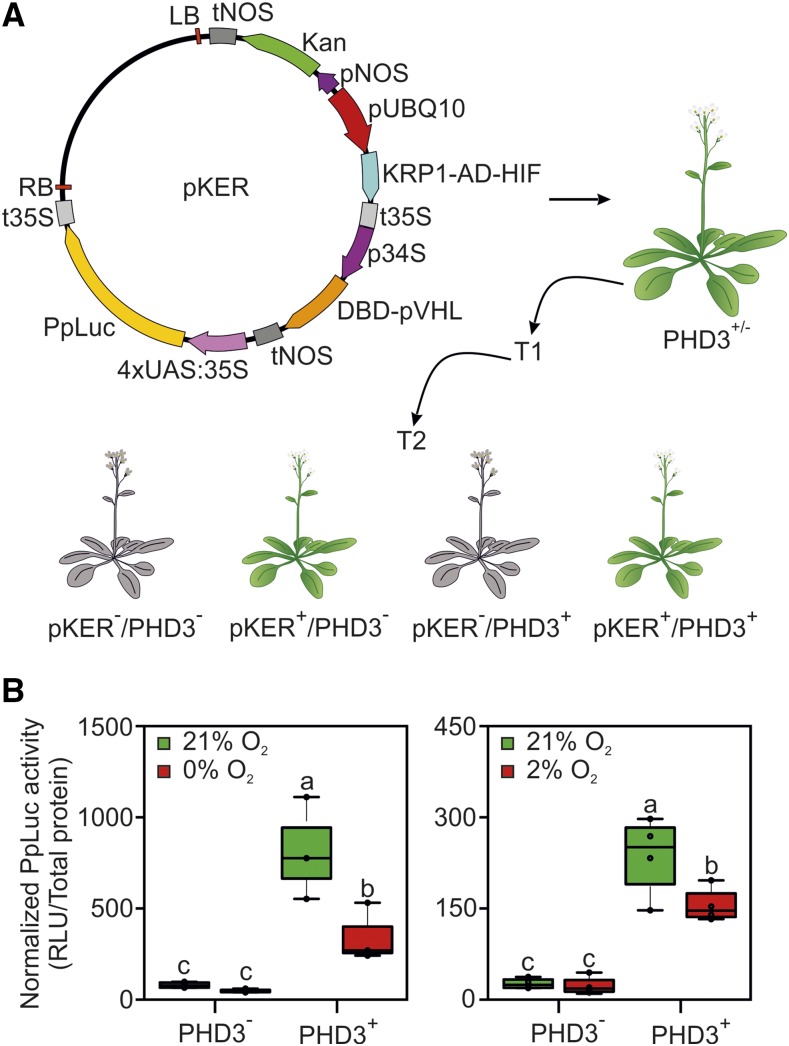
Although one potential endogenous target of the oxygen sensor effectors was found (the At1g65390 gene), its lower induction in the presence of PHD3 and its possible down-regulation by hypoxia make the use of a specifically designed output module still advisable. Therefore, a further construct, bearing the firefly luciferase gene under the control of the UAS promoter, was delivered into Arabidopsis for stable expression. To reduce the time required for the identification of plants carrying all four transgenes, or specific combinations of them depending on the purpose, we devised to generate multiple expression cassettes. In this new strategy, we incorporated three transcriptional units on a single T-DNA, for the expression of the effector modules (encoding KRP121-30-AD-HIF554-576 and DBD-pVHL63-157), and an output module expressing the luciferase enzyme (Fig. 7A). Since the UAS:FLuc reporter construct used for transient assays in protoplasts did not produce a detectable luciferase signal (Supplemental Fig. S3), we designed an alternative reporter consisting of four tandem repeats of UAS inserted between the A1 and B5 domains of the CaMV 35S promoter (Mazarei et al., 2008). The pKER T-DNA (for Kanamycin, Effector and Reporter module) then was delivered to heterozygous Arabidopsis plants carrying the PHD3 sensory module. The integration of three transgenes in a single locus enabled us to rapidly identify plants expressing the transcriptional components of the sensor, whereas the PHD3− control genotype was recovered thanks to the independent segregation of the PHD3-containing T-DNA (Fig. 7A). High transgene expression was measured in the lines selected for the subsequent experiments (Supplemental Fig. S4).
Figure 7.
Responsiveness of the synthetic oxygen sensor stably expressed in Arabidopsis. A, Map of the T-DNA used to link the KRP1-AD-HIF, DBD-pVHL, and 4xUAS:35S:Luc expression cassettes. The construct was transformed into heterozygous 35S:PHD3 plants (PHD3+/−), and first-generation transgenic plants (T1) were selected on kanamycin for the presence of pKER (in the hemizygous state). From their offspring (T2), two genotypes of interest were isolated upon the independent segregation of the PHD3- and pKER-containing T-DNAs: pKER (pKER+/PHD3−) and pKER/PHD (pKER+/PHD3+), corresponding to the colored Arabidopsis plants. Sensor performance in one of the T2 populations produced is shown. B, Normalized firefly activity in pKER seedlings, stably expressing the effector components of the oxygen-responsive device, in the presence or absence of PHD3. Sensor output was recorded after 12-h treatments under complete anoxia (left) or 2% (v/v) oxygen (right). In the box plots, dots represent single data points, the black line marks the median, and the box indicates the interquartile range (IQR). Whiskers extend to data points that are lower than 1.5× IQR away from the box extremities. Different letters indicate statistically significant differences among means (P ≤ 0.05) calculated from two-way ANOVA followed by Tukey’s posttest. RLU, Relative luminescence units.
PHD3+ and PHD3− pKER seedlings were grown on liquid medium and incubated under aerobic (21% [v/v] oxygen) or hypoxic (2% [v/v] oxygen) atmosphere for 12 h. After the treatment, total proteins were extracted, quantified, and subjected to luminescence quantification. PHD3+ aerobic and hypoxia-treated seedlings showed higher luciferase activity than PHD3− seedlings, indicating that the transcriptional complex is forming and inducing transcription of the reporter gene. Moreover, consistent and significant reduction of reporter activity was detected in hypoxic seedlings coming from plants carrying all the components, while no inhibition was observed in plants devoid of PHD3 (Fig. 7B). These data indicate that the device we examined in the transient test system retained full functionality when stably encoded in Arabidopsis plants.
The HIF-pVHL Device Exhibits Higher Hypoxia Specificity Compared with Marker Genes
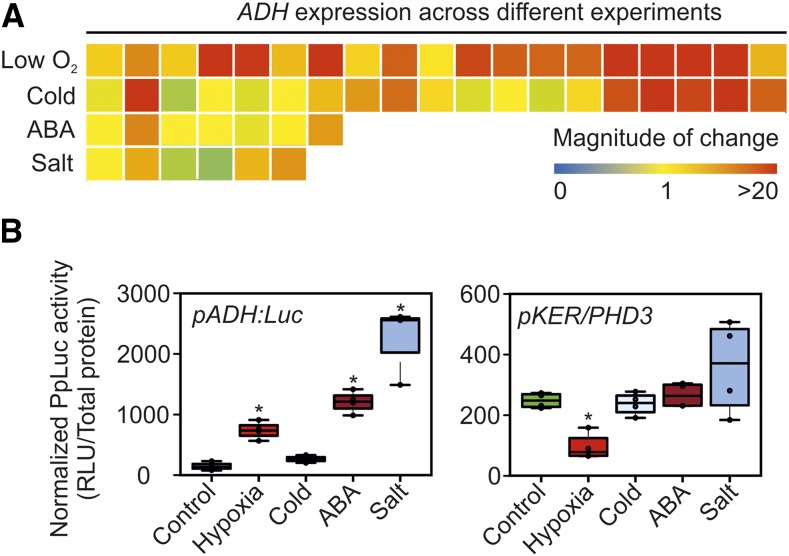
The occurrence of hypoxic conditions in plant tissues can be inferred on the basis of high expression of hypoxia-inducible transcripts, such as ALCOHOL DEHYDROGENASE (ADH; Paul et al., 2001; Gravot et al., 2016). However, an extensive survey of the changes in ADH mRNA under different environmental conditions revealed that this gene actually is controlled by many cues, including cold, high salinity, and abscisic acid (ABA; Fig. 8A; Supplemental File S2). Therefore, we tested whether the synthetic HIF-pVHL-based device responds more specifically to low-oxygen conditions than an ADH promoter-based reporter. To this end, we compared the luminescent output of pKER/PHD3 seedlings with that of plants at the same developmental stage bearing the firefly luciferase gene driven by the genomic region upstream of the ADH gene of Arabidopsis. This pADH:Luc transcriptional reporter showed higher activity in response to 10 μm ABA or 150 mm NaCl than after exposure to hypoxia (1% [v/v] oxygen) for 12 h, while low-temperature treatment of equal duration did not significantly stimulate luciferase accumulation (Fig. 8B). On the other hand, pKER/PHD3 seedlings only produced a significant output under hypoxia (Fig. 8B), demonstrating the superior specificity of the synthetic device over a plant promoter-based reporter.
Figure 8.
Comparative responses of pADH:Luc and PHD3-positive pKER seedlings to abiotic stresses. A, Changes in ADH mRNA across different experiments, gathered with the aid of the Genevestigator reference expression database (Hruz et al., 2008) and listed in Supplemental File S2. B, Luminescent output of pKER/PHD3 pADH:Luc seedlings in response to four different abiotic treatments. In the box plots, dots represent single data points, the black line marks the median, and the box indicates the interquartile range (IQR). Whiskers extend to data points that are lower than 1.5× IQR away from the box extremities. Asterisks indicate statistically significant differences (P ≤ 0.05) from the mean of the control, calculated from one-way ANOVA comparison followed by Fisher’s lsd test. RLU, Relative luminescence units.
DISCUSSION
In this work, we successfully exploited the oxygen-dependent interaction of the mammalian pVHL β-domain with the HIF-CODD to develop a molecular switch to drive gene expression in an oxygen-dependent manner in plant cells and whole seedlings (Figs. 3 and 7). This represents a valuable resource to control metabolism, growth, and development under low- or high-oxygen conditions.
Due to the absence of an efficient transport system for oxygen, its levels vary greatly in plant tissues, depending on environmental parameters and endogenous cues (Sasidharan and Voesenek, 2015). Indeed, the photosynthetic release of oxygen from water molecules and its consumption by mitochondrial respiration contribute to its concentration, while light, temperature, and, especially, exogenous oxygen availability impact both processes. To adjust cell metabolism, tissue growth, and developmental programs to the actual oxygen availability, plants have developed a series of direct and indirect sensing mechanisms of the oxic status. The mechanisms identified so far are tightly interconnected with other signaling pathways, including light perception, sugar sensing, nitric oxide, and reactive oxygen species responses. Moreover, the transcriptional network activated in this way has been shown to control a plethora of responses, including the induction of fermentation, inhibition of chlorophyll biosynthesis, defense against pathogens, and root and shoot development (Giuntoli and Perata, 2018).
For these reasons, genetic intervention to selectively modulate a subset of these processes in response to variations in oxygen availability represents a true challenge in plants, even accounting for the possibility of precise genome editing offered by emerging technologies. A valuable alternative is represented by synthetic sensory modules affected by oxygen but not by other cellular pathways (orthogonal) engineered to specifically control one or more processes. The HIF-pVHL device used here already has been proven to work in a heterologous chassis by Alcaide-German et al. (2008), who developed an in vivo assay for the study of PHD biochemistry in S. cerevisiae. Using a similar set of protein fusions between the pVHL β-domain and the HIF1α-CODD with the DBD and AD of the Gal4 transcription factor, respectively, we confirmed PHD3 activity and selectivity on HIF-CODD, together with the ability of the pVHL β-domain to interact with the hydroxylated form of HIF-CODD (HIFHyp) also in plant cells (Fig. 2B). Moreover, since no activation of the downstream reporter was observed in the absence of PHD3 (Fig. 2B), we concluded that the interaction of the two chimeric proteins is orthogonal to the host plant system. Indeed, we showed that our synthetic oxygen sensor responds more specifically to hypoxia than simple transcriptional reporters (Fig. 8B).
A first main challenge encountered during the optimization of the sensor consisted in expanding the extent of repression brought about by hypoxia (i.e. its dynamic range). Ideally, the device is expected to switch from an on state under normoxia to an off state under near anoxia, passing through a series of gradual states at intermediate oxygen levels. Indeed, this kind of continuous state transition has been observed for the endogenous HIF and Group VII Ethylene Response Factor systems in animals and plants, respectively (Ceradini et al., 2004; Kosmacz et al., 2015). These oxygen-sensing mechanisms are both based on proteasomal degradation and additionally integrated in a complex network of feed-back and feed-forward loops (Masson et al., 2012; Weits et al., 2014; Kietzmann et al., 2016) that likely ensure stepwise regulation. Relying on a single oxygen-dependent mechanism, instead, our luminescent reporter exhibited only marginal repression when environmental anoxia was imposed (Fig. 2C). By enhancing the turnover rate of the AD-HIF module via a KRP1 degron, we successfully achieved greater response by our oxygen sensor (Fig. 3).
Rational substitution of amino acids crucial for the HIF-pVHL interaction did not lead to any overall improvement of the dynamic range of our sensor. Among all mutations tested, only a Tyr-98Asn conversion on the pVHL fragment was associated with mildly (by 10%) increased luciferase activity between anoxia and normoxia, although at the same time decreasing reporter expression under aerobic conditions (Fig. 4D). Nonetheless, this approach allowed the unprecedented identification of a set of pVHL variants (Tyr-98Phe, Tyr-98Cys, and Ser-111Ala) whose binding to the HIF1α-CODD was not inhibited by anoxia (Fig. 4, C and D; Supplemental Fig. S2C). Future mutational strategies may be directed at displacing residues involved in weak interactions to modify the local conformation of the complex. An alternative strategy aimed at further reducing reporter expression under anoxia may consist in providing the DBD-pVHL with a repressor domain that is masked by the interaction with AD-HIF, thereby stimulating the competition between repressive VHL monomers and activating heterodimers in an oxygen-dependent manner.
A second limiting aspect of the proposed oxygen sensor is represented by the time scale required to observe substantial output changes after a reduction in oxygen availability. In both transiently and stably transformed cells, 12 h were necessary to observe a significant decrease in luminescence (Fig. 5), a time frame unsuitable to monitor oxygen dynamics within tissues on a daily basis. Attainment of the inactive state was comparable with that caused by translation inhibition and thereby in line with the time required for luciferase protein turnover, whereas more rapid activation by reoxygenation occurred in 1 h (Fig. 5B), compatible with the time required for de novo luciferase synthesis. Under this perspective, future improvements of this kind of transcriptional sensor are likely to be achieved with alternative reporters characterized by rapid turnover; for instance, a luciferase fusion with the KRP121-30 destabilization domain could be adopted.
Finally, a third crucial property of a molecular sensor is its sensitivity to the concentration of the analyte. In the case of our device, sensitivity is expected to be determined by PHD affinity for oxygen. In vitro assays revealed an apparent Km of 230 to 250 µm for HIF-PHDs (Flashman et al., 2008), low enough to be compatible with a sensing function in animal cells; indeed, in human cell cultures, PHD2 activity on a HIF1α556-574 peptide was observed to decrease upon a transition from 210 to 100 µm dissolved oxygen and drop in the range of 30 to 80 µm, with a half-maximum activity close to 50 µm oxygen (Berchner-Pfannschmidt et al., 2008). No in vivo kinetic data have been produced so far for the PHD3 isoform. When expressed transiently in Arabidopsis protoplasts, our PHD3-based sensor worked in response to a moderate decrease in oxygen concentration (5% [v/v] oxygen, corresponding to 67 µm dissolved oxygen; Fig. 3E), compatible with the behavior of PHD2. However, the response could not be pushed further at lower oxygen concentrations, at least in the same time frame. The inability to discriminate signals in the range of 0% to 5% (v/v) oxygen currently prevents the exploitation of this molecular switch to drive selective responses to severe oxygen deficiency, such as those arising in plant organs experiencing dark submergence stresses (Lee et al., 2011). Nonetheless, our device holds the potential to detect softer hypoxic conditions, such as the onset of endogenous oxygen gradients that generate inside plant tissues during growth and development (van Dongen and Licausi, 2015).
After correcting some pitfalls that currently limit the applicability of the molecular switch, multiple options for future exploitation can be envisioned. First, as just stated, it can be used to report the actual oxygen status of cells and tissues in plants. This requires equipping the device with a suitable reporter protein characterized by rapid synthesis and turnover as well as independence from the provision of exogenous substrates. To this end, fluorescent proteins are probably the preferable option, as demonstrated by their employment to build a plethora of synthetic sensors (Walia et al., 2018). Second, the PHD-dependent HIF-pVHL dimerization can be exploited to reconstitute transcription factors different from yeast Gal4, or even enzymes, and drive signaling, metabolic, or developmental processes in an oxygen-dependent manner in plants. This can be exploited to improve crop yield under challenging oxygen limitation, such as flooding or waterlogging. Alternatively, the artificial alteration of oxygen provision to plants or cell cultures can be used, through the HIF-pVHL switch, as a means to control biochemical pathways toward the production of specific molecules.
In summary, with this work, we demonstrated that the mammalian oxygen-sensing machinery can be harnessed to generate an orthogonal switch able to regulate gene expression in response to hypoxic conditions. It represents the first step for the introduction and enhancement of adaptive traits in response to hypoxic conditions, such as flooding and waterlogging.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Columbia-0 ecotype of Arabidopsis (Arabidopsis thaliana) was used as the wild-type background in all the experiments. Transgenic plants expressing the individual modules or the entire synthetic regulatory device were obtained by Agrobacterium tumefaciens-mediated transformation, applying the floral dip method (Clough and Bent, 1998). Transgenic seedlings were selected for resistance on the appropriate antibiotic or herbicide and verified subsequently by PCR for the presence of the desired transgenes.
For plant growth in soil, seeds were sown directly on a peat:perlite 3:1 mixture, vernalized at 4°C for 48 h in the dark, and then germinated at 22°C day/18°C night with a photoperiod of 12 h. For plant growth in sterility, seeds were sterilized using sequential washes in 70% (v/v) ethanol and 10% (v/v) commercial bleach solution and then rinsed five times with sterile distilled water. Seeds then were sown on either liquid or agarized (8 g L−1) Murashige and Skoog one-half-strength medium, vernalized, and grown as described above.
Abiotic Treatments
The plant material was subjected to low-oxygen atmospheres under darkness, using a hypoxic chamber glove box (COY Lab Products), for the time specified. ABA and NaCl were supplemented in the sterile medium at final concentrations of 10 μm and 150 mm, respectively. Cold stress was applied by incubating at 4°C in the dark. All treatments applied to seedlings lasted 12 h.
DNA Fragment Cloning and Assembly
Transcriptional units were either cloned from genomic DNA or cDNA or de novo synthesized by GeneArt (Thermo Fisher Scientific). Protein-coding sequences were codon optimized using the EMBOSS Backtranseq online tools (McWilliam et al., 2013). The KRP1-AD-HIF transcriptional unit was realized by overlapping PCR using KRP1-AD-Fw and KRP1-Fw in combination with HIF-Rv primers. The DBD-pVHL transcriptional unit was realized by overlapping PCR using DBD-Fw, DBD-pVHL-Fw, DBD-pVHL-Rv, and pVHL-Rv primers. Four tandem repeats of the UAS element were inserted, via overlapping PCR, 90 bp upstream of the transcriptional start unit of a 464-bp-long CaMV 35S promoter using 35S-Fw, 35S-UAS-Rv, 35S-UAS-Fw, UAS-35S-Fw, UAS-35S-Rv, and UAS-Rv primers. A full list of synthetic sequences used in this work is provided in Supplemental Information S1. The various DNA fragments were initially subcloned either in the pENTR/D-TOPO vector, to be recombined into destination vectors using Gateway LR Clonase II Enzyme mix (Thermo Fisher Scientific), or in the pCR2.1-TOPO-TA vector (Thermo Fisher Scientific), to be inserted into expression vectors via a restriction-ligation strategy. The resulting plasmids then were verified using restriction enzymes and, when needed, by sequencing. Plasmid maps were generated with Serial Cloner 2.6.1 (Serial Basic). A full list and description of expression plasmids is provided Supplemental Table S1.
The pKER plasmid containing three expression cassettes was obtained by assembling two promoter-terminator cassettes into a compatible SacI restriction site of the pKGWL7 plasmid (Karimi et al., 2002). The first cassette consisted of the Arabidopsis UBQ10 promoter (Grefen et al., 2010), flanked by the SacI and BstBI restriction sites, followed by the CaMV 35S terminator (Hirt et al., 1990), cloned in between the SfaAI and PacI recognition sequences. The second cassette comprised the viral fig mosaic virus 34S promoter (Sanger et al., 1990), separated from the NOS terminator (Depicker et al., 1982) by sequential AfeI and XmaJ1 restriction sites. The two sites present between the promoters and terminators were used to insert specific coding sequences. Specifically, BstBI and SfaAI were used to ligate the KRP1-AD-HIF effector in the first cassette and AfeI and XmaJ1 were used to insert the DBD-pVHL effector in the second cassette; these restriction extremities were incorporated in the primers used to subclone each coding sequence (BstbIKRP1-Fw and SfaAIKRP1-Rv primers for KRP1-AD-HIF and AfeIDBD-Fw and XmajDBD-Rv primers for DBD-pVHL). The resulting destination vector was finally recombined with an entry vector containing the 4xUAS:min35S sequence to generate the final expression vector pKER4xUAS, whose sequence is provided in Supplemental Information S1. The 5′ region upstream of the ADH gene (At1g77120) was PCR amplified using Phusion polymerase (Thermo Fisher Scientific) on Arabidopsis Columbia-0 genomic DNA and cloned into pENTR/D-TOPO vector (Thermo Fisher Scientific). Subsequently, the ADH promoter was recombined into pH7LWG by LR Clonase (Thermo Fisher Scientific). A full list of primers used in cloning procedures is provided in Supplemental Table S2. pVHL and HIF site-directed mutagenesis was performed by PCR. A full list of primers utilized for site-directed mutagenesis is provided in Supplemental Table S3.
Large-Scale Plasmid DNA Purification
Highly concentrated plasmid DNA, required for protoplast transformation, was extracted from a 100-mL bacterial culture (Luria-Bertani medium supplemented with the appropriate antibiotic) by resuspension in 2 mL of a buffer containing 50 mm Glc, 25 mm Tris-HCl, and 10 mm EDTA (pH 8), lysis in 3 mL of a buffer containing 200 mm sodium hydroxide and 1% (w/v) SDS, followed by neutralization with 4 mL of ice-cold 3 m potassium acetate in 11.5% (v/v) glacial acetic acid. Nucleic acids were precipitated with an equal volume of isopropanol, and plasmid DNA was further purified by RNase A (Sigma-Aldrich) treatment for 30 min at 37°C followed by polyethylene glycol (PEG) precipitation (13% [w/v] PEG 8000 dissolved in 1.6 m NaCl), phenol:chloroform extraction, and ethanol precipitation. Plasmid DNA was finally eluted in a suitable volume of nuclease-free water to achieve a concentration of 1 to 2 μg μL−1.
Protoplast Isolation and Transformation
Protoplast isolation and transformation were performed as reported by Wu et al. (2009) with some modifications. All solutions, with the exception of PEG, were filter sterilized before use. Leaves of 3-week-old plants were deprived of the lower epidermis using paper tape, immersed in an enzymatic solution (0.4 m mannitol, 20 mm KCl, 20 mm MES, pH 5.7, 10 mm CaCl2, 1% [w/v] cellulase, and 0.4% [w/v] macerozyme), and incubated at 22°C in the dark for 3 h. The released protoplasts were filtered and washed in W5 (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES). For transfection, protoplasts were centrifuged at 100g for 1 min and resuspended in 0.4 m mannitol, 15 mm MgCl2, and 5 mm MES to a final concentration of 5 × 105 cells mL−1. Then, 100 µL of protoplasts was mixed with 4 µg of each effector and reporter plasmid, plus an equal volume of freshly prepared solution of 40% (w/v) PEG 4000, 100 mm CaCl2, and 200 mm mannitol. The mixture was incubated for 20 min at room temperature in the dark. After incubation, 440 µL of W5 was added, and protoplasts were gently pelleted at 100g for 1 min and resuspended in 1 mL of WI (500 mm mannitol, 4 mm MES, and 20 mm KCl). Protoplasts were incubated on six-well plates at 22°C in the dark for a minimum time of 12 h before starting any treatment. CHX treatment was performed as reported by Giuntoli et al. (2017). For luciferase activity assays, protoplasts were collected and centrifuged at 700g for 2 min. WI was removed, and protoplasts were flash frozen in liquid nitrogen.
Luciferase Activity Quantification
Frozen protoplasts were lysed using 50 μL of Passive Lysis buffer (Promega). Firefly (Photinus pyralis) and sea pansy (Renilla reniformis) luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s instructions. Luciferase activity in transgenic seedlings was measured with the same method in total protein pools extracted with Passive Lysis buffer. Total proteins were quantified using the Bradford protein assay (Bio-Rad).
Confocal Imaging
Confocal investigations were performed using a Zeiss AiryScan confocal microscope. GFP fluorescence was excited at 488 nm and collected with a 497- to 554-nm long-pass emission filter. Chlorophyll autofluorescence was excited at 633 nm and collected at 650 to 750 nm.
Images were analyzed with the ZEN 2010 software (Zeiss). Nuclei were stained with 1 µg µL−1 4′,6-diamidino-2-phenylidone (Sigma-Aldrich), and fluorescence was excited at 405 nm and collected at 410 to 470 nm.
MD Simulations and MM-GBSA
Classical MD simulations were performed with the Amber14 package. The HIF1α-pVHL complex structure was extracted from the crystal structure of a hydroxylated HIF1α peptide bound to the pVHL/elongin-C/elongin-B complex (Protein Data Bank code 1LQB; Hon et al., 2002).
The water-solvated system was minimized, heated up to 300 K, and equilibrated by applying positional harmonic restraints (2 kcal mol−1 Å−2) on the protein residues. Finally, a production run of 35 ns in the constant temperature and pressure ensemble was conducted without any restraints. Technical details about the MD simulation are reported in Supplemental Methods S1. MM/GBSA calculations (Kollman et al., 2000; Onufriev et al., 2000; Wang et al., 2006; Hou et al., 2011) were performed to determine the binding free energy (ΔG) for each of the considered mutants. We employed the MMPBSA.py module of AmberTools (Miller et al., 2012). MM/GBSA calculations were run along short (1-ns) MD runs performed with strong positional restraints (25 kcal mol−1 Å−2) on all the protein residues outside a 20-Å radius from the Hyp-564 residue. This allows focusing on the contributions from the protein residues close to the binding pocket while neglecting the contributions from conformational changes in the flexible region of the protein. In short, 15 uncorrelated frames every 1 ns were extracted and the protein fragments were mutated, maintaining the coordinates of the wild-type backbone and the unchanged side chain atoms; subsequently, the geometry of the mutated residue was relaxed with 1,000 optimization steps, keeping the rest of the system frozen, and finally a short MD run was performed to extract 100 frames while skipping the first 10 ps to allow the system to equilibrate. For each frame, MM/GBSA calculations were carried out to obtain the free energy of HIF1α, pVHL, and the HIF1α-pVHL complex and to compute the binding free energy (further technical details are provided in Supplemental Methods S2).
RNA Extraction and Gene Expression Analysis
For microarray analysis, pools of 20 seedlings were harvested at the end of the day and total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma). Hybridization against the Arabidopsis Gene 1.0 ST Array and scanning procedures were performed by Atlas Biolabs. The raw microarray data were normalized and signal intensities were calculated using RobiNA (Lohse et al., 2012), applying the Robust MultiArray methodology. Differential expression analysis was carried out using the Limma R package (Ritchie et al., 2015).
For qPCR analysis, total RNA was extracted from 4-week-old plants as described by Kosmacz et al. (2015). cDNA was synthesized from 1 µg of total RNA using the Maxima Reverse Transcriptase kit (Life Technologies). qPCR amplification was performed on 12.5 ng of cDNA with the ABI Prism 7300 sequence detection system (Applied Biosystems), using the PowerUp SYBR Green Master Mix (Applied Biosystems). Ubiquitin10 (At4g053290) was exploited as the housekeeping gene. A full list of the primers used for qPCR is provided in Supplemental Table S4.
Statistical Analysis
Ordinary two-way and one-way ANOVA and multiple comparisons for statistical differences were performed with GraphPad Prism 7 for Windows 10.
Accession Numbers
Microarray data are deposited in the Gene Expression Omnibus database with the series number GSE118364.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of sensor output between the stable and destabilized AD-HIF variants in Arabidopsis protoplasts.
Supplemental Figure S2. In silico prediction and testing of pVHL variants.
Supplemental Figure S3. Basal activity of the exogenous 4xUAS promoter in plants.
Supplemental Figure S4. Overexpression of sensor modules in pKER plants.
Supplemental Table S1. List of plasmids generated in the frame of the study.
Supplemental Table S2. List of oligonucleotides used for cloning.
Supplemental Table S3. List of oligonucleotides used for site-directed mutagenesis.
Supplemental Table S4. List of qPCR primers used in this study.
Supplemental Information S1. Nucleotide sequences of the constructs used in the study.
Supplemental Methods S1. Extended description of the MD simulation procedure.
Supplemental Methods S2. Extended description of the MM/GBSA calculations.
Supplemental File S1. Microarray comparison of transgenic seedlings expressing two effector modules in the presence or absence of the PHD3 sensory module.
Supplemental File S2. Changes in ADH mRNA levels in response to different treatments.
Acknowledgments
We thank Daan A. Weits and Vinay Shukla for technical support with the confocal microscopy and Giacomo Novi for plant growth supervision.
References
- Abbas M, Berckhan S, Rooney DJ, Gibbs DJ, Vicente Conde J, Sousa Correia C, Bassel GW, Marín-de la Rosa N, León J, Alabadí D, et al. (2015) Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr Biol 25: 1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide-German ML, Vara-Vega A, Garcia-Fernandez LF, Landazuri MO, del Peso L (2008) A yeast three-hybrid system that reconstitutes mammalian hypoxia inducible factor regulatory machinery. BMC Cell Biol 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem 279: 38458–38465 [DOI] [PubMed] [Google Scholar]
- Armstrong W. (1980) Aeration in higher plants. Adv Bot Res 7: 225–332 [Google Scholar]
- Bailey-Serres J, Voesenek LACJ (2008) Flooding stress: Acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Lee SC, Brinton E (2012) Waterproofing crops: Effective flooding survival strategies. Plant Physiol 160: 1698–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Voytas DF (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33: 120–131 [DOI] [PubMed] [Google Scholar]
- Berchner-Pfannschmidt U, Tug S, Trinidad B, Oehme F, Yamac H, Wotzlaw C, Flamme I, Fandrey J (2008) Nuclear oxygen sensing: Induction of endogenous prolyl-hydroxylase 2 activity by hypoxia and nitric oxide. J Biol Chem 283: 31745–31753 [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, Capla JM, Galiano RD, Levine JP, Gurtner GC (2004) Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med 10: 858–864 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crane TA, Roncoli C, Hoogenboom G (2011) Adaptation to climate change and climate variability: The importance of understanding agriculture as performance. NJAS Wagening J Life Sci 57: 179–185 [Google Scholar]
- Cui X. (2012) Nutrient sensing in plants. Mol Plant 5: 1167–1169 [DOI] [PubMed] [Google Scholar]
- Depicker A, Stachel S, Dhaese P, Zambryski P, Goodman HM (1982) Nopaline synthase: Transcript mapping and DNA sequence. J Mol Appl Genet 1: 561–573 [PubMed] [Google Scholar]
- Depping R, Steinhoff A, Schindler SG, Friedrich B, Fagerlund R, Metzen E, Hartmann E, Köhler M (2008) Nuclear translocation of hypoxia-inducible factors (HIFs): Involvement of the classical importin α/β pathway. Biochim Biophys Acta 1783: 394–404 [DOI] [PubMed] [Google Scholar]
- Ehrismann D, Flashman E, Genn DNN, Mathioudakis N, Hewitson KSS, Ratcliffe PJJ, Schofield CJJ (2007) Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem J 401: 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y (1997) A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA 94: 4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein ACR, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Erb TJ, Zarzycki J (2016) Biochemical and synthetic biology approaches to improve photosynthetic CO2-fixation. Curr Opin Chem Biol 34: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman E, Bagg EAL, Chowdhury R, Mecinović J, Loenarz C, McDonough MA, Hewitson KS, Schofield CJJ (2008) Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases. J Biol Chem 283: 3808–3815 [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Isa NM, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP, et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B, Perata P (2018) Group VII ethylene response factors in Arabidopsis: Regulation and physiological roles. Plant Physiol 176: 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B, Shukla V, Maggiorelli F, Giorgi FM, Lombardi L, Perata P, Licausi F (2017) Age-dependent regulation of ERF-VII transcription factor activity in Arabidopsis thaliana. Plant Cell Environ 40: 2333–2346 [DOI] [PubMed] [Google Scholar]
- Gravot A, Richard G, Lime T, Lemarié S, Jubault M, Lariagon C, Lemoine J, Vicente J, Robert-Seilaniantz A, Holdsworth MJ, et al. (2016) Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot. BMC Plant Biol 16: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA (1998) Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr 7: 205–213 [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Yocum RR, Gifford P (1982) A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci USA 79: 7410–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem 278: 30772–30780 [DOI] [PubMed] [Google Scholar]
- Hirt H, Kögl M, Murbacher T, Heberle-Bors E (1990) Evolutionary conservation of transcriptional machinery between yeast and plants as shown by the efficient expression from the CaMV 35S promoter and 35S terminator. Curr Genet 17: 473–479 [DOI] [PubMed] [Google Scholar]
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY (2002) Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417: 975–978 [DOI] [PubMed] [Google Scholar]
- Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr (2001) HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. (2001) Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472 [DOI] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keegan L, Gill G, Ptashne M (1986) Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science 231: 699–704 [DOI] [PubMed] [Google Scholar]
- Kietzmann T, Mennerich D, Dimova EY (2016) Hypoxia-Inducible Factors (HIFs) and phosphorylation: Impact on stability, localization, and transactivity. Front Cell Dev Biol 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauth K, Bex C, Jemth P, Buchberger A (2006) Renal cell carcinoma risk in type 2 von Hippel-Lindau disease correlates with defects in pVHL stability and HIF-1alpha interactions. Oncogene 25: 370–377 [DOI] [PubMed] [Google Scholar]
- Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. (2000) Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res 33: 889–897 [DOI] [PubMed] [Google Scholar]
- Kosmacz M, Parlanti S, Schwarzländer M, Kragler F, Licausi F, Van Dongen JT (2015) The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ 38: 1094–1103 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Pelayo MAF, Trijatmiko KR, Gabunada LFM, Alam R, Jimenez R, Mendioro MS, Slamet-Loedin IH, Sreenivasulu N, Bailey-Serres J, et al. (2015) A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat Plants 1: 15124. [DOI] [PubMed] [Google Scholar]
- Lee SC, Mustroph A, Sasidharan R, Vashisht D, Pedersen O, Oosumi T, Voesenek LACJ, Bailey-Serres J (2011) Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol 190: 457–471 [DOI] [PubMed] [Google Scholar]
- Li Q, Shi X, Ye S, Wang S, Chan R, Harkness T, Wang H (2016) A short motif in Arabidopsis CDK inhibitor ICK1 decreases the protein level, probably through a ubiquitin-independent mechanism. Plant J 87: 617–628 [DOI] [PubMed] [Google Scholar]
- Licausi F, Perata P (2009) Low oxygen signaling and tolerance in plants. Adv Bot Res 50: 139–198 [Google Scholar]
- Liu W, Stewart CN Jr (2015) Plant synthetic biology. Trends Plant Sci 20: 309–317 [DOI] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B (2012) RobiNA: A user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40: W622–W627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Valeri MC, Novi G, Perata P (2018) Gene regulation and survival under hypoxia requires starch availability and metabolism. Plant Physiol 176: 1286–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ (2001) Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J 20: 5197–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Singleton RS, Sekirnik R, Trudgian DC, Ambrose LJ, Miranda MX, Tian YM, Kessler BM, Schofield CJ, Ratcliffe PJ (2012) The FIH hydroxylase is a cellular peroxide sensor that modulates HIF transcriptional activity. EMBO Rep 13: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275 [DOI] [PubMed] [Google Scholar]
- Mazarei M, Teplova I, Hajimorad MR, Stewart CN (2008) Pathogen phytosensing: Plants to report plant pathogens. Sensors (Basel) 8: 2628–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41: W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitha K, Agudelo-Romero P, Signorelli S, Gibbs DJ, Considine JA, Foyer CH, Considine MJ (2018) Developmental control of hypoxia during bud burst in grapevine. Plant Cell Environ 41: 1154–1170 [DOI] [PubMed] [Google Scholar]
- Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, Huang WQ, Wotzlaw C, Hellwig-Bürgel T, Jelkmann W, et al. (2003) Intracellular localisation of human HIF-1 alpha hydroxylases: Implications for oxygen sensing. J Cell Sci 116: 1319–1326 [DOI] [PubMed] [Google Scholar]
- Miller BR III, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA.py: An efficient program for end-state free energy calculations. J Chem Theory Comput 8: 3314–3321 [DOI] [PubMed] [Google Scholar]
- Min JH, Yang H, Ivan M, Gertler F, Kaelin WG Jr, Pavletich NP (2002) Structure of an HIF-1alpha-VHL complex: Hydroxyproline recognition in signaling. Science 296: 1886–1889 [DOI] [PubMed] [Google Scholar]
- Mommer L, Visser EJW (2005) Underwater photosynthesis in flooded terrestrial plants: A matter of leaf plasticity. Ann Bot 96: 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F, Crook MB, Garcia K, Garcia Costas A, Geddes BA, Kouri ED, Paramasivan P, Ryu MH, Oldroyd GED, Poole PS, et al. (2016) Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol 82: 3698–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GC, Valin H, Sands RD, Havlík P, Ahammad H, Deryng D, Elliott J, Fujimori S, Hasegawa T, Heyhoe E, et al. (2014) Climate change effects on agriculture: Economic responses to biophysical shocks. Proc Natl Acad Sci USA 111: 3274–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufriev A, Bashford D, Case DA (2000) Modification of the generalized born model suitable for macromolecules. J Phys Chem B 104: 3712–3720 [Google Scholar]
- Paul AL, Daugherty CJ, Bihn EA, Chapman DK, Norwood KL, Ferl RJ (2001) Transgene expression patterns indicate that spaceflight affects stress signal perception and transduction in Arabidopsis. Plant Physiol 126: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingali PL. (2012) Green revolution: Impacts, limits, and the path ahead. Proc Natl Acad Sci USA 109: 12302–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A, Fleck C, Hiltbrunner A (2014) Shedding (far-red) light on phytochrome mechanisms and responses in land plants. Plant Sci 217-218: 36–46 [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger M, Daubert S, Goodman RM (1990) Characteristics of a strong promoter from figwort mosaic virus: Comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14: 433–443 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Voesenek LACJ (2015) Ethylene-mediated acclimations to flooding stress. Plant Physiol 169: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG Jr, Pavletich NP (1999) Structure of the VHL-ElonginC-ElonginB complex: Implications for VHL tumor suppressor function. Science 284: 455–461 [DOI] [PubMed] [Google Scholar]
- van Dongen JT, Licausi F (2015) Oxygen sensing and signaling. Annu Rev Plant Biol 66: 345–367 [DOI] [PubMed] [Google Scholar]
- Walia A, Waadt R, Jones AM (2018) Genetically encoded biosensors in plants: Pathways to discovery. Annu Rev Plant Biol 69: 497–524 [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA 92: 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hou T, Xu X (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput Aided Drug Des 2: 287–306 [Google Scholar]
- Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten HM, Riegler H, Hoefgen R, Perata P, van Dongen JT, Licausi F (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun 5: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yang S, Vanderbeld B, Wan J, Huang Y (2010) Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol Plant 3: 469–490 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]