Parts-prospecting in prokaryotes and plants identified candidate enzymes to design and build a more energy-efficient thiamin synthesis pathway to boost crop yields.
Abstract
Plants synthesize the thiazole precursor of thiamin (cThz-P) via THIAMIN4 (THI4), a suicide enzyme that mediates one reaction cycle and must then be degraded and resynthesized. It has been estimated that this THI4 turnover consumes 2% to 12% of the maintenance energy budget and that installing an energy-efficient alternative pathway could substantially increase crop yield potential. Available data point to two natural alternatives to the suicidal THI4 pathway: (i) nonsuicidal prokaryotic THI4s that lack the active-site Cys residue on which suicide activity depends, and (ii) an uncharacterized thiazole synthesis pathway in flowers of the tropical arum lily Caladium bicolor that enables production and emission of large amounts of the cThz-P analog 4-methyl-5-vinylthiazole (MVT). We used functional complementation of an Escherichia coli ΔthiG strain to identify a nonsuicidal bacterial THI4 (from Thermovibrio ammonificans) that can function in conditions like those in plant cells. We explored whether C. bicolor synthesizes MVT de novo via a novel route, via a suicidal or a nonsuicidal THI4, or by catabolizing thiamin. Analysis of developmental changes in MVT emission, extractable MVT, thiamin level, and THI4 expression indicated that C. bicolor flowers make MVT de novo via a massively expressed THI4 and that thiamin is not involved. Functional complementation tests indicated that C. bicolor THI4, which has the active-site Cys needed to operate suicidally, may be capable of suicidal and – in hypoxic conditions – nonsuicidal operation. T. ammonificans and C. bicolor THI4s are thus candidate parts for rational redesign or directed evolution of efficient, nonsuicidal THI4s for use in crop improvement.
Searching nature for genes and proteins to develop into synthetic biology components (parts-prospecting) is a key activity that provides new parts to the engineering design-build-test-learn cycle and helps drive continuous innovation (Wang et al., 2013; Jensen and Keasling, 2015). Parts-prospecting is becoming increasingly central to progress in the synthetic biology of plant metabolism as the field runs out of targets of opportunity (i.e. well-established primary and secondary pathways) and moves on strategically to next-generation target pathways for which defined components are not yet available off-the-shelf (Erb et al., 2017; Pouvreau et al., 2018).
Some of the most promising next-generation targets are energy-inefficient metabolic pathways for which efficient alternatives exist in nature, or could theoretically exist (Erb et al., 2017). Using such energy-saving pathways to replace native ones has the potential to increase crop yields; a well-explored example is the Tyr shortcut to p-coumarate in lignin biosynthesis (Amthor, 2003; Maeda, 2016). Another target is the inefficient way that plants synthesize the thiazole moiety of thiamin (Fig. 1A), as explained briefly below and more fully elsewhere (Hanson et al., 2018). Thiamin, in its diphosphate form, is the cofactor for several central metabolic enzymes including the α-keto acid dehydrogenase complexes, transketolase, and acetolactate synthase (Goyer, 2010; Fitzpatrick and Thore, 2014).
Figure 1.
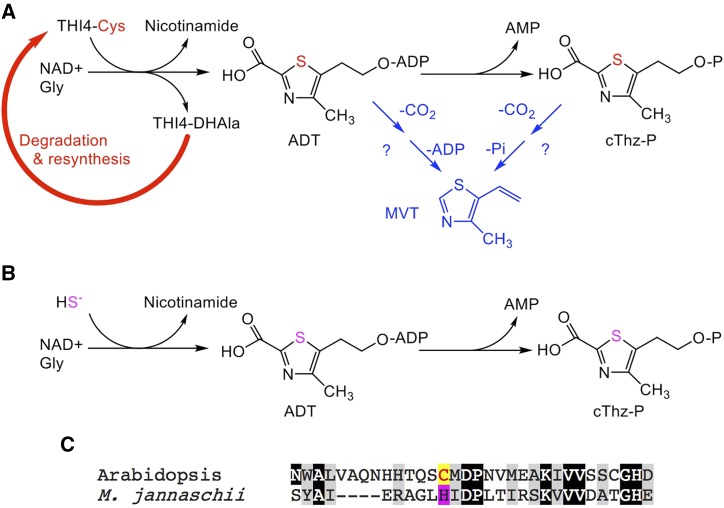
THI4-mediated synthesis of the thiamin thiazole precursors and their possible relationship to MVT. A, In the plant thiazole synthesis pathway, THI4 forms the adenylated thiazole precursor ADT from NAD+, Gly, and a sulfur atom stripped from an active-site Cys residue, which is changed to dehydro-Ala (DHAla). The inactive DHAla form of THI4 is degraded. ADT is cleaved to give cThz-P, the direct precursor of thiamin. MVT could theoretically be derived from ADT or cThz-P by decarboxylation and elimination of ADP or phosphate, respectively (hypothetical reactions shown in blue). B, In thiazole synthesis in Methanococci, sulfide (as HS-) serves as the sulfur donor, and the THI4 enzyme is not inactivated in the reaction. The pathway is otherwise the same as in plants. C, Partial sequence alignment showing the active site Cys in Arabidopsis THI4 and its replacement by His in Methanococcus jannaschii THI4.
Thiamin diphosphate can be destroyed during enzymatic catalysis (McCourt et al., 2006). Replacing the destroyed thiamin drives a high rate of thiamin synthesis, which in leaves can be estimated as ∼ 0.1 nmol h−1 g−1 fresh weight (Hanson et al., 2018). Plants, like yeast, make the thiazole moiety of thiamin via the thiazole synthase THIAMIN4 (THI4; Woodward et al., 2010; Chatterjee et al., 2011), which forms an adenylated thiazole derivative (ADT) from NAD+, Gly, and a sulfur atom taken from a Cys residue in its active site. The desulfurization of the Cys abolishes THI4 activity; THI4 is thus a suicide enzyme that catalyzes its reaction just once and must then be degraded and resynthesized (Fig. 1A). The combination of a high demand for thiazole synthesis with a suicide synthesis mechanism leads to a uniquely high turnover rate for the THI4 protein (Nelson et al., 2014; Li et al., 2017). Because this rapid turnover is estimated to consume 2-12% of the maintenance energy budget, suppressing it could substantially increase crop yields (Hanson et al., 2018). This makes building an energy-efficient alternative to the THI4-mediated thiazole pathway a rational target for synthetic biology.
There are two leads to potential parts for such an alternative pathway in the literature. The first is the discovery of nonsuicidal THI4s in the archaea Methanococcus jannaschii and M. igneus (Eser et al., 2016). These THI4s are true catalysts that use sulfide (as HS-) as the sulfur donor for the reaction and lack the active-site Cys of suicidal THI4s, having His instead (Fig. 1, B and C). However, M. jannaschii and M. igneus are oxygen-intolerant hyperthermophiles whose normal environments (hydrothermal vents) contain millimolar levels of sulfide (Liu et al., 2012). Their THI4s might not function in plants, in which temperatures and sulfide levels are low and oxygen is generally abundant.
The second lead is the report that inflorescences from wild populations of the tropical arum lily Caladium bicolor emit large amounts of 4-methyl-5-vinylthiazole (MVT; Maia et al., 2012). MVT is a structural analog of ADT and its metabolite cThz-P (the direct precursor of thiamin) and is theoretically derivable from either (Fig. 1A). MVT was emitted in a ∼ 4-h nocturnal burst at an average rate per inflorescence of 6.3 μmol h−1 (range 0.3-18 μmol h−1). Assuming an inflorescence weight of 2 g (Maia and Schlindwein, 2006), this average rate is ∼ 30,000-fold above the estimated rate of thiazole synthesis in leaves (see above). If MVT is synthesized only at the time of emission by a suicidal THI4, the MVT synthesis rate would require the impossibly high THI4 synthesis rate of 118 mg h−1 g−1 fresh weight (FW; arum lily inflorescences contain only ∼ 20 mg protein g−1 FW; Ap Rees et al., 1977). Even if, like other arum lily floral volatiles (Leguet et al., 2014), MVT is made throughout two weeks of flower development and stored until emission, the MVT synthesis rate would still have to be ∼ 400-fold more than the estimated thiazole synthesis rate in leaves and THI4 would have to be made at the extreme rate of ∼ 1.4 mg h−1 g−1 FW. These numbers suggest three possibilities for MVT production in C. bicolor: (1) a new MVT synthesis pathway not involving THI4; (2) a nonsuicidal THI4 plus enzymes to convert ADT or cThz-P to MVT; or (3) a hyperexpressed suicidal THI4, again plus additional enzymes. These alternatives do not exclude each other.
This study followed up these leads. The extremophile Methanococcus THI4 lead was pursued by analyzing the distribution of predicted nonsuicidal THI4s among prokaryotes from diverse habitats to identify candidates that operate under less extreme conditions. Selected candidates were then tested for function in Escherichia coli as a proxy for plants. To investigate the C. bicolor MVT lead, we first confirmed that cultivated C. bicolor plants produce MVT. To explore how MVT is made, we monitored extractable MVT pools, MVT emission, and thiamin pools throughout inflorescence development, cloned and sequenced C. bicolor THI4 cDNAs, and analyzed THI4 expression. Both leads identified potential parts for use in developing an energy-efficient plant thiazole synthesis pathway.
RESULTS AND DISCUSSION
Lead 1: Predicted Nonsuicidal THI4s Occur in Diverse Prokaryotes
The nonsuicidal THI4s from hyperthermophilic anaerobic archaea are a priori poorly adapted to function in plants because they have evolved to operate at ≥ 90°C in the presence of high sulfide levels and in the absence of oxygen (Vieille and Zeikus, 2001). The strictly anoxic normal environment of these enzymes is a special concern because of their dependence on an Fe(II) cofactor that could be oxidation-sensitive (Eser et al., 2016; Zhang et al., 2016). We therefore searched the SEED database (Overbeek et al., 2005) for predicted nonsuicidal THI4 homologs in prokaryotes from less extreme environments. The search criteria were an active-site His or other residue instead of Cys (Fig. 1C), and the presence in the genome of other thiamin synthesis genes, indicating that the THI4 homolog is likely to function in thiamin synthesis (Hanson et al., 2009).
Of > 11,000 prokaryote genomes in the SEED, only 106 (1%) encode a THI4 homolog. The other genomes either have canonical prokaryotic thiazole synthesis genes (including thiG, thiS, thiF, thiI, and thiH or thiO) or lack these genes, indicating that they are thiazole auxotrophs (Rodionov et al., 2002). The full results of this analysis are encoded in the SEED (http://pubseed.theseed.org/; see subsystem named ‘THI4’). Of the 106 THI4 homologs found, only ten have an active-site Cys; these THI4s are from aerobic archaea. Of the remaining 96, 92 have an active-site His and four have Leu or Met in the equivalent position. These 96 organisms are taxonomically and ecologically diverse; they belong to three archaeal and nine bacterial phyla, and include mesophiles and aerobes or microaerophiles as well as thermophiles and anaerobes.
Five representative bacteria with complete sets of thiamin synthesis genes were chosen as sources of THI4s to test for the capacity to function in E. coli (Table 1). These bacteria include two anaerobic thermophiles (Thermovibrio ammonificans and Hippea maritima) and three mesophiles, of which one is anaerobic (Pseudoramibacter alactolyticus), and two are aerobic or microaerophilic (Nitrospira defluvii and Poribacteria sp.; Willems and Collins, 1996; Miroshnichenko et al., 1999; Vetriani et al., 2004; Spieck et al., 2006; Siegl et al., 2011; Ushiki et al., 2013). The anaerobe T. ammonificans has a katG catalase gene (Table 1) and catalase activity, indicating a degree of aerotolerance (Vetriani et al., 2004). P. alactolyticus likewise has some aerotolerance (Cabral, 2010). The thermophiles are from sulfide-rich environments; the mesophiles are not. In place of Cys, two of the THI4s have His in the active site, two have Met, and one has Leu (Supplemental Fig. S1). The organisms adapted to, or able to survive in, oxygen-containing environments seemed likeliest to have THI4s that can function when oxygen is present.
Table 1. Characteristics of five diverse bacteria whose genomes encode THI4 homologs with no active-site Cys.
In addition to the gene encoding THI4, all five genomes have the other genes needed to synthesize thiamin and its diphosphate (thiC, thiD or thiD2, thiE, and thiL or thiN).
| Species (Phylum) | Temperature range (°C) | Aerobe or anaerobe | Catalase presenta | Natural habitat | ThiG route to thiazoleb | Residue replacing THI4 active-site Cys | ||
|---|---|---|---|---|---|---|---|---|
| Type | Sulfide level | Pressure (atm) | ||||||
| Thermovibrio ammonificans (Aquificae) | 60-80 | Anaerobe | Yes | Hydrothermal vent | High | 250 | No | Met |
| Hippea maritime (Proteobacteria) | 40-65 | Anaerobe | No | Hydrothermal vent | High | 4 | No | Leu |
| Nitrospira defluvii (Nitrospirae) | 22-34 | Aerobe | No | Sewage sludge | Low | 1 | Yes | His |
| Pseudoramibacter alactolyticus (Firmicutes) | 35-37 | Anaerobe | No | Abscesses | Low | 1 | No | Met |
| Poribacteria sp. WGA-A3 (unclassified) | 15-25 | Facultative aerobe | No | Sponge symbiont | Low | 1 | Yes | His |
Having a gene specifying a homolog of E. coli KatE or KatG, or Bacillus subtilis YdbD.
Having thiG and other genes encoding the canonical bacterial pathway to thiazole.
Prokaryotic THI4 Activity in Anaerobically and Aerobically Grown E. coli
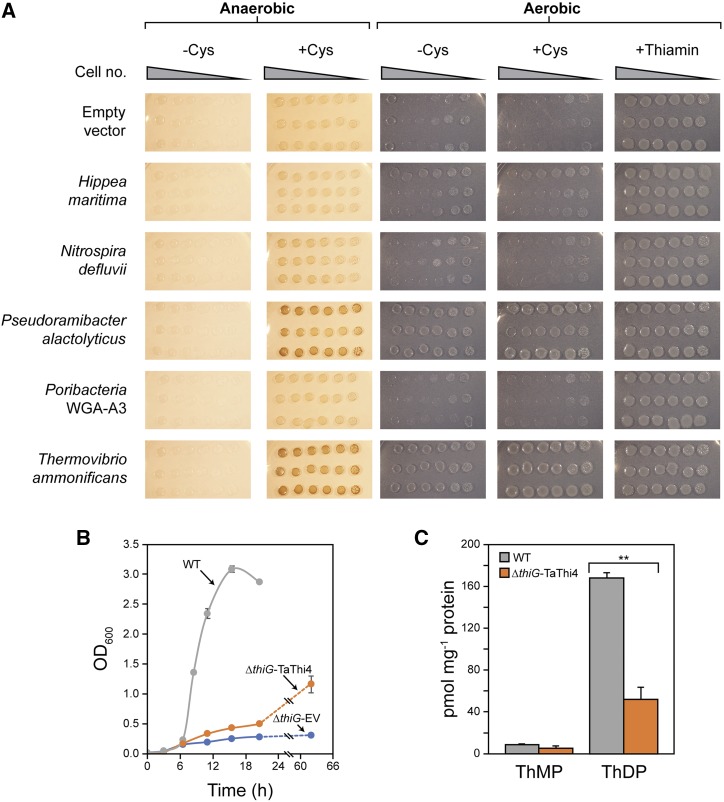
The five selected sequences were recoded for expression in E. coli and tested for their ability to complement a ΔthiG (thiazole-requiring) E. coli strain in anaerobic or aerobic conditions, minus or plus supplementary Cys to increase internal sulfide availability (Korshunov et al., 2016). [Complementation requires E. coli to hydrolyze ADT to cTHZ-P (Fig. 1A), e.g. via a nonspecific Nudix diphosphatase (Bessman et al., 1996).] T. ammonificans and P. alactolyticus THI4s gave complementation on Cys-supplemented plates in anaerobic and aerobic conditions, and weaker complementation without Cys in aerobic conditions (Fig. 2A). The other THI4s gave little or no complementation in any condition (Fig. 2A). T. ammonificans THI4, which performed slightly better than P. alactolyticus THI4, was selected for testing in aerobic liquid culture; complementing activity was again apparent (Fig. 2B). The complemented cells had thiamin diphosphate contents about one-third of those in wild type cells (Fig. 2C), but within the normal range for E. coli (∼ 50–180 pmol mg−1 protein; Leonardi and Roach, 2004). The essentially normal thiamin diphosphate level confirms that the complementing activity reflects restoration of thiazole synthesis.
Figure 2.
Tests of functional complementation of an E. coli ΔthiG strain by diverse bacterial THI4s. A, The ΔthiG strain harboring the pBAD24 vector alone or containing a THI4 gene was cultured in MOPS minimal medium containing 0.2% (w/v) glycerol and 0.02% (w/v) arabinose, plus or minus 1 mm Cys, in aerobic or anaerobic conditions. Aerobic controls supplemented with 100 nm thiamin were included. Anaerobic media contained 40 mm nitrate as the electron acceptor. Overnight liquid cultures of three independent clones for each construct were 10-fold serially diluted and spotted on the plates. Images were captured after incubation at 37°C for 7 d. B, Growth in liquid MOPS medium as above, plus 1 mm Cys, of wild type E. coli (WT) and the ΔthiG strain harboring pBAD24 alone (EV) or containing T. ammonificans THI4 (TaTHI4). Cultures were incubated in aerobic conditions at 37°C. C, Thiamin monophosphate (ThMP) and diphosphate (ThDP) contents of wild type and TaTHI4-complemented cells from B, harvested when OD600 reached 1.0–1.5; free thiamin was undetectable. Values in B and C are means and SE for three independent replicates. The difference in ThDP content between wild type and TaTHI4-complemented cells was significant at P < 0.001 (**), as determined by Student’s t test. Where no error bars appear in B they are smaller than the symbol.
The activity of the THI4 from T. ammonificans in aerobic conditions and the positive effect of giving Cys fit with this organism’s predicted aerotolerance and its provenance from a sulfide-rich environment (Vetriani et al., 2004). It is nonetheless striking that this enzyme functions in E. coli, given that T. ammonificans is a thermophile from a deep-sea (i.e. high pressure) environment (Table 1). That three of the THI4s tested gave little or no complementation could reflect poor THI4 expression rather than failure to operate in the conditions used; it is unlikely to be because THI4 expression is toxic because all strains grew when given thiamin (Fig. 2A).
The activity of T. ammonificans THI4 in aerobic conditions and the greater activity when Cys is supplied to drive up internal sulfide levels make this THI4 a rational candidate to replace the suicidal plant enzyme. Plant THI4 is localized in the chloroplast stroma (Goyer, 2010), where reported free sulfide concentrations (Krueger et al., 2009) exceed those estimated for aerobic E. coli cells (Korshunov et al., 2016). If this is the case, T. ammonificans THI4 might function better in chloroplasts than in E. coli. However, free sulfide is notoriously difficult to measure, and an indirect indicator of sulfide levels in vivo — the Km values for sulfide of the Cys synthetases from Arabidopsis thaliana chloroplasts and E. coli (3 and 6 μm, respectively) — suggest little difference (Mino et al., 2000; Wirtz et al., 2004). T. ammonificans THI4 might therefore need engineering to reduce the Km for sulfide; that it operates naturally at millimolar sulfide concentrations suggests that there is scope for such reduction.
Lead 2: MVT Production by C. bicolor Inflorescences
To confirm the observations made with wild populations of C. bicolor (Maia et al., 2012), we screened whole inflorescences of eight diverse C. bicolor cultivars for MVT emission (Supplemental Fig. S2A). All showed a burst of MVT emission in the female phase of anthesis, beginning in early evening and ending soon after midnight; cultivars differed slightly in the timing of emission but substantially (5-fold) in the total amount of MVT emitted (Supplemental Fig. S2B). The cultivar with the highest and least variable MVT emission, ‘Tapestry’, was used for most subsequent experiments. The identity of emitted MVT was confirmed by matches with authentic MVT with respect to gas chromatography-mass spectrometry (GC-MS) retention time and fragmentation pattern (Supplemental Fig. S3), and to LC-MS exact mass (m/z 126.0374, [M+H]+; theoretical = 126.0372).
The peak MVT emission rates (from cvs. ‘Tapestry’ and ‘Red Flash’) were about 0.4 μmol h−1 per inflorescence, which is the low end of the reported range (0.3-18 μmol h−1) and an order of magnitude below the average (6.3 μmol h−1; Maia et al., 2012) – but still extremely high relative to normal thiazole synthesis rates (Hanson et al., 2018). Genetics and/or environment may explain the divergence (wild C. bicolor populations in a Brazilian forest versus ornamental cultivars grown in a greenhouse).
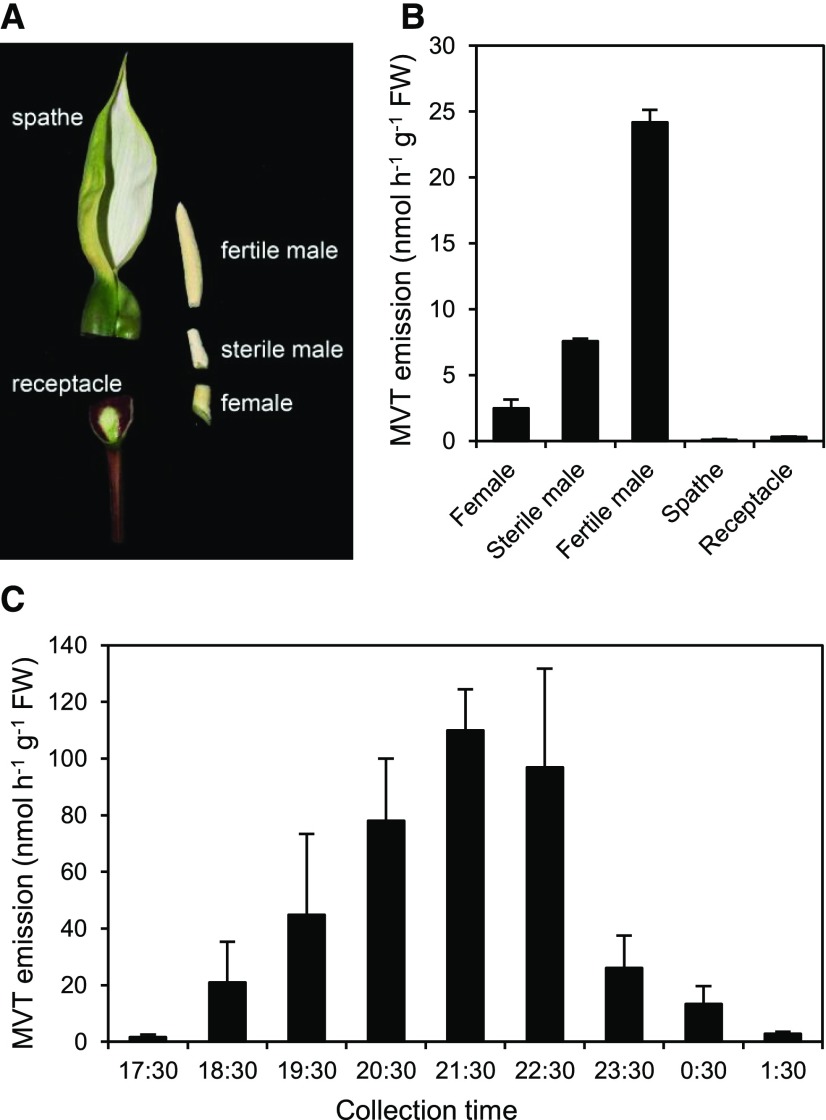
Most MVT Emission Comes from Male Flowers
Inflorescences were dissected into their five major parts (Fig. 3A) and assayed for MVT emission at its peak time. Virtually all the MVT came from the spadix, with 90% contributed by the sterile and fertile male flower regions (Fig. 3B). Subsequent work therefore used a pooled sterile and fertile male flower region. This dissected region reproduced the MVT emission peak observed in whole inflorescences (Fig. 3C; Supplemental Fig. S4, A and B). As reported (Maia et al., 2012), other volatiles were emitted, including methyl salicylate, linalool, (Z)-jasmone, and trimethoxybenzene (Supplemental Fig. S4C).
Figure 3.
MVT emission from C. bicolor inflorescences. A, Inflorescence dissection showing the parts tested for emissions. The female, sterile male, and fertile male flowers together comprise the spadix. B, MVT emission from inflorescence parts of cv ‘Pink Gem’. Data are means and SE for two biological replicates. Volatiles were collected for 2 h (19:30-21:30). C, Hourly MVT emission rates from sterile and fertile male flower tissue of cv ‘Tapestry’. Data are means and SE for three biological replicates. The x axis shows the start time for each 1-h collection period.
MVT Is Presynthesized during Flower Development
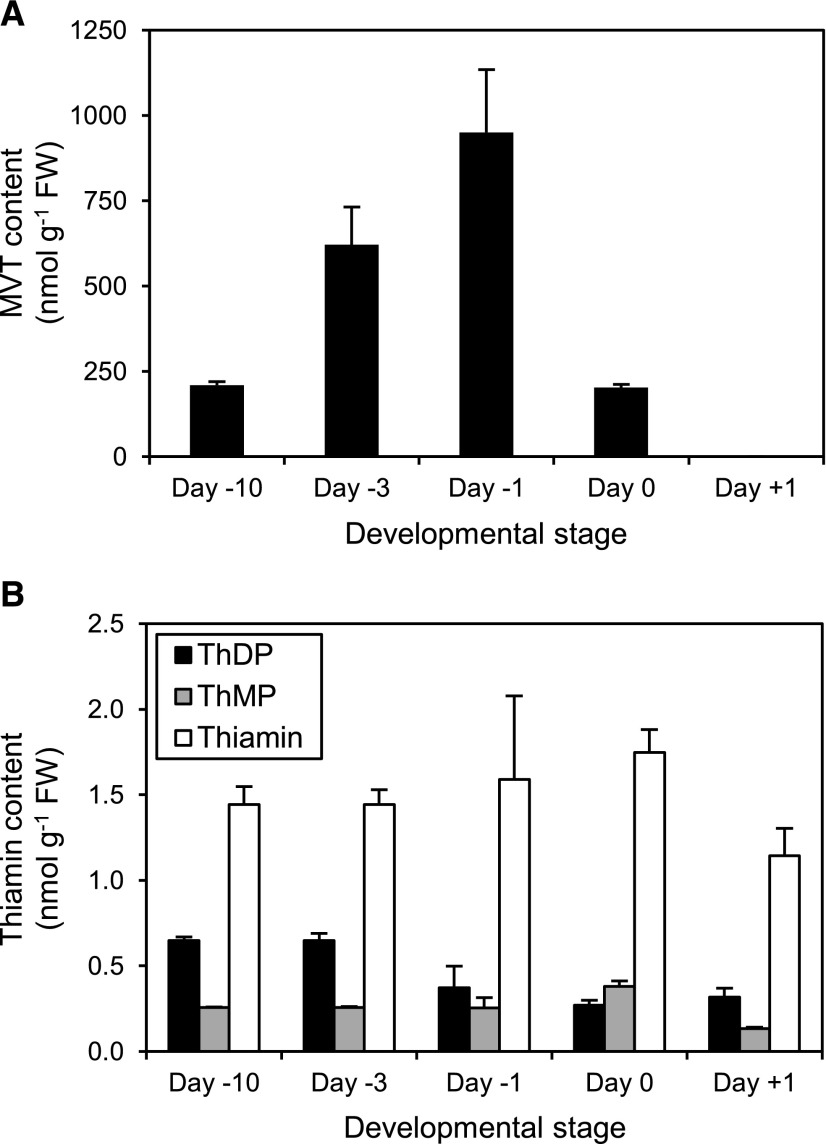
To determine when during flower development MVT is synthesized, male flowers of cv ‘Tapestry’ were harvested at various stages (from 10 d before to 1 d after female anthesis) and extracted with hexane. Analysis of the extracts showed that MVT, like other arum lily volatiles (Leguet et al., 2014), is synthesized throughout flower development and stored until it is emitted (Fig. 4A). The maximal MVT content on the day before emission (∼ 1 μmol g−1 FW) fell to zero on the day after the emission burst. This loss of extractable MVT is in acceptable agreement with the summed MVT emissions from ‘Tapestry’ male flowers (∼ 0.4 μmol g−1 FW; Supplemental Fig. S4B) when the large variation in MVT emission among individual plants (Fig. 3C; Supplemental Fig. S4A) and the incomplete recovery of emitted MVT (see “Materials and Methods”) are taken into account. Note that MVT emission and extractable MVT content necessarily cannot be measured on the same sample.
Figure 4.
Developmental time courses of the levels of extractable MVT (A) and of thiamin and its phosphorylated forms (B) in male flower tissue of cv ‘Tapestry’. Data are means and SE of three biological replicates. Tissue was harvested 1, 3, and 10 d before female anthesis, on day 0 (female anthesis), and 1 d after female anthesis. The day 0 harvest was at 20:30, when the MVT emission burst was in progress. ThDP, thiamin diphosphate; ThMP, thiamin monophosphate.
To test whether thiamin breakdown contributes thiazole moieties for conversion to MVT, thiamin and its phosphates were determined in male flowers of cv ‘Tapestry’ harvested at various developmental stages as detailed above (Fig. 4C). The levels of free thiamin and thiamin phosphates changed little with time and their total never exceeded 2.6 nmol g−1 FW, which is trivial compared to the maximal MVT content of ∼ 1 μmol g−1 FW (Fig. 4A). These data show that net thiamin degradation cannot be a significant source of thiazole for MVT synthesis.
Figure 4A indicates an average net MVT synthesis rate in male flowers of ∼ 80 nmol d−1 g−1 FW or ∼ 3 nmol h−1 g−1 FW, rising to ∼ 7 nmol h−1 g−1 FW in the two days before MVT emission starts. These rates are 30–70 times the estimated rate of thiazole synthesis in leaves (∼ 0.1 nmol h−1 g−1 FW; see above) and, if mediated by a suicidal THI4, would require THI4 turnover rates of 0.11–0.26 mg h−1 g−1 FW. These rates are high, but not inconceivably so. Therefore, our observed MVT synthesis rates could in principle reflect operation of a suicidal THI4 expressed at exceptional levels.
C. bicolor THI4s Have an Active-Site Cys and Are Massively Expressed in Male Flowers
To explore C. bicolor THI4 function and expression, full-length THI4 cDNAs were cloned from immature male flower tissue by touchdown reverse transcription PCR (RT-PCR) followed by RACE. Two cDNA sequences were obtained; they encode THI4s that have an active-site Cys residue and are 96% identical to each other and 72% identical to Arabidopsis THI4 (Supplemental Fig. S5). C. bicolor male flowers thus express canonical THI4s that are predicted to be suicide enzymes.
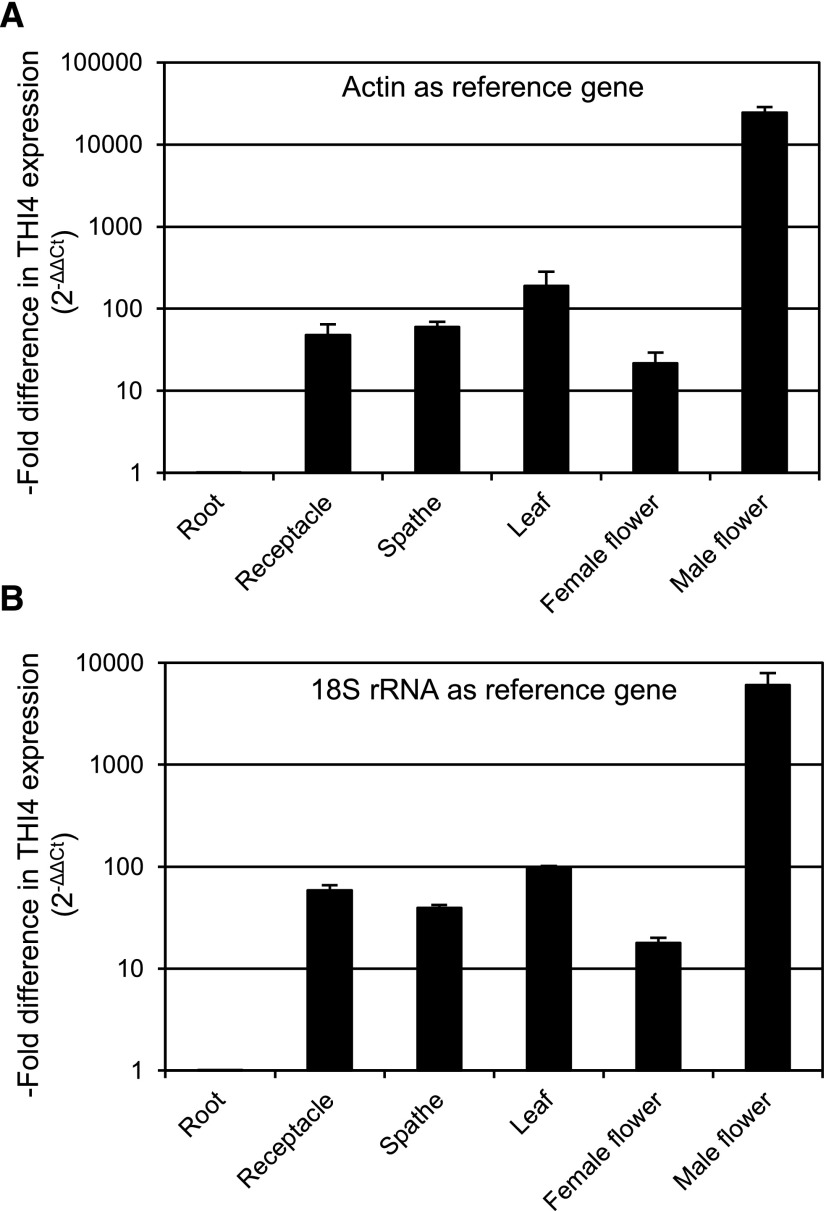
The summed expression of both THI4 mRNAs in various organs was assayed by reverse transcription quantitative PCR (RT-qPCR) using primers common to both sequences, and quantified by the 2-ΔΔCT method with actin or 18S rRNA as the reference gene (Fig. 5). THI4 expression in leaves was two orders of magnitude higher than in roots, which parallels the difference between Arabidopsis leaves and roots (Schmid et al., 2005). THI4 expression in male flowers, however, was two orders of magnitude higher still, i.e. ∼ 104-fold more than in roots and ∼ 100-fold more than in leaves.
Figure 5.
RT-qPCR assay of THI4 mRNA levels in various organs of C. bicolor cv ‘Tapestry’. THI4 mRNA was quantified by the 2-ΔΔCT method. Data are presented as the -fold difference in THI4 gene expression in each organ normalized to the reference gene (actin or 18S rRNA) and relative to roots (= 1.0). Inflorescence tissues were harvested on Day 0, just before the MVT emission burst started. Data are means and SE of three biological replicates. Note that the y axis scale is logarithmic.
C. bicolor THI4 May Retain Catalytic Activity After Loss of the Active-Site Cys
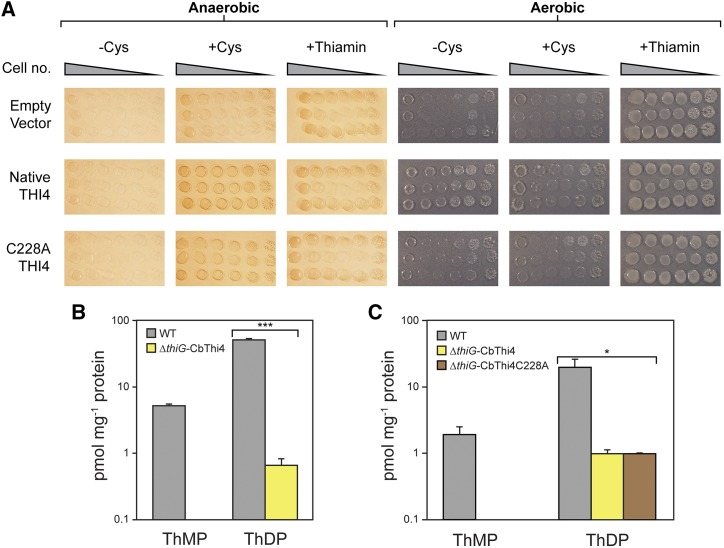
The ∼ 100-fold difference in THI4 expression between male flowers and leaves roughly matches the ∼ 30-70-fold difference in their thiazole synthesis rates estimated above. This correspondence suggests that extreme expression of a suicidal THI4 might, in itself, be enough to support the MVT production we observed. However, the total amounts of MVT produced in our experiments are far below the highest reported for C. bicolor (Maia et al., 2012). We therefore used complementation of the ΔthiG E. coli strain to test whether C. bicolor THI4 can, as can yeast THI4 in vitro (Eser et al., 2016), function catalytically using sulfide as the sulfur donor when the catalytic Cys residue (Cys 228, Supplemental Fig. S5) is replaced with Ala as a facsimile of the dehydro-Ala residue formed by the suicide reaction. (Note, however, that dehydro-Ala differs from Ala in having an electrophilic double bond.) The native THI4 complemented the ΔthiG strain under aerobic conditions in the absence of added Cys but the C228A mutant did not, nor did it do so in the presence of Cys (Fig. 6A). In anaerobic conditions, however, both native and C228A enzymes showed complementing activity when Cys was added (Fig. 6A). Complementation in aerobic and anaerobic conditions restored a low level of thiazole synthesis, giving a thiamin diphosphate level of ∼ 1 pmol mg−1 protein (i.e. 20–50 times less than in wild type cells in the same conditions; Fig. 6, B and C). This minimal thiamin diphosphate level corresponds to ∼ 100 molecules per cell, which is the threshold level for growth estimated from the response of an E. coli thiamin auxotroph to supplied thiamin (Tian et al., 2016). Growth at such low thiamin diphosphate concentrations implies extensive reconfiguration of the metabolic network to reduce dependence on thiamin diphosphate-dependent enzymes (Tian et al., 2016).
Figure 6.
Functional complementation of the E. coli ΔthiG strain by native and C228A mutant C. bicolor THI4. A, The ΔthiG strain harboring the pBAD24 vector alone or containing the native or C228A mutant form of C. bicolor THI4_1 was cultured in MOPS minimal medium containing 0.2% (w/v) glycerol and 0.02% (w/v) arabinose, plus or minus 1 mm Cys, in aerobic or anaerobic conditions. Controls supplemented with 100 nm thiamin were included. Anaerobic media contained 40 mm nitrate as the electron acceptor. Overnight liquid cultures of three independent clones for each construct were 10-fold serially diluted and spotted on the plates. Images were captured after incubation at 37°C for 7 d. B, Thiamin monophosphate (ThMP) and diphosphate (ThDP) contents (log scale) of E. coli ΔthiG cells complemented with native C. bicolor THI4_1, grown aerobically without added Cys; wild type cells grown in these conditions are included for comparison. C, Thiamin mono- and diphosphate contents (log scale) of E. coli ΔthiG cells complemented with native or C228A C. bicolor THI4_1, grown anaerobically plus 1 mm Cys; wild type cells grown in these conditions are included for comparison. Values in B and C are means and SE for three independent replicates. No thiamin monophosphate was detectable in cells complemented with native or C228A C. bicolor THI4_1. The ThDP content differences between wild type and complemented cells were significant at P < 0.0001 (***) in aerobic conditions or P < 0.05 (*) in anaerobic conditions, as determined by Student’s t test.
Assuming that Ala adequately mimics dehydro-Ala in the THI4 active site, these results indicate that C. bicolor THI4 could function catalytically in male flower tissue after an initial suicide reaction, provided that the oxygen level is low and the sulfide level is relatively high. Aroid spadices are known to become internally hypoxic at the flowering (thermogenic) stage (Seymour et al., 2015), and classical data on oxygen consumption rates indicate that they also may be hypoxic at earlier stages when tightly wrapped in the spathe (James and Beevers, 1950; Seymour et al., 1984). As noted above, sulfide levels in plant tissues may exceed those in E. coli. If C. bicolor THI4 can indeed function catalytically after a suicide reaction, such ‘zombie’ activity would amplify the thiazole synthesis potential conferred by extremely high THI4 expression.
CONCLUSIONS
Parts-prospecting for an efficient thiazole synthesis route produced two promising candidates for further development – a catalytic bacterial THI4 likely to work in planta, and a plant THI4 that may function catalytically after committing suicide, at least under low-oxygen, high-sulfide conditions. One way to develop these THI4s would be by continuous directed evolution, selecting a THI4-complemented thiamin auxotroph for growth rate in aerobic conditions. This strategy could be implemented in an E. coli ΔthiG strain using the EvolvR system (Halperin et al., 2018) or in a yeast thi4 mutant using the OrthoRep system (Ravikumar et al., 2018). The yeast option is supported by the finding that maize (Zea mays) or Arabidopsis THI4 cDNAs complement a thi4 disruptant (Belanger et al., 1995; Godoi et al., 2006).
Finally, the trajectory of this project illustrates a key point about bioprospecting: even if rationally targeted, it can spring surprises. Thus, although our two leads – catalytic archaeal THI4s and massive MVT emission from C. bicolor – were followed up rationally, in neither case was the outcome fully predictable. Thus, the most effective bacterial nonsuicidal THI4 came from a thermophilic anaerobe from a deep-sea, high-sulfide environment rather than a mesophilic aerobe from a surface, low-sulfide environment. Similarly, although the massive MVT emission from C. bicolor could be reconciled with a THI4-mediated pathway only by making extreme assumptions about THI4 synthesis rates, these extreme assumptions appear to be largely correct, with the added twist that C. bicolor THI4 could be a zombie enzyme that still operates in catalytic mode with sulfide as the sulfur donor after a single cycle of operation in suicide mode. In any case, our results demonstrate that a targeted search for novel parts to engineer a specific system can be fruitful, notwithstanding its vagaries.
MATERIALS AND METHODS
Bioinformatics Analysis
Comparative genomics analysis was carried out using the SEED database and its tools (Overbeek et al., 2005). Protein sequences were aligned with Multalin (http://multalin.toulouse.inra.fr/multalin/) and formatted with BoxShade (https://embnet.vital-it.ch/software/BOX_form.html). Nucleotide sequences were aligned with Clustal Omega (http://www.clustal.org/omega/). Plant THI4 sequences were taken from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/).
Functional Complementation Tests with Bacterial THI4s
THI4 genes were recoded for expression in E. coli (Supplemental Table S1) and synthesized by GenScript (Piscataway, NJ). Synthesized DNA fragments were digested with PciI/XbaI (P. alactolyticus), NcoI/XbaI (N. defluvii, Poribacteria sp. WGA-A3), BspHI/XbaI (H. maritima) or EcoRI/XbaI (T. ammonificans) and ligated into pBAD24 digested with NcoI/XbaI (P. alactolyticus, N. defluvii, Poribacteria sp. WGA-A3, H. maritima) or EcoRI/XbaI (T. ammonificans). The E. coli MG1655 ΔthiG strain was made by recombineering (Datsenko and Wanner, 2000) using the ΔthiG cassette from the corresponding Keio collection strain (Baba et al., 2006). Briefly, MG1655 was first transformed with pKD46 and grown in LB with 100 µg/mL ampicillin at 30°C. These cells were transformed with an amplicon containing the ΔthiG cassette and transformants were selected on LB with 50 µg/mL kanamycin at 37°C. The cells were then cured of pKD46 by growth at 42°C and the ΔthiG mutation was verified by PCR. Plasmids encoding bacterial THI4s were transformed into the MG1655 ΔthiG strain. Single colonies were used to inoculate 3 mL of MOPS medium (Stewart and Parales, 1988) containing 0.2% (w/v) glycerol, 100 nm thiamin, and 100 µg/mL ampicillin. Cultures were grown for 18 h at 37°C, after which cells were harvested by centrifugation, washed three times with thiamin-deplete MOPS medium, and resuspended in 400 µL of the same medium. Washed cells (10 μL of a suspension of OD600 1.0, and 10-fold serial dilutions) were spotted on MOPS plates containing 0.2% (w/v) glycerol, 0.02% (w/v) arabinose, and 1.5% (w/v) agar, minus and plus 1 mm Cys, and grown at 37°C. For anaerobic growth, the medium was supplemented with 40 mm sodium nitrate and plates were incubated in an airtight 10-L chamber flushed continuously with nitrogen at ∼ 1 L/min). Plates were imaged after one week. Liquid cultures of 25 mL of MOPS medium containing 0.2% (w/v) glycerol, 0.02% (w/v) arabinose, 100 µg/mL ampicillin, and 1 mm Cys were inoculated with washed cells to an OD600 of 0.05, grown at 37°C with shaking at 225 rpm, harvested by centrifugation at OD600 1.0-1.5, and flash frozen in liquid nitrogen. Cells complemented with C. bicolor THI4 constructs were harvested for thiamin analysis from plates, suspended in thiamin-deplete MOPS medium, pelleted, and frozen as above. Cys-containing plates were a light tan color after anaerobic incubation, and colonies grown thereon were brown; these plates were consequently photographed against a light background. The slight growth of vector-alone controls seen in our complementation experiments can be attributed to pico- or subpicomolar traces of thiamin or thiazole coming from the carbon source; such minute concentrations support limited growth of E. coli ΔthiG cells (Paerl et al., 2018).
C. bicolor Plant Material
Tubers of the following eight hybrid C. bicolor cultivars were obtained from commercial sources: ‘Pink Symphony’, ‘Tapestry’, ‘FL Sweetheart’, ‘Red Flash’, ‘Freida Hemple’, ‘Candidum’, ‘Pink Gem’, and ‘FL Fantasy’. Tubers were soaked for 16 h in a 600 ppm gibberellic acid solution (4% Pro-Gibb, Valent BioSciences Corporation, Libertyville, IL). The bulbs were individually potted in 1-gallon pots containing Sunshine Mix #4 substrate (Sun Gro Horticulture, Agawam, MA) with 10 g of Osmocote 14-14-14 fertilizer (ICL Specialty Fertilizers, Dublin, OH) and placed in a shade house. Two crops were grown in FL shade house conditions (USDA zone 8b) and moved to an environmentally controlled greenhouse (24°C) when temperatures exceeded 24°C. The first crop was grown from May 2017 to September 2017 and the second was grown from October 2017 to February 2018.
Growth stages were determined by measuring time until inflorescence opening, which was typically 16-17 d after the inflorescence first emerged. Day -10 samples were collected 7 d after inflorescences emerged, but the peduncle was not extended. Day -3 samples were collected 14 d after emergence, marked by a fully extended peduncle and a light green spathe that was starting to loosen to open. Day -1 samples were collected 15 d after emergence, marked by a pale green to cream cracked-open spathe, exposing the spadix. Day 0 samples were collected 17 d after emergence, marked by a fully opened, cream-colored spathe exposing the spadix. At this stage, female flowers in the floral chamber are covered in a sticky residue. Day +1 samples were collected 18 d after emergence, marked by a senescing spathe exposing the spadix. At this stage, the fertile male flowers on the spadix shed pollen and the female flowers are not receptive.
Volatile Collections and GC-MS Analysis
For whole inflorescence measurements, inflorescences were excised 10 cm beneath the receptacle and the peduncle was placed in a 2-mL tube containing 1.5 mL of sterile distilled water and sealed with Parafilm. The inflorescence and tube were quickly placed inside a glass tube (2.5 cm internal diameter, 61 cm long; 300 mL volume) and connected to a dynamic headspace volatile collection system. Volatiles were collected on a 50 mg HaySep 80–100 porous polymer adsorbent (Hayes Separations Inc., Bandera, TX) contained in a glass column. Elution of volatiles from the adsorbent was as described (Schmelz et al., 2001). For extended collections, columns were replaced hourly. For separate flower part measurements, the parts were dissected, and each part was quickly placed in a separate glass tube connected to the dynamic headspace volatile collection system as above. Volatiles were collected for 2 h (19:30-21:30) and eluted immediately after the collection.
Volatiles in eluates were quantified by GC-MS using an Agilent 6890 n gas chromatograph (carrier gas; He fixed at 13.56 psi; split 20:1 injector, temperature 220°C, injection volume 2 μl) with a DB-5 column ((5%-phenyl)-methylpolysiloxane, 30 m × 250 μm, 1 μm film thickness; Agilent Technologies, Santa Clara, CA, USA) coupled to an Agilent 5975 mass spectrometer (MS) (Agilent Technologies, Santa Clara, CA, USA). Oven temperatures were programmed from 40°C with a 0.5-min hold at 5°C/min to 250°C with a 4-min hold. Retention times were compared with authentic standards (Sigma-Aldrich) for identification (Schmelz et al., 2001). MS peaks were manually integrated with MS Quantitative Analysis software (Agilent Technologies, Santa Clara, CA) and compound identity was confirmed using NIST Mass Spectral Library software (National Institute of Standards and Technology, Gaithersburg, MD). Duplicate calibration curves for standards were run to obtain a response factor for each compound that was used to calculate volatile emission mass. Volatile mass calculations were based on individual compound peak area relative to the peak area of a standard nonyl acetate spike, added to each sample. Calculations were made with JMP Pro (Version 10, SAS Institute Inc., Cary, NC, USA). The recovery of emitted MVT from the trapping and elution procedure was found to be < 100% but could not be reliably measured due to the difficulty of mimicking slow MVT release over time in the presence of a complex mix of other volatiles (Supplemental Figure S4C).
MVT Identification by High-Resolution Mass Spectrometry
A 1 ng/μL MVT standard solution and a C. bicolor volatile eluate containing 1 ng/μL MVT were analyzed via reversed phase gradient HPLC/electrospray ionization high-resolution time-of-flight mass spectrometry using an Agilent 6220 instrument. One μL of the standard MVT or 2 μL of the C. bicolor sample were injected. Standard MVT eluted at 26.6 min and produced an abundant m/z 126.0374 [M+H]+ ion (1.6 ppm error from the theoretical m/z 126.0372). MVT in the C. bicolor samples was detected at 26.6 min and produced an m/z 126.0374 [M+H]+ ion (1.6 ppm error from the theoretical).
Hexane Extractions
Fertile and sterile male flower tissue was excised, frozen in liquid nitrogen, and stored at -80°C until extraction. Tissue was ground to a fine powder in a mortar cooled with liquid nitrogen, and 250-mg portions (three technical replicates per sample) were placed in 2-mL Eppendorf tubes. One mL of hexane (Sigma-Aldrich) was added to each tube and samples were centrifuged at 24,000g for 20 min. The supernatant was transferred to a 1.5-mL glass vial and analyzed by GC-MS as above. Pilot extractions showed that other inflorescence parts contained negligible quantities of MVT.
Extraction and Determination of Thiamin and Its Phosphates
Fertile and sterile male flower tissue from inflorescences at different developmental stages was harvested at 17:30, ground as above, and stored at -80°C. Portions (50-100 mg) of powder were extracted in 0.3 mL of 7.2% (v/v) perchloric acid by sonicating for 30 min and chilling on ice for 15 min. After centrifuging at 14,000g for 10 min, thiamin and its phosphates in the supernatant were oxidized to thiochrome derivatives using potassium ferricyanide and analyzed by HPLC with fluorometric detection (Kuznetsova et al., 2015). Briefly, 15 μL of methanol and 100 μL of freshly made 12.14 mm potassium ferricyanide dissolved in 3.35 m NaOH were mixed with 160 μL of sample or standard for 2 min. One hundred μL of 1.43 m phosphoric acid was added to neutralize the derivatized sample, followed by deproteinization using Amicon Ultra 0.5-mL 10K units. A 10-μL portion of the flow-through was separated on an Alltima HP C18 amide column (150 × 4.6 mm, 5 μm, 190 Å, Alltech). The mobile phase (1 mL/min) was a gradient of potassium phosphate (20 mM, pH 7.0), 12% (v/v) methanol (buffer A) and 70% (v/v) methanol (buffer B). The running protocol was: 0-10 min, 100% A to 50% A; 10-13 min, 50% A to 100% B; 13-14 min, 100% B; 14-17 min, 100% B to 100% A; 17-21 min, 100% A. Thiamin and its phosphates in E. coli cells harvested from liquid cultures or plates were extracted and analyzed essentially as above (Hanson et al., 2018). Comparisons with literature on E. coli thiamin contents assumed dry weight to be 23% of wet weight, protein content to be 55% of dry weight, cultures at OD600 = 1 to have 0.39 g L−1 dry weight of cells, and a cell to contain ∼ 0.15 pg protein (http://bionumbers.hms.harvard.edu). Student's t test was used to determine the significance of differences in thiamin diphosphate contents.
Cloning of C. bicolor THI4 cDNAs
C. bicolor cv ‘Tapestry’ sterile and fertile male flower tissue was harvested on day 0, frozen in liquid nitrogen, stored at -80°C, and ground as above. RNA samples were isolated using Plant/Fungi Total RNA Purification Kit (Norgen Biotech Corp., Thorold, Canada) followed by on-column DNase treatment using RNase-Free DNase I Kit (Norgen Biotech). Total RNA was quantified using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE), and stored at -20°C. RACE-ready cDNA was generated using a SMARTer RACE cDNA Amplification Kit (Clontech) according to the manufacturer’s protocol. RACE primers (Supplemental Table S2) were designed based on alignments of THI4 nucleotide sequences from Z. mays (NM_001112227.2), foxtail millet (Setaria italica) (XM_004965684.4), sugarcane (Saccharum) hybrid cv GT28 (JN591765.1) and cv R570 (KF184858.1:c139219-138052), soybean (Glycine max) (XM_003536502.3), and sunflower (Helianthus annuus) (XM_022137923.1). The sequences obtained were assembled into contigs using the Snapgene software package. The full cDNA sequences were amplified using Turbo Pfu DNA polymerase (Agilent Technologies) and cloned into pGEMT-easy (Promega, Madison, WI) as described (Colquhoun et al., 2011). The coding sequences were determined and confirmed at Genewiz Sanger sequencing service (South Plainfield, NJ).
THI4 Expression Analysis
C. bicolor ‘Tapestry’ root, receptacle, spathe, leaf, female flower, and male flower tissues (two biological replicates) were harvested on day 0 at 17:30, and frozen, stored, and ground as above. RNA was isolated and quantified as above; the samples were diluted to 50 ng RNA/μL and stored at -80°C. 18S rRNA and actin were selected as RT-qPCR reference genes. A homolog of actin 7 was identified using C. bicolor root transcriptome data (Cao and Deng, 2017). The Arabidopsis mitochondrial 18S rRNA (ATMG01390) was used to design the 18S rRNA RT-qPCR primers based on the conserved region (Hadziavdic et al., 2014). The THI4 sequence was obtained from cloning (see above). RT-qPCR primers (Supplemental Table S2) were designed with PREMIER 5.0 (PREMIER Biosoft International, Palo Alto, CA). Power SYBR Green RNA-to-Ct 1-Step kits (Applied Biosystems) were used to amplify and detect the products according to the manufacturer’s instructions. The qPCR reactions (10 μL) contained 5 μL 2×Power SYBR Green RT-PCR Mix, 0.2 μm each primer, 50 ng RNA, 0.08 μL RT Enzyme Mix, and nuclease-free water. The reaction mixture was held at 48°C for 30 min and 95°C for 10 min, and for 40 cycles of 15 s at 95°C and 1 min at 60°C. The final dissociation curve was obtained between 60 and 95°C to demonstrate gene specificity of the primers. The relative transcription of THI4 in different C. bicolor tissues was quantified by the 2-ΔΔCT method, normalizing to the reference 18S rRNA or actin gene, and the transcript level in root tissue. Assays included three technical replicates per biological replicate.
Functional Complementation Tests with C. bicolor THI4
A CbTHI4_1 open reading frame (encoding a protein 99% identical to the sequence in Supplemental Figure S5) was amplified with Phusion DNA polymerase (NEB) from cDNA, adenylated with Taq DNA polymerase (ThermoFisher Scientific), cloned into pGEM-T Easy (Promega), and sequence-verified. A C228A mutant was generated using the Q5 Site-Directed Mutagenesis Kit (NEB). Expression constructs minus the predicted plastid targeting peptide (Supplemental Figure S5) were made by amplifying wild-type and C228A mutant sequences, digesting the amplicon with PciI/XbaI, and ligating into NcoI/XbaI-cut pBAD24. The plasmids were transformed into E. coli MG1655 ΔthiG and tested for complementation as above.
Accession Numbers
C. bicolor THI4 sequences from this article can be found in the NCBI database under the accession numbers: THI4_1, MH796125; THI4_2, MH796126.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of the sequences of the five selected bacterial THI4s with that of the characterized nonsuicidal THI4 from Methanococcus jannaschii.
Supplemental Figure S2. Volatile emission from whole C. bicolor inflorescences.
Supplemental Figure S3. Mass spectral analysis of MVT.
Supplemental Figure S4. Volatile emission from C. bicolor inflorescence parts.
Supplemental Figure S5. Alignment of the THI4 sequences obtained from C. bicolor male flower tissue with that of Arabidopsis THI4.
Supplemental Table S1. Recoded nucleotide sequences of bacterial THI4 genes.
Supplemental Table S2. Oligonucleotide primers used in this study.
Acknowledgments
We thank Dr. Manasi Kamat and the University of Florida Department of Chemistry Mass Spectrometry Research and Education Center for high-resolution mass spectrometry analysis of MVT.
Footnotes
This work was supported by NSF award IOS-1444202, the UF/IFAS Plant Innovation Center, the American Floral Endowment, the USDA-ARS Floriculture and Nursery Research Initiative, and by an endowment from the C.V. Griffin Sr. Foundation. High-resolution mass spectrometry was supported by NIH Shared Instrumentation grant no. S10 OD021758-01A1.
Articles can be viewed without a subscription.
References
- Amthor JS. (2003) Efficiency of lignin biosynthesis: a quantitative analysis. Ann Bot 91: 673–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ap Rees T, Wright BW, Fuller WA (1977) Measurements of starch breakdown as estimates of glycolysis during thermogenesis by the spadix of Arum maculatum L. Planta 134: 53–56 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger FC, Leustek T, Chu B, Kriz AL (1995) Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Mol Biol 29: 809–821 [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O’Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271: 25059–25062 [DOI] [PubMed] [Google Scholar]
- Cabral JP. (2010) Water microbiology. Bacterial pathogens and water. Int J Environ Res Public Health 7: 3657–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Deng Z (2017) De novo assembly, annotation, and characterization of root transcriptomes of three Caladium cultivars with a focus on necrotrophic pathogen resistance/defense-related genes. Int J Mol Sci 18: 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP (2011) Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478: 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG (2011) PhMYB4 fine-tunes the floral volatile signature of Petunia x hybrida through PhC4H. J Exp Bot 62: 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb TJ, Jones PR, Bar-Even A (2017) Synthetic metabolism: metabolic engineering meets enzyme design. Curr Opin Chem Biol 37: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser BE, Zhang X, Chanani PK, Begley TP, Ealick SE (2016) From suicide enzyme to catalyst: the iron-dependent sulfide transfer in Methanococcus jannaschii thiamin thiazole biosynthesis. J Am Chem Soc 138: 3639–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Thore S (2014) Complex behavior: from cannibalism to suicide in the vitamin B1 biosynthesis world. Curr Opin Struct Biol 29: 34–43 [DOI] [PubMed] [Google Scholar]
- Godoi PH, Galhardo RS, Luche DD, Van Sluys MA, Menck CF, Oliva G (2006) Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana. J Biol Chem 281: 30957–30966 [DOI] [PubMed] [Google Scholar]
- Goyer A. (2010) Thiamine in plants: aspects of its metabolism and functions. Phytochemistry 71: 1615–1624 [DOI] [PubMed] [Google Scholar]
- Hadziavdic K, Lekang K, Lanzen A, Jonassen I, Thompson EM, Troedsson C (2014) Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One 9: e87624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer DV, Dueber JE (2018) CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560: 248–252 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Pribat A, Waller JC, de Crécy-Lagard V (2009) ‘Unknown’ proteins and ‘orphan’ enzymes: the missing half of the engineering parts list--and how to find it. Biochem J 425: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Amthor JS, Sun J, Niehaus TD, Gregory JF III, Bruner SD, Ding Y (2018) Redesigning thiamin synthesis: Prospects and potential payoffs. Plant Sci 273: 92–99 [DOI] [PubMed] [Google Scholar]
- James WO, Beevers H (1950) The respiration of arum spadix. A rapid respiration, resistant to cyanide. New Phytol 49: 352–374 [Google Scholar]
- Jensen MK, Keasling JD (2015) Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res 15: 1–10 [DOI] [PubMed] [Google Scholar]
- Korshunov S, Imlay KR, Imlay JA (2016) The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol Microbiol 101: 62–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S, Niehl A, Lopez Martin MC, Steinhauser D, Donath A, Hildebrandt T, Romero LC, Hoefgen R, Gotor C, Hesse H (2009) Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ 32: 349–367 [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Nocek B, Brown G, Makarova KS, Flick R, Wolf YI, Khusnutdinova A, Evdokimova E, Jin K, Tan K, Hanson AD, Hasnain G, et al. (2015) Functional diversity of Haloacid Dehalogenase superfamily phosphatases from Saccharomyces cerevisiae. J Biol Chem 290: 18678–18698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguet A, Gibernau M, Shintu L, Caldarelli S, Moja S, Baudino S, Caissard JC (2014) Evidence for early intracellular accumulation of volatile compounds during spadix development in Arum italicum L. and preliminary data on some tropical Aroids. Naturwissenschaften 101: 623–635 [DOI] [PubMed] [Google Scholar]
- Leonardi R, Roach PL (2004) Thiamine biosynthesis in Escherichia coli: in vitro reconstitution of the thiazole synthase activity. J Biol Chem 279: 17054–17062 [DOI] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH (2017) Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29: 207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beer LL, Whitman WB (2012) Methanogens: a window into ancient sulfur metabolism. Trends Microbiol 20: 251–258 [DOI] [PubMed] [Google Scholar]
- Maeda HA. (2016) Lignin biosynthesis: Tyrosine shortcut in grasses. Nat Plants 2: 16080. [DOI] [PubMed] [Google Scholar]
- Maia AC, Schlindwein C (2006) Caladium bicolor (Araceae) and Cyclocephala celata (Coleoptera, Dynastinae): a well-established pollination system in the Northern Atlantic rainforest of Pernambuco, Brazil. Plant Biol (Stuttg) 8: 529–534 [DOI] [PubMed] [Google Scholar]
- Maia AC, Dötterl S, Kaiser R, Silberbauer-Gottsberger I, Teichert H, Gibernau M, do Amaral Ferraz Navarro DM, Schlindwein C, Gottsberger G (2012) The key role of 4-methyl-5-vinylthiazole in the attraction of scarab beetle pollinators: a unique olfactory floral signal shared by Annonaceae and Araceae. J Chem Ecol 38: 1072–1080 [DOI] [PubMed] [Google Scholar]
- McCourt JA, Nixon PF, Duggleby RG (2006) Thiamin nutrition and catalysis-induced instability of thiamin diphosphate. Br J Nutr 96: 636–638 [PubMed] [Google Scholar]
- Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K (2000) Effects of bienzyme complex formation of cysteine synthetase from escherichia coli on some properties and kinetics. Biosci Biotechnol Biochem 64: 1628–1640 [DOI] [PubMed] [Google Scholar]
- Miroshnichenko ML, Rainey FA, Rhode M, Bonch-Osmolovskaya EA (1999) Hippea maritima gen. nov., sp. nov., a new genus of thermophilic, sulfur-reducing bacterium from submarine hot vents. Int J Syst Bacteriol 49: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Alexova R, Jacoby RP, Millar AH (2014) Proteins with high turnover rate in barley leaves estimated by proteome analysis combined with in planta isotope labeling. Plant Physiol 166: 91–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33: 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl RW, Bertrand EM, Rowland E, Schatt P, Mehiri M, Niehaus TD, Hanson AD, Riemann L, Bouget FY (2018) Carboxythiazole is a key microbial nutrient currency and critical component of thiamin biosynthesis. Sci Rep 8: 5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau B, Vanhercke T, Singh S (2018) From plant metabolic engineering to plant synthetic biology: The evolution of the design/build/test/learn cycle. Plant Sci 273: 3–12 [DOI] [PubMed] [Google Scholar]
- Ravikumar A, Arzumanyan GA, Obadi MK, Javanpour AA, Liu CC (2018) Scalable, continuous evolution of genes at mutation rates above genomic error thresholds. Cell 175: 1946–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS (2002) Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem 277: 48949–48959 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Alborn HT, Tumlinson JH (2001) The influence of intact-plant and excised-leaf bioassay designs on volicitin- and jasmonic acid-induced sesquiterpene volatile release in Zea mays. Planta 214: 171–179 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Seymour RS, Barnhart MC, Bartholomew GA (1984) Respiratory gas exchange during thermogenesis inPhilodendron selloum Koch. Planta 161: 229–232 [DOI] [PubMed] [Google Scholar]
- Seymour RS, Ito K, Umekawa Y, Matthews PD, Pirintsos SA (2015) The oxygen supply to thermogenic flowers. Plant Cell Environ 38: 827–837 [DOI] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C, Dandekar T, Hentschel U (2011) Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 5: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, Lipski A, Daims H (2006) Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol 8: 405–415 [DOI] [PubMed] [Google Scholar]
- Stewart V, Parales J Jr (1988) Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol 170: 1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K, Niu D, Liu X, Prior BA, Zhou L, Lu F, Singh S, Wang Z (2016) Limitation of thiamine pyrophosphate supply to growing Escherichia coli switches metabolism to efficient D-lactate formation. Biotechnol Bioeng 113: 182–188 [DOI] [PubMed] [Google Scholar]
- Ushiki N, Fujitani H, Aoi Y, Tsuneda S (2013) Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant. Microbes Environ 28: 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani C, Speck MD, Ellor SV, Lutz RA, Starovoytov V (2004) Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate-ammonifying bacterium from deep-sea hydrothermal vents. Int J Syst Evol Microbiol 54: 175–181 [DOI] [PubMed] [Google Scholar]
- Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65: 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Wei KY, Smolke CD (2013) Synthetic biology: advancing the design of diverse genetic systems. Annu Rev Chem Biomol Eng 4: 69–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A, Collins MD (1996) Phylogenetic relationships of the genera Acetobacterium and Eubacterium sensu stricto and reclassification of Eubacterium alactolyticum as Pseudoramibacter alactolyticus gen. nov., comb. nov. Int J Syst Bacteriol 46: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R (2004) O-acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55: 1785–1798 [DOI] [PubMed] [Google Scholar]
- Woodward JB, Abeydeera ND, Paul D, Phillips K, Rapala-Kozik M, Freeling M, Begley TP, Ealick SE, McSteen P, Scanlon MJ (2010) A maize thiamine auxotroph is defective in shoot meristem maintenance. Plant Cell 22: 3305–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Eser BE, Chanani PK, Begley TP, Ealick SE (2016) Structural basis for iron-mediated sulfur transfer in archael and yeast thiazole synthases. Biochemistry 55: 1826–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]