Recent technological developments narrow the existing complexity gap between bacterial and plastid synthetic biology.
Abstract
Building on recombinant DNA technology, leaps in synthesis, assembly, and analysis of DNA have revolutionized genetics and molecular biology over the past two decades (Kosuri and Church, 2014). These technological advances have accelerated the emergence of synthetic biology as a new discipline (Cameron et al., 2014). Synthetic biology is characterized by efforts targeted at the modification of existing and the design of novel biological systems based on principles adopted from information technology and engineering (Andrianantoandro et al., 2006; Khalil and Collins, 2010). As in more traditional engineering disciplines such as mechanical, electrical and civil engineering, synthetic biologists utilize abstraction, decoupling and standardization to make the design of biological systems more efficient and scalable. To facilitate the management of complexity, synthetic biology relies on an abstraction hierarchy composed of multiple levels (Endy, 2005): DNA as genetic material, “parts” as elements of DNA encoding basic biological functions (e.g. promoter, ribosome-binding site, terminator sequence), “devices” as any combination of parts implementing a human-defined function, and “systems” as any combination of devices fulfilling a predefined purpose. Parts are designated to perform predictable and modular functions in the context of higher-level devices or systems, which are successively refined through a cycle of designing, building, and testing.
Within the past two decades, the synthetic biology approach has produced several notable successes, especially in microbial systems. These include, for example, the design of a minimal bacterial genome (Hutchison et al., 2016) and a highly modified yeast genome (Richardson et al., 2017), as well as the metabolic engineering of yeast for the biosynthesis of the antimalarial drug precursor artemisinic acid (Ro et al., 2006) and the opioid compounds thebaine and hydrocodone (Galanie et al., 2015). Compared to synthetic biology in bacteria and yeast, synthetic biology in algae and plants is still lagging behind. While the potential of photoautotrophic organisms for environmentally sustainable bioproduction has long been recognized (Georgianna and Mayfield, 2012; Fesenko and Edwards, 2014; Liu and Stewart, 2015; Boehm et al., 2017), their relatively slow growth, scarcely available tools for genetic manipulation, and the physiological as well as genomic complexity of plant systems have delayed their widespread adoption as synthetic biology chassis. However, especially the small genome of the plastid (chloroplast) represents a highly promising platform for engineering the sophisticated metabolism and physiology of the eukaryotic cell it is embedded in (Fig. 1).
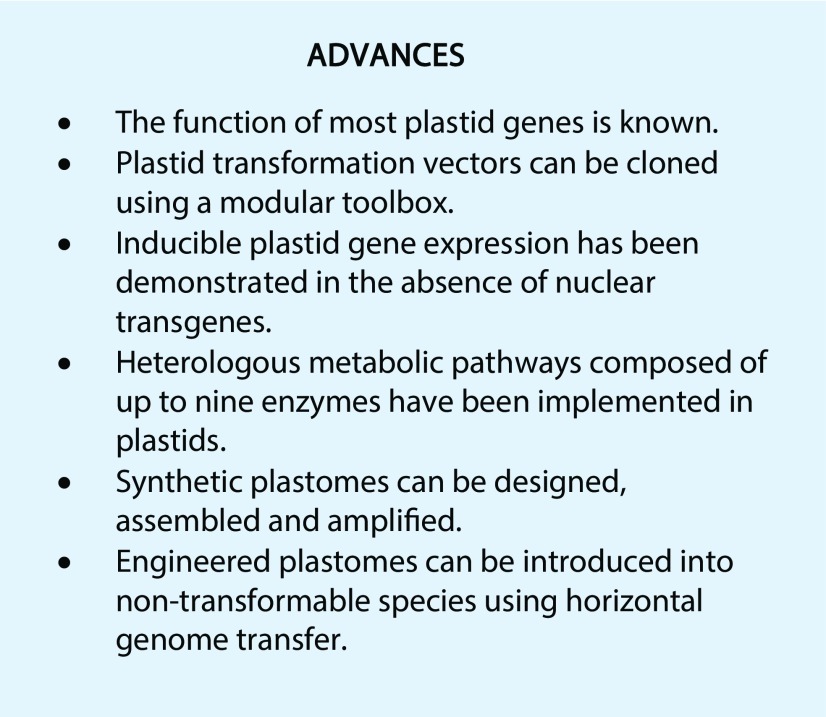
Figure 1.
Biological properties and existing technical capacities for synthetic biology of plastids compared to bacteria, yeast and the plant nucleus. The number of asterisks roughly illustrates the relative degree of (top) presence of a biological feature, (middle) availability of a tool or technique, and (bottom) current implementation of a type of application across the different chassis.
The chloroplast originated through the endosymbiotic uptake of a cyanobacterium by a heterotrophic eukaryote more than a billion years ago (Palmer, 2003). Following this event, the endosymbiont evolved mechanisms for facilitated exchange of metabolites with the host cell, underwent radical streamlining of its genome (by gene loss and large-scale transfer of genes to the host nuclear genome) and established an import machinery for the uptake of nucleus-encoded proteins. The resulting organelle serves as the major biosynthetic compartment in photoautotrophic organisms, and has been exploited as a platform for metabolic engineering and molecular farming since the successful development of transformation technologies in the late 1980s (Boynton et al., 1988; Svab et al., 1990). Compared to nuclear genetic engineering, plastid transformation offers several notable advantages relevant to plant biotechnology. These include (1) the high precision of genetic engineering enabled by efficient homologous recombination, (2) the possibility of transgene stacking in synthetic operons, (3) the potential for high-level expression of gene products, (4) the absence of epigenetic transgene silencing, and (5) the reduced risk of unwanted transgene transmission due to maternal inheritance of plastid DNA (Bock, 2015).
In this article, we provide an update on tools and technologies available for extending the synthetic biology approach to plastids and highlight key challenges to be addressed through future research. Guided by an abstraction hierarchy of biological design, we identify a scarcity of well-characterized genetic parts, tightly controlled expression devices, and quantitative knowledge of plastid gene expression as current key limitations to plastid synthetic biology. We highlight recent technological developments narrowing the existing complexity gap between bacterial and plastid synthetic biology and provide an outlook to the implementation of complex systems such as synthetic metabolic feedback loops, designer subcompartments and tailor-made genomes in chloroplasts.
Parts
The Registry of Standard Biological Parts (http://parts.igem.org) currently contains over 20,000 genetic elements which can be requested by researchers for use in synthetic biology applications. From this collection, approximately 100 parts each have been designed for use in the unicellular green alga Chlamydomonas reinhardtii and in multicellular plants (e.g. the seed plants Nicotiana tabacum and Arabidopsis thaliana, the moss Physcomitrella patens and the liverwort Marchantia polymorpha). The majority of these parts are designated for nuclear engineering, with only about two dozen suitable for gene expression from the chloroplast genome. One explanation for the relative paucity of plastid genetic elements in the Registry of Standard Biological Parts lies in the half-year timeframe of projects pursued as part of the international Genetically Engineered Machine (iGEM) competition (Smolke, 2009), which is barely compatible with the generation and characterization of stable plastid-engineered (transplastomic) organisms. Beyond iGEM, the repertoire of regulatory sequences routinely used for transgene expression in plastids has remained similarly small: it is comprised of a few preferred promoters (e.g. from the plastid rRNA operon, Prrn; the gene for the large subunit of Rubisco, PrbcL; and the gene for the D1 protein of photosystem II, PpsbA) and a handful of 5′-and 3′-UTRs (Jin and Daniell, 2015). In addition, the bacterial hybrid promoter Ptrc (Newell et al., 2003) and several bacteriophage-derived expression elements (McBride et al., 1994; Kuroda and Maliga, 2001; Yang et al., 2013) have been successfully used for plastid transgene expression. A greater variety of parts available for controlled expression of plastid transgenes is desirable for several reasons. First, multiple use of the same genetic element within the chloroplast genome is problematic due to the risk of unwanted homologous recombination between sequence stretches as short as 50 bp (Dauvillee et al., 2004; Rogalski et al., 2006). Second, synthetic genetic circuits commonly require precise tuning of the activity of their constitutive parts for optimal function (Brophy and Voigt, 2014).
For synthetic biology applications in plastids to catch up in versatility and complexity with those already demonstrated in bacteria, gene expression elements covering a wider activity range will be required. Natural plastid genomes represent an obvious source of such elements. The small size and low coding capacity of chloroplast genomes (in most seed plants, approximately 130 genes in an ∼ 150 kb genome) should allow refactoring of all coding and regulatory regions into standardized genetic parts. The sequences of over 800 chloroplast genomes have been determined (Daniell et al., 2016), and the functions of most of their (widely conserved) genes are known (Scharff and Bock, 2014). Plastid genetic elements contained within this wealth of sequence data can be domesticated according to a recently proposed common syntax for plant synthetic biology (Patron et al., 2015). This scheme promises to facilitate sharing of genetic resources among the community and, although developed for a eukaryotic system, is also compatible with GoldenBraid-based modular cloning of chloroplast transformation vectors (Vafaee et al., 2014). Plastid parts containing internal recognition sites for type IIS restriction enzymes (e.g. BsaI, BsmBI, BbsI) that cannot be synonymously changed (e.g. because they constitute essential sequence motifs in a promoter or UTR sequence) may alternatively be assembled using long-overlap-based methods such as Gibson Assembly (Gibson et al., 2009).
Gene Expression Devices
Gene expression devices send or receive signals in the form of levels of gene expression. A basic device of this kind may be composed of four parts: a promoter, a ribosome-binding site, a coding sequence and a terminator. This device architecture is commonly used for the quantification of part performance to inform the rational design of genetic circuits. Hundreds of prokaryotic gene-expression elements (including promoters, ribosome-binding sites and terminators) have been characterized in bacterial hosts using reporter gene-based assays (Salis et al., 2009; Cambray et al., 2013; Chen et al., 2013; Kosuri et al., 2013; Mutalik et al., 2013), and standards have been formulated for quantifying their activities (Canton et al., 2008; Kelly et al., 2009; Rudge et al., 2016). To reduce the context dependence of part activity, standardized flanking sequences (Mutalik et al., 2013), strong terminators (Chen et al., 2013) and enzymatic cleavage of UTRs (Lou et al., 2012; Qi et al., 2012) have been successfully employed as insulators in bacteria. In plastids, not more than two dozen combinations of regulatory elements (i.e. promoters, 5′-UTRs and 3′-UTRs) have been systematically characterized for their impact on transgene expression using GFP (Barnes et al., 2005; Caroca et al., 2013), GUS (Eibl et al., 1999; Herz et al., 2005; Gerasymenko et al., 2017) or other reporter proteins (Ruhlman et al., 2010; Zhang et al., 2012).
Compared to part characterization in microbes, that in plastids involves several notable challenges. First, relatively long timescales are required to generate transplastomic organisms ready for characterization. While only a few days are needed for transformation of the microbial models Escherichia coli or Saccharomyces cerevisiae by a genetic part, several months of selection are needed to recover homoplasmic plastid transformants (i.e. transplastomic cells or plants that are devoid of residual copies of the wild-type plastid genome). In theory, the establishment of homoplasmy could be accelerated through inducible expression of endonucleases that selectively target the wild-type chloroplast genome, but it remains to be tested how much time this approach can save. Alternatively, measurement fidelity can be traded for high-throughput, transient assays to quantify part performance within days of particle bombardment of algal cells or plant tissues. Such assays will require (1) high transient transformation frequencies, (2) high sensitivity, and (3) a robust way of normalizing the primary reporter signal to the copy number of transformed plastomes. The latter could be achieved by using a ratiometric approach (Rudge et al., 2016; Boehm et al., 2018). If a suitable reporter system can be developed, the activities of hundreds of plastid parts could rapidly be measured in algal cells or plant protoplasts using microtiter plate-based assays (Schaumberg et al., 2016) or microfluidic devices (Yu et al., 2018).
Second, the plastome exhibits abundant read-through transcription due to inefficient termination (Stern and Gruissem, 1987; Rott et al., 1996; Legen et al., 2002; Shi et al., 2016). Consequently, part behavior is, by default, poorly insulated from its specific genetic context: both upstream promoters and downstream antisense promoters may significantly affect the expression level of a target gene (Quesada-Vargas et al., 2005; Sharwood et al., 2011). However, some sequences such as the endogenous tRNA genes trnS and trnH (Stern and Gruissem, 1987) or the heterologous E. coli Thr attenuator (thra; Chen and Orozco, 1988) have been shown to terminate plastid transcription with at least 85% efficiency. Use of insulators based on these parts or new synthetic terminators can potentially enhance the robustness of gene expression levels generated by plastid synthetic biology devices.
Third, plastid transgene expression has been shown to be primarily determined by posttranscriptional control and protein stability rather than by the accumulation of mRNA (Eberhard et al., 2002; Birch-Machin et al., 2004; Bellucci et al., 2005; Kahlau and Bock, 2008; Valkov et al., 2009; Zoschke and Bock, 2018). Chloroplast transcripts are subject to a series of complex processing steps which include intercistronic cleavage, 5′-and 3′-end maturation, intron splicing and mRNA editing (Stern et al., 2010). These steps are largely mediated by nucleus-encoded and organelle-targeted factors, including a large family of modular proteins known as pentatricopeptide repeat (PPR) proteins that site-specifically bind to one or several premRNAs (Barkan and Small, 2014). Plastid gene expression levels can, therefore, vary considerably between different transgenes even if the same promoter and 3′-UTR are used, limiting the informative value of part characterization based on standard reporter protein assays. While the amino acid sequence of the N-terminus is thought to substantially influence protein stability in the chloroplast (Apel et al., 2010; De Marchis et al., 2012), our general knowledge of plastid proteostasis remains limited. A better understanding of the molecular determinants of plastid protein (in)stability may in the future allow the design of protective amino acid sequences (Elghabi et al., 2011) that level the stabilities of different plastid-expressed proteins and make transgene expression from the plastid genome more predictable.
Metabolic Devices
Metabolic devices send or receive signals in the form of levels of metabolites. Accordingly, a synthetic metabolic pathway represents a metabolic device carrying out a specific series of enzyme-catalyzed reactions. A variety of metabolic devices have been successfully implemented in plastids for the production of molecules such as polyhydroxybutyrate (Bohmert-Tatarev et al., 2011), carotenoids (Wurbs et al., 2007; Hasunuma et al., 2008; Apel and Bock, 2009), fatty acids (Madoka et al., 2002; Craig et al., 2008), artemisinic acid (Fuentes et al., 2016), vitamin E (Lu et al., 2013) and dhurrin (Gnanasekaran et al., 2016). These applications have been reviewed in more detail elsewhere (Bock, 2015; Fuentes et al., 2018). While heterologous pathways composed of 20 genes or more have been expressed in bacteria and yeast (Temme et al., 2012; Galanie et al., 2015; Li et al., 2018), no more than seven transgenes have to date been simultaneously expressed from the plastome (Krichevsky et al., 2010). The complexity of plastid-based metabolic devices has primarily been limited by a scarcity of available expression signals (see Gene Expression Devices) rather than by the physical size of the introduced DNA (Adachi et al., 2007). Recently, the complexity and number of pathway variants accessible to experimental interrogation has been expanded through combinatorial supertransformation of transplastomic recipient lines (COSTREL). Using this approach, an up to 77-fold increase in artemisinic acid production has been demonstrated in transplastomic tobacco plants combinatorially supertransformed by five additional nuclear transgenes (Fuentes et al., 2016). There is no in-principle limitation to the number of transgenes that can be simultaneously introduced into the plant nucleus using combinatorial transformation (Naqvi et al., 2009). However, handling hundreds to thousands of plants resulting from combinatorial transformation with several dozen transgenes will require an effective screening pipeline.
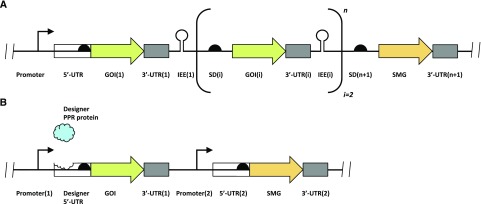
In plastid-based metabolic devices containing multicistronic operons, intercistronic expression elements (IEEs) can be used to facilitate correct processing of polycistronic transcripts into monocistronic mRNAs and their efficient translation (Fig. 2A; Zhou et al., 2007). To avoid defects in mRNA stabilization upon repeated use of the same IEE, more complex future metabolic devices may feature a variety of different such elements and/or additionally overexpress their cognate RNA-binding proteins (Legen et al., 2018).
Figure 2.
Design of plastid-based metabolic devices. A, Intercistronic expression elements (IEEs; Zhou et al., 2007) can be used to design synthetic operons composed of n genes of interest (GOIs) under the control of a single promoter. Alternatively, each transgene can be controlled by its own promoter. SD, Shine-Dalgarno sequence; SMG, selectable marker gene. B, Expression of a GOI can be controlled by a synthetic 5′-UTR that is specifically stabilized by a designer PPR protein (that recognizes a different binding sequence than all other RNA-binding proteins present in the plastid).
Genetic Circuits
Genetic circuits mimic logical functions commonly found in their electronic counterparts. A genetic circuit can be used to control the activity of other devices (such as the gene expression devices or metabolic devices discussed above) in response to external stimuli. A wide range of genetic circuits implementing Boolean logic functions such as yes, not, and, or, nand, nor, xor and n-imply has been reported for bacteria, yeast and mammalian cells (Miyamoto et al., 2013). In plastids, only the simplest logic function yes has been implemented in the form of chemically inducible transgene expression.
Chloroplast transcription is natively controlled by two different types of RNA polymerases in seed plants. The nucleus-encoded RNA polymerase (NEP) is a chloroplast-targeted bacteriophage-type single subunit enzyme, while the plastid-encoded RNA polymerase (PEP) is a eubacteria-type multisubunit enzyme (Barkan, 2011; Börner et al., 2015). The promoter specificity of PEP is modulated by nucleus-encoded and plastid-targeted sigma factors in response to light, hormones and biotic as well as abiotic stresses. However, due to their important role in plant growth, development and survival (and the pervasive transcription of essentially all plastid genes), NEP and PEP are poorly suited as stringent controllers of synthetic genetic circuits in plastids.
As an alternative to transgene control by the endogenous transcription machineries, plastid transgene expression has been controlled through nucleus-encoded and plastid-targeted bacteriophage RNA polymerases or processing factors that are responsive to chemical inducers such as salicylic acid (Magee et al., 2004), ethanol (Lössl et al., 2005), copper (Surzycki et al., 2007) or thiamine (Ramundo et al., 2013). To avoid (pollen-transmissible) nuclear transgenes and increase transgene containment, inducible expression systems encoded solely in the plastid genome are particularly desirable. Plastid-only inducible circuits responsive to isopropyl β-d-1-thiogalactopyranoside (IPTG; Mühlbauer and Koop, 2005) or theophylline (Verhounig et al., 2010; Emadpour et al., 2015) have been shown to be functional, yet fall short of binary behavior due to the pronounced transcriptional leakiness present in plastids (see Gene Expression Devices). To achieve a signal-to-noise ratio sufficient for the implementation of more complex logic gates, future plastid-based genetic circuits may employ synthetic RNA-binding proteins of the PPR class (see Gene Expression Devices; Coquille et al., 2014; Gully et al., 2015) to selectively control the maturation of target mRNAs in the chloroplast (Fig. 2B; Stern et al., 2010; Barkan and Small, 2014).
Systems
Beyond hard-wired logic gates, synthetic biologists have explored dynamic feedback mechanisms to enhance the efficiency of engineered metabolic pathways in bacteria and yeast (Venayak et al., 2015; Del Vecchio et al., 2016). Translation of this approach to plastids is currently hampered by our limited quantitative understanding of chloroplast gene expression, though new tools for analysis of the metabolic network shared between the chloroplast and its host cell are emerging (Gloaguen et al., 2017). Metabolic engineering in plastids may further be supported by expression of synthetic subcompartments for substrate concentration, metabolite channeling and the prevention of unwanted reactions between subcompartmentalized and endogenous plastid metabolites and enzymes (Winkel, 2004; Ort et al., 2015; Hanson et al., 2016). Synthetic subcompartments have already been introduced in bacteria and yeast (Bonacci et al., 2012; Lau et al., 2018), and carboxysomal shell proteins transiently expressed in leaves of Nicotiana benthamiana have been shown to be capable of assembling into carboxysome-like structures within chloroplasts (Lin et al., 2014), encouraging further efforts in this area.
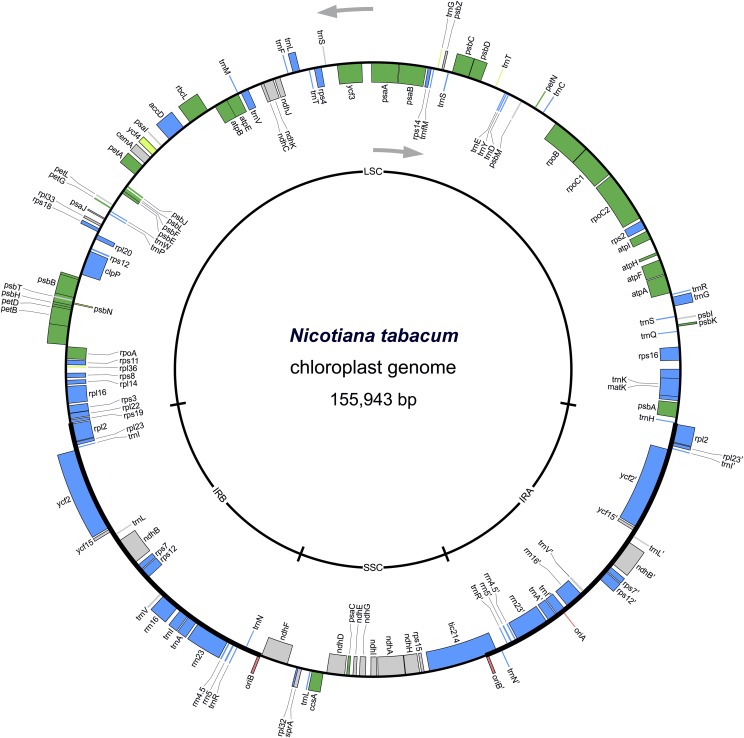
Among the most complex systems proposed for implementation in plastids are entire synthetic genomes, inspired by recent successes in microbial synthetic genomics (Hutchison et al., 2016; Richardson et al., 2017). A minimum-size plastid genome composed of the smallest possible number of components will be of great value for two reasons: it will advance our understanding of the regulatory network underlying plastid function, and it will serve as a template for engineering synthetic plastomes to be used in biotechnological applications. We have previously proposed a design for a synthetic minimal plastome of N. tabacum that is free of all genes nonessential under heterotrophic growth conditions (Fig. 3), intergenic spacers, introns, and isoaccepting tRNA genes that are dispensable or become dispensable after genome-wide modification of codon usage (Scharff and Bock, 2014). Such a synthetic chloroplast genome can be assembled from linear DNA fragments in yeast (O’Neill et al., 2012) and, prior to plant transformation, can be amplified in vitro using rolling circle amplification (Jansen et al., 2005). The major hurdle to the successful implementation of fully synthetic plastomes in planta is the high probability of homologous recombination between the (largely nonrecodeable) rRNA and tRNA genes and their counterparts in the resident plastid genome, leading to chimeric genomes of unpredictable structure and function (O’Neill et al., 2012). In addition, the effects of synthetic lethality (i.e. the combined knock-out of two nonessential genes being lethal; e.g. Ehrnthaler et al., 2014) cannot currently be excluded to occur in a synthetic minimal plastome.
Figure 3.
Physical map of the N. tabacum chloroplast genome with all genes classified by essentiality. Genes shown in blue are essential for both heterotrophic and autotrophic growth. Genes shown in green are essential for autotrophic growth only. Light green indicates borderline cases where knock-out plants survive under carefully controlled growth conditions. Genes shown in gray are nonessential under both heterotrophic and autotrophic growth conditions, in that their knock-out causes no or only a mild phenotype (Scharff and Bock, 2014). Origins of replication are highlighted in red. Gray arrows indicate the direction of transcription for the two DNA strands. The map was drawn using the OrganellarGenomeDRAW (OGDRAW) software (Lohse et al., 2013) based on the complete plastome sequence of N. tabacum (Shinozaki et al., 1986; GenBank accession number Z00044.2). LSC, large single-copy region; IRA, inverted repeat A; IRB, inverted repeat B; SSC, small single-copy region.
Despite numerous technical advances made over the past 30 years, the number of algal and plant species whose plastids can reliably be transformed has remained small (Bock, 2015). Transplantation of transgenic plastids from a species amenable to transformation to a species recalcitrant to transformation represents an attractive alternative to painstakingly developing specialized transformation protocols for the latter. Plastomes can be horizontally transferred across graft junctions with relative ease (Stegemann and Bock, 2009; Stegemann et al., 2012; Thyssen et al., 2012; for review, see Bock, 2017) and this process has been exploited for transplanting a plastid-encoded synthetic metabolic device into a currently nontransformable species (Lu et al., 2017). The graft-mediated horizontal transfer of transgenic plastid genomes may not be feasible between distantly related species due to the close coevolution of nuclear and plastid genomes, and the probability of nuclear-cytoplasmic incompatibilities that increases with phylogenetic distance and can cause deleterious phenotypes (Schmitz-Linneweber et al., 2005; Greiner and Bock, 2013). However, the transfer will certainly facilitate the expansion of transplastomic technologies from model species and cultivars used in research to related species and elite cultivars grown commercially.
Footnotes
Articles can be viewed without a subscription.
References
- Adachi T, Takase H, Tomizawa K (2007) Introduction of a 50 kbp DNA fragment into the plastid genome. Biosci Biotechnol Biochem 71: 2266–2273 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Basu S, Karig DK, Weiss R (2006) Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol 2: 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel W, Bock R (2009) Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol 151: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel W, Schulze WX, Bock R (2010) Identification of protein stability determinants in chloroplasts. Plant J 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011) Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Barnes D, Franklin S, Schultz J, Henry R, Brown E, Coragliotti A, Mayfield SP (2005) Contribution of 5′- and 3′-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Mol Genet Genomics 274: 625–636 [DOI] [PubMed] [Google Scholar]
- Bellucci M, De Marchis F, Mannucci R, Bock R, Arcioni S (2005) Cytoplasm and chloroplasts are not suitable subcellular locations for β-zein accumulation in transgenic plants. J Exp Bot 56: 1205–1212 [DOI] [PubMed] [Google Scholar]
- Birch-Machin I, Newell CA, Hibberd JM, Gray JC (2004) Accumulation of rotavirus VP6 protein in chloroplasts of transplastomic tobacco is limited by protein stability. Plant Biotechnol J 2: 261–270 [DOI] [PubMed] [Google Scholar]
- Bock R. (2015) Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu Rev Plant Biol 66: 211–241 [DOI] [PubMed] [Google Scholar]
- Bock R. (2017) Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu Rev Genet 51: 1–22 [DOI] [PubMed] [Google Scholar]
- Boehm CR, Pollak B, Purswani N, Patron N, Haseloff J (2017) Synthetic Botany. Cold Spring Harb Perspect Biol 9: 023887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm CR, Grant PK, Haseloff J (2018) Programmed hierarchical patterning of bacterial populations. Nat Commun 9: 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert-Tatarev K, McAvoy S, Daughtry S, Peoples OP, Snell KD (2011) High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol 155: 1690–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF (2012) Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci USA 109: 478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta - Bioenerg 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB, et al. (1988) Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538 [DOI] [PubMed] [Google Scholar]
- Brophy JAN, Voigt CA (2014) Principles of genetic circuit design. Nat Methods 11: 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray G, Guimaraes JC, Mutalik VK, Lam C, Mai QA, Thimmaiah T, Carothers JM, Arkin AP, Endy D (2013) Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res 41: 5139–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DE, Bashor CJ, Collins JJ (2014) A brief history of synthetic biology. Nat Rev Microbiol 12: 381–390 [DOI] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D (2008) Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol 26: 787–793 [DOI] [PubMed] [Google Scholar]
- Caroca R, Howell KA, Hasse C, Ruf S, Bock R (2013) Design of chimeric expression elements that confer high-level gene activity in chromoplasts. Plant J 73: 368–379 [DOI] [PubMed] [Google Scholar]
- Chen LJ, Orozco EM Jr (1988) Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res 16: 8411–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Liu P, Nielsen AA, Brophy JA, Clancy K, Peterson T, Voigt CA (2013) Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods 10: 659–664 [DOI] [PubMed] [Google Scholar]
- Coquille S, Filipovska A, Chia T, Rajappa L, Lingford JP, Razif MFM, Thore S, Rackham O (2014) An artificial PPR scaffold for programmable RNA recognition. Nat Commun 5: 5729. [DOI] [PubMed] [Google Scholar]
- Craig W, Lenzi P, Scotti N, De Palma M, Saggese P, Carbone V, McGrath Curran N, Magee AM, Medgyesy P, Kavanagh TA, Dix PJ, Grillo S, et al. (2008) Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res 17: 769–782 [DOI] [PubMed] [Google Scholar]
- Daniell H, Lin C-S, Yu M, Chang W-J (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillee D, Hilbig L, Preiss S, Johanningmeier U (2004) Minimal extent of sequence homology required for homologous recombination at the psbA locus in Chlamydomonas reinhardtii chloroplasts using PCR-generated DNA fragments. Photosynth Res 79: 219–224 [DOI] [PubMed] [Google Scholar]
- Del Vecchio D, Dy AJ, Qian Y (2016) Control theory meets synthetic biology. J R Soc Interface 13: 20160380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchis F, Pompa A, Bellucci M (2012) Plastid proteostasis and heterologous protein accumulation in transplastomic plants. Plant Physiol 160: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman F-A (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Ehrnthaler M, Scharff LB, Fleischmann TT, Hasse C, Ruf S, Bock R (2014) Synthetic lethality in the tobacco plastid ribosome and its rescue at elevated growth temperatures. Plant Cell 26: 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck a, Kim M, Mullet J, Koop H-U (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J 19: 333–345 [DOI] [PubMed] [Google Scholar]
- Elghabi Z, Karcher D, Zhou F, Ruf S, Bock R (2011) Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome. Plant Biotechnol J 9: 599–608 [DOI] [PubMed] [Google Scholar]
- Emadpour M, Karcher D, Bock R (2015) Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res 43: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy D. (2005) Foundations for engineering biology. Nature 438: 449–453 [DOI] [PubMed] [Google Scholar]
- Fesenko E, Edwards R (2014) Plant synthetic biology: a new platform for industrial biotechnology. J Exp Bot 65: 1927–1937 [DOI] [PubMed] [Google Scholar]
- Fuentes P, Zhou F, Erban A, Karcher D, Kopka J, Bock R (2016) A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. eLife 5: 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes P, Armarego-Marriott T, Bock R (2018) Plastid transformation and its application in metabolic engineering. Curr Opin Biotechnol 49: 10–15 [DOI] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgianna DR, Mayfield SP (2012) Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488: 329–335 [DOI] [PubMed] [Google Scholar]
- Gerasymenko IM, Sheludko YV, Klebanovych AA, Rudas VA, Shakhovsky AM, Klein TM, Kuchuk NV (2017) Comparison of effectiveness of 5′-regulatory sequences in transplastomic tobacco chloroplasts. Transgenic Res 26: 65–75 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gloaguen P, Bournais S, Alban C, Ravanel S, Seigneurin-Berny D, Matringe M, Tardif M, Kuntz M, Ferro M, Bruley C, Rolland N, Vandenbrouck Y, et al. (2017) ChloroKB: a web application for the integration of knowledge related to chloroplast metabolic network. Plant Physiol 174: 922–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekaran T, Karcher D, Nielsen AZ, Martens HJ, Ruf S, Kroop X, Olsen CE, Motawie MS, Pribil M, Møller BL, Bock R, Jensen PE (2016) Transfer of the cytochrome P450-dependent dhurrin pathway from Sorghum bicolor into Nicotiana tabacum chloroplasts for light-driven synthesis. J Exp Bot 67: 2495–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Bock R (2013) Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 35: 354–365 [DOI] [PubMed] [Google Scholar]
- Gully BS, Shah KR, Lee M, Shearston K, Smith NM, Sadowska A, Blythe AJ, Bernath-Levin K, Stanley WA, Small ID, Bond CS (2015) The design and structural characterization of a synthetic pentatricopeptide repeat protein. Acta Crystallogr D Biol Crystallogr 71: 196–208 [DOI] [PubMed] [Google Scholar]
- Hanson MR, Lin MT, Carmo-Silva AE, Parry MAJ (2016) Towards engineering carboxysomes into C3 plants. Plant J 87: 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T, Miyazawa S, Yoshimura S, Shinzaki Y, Tomizawa K, Shindo K, Choi S-K, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55: 857–868 [DOI] [PubMed] [Google Scholar]
- Herz S, Füssl M, Steiger S, Koop H-U (2005) Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic Res 14: 969–982 [DOI] [PubMed] [Google Scholar]
- Hutchison CA III, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, et al. (2016) Design and synthesis of a minimal bacterial genome. Science 351: aad6253–aad6253 [DOI] [PubMed] [Google Scholar]
- Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, Fourcade HM, Kuehl JV, et al. (2005) Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol 395: 348–384 [DOI] [PubMed] [Google Scholar]
- Jin S, Daniell H (2015) The engineered chloroplast genome just got smarter. Trends Plant Sci 20: 622–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, de Mora K, Glieberman AL, Monie DD, Endy D (2009) Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol Eng 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ (2010) Synthetic biology: applications come of age. Nat Rev Genet 11: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Church GM (2014) Large-scale de novo DNA synthesis: technologies and applications. Nat Methods 11: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM (2013) Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci USA 110: 14024–14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A, Meyers B, Vainstein A, Maliga P, Citovsky V (2010) Autoluminescent plants. PLoS One 5: e15461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2001) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res 29: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YH, Giessen TW, Altenburg WJ, Silver PA (2018) Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat Commun 9: 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Legen J, Ruf S, Kroop X, Wang G, Barkan A, Bock R, Schmitz-Linneweber C (2018) Stabilization and translation of synthetic operon-derived mRNAs in chloroplasts by sequences representing PPR protein-binding sites. Plant J 94: 8–21 [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Thodey K, Trenchard I, Cravens A, Smolke CD (2018) Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc Natl Acad Sci USA 115: E3922–E3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MAJ, Hanson MR (2014) β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Stewart CN Jr (2015) Plant synthetic biology. Trends Plant Sci 20: 309–317 [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW--a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41: W575-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lössl A, Bohmert K, Harloff H, Eibl C, Mühlbauer S, Koop H-U (2005) Inducible trans-activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol 46: 1462–1471 [DOI] [PubMed] [Google Scholar]
- Lou C, Stanton B, Chen Y-J, Munsky B, Voigt CA (2012) Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat Biotechnol 30: 1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Rijzaani H, Karcher D, Ruf S, Bock R (2013) Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc Natl Acad Sci USA 110: E623–E632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Stegemann S, Agrawal S, Karcher D, Ruf S, Bock R (2017) Horizontal transfer of a synthetic metabolic pathway between plant species. Curr Biol 27: 3034–3041 [DOI] [PubMed] [Google Scholar]
- Madoka Y, Tomizawa K, Mizoi J, Nishida I, Nagano Y, Sasaki Y (2002) Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol 43: 1518–1525 [DOI] [PubMed] [Google Scholar]
- Magee AM, Coyne S, Murphy D, Horvath EM, Medgyesy P, Kavanagh TA (2004) T7 RNA polymerase-directed expression of an antibody fragment transgene in plastids causes a semi-lethal pale-green seedling phenotype. Transgenic Res 13: 325–337 [DOI] [PubMed] [Google Scholar]
- McBride KE, Schaaf DJ, Daley M, Stalker DM (1994) Controlled expression of plastid transgenes in plants based on a nuclear DNA-encoded and plastid-targeted T7 RNA polymerase. Proc Natl Acad Sci USA 91: 7301–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Razavi S, DeRose R, Inoue T (2013) Synthesizing biomolecule-based Boolean logic gates. ACS Synth Biol 2: 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer SK, Koop H-U (2005) External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J 43: 941–946 [DOI] [PubMed] [Google Scholar]
- Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai QA, Tran AB, Paull M, Keasling JD, Arkin AP, Endy D (2013) Precise and reliable gene expression via standard transcription and translation initiation elements. Nat Methods 10: 354–360 [DOI] [PubMed] [Google Scholar]
- Naqvi S, Zhu C, Farre G, Ramessar K, Bassie L, Breitenbach J, Perez Conesa D, Ros G, Sandmann G, Capell T, Christou P (2009) Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Natl Acad Sci USA 106: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell CA, Birch-Machin I, Hibberd JM, Gray JC (2003) Expression of green fluorescent protein from bacterial and plastid promoters in tobacco chloroplasts. Transgenic Res 12: 631–634 [DOI] [PubMed] [Google Scholar]
- O’Neill BM, Mikkelson KL, Gutierrez NM, Cunningham JL, Wolff KL, Szyjka SJ, Yohn CB, Redding KE, Mendez MJ (2012) An exogenous chloroplast genome for complex sequence manipulation in algae. Nucleic Acids Res 40: 2782–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, Moore TA, Moroney J, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. (2003) The symbiotic birth and spread of plastids: How many times and whodunit? J Phycol 39: 4–11 [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, Smith A, Hibberd J, et al. (2015) Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol 208: 13–19 [DOI] [PubMed] [Google Scholar]
- Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP (2012) RNA processing enables predictable programming of gene expression. Nat Biotechnol 30: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, Daniell H (2005) Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, and translation. Plant Physiol 138: 1746–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix J-D (2013) Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, Lee D, Huang CLV, Chandrasegaran S, Cai Y, Boeke JD, Bader JS (2017) Design of a synthetic yeast genome. Science 355: 1040–1044 [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, et al. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943 [DOI] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott R, Drager RG, Stern DB, Schuster G (1996) The 3′ untranslated regions of chloroplast genes in Chlamydomonas reinhardtii do not serve as efficient transcriptional terminators. Mol Gen Genet 252: 676–683 [DOI] [PubMed] [Google Scholar]
- Rudge TJ, Brown JR, Federici F, Dalchau N, Phillips A, Ajioka JW, Haseloff J (2016) Characterization of intrinsic properties of promoters. ACS Synth Biol 5: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Verma D, Samson N, Daniell H (2010) The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol 152: 2088–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27: 946–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Bock R (2014) Synthetic biology in plastids. Plant J 78: 783–798 [DOI] [PubMed] [Google Scholar]
- Schaumberg KA, Antunes MS, Kassaw TK, Xu W, Zalewski CS, Medford JI, Prasad A (2016) Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat Methods 13: 94–100 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Kushnir S, Babiychuk E, Poltnigg P, Herrmann RG, Maier RM (2005) Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell 17: 1815–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, Halpert M, Luro S, Schuster G, Stern DB (2011) Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 17: 2165–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Wang S, Xia E-H, Jiang J-J, Zeng F-C, Gao L-Z (2016) Full transcription of the chloroplast genome in photosynthetic eukaryotes. Sci Rep 6: 30135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, et al. (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolke CD. (2009) Building outside of the box: iGEM and the BioBricks Foundation. Nat Biotechnol 27: 1099–1102 [DOI] [PubMed] [Google Scholar]
- Stegemann S, Bock R (2009) Exchange of genetic material between cells in plant tissue grafts. Science 324: 649–651 [DOI] [PubMed] [Google Scholar]
- Stegemann S, Keuthe M, Greiner S, Bock R (2012) Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA 109: 2434–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Gruissem W (1987) Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51: 1145–1157 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Surzycki R, Cournac L, Peltier G, Rochaix J-D (2007) Potential for hydrogen production with inducible chloroplast gene expression in Chlamydomonas. Proc Natl Acad Sci USA 104: 17548–17553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P (1990) Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA 87: 8526–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K, Zhao D, Voigt CA (2012) Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc Natl Acad Sci USA 109: 7085–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen G, Svab Z, Maliga P (2012) Cell-to-cell movement of plastids in plants. Proc Natl Acad Sci USA 109: 2439–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafaee Y, Staniek A, Mancheno-Solano M, Warzecha H (2014) A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS One 9: e110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkov VT, Scotti N, Kahlau S, Maclean D, Grillo S, Gray JC, Bock R, Cardi T (2009) Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol 150: 2030–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venayak N, Anesiadis N, Cluett WR, Mahadevan R (2015) Engineering metabolism through dynamic control. Curr Opin Biotechnol 34: 142–152 [DOI] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA 107: 6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel BSJ. (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55: 85–107 [DOI] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R (2007) Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J 49: 276–288 [DOI] [PubMed] [Google Scholar]
- Yang H, Gray BN, Ahner BA, Hanson MR (2013) Bacteriophage 5′ untranslated regions for control of plastid transgene expression. Planta 237: 517–527 [DOI] [PubMed] [Google Scholar]
- Yu Z, Boehm CR, Hibberd JM, Abell C, Haseloff J, Burgess SJ, Reyna-Llorens I (2018) Droplet-based microfluidic analysis and screening of single plant cells. PLoS One 13: e0196810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R (2012) Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J 72: 115–128 [DOI] [PubMed] [Google Scholar]
- Zhou F, Karcher D, Bock R (2007) Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J 52: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Bock R (2018) Chloroplast translation: structural and functional organization, operational control and regulation. Plant Cell 30: 745–770 [DOI] [PMC free article] [PubMed] [Google Scholar]