Abstract
Engineered proteins can be used to optimize desired traits in plants; even though recent advances have resulted in new application areas, certain methodological challenges remain.
Protein engineering and directed evolution are powerful technologies for probing protein sequence-function relationships. These methods have been used to engineer both plant-derived proteins and exogenous proteins heterologously expressed in plants. In this review, we aim to further increase the interdisciplinary crossover between the disciplines of protein engineering and plant biology by first introducing protein engineering in some detail. This introduction is key to understanding current limitations to protein engineering when applied to plants. Subsequently, we provide an overview of the recent methodological progress in, and novel applications of, protein engineering and directed evolution in plant research.
A PRIMER ON PROTEIN ENGINEERING
Proteins and Their Properties
Evolution has shaped the functions and properties of proteins found in nature such that they contribute to beneficial phenotype in living organisms. However, these functions and roles are just a fraction of those biologically possible. By modifying the sequence of individual proteins, one can go beyond what nature has evolved and gain completely new functions or properties (Brustad and Arnold, 2011). Such modified proteins can be used to improve the phenotype of living organisms or have industrial or medical applications (Kumar and Singh, 2013; Porter et al., 2016). The processes and frameworks for modifying protein sequences fall in the domain of protein engineering.
Protein Engineering
Protein engineering is the process by which a researcher modifies a protein sequence through substitution, insertion, or deletion of nucleotides in the encoding gene, with the goal of obtaining a modified protein that is more suitable for a particular application or purpose than the unmodified protein. The focus on application sets protein engineering apart from the broader term “targeted mutagenesis.” Targeted mutagenesis, or site-directed mutagenesis, is a method whereby a specific site within a gene sequence is altered (Hutchison et al., 1978). Such alterations can be performed for engineering purposes, as in protein engineering, or for examining the effect of specific mutations in a gene.
Directed protein evolution—a method that was awarded the Nobel prize in chemistry in 2018—is a specific conceptual and methodological approach within protein engineering (Chen and Arnold, 1993; Arnold, 1998). The conceptual approach recognizes that we have a limited capability to predict the impact of individual amino acid substitutions on protein properties, but measuring the effect of those same substitutions can be readily achieved. The methodological approach involves generating a large set of diverse protein sequences, with some representing a potential solution to the engineering goal, and then experimentally screening the resulting proteins for desirable properties and functions. In a striking parallel to mathematics, the problem in protein engineering resembles the P ≠ NP problem; whereby finding a solution to a problem is hard, but verifying the solution is easy (Pierce and Winfree, 2002).
In directed evolution, sequence diversification and screening are often repeated multiple times, with additional amino acid substitutions accumulating in each round and each round providing a protein sequence closer to that of the protein engineering target. Directed evolution relies on methods from molecular biology to perform sequence diversification and methods from biochemistry, analytical chemistry, and microbiology to screen the resulting proteins for desired properties.
Methods for Sequence Diversification
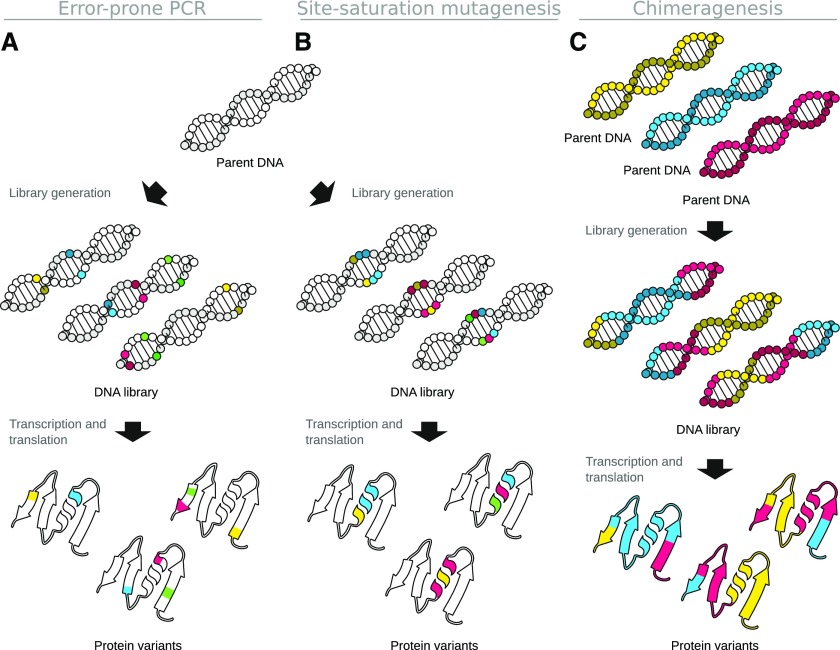
Many methods for DNA sequence diversification have been developed since directed evolution was first conceptualized (Fig. 1). Most of these methods fall into the following categories: error-prone PCR, site saturation mutagenesis, DNA shuffling or chimeragenesis, and random mutagenesis using chemical agents, physical agents, or hypermutator strains (Hiraga and Arnold, 2003; Wong et al., 2006; Labrou, 2010; Packer and Liu, 2015). In this update article, we only deal with the first three methods, since they enable the targeting of mutations to a specific locus. Regardless of the method used, the goal is to generate a sequence library, i.e. a large collection of diverse sequences, which include potential solutions to the engineering goal.
Figure 1.
Procedures for the diversification of genetic sequences. A limited number of random codon exchanges can be introduced via error-prone PCR (A). In this method, both the positions within the sequence and the nature of the amino acid modification are undefined. When employing site saturation mutagenesis (B), the positions of modification within the sequence are predetermined, and all possible amino acid modifications can be realized. A much higher rate of mutated amino acids relative to the original sequence can be achieved by utilizing chimeragenesis (C). In this method, several parental DNA sequences are being recombined, resulting in protein variants that contain different parts from different parents’ DNA sequences.
Error-Prone PCR
Error-prone PCR (Fig. 1A) relies on the introduction of random mutations throughout the amplified DNA sequence by means of DNA polymerase errors (Leung et al., 1989). The error rate in this method can be increased using specific polymerase mutants or by introducing low concentrations of MnCl2 into the PCR reaction. Error-prone PCR can be used to test the effect of mutations throughout the entire gene sequence (Fig. 1A). Consequently, improved variants identified from an error-prone PCR library often carry mutations at unexpected positions. Furthermore, the number of amino acid substitutions sampled at any given position of the sequence is limited by the inability to introduce concomitant random mutations at more than one base in a three-base codon sequence (Zhao et al., 2017). Improvements on the original method, such as sequence saturation mutagenesis, address some of these limitations (Wong et al., 2004).
Site Saturation Mutagenesis
Site saturation mutagenesis targets one or a few specific codons from the gene sequence (Fig. 1B) and introduce all possible amino acid substitutions at those sites (Zheng et al., 2004). This method is typically PCR based (Aiyar et al., 1996), but instead of relying on polymerase errors, the mutations are introduced using primers containing nucleotide mismatches at the targeted sites. Typically, pools of primers with the same binding site are used, with each individual primer encoding one specific amino acid substitution.
The advantage of using site saturation mutagenesis is that a small number of sites within a gene can be precisely targeted, and for these sites, all possible amino acid substitutions can be sampled. Hence, this method is suitable if one knows which positions in the amino acid sequence are important for a certain protein property (Zheng et al., 2004; Pedotti et al., 2009). Initial methods developed for site saturation mutagenesis introduced all 64 possible codons at a site, equivalent to using NNN of the International Union of Pure and Applied Chemistry standardized ambiguous nucleotide alphabet (Cornish-Bowden, 1985). Many of these codons are redundant and thus needlessly increase the extent of subsequent screening. To reduce this burden, many methods have been developed whereby a subset of codons are used (Reetz and Wu, 2008; Jochens and Bornscheuer, 2010), including computational tools for choosing codons for arbitrary selections of amino acids (Mena and Daugherty, 2005; Firth and Patrick, 2008; Engqvist and Nielsen, 2015).
Chimeragenesis
Chimeragenesis entails creating new protein sequences (chimeras) through concatenating parts of amino acid sequences derived from homologous proteins (Fig. 1C). This approach can be successful in combining desirable properties from two or more parent proteins, but also for generating proteins with properties not found in either parent (Hiraga and Arnold, 2003). Chimeragenesis implies a predefined reassembly of the genetic information and builds on the earlier approach of DNA shuffling (Stemmer, 1994a, 1994b). Chimeragenesis has been further developed to encompass computational methods for designing chimeric protein libraries through SCHEMA (Meyer et al., 2003; Silberg et al., 2004) and novel methods for recombination (Coco et al., 2001; Sun et al., 2003; Smith et al., 2013).
Methods for Screening
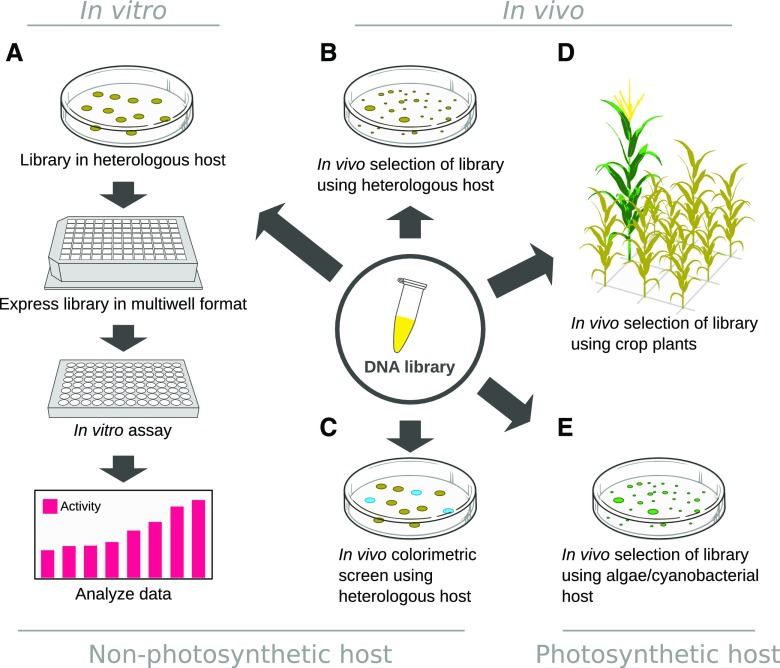
Regardless of the method used for sequence diversification, an efficient search for improved variants must be conducted using screening methods (Fig. 2). Screens can be performed using a wide array of methods that broadly fall into two categories: assaying protein properties in vitro or measuring protein effects in vivo. Both methods require an accurate and precise readout of the protein property one wishes to engineer. If the measurements are imprecise, improved variants will be overlooked (false negatives), and nonimproved variants will be incorrectly scored as improved (false positives). Such incorrect scoring will greatly increase the difficulty of finding improved variants that match the engineering goal.
Figure 2.
Procedures for library screening. Genetic libraries, generated via methods depicted in Figure 1, have to be analyzed in order to identify improved protein variants employing methods, which depend on the individual protein case and the screen available. As such, the library can be expressed in a heterologous host and analyzed in vitro, here exemplified by a screen in multiwell format (A). If screening in vivo in a nonphotosynthetic heterologous host, fitness or survival of the mutant strains (B) or a colorimetric detection of improved mutants (C) can be utilized. These two approaches can also be employed in the photosynthetic hosts (D and E).
In Vitro Methods
In vitro screens typically employ heterologous hosts, such as Escherichia coli or Saccharomyces cerevisiae or alternatively in vitro translation, to produce protein products from the sequence library (Fig. 2A). The protein products are assayed for improvement in the target property using colorimetric assays, analytical measurements of substrate consumption or product formation, target affinity assays, or a range of other methods (Aharoni et al., 2005). In addition to providing accurate readouts of the engineered protein property, such screens must ensure a link between data obtained from the screen and a genotype, allowing the identification of beneficial mutations. When performed in a multiwell format, the connection between phenotype and genotype is given by mapping the assay plate position to the plate position of the cells on which the assay was performed. In veritable high-throughput methods such as phage display (Smith, 1985), yeast surface display (Gai and Wittrup, 2007), or artificial microdroplet compartments (Tawfik and Griffiths, 1998), this connection is provided through physical colocalization of DNA and protein products.
In Vivo Methods
In vivo screens make use of a wide variety of phenotypes as a readout of the engineered protein properties and have to be carefully selected according to the function of the engineered protein (Fig. 2, B–E). Screening by selection is an elegant and powerful approach that establishes a connection between properties of the engineered protein and the survival of an organism (Fig. 2, B, D, and E). Selection has been used extensively, particularly in studying antibiotic resistance (Orencia et al., 2001). Other in vivo screens directly measure the color or fluorescence (Zlokarnik et al., 1998; McIsaac et al., 2014) of the engineered protein to identify improved variants (Fig. 2C). Yet other screens monitor the ability of an organism to consume or produce a specific compound, either colorimetrically (Zhang et al., 1997), fluorometrically (Jeschek et al., 2016), or analytically (Coelho et al., 2013). Cell surface display can be used to probe specific properties, such as the engineered affinity for a ligand (Xiao et al., 2015). A major benefit of in vivo screens is that they often lend themselves to extremely high throughout, allowing an investigator to screen large sequence libraries, particularly in cases where survival can be used as a readout or where cell sorting can be employed. In in vivo screens, the connection between the engineered target property and genotype is a natural consequence of physical colocalization of protein and DNA inside the organism, in a manner analogous to phage display.
METHODOLOGIES AND LIMITATIONS IN PLANT PROTEIN ENGINEERING
Review Scope
Protein engineering and directed evolution are methods; here, we review their use in plant research. We have considered literature relating to the engineering and use of plant proteins in nonplant organisms or for in vitro use and have selected different, prominent use cases. In addition to this, we have surveyed the literature relating to the engineering of proteins either derived from plant or nonplant organisms for the use in plants, algae, or cyanobacteria. Finally, we briefly explore recent methodological developments for performing protein engineering and directed evolution in planta.
To limit the scope of this review, we have chosen not to cover the introduction of a small number of targeted mutations but focus instead on approaches requiring the screening of many variants. Additionally, there is a large body of literature relating to directed evolution of enzymes to break down plant biomass (Álvarez et al., 2016; Kumar et al., 2016b), but these will not be covered here.
The protein engineering methodologies applied in plants and their outstanding problems differ widely depending on the area of application. To facilitate the structured discussion of these methods and difficulties, we divide the topic in two sections. This division is made based on which organism is used for the screening process. The first section deals with using heterologous hosts to screen protein variants for subsequent applications in plants. The second section deals specifically with using plants or algae to screen protein variants, without the use of nonphotosynthetic heterologous hosts. For both of these sections, we outline in what situation the approach may be applicable, give examples of past applications, and specify known difficulties and novel methodological advances.
The Use of Heterologous Hosts for Protein Engineering in Plant Biotechnology
Use Cases for Heterologous Hosts
Engineering proteins for applications in plants is a key method in plant biotechnology. However, much of this engineering has focused on improving a small number of plant traits, such as glyphosate resistance (Pollegioni et al., 2011) or Rubisco performance (Wilson and Whitney, 2017). Furthermore, this engineering is usually performed within heterologous hosts to leverage established microbial methods. According to current legislation, plants containing DNA that has been manipulated outside of the host are considered genetically modified organisms. Bringing genetically-modified-organism plants to market is coupled to a lengthy regulatory process and large capital investments (Bradford et al., 2005; Qaim, 2009). Combined, these two factors have resulted in companies targeting so-called “blockbuster traits” in crops. Blockbuster traits are those that have a very large market value, a necessary requirement to recoup the investment required to develop and deregulate the engineered plants. Below, we highlight approaches for engineering exogenous genes for crop pest and herbicide resistance, two of the most prominent blockbuster traits. In addition, we review the use of heterologous hosts to engineer Rubisco.
Engineering of Bacterial Toxins and Their Applications in Plants
Optimization of Bacillus thuringiensis toxin has been a focus of insect-specific pest control strategies. This optimization has been carried out in a variety of ways, for example, through truncation, domain swapping, peptide addition, and amino acid mutation (Deist et al., 2014). The most prevalent engineering goals addressed in these optimization approaches include increasing toxin potency to combat increasing pest resistance toward the toxins and expanding their applicability to a wider range of pests.
In a recent example, the toxin Cry1Ab, which does not significantly affect the insect pest Nilaparvata lugens (rice brown planthopper), was engineered to yield a variant that, when fed as a purified protein, increases the mortality of the pest (Shao et al., 2016). In this work, the key to increased pest mortality was to retain the toxin in the insect. To achieve this, the authors identified peptides that were reported to bind to the gut of the rice brown planthopper in an earlier study employing phage-display technology and randomized peptide libraries (Shao et al., 2013). When these gut-binding peptides were rationally introduced into loops on the surface of the Cry1Ab protein and the resulting proteins were fed to the pest, an increased mortality was observed because the toxin was able bind to and interact with Nilaparvata lugens, thus broadening the scope of affected pests.
Similarly, the toxin resistance of the insect pest Trichoplusia ni (Tn) was addressed (Badran et al., 2016). For this purpose, the authors chose the widely used Cry1Ac toxin and enabled its binding to protein receptors in the insect gut cell membrane that usually do not interact with the toxin. Specifically, they evolved the toxin to bind to TnCAD, an insect cell membrane cadherin-like receptor, employing a phage-based technology, which has recently been introduced and enables the continuous selection of efficient binding peptides (phage-assisted continuous evolution). Phage-assisted continuous evolution was used to continuously mutate and select variants of the Cry1Ac toxin that efficiently bind TnCAD. After 500 generations (approximately 22 d), several toxin variants with binding constants to TnCAD in the nanomolar range were identified, whereas for wild-type Cry1Ac, no binding was observed. When one of these protein variants was fed to Tn insects, a 335-fold increased rate of mortality was observed.
The two examples outlined above showcase efficient strategies to obtain early indications whether a protein evolution strategy is successful, without the need to express the proteins in plants. This is especially important if thousands of variants need to be tested, which is prohibitive in plants, due to low transformation rates. However, it is important to subsequently test promising protein variants in a plant system. A recent example of this was performed by Das et al. (2017). The authors found that feeding chickpea (Cicer arietinum) leaves expressing the Cry1Aabc protein, which had been generated by chimeragenesis, to larvae of the gram pod borer (Helicoverpa armigera Hubner) led to significantly increased mortality when compared to control leaves not expressing the protein. In this study, transformation efficiencies of 0.076% were reported, underscoring the benefit of initial studies outside of the plant to find promising protein candidates. This argument is further strengthened by the fact that not all transgenic plants carrying new genes also express the corresponding protein. For instance, in transgenic tobacco (Nicotiana tabacum NC89) protected against Cyr1Ac-resistant cotton bollworm, only 13%–38% of the regenerated plants expressed the target protein (Li et al., 2018a).
Engineering Enzymes for Glyphosate Tolerance and Their Applications in Plants
Glyphosate [N-(phosphonomethyl)Gly] is the best-selling herbicide to date, and much research has been devoted to generating transgenic plants that are not susceptible to its effects (Sammons and Gaines, 2014). Glyphosate inhibits the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), which is part of the essential shikimate pathway leading to the production of the aromatic amino acids Phe, Tyr, and Trp, and this inhibition results in plant death. Two main strategies have been employed to generate glyphosate-tolerant transgenic plants: engineering EPSPS to remain active in the presence of glyphosate or introducing genes encoding enzymes that remove glyphosate by breaking it down.
Engineering the EPSPS for activity in the presence of glyphosate typically involves screening large libraries of genetic variants. In plants, this approach is hampered by limited transformation efficiency. Therefore, initial libraries of EPSPS variants, generated by DNA shuffling (Tian et al., 2013) or error-prone PCR (Mao et al., 2017), are typically tested in E. coli by selecting for colony growth in the presence of glyphosate at inhibitory concentrations. Improved protein variants are then characterized and verified by generating transgenic plants, such as rice (Oryza sativa; Tian et al., 2013, 2015) and Arabidopsis (Arabidopsis thaliana; Tian et al., 2015; Mao et al., 2017).
An example for the removal of glyphosate is the use and engineering of bacterial Gly oxidases. Gly oxidases cleave the carbon-nitrogen bond in glyphosate, allowing libraries generated by error-prone PCR, site-directed mutagenesis, and DNA shuffling to be screened in E. coli using glyphosate as the sole nitrogen source (Zhan et al., 2013). Enzyme variants obtained in this approach showed up to a 160-fold increase in substrate affinity and a 326-fold enhancement in catalytic efficiency against glyphosate. Nicolia et al. (2014) have used a rational engineering approach employing site saturation and site-directed mutagenesis to show that transgenic alfalfa (Medicago sativa) plants do indeed show an increased tolerance to glyphosate when expressing optimized Gly oxidase variants.
Engineering Rubisco for Improved Carboxylation Properties
Rubisco catalyzes the incorporation of CO2 into ribulose-1,5-bisphosphate (RuBP), a key reaction in carbon fixation in plants. In a competing reaction, Rubisco also catalyzes the incorporation of O2 into RuBP. The metabolites produced in this competing reaction are salvaged through the photorespiratory pathway, a wasteful process in which CO2 is lost (Peterhansel and Maurino, 2011). Improving the carboxylation properties of Rubisco thus has the potential to significantly improve crop yield.
Research relating to this important enzyme has an almost 50-year history. In 1971, Rubisco was identified as the direct cause for photorespiration (Bowes et al., 1971), and already in 1980, the first attempts were made to engineer more efficient carbon fixation by screening for suppressor mutants in plants harboring a deficient photorespiratory pathway (Somerville and Ogren, 1980). Rubisco itself was first targeted through site-directed mutagenesis in 1984 (Gutteridge et al., 1984). The first report describing the directed evolution of Rubisco, using an in vivo Rhodoobacter capsulatus screening system, was published fifteen years later (Smith and Tabita, 2003). Subsequently, E. coli was developed as a Rubisco screening system (Parikh et al., 2006; Mueller-Cajar et al., 2007; Antonovsky et al., 2016).
E. coli screening systems typically depend on the heterologous expression of phosphoribulokinase, which produces RuBP. RuBP is toxic to bacteria, and Rubisco activity can therefore be used to alleviate the toxicity of this compound and ensure survival of the host organism. One issue in these screens is that false positives are obtained at high frequencies (Greene et al., 2007; Cai et al., 2014) due to natural transposon-mediated silencing of phosphoribulokinase (Wilson and Whitney, 2017). In a clever approach, the problem of false positives was combated by expressing a phosphoribulokinase-neomycin phosphotransferase fusion protein and including the additional selection pressure of antibiotic resistance (Wilson et al., 2018). In an approach similar to the one taken in E. coli, the soil bacterium Ralstonia eutropha has also been developed for in vivo screening of Rubisco variants (Satagopan and Tabita, 2016).
Even though these in vivo screening methods show great promise, their impact for improving crop yields remains to be realized. This may soon change, however, as it is now possible, using coexpression of five plant-derived chaperones, to obtain functional plant Rubisco in E. coli (Aigner et al., 2017). Heterologous expression of land plant Rubisco represents a major advance, as established mutagenesis procedures and selection systems can be leveraged for improving its catalytic properties. Improved Rubisco variants will subsequently need to be reintroduced into plants, a process for which a recent proof of principle was achieved using an improved nonphotosynthetic Rubisco from Methanococcoides burtonii (Wilson et al., 2016).
Difficulties and Recent Developments in Using Heterologous Hosts
There are several challenges when expressing plant proteins in heterologous hosts. Some of these challenges relate to problems with the RNA transcript sequence, such as poor codon usage (Chaney and Clark, 2015) and the formation of hairpin structures (Cambray et al., 2018). Further challenges relate to how other proteins interact with the target protein to improve folding, such as chaperones (Aigner et al., 2017) and enzymes performing posttranslational modifications on the protein product (Hou et al., 2012; Mattanovich et al., 2012). A protein that is poorly expressed or incorrectly folded is difficult or impossible to engineer.
Much work has been invested in solving the codon bias problem, which can lead to poor expression or incorrect folding of plant proteins in heterologous hosts. Living organisms have widely different usage preferences for codons that encode the same amino acids (Chaney and Clark, 2015). The specific preferences for each organism can be obtained by computationally analyzing their genome sequence (Athey et al., 2017). Codon optimization, a process where less-frequent codons in the coding sequence are replaced by more frequent synonymous codons, has long been used to address this issue (Burgess-Brown et al., 2008; Welch et al., 2009; Maertens et al., 2010). However, in many cases, codon optimization improves expression but fails to yield correctly folded protein. These failures may be due to the fact that some proteins require sections of less-frequent codons for translation to slow down, thereby allowing time for the emerging protein chain to fold properly (Marin, 2008; Zhang et al., 2009; Deane and Saunders, 2011; Zhang and Ignatova, 2011; Rosenblum et al., 2013).
Several recent computational approaches have been developed to improve codon optimization methods. Some of these approaches provide a tool without experimentally testing its efficacy (Rodriguez et al., 2018). In other cases, investigators do experimentally test their predictions, sometimes verifying the tool (Tian et al., 2017) and sometimes finding that the tool has more limited efficacy (Mignon et al., 2018). There are also experimental methods leveraging directed evolution to improve protein folding in vivo, as reviewed recently (Sachsenhauser and Bardwell, 2018).
The Use of Photosynthetic Organisms for Protein Engineering in Plant Biotechnology
Use Cases for In Planta Protein Engineering
Engineering proteins by screening sequence libraries directly in plants, instead of using heterologous hosts, is currently advisable only for a small set of use cases where specific circumstances make it necessary. Such circumstances typically involve plants having some property that is required to evaluate the engineered proteins’ performance—a property that is difficult to replicate in vitro or in a heterologous host. Examples for use cases where engineering in plants is preferable include engineering of plant signaling pathways, engineering enzymes acting on plant metabolites that are difficult to obtain, and engineering plant-microbe interactions. The use of in planta screening of variants of a single gene essentially provides a more focused and powerful approach for interrogating plant physiology through mutagenesis, as compared to genome-wide mutagenesis.
Engineering Plant Immune Effectors
Recently, there has been an increased interest in further understanding and modulating the innate immune response of plants (Bent and Mackey, 2007; Grant et al., 2013; Kourelis et al., 2016; Sun et al., 2017). A key part of the molecular system to defend against pathogens is the intracellular immune receptors, which belong to the nucleotide-binding Leu-rich-repeat-containing protein family. Nucleotide-binding Leu-rich-repeat-containing proteins bind to specific effectors found in pathogens and trigger a defense reaction. Random mutagenesis of these proteins can modify or broaden the spectrum of potential pathogens being detected, as has been demonstrated with initial diversification of the genes by error-prone PCR (Segretin et al., 2014; Steinbrenner et al., 2015; Sueldo et al., 2015) or site saturation mutagenesis (Helft et al., 2016) and subsequent transformation employing Argobacterium tumefaciens. Such studies can only be performed inside the plant host system, as the response (often cell death) can only be observed there.
Engineering Genes Encoded in the Plastid Genome
Genes-encoding proteins participating in both the photosynthetic dark and light reactions have been targeted by mutagenesis and screening in the photosynthetic unicellular alga Chlamydomonas reinhardtii. For engineering the dark reactions, Zhu and colleagues screened a DNA-shuffled C. reinhardtii Rubisco large subunit library through chloroplast transformation of a Rubisco large subunit-deficient C. reinhardtii strain (Zhu et al., 2010). A three-tiered selection/screening procedure was used, involving selection for autotrophic growth on minimal media, followed by selection by competitive growth and subsequent identification of improved variants. This allowed the investigators to identify multiple clones with increased carboxylase activity (Zhu et al., 2010). Some of the specific claims relating to the improved Rubisco properties have been challenged (Wilson and Whitney, 2017), but the study remains an important proof of principle for screening sequence libraries directly in chloroplasts.
In a similar approach, petD—the gene which encodes the core protein subunit of the cytochrome b6f complex—was engineered through the in vivo screening of an in vitro-generated error-prone PCR library. The library was integrated directly at the petD locus inside chloroplasts of a petD-deficient C. reinhardtii strain, followed by a screen for photoautotrophic growth (Dumas et al., 2018). In this study, the goal was not to engineer a more efficient protein but rather to probe the robustness and plasticity of this transmembrane complex subunit through mutagenesis and screening. In principle, it should also be possible to apply methods such as these to plastids of land plants (Dumas et al., 2018). The two approaches have important limitations, however. The first is that they rely on selection systems requiring mutant strains that are impaired in the functional phenotype being selected for. Such strains may be difficult to obtain. A second limitation is the low transformation efficiencies, which limits the number of distinct library sequences that can be introduced. For further reading regarding plastid synthetic biology, we refer interested readers to the companion paper by Boehm and Bock (2019) as well as the companion paper on engineering the photosynthetic light reactions by (Leister, 2019).
Difficulties and Recent Developments for In Planta Protein Engineering
A main difficulty for in planta protein engineering is low transformation rates. Improvements to current transformation methods or the development of novel ones (Altpeter et al., 2016) will be key for expanding the use of plants for protein engineering. Alternatively, in vivo mutagenesis could provide a viable option for species wherein transformation rates are low. The power of this approach comes from the fact that the sequence diversity is generated directly inside the target organism, thus negating constraints on transformation efficiency. For example, a nitrogen-regulated mutator strain has been developed in the cyanobacterium Synechococcus sp. to alleviate transformation bottlenecks (Emlyn-Jones et al., 2003). A drawback of current methods for in vivo mutagenesis is that mutations cannot be targeted to a specific locus. Novel methods to perform targeted in vivo mutagenesis in plants are needed. Indeed, in vivo site saturation mutagenesis has already been successfully performed in human (Homo sapiens) cell lines (Findlay et al., 2014; Ma et al., 2017). The key technological advance in these methods is to couple CRISPR/Cas9-induced double-strand breaks with multiplex homology-directed repair. Whether this approach can be adapted to plants is an open question.
Even though in vivo saturation mutagenesis has not yet been performed in plants, the use of CRISPR/Cas9 for plant genome editing was achieved as early as in 2013 (Li et al., 2013; Nekrasov et al., 2013; Shan et al., 2013). This technology has since been used in a wide variety of applications to improve crop plants. For example, CRISPR/Cas9 has been used for the domestication of new crop plants, as recently showcased in work on the orphan crop groundcherry (Physalis pruinosa; Lemmon et al., 2018) and wild tomato (Solanum pimpinellifolium; Li et al., 2018b; Zsögön et al., 2018). Similarly, CRISPR/Cas9 was used for simultaneously modifying different homeologous gene copies in Brassica napus to improve the agronomic trait shatter resistance (Braatz et al., 2017). In another important example, the CRISPR/Cas9 system was used to achieve nontransgenic mutations in perennial heterozygous plants. The method leverages agrobacterial transformation and transient expression to perform edits with an overall nontransgenic mutation rate of 8.2% (Chen et al., 2018). Genomes in leaf disks, shoots, roots, or cotyledons can be edited using this method. This represents an important advance, as plant regeneration from such tissues is established for most crop plants. For an overview on CRISPR/Cas9-mediated genome editing in crops, see several recent reviews (Songstad et al., 2017; Jaganathan et al., 2018; Jung et al., 2018). Perspectives on future application areas of CRISPR/Cas9 in plant breeding have recently been reviewed (Puchta, 2017; Scheben et al., 2017).
A transformative technology such as CRISPR/Cas9 raises important ethical and legal concerns. Encouragingly, some researchers have started forming interdisciplinary research teams to identify and analyze the ethical and legal implications of using these technologies (Nordberg et al., 2018).
CONCLUSION
Protein engineering and directed evolution are powerful technologies in biotechnology. However, these technologies have only been applied to a limited set of plant traits. Further developments in transformation technologies, the use of CRISPR/Cas9 for targeted mutagenesis, and possibly the development of technologies for in planta library generation are expected to yield more protein engineering approaches in plant biotechnology (see Outstanding Questions). However, any new technologies resulting from such developments must also be accompanied by favorable regulatory frameworks or they will likely result in limited use for plant improvement.
One underdeveloped application area for protein engineering lies in engineering plant-microbiome interactions. We believe that engineering such interactions will be a key component in the future of plant biotechnology. A holistic approach is needed, encompassing soil amendment, microbial engineering, and plant engineering, to sufficiently raise crop yields (Dessaux et al., 2016). Whereas protein engineering for plants has been the main focus of this review, plant-microbe interactions can also be modified using gene-editing and systems biology tools (Kumar et al., 2016a). Techniques to perform host-mediated microbiome engineering already exist (Mueller and Sachs, 2015), but protein engineering is not commonly used for this purpose. Using protein engineering to achieve these goals should not only focus on crop improvement and product development but also serve as a powerful tool to further understand the basis of plant-microbe interactions. We look forward to future developments in this area.
Acknowledgments
The authors would like to dedicate this article to Prof. Frances Arnold to mark the occasion of her winning the Nobel prize in chemistry in 2018 for her work on directed evolution of enzymes, as well as to show our gratitude for her mentorship and support. The authors declare no conflicts of interest.
Footnotes
M.K.M.E. acknowledges funding through the Chalmers University of Technology Life Science Engineering Area of Advance. K.S.R. acknowledges financial support by the Helmholtz program “BioInterfaces in Technology and Medicine.”
Articles can be viewed without a subscription.
References
- Aharoni A, Griffiths AD, Tawfik DS (2005) High-throughput screens and selections of enzyme-encoding genes. Curr Opin Chem Biol 9: 210–216 [DOI] [PubMed] [Google Scholar]
- Aigner H, Wilson RH, Bracher A, Calisse L, Bhat JY, Hartl FU, Hayer-Hartl M (2017) Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Aiyar A, Xiang Y, Leis J (1996) Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol 57: 177–191 [DOI] [PubMed] [Google Scholar]
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, et al. (2016) Advancing crop transformation in the era of genome editing. Plant Cell 28: 1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, Reyes-Sosa FM, Díez B (2016) Enzymatic hydrolysis of biomass from wood. Microb Biotechnol 9: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonovsky N, Gleizer S, Noor E, Zohar Y, Herz E, Barenholz U, Zelcbuch L, Amram S, Wides A, Tepper N, et al. (2016) Sugar synthesis from CO2 in Escherichia coli. Cell 166: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold FH. (1998) Design by directed evolution. Acc Chem Res 31: 125–131 [Google Scholar]
- Athey J, Alexaki A, Osipova E, Rostovtsev A, Santana-Quintero LV, Katneni U, Simonyan V, Kimchi-Sarfaty C (2017) A new and updated resource for codon usage tables. BMC Bioinformatics 18: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran AH, Guzov VM, Huai Q, Kemp MM, Vishwanath P, Kain W, Nance AM, Evdokimov A, Moshiri F, Turner KH, et al. (2016) Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 533: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Boehm CR, Bock R (2019) Recent advances and current challenges in synthetic biology of the plastid genetic system and metabolism. Plant Physiol 179: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes G, Ogren WL, Hageman RH (1971) Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem Biophys Res Commun 45: 716–722 [DOI] [PubMed] [Google Scholar]
- Braatz J, Harloff H-J, Mascher M, Stein N, Himmelbach A, Jung C (2017) CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol 174: 935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford KJ, Van Deynze A, Gutterson N, Parrott W, Strauss SH (2005) Regulating transgenic crops sensibly: Lessons from plant breeding, biotechnology and genomics. Nat Biotechnol 23: 439–444 [DOI] [PubMed] [Google Scholar]
- Brustad EM, Arnold FH (2011) Optimizing non-natural protein function with directed evolution. Curr Opin Chem Biol 15: 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Brown NA, Sharma S, Sobott F, Loenarz C, Oppermann U, Gileadi O (2008) Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif 59: 94–102 [DOI] [PubMed] [Google Scholar]
- Cai Z, Liu G, Zhang J, Li Y (2014) Development of an activity-directed selection system enabled significant improvement of the carboxylation efficiency of Rubisco. Protein Cell 5: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray G, Guimaraes JC, Arkin AP (2018) Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli. Nat Biotechnol 36: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Chaney JL, Clark PL (2015) Roles for synonymous codon usage in protein biogenesis. Annu Rev Biophys 44: 143–166 [DOI] [PubMed] [Google Scholar]
- Chen K, Arnold FH (1993) Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc Natl Acad Sci USA 90: 5618–5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li W, Katin-Grazzini L, Ding J, Gu X, Li Y, Gu T, Wang R, Lin X, Deng Z, et al. (2018) A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic Res 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH, Monticello DJ (2001) DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat Biotechnol 19: 354–359 [DOI] [PubMed] [Google Scholar]
- Coelho PS, Brustad EM, Kannan A, Arnold FH (2013) Olefin cyclopropanation via carbene transfer catalyzed by engineered cytochrome P450 enzymes. Science 339: 307–310 [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. (1985) Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res 13: 3021–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Datta S, Thakur S, Shukla A, Ansari J, Sujayanand GK, Chaturvedi SK, Kumar PA, Singh NP (2017) Expression of a chimeric gene encoding insecticidal crystal protein Cry1Aabc of Bacillus thuringiensis in chickpea (Cicer arietinum L.) confers resistance to gram pod borer (Helicoverpa armigera Hubner.). Front Plant Sci 8: 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane CM, Saunders R (2011) The imprint of codons on protein structure. Biotechnol J 6: 641–649 [DOI] [PubMed] [Google Scholar]
- Deist BR, Rausch MA, Fernandez-Luna MT, Adang MJ, Bonning BC (2014) Bt toxin modification for enhanced efficacy. Toxins (Basel) 6: 3005–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaux Y, Grandclément C, Faure D (2016) Engineering the rhizosphere. Trends Plant Sci 21: 266–278 [DOI] [PubMed] [Google Scholar]
- Dumas L, Zito F, Auroy P, Johnson X, Peltier G, Alric J (2018) Structure-function analysis of chloroplast proteins via random mutagenesis using error-prone PCR. Plant Physiol 177: 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlyn-Jones D, Price GD, Andrews TJ (2003) Nitrogen-regulated hypermutator strain of Synechococcus sp. for use in in vivo artificial evolution. Appl Environ Microbiol 69: 6427–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MKM, Nielsen J (2015) ANT: Software for generating and evaluating degenerate codons for natural and expanded genetic codes. ACS Synth Biol 4: 935–938 [DOI] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J (2014) Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513: 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Patrick WM (2008) GLUE-IT and PEDEL-AA: New programmes for analyzing protein diversity in randomized libraries. Nucleic Acids Res 36: W281–W285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai SA, Wittrup KD (2007) Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol 17: 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Kazan K, Manners JM (2013) Exploiting pathogens’ tricks of the trade for engineering of plant disease resistance: challenges and opportunities. Microb Biotechnol 6: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DN, Whitney SM, Matsumura I (2007) Artificially evolved Synechococcus PCC6301 Rubisco variants exhibit improvements in folding and catalytic efficiency. Biochem J 404: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S, Sigal I, Thomas B, Arentzen R, Cordova A, Lorimer G (1984) A site-specific mutation within the active site of ribulose-1,5-bisphosphate carboxylase of Rhodospirillum rubrum. EMBO J 3: 2737–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft L, Thompson M, Bent AF (2016) Directed evolution of FLS2 towards novel flagellin peptide recognition. PLoS One 11: e0157155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga K, Arnold FH (2003) General method for sequence-independent site-directed chimeragenesis. J Mol Biol 330: 287–296 [DOI] [PubMed] [Google Scholar]
- Hou J, Tyo KEJ, Liu Z, Petranovic D, Nielsen J (2012) Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res 12: 491–510 [DOI] [PubMed] [Google Scholar]
- Hutchison CA III, Phillips S, Edgell MH, Gillam S, Jahnke P, Smith M (1978) Mutagenesis at a specific position in a DNA sequence. J Biol Chem 253: 6551–6560 [PubMed] [Google Scholar]
- Jaganathan D, Ramasamy K, Sellamuthu G, Jayabalan S, Venkataraman G (2018) CRISPR for crop improvement: An update review. Front Plant Sci 9: 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschek M, Reuter R, Heinisch T, Trindler C, Klehr J, Panke S, Ward TR (2016) Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature 537: 661–665 [DOI] [PubMed] [Google Scholar]
- Jochens H, Bornscheuer UT (2010) Natural diversity to guide focused directed evolution. ChemBioChem 11: 1861–1866 [DOI] [PubMed] [Google Scholar]
- Jung C, Capistrano-Gossmann G, Braatz J, Sashidhar N, Melzer S (2018) Recent developments in genome editing and applications in plant breeding. Plant Breed 137: 1–9 [Google Scholar]
- Kourelis J, van der Hoorn RAL, Sueldo DJ (2016) Decoy engineering: the next step in resistance breeding. Trends Plant Sci 21: 371–373 [DOI] [PubMed] [Google Scholar]
- Kumar A, Singh S (2013) Directed evolution: tailoring biocatalysts for industrial applications. Crit Rev Biotechnol 33: 365–378 [DOI] [PubMed] [Google Scholar]
- Kumar V, Baweja M, Singh PK, Shukla P (2016a) Recent developments in systems biology and metabolic engineering of plant-microbe interactions. Front Plant Sci 7: 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Marín-Navarro J, Shukla P (2016b) Thermostable microbial xylanases for pulp and paper industries: Trends, applications and further perspectives. World J Microbiol Biotechnol 32: 34. [DOI] [PubMed] [Google Scholar]
- Labrou NE. (2010) Random mutagenesis methods for in vitro directed enzyme evolution. Curr Protein Pept Sci 11: 91–100 [DOI] [PubMed] [Google Scholar]
- Leister D. (2019) Genetic engineering, synthetic biology and the light reactions of photosynthesis. Plant Physiol 179: 778–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, Van Eck J, Lippman ZB (2018) Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plants 4: 766–770 [DOI] [PubMed] [Google Scholar]
- Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N (1989) Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246: 1306–1309 [DOI] [PubMed] [Google Scholar]
- Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31: 688–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang Z, Zhou Y, Li C, Wang G, Wang H, Zhang J, Liang G, Lang Z (2018a) Expression of cry2Ah1 and two domain II mutants in transgenic tobacco confers high resistance to susceptible and Cry1Ac-resistant cotton bollworm. Sci Rep 8: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C (2018b) Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol [DOI] [PubMed] [Google Scholar]
- Ma L, Boucher JI, Paulsen J, Matuszewski S, Eide CA, Ou J, Eickelberg G, Press RD, Zhu LJ, Druker BJ, et al. (2017) CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc Natl Acad Sci USA 114: 11751–11756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens B, Spriestersbach A, von Groll U, Roth U, Kubicek J, Gerrits M, Graf M, Liss M, Daubert D, Wagner R, et al. (2010) Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci 19: 1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Xie H, Chen S, Valverde BE, Qiang S (2017) Error-prone PCR mutation of Ls-EPSPS gene from Liriope spicata conferring to its enhanced glyphosate-resistance. Pestic Biochem Physiol 141: 90–95 [DOI] [PubMed] [Google Scholar]
- Marin M. (2008) Folding at the rhythm of the rare codon beat. Biotechnol J 3: 1047–1057 [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D (2012) Recombinant protein production in yeasts. In Lorence A, ed, Recombinant Gene Expression. Humana Press, Totowa, NJ, pp 329–358 [DOI] [PubMed] [Google Scholar]
- McIsaac RS, Engqvist MKM, Wannier T, Rosenthal AZ, Herwig L, Flytzanis NC, Imasheva ES, Lanyi JK, Balashov SP, Gradinaru V, et al. (2014) Directed evolution of a far-red fluorescent rhodopsin. Proc Natl Acad Sci USA 111: 13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena MA, Daugherty PS (2005) Automated design of degenerate codon libraries. Protein Eng Des Sel 18: 559–561 [DOI] [PubMed] [Google Scholar]
- Meyer MM, Silberg JJ, Voigt CA, Endelman JB, Mayo SL, Wang Z-G, Arnold FH (2003) Library analysis of SCHEMA-guided protein recombination. Protein Sci 12: 1686–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignon C, Mariano N, Stadthagen G, Lugari A, Lagoutte P, Donnat S, Chenavas S, Perot C, Sodoyer R, Werle B (2018) Codon harmonization: going beyond the speed limit for protein expression. FEBS Lett 592: 1554–1564 [DOI] [PubMed] [Google Scholar]
- Mueller UG, Sachs JL (2015) Engineering microbiomes to improve plant and animal health. Trends Microbiol 23: 606–617 [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Morell M, Whitney SM (2007) Directed evolution of rubisco in Escherichia coli reveals a specificity-determining hydrogen bond in the form II enzyme. Biochemistry 46: 14067–14074 [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 691–693 [DOI] [PubMed] [Google Scholar]
- Nicolia A, Ferradini N, Molla G, Biagetti E, Pollegioni L, Veronesi F, Rosellini D (2014) Expression of an evolved engineered variant of a bacterial glycine oxidase leads to glyphosate resistance in alfalfa. J Biotechnol 184: 201–208 [DOI] [PubMed] [Google Scholar]
- Nordberg A, Minssen T, Holm S, Horst M, Mortensen K, Møller BL (2018) Cutting edges and weaving threads in the gene editing (Я)evolution: reconciling scientific progress with legal, ethical, and social concerns. J Law Biosci 5: 35–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC (2001) Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol 8: 238–242 [DOI] [PubMed] [Google Scholar]
- Packer MS, Liu DR (2015) Methods for the directed evolution of proteins. Nat Rev Genet 16: 379–394 [DOI] [PubMed] [Google Scholar]
- Parikh MR, Greene DN, Woods KK, Matsumura I (2006) Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E.coli. Protein Eng Des Sel 19: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedotti M, Rosini E, Molla G, Moschetti T, Savino C, Vallone B, Pollegioni L (2009) Glyphosate resistance by engineering the flavoenzyme glycine oxidase. J Biol Chem 284: 36415–36423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterhansel C, Maurino VG (2011) Photorespiration redesigned. Plant Physiol 155: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce NA, Winfree E (2002) Protein design is NP-hard. Protein Eng 15: 779–782 [DOI] [PubMed] [Google Scholar]
- Pollegioni L, Schonbrunn E, Siehl D (2011) Molecular basis of glyphosate resistance-different approaches through protein engineering. FEBS J 278: 2753–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JL, Rusli RA, Ollis DL (2016) Directed evolution of enzymes for industrial biocatalysis. ChemBioChem 17: 197–203 [DOI] [PubMed] [Google Scholar]
- Puchta H. (2017) Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr Opin Plant Biol 36: 1–8 [DOI] [PubMed] [Google Scholar]
- Qaim M. (2009) The economics of genetically modified crops. Annu Rev Resour Economics 1: 665–694 [Google Scholar]
- Reetz MT, Wu S (2008) Greatly reduced amino acid alphabets in directed evolution: Making the right choice for saturation mutagenesis at homologous enzyme positions. Chem Commun (Camb) 43: 5499–5501 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Wright G, Emrich S, Clark PL (2018) %MinMax: a versatile tool for calculating and comparing synonymous codon usage and its impact on protein folding. Protein Sci 27: 356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum G, Chen C, Kaur J, Cui X, Zhang H, Asahara H, Chong S, Smilansky Z, Goldman YE, Cooperman BS (2013) Quantifying elongation rhythm during full-length protein synthesis. J Am Chem Soc 135: 11322–11329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachsenhauser V, Bardwell JC (2018) Directed evolution to improve protein folding in vivo. Curr Opin Struct Biol 48: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammons RD, Gaines TA (2014) Glyphosate resistance: State of knowledge. Pest Manag Sci 70: 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satagopan S, Tabita FR (2016) RubisCO selection using the vigorously aerobic and metabolically versatile bacterium Ralstonia eutropha. FEBS J 283: 2869–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheben A, Wolter F, Batley J, Puchta H, Edwards D (2017) Towards CRISPR/Cas crops: Bringing together genomics and genome editing. New Phytol 216: 682–698 [DOI] [PubMed] [Google Scholar]
- Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JIB, Banfield MJ, Kamoun S (2014) Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol Plant Microbe Interact 27: 624–637 [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu J-L, et al. (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31: 686–688 [DOI] [PubMed] [Google Scholar]
- Shao E, Zhuang H, Guan X (2013) Screening of peptides bound to brash border membrane vesicle of Nilaparvata lugens (Hemiptera: Delphacidae). Chin J Appl Environ Biol 19: 637–642 [Google Scholar]
- Shao E, Lin L, Chen C, Chen H, Zhuang H, Wu S, Sha L, Guan X, Huang Z (2016) Loop replacements with gut-binding peptides in Cry1Ab domain II enhanced toxicity against the brown planthopper, Nilaparvata lugens (Stål). Sci Rep 6: 20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg JJ, Endelman JB, Arnold FH (2004) SCHEMA-guided protein recombination. Methods Enzymol 388: 35–42 [DOI] [PubMed] [Google Scholar]
- Smith GP. (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228: 1315–1317 [DOI] [PubMed] [Google Scholar]
- Smith SA, Tabita FR (2003) Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1,5-bisphosphate carboxylase/oxygenase. J Mol Biol 331: 557–569 [DOI] [PubMed] [Google Scholar]
- Smith MA, Romero PA, Wu T, Brustad EM, Arnold FH (2013) Chimeragenesis of distantly-related proteins by noncontiguous recombination. Protein Sci 22: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1980) Photorespiration mutants of Arabidopsis thaliana deficient in serine-glyoxylate aminotransferase activity. Proc Natl Acad Sci USA 77: 2684–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad DD, Petolino JF, Voytas DF, Reichert NA (2017) Genome editing of plants. CRC Crit Rev Plant Sci 36: 1–23 [Google Scholar]
- Steinbrenner AD, Goritschnig S, Staskawicz BJ (2015) Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog 11: e1004665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WP. (1994a) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA 91: 10747–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WPC. (1994b) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370: 389–391 [DOI] [PubMed] [Google Scholar]
- Sueldo DJ, Shimels M, Spiridon LN, Caldararu O, Petrescu A-J, Joosten MHAJ, Tameling WIL (2015) Random mutagenesis of the nucleotide-binding domain of NRC1 (NB-LRR Required for Hypersensitive Response-Associated Cell Death-1), a downstream signalling nucleotide-binding, leucine-rich repeat (NB-LRR) protein, identifies gain-of-function mutations in the nucleotide-binding pocket. New Phytol 208: 210–223 [DOI] [PubMed] [Google Scholar]
- Sun J, Katzenellenbogen JA, Zhao H, Katzenellenbogen BS (2003) DNA shuffling method for generating estrogen receptor α and β chimeras in yeast. Biotechniques 34: 278–280, 282, 284 passim [DOI] [PubMed] [Google Scholar]
- Sun L, Qin J, Wang K, Zhang J (2017) Expansion of pathogen recognition specificity in plants using pattern recognition receptors and artificially designed decoys. Sci China Life Sci 60: 797–805 [DOI] [PubMed] [Google Scholar]
- Tawfik DS, Griffiths AD (1998) Man-made cell-like compartments for molecular evolution. Nat Biotechnol 16: 652–656 [DOI] [PubMed] [Google Scholar]
- Tian J, Yan Y, Yue Q, Liu X, Chu X, Wu N, Fan Y (2017) Predicting synonymous codon usage and optimizing the heterologous gene for expression in E. coli. Sci Rep 7: 9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y-S, Xu J, Peng R-H, Xiong A-S, Xu H, Zhao W, Fu X-Y, Han H-J, Yao Q-H (2013) Mutation by DNA shuffling of 5-enolpyruvylshikimate-3-phosphate synthase from Malus domestica for improved glyphosate resistance. Plant Biotechnol J 11: 829–838 [DOI] [PubMed] [Google Scholar]
- Tian Y-S, Xu J, Xing X-J, Zhao W, Fu X-Y, Peng R-H, Yao Q-H (2015) Improved glyphosate resistance of 5-enolpyruvylshikimate-3-phosphate synthase from Vitis vinifera in transgenic Arabidopsis and rice by DNA shuffling. Mol Breed 35: 148 [Google Scholar]
- Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C (2009) Design parameters to control synthetic gene expression in Escherichia coli. PLoS One 4: e7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RH, Whitney SM (2017) Improving CO2 fixation by enhancing Rubisco performance. In Alcade M, Directed Enzyme Evolution: Advances and Applications. Springer International, Cham, Switzerland, pp 101–126 [Google Scholar]
- Wilson RH, Alonso H, Whitney SM (2016) Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci Rep 6: 22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RH, Martin-Avila E, Conlan C, Whitney SM (2018) An improved Escherichia coli screen for Rubisco identifies a protein-protein interface that can enhance CO2-fixation kinetics. J Biol Chem 293: 18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TS, Tee KL, Hauer B, Schwaneberg U (2004) Sequence saturation mutagenesis (SeSaM): A novel method for directed evolution. Nucleic Acids Res 32: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TS, Zhurina D, Schwaneberg U (2006) The diversity challenge in directed protein evolution. Comb Chem High Throughput Screen 9: 271–288 [DOI] [PubMed] [Google Scholar]
- Xiao H, Bao Z, Zhao H (2015) High throughput screening and selection methods for directed enzyme evolution. Ind Eng Chem Res 54: 4011–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T, Zhang K, Chen Y, Lin Y, Wu G, Zhang L, Yao P, Shao Z, Liu Z (2013) Improving glyphosate oxidation activity of glycine oxidase from Bacillus cereus by directed evolution. PLoS One 8: e79175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z (2011) Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr Opin Struct Biol 21: 25–31 [DOI] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Dawes G, Stemmer WP (1997) Directed evolution of a fucosidase from a galactosidase by DNA shuffling and screening. Proc Natl Acad Sci USA 94: 4504–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Frauenkron-Machedjou VJ, Kardashliev T, Ruff AJ, Zhu L, Bocola M, Schwaneberg U (2017) Amino acid substitutions in random mutagenesis libraries: lessons from analyzing 3000 mutations. Appl Microbiol Biotechnol 101: 3177–3187 [DOI] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond J-L (2004) An efficient one-step site-directed and site saturation mutagenesis protocol. Nucleic Acids Res 32: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Kurek I, Liu L (2010) Engineering photosynthetic enzymes involved in CO2–assimilation by gene shuffling, Chapter 20. In Rebeiz CA, Benning C, Bohnert HJ, Daniell H, Hoober JK, Lochtenthaler HK, Portis AR, Tripathy BC, eds, Advances in Photosynthesis and Respiration: The Chloroplast, Springer Netherlands, Dordrecht, the Netherlands, pp 307–322 [Google Scholar]
- Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien RY (1998) Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279: 84–88 [DOI] [PubMed] [Google Scholar]
- Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36: 1211–1216 [DOI] [PubMed] [Google Scholar]