An importin β regulates nucleocytoplasmic transport of Arabidopsis transcriptional co-activators GIFs, and by doing so, it regulates ovule development.
Abstract
Ovule development is critical for seed development and plant reproduction. Multiple transcription factors (TFs) have been reported to mediate ovule development. However, it is not clear which intracellular components regulate these TFs during ovule development. After their synthesis, TFs are transported into the nucleus a process regulated by karyopherins commonly known as importin alpha and β. Around half of Arabidopsis (Arabidopsis thaliana) importin β-coding genes have been functionally characterized but only two with specific cargos have been identified. We report here that Arabidopsis IMPORTIN β4 (IMB4) regulates ovule development through nucleocytoplasmic transport of transcriptional coactivator growth regulating factors–interacting factors (GIFs). Mutations in IMB4 impaired ovule development by affecting integument growth. imb4 mutants were also defective in embryo sac development, leading to partial female sterility. IMB4 directly interacts with GIFs and is critical for the nucleocytoplasmic transport of GIF1. Finally, functional loss of GIFs resulted in ovule defects similar to those in imb4 mutants, whereas enhanced expression of GIF1 partially restored the fertility of imb4. The results presented here uncover a novel genetic pathway regulating ovule development and reveal the upstream regulator of GIFs.
Ovule development is critical for seed development and plant reproduction. Mature ovules contain sporophytic integuments and gametophytic embryo sacs and are formed through two processes, i.e. megasporogenesis and megagametogenesis (Schneitz et al., 1995, 1997; Drews et al., 1998). During megasporogenesis, ovules establish proximal-distal polarity. The distal part of developing ovules, i.e. the nucellus, forms megaspore mother cell (MMC), which produces a tetrad of haploid spores through meiotic division. The most proximal one survives to form the functional megaspore (FM), whereas the other three degenerate (Schneitz et al., 1995, 1997; Christensen et al., 1997; Drews et al., 1998). During megagametogenesis, FM eventually forms a seven-cell eight-nuclei female gametophyte, i.e. embryo sac (Christensen et al., 1997; Drews et al., 1998). In Arabidopsis (Arabidopsis thaliana), the growth of outer and inner integuments initiates and eventually envelops the embryo sac during megagametogenesis. Integuments grow asymmetrically, resulting in anatropy of mature ovules, i.e. mature ovules bend so that the micropyle is close to the funiculus (Schneitz et al., 1995, 1997; Drews et al., 1998). Because mutants defective in integument cells often result in defective formation of female gametophyte (Bencivenga et al., 2011; Chevalier et al., 2011; Wang et al., 2016), it is generally considered that integuments control embryo sac development through unknown signaling molecules.
Multiple transcription factors (TFs) were reported to mediate ovule development (Colombo et al., 2008). INNER NO OUTER (INO) regulates abaxial-adaxial patterning of Arabidopsis ovules (Villanueva et al., 1999). Mutations at ABERRANT TESTA SHAPE result in the formation of a single integument layer due to congenital fusion of the two integuments (McAbee et al., 2006). Functional loss of AINTEGUMENTA abolishes the growth of both integuments (Elliott et al., 1996; Klucher et al., 1996). Some homeodomain proteins such as BELL1 (Reiser et al., 1995), PHABULOSA (Sieber et al., 2004), and WUSHEL (Gross-Hardt et al., 2002; Lieber et al., 2011) are also mediators of integument growth and ovule development. Although extensive studies have been performed to TF regulation of ovule development, it is not yet clear which intracellular components regulate their targeting and activities.
TFs are transported into the nucleus through the formation of the nuclear pore complex (NPC), a process mediated by karyopherins, often called importins or exportins, depending on the direction of transport (Meier and Brkljacic, 2009; Tamura and Hara-Nishimura, 2014). There are two subfamilies of karyopherins: importin alpha and importin beta (Tamura and Hara-Nishimura, 2014). It is generally believed that importin alpha recognizes a short positively charged nuclear localization signal, whereas importin beta mediates their interaction with the NPC and Ran-GTP to complete cargo transport (Tamura and Hara-Nishimura, 2014). However, importin beta may play roles in cells other than merely cargo transport (Harel and Forbes, 2004; Tamura and Hara-Nishimura, 2014), such as mediation of microRNA activity as reported in plants (Wang et al., 2011; Cui et al., 2016).
The Arabidopsis genome encodes 18 importin βs (IMB), half of which were functionally characterized. IMB1/ATKPNB1 participates in responses to abscisic acid and drought (Luo et al., 2013); HASTY/XPO4 was involved in shoot meristem maintenance, flowering time, and fertility (Telfer and Poethig, 1998; Bollman et al., 2003); SAD2/EMA1/IPO8 not only mediates responses to abscisic acid and UV but also regulates trichome development (Zhao et al., 2007; Wang et al., 2011); XPO1A and XPO1B are essential for gametophytic fertility (Blanvillain et al., 2008), whereas XPO1A also mediates heat responses (Wu et al., 2010); XPOT/PAUSED regulates leaf development and meristem maintenance (Hunter et al., 2003; Li and Chen, 2003); TRANSPORTIN1 (TRN1) and KETCH1/IMP3 are both involved in leaf development and fertility (Cui et al., 2016; Zhang et al., 2017). Despite the extensive functional studies on importin β, only two have identified a cargo: one showed that SAD2 mediates the nucleocytoplasmic transport of MYB4 (Zhao et al., 2007), whereas another showed that KETCH1/IMP3 mediates that of hyponastic leaves 1 (Zhang et al., 2017).
Here we report that Arabidopsis IMB4, an importin β, regulates ovule development through nucleocytoplasmic transport of transcriptional coactivator growth regulating factors (GRF)-INTERACTING FACTORS (GIFs). Functional loss of IMB4 compromised ovule development by impairing integument growth. Consequently, embryo sacs were not properly developed in imb4 mutants, leading to partial female sterility. We showed that GIFs directly interact with IMB4. Furthermore, through performing cell fractionation and examining fluorescence distribution, we found that nucleocytoplasmic transport of GIF1 relies on IMB4. Finally, functional loss of GIFs resulted in ovule defects similar to those in imb mutants. Our results have uncovered a genetic pathway regulating ovule development and the upstream regulator of the important transcriptional coactivator GIFs.
RESULTS
Functional Loss of IMB4 Reduces Fertility
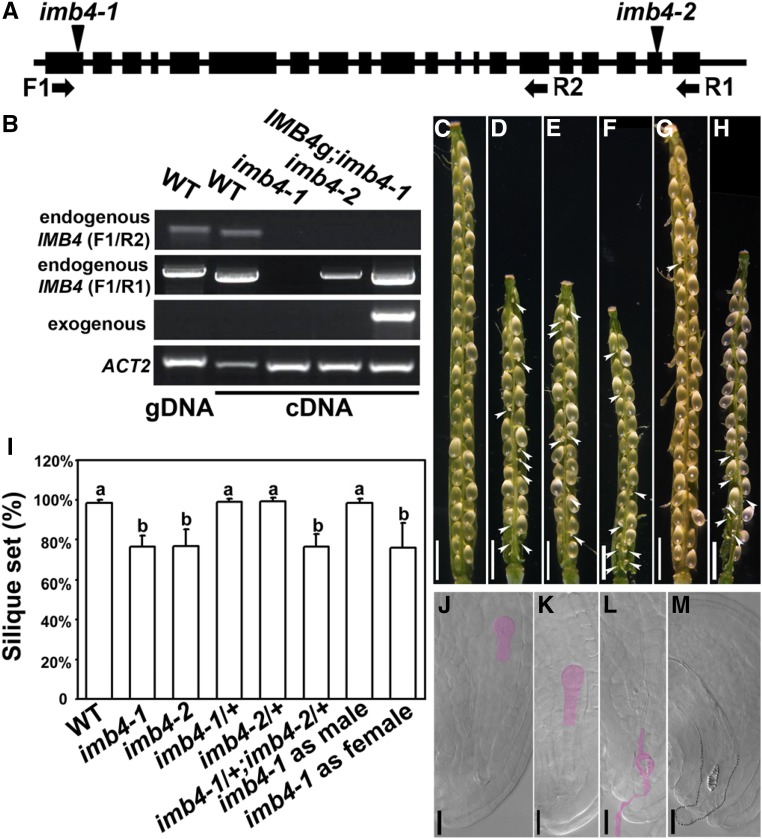
To identify novel components involved in fertility, we characterized Arabidopsis IMB4 (At4g27640), which encodes importin β, whose function has not yet been understood. Two T-DNA insertion mutant lines were isolated (Fig. 1A). Transcript analysis showed that no full-length transcript was detected in either mutant, although imb4-2 did express a partial transcript (Fig. 1B). The two mutants displayed similar pleiotropic developmental defects (Supplemental Fig. S1). In this study, we focused on their reduced fertility for further analysis (Fig. 1A, 1I). Unlike wild type in which siliques contained fully developed seeds (Fig. 1, C and I), imb4 siliques contained white and wrinkled ovules among developing seeds (Fig. 1, D and E). Embryogenesis started in wild type at 24 h after pollination (HAP), as judged from whole-mount clearing of pistils (Fig. 1J). However, although 66.8% ± 3.7% (means ± sd, n = 10) ovules were fertilized in imb4 pistils, as judged from the presence of developing embryos (Fig. 1K), 33.2% ± 8.6% ovules of imb4 showed no pollen tube entrance (Fig. 1M) or occasionally a nondischarging pollen tube (Fig. 1L), suggesting that the reduced fertility in imb4 was because of failed fertilization. Crosses between imb4-1 and imb4-2 yielded F1 plants with a reduced seed set comparable with either parent (Fig. 1, F and I), suggesting that imb4-1 and imb4-2 were allelic. The following results were mostly observed for imb4-1, but were also observed for imb4-2.
Figure 1.
Functional loss of IMB4 reduces fertility. A, Schematic illustration of the IMB4 genomic region. Boxes indicate exons; short lines indicate untranslated regions or introns. Triangles point at the insertion sites of two T-DNA mutants. Primer binding sites are indicated with arrows. B, Transcript analysis demonstrating that neither imb4-1 nor imb4-2 expresses the full-length IMB4 transcript, whereas imb4-2 expresses a partial IMB4 transcript. The complemented line, IMB4g-YFP imb4-1, expressed only exogenous IMB4. C to H, a representative silique from the wild type (C), imb4-1 (D), imb4-2 (E), imb4-1/+ imb4-2/+ (F), or from a cross between the wild type and imb4-1 when imb4-1 was used as the male (G) or as the female (H) parent. Arrowheads point at unfertilized ovules. I, Quantification of seed set from designated genetic background. Results shown are means ± sd (sd). Twenty siliques were analyzed for each genotype. Means with different letters are significantly different (one-way ANOVA, Tukey-Kramer test, P < 0.05). J to M, Whole-mount clearing of wild-type (J) or imb4-1 (K to M) ovules at 24 HAP. Pink-highlighted regions are developing embryos (J and K) or a nondischarging pollen tube (L). The dotted line illustrates a protruding embryo sac with tracheary element-like structure inside (M). Bars = 1 mm (C to H) and 20 µm (J to M).
Reciprocal crosses between the homozygous imb4-1 mutant and wild type indicated that the reduced seed set of imb4-1 was because of defects from the female side, whereas pollen of the imb4 mutants were not affected (Fig. 1, G–I). Indeed, pollen development was comparable between imb4 and the wild type (Supplemental Fig. S2). Next, we performed reciprocal crosses between the wild type and the heterozygous imb4 mutants to determine whether defects of imb4 were gametophytic. The segregation ratio showed that female and male transmission of imb4 was comparable with that of the wild type (Supplemental Table S1), indicating a sporophytic defect in female tissues due to IMB4 loss-of-function.
Functional Loss of IMB4 Compromises Ovule-Controlled Pollen Tube Guidance
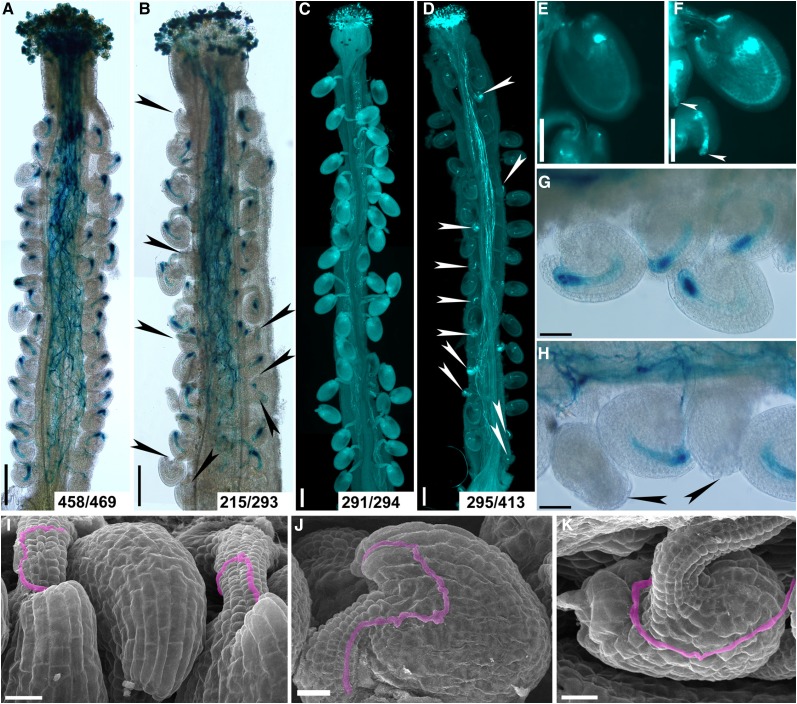
To determine the cause of failed fertilization when the imb4 mutants were used as the female parent, we first pollinated wild-type or imb4-1 pistils with ProLAT52:GUS pollen and performed histochemical β-glucuronidase (GUS) staining at 12 HAP. At 12 HAP, all wild-type ovules were targeted by a pollen tube (Fig. 2, A and G), indicating proper pollen tube guidance by wild-type ovules. By contrast, 78 out of 293 imb4-1 ovules failed to attract pollen tubes (Fig. 2, B and H). To determine whether failed pollen tube guidance led to reduced fertility in imb4-1, we pollinated imb4-1 pistils with wild-type pollen and analyzed pistils by aniline blue staining at 48 HAP when wild-type ovules were fertilized, and found rapid size increase (Fig. 2, C and E). Indeed, 118 out of 413 imb4-1 ovules did not grow as a consequence of failed fertilization (Fig. 2, D and F). To provide further evidence of defective pollen tube guidance in imb4-1, we analyzed wild-type and imb4-1 pistils pollinated by wild-type pollen at 12 HAP by scanning electron micrographs (SEMs). Pollen tubes efficiently targeted to the micropyle of wild-type ovules as expected (Fig. 2I). In comparison, imb4-1 pistils contained not only ovules normally targeted by a pollen tube (Fig. 2J), but those that failed to attract a pollen tube at the micropyle (Fig. 2K). These results all point to a key role of IMB4 in ovule-guided pollen tube targeting.
Figure 2.
Functional loss of IMB4 compromises female-controlled pollen tube guidance. A and B, Histochemical GUS staining of wild-type (A) or imb4-1 (B) pistils at 12 HAP with ProLAT52:GUS pollen. Arrowheads point at ovules that failed to attract pollen. Two to three overlapping high-magnification images were taken for one pistil. The images were then overlaid with Photoshop (Adobe) to show the whole pistil. Numbers at the bottom: ovules targeted by a ProLAT52:GUS pollen tube versus ovules examined. C and D, Aniline blue staining of wild-type (C) or imb4-1 (D) pistils at 48 HAP with wild-type pollen. Two to three overlapping high-magnification images were taken for one pistil. The images were then overlaid with Photoshop (Adobe) to show the whole pistil. Arrowheads point at ovules that did not develop as a result of fertilization failure. Numbers at the bottom: fertilized ovules versus total ovules examined. E and F, A close-up of aniline blue stained wild-type (E) or imb4-1 (F) pistils at 48 HAP with wild-type pollen. Arrowheads point at the unfertilized ovules. G and H, A close-up of histochemical GUS stained wild-type (G) or imb4-1 (H) pistils at 12 HAP with ProLAT52:GUS pollen. Arrowheads point at the micropyles that failed to attract pollen tubes. I to K, SEMs of wild-type (I) or imb4-1 (J-K) pistils at 12 HAP with wild-type pollen. Incoming pollen tubes are highlighted in pink. A pollen tube enters through the micropyle of a normal-developing ovule of imb4-1 (J) but not of a defective ovule of imb4-1 (K). Bars = 200 µm (A to D), 100 µm (E and F), 50 µm (G and H), and 20 µm (I to K).
Functional Loss of IMB4 Impairs Integument Growth
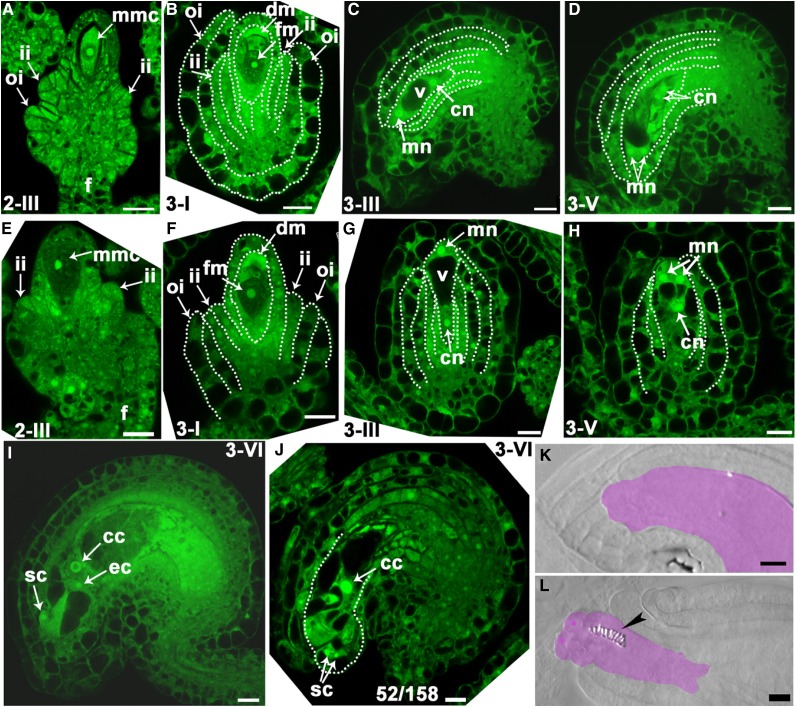
To determine how IMB4 loss-of-function caused ovule defects, we first performed SEMs on mature unpollinated ovules. In the wild type, mature ovules were asymmetric, whose abaxial-adaxial polarity led to ovule bending such that the micropyle was close to the funiculus (Supplemental Fig. S3), as well known (Schneitz et al., 1995, 1997). By contrast, 21.4% ± 3.7% of imb4-1 ovules were abnormal such that outer integuments did not fully extend to enclose the inner integument (Supplemental Fig. S3). As a consequence of the disrupted polarity, the micropyle of imb4-1 often was not clearly formed and perpendicular to the funiculus (Supplemental Fig. S3). These results suggested that integument development was impaired in imb4-1.
To determine the exact stage when imb4-1 showed defective ovule development, we performed optical sections of ovules over entire developmental stages. Initiation of outer and inner integument was comparable between the wild type and imb4-1 (Fig. 3, A and E). At these stages, FMs were formed, i.e. three progenies of megasporocytes degenerated and only the one proximal to the funiculus persisted (Schneitz et al., 1995, 1997; Christensen et al., 1997). At stage 3-I, when rapid differentiation and growth of the outer integument began in the wild type, especially at the abaxial side of ovules (Fig. 3B), the growth of the outer integuments in imb4-1 was much delayed and often symmetric (Fig. 3F). At stage 3-III, wild-type ovules established the abxial-adaxial polarity by bending toward the funiculus through differential growth of outer integuments (Fig. 3C). At this stage, a central vacuole in embryo sac separated the chalazal nucleus and the micropylar nucleus (Fig. 3C). In contrast, the outer integuments in imb4-1 showed symmetric growth and ovules did not show the classic anatropy (Fig. 3G).
Figure 3.
Functional loss of IMB4 impairs integument growth. A to J, Confocal laser scanning microscopy (CLSM) of wild-type (A to D, I) or imb4-1 (G, H, J) ovules at stage 2-III (A, E), 3-I (B, F), 3-III (C, G), 3-V (D, H), or 3-VI (I, J). Midoptical sections of ovules are shown. Only imb4-1 ovules with visible nuclei are documented. cc, Central cell; cn, chalazal nucleus; ec, egg cell; fm, functional megaspore; F, funiculus; ii, inner integument; oi, outer integument; mmc, microspore mother cell; mn, micropylar nucleus; sc, synergid cell; V, vacuole. Numbers at the bottom of (J): displayed ovules versus total ovules examined. K to L, Whole-mount ovule clearing of wild type (WT; K) or imb4-1 (L). Lilac-shaded areas indicate embryo sacs. Arrowhead points at tracheary element-like structure in the embryo sac of imb4-1. Bars = 10 µm.
Subsequently, wild-type ovules maintained the bending status and embryo sacs developed (Fig. 3, D and I), whereas in 63 out of 210 imb4-1 ovules, embryo sac development was defective: either no embryo sac structures could be seen or embryo sacs did not contain central cell, egg cell, and synergid cells (Fig. 3, H and J). Because of defective integument growth, the embryo sacs in imb4-1 were often protruding (Fig. 3J). To provide further evidence that embryo sac development was defective in imb4-1, we performed whole-mount ovule clearing on unfertilized mature pistils. In contrast with well-enclosed embryo sacs in wild-type ovules (Fig. 3K), 67 out of 239 imb4-1 ovules contained protruding embryo sacs, occasionally with tracheary element-like structures inside (Fig. 3L), suggesting that functional loss of IMB4 results in defective embryo sac development.
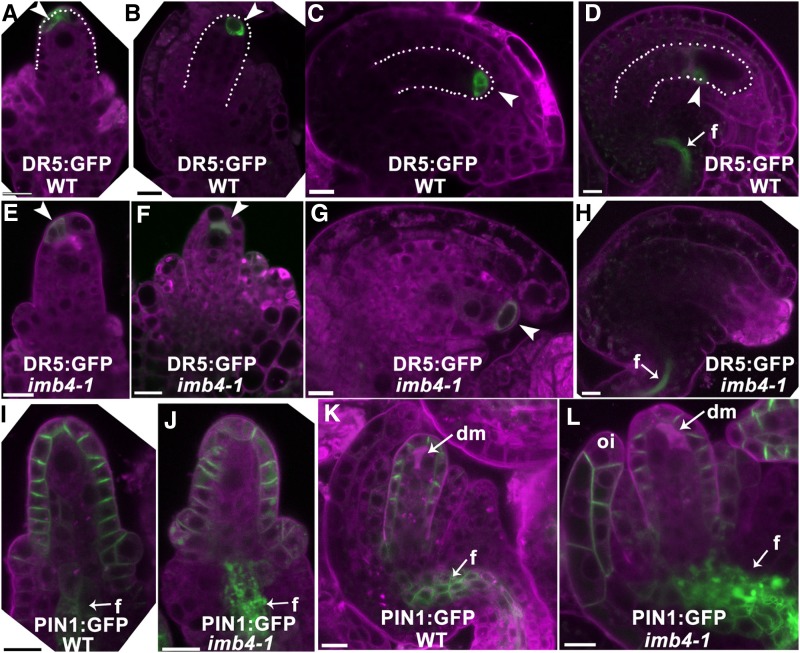
Functional Loss of IMB4 Compromises Auxin and Cytokinin Responses during Ovule Development
Because auxin and cytokinins (CK) play important roles in ovule development (Benková et al., 2003; Bencivenga et al., 2012; Kelley et al., 2012; Ceccato et al., 2013), we examined whether auxin and CK responses were compromised in imb4-1 by analyzing the distribution of their respective fluorescence reporters, DR5:GFP (Benková et al., 2003) and TCS:yellow fluorescence protein (YFP; Bencivenga et al., 2012). In the wild type, GFP signals showed a polar distribution in the micropylar end of ovules from stage 2-III to 3-III (Fig. 4, A–C), which was consistent with the polar distribution of PIN-FORMED1 (PIN1; Fig. 4, I and K), the auxin efflux carrier mediating auxin distribution during ovule development (Benková et al., 2003; Bencivenga et al., 2012; Ceccato et al., 2013). At maturation, strong GFP signals were detected only in the funiculus (Fig. 4D). By contrast, GFP signals were hardly detectable in DR5:GFP;imb4-1 ovules over various developmental stages (Fig. 4, E–H). Correspondingly, PIN1 was mis-targeted in imb4-1 ovules such that PIN1 showed irregular membrane distribution and was often detected in vacuoles or cytoplasmic aggregates both in the nucellus and in the funiculus (Fig. 4, J and L). Strong signals of PIN1 were also ectopically detected in the plasma membrane of outer integuments (Fig. 4L).
Figure 4.
Distribution of auxin maximum and PIN1 is compromised by IMB4 loss-of-function. A to H, CLSM of DR5:GFP;wild type (WT; A to D) or DR5:GFP;imb4-1 (E to H) at stage 2-III (A, E), 3-I (B, F), 3-III (C, G), or 3-V (D, H). I to L, CLSM of PIN1:GFP (I, K) or PIN1:GFP;imb4-1 (J, L) at stage 2-I (I, J) or 3-I (K, L). Arrowheads point at DR5:GFP signals. Arrows point at funiculus (f). Dotted lines highlight nucellus (A, B) or developing embryo sacs (C, D). dm, Degenerating megaspores; oi, outer integument. Cells were stained with Lysotracker red (magenta). Bars = 10 µm.
Because CK is another important phytohormone for ovule development and was reported to show a distribution reciprocal to that of auxin (Bencivenga et al., 2012), we introduced TCS:YFP in imb4-1 to determine CK responses in the mutant. Indeed, strong CK signals were detected at the chalazal ends of ovules during development (Supplemental Fig. S4), reciprocal to that of DR5:GFP (Fig. 4). By contrast, YFP signals were enhanced and expanded at early stages while being irregularly distributed at the micropylar end at maturation in imb4-1 (Supplemental Fig. S4). The enhanced and ectopic CK responses were consistent with reduced auxin signaling in imb4-1 ovules, providing further evidence that IMB4 is critical for ovule development.
IMB4 Is Expressed Constitutively during Ovule Development and its Protein Localizes in Both the Cytoplasm and the Nucleus
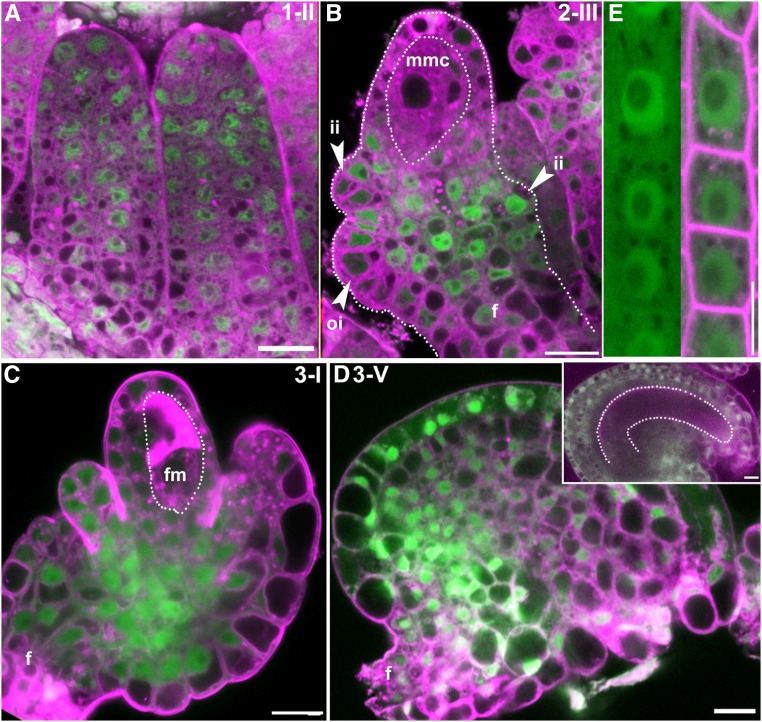
To determine the expression pattern and subcellular localization of IMB4, we generated IMB4g-YFP, in which the coding sequences of IMB4 fused with YFP-coding sequences were driven by the native promoter of IMB4, and introduced it into imb4-1 (Fig. 1). The transgene fully complemented the mutant defects of imb4-1, including seed set and the ability to attract pollen tubes (Supplemental Fig. S5), indicating that YFP fusion did not interfere with its functionality. Consistent with the observation of its mutant phenotype in ovules, IMB4 was expressed in all stages of ovule development (Fig. 5, A–D). YFP signals were detected in all cells during ovule primordial formation (Fig. 5A). At stage 2-III, when MMC were formed, IMB4 was detected in both outer and inner integument cells but was absent in MMC (Fig. 5B). At stage 3-I, when FM was established, IMB4 was also detected in outer and inner integuments but not in FM (Fig. 5C). In mature ovules, IMB4 was expressed in all sporophytic cells of ovules but not in the embryo sacs (Fig. 5D). Consistent with the pleiotropic defects of the imb4 mutants (Supplemental Fig. S1), IMB4 was detected in all tissues by reverse transcription-quantitative PCRs (RT-qPCRs; Supplemental Fig. S6). YFP signals were detected in both the cytoplasm and the nucleus in ovules and roots (Fig. 5, A–E), an observation in accord with its role as an importin.
Figure 5.
IMB4 is expressed constitutively during ovule development and protein localizes at both cytoplasm and the nucleus. A to D, CLSM of IMB4g-YFP imb4-1 ovule at developmental stage 1-II (A), 2-III (B), 3-I (C), or 3-V (D). D, inset is a stage 3-V ovule at a different angle showing the embryo sac (illustrated by a dotted line). F, funiculus; fm, functional megaspore; ii, inner integument; mmc, megaspore mother cell; oi, outer integument. E, CLSM of IMB4g-YFP imb4-1 root at 4 DAG. Lysotracker red (A to D) or FM4-64 (E) in magenta was used to show cell silhouette. Bars = 10 µm.
To exclude the possibility that compromised vegetative growth of imb4 (Supplemental Fig. S1) resulted in ovular defects, we expressed IMB4 specifically in outer integuments of ovules (Supplemental Fig. S7) by using the promoter of INO (Wang et al., 2016). Indeed, the expression of IMB4 in outer integuments was sufficient in complementing the ovular defects of the imb4 mutants (Supplemental Fig. S7). This result strongly supported a specific role of IMB4 in ovules, in addition to its roles during vegetative development.
IMB4 Interacts with GIF1/AN3
Because importins shuttle specific cargo proteins between the cytoplasm and the nucleus (Merkle, 2011; Tamura and Hara-Nishimura, 2014), we reasoned that the defects of imb4 were because of mis-localization of specific IMB4-cargos. Although a classic role of importin beta is to facilitate cargo transport indirectly, there were cases when importin beta directly mediates nucleocytoplasmic transport (Harel and Forbes, 2004; Tamura and Hara-Nishimura, 2014; Zhang et al., 2017). A study of high-throughput mass spectrometry reported that a transcriptional coactivator GIF1/ANGUSTIFOLIA3 (AN3) may interact with importin βs (Vercruyssen et al., 2014). Interestingly, a recent report showed that functional loss of Arabidopsis GIF1 and its two homologs GIF2, GIF3 (Kim et al., 2003; Kim and Kende, 2004; Omidbakhshfard et al., 2015) affects fertility (Lee et al., 2014, 2018).
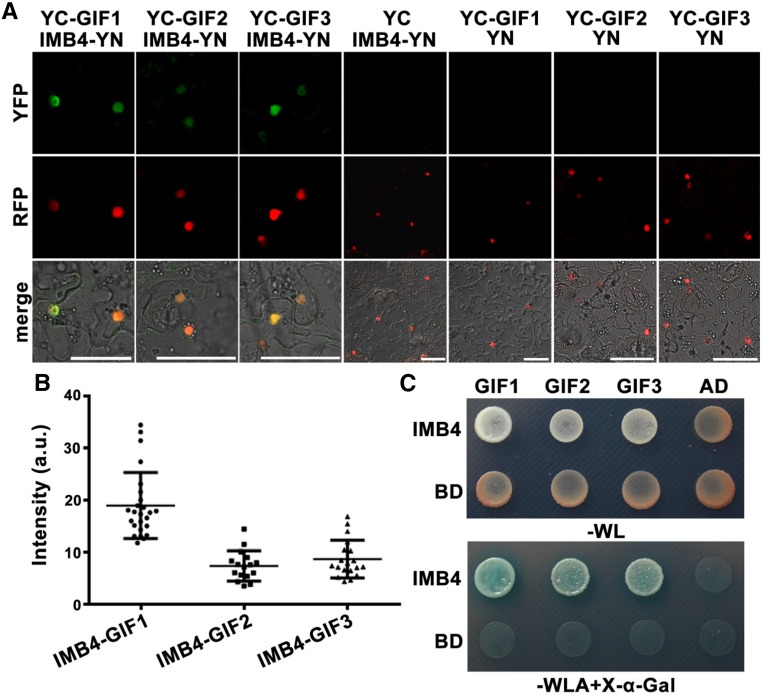
To test whether IMB4 interacted with GIFs directly, we applied bimolecular fluorescence complementation (BiFC) and yeast two-hybrid (Y2H) assays. IMB4 interacted with all three GIFs and most strongly with GIF1/AN3 (Fig. 6, A-C), suggesting that GIFs are cargos of IMB4.
Figure 6.
IMB4 interacts with the GIF1 family proteins. A, CLSM of BiFC assays showing the interaction between IMB4 and the GIF1 family proteins. Magenta signals indicate the nuclear-localized U1-70K-mCherry. The bottom shows merged images of the YFP, RFP, and transmission channels. B, Quantification of BiFC intensity. a.u., Arbitrary fluorescence units. Results are means ± sd. Sixteen to twenty-five cells were measured. No signals were detected for the negative controls. C, Representative yeast two-hybrid assays. Selection of interaction was performed on YSD medium lacking Trp (-W), Leu (-L), and Ade (-A), together with X-α-Gal. Bars = 50 µm.
Efficient Nucleus Import of GIF1/AN3 Requires IMB4
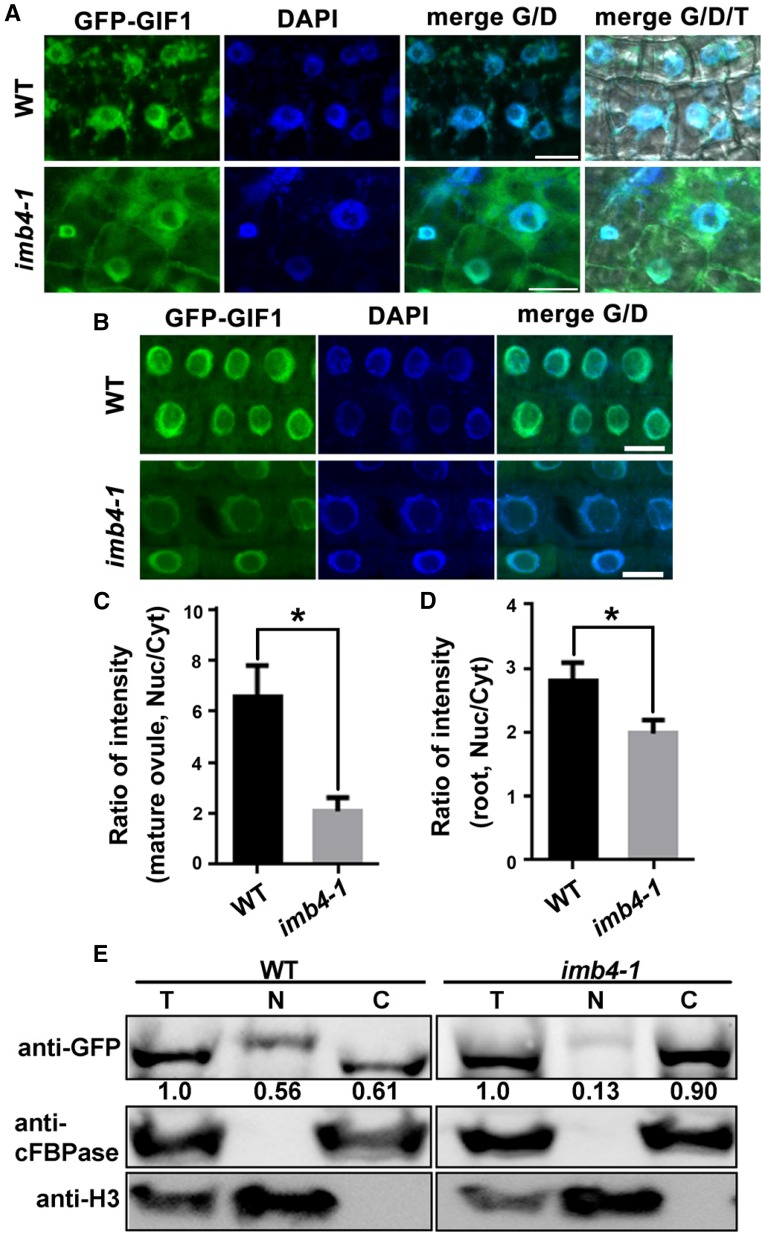
To examine whether nucleocytoplasmic transport of GIF1/AN3 depended on IMB4, we adopted two approaches. First, we introduced a GFP-GIF1/AN3 translational fusion into imb4-1 and analyzed its subcellular distribution in stable transgenic plants. By confocal laser scanning microscopy (CLSM), we detected strong nuclear signals of GFP-GIF1/AN3 in wild-type ovules but enhanced cytoplasmic signals in imb4-1 ovules (Fig. 7A). Quantitative analysis supported a significant difference on the ratio of nucleus-associated versus cytoplasmic-associated GFP-GIF1/AN3 distribution between wild type and imb4-1 (Fig. 7C). We also examined the nucleus-cytoplasmic distribution of GIF1 in root epidermal cells. The results were consistent such that the nuclear accumulation of GIF1 was reduced in imb4-1 (Fig. 7, B and D). Second, we performed cell fractionation on inflorescences from the ProUBQ10:GFP-GIF1/AN3 or ProUBQ10:GFP-GIF1/AN3;imb4-1 plants. The successful separation of nuclei and cytoplasm was demonstrated by using antihistone 3 (H3) and anticytosolic Fru-1,6-bisphosphatase (cFBPase; Fig. 7E). Indeed, the signals of anti-GFP in the nucleus fraction of imb4-1 were substantially reduced compared with that in the wild type (Fig. 7E). Thus, cell fractionation assays also support cytoplasmic retention of GIF1/AN3 in imb4-1. Interestingly, we detected a size increase in GIF1 in the nuclear fraction but not in the cytoplasmic or total fraction (Fig. 7E), implying that GIF1 might be modified posttranslationally in the nucleus and de-modified in the cytoplasm.
Figure 7.
Nucleocytoplasmic transport of GIF1/AN3 requires IMB4. A and B, CLSM of mature ovules (A) or root epidermal cells (B) from ProUBQ10:GFP-GIF1;wild type (WT) or ProUBQ10:GFP-GIF1;imb4-1 stained with DAPI. Overlays of fluorescent (G for GFP, D for DAPI) and transmission (T) images are shown at the right. C and D, Quantification of intensity ratio between nuclear- (Nuc) and cytoplasmic- (Cyt) associated signals in mature ovules (C) or in root epidermal cells (D) from ProUBQ10:GFP-GIF1;wild type or ProUBQ10:GFP-GIF1;imb4-1. Results are means ± sd (n = 30). Asterisk indicates significant difference (t test, P < 0.05). E, Cell fractionation and Western blot analysis. Inflorescences from ProUBQ10:GFP-GIF1;wild type or ProUBQ10:GFP-GIF1;imb4-1 were used for protein extraction, followed by subcellular fractionation and Western blotting. C, cytoplasm; N, nucleus; T, total protein extract. Histone H3 and cFBPase were used for the nuclei and cytoplasmic markers, respectively. Numbers at the bottom are the values of GFP-GIF1 in the nuclear fraction (N) or cytoplasmic fraction (C) relative to that in total protein fraction (T), which is set to 1.0. Main inflorescences from 30 plants of each genotype were collected for protein extraction used in one experiment. Results shown are representative of three biological replicates. Bars = 10 µm.
To further confirm that GIFs were cargos of IMB4 during ovule development, we examined the gif1;gif2;gif3 triple mutant based on the rationale that functional loss of GIFs would display ovule defects similar to those in imb4-1. Although the gif1;gif2;gif3 triple mutant was reported to be defective in both male and female reproductive development, integument growth during various developmental stages was not closely examined (Lee et al., 2014). Because we could not obtain the homozygous triple mutant probably due to its severe growth defects, we examined ovule development of the gif1;gif2;gif3/+ plants by CLSM of optical sections over different developmental stages. Indeed, the gif1;gif2;gif3/+ mutant showed integument growth defects similar to those in imb4-1, albeit more pronounced (Supplemental Fig. S8). At stage 3-I, outer integuments of the triple mutant developed slow and symmetric; at stage 3-III to 3-V, outer integument cells of the triple mutant proliferated only slightly; at maturation, i.e. stage 3-VI and above, outer integuments of the triple mutant could not enclose inner integuments, and no embryo sac structure could be detected in these ovules (Supplemental Fig. S8). On the other hand, overexpressing GIF1 in imb4-1 partially restored its fertility (Fig. 8). These results suggested genetic epistasis between IMB4 and GIF1 in ovule development.
Figure 8.
Enhanced expression of GIF1 partially restores the fertility of imb4-1. A, A representative silique from wild type (WT), imb4-1, two ProUBQ10:GFP-GIF1 transgenic plants, and two ProUBQ10:GFP-GIF1;imb4-1 transgenic plants that were generated by crossing the ProUBQ10:GFP-GIF1 transgenic lines with imb4-1. B, Quantification of seed set from designated genetic backgrounds. Results shown are means ± sd. Twenty-four siliques were analyzed for each genotype. C, Quantitative real-time PCRs showing the expression of GIF1 in designated genetic backgrounds. Results are means ± s.e.m. (ses, n = 3). Means with different letters in (B and C) are significantly different (one-way ANOVA, Tukey-Kramer test, P < 0.05). Bars = 1 mm.
Because the imb4 mutants also displayed defects in vegetative tissues, such as reduced primary roots and dwarfism (Supplemental Fig. S1), we wondered whether those defects were also because of reduced nuclear accumulation of GIF1. By examining ProUBQ10:GFP-GIF1;imb4-1 plants, we determined that leaf development, but not primary root growth or stem elongation, in imb4-1 was partially rescued by overexpressing GIF1 (Supplemental Fig. S9). Thus, the reduced nuclear accumulation of GIF1 explains some but not all developmental defects of imb4-1.
DISCUSSION
Although many studies on NPC components and importins have suggested the importance of nucleocytoplasmic transport in plant reproduction (Blanvillain et al., 2008; Tamura and Hara-Nishimura, 2014; Boruc et al., 2015; Cui et al., 2016), no mechanistic insights have been obtained. We show here that Arabidopsis IMB4, an importin β, is critical for ovule development through mediating the nucleocytoplasmic transport of GIFs. The imb4 mutants showed reduced seed set only as the female parents during crosses (Fig. 1), suggesting that the reduced fertility of imb4 mutants was because of female sporophytic defects. Indeed, optical sections during ovule development showed that integument growth in imb4 mutants was compromised (Fig. 3). As in most mutants defective in integument growth (Villanueva et al., 1999; McAbee et al., 2006; Chevalier et al., 2011; Vaddepalli et al., 2011; Kelley et al., 2012; Wang et al., 2016), embryo sac development in imb4 was impaired because of defects in the sporophytic integuments. It is supported by three lines of evidence: (1) female transmission of imb4 was not affected (Supplemental Table S1); (2) IMB4 was not expressed in FM or embryo sacs (Fig. 5); (3) heterozygous imb4 plants showed full seed set (Fig. 1). Ovule development defects in imb4 were not full penetrant (Fig. 1). It is likely that other importin βs play functionally redundant roles (Tamura and Hara-Nishimura, 2014). Indeed, functional loss of GIFs resulted in a much severe defect than that of IMB4 (Supplemental Fig. S7), suggesting redundancy by other importins.
According to classic thoughts, the role of importin beta in nucleocytoplasmic transport is indirect, through interaction with the NPC and RAN-GTP (Harel and Forbes, 2004; Merkle, 2011; Tamura and Hara-Nishimura, 2014). However, we show here that IMB4 directly interacts with GIFs and mediates the nucleocytoplasmic transport of GIF1 (Figs. 6 and 7). Using both fluorescence imaging and cell fractionation, we demonstrated that nuclear accumulation of GIF1 requires IMB4 (Fig. 7). In addition, functional loss of GIFs resulted in similar ovule developmental defects to imb4 (Supplemental Fig. S8). All these results supported a direct role of IMB4 in the nucleocytoplasmic transport of GIF1.
As transcriptional coactivators, GIFs function through other TFs, among which nine GRFs are the best characterized GIF interactors (Kim et al., 2003; Kim and Kende, 2004; Kim and Tsukaya, 2015). As transcriptional coactivators (Kim and Kende, 2004), GRFs participate in several developmental processes in plants such as leaf development and leaf primordial proliferation (Kim et al., 2003; Horiguchi et al., 2005). Although it is unclear whether GRFs mediate ovule development, overexpression of microRNA396, an upstream regulator of GRFs, affected carpel development and caused seed set reduction (Liang et al., 2014; Omidbakhshfard et al., 2015). It is thus likely that GRFs are interactors of GIFs during ovule development. Recently, it was reported that GIFs also interact with chromatin remodelers (Vercruyssen et al., 2014; Nelissen et al., 2015), suggesting that GIFs regulate chromatin dynamics for growth regulation.
In addition to ovule development defects, imb4 mutants also displayed pleiotropic defects (Supplemental Fig. S1). Some defects might have resulted from compromised nuclear transport of GIFs such as leaf size because mutations at GIFs resulted in similar defects (Kim et al., 2003; Kim and Kende, 2004; Kawade et al., 2013; Vercruyssen et al., 2014; Omidbakhshfard et al., 2015; Ercoli et al., 2018). However, other phenotypes of imb4, such as dwarfism (Supplemental Fig. S1), are not reported for mutants of GIFs, suggesting the presence of other IMB4 cargos regulating these processes. Indeed, enhancing the expression of GIF1 failed to restore primary root growth or stem elongation in imb4-1 (Supplemental Fig. S9), hinting at the presence of other IMB4 cargos whose defects in nucleocytoplasmic shuttling caused pleiotropic growth defects in imb4-1. On the other hand, the gif1;gif2;gif3 triple mutant failed to develop anthers (Lee et al., 2014, 2018), whereas imb4 has no anther developmental defects (Supplemental Fig. S2), indicating that other importins might mediate the nucleocytoplasmic transport of GIFs in tissues such as anthers.
We also showed that auxin and CK responses were altered in imb4-1 ovules (Fig. 4; Supplemental Fig. S4). Both auxin and CK are instrumental for ovule development (Benková et al., 2003; Bencivenga et al., 2012; Ceccato et al., 2013). Auxin responses were substantially reduced in imb4-1, which correlates with the disrupted polar distribution of PIN1 (Fig. 4). The abnormal auxin responses at the micropyle (Fig. 4) might explain defective embryo sac development in imb4-1 because auxin is critical for embryo sac patterning and gamete specification (Pagnussat et al., 2009). As a reciprocal phytohormone to auxin during ovule development (Bencivenga et al., 2012; Ceccato et al., 2013), CK signals were enhanced correspondingly (Supplemental Fig. S4). Because there is no evidence supporting a direct role of IMB4 on auxin and CK signaling, it is more likely that defective auxin and CK signaling in imb4-1 ovules is associated with abnormal ovule development. Alternatively, GIFs may mediate transcriptional changes through their interactors CYTOKININ RESPONSE FACTOR2 and ARABIDOPSIS RESPONSE REGULATOR4 (Vercruyssen et al., 2014) to influence hormonal responses during ovule development.
MATERIALS AND METHODS
Plant Growth and Transformation
The T-DNA insertion lines SALK_049564 (imb4-1) and SALK_078852 (imb4-2) were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus; http://www.Arabidopsis.org). Other materials, including ProLAT52:GUS (Li et al., 2013), DR5:GFP (Ulmasov et al., 1997), PIN1:GFP (Benková et al., 2003), TCS:YFP (Bencivenga et al., 2012), and gif1;gif2;gif3/+ (Lee et al., 2014), have been previously described. Columbia-0 was used as the wild type. Plant growth, transformation, and selection were as previously described (Zhou et al., 2013).
DNA Manipulation
All constructs were generated using the Gateway technology (Invitrogen) unless noted otherwise. Entry vectors were generated using pENTR/D/TOPO (Invitrogen). For plant-expressing vectors, ProIMB4 containing the upstream 1200-bp sequences before the start codon of IMB4 was amplified with the primer pair ZP3899/ZP3900. PCR fragments were digested with HindIII/SpeI and replaced the Pro35S of the destination vector Pro35S:GW-YFP (Karimi et al., 2002) to generate the destination vector ProIMB4:GW-YFP. The IMB4 genomic sequence containing 4963 bp from 5′-untranslated region to the nucleotide before stop codon was amplified with the primer pair ZP3605/ZP3549 and introduced into ProIMB4:GW-YFP to generate IMB4g-YFP. Coding sequences of IMB4, GIF1, GIF2, and GIF3 were amplified with the primer pair ZP3504/ZP4475, ZP6904/ZP6907, ZP6671/ZP6672, and ZP6673/ZP6674, respectively. ProUBQ10:GFP-GIF1 was generated by combining the GIF1 entry vector and ProUBQ10:GFP-GW (Zhang et al., 2015) in an LR reaction. ProINO:red fluorescence protein (RFP)-IMB4 was generated by combining the IMG genomic entry vector and ProINO:RFP-GW (Wang et al., 2016) in an LR reaction.
For vectors used in Y2H assays, entry vectors for IMB4, GIF1, GIF2, and GIF3 were used in LR reactions with the destination bait vector pDEST-GBKT7 or the destination prey vector pDEST-GADT7 (Clontech) to generate expression vectors pGBKT7-IMB4, pGADT7-GIF1, pGADT7-GIF2, and pGADT7-GIF3. For vectors used in BiFC assays, entry vectors were used in LR reactions with the destination vector pSITE::cEYFP-C1 or pSITE-nEYFP-C1 (Martin et al., 2009) to generate pSITE-nEYFP-IMB4, pSITE::cEYFP-GIF1, pSITE::cEYFP-GIF2, or pSITE::cEYFP-GIF3.
All PCR amplifications were performed using Phusion TM hot-start high-fidelity DNA polymerase, at the annealing temperature and extension times recommended by the manufacturer (Finnzyme). All entry vectors were sequenced, and sequences were analyzed using Vector NTI (Invitrogen). The Bioneer PCR purification kit and the Bioneer Spin miniprep kit were used for PCR product recovery and plasmid DNA extraction, respectively. All primers are listed in Supplemental Table S2.
RNA Extraction, RT-PCRs, and qPCRs
Genotyping PCRs of imb4-1 and imb4-2 were performed using the following primers: ZP4342/ZP4343 and ZP3917/ZP3918 for the wild-type copy of IMB4, ZP1/ZP4342 for imb4-1,ZP1/ZP3918 for imb4-2. Genotyping PCRs of the IMB4g-YFP;imb4-1 were performed using the following primers: ZP3915/ZP4438 for the endogenous IMB4 and ZP3915/ZP1848 for the transgene. ACT2 was amplified with the primer pair ZP16/ZP17.
For RT-qPCRs analyzing the expression pattern of IMB4 and for RT-PCRs, total RNAs were isolated from open flower, seedlings of 3 d after germination (DAG) and 7 DAG, leaves of 14 DAG, stems of 25 DAG, inflorescence, mature ovules, and mature pollen using a Qiagen RNeasy plant mini kit according to the manufacturer’s instructions. Oligo(dT)-primed cDNAs were synthesized using Superscript III reverse transcriptase with on-column DNase II digestion (Invitrogen).
Expression analysis of IMB4 by RT-qPCRs was performed with the Bio-Rad CFX96 real-time system using SYBR Green real-time PCR master mix (Toyobo) as described (Zhou et al., 2013). The specific primers used for IMB4, ACT2, and GAPDH were ZP4492/ZP4493, ZP313/ZP314, and ZP687/ZP688, respectively. All primers are listed in Supplemental Table S2.
Phenotypic Analysis
Pollen development by Alexander staining, 4’,6-diamino-phenylindole (DAPI) staining, SEM, examination of the pollen tube in vivo growth by histochemical GUS staining of ProLAT52:GUS-pollinated pistils and aniline blue staining were performed as previously described (Li et al., 2013). Whole-mount ovule clearing and CLSM of ovules were performed as described (Wang et al., 2016). For each assay on ovules, including the ProLAT52:GUS pollination assay, aniline blue staining, whole-mount ovule clearing, and optical sectioning by CLSM, 10 pistils were analyzed. Results are shown either as means ± sd, or shown as the number of given category/total number of ovules (n = 210 to 469).
Protein Interaction Assays
Y2H assays were performed as previously described (Park et al., 2014), with slight modifications. Briefly, different combinations of bait and prey vectors were cotransformed into the Y2HGold yeast strain (Clontech). Protein-protein interactions were determined based on the growth of transformants after 3 d on YSD-WLA containing 80 mg/l X-α-Gal. Agrobacterium infiltration of expression vectors used in BiFC was performed as described, in which a P19 protein was used to suppress gene silencing (Park et al., 2014). The nuclear-localized protein U1-70K-mCherry has been previously described (Wang et al., 2012). Confocal imaging was performed 48 h after infiltration.
Cell Fractionation and Western Blot
Cell fraction was performed according to a previously described method (Wierzbicki et al., 2008), with slight modifications. Specifically, two grams of inflorescences were ground into powder in liquid nitrogen, suspended in 20 ml of Honda buffer (20 mm HEPES-KOH [pH 7.4], 0.44 m Suc, 1.25% [v/v] ficoll, 2.5% [w/v] Dextran T40, 10 mm MgCl2, 0.5% [v/v] Triton X-100, 5 mM dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1% [w/v] plant protease inhibitors), and filtered through two layers of Miracloth and centrifuged at 5000 g for 20 min. The supernatant was used as the cytosolic fraction. The pellets were washed five times, each with 1 ml of Honda buffer. Total nuclear protein was obtained by adding 3 volumes of 1% (w/v) SDS directly to the nucleus pellet, followed by boiling at 95°C for 10 min. Equal volumes of each fraction were mixed with loading buffer, boiled, gel-separated, and subjected to protein gel blot analysis. Immunolabeling of GFP-GIF1/AN3 was performed using an anti-GFP antibody (Beyotime, 1:1000 dilution). Anti-H3 (Beyotime, 1:1000 dilution) and AnticFBPase (Agrisera, 1:5000 dilution) were used as the nuclear and cytoplasmic marker, respectively. Quantification of total proteins was performed using Gel Image System 4.1.2 (www.bio-tanon.com.cn).
Fluorescence Microscopy and Pharmacological Treatment
Lysotracker red staining or FM4-64 staining was used to show cell silhouettes as previously described (Wang et al., 2016). CLSM of fluorescence materials was performed with an LSM 880 (Zeiss) with the excitation and emission wavelengths set to 488 nm/505–550 nm for YFP and GFP signals and 561 nm/600 nm for RFP signals. For the quantification of fluorescence intensity between nuclear and cytoplasmic fractions in ovules, a region of interest of the same size was defined either in the nucleus or in cytoplasm within an outer integument cell. For the quantification of fluorescence intensity between nuclear and cytoplasmic fractions in roots, a region of interest of the same size was defined in either the nucleus or the cytoplasm within a root epidermal cell. The ratio of fluorescence intensity between the nuclear and cytoplasmic reactive oxygen species (Nucleus/Cytoplasm) was calculated using ImageJ (http://rsbweb.nih.gov/ij/).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At4g27640 for IMB4; At5g28640 for GIF1/AN3; At1g01160 for GIF2; At4g00850 for GIF3; At1g23420 for INO.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article.
Supplemental Figure S1. Functional loss of IMB4 causes pleiotropic developmental defects.
Supplemental Figure S2. Functional loss of IMB4 does not affect pollen development.
Supplemental Figure S3. Integument growth is compromised by functional loss of IMB4.
Supplemental Figure S4. CK signaling during ovular development is affected by functional loss of IMB4.
Supplemental Figure S5. Phenotypic defects in IMB4 loss-of-function are suppressed by IMB4g-YFP.
Supplemental Figure S6. IMB4 is constitutively expressed.
Supplemental Figure S7. Specific expression of IMB4 in outer integuments suppresses ovular defects of imb4-1.
Supplemental Figure S8. GIFs are critical for ovule development.
Supplemental Figure S9. Leaf development, but not primary root growth or stem elongation, is partially rescued by ProUBQ10:GFP-GIF1 in imb4-1.
Supplemental Table S1. IMB4 loss-of-function does not compromise male or female transmission.
Supplemental Table S2. Oligos used in this study.
Acknowledgments
We thank Prof. Xian Sheng Zhang for DR5:GFP, PIN1:GFP, and TCS:YFP. We thank Prof. Jeong Hoe Kim for the kind gift of the gif1;gif2;gif3 mutant.
Footnotes
This work was supported by the National Natural Science Foundation of China (NSFC) (31871422 and 31625003 to Y.Z. and 31771558 to S.L.), by Major Research Plant from the Ministry of Science and Technology of China (grant 2013CB945102), and by the Natural Science Foundation of Shandong Province (ZR2014CM027 to S.L.). Y.Z.’s laboratory is partially supported by the Tai-Shan Scholar Program of the Shandong Provincial Government.
Articles can be viewed without a subscription.
References
- Bencivenga S, Colombo L, Masiero S (2011) Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod 24: 113–121 [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benková E, Colombo L (2012) The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24: 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blanvillain R, Boavida LC, McCormick S, Ow DW (2008) Exportin1 genes are essential for development and function of the gametophytes in Arabidopsis thaliana. Genetics 180: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Boruc J, Griffis AH, Rodrigo-Peiris T, Zhou X, Tilford B, Van Damme D, Meier I (2015) GAP activity, but not subcellular targeting, is required for Arabidopsis RanGAP cellular and developmental functions. Plant Cell 27: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccato L, Masiero S, Sinha Roy D, Bencivenga S, Roig-Villanova I, Ditengou FA, Palme K, Simon R, Colombo L (2013) Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS One 8: e66148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier É, Loubert-Hudon A, Zimmerman EL, Matton DP (2011) Cell-cell communication and signalling pathways within the ovule: From its inception to fertilization. New Phytol 192: 13–28 [DOI] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Colombo L, Battaglia R, Kater MM (2008) Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci 13: 444–450 [DOI] [PubMed] [Google Scholar]
- Cui Y, Fang X, Qi Y (2016) TRANSPORTIN1 promotes the association of microRNA with ARGONAUTE1 in Arabidopsis. Plant Cell 28: 2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Lee D, Christensen CA (1998) Genetic analysis of female gametophyte development and function. Plant Cell 10: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR (1996) AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercoli MF, Ferela A, Debernardi JM, Perrone AP, Rodriguez RE, Palatnik JF (2018) GIF transcriptional co-regulators control root meristem homeostasis. Plant Cell 30: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Forbes DJ (2004) Importin beta: Conducting a much larger cellular symphony. Mol Cell 16: 319–330 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS (2003) PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol 132: 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H (2013) ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol 23: 788–792 [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS (2012) ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kende H (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Tsukaya H (2015) Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot 66: 6093–6107 [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL (1996) The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Wynn AN, Franks RG, Hwang YS, Lim J, Kim JH (2014) The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev Biol 386: 12–24 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Lee BH, Jung JH, Park SK, Song JT, Kim JH (2018) GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol 176: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen X (2003) PAUSED, a putative exportin-t, acts pleiotropically in Arabidopsis development but is dispensable for viability. Plant Physiol 132: 1913–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ge FR, Xu M, Zhao XY, Huang GQ, Zhou LZ, Wang JG, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Wang F, Yu D (2014) Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol 164: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber D, Lora J, Schrempp S, Lenhard M, Laux T (2011) Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr Biol 21: 1009–1017 [DOI] [PubMed] [Google Scholar]
- Luo Y, Wang Z, Ji H, Fang H, Wang S, Tian L, Li X (2013) An Arabidopsis homolog of importin β1 is required for ABA response and drought tolerance. Plant J 75: 377–389 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- McAbee JM, Hill TA, Skinner DJ, Izhaki A, Hauser BA, Meister RJ, Venugopala Reddy G, Meyerowitz EM, Bowman JL, Gasser CS (2006) ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J 46: 522–531 [DOI] [PubMed] [Google Scholar]
- Meier I, Brkljacic J (2009) The nuclear pore and plant development. Curr Opin Plant Biol 12: 87–95 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2011) Nucleo-cytoplasmic transport of proteins and RNA in plants. Plant Cell Rep 30: 153–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Eeckhout D, Demuynck K, Persiau G, Walton A, van Bel M, Vervoort M, Candaele J, De Block J, Aesaert S, et al. (2015) Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B (2015) Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol Plant 8: 998–1010 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V (2009) Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Tal L, Yichie Y, Gar O, Zamir D, Eshed Y, Lippman ZB (2014) Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet 46: 1337–1342 [DOI] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL (1995) The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83: 735–742 [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hülskamp M, Pruitt RE (1995) Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J 7: 731–749 [Google Scholar]
- Schneitz K, Hülskamp M, Kopczak SD, Pruitt RE (1997) Dissection of sexual organ ontogenesis: A genetic analysis of ovule development in Arabidopsis thaliana. Development 124: 1367–1376 [DOI] [PubMed] [Google Scholar]
- Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K (2004) Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol 273: 321–334 [DOI] [PubMed] [Google Scholar]
- Tamura K, Hara-Nishimura I (2014) Functional insights of nucleocytoplasmic transport in plants. Front Plant Sci 5: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Poethig RS (1998) HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125: 1889–1898 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P, Fulton L, Batoux M, Yadav RK, Schneitz K (2011) Structure-function analysis of STRUBBELIG, an Arabidopsis atypical receptor-like kinase involved in tissue morphogenesis. PLoS One 6: e19730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han SK, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. (2014) ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26: 210–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS (1999) INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev 13: 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JG, Feng C, Liu HH, Ge FR, Li S, Li HJ, Zhang Y (2016) HAPLESS13-mediated trafficking of STRUBBELIG is critical for ovule development in Arabidopsis. PLoS Genet 12: e1006269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y (2011) An importin β protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, et al. (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24: 3278–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Wang LC, Yeh CH, Lu CA, Wu SJ (2010) Isolation and characterization of the Arabidopsis heat-intolerant 2 (hit2) mutant reveal the essential role of the nuclear export receptor EXPORTIN1A (XPO1A) in plant heat tolerance. New Phytol 186: 833–842 [DOI] [PubMed] [Google Scholar]
- Zhang YL, Li E, Feng QN, Zhao XY, Ge FR, Zhang Y, Li S (2015) Protein palmitoylation is critical for the polar growth of root hairs in Arabidopsis. BMC Plant Biol 15: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Guo X, Ge C, Ma Z, Jiang M, Li T, Koiwa H, Yang SW, Zhang X (2017) KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc Natl Acad Sci USA 114: 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y (2007) SAD2, an importin β-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LZ, Li S, Feng QN, Zhang YL, Zhao X, Zeng YL, Wang H, Jiang L, Zhang Y (2013) Protein S-ACYL Transferase10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 25: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]