Extracellular ATP-responsive transcription interacts with other defense signaling pathways and relies on MYC and CAMTA transcription factors.
Abstract
ATP is not only an essential metabolite of cellular biochemistry but also acts as a signal in the extracellular milieu. In plants, extracellular ATP is monitored by the purinergic receptor P2K1. Recent studies have revealed that extracellular ATP acts as a damage-associated molecular pattern in plants, and its signaling through P2K1 is important for mounting an effective defense response against various pathogenic microorganisms. Biotrophic and necrotrophic pathogens attack plants using different strategies, to which plants respond accordingly with salicylate-based or jasmonate/ethylene-based defensive signaling, respectively. Interestingly, defense mediated by P2K1 is effective against pathogens of both lifestyles, raising the question of the level of interplay between extracellular ATP signaling and that of jasmonate, ethylene, and salicylate. To address this issue, we analyzed ATP-induced transcriptomes in wild-type Arabidopsis (Arabidopsis thaliana) seedlings and mutant seedlings defective in essential components in the signaling pathways of jasmonate, ethylene, and salicylate (classic defense hormones) as well as a mutant and an overexpression line of the P2K1 receptor. We found that P2K1 function is crucial for faithful ATP-induced transcriptional changes and that a subset of genes is more responsive in the P2K1 overexpression line. We also found that more than half of the ATP-responsive genes required signaling by one or more of the pathways for the classical defense hormones, with the jasmonate-based signaling being more critical than others. By contrast, the other ATP-responsive genes were unaffected by deficiencies in signaling for any of the classical defense hormones. These ATP-responsive genes were highly enriched for defense-related Gene Ontology terms. We further tested the ATP-induced genes in knockout mutants of transcription factors, demonstrating that MYCs acting downstream of the jasmonate receptor complex and calmodulin-binding transcription activators are nuclear transducers of P2K1-mediated extracellular ATP signaling.
ATP, a universal biological energy currency and genetic building block, is maintained at a high concentration (approximately millimolar) intracellularly. By contrast, the resting extracellular ATP concentration is roughly 6 orders of magnitude lower (Watt et al., 1998; Weerasinghe et al., 2009). This steep chemical gradient provides an elegant basis for cellular signaling (Verkhratsky and Burnstock, 2014). Transmembrane receptors monitor changes in extracellular ATP concentration to detect either uncontrolled ATP release caused by necrosis of nearby cells (Davalos et al., 2005; Song et al., 2006) or active ATP release induced after pathogen detection or treatment with, for example, salicylic acid (Chivasa et al., 2009; Chen et al., 2017). In this manner, extracellular ATP can be perceived as a sign of damaged self, or a damage-associated molecular pattern (DAMP; Tanaka et al., 2010a, 2014; Cho et al., 2017). For general discussions of the DAMP concept, see reviews by Heil (2009), Boller and Felix (2009), and Lotze et al. (2007). In animals, extracellular ATP has been well studied due to its significant medical implications, and its important roles in physiological processes and signal transduction mechanisms are documented in detail (Burnstock, 2017). In contrast, extracellular ATP signaling in plants is only beginning to be understood.
A plant extracellular ATP receptor, P2K1, originally identified by genetic screen as an Arabidopsis (Arabidopsis thaliana) mutant, does not respond to nucleotides1 (dorn1), is a legume-type lectin receptor kinase, LecRK-I.9 (Choi et al., 2014). ATP binding to the extracellular domain and kinase activity of the intracellular domain of P2K1 are essential for the plant response to extracellular ATP (Choi et al., 2014). P2K1 is expressed during the major stages of plant growth and development (Cho et al., 2017), suggesting that extracellular ATP is involved in various physiological processes, including seedling growth, stomata movement, pollen tube development, root hair growth, gravitropism, and abiotic and biotic stress responses.
Mechanisms of extracellular ATP signal transduction through P2K1 are largely unknown, although a growing body of circumstantial evidence indicates a role in plant defense against pathogens. For example, overexpression of P2K1 protects plants against the oomycete pathogen Phytophthora brassicae and the bacterial pathogen Pseudomonas syringae (Bouwmeester et al., 2011; Balagué et al., 2017). A recent study demonstrated that extracellular ATP-induced plant protection against the fungal pathogen Botrytis cinerea depended on a functional jasmonate signaling pathway but not jasmonate biosynthesis (Tripathi and Tanaka, 2018; Tripathi et al., 2018). Moreover, ATP-induced stomatal immunity against the bacterial pathogen P. syringae was caused by P2K1-mediated activation of the plasma membrane-localized NADPH oxidase RBOHD that produces reactive oxygen species (Chen et al., 2017). Lastly, application of extracellular ATP to lima bean (Phaseolus lunatus) induced an indirect defense, the secretion of extrafloral nectar (Heil et al., 2012). Therefore, extracellular ATP is likely involved in different resistance mechanisms.
Plant pathogens can be divided into biotrophs and necrotrophs based on their lifestyles. Plant defense against hemibiotrophs/biotrophs (e.g. P. syringae and P. brassicae) and necrotrophs (e.g. B. cinerea) generally relies on salicylate-based and jasmonate/ethylene-based defensive signaling pathways, respectively (Glazebrook, 2005). Interestingly, the above-mentioned ATP-induced resistances were effective against both types of pathogens (i.e. necrotrophs and biotrophs). This led us to hypothesize that the extracellular ATP signaling pathway interacts with other signaling pathways of classical defense hormones (i.e. jasmonate, ethylene, and salicylate).
To characterize the interaction between extracellular ATP signaling and that of these classical defense hormones, we investigated the ATP-induced transcriptomes in several mutants defective in signaling of jasmonate, ethylene, and salicylate (coi1, ein2, and npr1) as well as the dorn1 mutant and a P2K1 overexpression line. Our results reveal that (1) the ATP-responsive transcriptome depends on a functional P2K1 receptor, (2) many ATP-responsive genes depend on one or more of the signaling pathways of the classical defense hormones, (3) nearly 30% of ATP-responsive genes are solely dependent on jasmonate-mediated signaling, and (4) nearly half of ATP-responsive genes are unaffected by deficiencies in the classical defense signaling pathways of jasmonate, ethylene, and salicylate. These results suggest that extracellular ATP induces a number of defense-related genes, half of them independent of, but the other half acting cooperatively with, the classical defense hormones. Finally, we find that MYC transcription factors and a calmodulin-binding transcription activator (CAMTA3) are important signaling components for ATP-responsive gene expression.
RESULTS
Experimental Design and Validation of Transcriptome Profiling for Extracellular ATP-Induced Responses
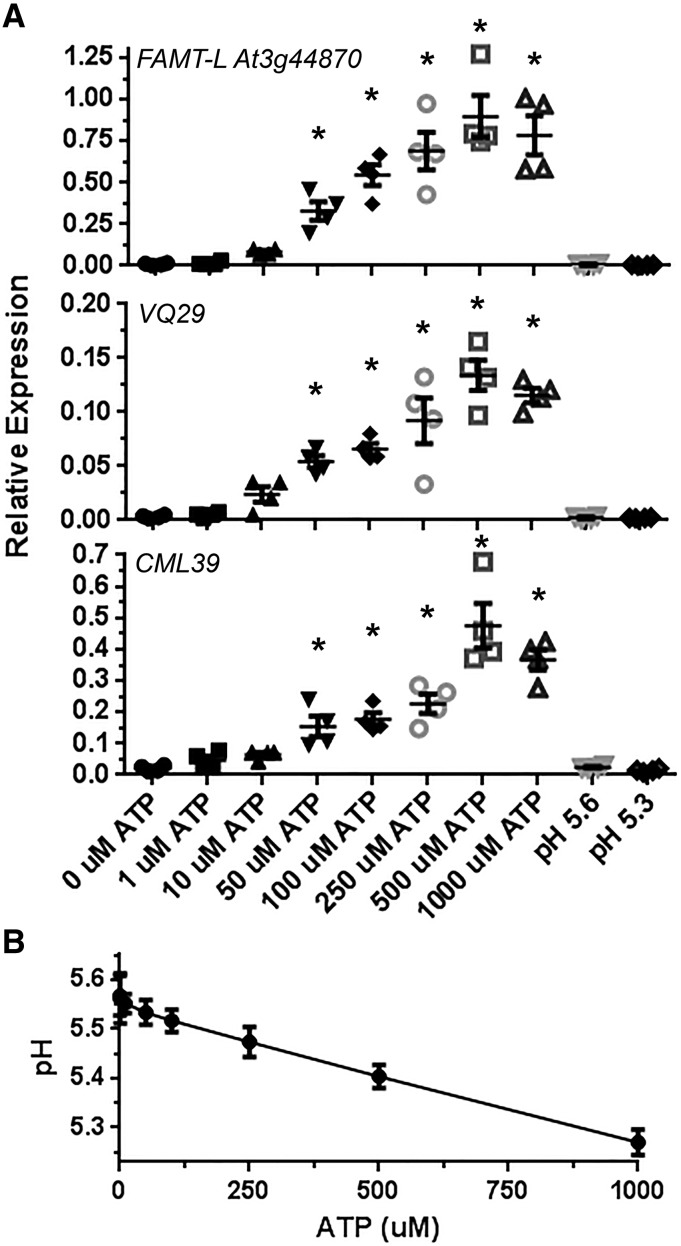
In a preliminary dose-response experiment, we found that treating seedlings with increasing ATP concentrations increased the induction of selected ATP-responsive genes, with 500 µm ATP treatment maximally stimulating expression, although significant gene induction could also be detected with treatments of ATP as low as 50 µm (Fig. 1A). We also tested whether addition of the polyvalent anion ATP to the test solution would alter the pH. As expected, even though its stock solution was prepared in MES buffer, ATP reduced the pH level in a dose-dependent manner, with 1 mm ATP lowering the pH to 5.3 (Fig. 1B). This result led us to further examine the effect of the pH drop on ATP-responsive gene expression. RT-qPCR demonstrated no changes in the gene expression tested even in the lowest pH condition (i.e. pH 5.3) in the absence of ATP (Fig. 1A). Therefore, we concluded that ATP has bona fide effects on plant physiology (gene expression in our case) regardless of its anionic effect in the treatment solution.
Figure 1.
Dose-dependent effect of ATP on gene expression in wild-type plants and pH in the treatment solution. A, Eight-day-old seedlings were treated with varying concentrations of ATP or with different pH levels for 30 min, and gene expression of the indicated genes was estimated by reverse transcription quantitative PCR (RT-qPCR; mean, individual replicate values and se are shown, n = 4). The expression of each gene was normalized by comparison with the internal control PP2A (Czechowski et al., 2005). Gene expression different from the non-ATP treatment is indicated by asterisks (P < 0.05). B, Addition of ATP to the test solution decreases the pH. Note that ATP treatment induces ATP-responsive gene expression but that pH decrease does not.
To define the interaction of ATP-responsive transcription with classical defense signaling pathways, we treated 8-d-old seedlings of wild-type and mutant lines with 500 µm ATP or vehicle solution for 30 min. The mutants used were coi1-30, defective in all known jasmonate responses (Xie et al., 1998; Yang et al., 2012); ein2-1, defective in most known ethylene responses (Alonso et al., 1999); npr1-3, deficient in all known salicylate responses (Cao et al., 1997); and dorn1-3, defective in ATP-induced cytosolic calcium transients and ATP-induced pathogen resistance (Choi et al., 2014; Tripathi et al., 2018). In addition, we examined OxP2K1, an overexpression line of the extracellular ATP receptor (Choi et al., 2014). The experiment was replicated six times in order to have the statistical resolving power needed to determine quantitative differences between the treatments. Seventy-two RNA samples were used in mRNA sequencing (mRNA-seq) analysis, using 1 × 100-bp Illumina sequencing. The mRNA-seq libraries were prepared from total RNA using poly(A) enrichment of the mRNA. Hierarchical clustering and principal component analysis indicated a high level of data quality, with different samples of treatment/genotype combinations clustering together (Supplemental Figs. S1 and S2). Moreover, RT-qPCR analysis of a subset of these genes indicated a high concordance with the sequencing by synthesis-based gene expression estimates (R2 = 0.98; Supplemental Fig. S3), which confirmed the validity of the mRNA-seq data in this study.
Extracellular ATP-Responsive Gene Expression in the Wild Type, the P2K1 Overexpression Line, and the dorn1 Mutant
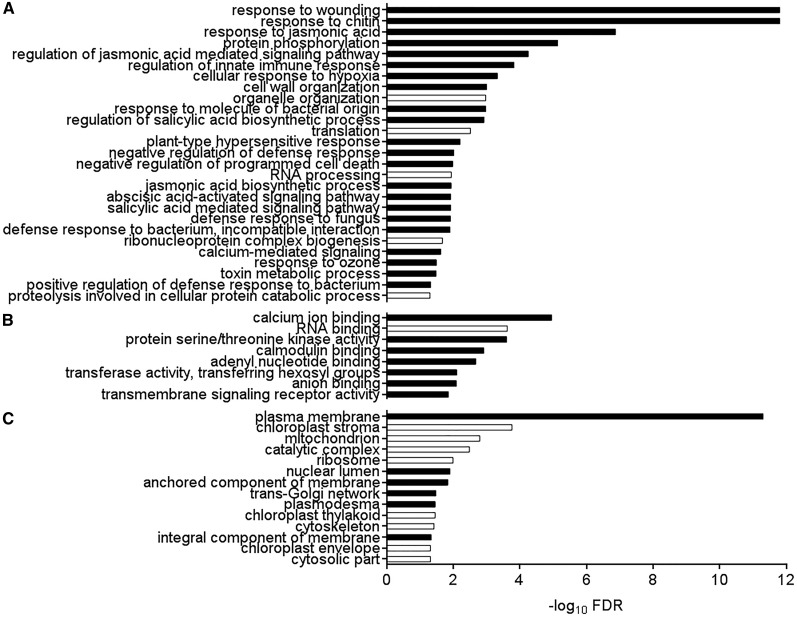
The response to extracellular ATP in wild-type seedlings led to the induction of 500 genes and the repression of 43 genes, with a cutoff at false discovery rate (FDR) < 0.05 and fold change (FC) > 1.5. All gene expression changes after ATP treatment are shown in Supplemental Data Set S1. The ATP-responsive genes were significantly enriched in Gene Ontology (GO) biological processes (Fig. 2A) related to response to wounding (FDR = 2.89e-12) and response to chitin (FDR = 2.96e-12), similar to what was described for ATP response in root tissue (Cao et al., 2014; Choi et al., 2014). In addition, protein phosphorylation (FDR = 8.52e-7), response to jasmonic acid (FDR = 1.35e-7), regulation of innate immune response (FDR = 1.55e-4), salicylic acid biosynthesis (FDR = 1.22e-3), and abscisic acid-activated signaling pathway (FDR = 8.21e-3) were also highly enriched biological process GO terms (Fig. 2A). We further performed MapMan mapping in the metabolism overview pathway, demonstrating that the ATP-induced transcriptome was highly enriched in the category biotic stress (Supplemental Fig. S4). With regard to molecular function (Fig. 2B), the ATP-responsive genes were enriched for calcium ion binding (FDR = 1.13e-5), calmodulin binding (FDR = 3.12e-5), and protein Ser/Thr kinase activity (FDR = 1.08e-4). Genes responsive to ATP were highly enriched for plasma membrane cellular components (FDR = 9.10e-12; Fig. 2C). In summary, ATP-responsive genes in wild-type seedlings tend to encode plasma membrane-localized defense-related proteins with kinase or calcium-binding activity.
Figure 2.
GO term enrichment for ATP-responsive gene expression in the wild type. Biological processes (A), molecular functions (B), and cellular components (C) significantly enriched (FDR < 0.05) in 543 ATP-responsive genes in wild-type seedlings are shown. Black and white bars indicate enrichment and depletion, respectively. Note that, perhaps because of the low number of ATP-repressed genes, no functional categories were detected to be enriched in ATP-repressed genes, so these GO terms represent all 543 ATP-responsive genes whether repressed or induced.
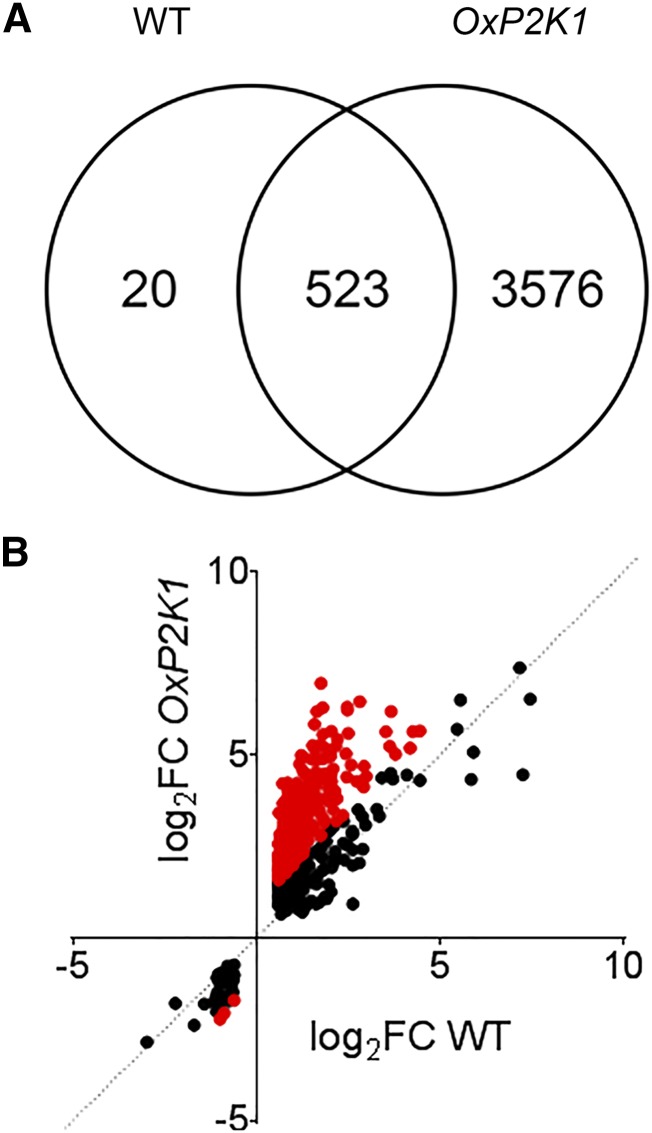
We further compared the ATP-induced transcriptome in the wild type with that in OxP2K1. The data indicated that there was substantial overlap between ATP-responsive gene expression in the wild type and OxP2K1, with 96% of genes identified in the wild type also being identified in the P2K1-overexpressing line (523 of 543; Fig. 3A). A total of 329 of the 523 ATP-responsive genes in common between the wild type and OxP2K1 were greater than 2-fold more responsive in the OxP2K1 line compared with the wild type (red dots in Fig. 3B; P = 7.54e-79, two-tailed heteroscedastic Student’s t test), indicating that most of the ATP-responsive genes are regulated in correlation with the amount of the receptor, P2K1. Those 329 genes were composed of the same ontological components based on GO analysis (Supplemental Fig. S5). In parallel, 3,576 genes were identified as ATP responsive in OxP2K1 that were not differentially expressed in the wild type. The genes identified in OxP2K1 but not in the wild type were enriched for biological processes also noted in the wild type (Supplemental Fig. S6), such as response to chitin (FDR = 1.42e-20), response to wounding (FDR = 3.47e-4), response to jasmonic acid (FDR = 1.08e-3), regulation of innate immune response (FDR = 5.97e-3), and plasma membrane localization (FDR = .72e-17). In addition, DNA-binding transcription factor activity (FDR = 3.83e-20), ethylene-activated signaling pathway (FDR = 1.50e-4), response to gibberellin (FDR = 2.29e-3), and response to karrikin (FDR = 5.10e-3) were included.
Figure 3.
Comparison of gene expression between OxP2K1 and the wild type. A, Venn diagram showing overlap between genes responsive to ATP in the wild type (WT) and OxP2K1. B, The log2FCs (ATP/mock) were compared for the 523 genes differentially expressed in both the wild type and OxP2K1. Red dots represent the 329 genes whose expression was greater than 2-fold more highly responsive in OxP2K1 than in the wild type.
In contrast, only a single gene (AT1G80240; DUF642) was identified as ATP responsive in the dorn1-3 mutant (log2FC = 1.45, FDR = 0.038). The expression of this gene of unknown function was also induced by ATP in a similar manner in the wild type (log2FC = 1.28, FDR = 6.36e-4) and OxP2K1 seedlings (log2FC = 1.33, FDR = 1.85e-5), perhaps indicating the existence of another signaling pathway that mediates extracellular ATP-responsive gene expression irrespective of P2K1 function. However, the vast majority (99.8%) of ATP-responsive genes identified here depended on P2K1.
To identify conserved promoter elements that may be responsible for ATP-responsive transcription, we submitted the 1,000 bp upstream of the transcription start site of the 543 ATP-responsive genes to the regulatory sequence analysis tools (RSAT) server to search de novo for nucleotide sequences enriched relative to the promoters of the Arabidopsis genome (Defrance et al., 2008; Nguyen et al., 2018). Among the 33 significantly enriched 8-mer sequences (P < 3e-5; Supplemental Table S1), 23 contained a CGCG motif, the core motif of an element referred to as a CAM-box and known to be bound by so-called CAMTAs (Benn et al., 2014). Eight of the 33 sequences contained a TGAC element, the core motif of the WRKY transcription factor-bound W-box (Eulgem et al., 1999); one was composed of GAAAATTC, a portion of the minimal heat shock element (HSE) bound by heat shock transcription factors (Kumar et al., 2009); and one was a pyrimidine-rich sequence (CCTCCTCC), which may represent enrichment of the Y patch, which has previously been proposed to be a general component of core plant promoters and for which a binding factor is unknown (Yamamoto et al., 2007). To directly evaluate the enrichment of the CAM-box, the W-box, and the HSE in these promoters, we scored the presence of these elements in ATP-responsive gene promoters and in the promoters of the Arabidopsis genome, evaluating significance by hypergeometric distribution. Given the apparent role for extracellular ATP signaling in pathogen defense, we also tested for the enrichment of elements previously implicated in defense-related gene expression. These additional elements are the G-box (CACGTG), implicated in jasmonate-responsive expression (Fernández-Calvo et al., 2011; Figueroa and Browse, 2012); the abscisic acid response element (ABRE; Anderson et al., 2004; Adie et al., 2007); and the GCC-box (GCCGCC), implicated in ethylene-responsive gene expression (Ohme-Takagi and Shinshi, 1995; Solano et al., 1998). These analyses are summarized in Table 1. Notably, the CAM-box was highly enriched (2-fold, P = 5.1e-26) in ATP-responsive promoters in the wild type. Significant enrichment was also detected for the W-box (P = 1.1e-9), the HSE (P = 3.7e-4), the G-box (P = 4.1e-3), and the ABRE (P = 0.017). The GCC-box was marginally significantly (P = 0.058, fold enrichment = 0.94) underrepresented in these promoters.
Table 1. Enrichment of defense-related cis-elements in promoters of ATP-responsive genes in wild-type seedlings.
The presence or absence of the indicated promoter elements in the presumptive promoters (−1,000 bp upstream of the transcriptional start site) was scored in 543 ATP-responsive genes and 33,693 genes of the Arabidopsis nuclear genome, and significant enrichment was tested by hypergeometric test. Italicized values indicate that the element was not significantly enriched (P > 0.05). Fold enrichment is calculated as (percentage presence in ATP-responsive genes)/(percentage presence in all Arabidopsis genes).
| Element | Sequence | Presence | Fold Enrichment | P |
|---|---|---|---|---|
| CAM-box | VCGCGB | 206 | 2.01 | 5.1e-26 |
| W-box | TTGACY | 428 | 1.17 | 1.1e-9 |
| HSE | GAANNTTC | 215 | 1.18 | 3.7e-4 |
| G-box | CACGTG | 93 | 1.24 | 4.1e-3 |
| ABRE | ACGTGKC | 82 | 1.16 | 0.017 |
| GCC-box | GCCGCC | 43 | 0.94 | 0.058 |
We similarly analyzed the enrichment of these promoter elements in the 4,099 genes responsive to ATP in OxP2K1 seedlings, where broadly comparable results were seen (Supplemental Table S2), although here the GCC-box was significantly, although subtly, underrepresented (P = 0.019, fold enrichment = 0.977).
ATP-Responsive Gene Expression in Mutant Seedlings Defective in Jasmonate, Ethylene, and Salicylate Signaling
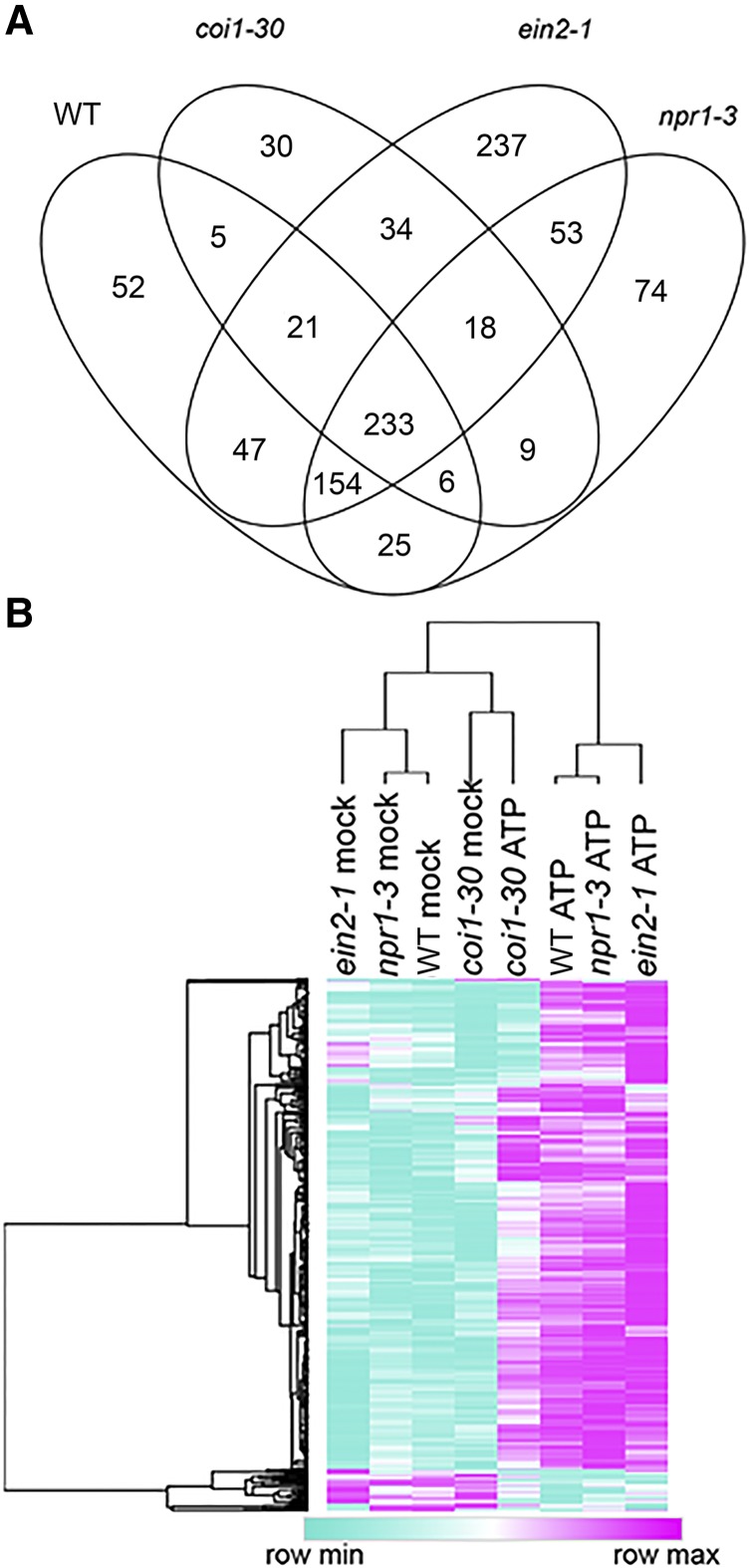
Next, we compared ATP-responsive gene expression in wild-type seedlings with that of the coi1-30, ein2-1, and npr1-3 mutants. Similar numbers of genes were responsive to ATP in these mutants and wild-type plants (Supplemental Table S3). However, hierarchical clustering of gene expression in the different genotype/treatment combinations revealed a clear disparity between ATP-responsive gene expression in coi1-30 and the three other genotypes; both ATP- and mock-treated samples of coi1-30 were more similar to mock-treated than ATP-treated samples of the other genotypes (Fig. 4B). On average, genes responding to ATP in the wild type were 67% less responsive to ATP in coi1-30 (P = 4.8e-20, two-tailed heteroscedastic Student’s t test), where no such differences were observed in comparisons between wild-type seedlings and ein2-1 or npr1-3 (P = 0.58 and 0.14, respectively).
Figure 4.
Overview of ATP-responsive gene expression in the wild type and coi1-30, npr1-3, and ein2-1 mutants. A, Overlap between ATP-responsive gene expression in the indicated genotypes 30 min after treatment with 500 µm ATP. B, Hierarchical clustering of average expression [log2(cpm+1)] for 543 genes identified as ATP responsive in the wild type (WT). Rows and columns were clustered by 1 − Pearson correlation using the Morpheus online tool (https://software.broadinstitute.org/morpheus), and the tree was drawn using average linkage.
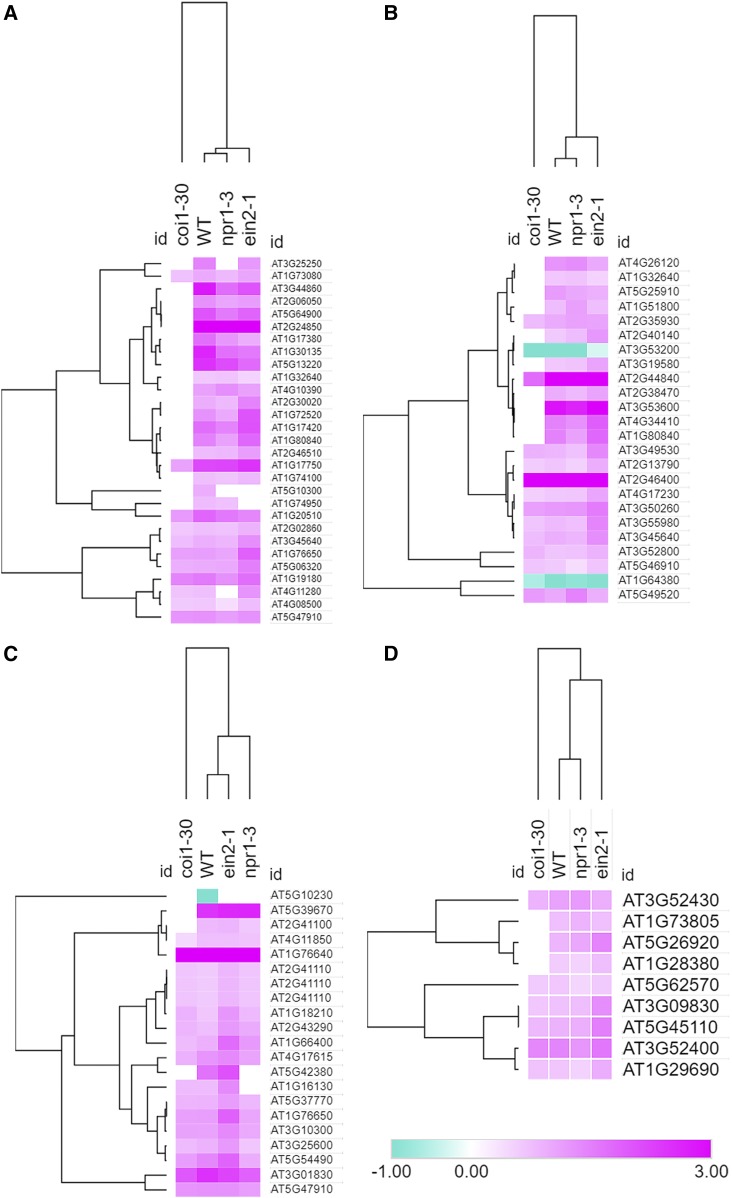
The unique transcriptomic response to ATP in the coi1-30 mutant was also evident in heat maps comparing the responses of these four genotypes for classes of GO terms enriched in the wild type. Hierarchical clustering of these data revealed that wild-type, npr1-3, and ein2-1 responses were more similar to each other in comparison with coi1-30 in GO classes including response to wounding, response to chitin, regulation of salicylic acid biosynthesis, calcium ion binding (Fig. 5), and protein phosphorylation and compared with genes encoding plasma membrane-localized proteins (Supplemental Fig. S6).
Figure 5.
Regardless of the biological process, molecular function, or cellular component, coi1-30 shows a markedly different transcriptional response to ATP stimulus. Log2FC for genes responsive to ATP in the wild type (WT) in selected GO categories, response to wounding (A), response to chitin (B), regulation of salicylate biosynthesis (C), and calcium ion binding (D), is shown. If a given gene had FDR > 0.1 in a given genotype, the raw FC value was set to 1, and log2FC and genotype were clustered using Morpheus.
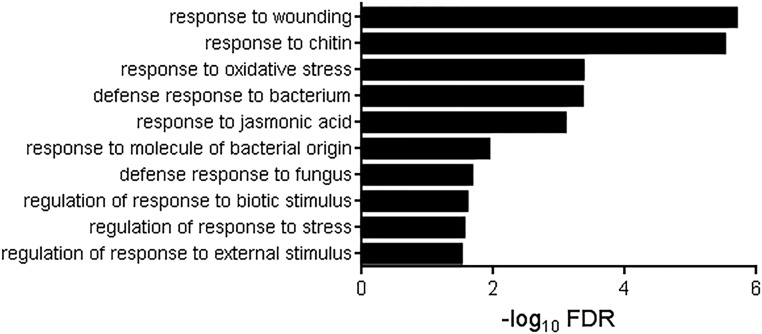
There were 154 genes responsive to ATP in the wild type that were solely dependent on intact COI1 but at the same time unaffected by the NPR1 and EIN2 functions. In contrast, there were only six and 21 genes dependent solely on EIN2 (not COI1 and NPR1) and NPR1 (not COI1 and EIN2), respectively (FDR < 0.05 and absolute fold change > 1.5; Fig. 4A). Interestingly, 129 genes depended on two or more of these pathways. With regard to GO enrichment, the 154 COI1-dependent ATP-responsive genes were significantly enriched for eight biological processes (Fig. 6), in contrast to NPR1-dependent and EIN2-dependent genes in which no enrichment for any GO term was detected. COI1-dependent ATP-responsive genes were enriched in expected GO biological processes such as response to wounding (FDR = 3.45e-9), response to chitin (FDR = 6.33e-9), and response to jasmonic acid (FDR = 3.05e-6).
Figure 6.
GO term enrichment for ATP-responsive gene expression dependent on COI1. Biological processes significantly enriched in 154 genes solely dependent on COI1 for ATP responsiveness are shown. Note that no functional categories were detected in 21 NPR1-dependent and six EIN2-dependent genes (see text for details).
Finally, we examined the promoters of these genes dependent on COI1, NPR1, or EIN2 to look for elements enriched in putative promoters of genes responsive to ATP in wild-type seedlings (Table 2). As observed for ATP-responsive gene promoters in wild-type plants, the CAM-box was significantly enriched in promoters of COI1-dependent genes (P = 1.59e-4). Although G-box-binding basic helix-loop-helix transcription factors such as MYC2, MYC3, and MYC4 are important primary signal transducers in COI1-dependent signaling (Chini et al., 2007; Fernández-Calvo et al., 2011; Niu et al., 2011), we observed that G-box elements were not significantly enriched in COI1-dependent ATP response genes (P = 0.09) but rather were found 2-fold enriched in NPR1-dependent genes (P = 0.04). Interestingly, the W-box, long known as a salicylate-responsive element, was enriched in COI1-dependent ATP-responsive promoters (P = 1.20e-5) but not found enriched in NPR1-dependent promoters (P = 0.137). Furthermore, to our knowledge, the HSE has not been identified previously as enriched in COI1-dependent promoters, yet we found it enriched in COI1-dependent ATP-responsive genes (P = 8.93e-4).
Table 2. Enrichment of defense-related cis-elements in COI1-, NPR1-, and EIN2-dependent ATP-responsive promoters and ATP-specific responsive promoters.
The presence or absence of the indicated promoter elements in the presumptive promoters (−1,000 bp upstream of transcriptional start site) was scored in 154 COI1-dependent, 21 NPR1-dependent, and six EIN2-dependent ATP-responsive genes as well as in 233 ATP-specific genes (Fig. 3) and 33,693 genes of the Arabidopsis nuclear genome, and significant enrichment was tested by hypergeometric test. Italicized values indicate that the element was not significantly enriched (P > 0.05). Fold enrichment is calculated as (percentage presence in ATP-responsive genes)/(presence in all Arabidopsis genes).
| Element | Sequence | P (Fold Enrichment) | |||
|---|---|---|---|---|---|
| COI1 Dependent | NPR1 Dependent | EIN2 Dependent | ATP Specific | ||
| CAM-box | VCGCGB | 1.59e-4 (1.62) | 0.171 (1.26) | 0.286 (0) | 3.60e-26 (2.62) |
| W-box | TTGACY | 1.20e-5 (1.22) | 0.137 (1.13) | 0.273 (1.24) | 1.60e-3 (1.12) |
| G-box | CACGTG | 0.090 (1.04) | 0.040 (2.07) | 0.394 (1.21) | 1.80-e3 (1.46) |
| ABRE | ACGTGKC | 0.049 (0.748) | 0.033 (2.20) | 0.433 (0) | 4.20e-3 (1.42) |
| HSE | GAANNTTC | 8.93e-4 (1.34) | 0.169 (0.854) | 0.262 (0.498) | 0.021 (1.13) |
| GCC-box | GCCGCC | 0.105 (0.848) | 0.280 (1.13) | 0.590 (0) | 0.091 (0.94) |
ATP-Responsive Gene Expression Independent of COI1, NPR1, and EIN2
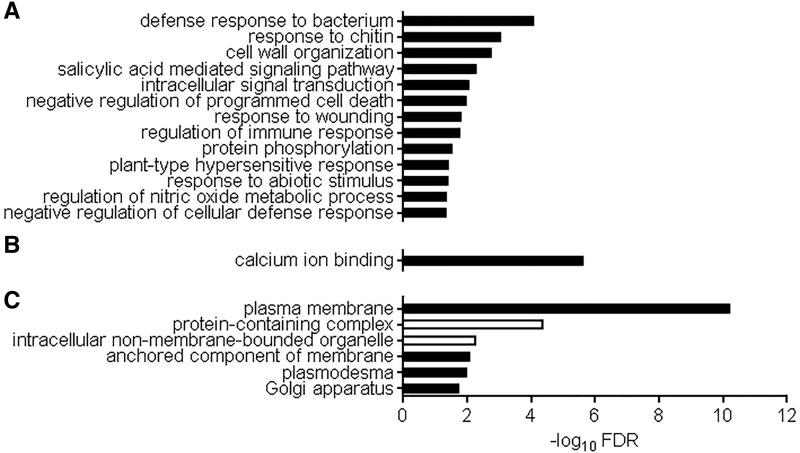
In total, we found 233 genes induced by extracellular ATP in the wild type that were also responsive in coi1-30, ein2-1, and npr1-3, indicating that these genes are regulated independently of the functioning of any signaling pathways of classical defense hormones (i.e. jasmonate, ethylene, and salicylate; Fig. 4A). These 233 genes were termed as putative ATP-specific genes for convenience. As shown in Figure 7, GO term enrichment in the putative ATP-specific genes presented defense response to bacterium (FDR = 3.43e-7), response to chitin (FDR = 3.33e-6), salicylic-acid-mediated signaling pathway (FDR = 8.79e-6), calcium and calmodulin binding (FDR = 7.96e-10 and 1.16e-6, respectively), and genes encoding plasma membrane-localized proteins (FDR = 1.73e-13).
Figure 7.
GO term enrichment for ATP-specific genes that require neither COI1, NPR1, nor EIN2 for expression. Biological processes (A), molecular function (B), and cellular components (C) for proteins significantly enriched in ATP-specific genes are shown (FDR < 0.05). Black and white bars indicate enrichment and depletion, respectively.
As for genes responsive to ATP in wild-type seedlings, we submitted the presumptive promoters for these genes to the RSAT server to search de novo for enriched sequence elements. No novel elements were identified. Rather, in 23 identified 8-mer sequences, 21 contained a CGCG CAM-box and two contained a W-box (Supplemental Table S4). Indeed, the VCGCGB CAM-box is highly enriched in these promoters relative to Arabidopsis nuclear genome promoters (2.6-fold, P = 3.6e-26) and is more enriched in these promoters than in the 543 ATP-responsive genes in the wild type. The W-box, G-box, ABRE, and HSE are also significantly enriched in these ATP-specific promoters (Table 2).
Contribution of MYC Transcription Factors to ATP-Specific Gene Expression
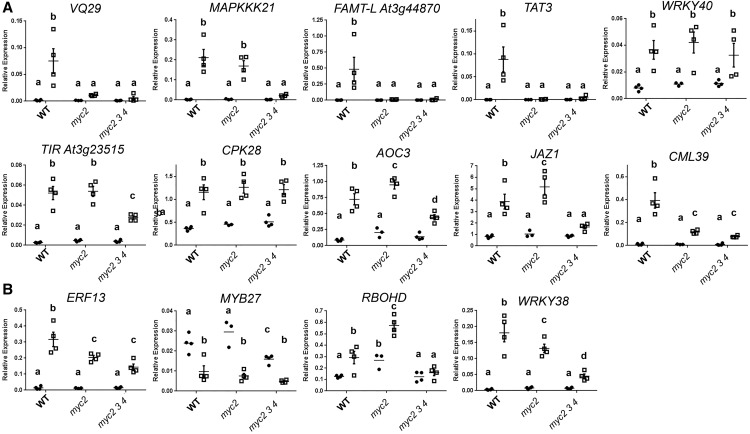
Given the larger number of genes dependent on COI1 for ATP-responsive expression, and given the importance of MYC transcription factors for jasmonate signaling (Fernández-Calvo et al., 2011; Niu et al., 2011), we investigated the contribution of MYC transcription factors to ATP-responsive transcription. We treated 8-d-old wild type, the myc2-1 mutant (Boter et al., 2004), and the myc2-1 myc3-1 myc4-1 triple mutant (Major et al., 2017) for 30 min with 500 µm ATP or vehicle solution and performed RT-qPCR analysis on COI1-dependent or ATP-specific transcripts. We chose 14 ATP-responsive genes for analysis: five dependent on COI1 and nine ATP specific. As shown in Figure 8A, genes dependent on COI1 for induction by ATP were, to differing degrees, dependent on MYC function for ATP response. TAT3, well known as a COI1- and MYC2-dependent jasmonate response marker gene (Titarenko et al., 1997; Jung et al., 2015), and FAMT-L (AT3G44870), an uncharacterized gene encoding a protein 93% identical to a farnesoic acid methyl transferase (Yang et al., 2006), were induced by ATP treatment in the wild type but not in myc2 or the myc2 myc3 myc4 triple mutant. VQ29, a gene encoding a VQ motif protein dependent on JAZ3 degradation for jasmonate-responsive induction (Chini et al., 2007), was more subtly perturbed in ATP response, with significantly reduced induction only in the triple mutant. Among tested ATP-specific genes, ERF13, encoding an ethylene-responsive element-binding factor, was induced similarly, irrespective of MYC mutation (Fig. 8B). WRKY38 and the uncharacterized gene AT4G23515, encoding a Toll/IL-1 receptor homology domain-containing protein, were similarly responsive to ATP treatment in myc2 and the wild type, and substantially reduced induction was observed in the triple mutant.
Figure 8.
Involvement of MYC transcription factors in ATP-responsive transcription. Wild-type (WT) seedlings, the myc2-1 single mutant, and the myc2-1 myc3-1 myc4-1 triple mutant were treated with 500 µm ATP or vehicle solution for 30 min. Expression of COI1-dependent genes (A) and ATP-specific genes (B) in the indicated genotypes is shown. The expression of each gene was normalized by comparison with the expression of PP2A. Data indicate means, individual replicate values and se (n = 3–4). In a subpanel, columns with different letters are significantly different (P < 0.05, Fisher’s lsd).
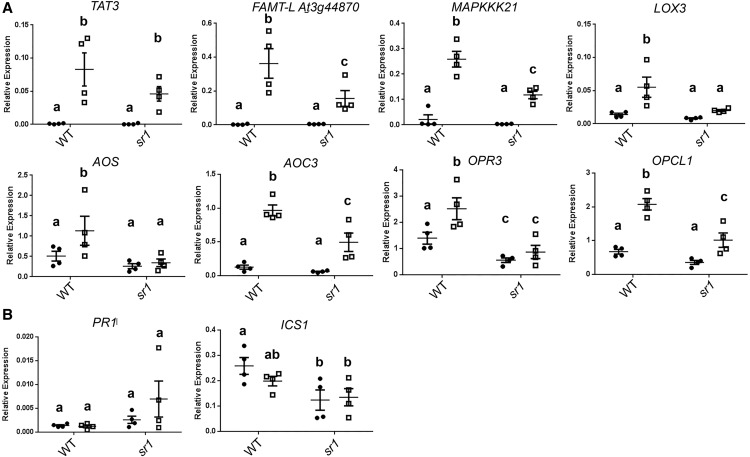
Contribution of CAMTA3 to ATP-Responsive Gene Expression
Since the CAM-box is highly enriched in ATP-responsive genes and Arabidopsis CAM-box-binding factors are known, we compared ATP-responsive gene expression in 8-d-old seedlings of wild-type Arabidopsis and the atsr1-1 mutant, a null mutation of the CAM-box-binding transcription factor CAMTA3 (Du et al., 2009). Seven of the tested ATP-responsive genes had significantly lower expression after ATP treatment in the atsr1 mutant than in wild-type seedlings (P < 0.05, Fisher’s lsd; Fig. 9A). Because a recent study indicated that this mutant exhibits an autoimmunity phenotype due to constitutive activation of nucleotide-binding, Leu-rich repeat guard proteins (Lolle et al., 2017), and this immune activation could potentially alter the transcriptional response to extracellular ATP, we also tested the expression of two immune markers reported to be highly expressed in mature atsr1-1 plants, PATHOGENESIS-RELATED1 (PR1) and ISOCHORISMATE SYNTHASE1 (ICS1; Du et al., 2009; Kim et al., 2017; Lolle et al., 2017). As shown in Figure 9B, PR1 and ICS1 were not significantly accumulated under mock conditions in atsr1; in fact, ICS1 was slightly less abundant (P = 0.012, Fisher’s lsd). Based on this result, we conclude that CAMTA3 function is required for proper gene induction in response to extracellular ATP. Furthermore, lack of immunity marker gene expression suggests that autoimmunity due to loss of CAMTA3 function is not likely to be responsible for the perturbed ATP-responsive expression.
Figure 9.
Involvement of CAMTA3 in ATP-responsive transcription. Wild-type (WT) seedlings and the atsr1-1 (camta3) mutants were treated with 500 µm ATP or vehicle solution for 30 min. Expression of ATP-responsive genes (A) and autoimmunity marker genes, PR1 and ICS1 (B), in the wild type and atsr1-1 is shown. The expression of each gene was normalized by comparison with the expression of PP2A. Data indicate means, individual replicate values and se (n = 3–4). In a subpanel, columns with different letters are significantly different (P < 0.05, Fisher’s lsd).
DISCUSSION
ATP, a fundamental building block of life on Earth, is an extracellular signaling molecule in animals (Verkhratsky and Burnstock, 2014) and is recently coming to be considered an extracellular signaling molecule in plants as well (Tanaka et al., 2010a). Although the physiological function and signal transduction downstream of plant extracellular ATP recognition remain poorly understood, our study provides several insights from mRNA-seq analysis, as discussed below, and demonstrates that extracellular ATP treatment causes transcriptional reprogramming of defense-related genes that fully depends on the functional P2K1 receptor.
Extracellular ATP Orchestrates Plant Defense Responses Independent of, But in Parallel with, the Classical Defense Hormones
Several studies have indicated that extracellular ATP plays an important role as a DAMP that contributes to successful plant defense against hemibiotrophic and necrotrophic pathogens (Bouwmeester et al., 2011; Balagué et al., 2017; Chen et al., 2017; Tripathi et al., 2018). Plant defense responses depend to varying degrees on signaling of the classical defense hormones salicylate, ethylene, and jasmonate (Glazebrook, 2005; Spoel et al., 2007; Hillmer et al., 2017).
Our transcriptomics experiments demonstrated that more than half (57%, 310 of 543) of ATP-responsive genes required signaling by one or more of the pathways of the classical defense hormones. This result is in line with a recent report in which 65% (5,189 of 7,918) of flg22-responsive transcriptional changes depended on one or more of the pathways of the classical defense hormones (Hillmer et al., 2017). Moreover, our data indicated that GO biological processes for response to jasmonic acid and for salicylic acid-mediated signaling are both enriched in the set of genes responsive to extracellular ATP treatment in the wild type. Indeed, the genes induced include primary signal transduction components of both salicylic acid and jasmonate signaling pathways, such as several JAZ genes, MYC2, JAM1, and NPR3 (Sasaki-Sekimoto et al., 2013; Ding et al., 2018). In addition, there were genes involved in the regulation of biosynthesis of salicylic acid, such as SARD1, PAD4, CBP60G, and CBP60A (Zhou et al., 1998; Zhang et al., 2010), and nearly all of the genes encoding jasmonate biosynthetic enzymes. Although ethylene signaling and biosynthesis were not identified as enriched processes in ATP-responsive genes in the wild type, the ethylene-activated signaling pathway was found to be enriched in OxP2K1. Moreover, several ERFs were induced in the wild type, as was AMINOCYCLOPROPANE CARBOXYLIC ACID SYNTHASE6, which is required for ethylene biosynthesis induced by MAPK6 activation (Liu and Zhang, 2004). Our results suggest one way that extracellular ATP confers resistance against different types of pathogens is by direct enhancement of the downstream signaling pathways activated by these classic defense hormones, with later pathogen challenge providing specificity.
Other evidence supporting the idea that extracellular ATP enhances defensive adaptation nonspecifically is found in the induction of many genes involved in the detection or response to pathogens. These include the induction of 29 genes encoding proteins containing a Toll/IL-1 receptor homology domain or a nucleotide-binding, Apaf1, Resistance, CED4 domain. Proteins containing these domains are associated with the recognition of microbe-derived molecules (i.e. effectors) or with microbe-modified host proteins (Jones et al., 2016). This result suggests that extracellular ATP induces the capacity for recognition of a broad range of danger-associated molecules, regardless of their source.
In addition, a number of ATP-responsive genes (233 of 543) were not disrupted in the coi1, ein2, and npr1 mutant backgrounds; we term these genes putative ATP-specific genes. These genes are highly enriched in defense-related processes, including response to bacteria, response to chitin, and salicylic acid-mediated signaling, and include NPR3, JAZ1, several defense-related ERF and WRKY transcription factors, jasmonate biosynthetic genes, the two PEP receptors, AtPEPR1 and AtPEPR2, and the PEP precursors PROPEP3 and PROPEP4 (Yamaguchi et al., 2010). The enrichment of defense-related genes among ATP-specific genes again highlights the importance of extracellular ATP signaling in fortification against pathogen attack, as has been discussed elsewhere (Tanaka et al., 2014). Our results highlight the role for extracellular ATP as a cue for the adoption of a general defensive posture.
Defense-Related Transcription Factors Play an Important Role in the ATP-Induced Transcriptome
The enrichment in ATP-responsive promoters also highlights the indiscriminate induction of defense platforms. For example, the G-box and W-box are both enriched in the ATP-responsive promoters. The W-box is a salicylic acid- and pathogen elicitor-responsive promoter element bound by WRKY and TGA transcription factors (Katagiri et al., 1989; Rushton et al., 1996; Yu et al., 2001), while the G-box, depending on the promoter context, can act as a jasmonate-inducible element bound by basic helix-loop-helix transcription factors (e.g. MYCs; Fernández-Calvo et al., 2011; Figueroa and Browse, 2012). Another enriched element in the ATP-responsive promoters was ABRE, which has long been known as an abiotic stress and abscisic acid-responsive element bound by bZIP transcription factors such as ABI5 (Guiltinan et al., 1990; Skriver et al., 1991; Uno et al., 2000). Abscisic acid signaling is more recently becoming recognized as important for pathogen defense (Anderson et al., 2004; Adie et al., 2007). The most enriched promoter element in ATP-responsive promoters is the CAM-box (Whalley et al., 2011), bound by CAMTAs that have been implicated in defense against pathogens and insects as well as response to abiotic stress (Bouché et al., 2002; Yang and Poovaiah, 2002; Du et al., 2009; Qiu et al., 2012; Benn et al., 2014). The analysis of promoter element enrichment suggests the ATP-responsive activation of multiple defense-responsive pathways.
Having delineated the importance of COI1 for ATP-responsive gene expression, we examined a subset of ATP-responsive genes in myc2 and the myc2 myc3 myc4 triple mutant. Among five COI1-dependent genes tested by RT-qPCR, four depended on MYC transcription factors. We also examined ATP-specific genes (i.e. expression was independent of COI1, EIN2, and NPR1 functions) and found that most of the tested genes showed a clearly reduced induction by ATP in the myc2 or myc2 myc3 myc4 background. As COI1 controls the activity of these MYC transcription factors by regulating the abundance of the JAZ corepressors (Chini et al., 2007), it is somewhat unexpected that COI1-independent genes (although those are induced by ATP) were dependent on the MYCs for activation, although we can speculate that MYC factors control the activity of transcriptional repressors or that known protein-protein interactions of MYC could explain this phenomenon (Jaspers et al., 2009; Hong et al., 2012; Shin et al., 2012). Indeed, several of the MYC-dependent genes lack the MYC-regulated G-box in their presumptive promoters, so ATP-responsive MYC regulation of these genes is likely indirect.
Since extracellular ATP is known to induce cytosolic calcium elevation in a P2K1-dependent manner, it is exciting to find significant enrichment for the CAM-box-containing sequences. The CAM-box is bound by calcium-regulated CAMTA transcription factors (da Costa e Silva, 1994; Yang and Poovaiah, 2002; Choi et al., 2005; Doherty et al., 2009; Nie et al., 2012; O’Malley et al., 2016), and this element is transcriptionally activated by CAMTA3 in a tobacco (Nicotiana tabacum) transient expression system (Benn et al., 2014). Given that extracellular ATP treatment induces an increase in cytosolic calcium content (Tanaka et al., 2010b), it is attractive to hypothesize that the cytosolic calcium increases upon ATP perception promote CAMTA-mediated transcriptional activation of ATP-responsive genes. Our RT-qPCR analysis in a CAMTA3 loss-of-function mutant, atsr1, demonstrated that most tested genes showed reduced ATP-induced response compared with the wild type. Importantly, the expression of two autoimmune markers, PR1 and ICS1, was not induced in the mutant, suggesting that the autoimmune phenotype of the CAMTA3 mutant is not manifested in the plants under our growth conditions and is unlikely to disturb the ATP response, although the possibility cannot be discounted entirely. Other studies found high expression of the autoimmune marker genes in soil-grown atsr1 mutant plants aged from 3 to 6 weeks (Du et al., 2009; Kim et al., 2017; Lolle et al., 2017), while our axenically grown seedlings were 8 d old, which could explain the difference in immune marker expression. Our study suggests an important role for extracellular ATP in modulating the calcium-dependent cellular response, where the CAMTA-mediated ATP signaling can be responsible for regulating the ATP-specific genes, which contribute to GO for calcium- and calmodulin-binding molecular functions such as calmodulin and calmodulin-like proteins, calcium-dependent protein kinases, EF-hand domain-containing proteins, and putative calcium channels.
CONCLUSION
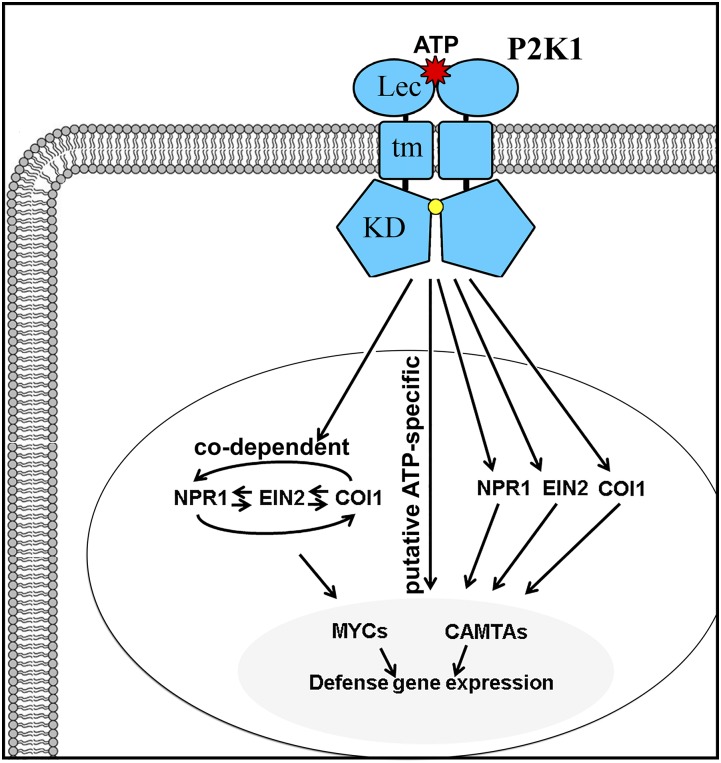
In conclusion, we present a model (Fig. 10) in which ATP binding to P2K1 results in defense gene activation, where more than half of ATP-responsive genes required signaling by one or more of the pathways through EIN2, NPR1, and COI1, but with a more substantial reliance on COI1. MYC transcription factors participate in some level of COI1-independent activation of ATP response. Cytosolic calcium transients induce some proportion of COI1/EIN2/NPR1-independent genes, likely with the participation of CAMTA transcription factors.
Figure 10.
Proposed model of extracellular ATP signaling pathway to the defense-related transcriptome. Once extracellular ATP binds to the P2K1 receptor on the plasma membrane, which induces the autophosphorylation of the receptor and transphosphorylation of downstream targets (Chen et al., 2017), the activated P2K1 receptor results in transcriptional reprogramming of 542 genes. More than half of the ATP-responsive genes required signaling by one or more of the pathways through EIN2, NPR1, and COI1, for example, 154 genes in the COI1-dependent pathway, 21 genes in the NPR1-dependent pathway, six genes in the EIN2-dependent pathway, and 129 genes in the codependent signaling pathway. The other half of the ATP-responsive genes (233 genes) are independent of COI1/NPR1/EIN2-mediated pathways, the putative ATP-specific pathway. Many of the ATP-responsive genes are regulated under the participation of MYC and CAMTA transcription factors. KD, Kinase domain; Lec, lectin domain; tm, transmembrane domain.
MATERIALS AND METHODS
Plant Materials and Treatments
Arabidopsis (Arabidopsis thaliana) OxP2K1, dorn1-3, ein2-1, npr1-3, and coi1-30 were described previously (Cao et al., 1997; Alonso et al., 1999; Yang et al., 2012; Choi et al., 2014). The wild type (Columbia-0; CS70000), coi1-30 (Salk_035548), ein2-1 (CS3071), and npr1-3 (CS3802) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. Seeds of myc2-1 (Niu et al., 2011) and myc2-1 myc3-1 myc4-1 (Major et al., 2017) were provided by John Browse. Seeds of atsr1-1 were from Joe Poovaiah (Du et al., 2009). The genotype of mutant parental lines was verified as follows. For coi1-30, genomic DNA was used as a template in a three-primer PCR using primers shown in Supplemental Table S5, and hemizygous plants were selected. Genomic DNA of parental lines of ein2-1 and npr1-3 was amplified using primers shown in Supplemental Table S5, and the products were sequenced to verify that they carried the causative mutation. The following lines are T-DNA insertional mutants that lack expression of full-length transcripts of the respective mutant genes: coi1-30, dorn1-3, myc2-1, myc2-1 myc3-1 myc4-1, and atsr1-1 (Du et al., 2009; Niu et al., 2011; Yang et al., 2012; Choi et al., 2014; Major et al., 2017). The EIN2 gene in the ein2-1 mutant contains an ethyl methanesulfonate-induced mutation that introduces a premature stop codon prior to the 3′ region encoding 839 amino acids of the C-terminal domain essential for ethylene-induced signal transduction (Alonso et al., 1999). Likewise, the npr1-3 allele contains a premature stop codon in the NPR1 coding sequence that should result in a truncated protein lacking the C-terminal 194 amino acids (Cao et al., 1997).
Seeds were surface sterilized, sown on rectangular plates of one-half-strength Murashige and Skoog medium, 1% (w/v) Suc, 0.05% (w/v) MES, pH 5.7, and 1% (w/v) agar, and stratified in darkness at 4°C for 3 d. Homozygous mutants of coi1-30 were grown with inclusion of 20 µm methyl jasmonate (Bedoukian Research) in the medium and selected based on their obvious long-root phenotype. After seed sterilization and sowing, plates were transferred to a 22°C growth chamber (Conviron) with 12 h of light of 100 to 120 µmol photons m−2 s−1 and grown vertically for 7 d. On day 7, 20 seedlings of each genotype were transferred to six-well plates, each well containing 2 mL of one-half-strength Murashige and Skoog medium as above but lacking agar (liquid medium). All media were sterilized by autoclaving. On day 8, 2 mL of 1 mm ATP in liquid medium (ATP treatment) or 2 mL of liquid medium (mock treatment) was added to each well, and the plates were returned to the growth chamber. After 30 min, the seedlings were removed from the wells, gently blotted dry, and snap frozen in liquid nitrogen. Each genotype and treatment was a true biological replicate sampled on different days.
Preparation of ATP Solution
ATP was purchased from Sigma-Aldrich. Stock solution of ATP was prepared in 2 mm MES buffer at 0.1 m, buffered to pH 5.7, and filter sterilized. The stock solution was stored at −20°C until use.
RNA-Seq Library Construction, Sequencing, and Analysis
After tissue disruption using a bead beater, total RNA was isolated using a quick-RNA miniprep kit with on-column DNase treatment per the manufacturer’s instructions (Zymo Research). Using the TruSeq RNA Sample Preparation Kit v2 (Illumina; catalog no. RS-122-2002), the poly(A) RNA was isolated from 2 μg of the total RNA from each treatment sample using magnetic oligo(dT) beads. Following purification, the mRNA was fragmented by zinc treatment at 94°C for 5 min and reverse transcribed to synthesize first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) and random primers. Second-strand cDNA synthesis was performed, and the products were then subjected to end repair and phosphorylation, and an A base was added to the 3′ end of the blunt phosphorylated DNA fragments. Illumina multiple indexing adapters were ligated to the fragments, as described by Illumina’s TruSeq RNA Sample Preparation V2 Guide. The cDNA fragments flanked by Illumina PE adapters were selected and purified by AMPure XP beads for downstream enrichment. The cDNA fragments were amplified by PCR Primers PE 1.0 and PE 2.0 (Illumina) that anneal to the ends of the adapters, using the PCR program of 30 s at 98°C followed by 15 cycles of 10 s at 98°C, 30 s at 60°C, 30 s at 72°C, and a final elongation step of 5 min at 72°C. The products were purified using AMPure XP beads to create an Illumina paired-end library. Library quality control was performed with a Bioanalyzer DNA 1000 Chip Series II (Agilent). A qPCR method was employed for quantifying libraries in advance of generating clusters. The libraries were diluted to a final concentration of 10 nm. The paired-end libraries were applied for cluster generation at a concentration of 10 pm on a flowcell in a cBOT (Illumina). Sequencing was performed on an Illumina HiSeq 2500 platform by Macrogen with one lane of 12 pooled libraries to generate 100-bp single-end reads. The base-calling and quality-value calculations were performed by the Illumina data-processing pipelines CASAVA v1.8.2 and v1.7.0, respectively. Various quality controls, including removal of reads containing primer/adaptor sequences, trimming of read length, and filtering of high-quality reads based on the score value, were performed using Illumina CASAVA v1.7.0. After trimming and filtering, reads were aligned to the Araport11 annotation of the Arabidopsis genome (Cheng et al., 2017) using TopHat version 2.1.1 (Kim et al., 2013) on the Kamiak High Performance Computing Cluster at Washington State University. Transcript reads were counted on Kamiak using HTSeq-0.7.2 (Anders et al., 2015). Significant differences in gene expression between mock and ATP-treated samples were calculated using edgeR release 3.7 (Robinson et al., 2010) with a cutoff for FDR < 0.05 and absolute FC > 1.5. Enrichment of GO terms in a given set of transcripts was determined by accessing the PANTHER classification system through TAIR (https://www.arabidopsis.org/; Lamesch et al., 2012). The PANTHER overrepresentation test version 13.1 was used (Fisher’s exact test with FDR multiple test correction) by comparison with GO database release 2018-08-09 (Thomas et al., 2003). All gene expression changes after ATP treatment are shown in Supplemental Data Set S1.
RT-qPCR
For RT-qPCR, seedlings were grown and treated, and RNA was prepared as above for the RNA-seq experiments. After isolation of RNA, 1 µg of RNA was used in a 20-µL reverse transcription reaction according to the manufacturer’s instructions (iScript; Bio-Rad). After reverse transcription, the products were diluted 5-fold with water, and 2 µL was used in a 20-µL reaction with a SYBR Green dye/polymerase mix (SsoAdvanced; Bio-Rad) in a CFX96 thermocycler (Bio-Rad). Genes of interest were normalized to the PP2A (AT1G13320) reference gene as described (Czechowski et al., 2005; Rieu and Powers, 2009). Primers used are listed in Supplemental Table S5.
Promoter Analysis
For de novo identification of enriched promoter elements in a given list of genes, promoter sequences 1,000 bases upstream of the transcriptional start site as annotated in Araport11 were accessed via Araport (www.araport.org) and submitted to the oligoanalysis tool (van Helden et al., 1998) using the default settings as implemented in RSAT (Defrance et al., 2008; http://rsat.eead.csic.es/plants). To determine significant enrichment of a particular element in promoters of a list of genes, presence or absence was scored in promoters of the genes of interest and in the Araport11 Arabidopsis nuclear genome using the RSAT dna-pattern tool. Significance was determined using the hypergeometric distribution function in Microsoft Excel 2016.
Accession Numbers
Quality-filtered mRNA sequencing data are available at National Center for Biotechnology Information BioProject PRJNA494862 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA494862).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. High between-replicate concordance of ATP-responsive gene expression in this study: hierarchical clustering.
Supplemental Figure S2. High between-replicate concordance of ATP-responsive gene expression in this study: principal component analysis.
Supplemental Figure S3. Reliability of RNA-seq-based estimates of gene expression in this data set.
Supplemental Figure S4. MapMan enrichment analysis of cellular function overview and biotic stress.
Supplemental Figure S5. GO term enrichment for ATP-responsive genes common to OxP2K1 and the wild type.
Supplemental Figure S6. GO term enrichment for ATP-responsive gene expression in OxP2K1.
Supplemental Table S1. Promoter element analysis for ATP-responsive gene expression in wild-type plants.
Supplemental Table S2. Enrichment of defense-related cis-elements in promoters of ATP-responsive genes in OxP2K1 seedlings.
Supplemental Table S3. Number of ATP-responsive genes in each genotype.
Supplemental Table S4. Promoter element analysis for ATP-specific gene expression.
Supplemental Table S5. Primer sequences used in this study.
Supplemental Dataset S1. Table of gene expression changes after 30 min 500 μM ATP treatment.
Acknowledgments
We thank Dr. John Browse at Washington State University for providing Arabidopsis mutant lines, myc2 and myc2 myc3 myc4, and Dr. Joe Poovaiah at Washington State University for the gift of atsr1-1 seeds. Special thanks to Washington State University Honors College student Misumi Sano for technical assistance with RT-qPCR.
Footnotes
This work was supported by the National Science Foundation (grant no. IOS-1557813) and the USDA National Institute of Food and Agriculture (Hatch Project WNP00008).
Articles can be viewed without a subscription.
References
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagué C, Gouget A, Bouchez O, Souriac C, Haget N, Boutet-Mercey S, Govers F, Roby D, Canut H (2017) The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol Plant Pathol 18: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn G, Wang CQ, Hicks DR, Stein J, Guthrie C, Dehesh K (2014) A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J 80: 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Scharlat A, Snedden W, Bouchez D, Fromm H (2002) A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem 277: 21851–21861 [DOI] [PubMed] [Google Scholar]
- Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F (2011) The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog 7: e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. (2017) Purinergic signalling: Therapeutic developments. Front Pharmacol 8: 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cao Y, Tanaka K, Nguyen CT, Stacey G (2014) Extracellular ATP is a central signaling molecule in plant stress responses. Curr Opin Plant Biol 20: 82–87 [DOI] [PubMed] [Google Scholar]
- Chen D, Cao Y, Li H, Kim D, Ahsan N, Thelen J, Stacey G (2017) Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat Commun 8: 2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chivasa S, Murphy AM, Hamilton JM, Lindsey K, Carr JP, Slabas AR (2009) Extracellular ATP is a regulator of pathogen defence in plants. Plant J 60: 436–448 [DOI] [PubMed] [Google Scholar]
- Cho SH, Nguyen CT, Choi J, Stacey G (2017) Molecular mechanism of plant recognition of extracellular ATP. Adv Exp Med Biol 1051: 233–253 [DOI] [PubMed] [Google Scholar]
- Choi J, Tanaka K, Cao Y, Qi Y, Qiu J, Liang Y, Lee SY, Stacey G (2014) Identification of a plant receptor for extracellular ATP. Science 343: 290–294 [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim MC, Yoo JH, Moon BC, Koo SC, Park BO, Lee JH, Koo YD, Han HJ, Lee SY, et al. (2005) Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J Biol Chem 280: 40820–40831 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa e Silva O. (1994) CG-1, a parsley light-induced DNA-binding protein. Plant Mol Biol 25: 921–924 [DOI] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan WB (2005) ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8: 752–758 [DOI] [PubMed] [Google Scholar]
- Defrance M, Janky R, Sand O, van Helden J (2008) Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. Nat Protoc 3: 1589–1603 [DOI] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467.e15 [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Ali GS, Simons KA, Hou J, Yang T, Reddy AS, Poovaiah BW (2009) Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J 18: 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerink J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Browse J (2012) The Arabidopsis JAZ2 promoter contains a G-box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol 53: 330–343 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR Jr, Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 [DOI] [PubMed] [Google Scholar]
- Heil M. (2009) Damaged-self recognition in plant herbivore defence. Trends Plant Sci 14: 356–363 [DOI] [PubMed] [Google Scholar]
- Heil M, Ibarra-Laclette E, Adame-Álvarez RM, Martínez O, Ramirez-Chávez E, Molina-Torres J, Herrera-Estrella L (2012) How plants sense wounds: Damaged-self recognition is based on plant-derived elicitors and induces octadecanoid signaling. PLoS ONE 7: e30537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer RA, Tsuda K, Rallapalli G, Asai S, Truman W, Papke MD, Sakakibara H, Jones JDG, Myers CL, Katagiri F (2017) The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet 13: e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong GJ, Xue XY, Mao YB, Wang LJ, Chen XY (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, et al. (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Jung C, Zhao P, Seo JS, Mitsuda N, Deng S, Chua NH (2015) PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell 27: 2016–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Lam E, Chua NH (1989) Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340: 727–730 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, An C, Park S, Gilmour SJ, Wang L, Renna L, Brandizzi F, Grumet R, Thomashow MF (2017) CAMTA-mediated regulation of salicylic acid immunity pathway genes in Arabidopsis exposed to low temperature and pathogen infection. Plant Cell 29: 2465–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F (2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant 2: 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle S, Greeff C, Petersen K, Roux M, Jensen MK, Bressendorff S, Rodriguez E, Sømark K, Mundy J, Petersen M (2017) Matching NLR immune receptors to autoimmunity in camta3 mutants using antimorphic NLR alleles. Cell Host Microbe 21: 518–529.e4 [DOI] [PubMed] [Google Scholar]
- Lotze MT, Zeh HJ, Rubartelli A, Sparvero LJ, Amoscato AA, Washburn NR, Devera ME, Liang X, Tör M, Billiar T (2007) The grateful dead: Damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol Rev 220: 60–81 [DOI] [PubMed] [Google Scholar]
- Major IT, Yoshida Y, Campos ML, Kapali G, Xin XF, Sugimoto K, de Oliveira Ferreira D, He SY, Howe GA (2017) Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol 215: 1533–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NTT, Contreras-Moreira B, Castro-Mondragon JA, Santana-Garcia W, Ossio R, Robles-Espinoza CD, Bahin M, Collombet S, Vincens P, Thieffry D, et al. (2018) RSAT 2018: Regulatory sequence analysis tools 20th anniversary. Nucleic Acids Res 46: W209–W214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H, Zhao C, Wu G, Wu Y, Chen Y, Tang D (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158: 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J (2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Xi J, Du L, Suttle JC, Poovaiah BW (2012) Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol Biol 79: 89–99 [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ (2009) Real-time quantitative RT-PCR: Design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K (2013) Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol 163: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ (2012) TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J (1991) Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88: 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CJ, Steinebrunner I, Wang X, Stout SC, Roux SJ (2006) Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in Arabidopsis. Plant Physiol 140: 1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Gilroy S, Jones AM, Stacey G (2010a) Extracellular ATP signaling in plants. Trends Cell Biol 20: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Swanson SJ, Gilroy S, Stacey G (2010b) Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol 154: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Choi J, Cao Y, Stacey G (2014) Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front Plant Sci 5: 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A (2003) PANTHER: A library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, León J, Sánchez-Serrano JJ (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi D, Tanaka K (2018) A crosstalk between extracellular ATP and jasmonate signaling pathways for plant defense. Plant Signal Behav 13: e1432229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi D, Zhang T, Koo AJ, Stacey G, Tanaka K (2018) Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol 176: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, André B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827–842 [DOI] [PubMed] [Google Scholar]
- Verkhratsky A, Burnstock G (2014) Biology of purinergic signalling: Its ancient evolutionary roots, its omnipresence and its multiple functional significance. BioEssays 36: 697–705 [DOI] [PubMed] [Google Scholar]
- Watt WC, Lazarowski ER, Boucher RC (1998) Cystic fibrosis transmembrane regulator-independent release of ATP: Its implications for the regulation of P2Y2 receptors in airway epithelia. J Biol Chem 273: 14053–14058 [DOI] [PubMed] [Google Scholar]
- Weerasinghe RR, Swanson SJ, Okada SF, Garrett MB, Kim SY, Stacey G, Boucher RC, Gilroy S, Jones AM (2009) Touch induces ATP release in Arabidopsis roots that is modulated by the heterotrimeric G-protein complex. FEBS Lett 583: 2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley HJ, Sargeant AW, Steele JF, Lacoere T, Lamb R, Saunders NJ, Knight H, Knight MR (2011) Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 23: 4079–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T (2007) Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics 8: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yuan JS, Ross J, Noel JP, Pichersky E, Chen F (2006) An Arabidopsis thaliana methyltransferase capable of methylating farnesoic acid. Arch Biochem Biophys 448: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu S, Ding P, Wang D, Cheng YT, He J, Gao M, Xu F, Li Y, Zhu Z, et al. (2010) Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc Natl Acad Sci USA 107: 18220–18225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]