Abstract
An overview of common challenges and strategies underlying efforts to reconstruct plant isoprenoid, alkaloid, phenylpropanoid, and polyketide biosynthetic pathways in microbial systems.
Secondary metabolites are broadly defined as natural products synthesized by an organism that are not essential to support growth and life. The plant kingdom manufactures over 200,000 distinct chemical compounds, most of which arise from specialized metabolism. While these compounds play important roles in interspecies competition and defense, many plant natural products have been exploited for use as medicines, fragrances, flavors, nutrients, repellants, and colorants.
Despite this vast chemical diversity, many secondary metabolites are present at very low concentrations in planta, eliminating crop-based manufacturing as a means of attaining these important products. The structural and stereochemical complexity of specialized metabolites hinders most attempts to access these compounds using chemical synthesis. Although native plants can be engineered to accumulate target pathway metabolites (Zhou et al., 2009; Glenn et al., 2013; Lange and Ahkami, 2013; Wilson and Roberts, 2014; Tatsis and O’Connor, 2016), metabolic engineering is technically more challenging in plants than in microbes.
Advancements in synthetic biology have stimulated the synthesis of valuable natural products in tractable laboratory microbes by interfacing plant secondary pathways with core host metabolism. Microbial synthesis overcomes many of the obstacles hindering traditional chemical synthesis and plant metabolic engineering, thus providing an alternative avenue for exploring plant specialized pathways.
This Update provides a brief overview of engineering plant secondary metabolism in microbial systems. We briefly outline biosynthetic pathways mediating formation of the major classes of natural products with an emphasis on high-value terpenoids, alkaloids, phenylpropanoids, and polyketides. We also highlight common themes, strategies, and challenges underlying efforts to reconstruct and engineer these pathways in microbial hosts. We focus chiefly on de novo biosynthetic approaches in which plant specialized metabolites are synthesized directly from sugar feedstocks rather than supplemented precursors or intermediates. Readers are directed to a selection of pioneering supplementation studies within the context of microbially sourced plant natural products (Becker et al., 2003; Kaneko et al., 2003; Yan et al., 2005; Watts et al., 2006; Leonard et al., 2007, 2008; Hawkins and Smolke, 2008; Fossati et al., 2014, 2015).
OVERVIEW OF KEY PLANT SECONDARY METABOLIC PATHWAYS
Terpenoids
Terpenoids (also called isoprenoids) are the largest class of plant secondary metabolites, comprising more than 50,000 natural products (Connolly and Hill, 1991). Whereas the terpene classification refers strictly to hydrocarbons, terpenoids possess a range of chemical functionalities. The central precursors geranyl pyrophosphate (GPP; C10), farnesyl pyrophosphate (FPP; C15), and geranylgeranylpyrophosphate (GGPP; C20) form the structural basis of most higher order terpenoids (Fig. 1). Plant terpene synthases convert these pyrophosphate intermediates to terpenes, which are then functionalized in downstream reactions.
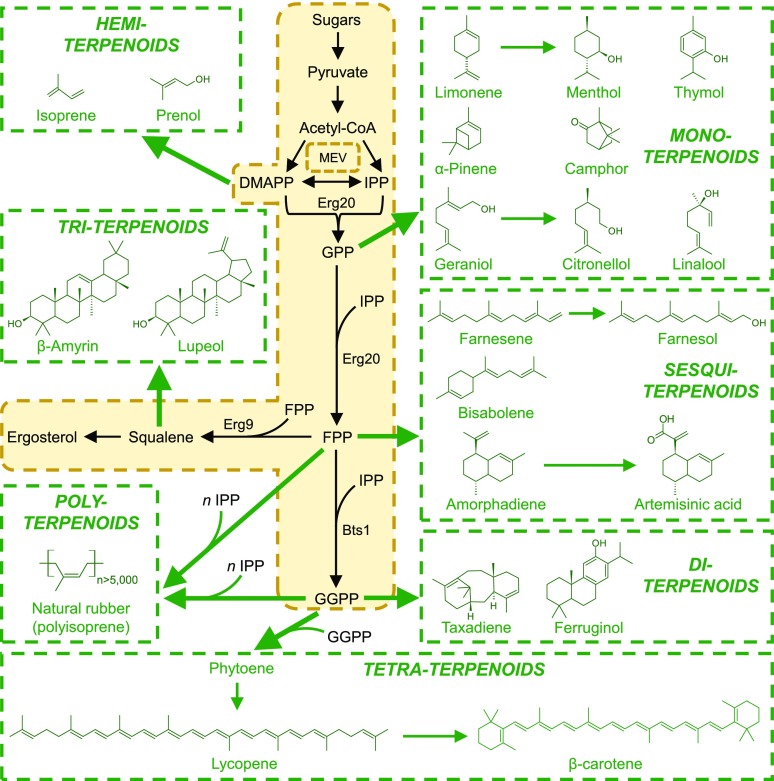
Figure 1.
Interfacing plant terpenoid secondary metabolic pathways with microbial metabolism. Plant secondary metabolic reactions and pathways (green) are shown linked to core microbial metabolism (beige shading; black font). S. cerevisiae is shown as the prospective host species. Key yeast enzymes discussed in the main text are shown. Abbreviations: Bts1, geranylgeranyl diphosphate synthase; DMAPP, dimethylallyl pyrophosphate; Erg9, squalene synthase; Erg20, farnesyl pyrophosphate synthetase; FPP, farnesyl pyrophosphate; GPP, geranyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate; IPP, isopentenyl pyrophosphate; MEV, mevalonate pathway.
GPP forms the backbone of most monoterpenoids (C10), including linear (geraniol and linalool) and cyclic (camphor and eucalyptol) terpenoids, as well as monoterpenes (limonene and pinene). Geraniol can be modified to the pest repellant citronellol, while limonene gives rise to menthol, a flavoring agent and decongestant. GPP also supplies the prenyl group in the biosynthesis of cannabinoids (Ahmed et al., 2015; Vickery et al., 2016), a class of natural products with promising pharmaceutical properties (Aizpurua-Olaizola et al., 2016). Sesquiterpenes (C15) are derived from FPP, itself generated through condensation of GPP and isopentenyl pyrophosphate (IPP) in yeast (Saccharomyces cerevisiae) or directly from IPP and dimethylallyl pyrophosphate units in plants. The sesquiterpene amorphadiene gives rise to the antimalarial drug precursor artemisinic acid. The addition of another IPP unit to FPP in yeast gives rise to GGPP, which forms the scaffold of the diterpenes and diterpenoids (C20), such as taxadiene, a precursor to the taxane family of chemotherapeutics. Condensation of two FPP units yields the linear triterpene squalene (C30), which serves as the universal building block of all sterols, including ergosterol in yeast. In plants, squalene gives rise to a number of pharmacologically active triterpenoids, such as β-amyrin and lupeol. Tetraterpenes (C40) are produced through condensation of two molecules of GGPP, yielding phytoene, the precursor to the carotenoids, such as lycopene and β-carotene. Polyterpenes such as natural rubber (cis-polyisoprene) comprise thousands of isoprene units.
Alkaloids
In the broadest sense, alkaloids are defined as low-molecular-weight metabolites containing heterocyclic (true alkaloids) or exocyclic (protoalkaloids, amines, and polyamines) nitrogen atoms. Approximately 20,000 natural alkaloids are known, many of which exhibit analgesic (morphine), stimulant (caffeine and ephedrine), psychotropic (mescaline and cocaine), antibacterial (sanguinarine), anticancer (vinblastine and vincristine), antitussive (codeine), anti-inflammatory (berberine), antispasmodic (papaverine), or antimalarial (quinine) activities.
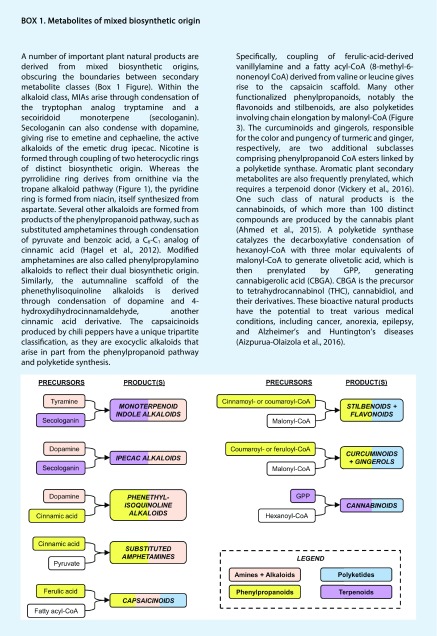
Phe, Tyr, and their derivatives (e.g. phenethylamine, tyramine, and dopamine) are the source of a tremendous number of alkaloids, including the benzylisoquinoline alkaloids (BIAs) and the phenethylisoquinoline alkaloids (Fig. 2). BIAs are a large class of roughly 2,500 metabolites that includes berberine, noscapine, sanguinarine, and morphine. These important medicines are derived from the condensation of dopamine and 4-hydroxyphenylacetaldehyde, both of which are synthesized from Tyr. Dopamine can also condense with derivatives of Phe (4-hydroxydihydrocinnamaldehyde), yielding the phenethylisoquinoline class of alkaloids. Phe and Tyr form the basis of some simpler protoalkaloids and catecholamines, including modified amphetamines (ephedrine and cathinone), dopamine, mescaline, and adrenaline. Owing to the ubiquity of the indole group in nature, Trp also forms the basis of many important alkaloids, such as simple indoles, β-carbolines (serotonin and harmine), and the monoterpenoid indole alkaloids (MIAs).
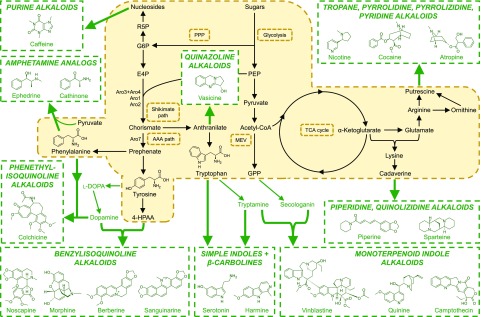
Figure 2.
Interfacing plant alkaloid secondary metabolic pathways with microbial metabolism. S. cerevisiae is shown as the prospective host species. Refer to Figure 1 for abbreviations and color coding. Additional abbreviations: AAA path, aromatic amino acid pathway; Aro1, pentafunctional arom protein; Aro2, chorismate synthase and Flavin reductase; Aro3+Aro4, 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) synthase isoenzymes; Aro7, chorismate mutase; E4P, erythrose 4-phosphate; 4-HPAA, 4-hydroxyphenylacetaldehyde; PPP, pentose phosphate pathway; PEP, phosphoenolpyruvate; R5P, ribose 5-phosphate; TCA cycle, tricarboxylic acid cycle.
MIAs, derived from the condensation of the Trp analog tryptamine and a monoterpenoid (secologanin), are one of the largest and most complex classes of alkaloids, giving rise to more than 3,000 structures. The Trp precursor anthranilate also serves as a precursor to several alkaloid subclasses, including the quinazolines. Beyond the aromatic amino acids, Arg and Orn form the basis of the tropane, pyrrolidine, pyrrolizidine, and pyridine alkaloids, whereas Lys is the precursor to the piperidines and quinolizidines. Nucleosides also form the basis of some alkaloids, such as caffeine and theobromine.
Phenylpropanoids and Polyketides
The phenylpropanoids represent a class of more than 8,000 plant phenolics derived from Tyr and Phe via the phenylpropanoid pathway (Wu and Chappell, 2008). The name phenylpropanoid refers to the distinctive C6-C3 structure of metabolites within this pathway. The key phenylpropanoid branch point intermediate is p-coumaroyl-CoA (Fig. 3), which forms the basis of the flavonoids, stilbenoids, coumarins, lignans, catechins, and aurones. In addition to the C6-C3 skeleton, the phenylpropanoid pathway also diverts at the level of cinnamic or ferulic acid to yield an array of C6-C1 benzoates, such as vanillin, benzaldehyde, and gallic acid (Vogt, 2010; Kallscheuer et al., 2018). Although the stilbenoids and flavonoids originate from the phenylpropanoid pathway, they are elongated by type III plant polyketide synthases (PKSs), underscoring the mixed biosynthetic nature of these specialized metabolites (Box 1). PKSs accept CoA-bound substrates (Yu et al., 2012), most often p-coumaroyl-CoA, although cinnamoyl- and feruloyl-CoA form the basis of some phenylpropanoid polyketides, such as pinosylvin and curcumin (Preisig-Müller et al., 1999; Kita et al., 2008). Once loaded with a starter molecule, malonyl-CoA moieties are incorporated into the growing polyketide chain.
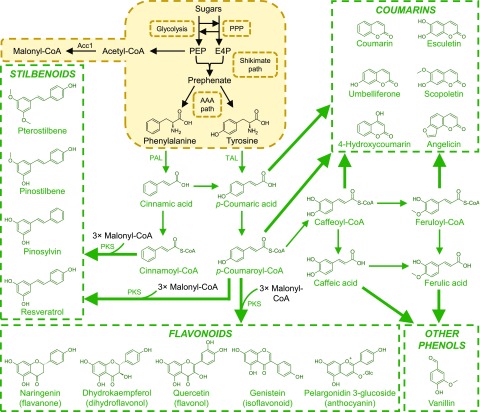
Figure 3.
Interfacing plant phenylpropanoid and polyketide secondary metabolic pathways with microbial metabolism. A selection of natural plant pathways in a prospective S. cerevisiae host is shown. More extensive networks of natural and synthetic phenylpropanoid routes have been shown previously in Wang et al. (2015) and Zhao et al. (2015). Refer to Figures 1 and 2 for abbreviations and color coding. Additional abbreviations: Acc1, acetyl-CoA carboxylase; PAL, Phe ammonia-lyase; PKS, polyketide synthase; TAL, Tyr ammonia-lyase.
Several different polyketide backbones can be produced, though the most heavily targeted for metabolic engineering are the naringenin chalcone and resveratrol scaffolds, which give rise to the respective flavonoid and stilbenoid subclasses (Lussier et al., 2013). These compounds are functionalized in downstream reactions including aromatic hydroxylation, NADP (NADPH)-dependent reduction, O-methylation, and glycosylation. The flavonoids alone encompass more than 6,000 natural products, including chalcones, aurones, catechins, flavanones, flavanols, isoflavonoids, and anthocyanins (Yu et al., 2012). In contrast to the flavonoids and stilbenoids, coumarins are not polyketides, as their lactone ring structure is derived from hydroxylation, isomerization, and lactonization of phenolic acids (cinnamic, p-coumaric, caffeic, or ferulic acid) or the corresponding phenolic acyl-CoA esters (Kai et al., 2008; Yao et al., 2017).
PIONEERING STUDIES IN THE MICROBIAL SYNTHESIS OF PLANT METABOLITES
The most famous application of synthetic biology for the synthesis of valuable plant products is semisynthetic artemisinin, an antimalarial terpenoid natively produced by sweet wormwood (Artemisia annua). Over a period of more than a decade, artemisinic acid production has reached titers of 25 g/L in yeast (Paddon et al., 2013), whereas Escherichia coli has been engineered to produce 27 g/L of the amorphadiene precursor (Tsuruta et al., 2009). The yeast artemisinic acid production strain has been repurposed as a sesquiterpenoid platform, facilitating industrial-scale production of more than 130 g/L farnesene (Meadows et al., 2016). Paclitaxel is another terpenoid medicine and microbes have been engineered to produce up to 1 g/L of the taxadiene scaffold (Ajikumar et al., 2010; Ding et al., 2014; Zhou et al., 2015). Other notable terpenoid pathway reconstructions include limonene, pinene, geraniol, eucalyptol, humulene, β-carophyllene, patchoulol, santalene, and bisabolene (Table 1).
Table 1. Selection of de novo plant secondary pathway reconstructions in microbial systems.
ND, no data available; NR, titer not reported.
| Metabolite class | Metabolite | Titer (mg/L) | Reference(s) | |
|---|---|---|---|---|
| S. cerevisiae | E. coli | |||
| PRECURSORS | Tyr | 1,930a | 55,000 | (Patnaik et al., 2008) |
| p-coumaric acid | 1,930 | 2,510 | (Rodriguez et al., 2015; Jones et al., 2017) | |
| dopamine | 24 | 2,150 | (Nakagawa et al., 2014; DeLoache et al., 2015) | |
| TERPENOIDS | Monoterpenoids | |||
| geraniol | 1,680 | 2,000 | (Liu et al., 2016; Jiang et al., 2017) | |
| pinene | ND | 970 | (Yang et al., 2013) | |
| limonene | 0.49 | 2,700 | (Willrodt et al., 2014; Jongedijk et al., 2015) | |
| eucalyptol | 400 | 653 | (Ignea et al., 2011; Mendez-Perez et al., 2017) | |
| linalool | 0.75 | 505 | (Mendez-Perez et al., 2017; Camesasca et al., 2018) | |
| Sesquiterpenoids | ||||
| β-farnesene | 130,000 | 8,700 | (Meadows et al., 2016; You et al., 2017) | |
| bisabolene | 5,200 | 912 | (Peralta-Yahya et al., 2011; Özaydin et al., 2013) | |
| α-humulene | 1,300 | ND | (Zhang et al., 2018b) | |
| β-caryophyllene | ND | 1,520 | (Yang et al., 2016) | |
| amorphadiene | 37,000 | 27,400 | (Tsuruta et al., 2009; Westfall et al., 2012) | |
| artemisinic acid | 25,000 | ND | (Paddon et al., 2013) | |
| santalene | 163 | ND | (Tippmann et al., 2016) | |
| patchoulol | 40.9 | ND | (Albertsen et al., 2011) | |
| Diterpenoids | ||||
| jolkinol C | 800 | ND | (Wong et al., 2018) | |
| taxadiene | 72.8 | 1,020 | (Ajikumar et al., 2010; Ding et al., 2014) | |
| Triterpenoids | ||||
| β-amyrin | 108.1 | NR | (Takemura et al., 2017; Zhu et al., 2018) | |
| Tetraterpenoids | ||||
| β-carotene | 162.1 | 3,200 | (Yang and Guo, 2014; Wang et al., 2016) | |
| lycopene | 1,650 | 3,520 | (Sun et al., 2014; Chen et al., 2016) | |
| ALKALOIDS | Benzylisoquinoline alkaloids | |||
| (S)-reticuline | 0.082 | 160 | (DeLoache et al., 2015; Matsumura et al., 2018) | |
| thebaine | 0.0064 | 2.1 | (Galanie et al., 2015; Nakagawa et al., 2016) | |
| hydrocodone | 0.0003 | 0.36 | (Galanie et al., 2015; Nakagawa et al., 2016) | |
| noscapine | 2.2 | ND | (Li et al., 2018) | |
| Monoterpene indole alkaloids | ||||
| strictosidine | 0.5 | ND | (Brown et al., 2015) | |
| PHENYLPROPANOIDS | Stilbenoids | |||
| resveratrol | >5,000 | 51.8 | (Katz et al., 2013; Yang et al., 2018) | |
| pinostilbene | 5.52 | ∼2.5 | (Kang et al., 2014; Li et al., 2016b) | |
| pterostilbene | 34.93 | 33.6 | (Li et al., 2016b; Heo et al., 2017) | |
| pinosylvin | 130.02 | 281 | (Katz et al., 2008; Wu et al., 2017) | |
| Flavonoids | ||||
| naringenin | >200 | 103.8 | (Lehka et al., 2017; Yang et al., 2018) | |
| scutellarin | 108 | ND | (Liu et al., 2018) | |
| genistein | 7.7 | ND | (Trantas et al., 2009) | |
| pinocembrin | 2.6 | 525.8 | (Wu et al., 2016b; Eichenberger et al., 2017) | |
| quercetin | 20.38 | ND | (Rodriguez et al., 2017) | |
| kaempferol | 66.29 | 57b | (Yang et al., 2014; Duan et al., 2017) | |
| dihydrokaempferol | 44 | ND | (Levisson et al., 2018) | |
| pelargonidin-3-O-glucoside | 0.02 | 9.5 | (Jones et al., 2017; Levisson et al., 2018) | |
| phloretin | 42.7 | ND | (Eichenberger et al., 2017) | |
| Coumarins | ||||
| umbelliferone | ND | 66.1 | (Yang et al., 2015a) | |
| esculetin | ND | 61.4 | (Yang et al., 2015a) | |
| 4-hydroxycoumarin | ND | 483.1 | (Lin et al., 2013) | |
Titer reported for the downstream metabolite p-coumaric acid.
Titer reported for kaempferol 3-O-rhamnoside.
Because geraniol forms half of the MIA scaffold, strategies successfully used to produce monoterpenoids were exploited to synthesize MIA intermediates in yeast, such as nepetalactol (Campbell et al., 2016; Billingsley et al., 2017) and strictosidine (Brown et al., 2015). These efforts have yet to be extended to the de novo synthesis of functionalized MIAs in a microbial system, although impressive partial pathway reconstructions have been reported (Qu et al., 2015). The BIA class has also recently experienced several breakthrough microbial pathway reconstitutions. As a result of successes in resolving upstream pathways involving conversion of Tyr to dopamine, de novo formation of (S)-reticuline was demonstrated for the first time (Nakagawa et al., 2011; DeLoache et al., 2015). This pathway was later extended for de novo biosynthesis of morphinan alkaloids in E. coli and yeast (Galanie et al., 2015; Nakagawa et al., 2016), as well as noscapine in yeast (Li et al., 2018). Titers of (S)-reticuline have recently been increased to 160 mg/L in E. coli (Matsumura et al., 2018). Stilbenoids have been the target of several microbial synthesis efforts and the de novo titer of resveratrol has reached 5 g/L in yeast (Katz et al., 2013; Li et al., 2016b). Other stilbenoids produced in microbes include pinostilbene, pterostilbene, pinosylvin, and piceatannol (Table 1). Similarly, the branch point flavonoid naringenin has been produced from glucose at titers above 200 mg/L (Lehka et al., 2017). The flavonoid pathway has been diversified in E. coli and yeast to yield the dihydrochalcone phloretin and its derivatives, the flavanone liquiritigenin, and the flavonols kaempferol, quercetin, and fisetin, among others (Table 1). This pathway was extended to produce anthocyanin pigments using E. coli polycultures (Jones et al., 2017) and a single engineered yeast strain (Eichenberger et al., 2018; Levisson et al., 2018).
Not all natural product syntheses involve straightforward maximization of product titer, because some applications require careful tuning of product concentrations. One such study involved combinatorial engineering of industrial brewing yeasts (S. cerevisiae) to generate strains with a diverse range of geraniol and linalool production profiles, culminating in the formulation of beers with a more desirable hops flavor compared with commercial varieties (Denby et al., 2018). Studies such as these highlight the wide range of applications afforded through the manipulation of plant metabolic pathways in microbial systems.
ELUCIDATION OF PLANT METABOLIC PATHWAYS
In Planta
Traditionally, the elucidation of metabolic pathways in source plants has been the driving force and often limiting factor of microbial biosynthesis. Owing to the complexity of plant metabolism, gaps in pathways are common, particularly within downstream reactions mediating the formation of functionalized products. Pathway gaps in the context of microbial synthesis refer to a lack of enzyme identification, as the biochemical nature of metabolic transformations is typically known.
To bypass gaps in enzyme elucidation, pathway intermediates are often supplied to cells exogenously, granted they are available and an uptake system exists. Although enzyme gaps have plagued many natural product pathways, several important pathways have been completed. While it was known that production of the nepetalactol backbone within the MIA pathway involved reductive cyclization of 10-oxogeranial, the enzyme mediating this transformation was finally discovered in 2012 (Geu-Flores et al., 2012). Critical to this success was the search of source plant transcriptomes for redox enzymes coexpressed with MIA pathway transcripts. Other key enzymes within the early MIA pathway were discovered in 2014 (Miettinen et al., 2014), prompting the synthesis of the key branch point MIA strictosidine in yeast soon thereafter (Brown et al., 2015).
The MIA pathway leading to vinblastine was fully elucidated (Caputi et al., 2018); thus, it is only a matter of time before the de novo synthesis of this potent anticancer medicine is achieved in a microbial system. Similarly, the final enzymes involved in the formation of substituted amphetamines, such as ephedrine and pseudoephedrine (Hagel et al., 2012), were also identified (Morris et al., 2018), providing rationale for their synthesis in a microbial system. The opium poppy (Papaver somniferum) enzyme responsible for the stereochemical inversion of (S)- to (R)-reticuline eluded identification for many years, limiting morphine pathway reconstructions to upstream [Glc to (S)-reticuline] and downstream [(R)-reticuline to morphine] modules. Eventual identification of the fusion enzyme by three research groups led to the reconstruction of the de novo morphinan pathway within a microbial host (Galanie et al., 2015). Candidate epimerase genes were identified using transcriptome database mining, and candidates were silenced in opium poppy plants sequenced from mutants that accumulate (S)-reticuline, or cloned in S. cerevisiae for functional analysis (Farrow et al., 2015; Galanie et al., 2015; Winzer et al., 2015). While this sequence of events permeates the field of microbial synthesis, in which pathway elucidation in planta precedes pathway reconstruction in microbes, a paradigm has arisen in response to this laborious and time-consuming process.
In Microbes
Due to the declining cost of DNA synthesis, rapidly advancing bioinformatics tools, and expanding omics databases, it has become possible to resolve single and multienzyme gaps within a heterologous microbial host. Using this synthetic biology approach, libraries of candidate gap-filling enzymes are compiled by querying plant transcriptome databases, such as 1000 Plants (Matasci et al., 2014) or PhytoMetaSyn (Xiao et al., 2013) Projects. The corresponding genes are then codon-optimized, synthesized, and expressed in a laboratory microbe. This workflow facilitates the resolution of enzyme gaps in plant metabolic pathways without having to obtain physical plant material or cDNAs (Narcross et al., 2016a). In this regard, synthetic biologists do not need to wait for gap-filling enzymes to be isolated and characterized from natural sources to reconstitute target pathways in a microbial species. One of the groups involved in identifying the aforementioned reticuline epimerase gene employed this strategy by querying plant transcriptome databases and integrating synthetic genes into a yeast strain harboring a downstream module for thebaine synthesis (Galanie et al., 2015). An ambitious combinatorial variation of this technique was attempted to fill a seven-enzyme gap within the vinblastine MIA pathway (Casini et al., 2018). While Casini et al. (2018) were unable to resolve this intricate pathway, they succeeded in building 74 strains, each possessing a distinct combination of the seven gene candidates. This study illustrates the immense potential of synthetic biology for elucidating and reconstructing complex biochemical pathways in microbial systems.
HOST SELECTION
Host selection is a fundamental facet of microbial synthesis and a key determinant of pathway performance. Because it is virtually impossible to forecast the ideal host for synthesizing a target metabolite, numerous factors must be weighed, including access to a complete genome sequence and the availability of genetic tools. Precursor availability is perhaps the most important criterion of host selection, as an abundant precursor pool, such as acetyl-CoA or shikimate, sidesteps the need to rewire core metabolism. Host selection is further complicated by the existence of phenotypically distinct strains within a given species, for instance the E. coli K and B lineages or the S288C and CEN.PK strains of S. cerevisiae. In a dramatic instance, use of E. coli DH1 improved oxidation of amorphadiene by a factor of 1000 relative to related strains (Chang et al., 2007). Screening of E. coli lineages for polyketide synthesis has revealed not only strain-specific differences in baseline polyketide production but also differences in response to the same genetic manipulations (Yang et al., 2018). Due to differences in amino acid biosynthesis between S. cerevisiae (Canelas et al., 2010), production of vanillin-β-glucoside was significantly higher in S288C than in CEN.PK (Strucko et al., 2015). Similar yeast-specific differences were observed for the synthesis of the polyketide triacetic acid lactone (Saunders et al., 2015). These studies highlight the often overlooked task of screening strains to identify optimal producers prior to comprehensively engineering a production strain.
Workhorses and Specialized Hosts
The overwhelming majority of microbial syntheses have employed E. coli or S. cerevisiae owing to their ease of genetic and metabolic manipulation, a comprehensive understanding of their genetics, metabolism, and physiology, and their highly active central metabolic pathways. Several critical differences between these industry-proven hosts inform many microbial synthesis campaigns (Box 2).
Recently, the diversity of microbial hosts exploited for the synthesis of plant natural products has expanded dramatically. Some microorganisms naturally synthesize aromatic amino acids at levels well above those of E. coli and S. cerevisiae, indicating a major potential of these hosts for the production of alkaloids and phenylpropanoids. Of particular importance is Corynebacterium glutamicum, an industrial bacterium with a tremendous capacity to synthesize glucose and Lys, as well as aromatic amino acids (Azuma et al., 1993; Ikeda et al., 1993). Lactococcus lactis is another industry-proven bacterium with a demonstrated capacity to synthesize plant terpenoids and phenylpropanoids (Gaspar et al., 2013; Dudnik et al., 2018). The yeast Scheffersomyces stipitis has gained interest for its ability to use C5 sugars, suggesting that this organism has a more active pentose phosphate pathway than does S. cerevisiae. Indeed, the highest shikimate titer reported to date in yeast was achieved using S. stipites (Gao et al., 2017).
Within the polyketide class of natural products, bacteria from the genus Streptomyces are major natural producers, providing rationale for diverting these native pathways to plant-derived polyketides (Baltz, 2010). The oleaginous yeast Yarrowia lipolytica has also been demonstrated to be a promising host for terpenoids, polyketides, and other compounds derived from acetyl-CoA (Abdel-Mawgoud et al., 2018). Other hosts are desirable for superior growth characteristics, such as Pichia pastoris, a yeast that attains higher cell densities than S. cerevisiae (Wriessnegger et al., 2014).
RECONSTITUTION AND OPTIMIZATION OF PLANT PATHWAYS IN MICROBIAL SYSTEMS
Pathway Assembly and Delivery
Pathway assembly refers to the design and organization of genetic parts required to reconstruct heterologous pathways in microbes. This endeavor begins by selecting and synthesizing gene variants responsible for mediating the synthesis of the target product. The selected candidates are placed within transcriptional units containing a promoter and terminator. Traditionally, heterologous genes and metabolic pathways have been delivered and expressed from plasmids. While this technique has certain merits, such as facilitating the rapid screening of prospective hosts (Casini et al., 2018) and easily assessing the performance of pathway modules (Fossati et al., 2014), access to efficient genome editing tools has led to the adoption of chromosomal expression systems (Horwitz et al., 2015; Jakočiūnas et al., 2015). Furthermore, plasmid-borne expression leads to significant variability in gene expression and copy number between cells (Ryan et al., 2014).
With respect to transcriptional control, most pathways are assembled by placing all genes under control of constitutive promoters. For instance, amorphadiene and artemisinic acid production strains were constructed using Gal induction, which was later converted to constitutive expression without affecting production or fitness (Westfall et al., 2012; Paddon et al., 2013). Because even “constitutive” promoters exhibit distinct transcriptional profiles, which vary based on carbon source or medium formulation (Reider Apel et al., 2017), major efforts have focused on comprehensively characterizing both promoter and terminator elements (Yamanishi et al., 2013; Lee et al., 2015). These toolkits facilitate the exploration of vast pathway design spaces comprised of a staggering number of possible gene and expression combinations.
Product titer, rate, and yield following initial pathway reconstitution are often very poor, necessitating many design-build-test engineering cycles to reach commercially viable levels. Historically, this process lasts six to eight years and costs more than $50 million (Nielsen and Keasling, 2016). These metrics are declining at a dramatic rate as a result of the standardization of synthetic biology and the advent of high-throughput strain engineering facilities (“biofoundries”), which aim to miniaturize and automate strain construction workflows (Chao et al., 2017; Casini et al., 2018).
Expression of Plant Genes in Microbial Species
The functional expression of plant genes in microbial species poses technical challenges. Many plant enzymes possess localization signals for targeting to specific organelles, such as the chloroplast. Implementation of these plant genes in microbial hosts typically leads to poor gene expression or insolubility of the corresponding protein (Williams et al., 1998). Most terpene synthases are plastid-localized and thus N-terminal truncation is an effective strategy for improving activity (Alonso-Gutierrez et al., 2013; Zhao et al., 2016). Production of BIAs is also improved through N-terminal truncation of norcoclaurine synthase (Li et al., 2016a). While software tools exist for predicting optimal signal peptide cleavage sites (Petersen et al., 2011), assessing various-sized truncations affords a range of improvements (Nishihachijo et al., 2014; Jiang et al., 2017).
Perhaps the most challenging facet of synthesizing plant-derived metabolites in microbial hosts is the functional expression of cytochrome P450 (CYP) enzymes. This obstacle provides much of the rationale for using eukaryotic expression systems over bacteria (Box 2). CYPs are membrane-bound enzymes that carry out many of the complex functionalizations in secondary metabolic pathways, including hydroxylations, double bond epoxidations, and dealkylations (Meunier et al., 2004). Because CYPs mediate oxidation reactions, catalysis is coupled to reduction of NADPH, which in turn requires a cytochrome P450 reductase (CPR) partner to shuttle electrons between NADPH and CYP. Additional components can also be involved, such as cytochrome b5 and a cytochrome b5 reductase (Porter, 2002; Paddon et al., 2013; Li et al., 2016b).
Some species of plant contain hundreds of CYPs and multiple CPRs, leading to immense challenges in optimizing heterologous CYP-CPR pairings. Although yeast contains a native CPR, most plant CYPs exhibit greater activity when paired with CPRs from the same plant species (Fossati et al., 2014). Production of highly functionalized secondary metabolites, such as paclitaxel and hydrocodone, involves numerous CYP-catalyzed reactions, requiring the coordinated expression of multiple CPR genes. To overcome this hurdle, most microbial synthesis studies employ a “one-size-fits-all” strategy by pairing multiple CYPs with a single CPR. CYP and CPR expression must be carefully balanced to ensure efficient shuttling of electrons and avoid the production of toxic reactive oxygen species. Optimization of CYP-CPR pairing was paramount to the success of the semisynthetic artemisinin process (Paddon et al., 2013) and enabled production of 500 mg/L oxygenated taxadiene in E. coli (Biggs et al., 2016).
The most efficient natural CYP is a primitive bacterial CYP-CPR fusion from Bacillus megaterium (Munro et al., 2002). This architecture has been exploited for the expression of plant CYPs in E. coli, in which the N-terminal membrane-binding domain is cleaved or modified for more efficient insertion within bacterial membranes (Hausjell et al., 2018). Conversely, it has been shown that a direct CYP-CPR fusion in E. coli was less active than carefully balanced expression of both components for oxygenated taxadiene synthesis (Biggs et al., 2016). Biggs et al. (2016) suggest that because plant CYPs and CPRs do not interact in a 1:1 ratio, a direct fusion approach is not appropriate.
Precursor Supply
Engineering microbial systems for the production of phenylpropanoids and most alkaloids demands an abundant supply of aromatic amino acid precursors (Figs. 2 and 3). Tremendous strides have been made in engineering platform strains producing high levels of Tyr, Phe, Trp, and their derivatives (Table 1).
Deregulating host precursor pathways is regarded as the chief hurdle to the production of heterologous metabolites (Nielsen and Keasling, 2016). The yeast shikimate and aromatic amino acid pathways are subject to multiple layers of transcriptional and posttranscriptional control. The entry point to the shikimate pathway involving condensation of E4P and PEP is catalyzed by 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase, for which two isoenzymes exist in yeast (Aro3 and Aro4). Allosteric inhibition by Tyr can be overcome in both enzymes using analogous mutations (Aro3K222L and Aro4K229L; Luttik et al., 2008; Brückner et al., 2018). Similarly, chorismate mutase (Aro7) is feedback-inhibited by Tyr, prompting discovery of the Aro7G141S allele for deregulating Tyr biosynthesis (Luttik et al., 2008). Implementation of Aro4K229L and Aro7G141S in the same host leads to a 200-fold increase in extracellular levels of aromatic compounds.
The terpenoid branch of secondary metabolism has also been the focal point of engineering precursor supply. The rate-limiting step of the mevalonate pathway catalyzed by HMG-CoA reductase (HMGR) can be improved by implementing a truncated mutant (tHMGR). Owing to the importance of this enzymatic step, studies achieving high-level terpenoid or MIA production have incorporated up to four gene copies of tHMGR (Paddon et al., 2013; Brown et al., 2015).
The production of GPP-derived monoterpenoids poses a unique engineering challenge pertaining to the dual nature of the yeast Erg20 protein. This enzyme catalyzes the synthesis of GPP from IPP and dimethylallyl pyrophosphate, as well as the subsequent condensation of GPP and an additional unit of IPP to give FPP (Fig. 1). Owing to the central role of FPP in ergosterol biosynthesis, Erg20 is an essential enzyme, signifying that its FPP synthase activity cannot be abolished. Instead, Erg20 variants with reduced GPP-binding properties have been described (Chambon et al., 1990) and exploited for the production of various monoterpenoids and MIAs (Brown et al., 2015; Campbell et al., 2016). Relatedly, the essential squalene synthase enzyme (Erg9) generates squalene from two FPP units, thus placing constraints on the production of all terpenoids deriving from FPP (Fig. 1).
The most fundamental means of circumventing host regulation is replacing native precursor pathways with a heterologous counterpart. This approach was used in early efforts to produce amorphadiene in E. coli, wherein the native MEP pathway was replaced by the yeast mevalonate pathway (Martin et al., 2003). Reconstructing the yeast pathway within a bacterial operon eliminates both E. coli and S. cerevisiae modes of regulation, facilitating constitutive production of terpenoids.
Acetyl-CoA plays a pivotal role in the biosynthesis of terpenoids, polyketides, phenylpropanoids, and cannabinoids (Figs. 1 and 3), and its overproduction has also been a target for improvement. In yeast, multiple acetyl-CoA synthesis pathways exist in different subcellular compartments that acetyl-CoA cannot directly cross (Strijbis and Distel, 2010). This provides options for strain engineering based on the pathway that is most suited for a particular heterologous product (van Rossum et al., 2016). Alternatively, heterologous cytosolic acetyl-CoA pathways are of great interest (Kozak et al., 2014a, 2014b, 2016; Zhang et al., 2015; Cardenas and Da Silva, 2016; Meadows et al., 2016; Rodriguez et al., 2016). The highest reported titer of a heterologous product derived from cytosolic acetyl-CoA is the synthesis of 130 g/L of the sesquiterpene farnesene under fermentative conditions (Meadows et al., 2016). This approach used multiple alternative cytosolic acetyl-CoA pathways to overproduce farnesene.
In prokaryotes, all routes to acetyl-CoA are equally accessible, reducing the necessity for alternative pathways, although they have been demonstrated (Wang et al., 2018). However, buildup of acetyl-CoA results in its conversion to acetate, which is an unproductive carbon sink. In E. coli, native acetate assimilation enzymes can be overexpressed to convert acetate back to acetyl-CoA, resulting in improved flavanone biosynthesis (Leonard et al., 2007; Zha et al., 2009). Gene knockouts have been identified that prevent acetate synthesis and improve heterologous production without reducing cellular fitness (Zha et al., 2009; Liu et al., 2017a). Eliminating pathway bottlenecks to improve flux through acetyl-CoA can also reduce acetate overflow (Li et al., 2015; King et al., 2017; Wang et al., 2018). Relatively little attention has been placed on the availability of free Coenzyme A to accept increased carbon flux. Supplementation of the Coenzyme A precursor pantothenate can improve the yield of heterologous products requiring acetyl-CoA. Several studies have demonstrated that overexpression of the rate-limiting step of Coenzyme A synthesis, pantothenate kinase, improves product synthesis 2-fold (Fowler et al., 2009; Schadeweg and Boles, 2016; Liu et al., 2017b).
Phenylpropanoid, polyketide, and cannabinoid synthesis require malonyl-CoA, which is used for fatty acid biosynthesis in E. coli and S. cerevisiae. In these organisms, fatty acid synthesis is tightly regulated (Davis et al., 2000). Improving synthesis and reducing off-target consumption of malonyl-CoA are the major areas for overproduction. Acetyl-CoA is converted to malonyl-CoA by acetyl-CoA carboxylase (Acc), and improvements in this activity have been achieved by overexpression (Zha et al., 2009; Xu et al., 2011; Yang et al., 2018), ablating posttranslational modification sites (Choi and Da Silva, 2014; Shi et al., 2014; Chen et al., 2018), and improving the availability of its biotin cofactor (Leonard et al., 2007). Acc-independent malonyl-CoA synthesis has also been demonstrated (Leonard et al., 2008), though the pathway requires supplemented malonate. Reducing the diversion of malonyl-CoA to essential pathways is a challenge, because fatty acids are required for biomass production. In yeast, knockout of genes regulating fatty acid biosynthesis improved titers of malonyl-CoA-derived products (Zha et al., 2009; Chen et al., 2017), yet the strain was auxotrophic for inositol. More promisingly, biomass accumulation prior to conditional gene knockdown has been demonstrated to improve polyketide titers in E. coli (Yang et al., 2015b, 2018; Liang et al., 2016; Wu et al., 2016a).
As a ubiquitous cofactor and the source of reductant for plant CYPs, NADPH is an important engineering target for the synthesis of natural products. A proven strategy for enhancing NADPH availability is the inactivation of native NADPH-consuming reactions. A caveat of this approach pertains to the targeting of enzymes involved in central carbon metabolism, such as Glc-6-phosphate isomerase and phosphoenolpyruvate carboxylase in E. coli (Chemler et al., 2010) or Glc-6-phosphate dehydrogenase in S. cerevisiae (Gold et al., 2015). Glucose dehydrogenase is a promising candidate in both species (Asadollahi et al., 2009; Chemler et al., 2010). Inactivation of these central targets often has deleterious effects on strain fitness, including the generation of amino acid auxotrophy (Gold et al., 2015), demanding a delicate balancing of core pathways and sophisticated metabolic models for predicting combinatorial modifications (Chemler et al., 2010).
The counter approach of up-regulating NADPH-generating enzymes is a more viable avenue for engineering NADPH supply. In yeast, these enzymes include prephenate dehydrogenase (Tyr-1) and cytosolic aldehyde dehydrogenase (Li et al., 2018). Curiously, both overexpression and deletion of Glc-6-phosphate dehydrogenase have been shown to enhance formation of Tyr products (Gold et al., 2015; Li et al., 2018), underscoring the complexity of yeast central metabolism and redox homeostasis.
Improving Flux to Heterologous Pathways
Flux improvement involves iterative rounds of strain engineering with the aim of elevating product titer, rate, and yield. Efforts to improve flux through a target pathway begin with the identification of rate-limiting enzymatic conversions, which is diagnosed by monitoring levels of pathway intermediates. The most fundamental means of relieving metabolic bottlenecks is to increase levels of rate-limiting enzymes by increasing gene expression or copy number. In addition to high-level expression of the heterologous pathway, the entire eight-gene yeast pathway from acetyl-CoA to FPP was overexpressed in the industrial artemisinic acid strain (Paddon et al., 2013).
Poor pathway flux can also arise from the diversion of pathway intermediates to off-target routes, either by host activities or promiscuous plant enzymes. Because phenylpropanoid biosynthesis derives from Phe or Tyr, the yeast pathway for aromatic amino acid degradation (Ehrlich pathway) is typically inactivated through deletion of amino acid decarboxylase genes (ARO10 and PDC5; Rodriguez et al., 2015). Yeast BIA biosynthesis involves a more intricate balancing of Tyr pathways, as the dopamine and 4-hydroxyphenylacetaldehyde precursors are derived from its biosynthesis and degradation, respectively (Narcross et al., 2016b). The physiological fate of 4-HPAA in yeast involves conversion to tyrosol or 4-hydroxyphenylacetate, which can be mediated by more than 20 candidate enzymes (Hazelwood et al., 2008; DeLoache et al., 2015). These enzymes have also been implicated in the irreversible reduction of aldehyde intermediates within the MIA pathway (Billingsley et al., 2017), suggesting that any pathway possessing carbonyl intermediates is subject to diversion by host activities. In a similar scenario, a yeast reductase was shown to divert p-coumaroyl-CoA within the phenylpropanoid pathway to an unwanted side product (Lehka et al., 2017).
Pathway flux can also be diverted to off-target routes by heterologous pathway enzymes. O- and N-methyltransferases within the core BIA pathway have been shown to exhibit a wide substrate range (Frick et al., 2001; Ounaroon et al., 2003; Fossati et al., 2014), which was circumvented by screening of plant homologs for variants exhibiting a more stringent substrate specificity (Narcross et al., 2016a). In addition to catalyzing the reductive cyclization of 10-oxogeranial, iridoid synthase within the MIA pathway was found to exhibit activity on other intermediates (Campbell et al., 2016). In this case, enzyme activity was higher on the unwanted substrates than the physiological substrate. Enzyme promiscuity may be avoided in cocultures, in which a pathway is split between strains such that promiscuous enzymes are sequestered from pathway intermediates (Nakagawa et al., 2014). Loss of intermediates is also mitigated through enzyme fusion, as in the cases of geraniol synthase or patchoulol synthase through fusion to yeast farnesyl diphosphate synthase (Erg20; Albertsen et al., 2011; Jiang et al., 2017). Fusions have the potential to facilitate more efficient substrate channeling by maintaining close physical proximity of sequential pathway enzymes, thus limiting competition from host enzymes. In addition to terpenoid systems, fusion of resveratrol biosynthetic enzymes facilitated a 15-fold improvement in titer in engineered S. cerevisiae (Zhang et al., 2006). Enzyme fusions have the added potential to improve protein stability or solubility, as demonstrated through fusions of taxadiene synthase with the soluble E. coli maltose binding protein, yielding a 25-fold improvement in taxadiene titer (Reider Apel et al., 2017). Fusions with maltose-binding protein also improved production of α-ionone up to 50-fold (Zhang et al., 2018a).
EXPANDING THE PLANT SECONDARY METABOLIC SPACE IN MICROBIAL SYSTEMS
An emerging application of synthetic biology aims to exploit microbial systems for drug discovery. In this manner, plant enzyme promiscuity and combinatorial biochemistry are exploited to synthesize structural scaffolds and functionalizations not found in nature.
A common chemical modification within the pharmaceutical industry is halogenation, in which fluoro or chloro moieties are introduced to drug candidates to modify the physicochemical properties of the compound. Because many natural products derive from amino acids, halogenated analogs can be supplemented to engineered microbes leading to the formation of halogenated products. This strategy was used to feed yeast fluoro- or chloro-Tyr, resulting in the synthesis of halogenated BIA pathway intermediates (Li et al., 2018). Halogenated end products were not detected, suggesting a more stringent substrate specificity of downstream BIA enzymes. Moreover, this method relies on the supplementation of costly amino acid analogs and could be overcome by engineering de novo production of halogenated BIAs.
O-sulfated reticuline has been produced de novo by implementing a human sulphotransferase into E. coli engineered for reticuline biosynthesis (Matsumura et al., 2018). Promiscuity of BIA pathway enzymes has also been exploited for the production of innovative scaffolds (such as disubstituted- and spiro-tetrahydroisoquinolines) by harnessing the capacity of norcoclaurine synthase to accept an exceptional range of carbonyl substrates (Lichman et al., 2017). Again, this strategy relies on the supplementation of costly substrates, in this case carbonyl species that are not synthesized by microorganisms. Moving these in vitro assays to robust de novo biosynthetic processes presents a formidable challenge.
CONCLUSION
Microbial synthesis provides a fresh avenue for the production of natural plant pharmaceuticals, fragrances, nutrients, and colorants. Although this field is in its infancy and faces many challenges (see Outstanding Questions), the wealth of successes in recent years has solidified microbial biosynthetic production as a viable alternative to natural product extraction and total chemical synthesis. The elucidation of increasingly complex plant secondary pathways continues to drive the reconstruction of longer and more challenging routes in microbial systems. Indeed, the discovery of missing enzymes in several highly sought-after plant-specialized pathways has primed a number of untapped natural products for de novo biosynthesis in a microbial species. Several microbially derived natural products have reached commercialization, and these successes will continue to pave the way for opportunities. The exploitation of microbial systems for drug discovery promises to deliver important new structural scaffolds and chemical functionalizations that have thus far not been observed in nature.
Acknowledgments
Many thanks to NSERC-Industrial Biocatalysis Network (IBN) and NSERC Discovery for financial support. M.P. is grateful for a NSERC postdoctoral fellowship. L.N. is appreciative of support from a FQRNT DE Doctoral Research Scholarship for Foreign Students. V.M. is thankful to Concordia University for their support of his role as Research Chair.
Footnotes
This work was supported financially by a Natural Sciences and Engineering Research Council of Canada (NSERC) Industrial Biocatalysis Network (IBN) grant, and a NSERC Discovery grant. M.P. was aided by a NSERC postdoctoral fellowship, and L.N. was assisted by a FQRNT DE Doctoral Research Scholarship for Foreign Students. V.M. is supported by Concordia University in his position as Research Chair.
Articles can be viewed without a subscription.
References
- Abdel-Mawgoud AM, Markham KA, Palmer CM, Liu N, Stephanopoulos G, Alper HS (2018) Metabolic engineering in the host Yarrowia lipolytica. Metab Eng 50: 192–208 [DOI] [PubMed] [Google Scholar]
- Ahmed SA, Ross SA, Slade D, Radwan MM, Khan IA, ElSohly MA (2015) Minor oxygenated cannabinoids from high potency Cannabis sativa L. Phytochemistry 117: 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizpurua-Olaizola O, Soydaner U, Öztürk E, Schibano D, Simsir Y, Navarro P, Etxebarria N, Usobiaga A (2016) Evolution of the cannabinoid and terpene content during the growth of Cannabis sativa plants from different chemotypes. J Nat Prod 79: 324–331 [DOI] [PubMed] [Google Scholar]
- Ajikumar PK, Xiao W-H, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330: 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, Brix S, Nielsen J, Mortensen UH (2011) Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol 77: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS (2013) Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng 19: 33–41 [DOI] [PubMed] [Google Scholar]
- Asadollahi MA, Maury J, Patil KR, Schalk M, Clark A, Nielsen J (2009) Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng 11: 328–334 [DOI] [PubMed] [Google Scholar]
- Azuma S, Tsunekawa H, Okabe M, Okamoto R, Aiba S (1993) Hyper-production of L-trytophan via fermentation with crystallization. Appl Microbiol Biotechnol 39: 471–476 [Google Scholar]
- Baltz RH. (2010) Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37: 759–772 [DOI] [PubMed] [Google Scholar]
- Becker JV, Armstrong GO, van der Merwe MJ, Lambrechts MG, Vivier MA, Pretorius IS (2003) Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res 4: 79–85 [DOI] [PubMed] [Google Scholar]
- Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, Ajikumar PK (2016) Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 113: 3209–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingsley JM, DeNicola AB, Barber JS, Tang M-C, Horecka J, Chu A, Garg NK, Tang Y (2017) Engineering the biocatalytic selectivity of iridoid production in Saccharomyces cerevisiae. Metab Eng 44: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Clastre M, Courdavault V, O’Connor SE (2015) De novo production of the plant-derived alkaloid strictosidine in yeast. Proc Natl Acad Sci USA 112: 3205–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner C, Oreb M, Kunze G, Boles E, Tripp J (2018) An expanded enzyme toolbox for production of cis, cis-muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisiae. FEMS Yeast Res 18: foy017. [DOI] [PubMed] [Google Scholar]
- Camesasca L, Minteguiaga M, Fariña L, Salzman V, Aguilar PS, Gaggero C, Carrau F (2018) Overproduction of isoprenoids by Saccharomyces cerevisiae in a synthetic grape juice medium in the absence of plant genes. Int J Food Microbiol 282: 42–48 [DOI] [PubMed] [Google Scholar]
- Campbell A, Bauchart P, Gold ND, Zhu Y, De Luca V, Martin VJ (2016) Engineering of a nepetalactol-producing platform strain of Saccharomyces cerevisiae for the production of plant seco-iridoids. ACS Synth Biol 5: 405–414 [DOI] [PubMed] [Google Scholar]
- Canelas AB, Harrison N, Fazio A, Zhang J, Pitkänen J-P, van den Brink J, Bakker BM, Bogner L, Bouwman J, Castrillo JI, et al. (2010) Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat Commun 1: 145. [DOI] [PubMed] [Google Scholar]
- Caputi L, Franke J, Farrow SC, Chung K, Payne RME, Nguyen T-D, Dang T-TT, Soares Teto Carqueijeiro I, Koudounas K, Dugé de Bernonville T, et al. (2018) Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 360: 1235–1239 [DOI] [PubMed] [Google Scholar]
- Cardenas J, Da Silva NA (2016) Engineering cofactor and transport mechanisms in Saccharomyces cerevisiae for enhanced acetyl-CoA and polyketide biosynthesis. Metab Eng 36: 80–89 [DOI] [PubMed] [Google Scholar]
- Casini A, Chang F-Y, Eluere R, King AM, Young EM, Dudley QM, Karim A, Pratt K, Bristol C, Forget A, et al. (2018) A pressure test to make 10 molecules in 90 days: External evaluation of methods to engineer biology. J Am Chem Soc 140: 4302–4316 [DOI] [PubMed] [Google Scholar]
- Chambon C, Ladeveze V, Oulmouden A, Servouse M, Karst F (1990) Isolation and properties of yeast mutants affected in farnesyl diphosphate synthetase. Curr Genet 18: 41–46 [DOI] [PubMed] [Google Scholar]
- Chang MC, Eachus RA, Trieu W, Ro D-K, Keasling JD (2007) Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol 3: 274–277 [DOI] [PubMed] [Google Scholar]
- Chao R, Mishra S, Si T, Zhao H (2017) Engineering biological systems using automated biofoundries. Metab Eng 42: 98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemler JA, Fowler ZL, McHugh KP, Koffas MA (2010) Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng 12: 96–104 [DOI] [PubMed] [Google Scholar]
- Chen X, Yang X, Shen Y, Hou J, Bao X (2017) Increasing malonyl-CoA derived product through controlling the transcription regulators of phospholipid synthesis in Saccharomyces cerevisiae. ACS Synth Biol 6: 905–912 [DOI] [PubMed] [Google Scholar]
- Chen X, Yang X, Shen Y, Hou J, Bao X (2018) Screening phosphorylation site mutations in yeast acetyl-CoA carboxylase using malonyl-CoA sensor to improve malonyl-CoA-derived product. Front Microbiol 9: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xiao W, Wang Y, Liu H, Li X, Yuan Y (2016) Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb Cell Fact 15: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Da Silva NA (2014) Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J Biotechnol 187: 56–59 [DOI] [PubMed] [Google Scholar]
- Connolly JD, Hill RA (1991) Dictionary of terpenoids. New York: Chapman and Hall [Google Scholar]
- Davis MS, Solbiati J, Cronan JE Jr (2000) Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem 275: 28593–28598 [DOI] [PubMed] [Google Scholar]
- DeLoache WC, Russ ZN, Narcross L, Gonzales AM, Martin VJ, Dueber JE (2015) An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat Chem Biol 11: 465–471 [DOI] [PubMed] [Google Scholar]
- Denby CM, Li RA, Vu VT, Costello Z, Lin W, Chan LJG, Williams J, Donaldson B, Bamforth CW, Petzold CJ, et al. (2018) Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat Commun 9: 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding MZ, Yan HF, Li LF, Zhai F, Shang LQ, Yin Z, Yuan YJ (2014) Biosynthesis of taxadiene in Saccharomyces cerevisiae: Selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy. PLoS One 9: e109348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Ding W, Liu X, Cheng X, Cai J, Hua E, Jiang H (2017) Biosynthesis and engineering of kaempferol in Saccharomyces cerevisiae. Microb Cell Fact 16: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudnik A, Almeida AF, Andrade R, Avila B, Bañados P, Barbay D, Bassard J-E, Benkoulouche M, Bott M, Braga A et al. (2018) BacHBerry: BACterial hosts for production of bioactive phenolics from bERRY fruits. Phytochem Rev 17: 291–326 [Google Scholar]
- Eichenberger M, Lehka BJ, Folly C, Fischer D, Martens S, Simón E, Naesby M (2017) Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab Eng 39: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger M, Hansson A, Fischer D, Dürr L, Naesby M (2018) De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res 18: foy046. [DOI] [PubMed] [Google Scholar]
- Farrow SC, Hagel JM, Beaudoin GA, Burns DC, Facchini PJ (2015) Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat Chem Biol 11: 728–732 [DOI] [PubMed] [Google Scholar]
- Fossati E, Ekins A, Narcross L, Zhu Y, Falgueyret J-P, Beaudoin GA, Facchini PJ, Martin VJ (2014) Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat Commun 5: 3283. [DOI] [PubMed] [Google Scholar]
- Fossati E, Narcross L, Ekins A, Falgueyret J-P, Martin VJ (2015) Synthesis of morphinan alkaloids in Saccharomyces cerevisiae. PLoS One 10: e0124459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler ZL, Gikandi WW, Koffas MA (2009) Increased malonyl coenzyme A biosynthesis by tuning the Escherichia coli metabolic network and its application to flavanone production. Appl Environ Microbiol 75: 5831–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick S, Ounaroon A, Kutchan TM (2001) Combinatorial biochemistry in plants: The case of O-methyltransferases. Phytochemistry 56: 1–4 [DOI] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Cao M, Suástegui M, Walker J, Rodriguez Quiroz N, Wu Y, Tribby D, Okerlund A, Stanley L, Shanks JV, et al. (2017) Innovating a nonconventional yeast platform for producing shikimate as the building block of high-value aromatics. ACS Synth Biol 6: 29–38 [DOI] [PubMed] [Google Scholar]
- Gaspar P, Carvalho AL, Vinga S, Santos H, Neves AR (2013) From physiology to systems metabolic engineering for the production of biochemicals by lactic acid bacteria. Biotechnol Adv 31: 764–788 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui Y, O’Connor SE (2012) An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature 492: 138–142 [DOI] [PubMed] [Google Scholar]
- Glenn WS, Runguphan W, O’Connor SE (2013) Recent progress in the metabolic engineering of alkaloids in plant systems. Curr Opin Biotechnol 24: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold ND, Gowen CM, Lussier F-X, Cautha SC, Mahadevan R, Martin VJ (2015) Metabolic engineering of a tyrosine-overproducing yeast platform using targeted metabolomics. Microb Cell Fact 14: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Krizevski R, Marsolais F, Lewinsohn E, Facchini PJ (2012) Biosynthesis of amphetamine analogs in plants. Trends Plant Sci 17: 404–412 [DOI] [PubMed] [Google Scholar]
- Hausjell J, Halbwirth H, Spadiut O (2018) Recombinant production of eukaryotic cytochrome P450s in microbial cell factories. Biosci Rep 38: BSR20171290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins KM, Smolke CD (2008) Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat Chem Biol 4: 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelwood LA, Daran J-M, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74: 2259–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo KT, Kang S-Y, Hong Y-S (2017) De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis. Microb Cell Fact 16: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz AA, Walter JM, Schubert MG, Kung SH, Hawkins K, Platt DM, Hernday AD, Mahatdejkul-Meadows T, Szeto W, Chandran SS, et al. (2015) Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst 1: 88–96 [DOI] [PubMed] [Google Scholar]
- Ignea C, Cvetkovic I, Loupassaki S, Kefalas P, Johnson CB, Kampranis SC, Makris AM (2011) Improving yeast strains using recyclable integration cassettes, for the production of plant terpenoids. Microb Cell Fact 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ozaki A, Katsumata R (1993) Phenylalanine production by metabolically engineered Corynebacterium glutamicum with the pheA gene of Escherichia coli. Appl Microbiol Biotechnol 39: 318–323 [DOI] [PubMed] [Google Scholar]
- Jakočiūnas T, Bonde I, Herrgård M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28: 213–222 [DOI] [PubMed] [Google Scholar]
- Jiang G-Z, Yao M-D, Wang Y, Zhou L, Song T-Q, Liu H, Xiao W-H, Yuan Y-J (2017) Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metab Eng 41: 57–66 [DOI] [PubMed] [Google Scholar]
- Jones J, Vernacchio V, Collins S, Shirke A, Xiu Y, Englaender J, Cress B, McCutcheon C, Linhardt R, Gross R (2017) Complete biosynthesis of anthocyanins using E. coli polycultures. American Society for Microbiology 8: e00621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongedijk E, Cankar K, Ranzijn J, van der Krol S, Bouwmeester H, Beekwilder J (2015) Capturing of the monoterpene olefin limonene produced in Saccharomyces cerevisiae. Yeast 32: 159–171 [DOI] [PubMed] [Google Scholar]
- Kai K, Mizutani M, Kawamura N, Yamamoto R, Tamai M, Yamaguchi H, Sakata K, Shimizu B (2008) Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J 55: 989–999 [DOI] [PubMed] [Google Scholar]
- Kallscheuer N, Classen T, Drepper T, Marienhagen J (2018) Production of plant metabolites with applications in the food industry using engineered microorganisms. Curr Opin Biotechnol 56: 7–17 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hwang EI, Ohnishi Y, Horinouchi S (2003) Heterologous production of flavanones in Escherichia coli: Potential for combinatorial biosynthesis of flavonoids in bacteria. J Ind Microbiol Biotechnol 30: 456–461 [DOI] [PubMed] [Google Scholar]
- Kang S-Y, Lee JK, Choi O, Kim CY, Jang J-H, Hwang BY, Hong Y-S (2014) Biosynthesis of methylated resveratrol analogs through the construction of an artificial biosynthetic pathway in E. coli. BMC Biotechnol 14: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Forster J, David H, Schmidt HP, Sendelius M, Bjorn SP, Durhuus TT (2008) Metabolically engineered cells for the production of pinosylvin. World Intellectual Property Organization Patent WO 2008/009728 A1. Patent Cooperation Treaty Gazette, PCT/EP/2007/057484. Google Patents, https://patentimages.storage.googleapis.com/73/83/96/dbecb827d803ab/WO2008009728A1.pdf
- Katz MP, Durhuus T, Smits HP, Förster J (2013) Production of metabolites. U.S. Patent US 20,130,209,613 A1. Patent Cooperation Treaty Gazette, PCT/EP/2011/058447. Google Patents, https://patentimages.storage.googleapis.com/03/22/9c/963bb4b4beea1f/US20130209613A1.pdf
- King JR, Woolston BM, Stephanopoulos G (2017) Designing a new entry point into isoprenoid metabolism by exploiting fructose-6-phosphate aldolase side reactivity of Escherichia coli. ACS Synth Biol 6: 1416–1426 [DOI] [PubMed] [Google Scholar]
- Kita T, Imai S, Sawada H, Kumagai H, Seto H (2008) The biosynthetic pathway of curcuminoid in turmeric (Curcuma longa) as revealed by 13C-labeled precursors. Biosci Biotechnol Biochem 72: 1789–1798 [DOI] [PubMed] [Google Scholar]
- Kozak BU, van Rossum HM, Benjamin KR, Wu L, Daran J-MG, Pronk JT, van Maris AJ (2014a) Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab Eng 21: 46–59 [DOI] [PubMed] [Google Scholar]
- Kozak BU, van Rossum HM, Luttik MA, Akeroyd M, Benjamin KR, Wu L, de Vries S, Daran J-M, Pronk JT, van Maris AJ (2014b) Engineering acetyl coenzyme A supply: Functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. MBio 5: e01696–e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak BU, van Rossum HM, Niemeijer MS, van Dijk M, Benjamin K, Wu L, Daran J-MG, Pronk JT, van Maris AJ (2016) Replacement of the initial steps of ethanol metabolism in Saccharomyces cerevisiae by ATP-independent acetylating acetaldehyde dehydrogenase. FEMS Yeast Res 16: fow006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ahkami A (2013) Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes—Current status and future opportunities. Plant Biotechnol J 11: 169–196 [DOI] [PubMed] [Google Scholar]
- Lee ME, DeLoache WC, Cervantes B, Dueber JE (2015) A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth Biol 4: 975–986 [DOI] [PubMed] [Google Scholar]
- Lehka BJ, Eichenberger M, Bjørn-Yoshimoto WE, Vanegas KG, Buijs N, Jensen NB, Dyekjær JD, Jenssen H, Simon E, Naesby M (2017) Improving heterologous production of phenylpropanoids in Saccharomyces cerevisiae by tackling an unwanted side reaction of Tsc13, an endogenous double-bond reductase. FEMS Yeast Res 17: fox004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Lim K-H, Saw P-N, Koffas MA (2007) Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol 73: 3877–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Fowler ZL, Li Z, Lim C-G, Lim K-H, Koffas MA (2008) Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm 5: 257–265 [DOI] [PubMed] [Google Scholar]
- Levisson M, Patinios C, Hein S, de Groot PA, Daran J-M, Hall RD, Martens S, Beekwilder J (2018) Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb Cell Fact 17: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ying L-Q, Zhang S-S, Chen N, Liu W-F, Tao Y (2015) Modification of targets related to the Entner-Doudoroff/pentose phosphate pathway route for methyl-D-erythritol 4-phosphate-dependent carotenoid biosynthesis in Escherichia coli. Microb Cell Fact 14: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee E-J, Chang L, Facchini PJ (2016a) Genes encoding norcoclaurine synthase occur as tandem fusions in the Papaveraceae. Sci Rep 6: 39256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schneider K, Kristensen M, Borodina I, Nielsen J (2016b) Engineering yeast for high-level production of stilbenoid antioxidants. Sci Rep 6: 36827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Thodey K, Trenchard I, Cravens A, Smolke CD (2018) Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc Natl Acad Sci USA 115: E3922–E3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JL, Guo LQ, Lin JF, He ZQ, Cai FJ, Chen JF (2016) A novel process for obtaining pinosylvin using combinatorial bioengineering in Escherichia coli. World J Microbiol Biotechnol 32: 102. [DOI] [PubMed] [Google Scholar]
- Lichman BR, Zhao J, Hailes HC, Ward JM (2017) Enzyme catalysed Pictet-Spengler formation of chiral 1,1′-disubstituted- and spiro-tetrahydroisoquinolines. Nat Commun 8: 14883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Shen X, Yuan Q, Yan Y (2013) Microbial biosynthesis of the anticoagulant precursor 4-hydroxycoumarin. Nat Commun 4: 2603. [DOI] [PubMed] [Google Scholar]
- Liu M, Ding Y, Chen H, Zhao Z, Liu H, Xian M, Zhao G (2017a) Improving the production of acetyl-CoA-derived chemicals in Escherichia coli BL21(DE3) through iclR and arcA deletion. BMC Microbiol 17: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xu X, Zhang R, Cheng T, Cao Y, Li X, Guo J, Liu H, Xian M (2016) Engineering Escherichia coli for high-yield geraniol production with biotransformation of geranyl acetate to geraniol under fed-batch culture. Biotechnol Biofuels 9: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhang B, Jiang R (2017b) Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol Biofuels 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cheng J, Zhang G, Ding W, Duan L, Yang J, Kui L, Cheng X, Ruan J, Fan W, et al. (2018) Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun 9: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier F-X, Colatriano D, Wiltshire Z, Page JE, Martin VJ (2013) Engineering microbes for plant polyketide biosynthesis. Comput Struct Biotechnol J 3: e201210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik MA, Vuralhan Z, Suir E, Braus GH, Pronk JT, Daran JM (2008) Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: Quantification of metabolic impact. Metab Eng 10: 141–153 [DOI] [PubMed] [Google Scholar]
- Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21: 796–802 [DOI] [PubMed] [Google Scholar]
- Matasci N, Hung L-H, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. (2014) Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura E, Nakagawa A, Tomabechi Y, Ikushiro S, Sakaki T, Katayama T, Yamamoto K, Kumagai H, Sato F, Minami H (2018) Microbial production of novel sulphated alkaloids for drug discovery. Sci Rep 8: 7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows AL, Hawkins KM, Tsegaye Y, Antipov E, Kim Y, Raetz L, Dahl RH, Tai A, Mahatdejkul-Meadows T, Xu L, et al. (2016) Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537: 694–697 [DOI] [PubMed] [Google Scholar]
- Mendez-Perez D, Alonso-Gutierrez J, Hu Q, Molinas M, Baidoo EEK, Wang G, Chan LJG, Adams PD, Petzold CJ, Keasling JD, et al. (2017) Production of jet fuel precursor monoterpenoids from engineered Escherichia coli. Biotechnol Bioeng 114: 1703–1712 [DOI] [PubMed] [Google Scholar]
- Meunier B, de Visser SP, Shaik S (2004) Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev 104: 3947–3980 [DOI] [PubMed] [Google Scholar]
- Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, et al. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Groves RA, Hagel JM, Facchini PJ (2018) An N-methyltransferase from Ephedra sinica catalyzing the formation of ephedrine and pseudoephedrine enables microbial phenylalkylamine production. J Biol Chem 293:13364-13376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro AW, Leys DG, McLean KJ, Marshall KR, Ost TW, Daff S, Miles CS, Chapman SK, Lysek DA, Moser CC, et al. (2002) P450 BM3: The very model of a modern flavocytochrome. Trends Biochem Sci 27: 250–257 [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Minami H, Kim J-S, Koyanagi T, Katayama T, Sato F, Kumagai H (2011) A bacterial platform for fermentative production of plant alkaloids. Nat Commun 2: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Matsuzaki C, Matsumura E, Koyanagi T, Katayama T, Yamamoto K, Sato F, Kumagai H, Minami H (2014) (R,S)-tetrahydropapaveroline production by stepwise fermentation using engineered Escherichia coli. Sci Rep 4: 6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Matsumura E, Koyanagi T, Katayama T, Kawano N, Yoshimatsu K, Yamamoto K, Kumagai H, Sato F, Minami H (2016) Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat Commun 7: 10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narcross L, Bourgeois L, Fossati E, Burton E, Martin VJ (2016a) Mining enzyme diversity of transcriptome libraries through DNA synthesis for benzylisoquinoline alkaloid pathway optimization in yeast. ACS Synth Biol 5: 1505–1518 [DOI] [PubMed] [Google Scholar]
- Narcross L, Fossati E, Bourgeois L, Dueber JE, Martin VJJ (2016b) Microbial factories for the production of benzylisoquinoline alkaloids. Trends Biotechnol 34: 228–241 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Keasling JD (2016) Engineering cellular metabolism. Cell 164: 1185–1197 [DOI] [PubMed] [Google Scholar]
- Nishihachijo M, Hirai Y, Kawano S, Nishiyama A, Minami H, Katayama T, Yasohara Y, Sato F, Kumagai H (2014) Asymmetric synthesis of tetrahydroisoquinolines by enzymatic Pictet-Spengler reaction. Biosci Biotechnol Biochem 78: 701–707 [DOI] [PubMed] [Google Scholar]
- Ounaroon A, Decker G, Schmidt J, Lottspeich F, Kutchan TM (2003) (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum —cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J 36: 808–819 [DOI] [PubMed] [Google Scholar]
- Özaydin B, Burd H, Lee TS, Keasling JD (2013) Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab Eng 15: 174–183 [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532 [DOI] [PubMed] [Google Scholar]
- Patnaik R, Zolandz RR, Green DA, Kraynie DF (2008) L-tyrosine production by recombinant Escherichia coli: Fermentation optimization and recovery. Biotechnol Bioeng 99: 741–752 [DOI] [PubMed] [Google Scholar]
- Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS (2011) Identification and microbial production of a terpene-based advanced biofuel. Nat Commun 2: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Porter TD. (2002) The roles of cytochrome b5 in cytochrome P450 reactions. J Biochem Mol Toxicol 16: 311–316 [DOI] [PubMed] [Google Scholar]
- Preisig-Müller R, Schwekendiek A, Brehm I, Reif H-J, Kindl H (1999) Characterization of a pine multigene family containing elicitor-responsive stilbene synthase genes. Plant Mol Biol 39: 221–229 [DOI] [PubMed] [Google Scholar]
- Qu Y, Easson ML, Froese J, Simionescu R, Hudlicky T, De Luca V (2015) Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc Natl Acad Sci USA 112: 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider Apel A, d’Espaux L, Wehrs M, Sachs D, Li RA, Tong GJ, Garber M, Nnadi O, Zhuang W, Hillson NJ,et al. (2017) A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res 45: 496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Kildegaard KR, Li M, Borodina I, Nielsen J (2015) Establishment of a yeast platform strain for production of p-coumaric acid through metabolic engineering of aromatic amino acid biosynthesis. Metab Eng 31: 181–188 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Strucko T, Stahlhut SG, Kristensen M, Svenssen DK, Forster J, Nielsen J, Borodina I (2017) Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour Technol 245(Pt B): 1645–1654 [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Denby CM, Van Vu T, Baidoo EE, Wang G, Keasling JD (2016) ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microb Cell Fact 15: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, et al. (2014) Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife 3: e03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LP, Bowman MJ, Mertens JA, Da Silva NA, Hector RE (2015) Triacetic acid lactone production in industrial Saccharomyces yeast strains. J Ind Microbiol Biotechnol 42: 711–721 [DOI] [PubMed] [Google Scholar]
- Schadeweg V, Boles E (2016) N-butanol production in Saccharomyces cerevisiae is limited by the availability of coenzyme A and cytosolic acetyl-CoA. Biotechnol Biofuels 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Chen Y, Siewers V, Nielsen J (2014) Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. MBio 5: e01130–e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K, Distel B (2010) Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot Cell 9: 1809–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strucko T, Magdenoska O, Mortensen UH (2015) Benchmarking two commonly used Saccharomyces cerevisiae strains for heterologous vanillin-β-glucoside production. Metab Eng Commun 2: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Miao L, Li Q, Dai G, Lu F, Liu T, Zhang X, Ma Y (2014) Production of lycopene by metabolically-engineered Escherichia coli. Biotechnol Lett 36: 1515–1522 [DOI] [PubMed] [Google Scholar]
- Takemura M, Tanaka R, Misawa N (2017) Pathway engineering for the production of β-amyrin and cycloartenol in Escherichia coli—a method to biosynthesize plant-derived triterpene skeletons in E. coli. Appl Microbiol Biotechnol 101: 6615–6625 [DOI] [PubMed] [Google Scholar]
- Tatsis EC, O’Connor SE (2016) New developments in engineering plant metabolic pathways. Curr Opin Biotechnol 42: 126–132 [DOI] [PubMed] [Google Scholar]
- Tippmann S, Scalcinati G, Siewers V, Nielsen J (2016) Production of farnesene and santalene by Saccharomyces cerevisiae using fed-batch cultivations with RQ-controlled feed. Biotechnol Bioeng 113: 72–81 [DOI] [PubMed] [Google Scholar]
- Trantas E, Panopoulos N, Ververidis F (2009) Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng 11: 355–366 [DOI] [PubMed] [Google Scholar]
- Tsuruta H, Paddon CJ, Eng D, Lenihan JR, Horning T, Anthony LC, Regentin R, Keasling JD, Renninger NS, Newman JD (2009) High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS One 4: e4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum HM, Kozak BU, Pronk JT, van Maris AJA (2016) Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: Pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab Eng 36: 99–115 [DOI] [PubMed] [Google Scholar]
- Vickery CR, La Clair JJ, Burkart MD, Noel JP (2016) Harvesting the biosynthetic machineries that cultivate a variety of indispensable plant natural products. Curr Opin Chem Biol 31: 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wang Q, Xu J, Sun Z, Luan Y, Li Y, Wang J, Liang Q, Qi Q (2018) Engineering an in vivo EP-bifido pathway in Escherichia coli for high-yield acetyl-CoA generation with low CO2 emission. Metab Eng 51: 79–87 [DOI] [PubMed] [Google Scholar]
- Wang R-z, Pan C-h, Wang Y, Xiao W-h, Yuan Y-j (2016) Design and construction of high β-carotene producing Saccharomyces cerevisiae. Zhongguo Shengwu Gongcheng Zazhi 36: 83–91 [Google Scholar]
- Wang S, Zhang S, Xiao A, Rasmussen M, Skidmore C, Zhan J (2015) Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab Eng 29: 153–159 [DOI] [PubMed] [Google Scholar]
- Watts KT, Lee PC, Schmidt-Dannert C (2006) Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, et al. (2012) Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA 109: E111–E118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DC, McGarvey DJ, Katahira EJ, Croteau R (1998) Truncation of limonene synthase preprotein provides a fully active “pseudomature” form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37: 12213–12220 [DOI] [PubMed] [Google Scholar]
- Willrodt C, David C, Cornelissen S, Bühler B, Julsing MK, Schmid A (2014) Engineering the productivity of recombinant Escherichia coli for limonene formation from glycerol in minimal media. Biotechnol J 9: 1000–1012 [DOI] [PubMed] [Google Scholar]
- Wilson SA, Roberts SC (2014) Metabolic engineering approaches for production of biochemicals in food and medicinal plants. Curr Opin Biotechnol 26: 174–182 [DOI] [PubMed] [Google Scholar]
- Winzer T, Kern M, King AJ, Larson TR, Teodor RI, Donninger SL, Li Y, Dowle AA, Cartwright J, Bates R, et al. (2015) Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349: 309–312 [DOI] [PubMed] [Google Scholar]
- Wong J, de Rond T, d’Espaux L, van der Horst C, Dev I, Rios-Solis L, Kirby J, Scheller H, Keasling J (2018) High-titer production of lathyrane diterpenoids from sugar by engineered Saccharomyces cerevisiae. Metab Eng 45: 142–148 [DOI] [PubMed] [Google Scholar]
- Wriessnegger T, Augustin P, Engleder M, Leitner E, Müller M, Kaluzna I, Schürmann M, Mink D, Zellnig G, Schwab H, et al. (2014) Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab Eng 24: 18–29 [DOI] [PubMed] [Google Scholar]
- Wu S, Chappell J (2008) Metabolic engineering of natural products in plants; tools of the trade and challenges for the future. Curr Opin Biotechnol 19: 145–152 [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang X, Dong M, Zhou J (2016a) Stepwise modular pathway engineering of Escherichia coli for efficient one-step production of (2S)-pinocembrin. J Biotechnol 231: 183–192 [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang X, Zhou J, Dong M (2016b) Efficient biosynthesis of (2S)-pinocembrin from d-glucose by integrating engineering central metabolic pathways with a pH-shift control strategy. Bioresour Technol 218: 999–1007 [DOI] [PubMed] [Google Scholar]
- Wu J, Zhang X, Zhu Y, Tan Q, He J, Dong M (2017) Rational modular design of metabolic network for efficient production of plant polyphenol pinosylvin. Sci Rep 7: 1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Zhang Y, Chen X, Lee E-J, Barber CJ, Chakrabarty R, Desgagné-Penix I, Haslam TM, Kim Y-B, Liu E, et al. (2013) Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol 166: 122–134 [DOI] [PubMed] [Google Scholar]
- Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA (2011) Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng 13: 578–587 [DOI] [PubMed] [Google Scholar]
- Yamanishi M, Ito Y, Kintaka R, Imamura C, Katahira S, Ikeuchi A, Moriya H, Matsuyama T (2013) A genome-wide activity assessment of terminator regions in Saccharomyces cerevisiae provides a ″terminatome″ toolbox. ACS Synth Biol 2: 337–347 [DOI] [PubMed] [Google Scholar]
- Yan Y, Kohli A, Koffas MA (2005) Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl Environ Microbiol 71: 5610–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Guo L (2014) Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb Cell Fact 13: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Kim WJ, Yoo SM, Choi JH, Ha SH, Lee MH, Lee SY (2018) Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc Natl Acad Sci U S A 115: 9835–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Nie Q, Ren M, Feng H, Jiang X, Zheng Y, Liu M, Zhang H, Xian M (2013) Metabolic engineering of Escherichia coli for the biosynthesis of alpha-pinene. Biotechnol Biofuels 6: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Li Z, Guo L, Du J, Bae H-J (2016) Biosynthesis of β-caryophyllene, a novel terpene-based high-density biofuel precursor, using engineered Escherichia coli. Renewable Energy 99: 216–223 [Google Scholar]
- Yang S-M, Han SH, Kim B-G, Ahn J-H (2014) Production of kaempferol 3-O-rhamnoside from glucose using engineered Escherichia coli. J Ind Microbiol Biotechnol 41: 1311–1318 [DOI] [PubMed] [Google Scholar]
- Yang S-M, Shim GY, Kim B-G, Ahn J-H (2015a) Biological synthesis of coumarins in Escherichia coli. Microb Cell Fact 14: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lin Y, Li L, Linhardt RJ, Yan Y (2015b) Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab Eng 29: 217–226 [DOI] [PubMed] [Google Scholar]
- Yao R, Zhao Y, Liu T, Huang C, Xu S, Sui Z, Luo J, Kong L (2017) Identification and functional characterization of a p-coumaroyl CoA 2′-hydroxylase involved in the biosynthesis of coumarin skeleton from Peucedanum praeruptorum Dunn. Plant Mol Biol 95: 199–213 [DOI] [PubMed] [Google Scholar]
- You S, Yin Q, Zhang J, Zhang C, Qi W, Gao L, Tao Z, Su R, He Z (2017) Utilization of biodiesel by-product as substrate for high-production of β-farnesene via relatively balanced mevalonate pathway in Escherichia coli. Bioresour Technol 243: 228–236 [DOI] [PubMed] [Google Scholar]
- Yu D, Xu F, Zeng J, Zhan J (2012) Type III polyketide synthases in natural product biosynthesis. IUBMB Life 64: 285–295 [DOI] [PubMed] [Google Scholar]
- Zha W, Rubin-Pitel SB, Shao Z, Zhao H (2009) Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab Eng 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen X, Lindley ND, Too HP (2018a) A “plug-n-play” modular metabolic system for the production of apocarotenoids. Biotechnol Bioeng 115: 174–183 [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu J, Zhao F, Lu C, Zhao G-R, Lu W (2018b) Production of sesquiterpenoid zerumbone from metabolic engineered Saccharomyces cerevisiae. Metab Eng 49: 28–35 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li S-Z, Li J, Pan X, Cahoon RE, Jaworski JG, Wang X, Jez JM, Chen F, Yu O (2006) Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J Am Chem Soc 128: 13030–13031 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dai Z, Krivoruchko A, Chen Y, Siewers V, Nielsen J (2015) Functional pyruvate formate lyase pathway expressed with two different electron donors in Saccharomyces cerevisiae at aerobic growth. FEMS Yeast Res 15: fov024. [DOI] [PubMed] [Google Scholar]
- Zhao J, Bao X, Li C, Shen Y, Hou J (2016) Improving monoterpene geraniol production through geranyl diphosphate synthesis regulation in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 100: 4561–4571 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Liu T, Luo J, Zhang Q, Xu S, Han C, Xu J, Chen M, Chen Y, Kong L (2015) Integration of a decrescent transcriptome and metabolomics dataset of Peucedanum praeruptorum to investigate the CYP450 and MDR genes involved in coumarins biosynthesis and transport. Front Plant Sci 6: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Qiao K, Edgar S, Stephanopoulos G (2015) Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol 33: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M-L, Shao J-R, Tang Y-X (2009) Production and metabolic engineering of terpenoid indole alkaloids in cell cultures of the medicinal plant Catharanthus roseus (L.) G. Don (Madagascar periwinkle). Biotechnol Appl Biochem 52: 313–323 [DOI] [PubMed] [Google Scholar]
- Zhu M, Wang C, Sun W, Zhou A, Wang Y, Zhang G, Zhou X, Huo Y, Li C (2018) Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab Eng 45: 43–50 [DOI] [PubMed] [Google Scholar]