Abstract
Argonaute1 activity is not limited to the cytoplasm and has been found to be associated with the regulation of gene expression in the nucleus and to be tightly associated with chromatin and transcription.
Argonaute (AGO) proteins, key factors in the RNA interference (RNAi) pathway, are involved in the regulation of gene expression at the transcriptional, posttranscriptional, and translational levels and play roles in genome integrity maintenance (Hutvagner and Simard, 2008; Meister, 2013; Kalantari et al., 2016; Carbonell, 2017; Ma et al., 2018). AGO proteins, found in all eukaryotes (excluding Saccharomyces cerevisiae) as well as in bacteria and archaea (Drinnenberg et al., 2009; Meister, 2013; Swarts et al., 2014), are classified into three paralogous groups: (1) AGO-like proteins, which account for most AGO proteins and are similar to Arabidopsis (Arabidopsis thaliana) AGO1; (2) P-element induced WImpy testis (PIWI)-like proteins, homologous to Drosophila melanogaster PIWI and exclusively expressed in animal germ cells; and (3) AGOs expressed in only Caenorhabditis elegans (Hutvagner and Simard, 2008; Zhang et al., 2015; Ma et al., 2018). The number of genes encoding AGO proteins varies among species, ranging from one in Schizosaccharomyces pombe to 27 in C. elegans (Carmell et al., 2002). All AGO proteins are evolutionarily conserved in structure and function (Höck and Meister, 2008), which emphasizes their importance in cellular metabolism and development.
In eukaryotes, AGO family proteins possess four characteristic domains: N-terminal, PAZ (PIWI-AGO-Zwille), MID, and PIWI (Hutvagner and Simard, 2008; Zhang et al., 2014). The PAZ domain anchors the 3′ end of the guide strand of a small RNA (sRNA) duplex (Ma et al., 2004), whereas the MID-PIWI domain creates a binding pocket for the 5′ end of the sRNA duplex guide strand (Boland et al., 2011). Moreover, the PIWI domain shows similarity to RNase H and is essential for target cleavage (Song et al., 2004; Parker et al., 2005). Finally, the N-terminal domain fragment (DUF1785) was recently described as being important for RNA duplex unwinding (Derrien et al., 2018).
The AGO proteins mediate posttranscriptional gene silencing (PTGS). Based on microRNA (miRNA) or small interfering RNA (siRNA) sequence complementarity, RNA-induced silencing complex (RISC) is guided to a target RNA, which leads to its cleavage (predominantly in plants; Baumberger and Baulcombe, 2005) or translation inhibition (Brodersen et al., 2008; Guo et al., 2010). However, AGO proteins also exhibit an sRNA-independent target recognition mechanism, as suggested for C. elegans, mammalian cells (Leung et al., 2011; Friend et al., 2012), and D. melanogaster (Pinder and Smibert, 2013). In Arabidopsis, ten AGO family members are expressed (Vaucheret, 2008; Borges and Martienssen, 2015), with the most prominent being AGO1, the main component of the miRNA-guided cytoplasmic RISC.
Other members of the AGO family were found to be associated with chromatin and involved in histone modification and DNA methylation in transcriptional gene silencing (reviewed in Castel and Martienssen, 2013; Martienssen and Moazed, 2015). The nuclear functions of plant AGO proteins were first described for AGO4 (Zilberman et al., 2003) and AGO6 (Zheng et al., 2007; McCue et al., 2015). Arabidopsis AGO4 binds 24-nt siRNAs, which are most likely generated in the nucleus by DCL3 in the RNA polymerase IV (RNAPIV) and RNA-dependent RNA polymerase 2 (RDR2)-dependent pathway (for review, see Wendte and Pikaard, 2017). Binding of the siRNA-AGO4 complex to RNA polymerase V (RNAPV) nascent transcripts leads to recruitment of the DNA methyltransferase DRM2 and induces de novo DNA methylation (m5C) at specific loci (Wierzbicki et al., 2008; Böhmdorfer et al., 2014; Zhong et al., 2014), leading to transcriptional silencing. The introduction of a methyl group at cytosine residues requires assistance from the SWI/SNF chromatin remodeling complex (Zhu et al., 2013) and is accompanied by histone modifications, such as H3 and H4 deacetylation, H3K9 methylation (H3K9me), and H3K4 demethylation (for review, see Matzke and Mosher, 2014). Similarly, Arabidopsis AGO6 is related to the RNA-directed DNA methylation pathway and may act redundantly (Zheng et al., 2007) or interdependently with (Duan et al., 2015) AGO4.
Moreover, Arabidopsis AGO6 exhibits a specific expression pattern (shoot and root meristem; Havecker et al., 2010; Eun et al., 2011) and participates in the methylation of transcriptionally active transposons (McCue et al., 2015). In S. pombe, the AGO-containing RNA-induced transcriptional silencing complex is required for the formation of constitutive heterochromatin at pericentromeric regions, stimulating histone H3K9me (Partridge et al., 2002). In C. elegans, the nuclear RNAi pathway is also involved in transcription silencing by promoting H3K9me (Gu et al., 2012), which protects germline cells from transposon activity (Vastenhouw and Plasterk, 2004).
In worms, AGO family proteins are involved in not only transcriptional suppression but also positive regulation of the transcription of target genes required for spermatogenesis (Conine et al., 2013; Seth et al., 2013), and in D. melanogaster, heterochromatin formation is dependent on RNAi and AGO proteins (Pal-Bhadra et al., 2002, 2004). High-throughput RNA-seq and ChIP-seq studies on flies revealed that AGO2 represses transcription by binding to chromatin, but this mechanism could be independent of its catalytic activity (Taliaferro et al., 2013). Moreover, Drosophila AGO2 was found to correlate with transcriptionally active regions of the genome and thought to be involved in the control of RNA polymerase II (RNAPII) processivity (Cernilogar et al., 2011). In human cells, AGO1 directs transcriptional gene silencing by associating with targeted promoters (Kim et al., 2006). However, it is also involved in the initiation of transcription and interacts directly with RNAPII (Huang et al., 2013).
Recent studies show that the action of Arabidopsis AGO1 is not limited to the cytoplasm, because it is also involved in the regulation of gene expression in the nucleus, where it is tightly associated with chromatin (Dolata et al., 2016; Schalk et al., 2017; Liu et al., 2018). Here, we summarize recent findings on the nuclear function of Arabidopsis AGO1 and provide ideas and directions for future studies (Fig. 1).
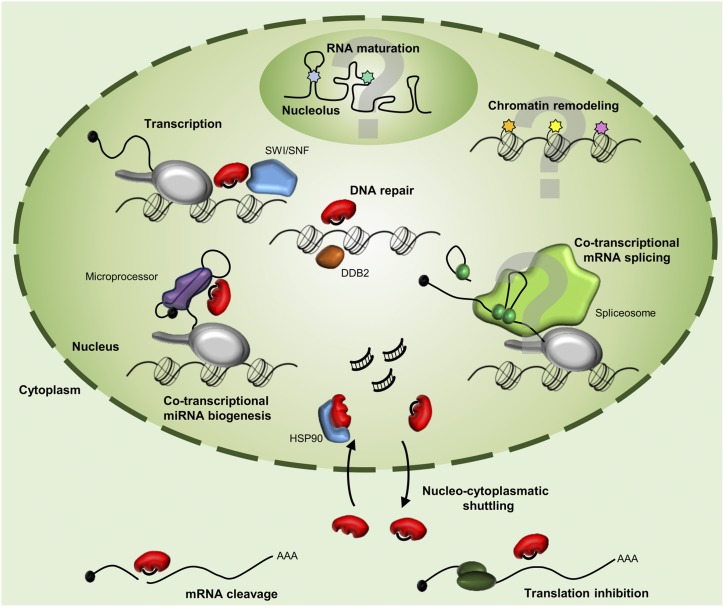
Figure 1.
Diverse functions of ARGONAUTE1 in Arabidopsis thaliana. In addition to its cytoplasmic role in mRNA target cleavage and translation inhibition, AGO1 is associated with chromatin and plays an important role in the regulation of transcription, cotranscriptional miRNA biogenesis, and DNA repair. It is highly probable that AGO1 is also involved in chromatin remodeling and cotranscriptional splicing regulation as well as RNA maturation in the nucleolus.
miRNA Loading into AGO1 in the Nucleus
In contrast with animal cells, wherein miRNA biogenesis occurs in both the nucleus and the cytoplasm, in plant cells, miRNA production occurs entirely in the nucleus (Stepien et al., 2017). miRNA/miRNA* duplexes are generated in two cleavage reactions, both of which are catalyzed by the endonuclease DICER-LIKE 1 (DCL1; Kurihara and Watanabe, 2004). The 2’ OH groups of the 3′ terminal nucleotides of both duplex strands generated in these reactions are methylated by the methylase hua enhancer 1 (Park et al., 2002), which protects them from uridylation and subsequent degradation (Yu et al., 2005). Interestingly, miRNA and miRNA* accumulate asymmetrically, because one strand remains more abundant than the other, and this effect is correlated with the formation of a functional RISC, initiated by AGO1 loading (traditionally thought to occur in the cytoplasm) in which the guide strand (miRNA) is retained inside this effector ribonucleoprotein complex, whereas the passenger strand (miRNA*) is removed by unwinding and degradation (Iki et al., 2010).
Until recently, very little was known about the exact site at which plant sRNA duplex loading occurs. According to previously published data, miRNA/miRNA* duplexes are exported to the cytoplasm by HASTY (HST; Papp et al., 2003; Park et al., 2005; Pontes et al., 2013), an ortholog of human Exportin 5 (Exp5; Yi et al., 2005) after completing biogenesis in the nucleus. However, this is not a general mechanism because HST has been reported to be required for the export of only some specific miRNAs (Park et al., 2005). Additionally, in contradiction to the current model of miRNA biogenesis in plants, data showing that the miRNA biogenesis machinery is necessary for miRNA/miRNA* duplex formation and RISC loading is already present in the nucleus have been reported; Fang and Spector showed that AGO1 and HYL1 (HYPONASTIC LEAVES 1), a double-stranded RNA-binding protein involved in primary transcript (pri-miRNA) processing and loading AGO1 with miRNAs (Vazquez et al., 2004) are present in Dicing bodies (d-bodies; Fang and Spector, 2007), nuclear structures involved in miRNA biogenesis in plants. HYL1 is an important factor for strand selection during AGO1 loading with miRNAs (Eamens et al., 2009). These observations are considered the first clue indicating that miRNA is loaded to AGO1 in the nucleus. Moreover, Pontes and colleagues showed that DCL1 colocalizes in nuclear d-bodies with hua enhancer 1 (Pontes et al., 2013).
Recently published data reshaped the miRNA biogenesis model and shed new light on the RISC-loading process in plants. Fascinatingly, a conserved coiled structure, termed an N-coil, deemed to have an important function, was observed at the AGO1 N-terminal domain. The N- and C-terminal regions of the N-coil possess both a nuclear localization signal (NLS) and a nuclear export signal (NES), indicating that AGO1 is a nuclear-cytoplasmic shuttling protein (Bologna et al., 2018). The prevalent presence of AGO1 in the cytoplasm results from the dominance of the NES over the NLS, which is recognized in the nucleus by the evolutionarily conserved CRM1/EXPORTIN1 protein (Kudo et al., 1999). However, according to Bologna and coworkers (Bologna et al., 2018), the nuclear localization of AGO1 is necessary for the loading and translocation of most methylated, mature miRNAs from the nucleus to the cytoplasm. In the cytoplasm, the NES element on the surface of unloaded AGO1 is unrecognizable, and the exposed NLS results in the translocation of AGO1 to the nucleus. Upon sRNA duplex binding in the nucleus, the NES element is exposed and begins to dominate over the NLS, resulting in the translocation of loaded AGO1 back to the cytoplasm.
When present in the nucleus, AGO1 undergoes two major conformational changes during RISC formation. One change occurs before the loading of miRNA/miRNA* duplexes and requires the molecular chaperone Hsp90, which binds to AGO1 in the form of a dimer, and it is a common feature of plant and animal RISC formation process (Iwasaki et al., 2010). In the presence of ATP, Hsp90 mediates the conformational opening of AGO1, enabling the fit of sRNA duplexes. In Arabidopsis and tobacco (Nicotiana tabacum), cyclophilin 40/SQUINT cochaperones assist Hsp90 in the loading of the siRNA duplex into AGO (Iki et al., 2012). Additionally, unlike in animals, in plants, the RNA duplex binding does not require energy generated from ATP hydrolysis, but hydrolysis is still required afterward for the dissociation of AGO1 from the Hsp90 dimer. Dissociation enforces a second conformational change that promotes unwinding and passenger strand ejection (Röhl et al., 2013).
Alteration of the NES element accessibility in AGO1 highly resembles the regulation of AGO4 nuclear localization in the RNA-directed DNA methylation pathway in plants (Ye et al., 2012). siRNAs must be loaded into the AGO4 protein to form AGO4/siRNA complexes, which occurs in the cytoplasm and triggers conformational changes in AGO4, exposing an initially hidden NLS and consequently allowing translocation to the nucleus. Assembly of the AGO4 complex before entry into the nucleus and selective transport may function as a quality control step before the nuclear RNAi process occurs (Ye et al., 2012), and a similar process could be applicable to the loading of miRNA into AGO1.
Bologna and colleagues have shown the importance of the NES for AGO1 function (AGO1 with mutated NES could not be transported to the cytoplasm). AGO1-mNES could not rescue the defects in ago1 mutants (Bologna et al., 2018), an observation similar to that made for cleavage activity mutants (Carbonell et al., 2012). However, the significance of AGO1 NLS remains unclear since AGO-mNLS shows only reduced nuclear localization (Bologna et al., 2018). The molecular phenotype of nuclear AGO1-depleted plants is not known and requires further investigation.
Argonaute1 Regulates Protein-Coding Gene Transcription
Arabidopsis AGO1 has been predominantly studied for its function in the cytoplasm in the PTGS pathway (Morel et al., 2002; Baumberger and Baulcombe, 2005; Brodersen et al., 2008; Li et al., 2013; Arribas-Hernández et al., 2016). Although AGO1 nuclear localization in Arabidopsis was reported several years ago (Fang and Spector, 2007; Wang et al., 2011), a function of chromatin-associated AGO1 was described much later (Dolata et al., 2016). Data recently published by Liu and colleagues (Liu et al., 2018) provide a model for AGO1 functioning as a positive regulator of transcription in Arabidopsis. Using the Chip-seq approach, the authors identified more than 900 AGO1-enriched sites in the Arabidopsis genome, which correspond mostly to genomic regions containing transcription start sites and transcription termination sites.
This AGO1 enrichment corresponds to the known total RNAPII distribution profile (Adelman and Lis, 2012; Hajheidari et al., 2013). Arabidopsis AGO1 was shown to coimmunoprecipitate with RNAPII complex subunits, and AGO1 depletion in the ago1-36 mutant induced a genome-wide decreased chromatin-associated accumulation of total RNAPII as well as that of its phosphorylated forms (P-Ser-5 and P-Ser-2), whereas the overall abundance of RNAPII in the cell did not decrease (Liu et al., 2018). These results support previously published data showing a reduced RNAPII occupancy at some MIR genes of the ago1-11 mutant (Dolata et al., 2016). Liu and colleagues suggest that in contrast with the cytoplasmic AGO1 pool, chromatin-associated AGO1 stimulates gene expression by assisting in the recruitment of RNAPII, and the presence of AGO1 on chromatin positively correlates with the expression levels of target genes. Moreover, the levels of nascent and mature transcripts encoded by AGO1-bound genes are predominantly reduced in the ago1-36 mutant (Liu et al., 2018).
For more than 50% of AGO1-bound genes, corresponding sRNAs associated with nuclear AGO1 have been identified (Liu et al., 2018). These sRNAs are 21 nucleotides in length and contain the 5′ uridine, which is consistent with the known AGO1-binding preference (Mi et al., 2008) and most likely produced by DCL1 with assistance from HYL1 (Liu et al., 2018). The association of AGO1 with chromatin was previously shown to be impaired in the HYL1-knockout mutant (hyl1-2) or upon alteration of the HYL1 phosphorylation status (in the cpl1-7 mutant; Dolata et al., 2016). Interestingly, the nuclear fraction of sRNAs bound to AGO1 is depleted in miRNAs and enriched in transacting siRNAs (Liu et al., 2018), which coincides with previous observations of AGO1 being detected on TAS1 and TAS2 loci but not on the TAS3 gene (Dolata et al., 2016). Finally, both sRNA binding and the RNase activity of AGO1 have been shown to be required for its interaction with chromatin (Liu et al., 2018), which contrasts with the results of studies on flies (Taliaferro et al., 2013).
In human cells, nuclear AGO1/AGO2 copurifies with many RNA-binding proteins, splicing factors, histones, and chromatin-related proteins (Ameyar-Zazoua et al., 2012; Batsché and Ameyar-Zazoua, 2015) and interacts with RNAPII (Huang et al., 2013). In Arabidopsis, FLAG-AGO1 coimmunoprecipitates with GFP-tagged SWI/SNF complex subunits (SWI3B, SWI3D, and BSH; Liu et al., 2018). Similar results showing the interaction of AGO2 with SWI/SNF components in an DNA/RNA-independent manner have been reported in HeLa cells (Carissimi et al., 2015). Liu and colleagues have demonstrated that the binding of AGO1 to its chromatin targets is affected in Arabidopsis SWI/SNF subunit mutants, whereas SWI3B recruitment to chromatin is not changed upon depletion of AGO1. These data suggest that SWI/SNF components are required for the AGO1-chromatin association (Liu et al., 2018). However, recent data show that the main ATPase of SWI/SNF, BRAHMA, CHR2, cotranscriptionally inhibits pri-miRNA processing (Wang et al., 2018b). These observations raise the question of whether impaired AGO1/chromatin binding in the brm-4 mutant (Liu et al., 2018) could be the result of misregulated sRNA production.
AGO1/chromatin binding was shown to be enhanced under biotic and abiotic stresses (Dolata et al., 2016; Liu et al., 2018), and a binding preference to stress-related genes was found among AGO1 chromatin targets (Liu et al., 2018). Interestingly, studies on the influence of the jasmonic acid derivative methyl jasmonate on root growth have shown that its inhibitory effect is reduced in ago1 mutants. Moreover, AGO1 binding to chromatin is induced by methyl jasmonate and correlates with elevated target gene expression (Liu et al., 2018).
Cotranscriptional Regulation of miRNA Biogenesis by Argonaute1
Although RNA processing and maturation are known to occur cotranscriptionally in eukaryotic cells (Bentley, 2014; Herzel et al., 2017; Grasser and Grasser, 2018), knowledge of cotranscriptional processes in plants is still limited. Several studies have shown functional correlations among mRNA transcription, splicing, and 5′- and 3′-end processing events in both animals and plants (Geraldo et al., 2009; Liu et al., 2012a; Dolata et al., 2015; Li et al., 2016). Similarly, the biogenesis of noncoding RNAs, especially miRNAs, is known to occur cotranscriptionally in both human and animal cells (Morlando et al., 2012; Dhir et al., 2015; Yin et al., 2015; Church et al., 2017). In plants, this field is rather underinvestigated and requires further study. In Arabidopsis, miRNA biogenesis processes, including (1) the amount and activity of various proteins involved in miRNA biogenesis (for review: Rogers and Chen, 2013; Achkar et al., 2016; Dolata et al., 2018), (2) the influence of splicing and polyadenylation (Bielewicz et al., 2013; Knop et al., 2017; Stepien et al., 2017), (3) alternative stem-loop structure formation (Iki et al., 2018), (4) miRNA modifications (Bhat et al., 2016), and (5) miRNA stability (Zhao et al., 2012), are highly regulated. Modulation of pri-miRNA processing based on secondary structure has also been recently shown (Wang et al., 2018b). Demonstrating a role of CHR2 in unwinding pri-miRNA stem and loop structures in association with Serrate (SE), Wang and colleagues provided evidence of cotranscriptional pri-miRNA processing, because CHR2 also plays a role in SE-independent transcription.
Upon the application of different environmental stresses, which alter these regulatory processes, the levels of mature miRNA expression are barely correlated with the expression levels of MIR pri-miRNAs (Barciszewska-Pacak et al., 2015). The role of AGO1 in miRNA biogenesis, especially under stress conditions, was investigated by Dolata and colleagues (Dolata et al., 2016), who identified some MIR genes (MIR161 and MIR173) that had decreased pri-miRNA levels and increased miRNA accumulation under salinity stress. The increased levels of miRNA161 and miRNA173 (but not the corresponding miRNAs*) were shown to result from AGO1-mediated stabilization, which is consistent with previously published data (Vaucheret et al., 2006; Vaucheret, 2009). Although the exact mechanism by which specific miRNAs are preferentially bound and stabilized by RISC under stress conditions is currently unknown, Iki and colleagues (Iki et al., 2018) may have shed some light on this phenomenon, because they showed that the structural flexibility of the pre-miRNA domain within Arabidopsis pri-miR168a could generate different miRNA/miRNA* duplexes with different AGO1-binding preferences.
Although loading could be one aspect underlying the enrichment of some miRNAs under stress conditions, the degradation of other miRNAs could also contribute to this selectivity. Recently, an association between the miRNA/miRNA* degradation pathway and AGO1 was also established. A 3′-5′ exoribonuclease, ATRM2, was shown to selectively degrade miRNA/miRNA* duplexes and partially colocalize with AGO1 in the cytoplasm, and this colocalization was enhanced under heat stress (Wang et al., 2018a). The authors suggested that the association with AGO1 may be important for strand selection during degradation and could play a role in modulating miRNA biogenesis in response to environmental conditions. Both miRNA161 and miRNA173 were previously shown to respond to various stresses, including viral infection and drought (Hajdarpašić and Ruggenthaler, 2012). Moreover, in addition to targeting protein-coding transcripts, both miRNAs have been shown to be involved in the generation of secondary siRNAs, including TAS1a/b/c- and TAS2-derived transacting siRNAs (Rhoades et al., 2002; Allen et al., 2005; Khraiwesh et al., 2012), indicating their importance and potentially explaining the precise and complicated pathways underlying their regulation.
Although stabilization or lack thereof can explain the up- or down-regulation of mature miRNAs, changes in pri-miRNA levels potentially suggest a role of AGO1 in transcription. This transcriptional/cotranscriptional role of AGO1 in Arabidopsis was suggested based on an observation that AGO1 binds to chromatin in close proximity of the MIR161 and MIR173 genes (Dolata et al., 2016). AGO1 enrichment on these two MIR genes was observed upon salt stress treatment and correlated with decreased RNAPII occupancy. However, reduced RNAPII levels were limited to MIR gene body regions and not to their promoters. These results together with those of studies on MIR promoter activity indicate that this regulation likely occurs at the elongation/termination phase of transcription. Moreover, the overaccumulation of nonpolyadenylated MIR transcripts indicates that the presence of AGO1 on the gene body causes RNAPII to pause and/or terminate transcription prematurely. Changes in the RNAPII distribution at MIR loci and alterations in pri-miRNA levels under salt stress conditions were far smaller in the ago1-11 background than in the wild-type background, suggesting that AGO1 plays a negative role in cotranscriptional miRNA biogenesis under salt stress. These results contrasted with those indicating the transcriptional role of AGO1, as demonstrated by Liu and colleagues (Liu et al., 2018).
AGO-dependent posttranscriptional processing of miRNA precursors has also been shown in human cells, albeit with AGO2 as the main player. AGO2-cleaved precursor miRNAs are products of AGO2-mediated premiRNA cleavage and endogenous products of miRNA biogenesis (Diederichs and Haber, 2007). On the other hand, certain Arabidopsis pri-miRNAs could be the targets of their own miRNAs (German et al., 2008). German and colleagues suggest that the interplay between polyadenylation and degradation machinery could lead to several different mechanisms by which pri-miRNAs are processed. Recent studies revealed that the binding of AGO1 to MIR genes is RNA dependent (Dolata et al., 2016). To test whether the association of AGO1 with chromatin requires sRNAs, the Arabidopsis hyl1-2 mutant was analyzed, in which the loading of miRNAs onto AGO1 is known to be hampered (Eamens et al., 2009). As expected, under salt stress, the association of AGO1 with chromatin was weaker in hyl1-2 mutant plants compared with that in wild-type plants. This information in combination with previous results showing colocalization of AGO1 with HYL1 in d-bodies (Fang and Spector, 2007) also indicates that AGO1 could act during early stages of miRNA biogenesis.
Role of Argonaute1 in RNA-Mediated DNA Repair
DNA double-stranded breaks (DSBs) are caused by reactive oxygen species, ionizing radiation, or failed DNA replication and underlie serious DNA damage, because they cause mutations and genome instability and may lead to cell death. Therefore, quick repair of DSBs is important for the maintenance of cellular genome integrity and excludes harmful mutations (Iyama and Wilson, 2013). In eukaryotes, DSBs are repaired by two major pathways: homologous recombination (HR) and nonhomologous end-joining (NHEJ). In the HR pathway, DNA is repaired using sister chromatids as a template, which accurately restores the DNA sequence. NHEJ involves joining of two double-stranded DNA strains by DNA synthesis and ligation (Iyama and Wilson, 2013). Wei and colleagues (Wei et al., 2012) elucidated the role of sRNAs in DSB repair via the HR pathway in both HeLa cells and Arabidopsis and showed that knocking down Dicer and AGO2 in human cells reduced the DSB repair efficiency (Wei et al., 2012). The main sRNA-processing enzymes Dicer and DROSHA have also been shown to be involved in activating a response to DNA damage upon genotoxic stress in fly and human cells (Francia et al., 2012; Michalik et al., 2012). Wei and colleagues (Wei et al., 2012) showed that RNAPIV, RDRs, and Dicer-like proteins (DCL2, DCL3, DCL4) are required for the proper biogenesis of DSB-induced sRNAs (diRNAs) in Arabidopsis.
These diRNAs are incorporated into AGO2 and interact with RNAPV scaffold transcripts to recruit DSB repair proteins to the DSB sites. Therefore, Arabidopsis mutants of proteins involved in diRNA biogenesis reduce the efficiency of DSB repair (Wei et al., 2012). Human AGO2 was later shown to interact with the Rad51 recombinase (Gao et al., 2014), and Rad51 mediates double-stranded DNA ligation during HR (San Filippo et al., 2008). Moreover, AGO2 together with sRNA recruit Rad51 to DSB sites, and the efficiency of HR depends on the association of AGO2 with sRNA (Gao et al., 2014). Further genetic studies have provided evidence that diRNAs are involved in not only HR but also the NHEJ repair pathway in plants (Qi et al., 2016), and Oliver and colleagues showed that Arabidopsis AGO9 is involved in somatic DNA repair (Oliver et al., 2014). The role of AGO9 in the repair of damaged DNA remains unknown, but it very likely acts by associating with sRNAs. In addition to Arabidopsis AGO2 and AGO9, AGO1 was recently shown to be involved in the RNA-mediated DNA repair pathway by forming a complex with DNA damage binding protein 2 (DDB2), the stability of which is mediated by DNA and sRNAs (Schalk et al., 2017). Using DDB2 with a mutated DNA-binding domain, the authors first showed that AGO1-DDB2 did not form, and this complex was shown to be less stable in RNAPV and DCL4 mutants, suggesting that the interactions between AGO1 and DDB2 are mediated by sRNAs.
Further experiments have revealed that the AGO1-DDB2 complex is involved in DNA repair after UV-C irradiation. Cyclobutane pyrimidine dimers (CPDs) are a major form of UV-induced DNA lesions that alter the structure of DNA and inhibit transcription and replication. Schalk and colleagues described plant UV-induced sRNAs (uviRNAs). Most uviRNAs associated with the AGO1-DDB2 complex are complementary to the DNA strand enriched in CPDs, and the biogenesis of uviRNAs requires transcription to be carried out by RNAPIV. RDR2 produces double-stranded RNAs, which are processed by DCL4 into 21-nt RNAs (Schalk et al., 2017). These data confirm that DNA repair strongly relies on sRNAs produced by PTGS and RNA-dependent DNA repair pathway (Wei et al., 2012; Oliver et al., 2014). DDB2 is a key factor for the recognition of DNA damage in the global genome repair pathway and for the repair of DNA lesions via nucleotide excision repair (Schärer, 2013; Schalk et al., 2017).
Similar to the observed accumulation of AGO1 upon salt stress (Dolata et al., 2016), accumulation of AGO1 together with DDB2 in the nucleus has been associated with UV-C irradiation (Schalk et al., 2017) and observed in the chromatin fraction. However, the levels of both proteins decrease very quickly, suggesting that the AGO1-DDB2 complex is transient and short-lived, existing for only a short period after UV-C stress. The ATR (Ataxia telangiectasia-mutated and RAD3-like) protein is important for transmitting DNA damage signals to downstream elements, and the AGO1-DDB2 complex stability has been shown to be ATR dependent. Furthermore, in an atr loss-of-function mutant, this complex occupies chromatin for a longer time, demonstrating that the ATR-mediated signaling of DNA damage is important for release of the AGO1-DDB2 complex from chromatin (Schalk et al., 2017). After detection of DNA damage, DDB2 must be released from chromatin, allowing downstream players to bind and proceed to the next step of DNA repair in the global genome repair pathway (Chen et al., 2001; Molinier et al., 2008).
Finally, the role of AGO1 in DNA repair in the response to UV stress is detectable at the morphological level. The ago1-27 mutant is more sensitive to UV-C during a period of recovery in the dark, which manifests as reduced root growth after UV-C treatment. Moreover, UV irradiation has a positive effect on recruitment of the AGO1-DDB2 complex to CPD-enriched DNA damage sites (Schalk et al., 2017). Therefore, it is possible that after UV stress, numerous uviRNAs are produced by RNAPV, RDR2, and DCL4, and these uviRNAs increase the stabilization of AGO1 and thus lead to the increased occupancy of AGO1-DDB2 complexes on many damaged chromatin sites.
Future Perspectives
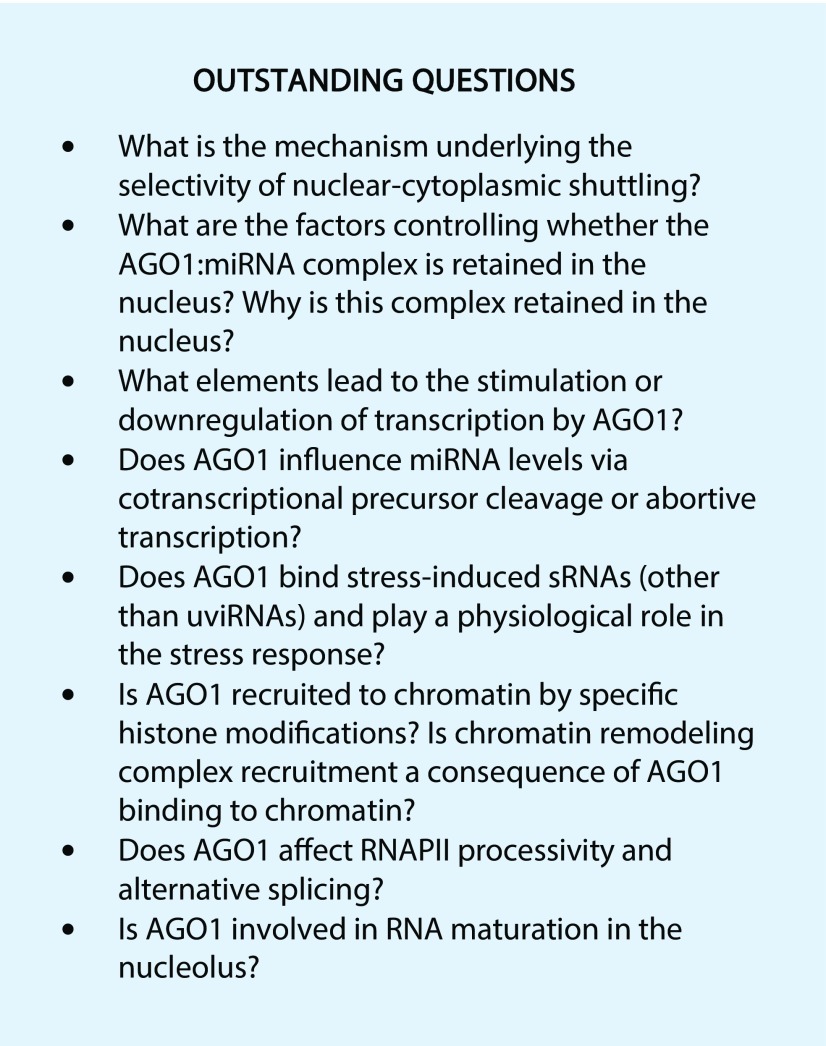
Studies published in the past few years decisively highlight the various important roles of AGO1 in the plant cell nucleus, but these data indicate only the outline of possible mechanisms. Nevertheless, many efforts have been exerted to describe the pathways driven by chromatin-associated AGO1 in Arabidopsis in detail. The biological relevance of AGO1 in the nucleus is still an open question, because published data point to certain (stress) conditions for its action (see Outstanding Questions). It is possible that nuclear AGO1 partially overlaps in function with its strictly nuclear homologs. However, severe phenotypes of ago1 knockout mutants in comparison with mutants of proteins acting upstream in the PTGS pathway suggest a much wider role for AGO1. Moreover, available data are based on studies in vegetative tissues and might be the tip of the iceberg. Alternatively, nuclear AGO1 functions potentially could be much more prominent in meristematic (Kidner and Martienssen, 2005) tissues, in generative tissues, or even specialized cells (Grant-Downton et al., 2009; Borges et al., 2011), where small RNA pathways are highly active and play distinct but very important roles (Slotkin et al., 2009; Calarco et al., 2012; Borges et al., 2018).
An important question that remains unanswered is how AGO1 is recruited to chromatin. Several possibilities for this phenomenon must be considered, beginning with sequence-specific site recognition in the native transcripts via AGO1-associated sRNAs (Alló et al., 2009; Liu et al., 2012b) and/or recruitment to specific histone modifications or chromatin-binding proteins (Ameyar-Zazoua et al., 2012; Taliaferro et al., 2013), as shown in animals. Furthermore, it is important to know how AGO1 binding affects transcription, because it may stimulate specific chromatin modifications and affect the ability of RNAPII to bind to promoter regions (Chu et al., 2010; Hu et al., 2012; Huang et al., 2013). Moreover, local changes in histone markers may result in RNAPII elongation rate alterations and alternative splice site selection (Alló et al., 2009; Ameyar-Zazoua et al., 2012).
The presence of sRNA-loaded AGO1 in the nucleus raises the question of which factors are responsible for deciding whether this complex is exported to the cytoplasm or retained in the nucleus. Until now, no sRNA sequence-based preferences have been found in plants (Bologna et al., 2018; Liu et al., 2018), although these preferences have been elucidated in human cells (Hwang et al., 2007). It is possible that retainment of AGO1 in the nucleus requires the binding of some additional factors or specific sRNA modifications. The list of potential AGO1/AGO2 nuclear interactors in mammalian cells is broad, suggesting the existence of such interactions in the plant nucleus as well (Ameyar-Zazoua et al., 2012).
Interestingly, in Arabidopsis germ line cells, several miRNA-targeted transcripts derived from reactivated transposable elements that trigger epigenetically activated sRNA production have been observed (Creasey et al., 2014; Borges et al., 2018), suggesting that nuclear AGO1 is also involved in protecting the genome from transposable elements–mediated epigenomic instability. Finally, the potential presence of AGO1 in the nucleolus and its association with small nucleolar RNA-derived sRNAs (Taft et al., 2009; González et al., 2010) raises many questions about its involvement in noncoding RNA maturation.
Footnotes
This work was supported by the Narodowe Centrum Nauki (Polish National Science Centre) (UMO-2014/13/N/NZ1/00049 to M.B., UMO-2017/27/N/NZ1/00202 to S.S.B., UMO-2015/19/N/NZ1/01997 to L.S., UMO-2016/23/B/NZ9/00862 to Z.S.-K., UMO-2013/10/A/NZ1/00557 to A.J., and UMO-2017/25/B/NZ1/00603 to J.D.) and the KNOW Poznan RNA Centre (01/KNOW2/2014).
Articles can be viewed without a subscription.
References
- Achkar NP, Cambiagno DA, Manavella PA (2016) miRNA biogenesis: A dynamic pathway. Trends Plant Sci 21: 1034–1044 [DOI] [PubMed] [Google Scholar]
- Adelman K, Lis JT (2012) Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet 13: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. (2009) Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. (2012) Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 [DOI] [PubMed] [Google Scholar]
- Arribas-Hernández L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, Meister G, Brodersen P (2016) The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28: 1563–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barciszewska-Pacak M, Milanowska K, Knop K, Bielewicz D, Nuc P, Plewka P, Pacak AM, Vazquez F, Karlowski W, Jarmolowski A, et al. (2015) Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front Plant Sci 6: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsché E, Ameyar-Zazoua M (2015) The influence of Argonaute proteins on alternative RNA splicing. Wiley Interdiscip Rev RNA 6: 141–156 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. (2014) Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SS, Jarmolowski A, Szweykowska-Kulińska Z (2016) MicroRNA biogenesis: Epigenetic modifications as another layer of complexity in the microRNA expression regulation. Acta Biochim Pol 63: 717–723 [DOI] [PubMed] [Google Scholar]
- Bielewicz D, Kalak M, Kalyna M, Windels D, Barta A, Vazquez F, Szweykowska-Kulinska Z, Jarmolowski A (2013) Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep 14: 622–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmdorfer G, Rowley MJ, Kuciński J, Zhu Y, Amies I, Wierzbicki AT (2014) RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J 79: 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O (2011) Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci USA 108: 10466–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Iselin R, Abriata LA, Sarazin A, Pumplin N, Jay F, Grentzinger T, Dal Peraro M, Voinnet O (2018) Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant MicroRNA pathway. Mol Cell 69: 709–719.e5 [DOI] [PubMed] [Google Scholar]
- Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD (2011) MicroRNA activity in the Arabidopsis male germline. J Exp Bot 62: 1611–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Parent JS, van Ex F, Wolff P, Martínez G, Köhler C, Martienssen RA (2018) Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat Genet 50: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó JA, Becker JD, et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A. (2017) Plant ARGONAUTEs: Features, functions, and unknowns. Methods Mol Biol 1640: 1–21 [DOI] [PubMed] [Google Scholar]
- Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC (2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24: 3613–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carissimi C, Laudadio I, Cipolletta E, Gioiosa S, Mihailovich M, Bonaldi T, Macino G, Fulci V (2015) ARGONAUTE2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human Transcription Start Sites. Nucleic Acids Res 43: 1498–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. (2011) Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480: 391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang Y, Douglas L, Zhou P (2001) UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem 276: 48175–48182 [DOI] [PubMed] [Google Scholar]
- Chu Y, Yue X, Younger ST, Janowski BA, Corey DR (2010) Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res 38: 7736–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church VA, Pressman S, Isaji M, Truscott M, Cizmecioglu NT, Buratowski S, Frolov MV, Carthew RW (2017) Microprocessor recruitment to elongating RNA polymerase II is required for differential expression of microRNAs. Cell Reports 20: 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D Jr., Yates JR III, Mello CC (2013) Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 155: 1532–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA (2014) miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B, Clavel M, Baumberger N, Iki T, Sarazin A, Hacquard T, Ponce MR, Ziegler-Graff V, Vaucheret H, Micol JL, et al. (2018) A suppressor screen for AGO1 degradation by the viral F-Box P0 protein uncovers a role for AGO DUF1785 in sRNA duplex unwinding. Plant Cell 30: 1353–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Proudfoot NJ, Jopling CL (2015) Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol 22: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA (2007) Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Dolata J, Guo Y, Kołowerzo A, Smoliński D, Brzyżek G, Jarmołowski A, Świeżewski S (2015) NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J 34: 544–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolata J, Bajczyk M, Bielewicz D, Niedojadlo K, Niedojadlo J, Pietrykowska H, Walczak W, Szweykowska-Kulinska Z, Jarmolowski A (2016) Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol 172: 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolata J, Taube M, Bajczyk M, Jarmolowski A, Szweykowska-Kulinska Z, Bielewicz D (2018) Regulation of plant microprocessor function in shaping microRNA landscape. Front Plant Sci 9: 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP (2009) RNAi in budding yeast. Science 326: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CG, Zhang H, Tang K, Zhu X, Qian W, Hou YJ, Wang B, Lang Z, Zhao Y, Wang X, et al. (2015) Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J 34: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM (2009) The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun C, Lorkovic ZJ, Naumann U, Long Q, Havecker ER, Simon SA, Meyers BC, Matzke AJ, Matzke M (2011) AGO6 functions in RNA-mediated transcriptional gene silencing in shoot and root meristems in Arabidopsis thaliana. PLoS One 6: e25730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F (2012) Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J (2012) A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol 19: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wei W, Li MM, Wu YS, Ba Z, Jin KX, Li MM, Liao YQ, Adhikari S, Chong Z, et al. (2014) Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res 24: 532–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo N, Bäurle I, Kidou S, Hu X, Dean C (2009) FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26: 941–946 [DOI] [PubMed] [Google Scholar]
- González I, Martínez L, Rakitina DV, Lewsey MG, Atencio FA, Llave C, Kalinina NO, Carr JP, Palukaitis P, Canto T (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: Their significance to RNA silencing suppressor activity. Mol Plant Microbe Interact 23: 294–303 [DOI] [PubMed] [Google Scholar]
- Grant-Downton R, Hafidh S, Twell D, Dickinson HG (2009) Small RNA pathways are present and functional in the angiosperm male gametophyte. Mol Plant 2: 500–512 [DOI] [PubMed] [Google Scholar]
- Grasser M, Grasser KD (2018) The plant RNA polymerase II elongation complex: A hub coordinating transcript elongation and mRNA processing. Transcription 9: 117–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A (2012) Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet 44: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdarpašić A, Ruggenthaler P (2012) Analysis of miRNA expression under stress in Arabidopsis thaliana. Bosn J Basic Med Sci 12: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M, Koncz C, Eick D (2013) Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci 18: 633–643 [DOI] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L, Ottoz DSM, Alpert T, Neugebauer KM (2017) Splicing and transcription touch base: Co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol 18: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Meister G (2008) The Argonaute protein family. Genome Biol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Chen Z, Xia D, Wu J, Xu H, Ye ZQ (2012) Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem J 447: 407–416 [DOI] [PubMed] [Google Scholar]
- Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC (2013) Ago1 interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet 9: e1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ (2008) Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT (2007) A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M (2010) In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Meshi T, Ishikawa M (2012) Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J 31: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T, Cléry A, Bologna NG, Sarazin A, Brosnan CA, Pumplin N, Allain FHT, Voinnet O (2018) Structural flexibility enables alternative maturation, ARGONAUTE sorting and activities of miR168, a global gene silencing regulator in plants. Mol Plant 11: 1008–1023 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y (2010) Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Iyama T, Wilson DM III (2013) DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 12: 620–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari R, Chiang CM, Corey DR (2016) Regulation of mammalian transcription and splicing by Nuclear RNAi. Nucleic Acids Res 44: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev Biol 280: 504–517 [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ (2006) Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13: 793–797 [DOI] [PubMed] [Google Scholar]
- Knop K, Stepien A, Barciszewska-Pacak M, Taube M, Bielewicz D, Michalak M, Borst JW, Jarmolowski A, Szweykowska-Kulinska Z (2017) Active 5’ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res 45: 2757–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA 96: 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA (2011) Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 18: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang D, Fu X, Luo X, Liu R, He Y (2016) Coupling of histone methylation and RNA processing by the nuclear mRNA cap-binding complex. Nat Plants 2: 16015. [DOI] [PubMed] [Google Scholar]
- Liu C, Xin Y, Xu L, Cai Z, Xue Y, Liu Y, Xie D, Liu Y, Qi Y (2018) Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev Cell 44: 348–361.e7 [DOI] [PubMed] [Google Scholar]
- Liu F, Bakht S, Dean C (2012a) Cotranscriptional role for Arabidopsis DICER-LIKE 4 in transcription termination. Science 335: 1621–1623 [DOI] [PubMed] [Google Scholar]
- Liu J, Hu J, Corey DR (2012b) Expanding the action of duplex RNAs into the nucleus: Redirecting alternative splicing. Nucleic Acids Res 40: 1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ (2004) Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zuo Z, Shao W, Jin Y, Meng Y (2018) The expanding roles of Argonautes: RNA interference, splicing and beyond. Brief Funct Genomic 17:191–197 [DOI] [PubMed] [Google Scholar]
- Martienssen R, Moazed D (2015) RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 7: a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- McCue AD, Panda K, Nuthikattu S, Choudury SG, Thomas EN, Slotkin RK (2015) ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J 34: 20–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. (2013) Argonaute proteins: Functional insights and emerging roles. Nat Rev Genet 14: 447–459 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik KM, Böttcher R, Förstemann K (2012) A small RNA response at DNA ends in Drosophila. Nucleic Acids Res 40: 9596–9603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J, Lechner E, Dumbliauskas E, Genschik P (2008) Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet 4: e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J-B, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I (2012) FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J 31: 4502–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C, Santos JL, Pradillo M (2014) On the role of some ARGONAUTE proteins in meiosis and DNA repair in Arabidopsis thaliana. Front Plant Sci 5: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA (2002) RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, Birchler JA, Elgin SC (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672; erratum Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, Birchler JA, Elgin SC (2018) Science 17:191–197 [DOI] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ (2003) Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS (2005) Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D (2005) Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Pinder BD, Smibert CA (2013) microRNA-independent recruitment of Argonaute 1 to nanos mRNA through the Smaug RNA-binding protein. EMBO Rep 14: 80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Vitins A, Ream TS, Hong E, Pikaard CS, Costa-Nunes P (2013) Intersection of small RNA pathways in Arabidopsis thaliana sub-nuclear domains. PLoS One 8: e65652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhang Y, Baller JA, Voytas DF (2016) Histone H2AX and the small RNA pathway modulate both non-homologous end-joining and homologous recombination in plants. Mutat Res 783: 9–14 [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A, Rohrberg J, Buchner J (2013) The chaperone Hsp90: Changing partners for demanding clients. Trends Biochem Sci 38: 253–262 [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Schalk C, Cognat V, Graindorge S, Vincent T, Voinnet O, Molinier J (2017) Small RNA-mediated repair of UV-induced DNA lesions by the DNA damage-binding protein 2 and ARGONAUTE 1. Proc Natl Acad Sci USA 114: E2965–E2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer OD. (2013) Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 5: a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D Jr., Mello CC (2013) The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 27: 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Stepien A, Knop K, Dolata J, Taube M, Bajczyk M, Barciszewska-Pacak M, Pacak A, Jarmolowski A, Szweykowska-Kulinska Z (2017) Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip Rev RNA 8: e1403. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J (2014) The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol 21: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS (2009) Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC (2013) Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: Alternative pre-mRNA splicing and transcriptional repression. Genes Dev 27: 378–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Plasterk RH (2004) RNAi protects the Caenorhabditis elegans germline against transposition. Trends Genet 20: 314–319 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2009) AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One 4: e6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y (2011) An importin β protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Dou Y, Chen L, Wang J, Jiang N, Guo C, Yao Q, Wang C, Liu L, et al. (2018a) Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3′ to 5′ exoribonuclease Atrimmer 2 in Arabidopsis. Proc Natl Acad Sci U S A. 115: E6659–E6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma Z, Castillo-González C, Sun D, Li Y, Yu B, Zhao B, Li P, Zhang X (2018b) SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557: 516–521 [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y (2012) A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112 [DOI] [PubMed] [Google Scholar]
- Wendte JM, Pikaard CS (2017) The RNAs of RNA-directed DNA methylation. Biochim Biophys Acta Gene Regul Mech 1860: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 46: 859–870 [DOI] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR (2005) Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Yu Y, Reed R (2015) Primary microRNA processing is functionally coupled to RNAP II transcription in vitro. Sci Rep 5: 11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xia R, Meyers BC, Walbot V (2015) Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol 27: 84–90 [DOI] [PubMed] [Google Scholar]
- Zhang X, Niu D, Carbonell A, Wang A, Lee A, Tun V, Wang Z, Carrington JC, Chang CE, Jin H (2014) ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat Commun 5: 5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Mo B, Chen X (2012) Mechanisms that impact microRNA stability in plants. RNA Biol 9: 1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK (2007) Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J 26: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE (2014) Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT (2013) A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell 49: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]