Abstract
Inefficiencies in photosynthetic carbon assimilation can be overcome by synthetic biology strategies.
Photosynthetic carbon assimilation (PCA) by plants has provided the basis for human civilization since the origin of agriculture. Transformative changes in agriculture, such as mechanization in the 19th century and the development of synthetic fertilizers, hybrid breeding, and the Green Revolution in the 20th century, have helped to support an almost 10-fold growth in the human population since 1800. Yield increases continued to grow in a linear manner since the 1960s, at an average annual rate of approximately 1.7% (Long et al., 2015). However, several recent studies predict that crop yield will have to double by 2050 to keep pace with the demand of population growth while accounting for changes in consumption patterns resulting from increased wealth and urbanization as well as increased use of agricultural resources for biorefineries (Tilman et al., 2011; Walker et al., 2016). Doubling agricultural yield within the next 30 years would require an annual yield increase of 2.2%, which exceeds the average annual increase witnessed over the past 50 years. Such yield increase cannot come from expanding agricultural land use since reserves in arable land are limited and further utilization of land for agricultural use come at the expense of natural habitats and therefore erodes biodiversity (Foley et al., 2011; Clark and Tilman, 2017). Hence, yield must be increased per unit area land, either by increasing planting density or by increasing the performance of individuals within a crop canopy. Planting density in intensely managed cropping systems is approaching a ceiling (Mansfield and Mumm, 2014), which defines increased performance of individual plants within a canopy as a prime target (Long et al., 2006).
Yield is a complex trait to which many intrinsic and extrinsic factors contribute, such as biotic and abiotic stresses, the efficiency of light energy capture by the photosynthetic light reactions, the efficiency of the conversion of light energy into biomass, and the harvest index (fraction of total energy in plant biomass contained in the harvestable organs). As the efficiency of light energy capture and the harvest index seem to reach their practical limit, improvement of the currently low conversion efficiency of light to biomass (∼2%) has received considerable attention (Zhu et al., 2010). This efficiency can be increased by relieving constraints imposed by individual components, as demonstrated by overexpression of SBPase and/or Gly decarboxylase, the activity of which limits carbon fixation and related processes (Timm et al., 2012; Simkin et al., 2015, 2017).
Instead of modulating individual components, a synthetic biology approach can be used, where entire processes are redesigned and engineered to overcome key barriers that cannot be easily addressed by tinkering with existing systems. This Update focuses on recent progress made by synthetic biology undertakings that aim to breach the boundaries of plant carbon fixation. We discuss in detail the concepts behind these projects rather than the specific technologies used for their implementation, for which we refer the readers to other recently published reviews (Engler et al., 2014; Patron et al., 2015).
LIMITATIONS IN PHOTOSYNTHETIC CARBON USE EFFICIENCY
The vast majority of organic carbon in the biosphere is derived from inorganic carbon captured by the Calvin-Benson cycle (CBC) in plants. The CBC consumes NADPH and ATP, which are regenerated by the light reactions. Hence, the light and carbon reactions of photosynthesis are tightly coupled via the production and consumption of the key cellular redox and energy carriers.
In the CBC, CO2 is first attached to the five-carbon acceptor molecule ribulose 1,5-bisphosphate (RuBP) by the enzyme Rubisco, yielding an unstable C6 intermediate that spontaneously and irreversibly hydrolyzes to yield two molecules of 3-phosphoglyceric acid (3PGA). 3PGA is then activated by phosphoglycerate kinase to yield 1,3-bisphosphoglycerate while consuming ATP. The latter intermediate is reduced, using NADPH as an electron donor, to give glyceraldehyde 3-phosphate (GAP), as catalyzed by glyceraldehyde phosphate dehydrogenase. ATP is also consumed during regeneration of the CO2 acceptor RuBP from GAP in the regenerative phase of the CBC. Overall, three molecules of ATP and two molecules of NADPH are consumed per assimilated CO2, without further considering the cost involved in making, maintaining, and degrading pathway enzymes as well as plant structures, such as chloroplasts, cell walls, etc. (see Amthor [2010] for a comprehensive review on the conversion of solar energy to plant phytomass and Williams [2016] for a general introduction to PCA).
The above calculation holds true only under the assumption that all reactions catalyzed by Rubisco are carboxylation reactions. In reality, however, under current atmospheric conditions in C3 plants at a leaf temperature of 25°C, approximately 25% of the reactions of Rubisco use O2 instead of CO2, which leads to the oxygenation of RuBP and the production of one molecule of 3PGA and one molecule of 2-phosphoglycolic acid (2PG). 2PG is a dead-end metabolite that must be recycled to GAP by an elaborate series of enzymic reactions and transport steps that are distributed over the chloroplasts, peroxisomes, mitochondria, and cytoplasm of the plant cell. This metabolic recycling process is called photorespiration (PR; Bauwe et al., 2010; Walker et al., 2016).
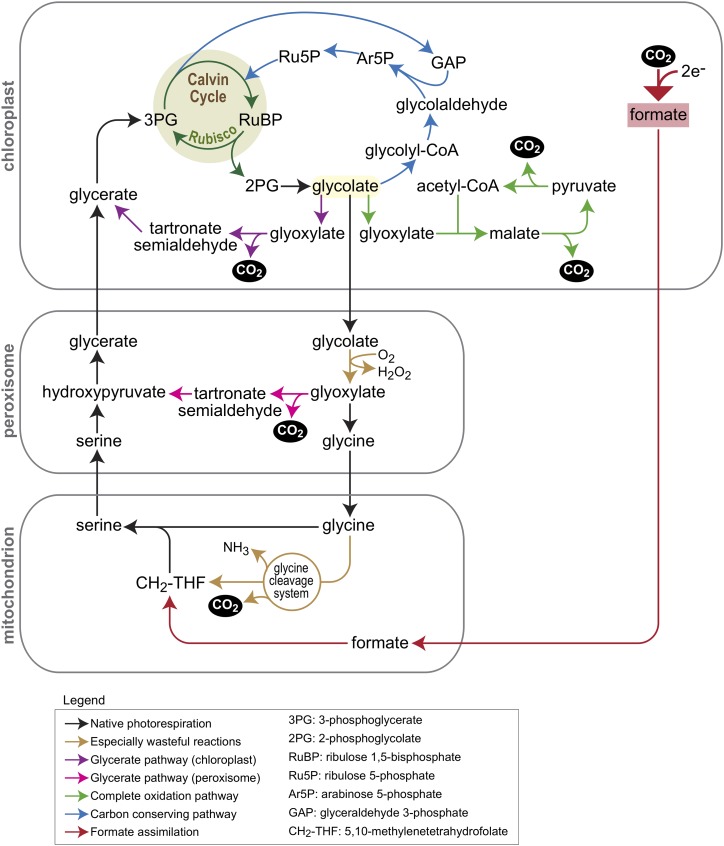
PR is one of the major factors contributing to inefficiency of photosynthetic energy conversion (Fig. 1). First and most importantly, PR recovers only 75% of the carbon contained in 2PG, whereas 25% is lost as CO2, directly counteracting carbon fixation by the CBC. Further, ammonia is released during this process (one molecule per two 2PG molecules assimilated), which needs to be reassimilated at the expense of one molecule of ATP and two reduced ferredoxins per molecule of ammonia. Moreover, redox power is dissipated in the oxidation of glycolate using molecular oxygen. Finally, one ATP is required to convert glycerate into 3PGA by glycerate kinase, the final step in the PR pathway. This brings the tally to 3.5 ATP and two NADPH for the recycling of two molecules of 2PG into one 3PGA. Hence, at 25°C and current atmospheric conditions, approximately one-third of the total ATP and NADPH consumed by a photosynthesizing leaf of a C3 plant is dedicated to the process of PR (see Walker et al. [2016] for details and tools to model energy consumption by PR under different environmental conditions).
Figure 1.
Schematic of native PR and of five synthetic bypasses to this pathway. Native PR is shown by black arrows, where brown arrows highlight especially wasteful reactions. The glycerate pathway (chloroplastic version shown in purple and peroxisomal version in pink) and the glycolate complete oxidation pathways (green arrows) have already been tested in plants, but all result in the release of CO2. The arabinose 5-phospate pathway (blue arrows) is a carbon-conserving route, assimilating glycolate to the CBC without the loss of CO2. Red arrows show a potential pathway that harnesses the low reduction potential in the chloroplast to reduce CO2 to formate, which is then assimilated into the photorespiratory pathway, transforming it into a carbon-fixing route.
Under conditions of low CO2 availability (e.g. when stomates close in the light due to low water potential) or at high temperatures, oxygenation rates of Rubisco and hence energy consumption by PR can be much higher. Even in conditions in which energy is not limiting, the oxygenation reaction and the release of CO2 during PR substantially limits the rate of carbon fixation. Thus, although PR performs important functions, such as provision of the amino acid Ser (Benstein et al., 2013) and dissipation of excess excitation energy from chloroplasts to mitochondria (Eisenhut et al., 2017), it also represents a major source of inefficiency that substantially reduces the yield potential of C3 crops (Betti et al., 2016; Walker et al., 2016).
It is hence not surprising that many research groups and consortia worldwide set out to improve the efficiency of photosynthetic carbon conversion by attempting to reduce the occurrence of the Rubisco oxygenation reaction or bypass the inefficiencies of PR. Multiple strategies are followed to this end: some are inspired by the diversity of naturally existing carbon concentration mechanisms (CCMs) in plants, algae, and cyanobacteria, while others aim to implement synthetic metabolic routes to supplement or replace the canonical PR pathway (Bar-Even, 2018; South et al., 2018).
INCREASING THE CO2 CONCENTRATION AT THE SITE OF RUBISCO
Oxygen competes for the enediolate carbanion of RuBP bound to Rubisco. Hence, increasing CO2 concentration at the site of Rubisco would effectively reduce the rate of oxygenation by increasing the rate of carboxylation of the enediolate carbanion (Tcherkez et al., 2006; Savir et al., 2010). Indeed, cyanobacteria, algae, and land plants have independently evolved mechanisms to concentrate CO2 at the site of Rubisco and thereby reduce oxygenation. Cyanobacteria and green algae have evolved a combination of bicarbonate pumps and physical confinement of Rubisco with carbonic anhydrase in close proximity. In angiosperms, a biochemical CO2 pump, so-called C4 photosynthesis, has evolved independently at least 66 times from the ancestral C3 state (Sage et al., 2012). To reduce the rate of the Rubisco oxygenation reaction and thereby the inefficiencies associated with PR, it would be desirable to engineer such CCMs into crops to increase their yield potential.
C4 PHOTOSYNTHESIS
In C4 photosynthesis, the photosynthetic pathway loses cell autonomy (with few exceptions) and becomes distributed over two distinct cell types, one functioning as a PCA unit and the other as a photosynthetic carbon reduction (PCR) unit. For C4 photosynthesis to function, the PCA and PCR cells are arranged as concentric layers, with each PCA unit having direct cell-to-cell contact with a PCR unit. Together, these two cell types form functional photosynthetic units. The mandatory interaction between PCA and PCR cells requires a characteristic leaf morphology, called Kranz anatomy, in which the vascular bundles (VBs) in leaves are surrounded by concentric layers of bundle sheath and mesophyll cells. Because each PCA cell requires contact with a PCR cell, only four cell layers fit between two VBs, leading to a stereotypic VB-PCR-PCA-PCA-PCR-VB pattern.
PCA cells contain little Rubisco activity. Instead, they exhibit high activity of phosphoenolpyruvate carboxylase (PEPC), an enzyme that condenses phosphoenolpyruvate with inorganic carbon to give the C4 acid oxaloacetate, the name-giving initial product of carbon fixation in C4 plants. PEPC reacts with the anion HCO3−, not CO2. In contrast to Rubisco, this reaction is not sensitive to oxygen. Oxaloacetate is then converted to malate and/or Asp, which diffuse along their concentration gradients to PCR cells, where they are decarboxylated. The decarboxylation of C4 acids in PCR cells increases the local CO2 concentration by approximately 10-fold over the concentration in PCA cells, which is sufficiently high to strongly reduce the rate of PR. Rubisco, which is mostly confined to PCR cells in C4 plants, hence operates under high CO2 concentration, which increases its efficiency and allows for reduced investment into this key protein. The C3 decarboxylation products pyruvate and/or Ala diffuse back to PCA cells, where they are converted to phosphoenolpyruvate, and a new cycle of CO2 transport to PCR cells by C4 acids can begin.
The C4 CCM is associated with an increased cost of two ATP per molecule of CO2 assimilated (five ATP and two NADPH per CO2). Hence, the pathway only pays off if the energy costs of PR exceed the extra ATP requirement for running the C4 pump. Further, evolving C4 photosynthesis, as outlined above, requires a special leaf anatomy. Some plant species, for example, the BEP-clade of grasses that contains many important crop species, such as rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare), may lack the genetic prerequisites to evolve the corresponding leaf anatomy (Christin et al., 2013). Thus, introduction of the C4 trait into plant species that may lack the ability to naturally evolve it is a challenging task (Denton et al., 2013; Schuler et al., 2016).
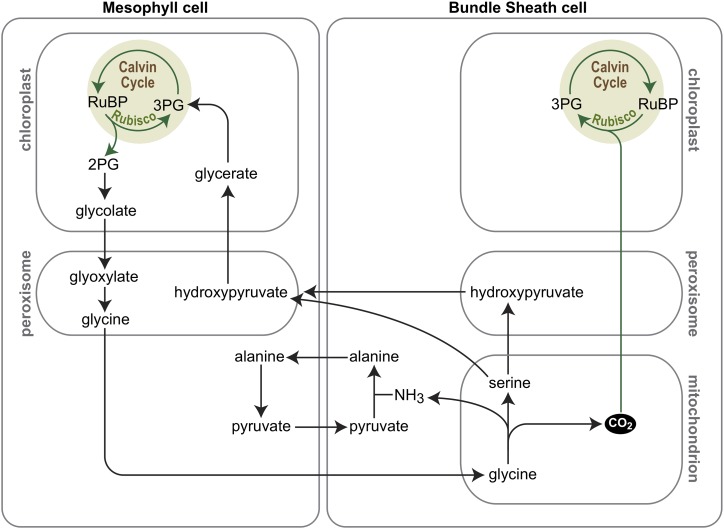
Efforts to engineer C4 photosynthesis into C3 plant species were hampered by an incomplete list of genes and gene functions required to support the trait. The prospects massively improved with the advent of next-generation sequencing technologies, which enabled the comparison of gene expression patterns between related C3, C3-C4 intermediate, and C4 species, thereby generating lists of candidate genes involved in setting up C4 leaf anatomy and completing the inventory of genes encoding the metabolic enzymes and solute transporters required to run C4 biochemistry (Wang et al., 2013, 2016; Fouracre et al., 2014; Burgess and Hibberd, 2015; Weber, 2015). Also, computational modeling of the evolutionary trajectory from C3 to C4 photosynthesis provided guidance for the implementation of the C4 trait in C3 backgrounds (Heckmann et al., 2013; Williams et al., 2013; Li et al., 2017). This recent work has indicated that achieving a C3-C4 intermediate state in which PR serves as a mechanism that increases the CO2 concentration in bundle sheath cells (Fig. 2) represents an important stepping stone during C4 evolution, which informs efforts aimed at synthetic experimental evolution of the pathway (Schuler et al., 2016).
Figure 2.
C3-C4 intermediate photosynthesis as a CCM. In C3-C4 intermediate photosynthesis, leaf mesophyll cells lose the activity of the mitochondrial glycine decarboxylase complex (GDC) while this activity is maintained in the bundle sheath cells. Hence, Gly produced as a consequence of PR in mesophyll cells is transported to bundle sheath cells, where it is decarboxylated by GDC in mitochondria. This locally increases the CO2 concentration in bundle sheath cells and promotes efficient fixation by Rubisco in bundle sheath chloroplasts. Ser and/or hydroxypyruvate are transported back to mesophyll cells for completion of the photorespiratory pathway.
A recent breakthrough en route to establishing C4 photosynthesis in the C3 crop rice was achieved by ectopic expression of Golden-like (GLK) transcription factor genes from maize (Zea mays; Wang et al., 2017). Transgenic rice lines expressing either ZmGLK1 or ZmG2 displayed increased numbers of differentiated chloroplasts in bundle sheath and mestome sheath cells of rice leaf VBs. Excitingly, increased chloroplast volume was accompanied by increased mitochondrial volume and increased amounts of plasmodesmata in these cell types. This indicates that proliferation of organelle volume and plasmodesmata numbers are emergent properties, triggered by the promotion of chloroplast differentiation through GLK transcription factors. Interestingly, transgenic rice calli that ectopically express ZmGLK1 or ZmG2 accumulate chlorophyll without the addition of cytokinin, indicating that the role of the GATA transcription factors GNC and GNL in regulating chlorophyll biosynthesis and plastid differentiation (Behringer and Schwechheimer, 2015; Cortleven and Schmülling, 2015) could be bypassed by expression of GLK1 or G2.
Increased organelle volume in cells surrounding the vasculature is characteristic of proto-Kranz anatomy, which is observed in several plant families that have evolved C3-C4 intermediate and C4 photosynthesis, such as Flaveria or Heliotropium, and is considered an important stepping stone on the evolutionary trajectory from C3 to C4 photosynthesis via intermediary states (Sage et al., 2012; Schlüter et al., 2017). However, to date, proto-Kranz anatomy has not been observed in BEP-clade grasses (Khoshravesh et al., 2016), hence the proto-Kranz-like anatomy observed in transgenic rice lines expressing ZmGLK1 or ZmG2 can be considered an important foundation and anatomical enabler for further engineering of rice toward C4. The next challenges on the way to C4 rice are now the implementation of a C4-like venation pattern (see Sedelnikova et al. [2018] for a recent review) and increasing the activity of enzymes and transporters mediating the C4 biochemistry in a cell-specific manner.
CARBOXYSOMES AND PYRENOIDS
Cyanobacteria and algae have evolved CCMs that rely on bringing Rubisco in close physical proximity with carbonic anhydrase and on increasing the cellular bicarbonate concentration through bicarbonate and/or CO2 transporters (Kerfeld and Melnicki, 2016; Meyer et al., 2016). In contrast to the C4 photosynthetic CCM outlined above, carboxysomes and pyrenoids do not depend on a specialized leaf anatomy or distribution of the CCM over specialized cell types and should hence be more straightforward to engineer into plant chloroplasts (Long et al., 2016).
The understanding of the molecular architecture of the pyrenoid from Chlamydomonas reinhardtii has seen dramatic progress during the past 3 years. Using high-throughput fluorescent protein tagging and affinity purification approaches, together with earlier proteomics work, 86 components of the C. reinhardtii pyrenoid have been identified (Mackinder et al., 2016, 2017). One key finding of this work was the discovery of the protein Essential Pyrenoid Component1, which occurs in equal abundance with Rubisco in the pyrenoid and likely serves to bind Rubisco via four interaction domains, therefore providing a scaffold for the assembly of Rubisco into the pyrenoid lattice. The C. reinhardtii pyrenoid does not behave like a crystalline structure, but is rather like a lipid and is a highly dynamic structure that de novo assembles and disassembles during cell cycle progression (Freeman Rosenzweig et al., 2017). While expression of individual components of the C. reinhardtii CCM did not enhance the growth of Arabidopsis (Arabidopsis thaliana) plants, these recent findings now set the stage for attempts to engineer a functional pyrenoid in plant cells (Atkinson et al., 2016).
Cyanobacterial CO2 and bicarbonate transporters increase the concentration of inorganic carbon inside the cytoplasm by 3 orders of magnitude over the external concentration (Mangan and Brenner, 2014). Bicarbonate then diffuses into carboxysomes, self-assembling proteinaceous microcompartments packed with Rubisco and carbonic anhydrase, where it is dehydrated to generate CO2. The low permeability of carboxysomes toward CO2 and O2 further ensures high CO2 concentration and low O2 concentration at the vicinity of Rubisco, thereby reducing and almost eliminating the oxygenation reaction. Using a diffusion-reaction model, it was recently shown that implementation of a carboxysome-type CCM in plant chloroplasts could potentially increase yield by 36% to 60% (McGrath and Long, 2014). Using simulations with individual CCM components, the model predicts a minimum set of components for a carboxysome to be effective in plant chloroplasts. These include knockout of the endogenous plastid-stroma carbonic anhydrase, plus the addition of the cyanobacterial BicA or BCT1 transporters, and/or the NAD(P) dehydrogenase complex1,3. Already the addition of the transporters BicA, BCT1, or SbtA to the plastid envelope membrane alone was predicted to increase the CO2 uptake under light saturation (McGrath and Long, 2014). Targeting of BicA and SbtA to the chloroplast envelope membrane was recently achieved in transiently transformed Nicotiana benthamiana cells (Rolland et al., 2016). Furthermore, production of a Rubisco-containing carboxysome in tobacco (Nicotiana tabacum) chloroplasts that replaces the endogenous tobacco Rubisco was recently achieved (Long et al., 2018), albeit the resulting transgenic plants displayed low growth rates even at 2% CO2, as expected from the modeling approach outlined above (McGrath and Long, 2014). The next challenge will now be to combine the individual components into a functional CCM.
BYPASSES TO THE CANONICAL PR PATHWAY
The first step of the PR pathway inside the chloroplast stroma is the dephosphorylation of 2PG to generate glycolate. Glycolate is then exported from the chloroplast by a glycolate transporter and imported into the peroxisomes, where it is oxidized to glyoxylate by glycolate oxidase (GOX). Many autotrophic and heterotrophic bacteria possess a dedicated pathway for the metabolism of glyoxylate to 3PGA (Eisenhut et al., 2006). This glycerate pathway proceeds via two reactions, the first condensing two molecules of glyoxylate to tartronate semialdehyde (by glyoxylate carboxylase; EC 4.1.1.47) while releasing CO2, and the second reducing tartronate semialdehyde to glycerate (by tartronate reductase; EC 1.1.1.60). Glycerate is then converted to 3PGA by glycerate kinase. This bacterial glycerate pathway has inspired plant researchers to implement it as a shortcut to the canonical PR pathway. This route prevents the wasteful release of ammonia in mitochondria by the GDC and hence increases the energy efficiency of the pathway (Peterhansel and Maurino, 2011). Furthermore, CO2 is released inside the chloroplasts and not in the mitochondria, thereby increasing local CO2 concentration in proximity to Rubisco and suppressing further oxygenation.
The implementation of the glycerate pathway in the chloroplast stroma requires the initial oxidation of glycolate to glyoxylate (Fig. 1, purple arrows). Hence, at least three enzymatic activities are needed inside the chloroplast stroma: GOX, glyoxylate carboxylase, and tartronate reductase. The GOX reaction releases H2O2, which needs to be detoxified, hence a fourth enzyme in the stroma is needed: catalase. Alternatively, glycolate can be converted to glyoxylate by a bacterial glycolate dehydrogenase, which does not generate H2O2. However, the bacterial enzyme (using a still unknown electron acceptor) consists of three subunits, which complicates the engineering task. In a first ground-breaking study, the bacterial glycolate dehydrogenase complex was introduced together with glyoxylate carboxylase and tartronate reductase into Arabidopsis chloroplasts. The resulting lines produced more biomass than the wild type, at least under short-day conditions (Kebeish et al., 2007). In another study, expression of the pathway in the chloroplasts of the biofuel crop Camelina sativa increased vegetative biomass and seed yields by more than 50% and supported faster development (Dalal et al., 2015).
Another bypass that completely oxidizes glycolate inside the chloroplast was developed by Maier et al. (2012)(Fig. 1, green arrows). In this pathway, glycolate is oxidized to glyoxylate by GOX and the resulting H2O2 is detoxified by expression of a plastid-targeted catalase. Glyoxylate is then condensed with acetyl-CoA to give malate (by malate synthase, as in the glyoxylate cycle). Finally, malate is oxidized to regenerate acetyl-CoA by the consecutive activities of NADP-dependent malic enzyme and pyruvate dehydrogenase, both of which are present in C3 plant chloroplasts at sufficient amounts to drive this pathway. Overall, this pathway converts all the carbon atoms of glycolate to CO2, which is released inside the chloroplast and reassimilated by Rubisco.
Despite being energetically less favorable than the wild-type pathway (12 ATP and eight NADPH needed to reassimilate the four molecules of CO2 released from two molecules of glycolate formed by two oxygenation reactions of Rubisco), Arabidopsis plants expressing this glycolate oxidation pathway displayed increased biomass production under short-day conditions. This is surprising since computational modeling predicted that the photosynthetic rate of plants expressing this pathway would be 31% lower than in wild-type plants (Xin et al., 2015).
One explanation for the discrepancy between the model and experimental results is that increased metabolism of glycolate relieves inhibition of Rubisco by this metabolite. However, Nölke et al. (2014) renders this explanation unlikely because lines displaying the highest yield gains also have higher glycolate concentrations than the wild type. In this work, only the three subunits of the Escherichia coli glycolate dehydrogenase (D, E, and F) were expressed in transgenic potato (Solanum tuberosum) plants, leading to increased conversion of glycolate to glyoxylate in chloroplasts. Albeit no enzyme activities for further conversion of glyoxylate were introduced, photosynthetic capacity was enhanced (increased Amax at 400 μL L−1 CO2) and tuber yield was increased by 2.3-fold (Nölke et al., 2014). The mechanism underpinning this increased yield remains to be determined. However, it seems that glycolate conversion to glyoxylate by itself suffices to enhance carbon fixation. Supporting this notion, fixation of 14CO2 was doubled in tobacco leaf discs exposed to 5 to 25 mm glyoxylate, indicating that Rubisco’s oxygenation is repressed by glyoxylate (Oliver and Zelitch, 1977). Similarly, CO2 fixation in soybean (Glycine max) mesophyll cells was increased by 150% after incubation with glyoxylate (Oliver, 1980).
As an alternative explanation, the growth benefits of the PR bypasses might result from pleiotropic effects, such as a decreased need for Rubisco protein due to higher plastidial CO2 concentration, which would reduce costs for de novo nitrogen assimilation and protein maintenance.
In all of these pathways, the metabolism of glycolate inside the chloroplast competes with export of glycolate from chloroplasts by the glycolate exporter PLGG1. As suggested by Weber and Bräutigam (2013), repression of this transporter should increase the flux into the bypasses and thereby increase their efficiency. Indeed, recent work by South et al. (2019) combined the above-described pathways, as well as another novel bypass, with the repression of PLGG1 expression using an antisense RNA in transgenic tobacco lines. They found that this combination reduced export of glycolate from the chloroplast and increased biomass gain as compared with plants in which a bypass was established without suppressing glycolate transport. The most efficient bypass found in this work was a variant of the glycolate oxidation pathway (Maier et al., 2012), in which the H2O2-producing GOX was replaced with a glycolate dehydrogenase from C. reinhardtii mitochondria (South et al., 2019). Hence, this pathway fully decarboxylates photorespiratory glycolate via glyoxylate, malate synthase, and NADP-malic enzyme. Importantly, as noted above, introduction of the photorespiratory bypasses suppressed the photorespiratory phenotype caused by reduced expression of PLGG1. This constitutes strong evidence for the concept that photorespiratory bypasses render the canonical photorespiratory pathway at least partially dispensable. This further opens new avenues for assessing assumptions on the roles of canonical PR beyond detoxification of 2-phosphoglycolate and salvage of CBC intermediates, such as production of Ser (Benstein et al., 2013) or dissipation of excess excitation energy (Eisenhut et al., 2017).
We conclude this section by emphasizing that much is yet unknown about the mechanisms that underlie the growth benefits obtained by the PR bypasses and whether they are truly related to the more efficient metabolism of glycolate. It is worth noting that a similar alternative PR route, aimed at bypassing downstream PR by converting peroxisomal glyoxylate to tartronate semialdehyde and hydroxypyruvate (Fig. 1, pink arrows), did not enhance photosynthetic productivity (Carvalho et al., 2011). This casts doubt on the validity of traditional explanations for growth enhancement via PR bypasses.
BYPASSING PR WITHOUT CO2 RELEASE
A bolder approach to the problem of PR is to try to abolish CO2 release altogether. For example, it was suggested that glyoxylate could be recycled into central metabolism via the activity of the downstream cycle of the prokaryotic 3-hydroxypropionate bicycle (Zarzycki et al., 2009; Shih et al., 2014). The net reaction of this cycle is the conversion of glyoxylate to pyruvate, a key cellular intermediate that can be further metabolized to various compounds. However, pyruvate cannot be easily reassimilated to the CBC because phosphoglycerate mutase and enolase, key enzymes of glycolysis/gluconeogenesis, are usually not expressed in chloroplasts (Prabhakar et al., 2009; Fukayama et al., 2015). While pyruvate can potentially be exported to the cytosol, converted to 3PGA or triose phosphate, and imported back into the chloroplast, the complexity of this route renders it quite unlikely to be useful. Another recent study put forward the synthetic malyl-CoA-glycerate cycle as a way to convert glycolate to acetyl-CoA (Yu et al., 2018). While this pathway could be useful for the biosynthesis of chemicals that originate from acetyl-CoA, it cannot be considered a true PR bypass as acetyl-CoA cannot be easily reassimilated to the CBC.
A different photorespiratory bypass was suggested to involve the reduction of 2PG to 2-phosphoglycolaldehyde, which is then condensed with dihydroxyacetone phosphate to give xylulose 1,5-bisphosphate (Ort et al., 2015). The latter intermediate is then dephosphorylated to give the CBC intermediate xylulose 5-phosphate. While the particular metabolism of this route might not be ideal, 2PG is unlikely to accumulate to high enough levels as to enable its reduction and xylulose 1,5-bisphosphate is a potent inhibitor of Rubisco (Bracher et al., 2015), the general structure it suggests could be very efficient. In particular, reduction of glycolate, the cellular concentration of which is in the millimolar range, to glycolaldehyde could be followed by aldol condensation with a phosphor-sugar of the CBC to produce a longer chain phosphor-sugar that is reassimilated to the cycle without carbon loss (Bar-Even, 2018).
Recently, a pathway based on the reduction of glycolate to glycolaldehyde was demonstrated in vitro (Trudeau et al., 2018). In this study, the substrate specificities of two enzymes were engineered to enable this reduction: acetyl-CoA synthetase was engineered to efficiently accept glycolate and generate glycolyl-CoA, and propionyl-CoA reductase was engineered to accept glycolyl-CoA as well as NADPH (the native enzyme prefers NADH). These engineered enzymes, which jointly convert glycolate to glycolaldehyde, were then combined with existing enzymes: an aldolase that condenses glycolaldehyde with GAP to give arabinose 5-phosphate, an isomerase that converts arabinose 5-phosphate to ribulose 5-phosphate, and the native kinase that generates RuBP. Together, this enzymatic sequence converted glycolate to Rubisco’s substrate RuBP without the release of CO2 and ammonia (Fig. 1, blue arrows). While the function of this metabolic sequence has been demonstrated in vitro, it awaits testing in plants.
RADICAL REDESIGN OF PCA WITHOUT RUBISCO AND THE CBC
Instead of fixing PR, other studies suggested replacing Rubisco and the CBC. The first of these studies used a computational approach to systematically identify all pathways that can be established using mix-and-match of existing enzymes from different sources (Bar-Even et al., 2010). Multiple candidate routes were uncovered this way, and one group of pathways, utilizing the most efficient carboxylating enzyme, PEPC (Cotton et al., 2018), was analyzed in detail and predicted to support a higher carbon fixation rate than the CBC. A recent study took a more compelling step toward synthetic carbon fixation, realizing in vitro a synthetic pathway (CETCH pathway) that converts CO2 into the C2 intermediate glyoxylate (Schwander et al., 2016). This pathway was based on the reductive carboxylation of organic acids carrying unsaturated bonds (i.e. crotonyl-CoA and acrylyl-CoA). Yet, while such synthetic carbon fixation routes provide an exciting avenue for further research, it is highly unlikely that we could use one of them to replace the CBC. The CBC seems to be too central in plant metabolism to be replaced, and the establishment of a long cyclic pathway in a plant host is highly challenging.
As an alternative strategy, instead of replacing the CBC, we can install a complementary route to assist its activity. This was suggested, for example, for the malyl-CoA-glycerate cycle mentioned above. A particularly interesting option is to use synthetic routes that are based on CO2 reduction rather than carboxylation (Cotton et al., 2018). Specifically, the low reduction potential that persists in the chloroplast could be harnessed to reduce CO2 into formate. Formate can be then assimilated to central metabolism via numerous routes (Bar-Even, 2016), directly contributing to carbon fixation. The advantages of this strategy are its very high energetic efficiency, its minimal overlap with central metabolism, and an implementation that requires a relatively small number of enzymes (Bar-Even, 2018; Cotton et al., 2018). An especially interesting formate assimilation strategy involves attaching this C1 intermediate to tetrahydrofolate and reducing it to 5,10-methylenetetrahydrofolate, which can be condensed with photorespiratory Gly to make Ser (Fig. 1, red arrows). This would avoid Gly decarboxylation and transform PR from being a carbon-negative route into a carbon-positive process (Bar-Even, 2018).
CONCLUSION
Clearly, reducing the rate of PR by increasing the concentration of CO2 at the site of Rubisco, and rerouting photorespiratory metabolism by introduction of synthetic bypasses and novel pathways that turn PR into a carbon-positive process, hold huge potential for increased yield, without increasing the need for arable land. Even higher yield gains should become possible by combining the engineering of photosynthetic carbon metabolism with recent approaches aimed at more rapid adaptation to fluctuating light conditions by reducing the loss of excitation energy due to overprotection of photosystems (Kromdijk et al., 2016; Slattery et al., 2018). Quoting Norman Borlaug (“Then I wake up and become disillusioned to find that mutation genetics programs are still engaged mostly in such minutiae as putting beards on wheat plants and taking off the hairs.”), we should leave the minutiae behind and reap the full potential of synthetic biology to overcome one of the major challenges of the 21st century, sustainably feeding a growing population without destroying the environment.
Footnotes
This work was supported by the German Federal Ministry of Research (031B0205 and 031B0194), the EU H2020 (CSA CropBooster-P), the Deutsche Forschungsgemeinschaft (EXC 1028), and the Max Planck Society (to A.B.-E.).
Articles can be viewed without a subscription.
References
- Amthor JS. (2010) From sunlight to phytomass: On the potential efficiency of converting solar radiation to phyto-energy. New Phytol 188: 939–959 [DOI] [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LCM, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ (2016) Introducing an algal carbon-concentrating mechanism into higher plants: Location and incorporation of key components. Plant Biotechnol J 14: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A. (2016) Formate assimilation: The metabolic architecture of natural and synthetic pathways. Biochemistry 55: 3851–3863 [DOI] [PubMed] [Google Scholar]
- Bar-Even A. (2018) Daring metabolic designs for enhanced plant carbon fixation. Plant Sci 273: 71–83 [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Noor E, Lewis NE, Milo R (2010) Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci USA 107: 8889–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR (2010) Photorespiration: Players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Behringer C, Schwechheimer C (2015) B-GATA transcription factors: Insights into their structure, regulation, and role in plant development. Front Plant Sci 6: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flügge UI, Krueger S (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25: 5011–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flügge U-I, Krueger S (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25: 5011–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betti M, Bauwe H, Busch FA, Fernie AR, Keech O, Levey M, Ort DR, Parry MAJ, Sage R, Timm S, et al. (2016) Manipulating photorespiration to increase plant productivity: Recent advances and perspectives for crop improvement. J Exp Bot 67: 2977–2988 [DOI] [PubMed] [Google Scholar]
- Bracher A, Sharma A, Starling-Windhof A, Hartl FU, Hayer-Hartl M (2015) Degradation of potent Rubisco inhibitor by selective sugar phosphatase. Nat Plants 1: 14002. [DOI] [PubMed] [Google Scholar]
- Burgess SJ, Hibberd JM (2015) Insights into C4 metabolism from comparative deep sequencing. Curr Opin Plant Biol 25: 138–144 [DOI] [PubMed] [Google Scholar]
- Carvalho JdeF, Madgwick PJ, Powers SJ, Keys AJ, Lea PJ, Parry MAJ (2011) An engineered pathway for glyoxylate metabolism in tobacco plants aimed to avoid the release of ammonia in photorespiration. BMC Biotechnol 11: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Osborne CP, Chatelet DS, Columbus JT, Besnard G, Hodkinson TR, Garrison LM, Vorontsova MS, Edwards EJ (2013) Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc Natl Acad Sci USA 110: 1381–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M, Tilman D (2017) Comparative analysis of environmental impacts of agricultural production systems, agricultural input efficiency, and food choice. Environ Res Lett 12: 064016 [Google Scholar]
- Cortleven A, Schmülling T (2015) Regulation of chloroplast development and function by cytokinin. J Exp Bot 66: 4999–5013 [DOI] [PubMed] [Google Scholar]
- Cotton CA, Edlich-Muth C, Bar-Even A (2018) Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation. Curr Opin Biotechnol 49: 49–56 [DOI] [PubMed] [Google Scholar]
- Dalal J, Lopez H, Vasani NB, Hu Z, Swift JE, Yalamanchili R, Dvora M, Lin X, Xie D, Qu R, et al. (2015) A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa. Biotechnol Biofuels 8: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton AK, Simon R, Weber APM (2013) C4 photosynthesis: From evolutionary analyses to strategies for synthetic reconstruction of the trait. Curr Opin Plant Biol 16: 315–321 [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Kahlon S, Hasse D, Ewald R, Lieman-Hurwitz J, Ogawa T, Ruth W, Bauwe H, Kaplan A, Hagemann M (2006) The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol 142: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Bräutigam A, Timm S, Florian A, Tohge T, Fernie AR, Bauwe H, Weber APM (2017) Photorespiration is crucial for dynamic response of photosynthetic metabolism and stomatal movement to altered CO2 availability. Mol Plant 10: 47–61 [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JDG, Patron NJ, Marillonnet S (2014) A Golden Gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Foley JA, Ramankutty N, Brauman KA, Cassidy ES, Gerber JS, Johnston M, Mueller ND, O’Connell C, Ray DK, West PC, et al. (2011) Solutions for a cultivated planet. Nature 478: 337–342 [DOI] [PubMed] [Google Scholar]
- Fouracre JP, Ando S, Langdale JA (2014) Cracking the Kranz enigma with systems biology. J Exp Bot 65: 3327–3339 [DOI] [PubMed] [Google Scholar]
- Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P, Plitzko JM, Förster F, et al. (2017) The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171: 148–162.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukayama H, Masumoto C, Taniguchi Y, Baba-Kasai A, Katoh Y, Ohkawa H, Miyao M (2015) Characterization and expression analyses of two plastidic enolase genes in rice. Biosci Biotechnol Biochem 79: 402–409 [DOI] [PubMed] [Google Scholar]
- Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber APM, Lercher MJ (2013) Predicting C4 photosynthesis evolution: Modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153: 1579–1588 [DOI] [PubMed] [Google Scholar]
- Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stäbler N, Schönfeld B, Kreuzaler F, Peterhänsel C (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599 [DOI] [PubMed] [Google Scholar]
- Kerfeld CA, Melnicki MR (2016) Assembly, function and evolution of cyanobacterial carboxysomes. Curr Opin Plant Biol 31: 66–75 [DOI] [PubMed] [Google Scholar]
- Khoshravesh R, Stinson CR, Stata M, Busch FA, Sage RF, Ludwig M, Sage TL (2016) C3-C4 intermediacy in grasses: Organelle enrichment and distribution, glycine decarboxylase expression, and the rise of C2 photosynthesis. J Exp Bot 67: 3065–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP (2016) Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354: 857–861 [DOI] [PubMed] [Google Scholar]
- Li Y, Heckmann D, Lercher MJ, Maurino VG (2017) Combining genetic and evolutionary engineering to establish C4 metabolism in C3 plants. J Exp Bot 68: 117–125 [DOI] [PubMed] [Google Scholar]
- Long BM, Rae BD, Rolland V, Förster B, Price GD (2016) Cyanobacterial CO2-concentrating mechanism components: Function and prospects for plant metabolic engineering. Curr Opin Plant Biol 31: 1–8 [DOI] [PubMed] [Google Scholar]
- Long BM, Hee WY, Sharwood RE, Rae BD, Kaines S, Lim YL, Nguyen ND, Massey B, Bala S, von Caemmerer S, et al. (2018) Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat Commun 9: 3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29: 315–330 [DOI] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu XG (2015) Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161: 56–66 [DOI] [PubMed] [Google Scholar]
- Mackinder LCM, Meyer MT, Mettler-Altmann T, Chen VK, Mitchell MC, Caspari O, Freeman Rosenzweig ES, Pallesen L, Reeves G, Itakura A, et al. (2016) A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc Natl Acad Sci USA 113: 5958–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, Jonikas MC (2017) A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171: 133–147.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Fahnenstich H, von Caemmerer S, Engqvist MK, Weber AP, Flügge UI, Maurino VG (2012) Transgenic introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front Plant Sci 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan N, Brenner M (2014) Systems analysis of the CO2 concentrating mechanism in cyanobacteria. eLife 3: e02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield BD, Mumm RH (2014) Survey of plant density tolerance in U.S. maize germplasm. Crop Sci 54: 157–173 [Google Scholar]
- McGrath JM, Long SP (2014) Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164: 2247–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, McCormick AJ, Griffiths H (2016) Will an algal CO2-concentrating mechanism work in higher plants? Curr Opin Plant Biol 31: 181–188 [DOI] [PubMed] [Google Scholar]
- Nölke G, Houdelet M, Kreuzaler F, Peterhänsel C, Schillberg S (2014) The expression of a recombinant glycolate dehydrogenase polyprotein in potato (Solanum tuberosum) plastids strongly enhances photosynthesis and tuber yield. Plant Biotechnol J 12: 734–742 [DOI] [PubMed] [Google Scholar]
- Oliver DJ. (1980) The effect of glyoxylate on photosynthesis and photorespiration by isolated soybean mesophyll cells. Plant Physiol 65: 888–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DJ, Zelitch I (1977) Increasing photosynthesis by inhibiting photorespiration with glyoxylate. Science 196: 1450–1451 [DOI] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, et al. (2015) Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol 208: 13–19 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Maurino VG (2011) Photorespiration redesigned. Plant Physiol 155: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar V, Löttgert T, Gigolashvili T, Bell K, Flügge UI, Häusler RE (2009) Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett 583: 983–991 [DOI] [PubMed] [Google Scholar]
- Rolland V, Badger MR, Price GD (2016) Redirecting the cyanobacterial bicarbonate transporters BicA and SbtA to the chloroplast envelope: Soluble and membrane cargos need different chloroplast targeting signals in plants. Front Plant Sci 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F (2012) Photorespiration and the evolution of C4 photosynthesis. Annu Rev Plant Biol 63: 19–47 [DOI] [PubMed] [Google Scholar]
- Savir Y, Noor E, Milo R, Tlusty T (2010) Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc Natl Acad Sci USA 107: 3475–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter U, Bräutigam A, Gowik U, Melzer M, Christin PA, Kurz S, Mettler-Altmann T, Weber APM (2017) Photosynthesis in C3-C4 intermediate Moricandia species. J Exp Bot 68: 191–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler ML, Mantegazza O, Weber APM (2016) Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J 87: 51–65 [DOI] [PubMed] [Google Scholar]
- Schwander T, Schada von Borzyskowski L, Burgener S, Cortina NS, Erb TJ (2016) A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354: 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OV, Hughes TE, Langdale JA (2018) Understanding the genetic basis of C4 Kranz anatomy with a view to engineering C3 crops. Annu Rev Genet 52: 249–270 [DOI] [PubMed] [Google Scholar]
- Shih PM, Zarzycki J, Niyogi KK, Kerfeld CA (2014) Introduction of a synthetic CO2-fixing photorespiratory bypass into a cyanobacterium. J Biol Chem 289: 9493–9500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, McAusland L, Headland LR, Lawson T, Raines CA (2015) Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. J Exp Bot 66: 4075–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Lopez-Calcagno PE, Davey PA, Headland LR, Lawson T, Timm S, Bauwe H, Raines CA (2017) Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnol J 15: 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery RA, Walker BJ, Weber APM, Ort DR (2018) The impacts of fluctuating light on crop performance. Plant Physiol 176: 990–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Lopez-Calcagno PE, Raines CA, Ort DR (2018) Optimizing photorespiration for improved crop productivity. J Integr Plant Biol 60: 1217–1230 [DOI] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR (2019) Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 362: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez GGB, Farquhar GD, Andrews TJ (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA 103: 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108: 20260–20264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Florian A, Arrivault S, Stitt M, Fernie AR, Bauwe H (2012) Glycine decarboxylase controls photosynthesis and plant growth. FEBS Lett 586: 3692–3697 [DOI] [PubMed] [Google Scholar]
- Trudeau DL, Edlich-Muth C, Zarzycki J, Scheffen M, Goldsmith M, Khersonsky O, Avizemer Z, Fleishman SJ, Cotton CAR, Erb TJ, et al. (2018) Design and in vitro realization of carbon-conserving photorespiration. Proc Natl Acad Sci USA 115: E11455–E11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR (2016) The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol 67: 107–129 [DOI] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA (2013) Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J 75: 656–670 [DOI] [PubMed] [Google Scholar]
- Wang P, Vlad D, Langdale JA (2016) Finding the genes to build C4 rice. Curr Opin Plant Biol 31: 44–50 [DOI] [PubMed] [Google Scholar]
- Wang P, Khoshravesh R, Karki S, Tapia R, Balahadia CP, Bandyopadhyay A, Quick WP, Furbank R, Sage TL, Langdale JA (2017) Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr Biol 27: 3278–3287.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM. (2015) Discovering new biology through sequencing of RNA. Plant Physiol 169: 1524–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, Bräutigam A (2013) The role of membrane transport in metabolic engineering of plant primary metabolism. Curr Opin Biotechnol 24: 256–262 [DOI] [PubMed] [Google Scholar]
- Williams ME. (2016) Carbon-fixing reactions of photosynthesis. Plant Cell 78: tpc.116.tt0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Johnston IG, Covshoff S, Hibberd JM (2013) Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2: e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin C-P, Tholen D, Devloo V, Zhu X-G (2015) The benefits of photorespiratory bypasses: How can they work?. Plant Physiol 167: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Li X, Duchoud F, Chuang DS, Liao JC (2018) Augmenting the Calvin-Benson-Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway. Nat Commun 9: 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzycki J, Brecht V, Müller M, Fuchs G (2009) Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc Natl Acad Sci USA 106: 21317–21322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR (2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]