Abstract
Aims:
Describe modifications to technical genomic terminology made by interpreters during disclosure of whole exome sequencing (WES) results.
Patients & Methods:
Using discourse analysis, we identified and categorized interpretations of genomic terminology in 42 disclosure sessions where Spanish-speaking parents received their child’s WES results either from a clinician using a medical interpreter, or directly from a bilingual physician.
Results:
Overall, 76% of genomic terms were interpreted accordantly, 11% were misinterpreted, and 13% were omitted. Misinterpretations made by interpreters and bilingual physicians included using literal and non-medical terminology to interpret genomic concepts.
Conclusions:
Modifications to genomic terminology made during interpretation highlight the need to standardize bilingual genomic lexicons. We recommend Spanish terms that can be used to refer to genomic concepts.
Keywords: medical interpreters, interpretation accuracy, genomic terminology, communication barriers, genomics, Spanish-speakers
INTRODUCTION
Language discordance between physicians and their patients has been shown to negatively affect patient comprehension of clinical information [1, 2]. Miscommunication resulting from language barriers in clinical settings can lead to adverse outcomes and even fatal medical errors [1, 3, 4]. The use of medical interpreters in language discordant encounters can bridge these linguistic gaps [5, 6] and improve clinical care and outcomes for limited English proficient (LEP) patients [7]. Utilizing professional medical interpreters instead of ad hoc interpreters, such as nurses or family members, results in interpretation that is both more effective and less likely to result in errors that have serious clinical consequences for patients [7, 8, 9, 10, 11].
While professional interpreters can lessen communication barriers between physician and patient, studies of medical interpretation in clinical and research settings have shown that interpreters also omit and modify clinicians’ words and phrases during interpretation [10, 12, 13], and that technical language can lead to increased misinterpretations [14]. As clinical genomic sequencing (GS) tests become more prevalent [15], modifications made during interpretation may be exacerbated by the lack of widely-accepted or standardized genomic lexicons in other languages and the absence of national training or certification standards for medical interpreters in the United States [12, 16, 17]. For example, an observational study of BRCA test result genetic counseling sessions with low-income Latina immigrants found professional interpreters were sometimes unfamiliar with the technical language and probabilistic nature of genetic information [18]. As broader GS tests, such as whole exome sequencing (WES), are utilized, it is critical to examine how medical interpreters interpret technical GS information for LEP families, what terms they use, and what implications interpretation modifications may have for the meaning of the clinical information presented.
We provide a first look into how WES terminology is being interpreted to Spanish-speaking families in order to begin addressing this gap in the literature of how technical GS information is interpreted for LEP populations. The goals of this manuscript are to describe the modifications to GS terminology made by hospital-based medical interpreters during disclosure of WES results for Spanish-speaking families of pediatric cancer patients and to discuss the implications of those modifications for the meaning of clinical information. Although some studies have described the types of linguistic modifications made on a semantic level during medical interpretation [10, 12, 13, 14, 18], this study is the first to do so with regard to technical GS test result terminology.
PATIENTS AND METHODS
The BASIC3 Study
As a Clinical Sequencing Exploratory Research (CSER) program project supported by the National Institutes of Health (NIH), the Baylor Advancing Sequencing in Childhood Cancer Care (BASIC3) study evaluates the incorporation of clinical WES results into the care of children with newly-diagnosed solid tumors. The study was approved by the Institutional Review Board (IRB) of Baylor College of Medicine, which also serves as the IRB for Texas Children’s Hospital (TCH). The study has recently completed enrollment of its primary study cohort at Texas Children’s Cancer Center (TCCC) with 287 patients as well as their parent(s)/legal guardian(s). Twenty pediatric oncologists were also enrolled as subjects in the study and completed their own informed consent procedure. As part of the BASIC3 study, germline and tumor (if available) WES results are returned to patients/families by their child’s primary pediatric oncologist and a study genetic counselor and entered into the child’s electronic health record [19, 20]. As a part of their participation in the BASIC3 study, pediatric oncologists participated in two genetics-specific training sessions and also had a medical geneticist and pediatric oncologist expert in genomics available as resources prior to each disclosure.
Hospital-Based Medical Interpreters
Hospital-based medical interpreters employed by TCH are requested to participate in the disclosure of WES results to families who self-identified as primarily Spanish-speaking, (53/287, 18%) per standard clinical care at TCH. According to TCH Language Services, interpreters must have at least two years of experience interpreting in a medical setting and must also pass a healthcare interpreter assessment exam administered by an outside partner organization in order to be employed full-time or part-time by TCH. While TCH does not require interpreters to be certified by an outside entity, most TCH interpreters do have some form of outside certification and generally have an average of five to ten years of experience interpreting in a medical setting. As interpreters were hospital-based and participated in disclosure sessions as part of routine clinical care, they were not enrolled into the BASIC3 study and did not receive training specific to the study or GS terminology. Since interpreters were performing their routine clinical care duties and not asked to participate in any additional study procedures, we received a waiver of consent from the BCM IRB to analyze interpreters’ statements during BASIC3 disclosure sessions.
Procedures
The hospital-based interpreters assisted in the BASIC3 disclosure sessions in the pediatric oncology clinic as part of their regular clinical duties. Although all consent documents were translated into Spanish for families, the actual WES test reports and a genetic counseling letter summarizing the germline results in lay language were provided to all families in English at the disclosure visit.
WES results disclosure sessions were audio-recorded and sessions with Spanish-speaking families were transcribed verbatim, including all English and Spanish as presented in the audio-recording. Transcripts of sessions were included in analysis if the patient’s primary parent/legal guardian, primary oncologist and/or study genetic counselor, and one of 16 hospital-based interpreters were present. The patient and/or other family members may also have been present. We also analyzed sessions in which a hospital-based interpreter was not present because the primary oncologist was bilingual and served as the session interpreter.
Discourse Analysis
Using discourse analysis [21, 22], we compared Spanish interpretations of GS terms to clinicians’ original statements in the interpreter-mediated disclosure sessions. Two bilingual (English-Spanish) investigators identified and compiled all sentences containing clinicians’ mentions of GS terminology and the interpreters’ corresponding Spanish interpretation. All Spanish interpretations of the aforementioned instances were translated into English as agreed upon by the bilingual investigators for the investigators who were not Spanish-speaking. The research team, including the two bilingual investigators and two study genetic counselors, met to assess each sentence spoken by the clinician in English that contained at least one GS term compared to the interpretation of that sentence into Spanish by the interpreter on a linguistic and semantic level to determine if the intended meaning was clearly conveyed. We categorized each interpretation as one of the following: (1) accordant, if either a Spanish term with an equivalent meaning was used or the meaning of the term was correctly communicated; (2) misinterpreted, if either an nonequivalent Spanish term was used or the meaning of the term was incorrectly communicated; or (3) omitted, if an equivalent Spanish term was not used and the meaning of the term was not communicated. Categorization of each sentence was discussed until consensus was reached. We then calculated the frequencies of modifications to GS terms. Although only excerpts containing a GS term were analyzed, events of misinterpretation were considered within the full context of the disclosure discussion to ensure the appropriate clinical message was ultimately conveyed. We also observed the use of Spanish GS terminology by two bilingual study oncologists who directly disclosed results to families in disclosure sessions and compared these to interpreter-mediated sessions. As we defined the unit of analysis in both types of sessions as a sentence in which the clinician mentions at least one GS term, we did not capture instances in which clinicians chose to describe a term in place of uttering the exact term.
RESULTS
Overview
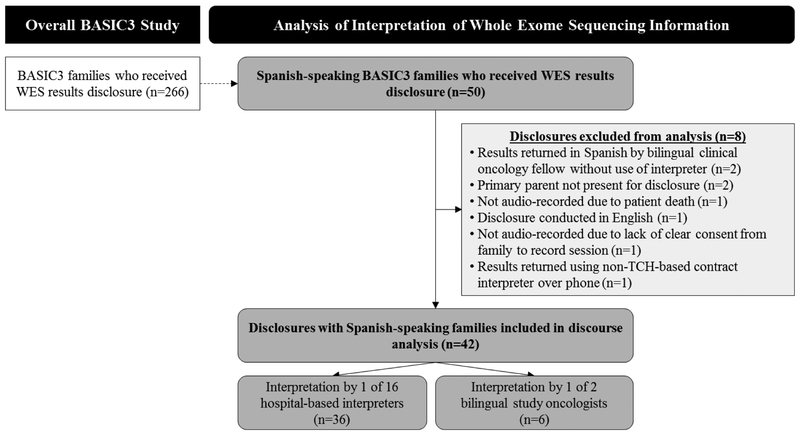
Of the 53 Spanish-speaking families enrolled, 50 disclosure sessions had been conducted at the time of analysis in May 2017 (Figure 1). Eight of these sessions were excluded from analysis for the following reasons: a bilingual clinical oncology fellow not enrolled as a physician subject in the study returned the results in Spanish without the use of an interpreter (n=2), the primary parent/guardian was not present at disclosure (n=2), a session was not audio-recorded due to patient death before disclosure (n=1), a session was not audio-recorded due to lack of clear consent from the family to audio-record the session (n=1), a session was conducted in English per parental preference with an interpreter present (n=1), and a session was conducted using a non-TCH-based contract interpreter over the phone (n=1). The remaining 42 disclosures were analyzed for this study. In 36 of the 42 analyzed disclosure sessions, one of 16 hospital-based interpreters, one of twelve English-speaking study oncologists and/or one of two study genetic counselors participated. In the other six disclosure sessions, one of two bilingual study oncologists provided the Spanish interpretation without a medical interpreter.
Figure 1.
BASIC3 interpreter discourse analysis flow diagram.
Parent demographic information is shown in Table 1. All parents self-identified as Hispanic or Latino and both bilingual oncologists self-identified as Hispanic or Latino. As interpreters were not enrolled into the BASIC3 study, their demographic information was not collected.
Table 1.
Parent demographics.
| Demographic characteristic, (n=42) | N (%) |
|---|---|
| Gender | |
| Female | 36 (86) |
| Ethnicity | |
| Hispanic or Latino | 42 (100) |
| Race | |
| White | 10 (24) |
| Native American or Alaska Native | 3 (7) |
| Declined or Not Specified | 29 (69) |
Interpreters participated in an average of two disclosure sessions, with a range of one to four sessions and a median of two sessions. The disclosure sessions analyzed with an interpreter present averaged 26 minutes in length (range: 8 minutes to 1 hour 13 minutes), compared to an average length of 21 minutes for disclosures with English-speaking families (range 5 minutes to 1 hour 7 minutes). The six disclosure sessions in which a bilingual oncologist returned the results directly to the family without the use of an interpreter averaged 16 minutes in length (range 9 minutes to 26 minutes).
Interpretation Accuracy of Technical Genomic Terms
We tracked mentions of all technical GS terminology by clinicians over the 36 transcripts in which an interpreter was present and analyzed the frequency of interpretation modifications. The GS terms and concepts mentioned included: “actionable,” “carrier/carrier status/carry,” “deleterious mutation,” “pathway,” “risk(s)/at risk/increased risk,” “sequencing/whole exome sequencing,” and “variants of uncertain/unknown significance.” Definitions of these terms are provided in Table 2.
Table 2.
Definitions of genomic sequencing terms observed in BASIC3 disclosure sessions.
| Term | Definition |
|---|---|
| Actionable | Pathogenic variants that suggest medical intervention is necessary, such as those found in genes listed in the American College of Medical Genetics and Genomics (ACMG) 2016 policy statement regarding reporting of secondary findings [23]. |
| Carrier / Carrier status | A patient with a single pathogenic variant in a gene known to be associated with conditions inherited in a recessive manner. A carrier is not at risk to have the condition, but could pass the pathogenic variant on to their children. |
| At risk | Average population risk to develop a condition. |
| Example: The general population risk for a woman to develop breast cancer is 12%. All women are at risk to develop breast cancer. | |
| Increased risk | Risk to develop a condition increased beyond the average population risk. |
| Example: A woman with a BRCA1 pathogenic variant has a 65% chance to develop breast cancer by age 70 [24]. She is at increased risk to develop breast cancer. | |
| Pathogenic variant / Deleterious mutation | A change in a gene sequence that can cause disease. This is a classification made by the clinical testing laboratory, consistent with the terminology recommended by ACMG in 2015 [25]. |
| Pathway | A sequence of gene or protein interactions. |
| Sequencing | Laboratory test used to identify changes in a gene’s nucleotide code. |
| Variants of uncertain / unknown significance | Novel or rare changes in a gene sequence, for which the clinical implications for the patient are unknown. This is a classification made by the clinical testing laboratory consistent with the 2015 ACMG guidelines, and may change as additional sequencing data becomes available [25]. |
Of note, as the focus of this discourse analysis is to describe what Spanish terms were used to describe the common English genomic terms in Table 2, we captured only interpretations of instances in which a clinician uttered one of these exact GS terms. The frequencies we report do not capture instances in which clinicians described a GS concept (e.g., “gene changes that we haven’t seen before and don’t know what they mean yet”) in place of uttering the exact term (e.g., “variants of uncertain significance”). This discrepancy affects all terms but most notably affects the frequency of “variants of uncertain significance” (VUS), as we only report seven instances of the term VUS. As a category of results, VUS was described in all disclosure sessions, although not all patients received VUS results.
Table 3 shows the total frequencies of accordant interpretations, misinterpretations, and omissions to GS terms in all 36 disclosure sessions. Overall, 76% of GS terms were interpreted accordantly, 11% of GS terms were misinterpreted, including incorrect/nonequivalent and literal interpretations, and 13% of GS terms were omitted completely during interpretation.
Table 3.
Frequency of interpretation modifications made to each genomic sequencing term.
| Genomic Sequencing Term* | Accordant Interpretation(s) (%) |
Misinterpretation(s) (%) |
Omission(s) (%) |
|---|---|---|---|
| Carrier / Carrier status / Carry (n=333) | 249 (75) | 38 (11) | 46 (14) |
| At risk / Increased risk (n=118) | 98 (83) | 7 (6) | 13 (11) |
| Variants of uncertain significance (n=7) | 6 (86) | 1 (14) | 0 |
| Actionable (n=3) | 2 (67) | 1 (33) | 0 |
| Pathway (n=3) | 1 (33) | 2 (67) | 0 |
| Deleterious mutation (n=2) | 1 (50) | 1 (50) | 0 |
| Sequencing (n=2) | 1 (50) | 1 (50) | 0 |
| Total (n=468) | 358 (76) | 51 (11) | 59 (13) |
The frequency of total mentions of each term is represented by “n”.
The majority (n=29, 81%) of transcripts had at least one omission or misinterpretation. Nineteen transcripts (53%) had at least one misinterpretation and 23 transcripts (64%) had at least one omission. While some interpreters were more accurate in their interpretations of these terms than others, all 16 interpreters made at least one of these two types of modifications. Most interpreters made at least one misinterpretation (n=12, 75%) and one omission (n=15, 94%).
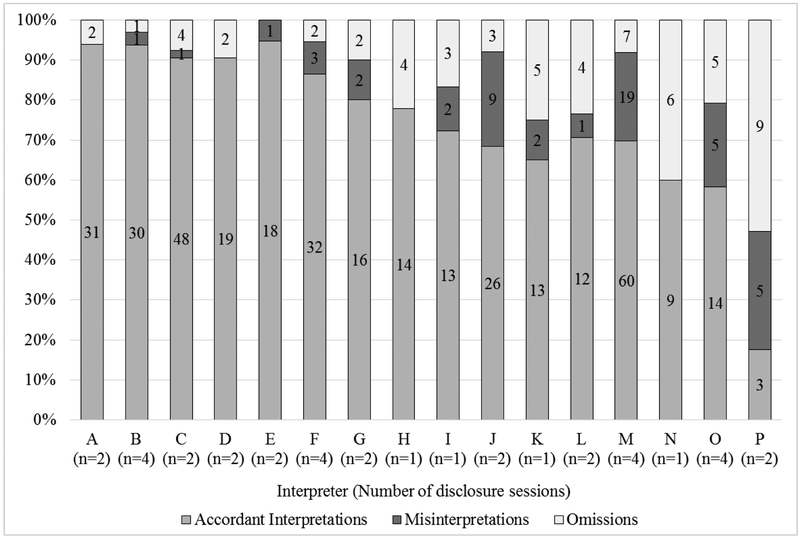
Figure 2 illustrates considerable variability in the frequencies of modifications made by each interpreter as well as the percentage of GS terms that each interpreter accordantly interpreted, misinterpreted, and omitted out of the total mentions of GS terms in all disclosure sessions in which that interpreter participated.
Figure 2.
Frequency and percentage of all genomic sequencing term modifications made by each interpreter.
*The number of disclosure sessions in which each interpreter participated is represented by “n” in the figure.
Misinterpretations of Technical Genomic Terms
We identified interpretations of GS terms as misinterpreted when interpreters used incorrect/nonequivalent Spanish terms, and when they interpreted information using literal non-medical terms instead of using precise genomic terms. The terms “carrier” and “carrier status” were interpreted literally 38 times across 13 transcripts by eight different interpreters. Instead of utilizing the correct medical Spanish term meaning ‘to carry a gene’ (“portar”), interpreters used more literal variations of the term “carry” including “cargar,” “llevar,” and “traer” which mean ‘to carry an object,’ ‘to take,’ ‘to hold,’ or ‘to bring.’ These literal interpretations of “carry” resulted in stating that the child ‘holds’ or ‘takes’ a gene mutation. Additionally, instead of using the correct medical term for genetic carrier (“portador”), some interpreters used the term “acarreador,” which translates to ‘a porter’ or ‘someone who transports items.’ Seven times in one disclosure session, an interpreter incorrectly interpreted the term “carrier” as “portero,” which translates to ‘a goalkeeper’ or ‘a doorman.’ It is important to note that these seven instances were counted as separate misinterpretations because the interpreter did not utilize the same misinterpretation throughout the session, but rather switched between accordant and discordant interpretations of “carrier.” These literal interpretations of “carrier” resulted in interpreters referring to patients as a ‘doorman’ or ‘porter’ of a gene. While these literal interpretations may be generally intelligible, they may have resulted in changing the medical meaning of the GS test result information being communicated to families.
Omissions of Technical Genomic Terms
Terms that were identified as omissions also potentially changed the meaning of the information presented by resulting in incomplete statements or statements that were missing crucial clinical information. For example, in the exchange below omission of the term “increased risk” resulted in a statement that may not have conveyed the genetic counselor’s point that the child did not have higher risk of developing the tumor:
Genetic counselor: “So we didn’t see that she was at any increased risk to develop the tumor.”
Interpreter: “Así que no vemos ningún aumento creciente de que se le desarrolle el tumor.” [So we do not see any growing increase from which the tumor can develop.] (Disclosure 393)
Additionally, omissions of certain GS terms might have resulted in presenting misleading clinical information to families. In the following example, omission of the term “carrier” resulted in a statement suggesting that the child ‘has’ five conditions rather than being a carrier for them:
Genetic counselor: “[The child] is a carrier for five different things”
Interpreter: “Ella tiene cinco diferentes cosas que tiene.” [She has five different things that she has.] (Disclosure 402)
Similarly, in the following exchange omission of the term “carrier” resulted in a statement suggesting that a future partner could also ‘have this condition’:
Genetic counselor: “So the chance of a partner being a carrier is also rare.” Interpreter: “Pero las posibilidades de que la pareja que ella escoja, también es raro que vaya a tener esta condición.” [But the possibilities that the partner that she chooses, it is also rare that s/he is going to have this condition.] (Disclosure 338)
Comparison of Interpretation by Hospital-Based Interpreters versus Bilingual Oncologists
We observed similarities between the 36 disclosure sessions with a hospital-based interpreter and the six disclosure sessions in which a bilingual oncologist directly returned study results to the family. We found that even though the two study oncologists had more medical and genetic knowledge than the interpreters, they made similar modifications as interpreters when presenting GS terminology in Spanish. In 13 instances during three disclosures, oncologists used the same variations of literal non-medical terms for “carrier” as did interpreters, including the terms “cargar” and “llevar.”
Additionally, we found that oncologists often avoided using the term “variants of uncertain significance.” In all six disclosures, both oncologists preferred to use the meaning or definition of VUS rather than the actual term “variants of uncertain significance” when discussing these results with families. Oncologists more accurately interpreted language about “risk” compared to interpreters as they used equivalent Spanish GS terms in all 17 mentions of “risk.”
DISCUSSION
To our knowledge, this study is the first to explore hospital-based medical interpreters’ utilization of specific technical GS terminology during disclosure of WES results for Spanish-speaking families. While interpreters mostly interpreted information accordantly, we found that all interpreters in this study either misinterpreted or omitted GS terms in the majority of WES disclosure sessions. These results are consistent with findings from previous studies on interpreter accuracy [10, 12, 13, 14, 18], however we expand upon the existing literature by identifying a trend in the way interpreters and bilingual oncologists misinterpreted technical GS terms. The misinterpretations we observed included using literal non-medical language, as well as using nonequivalent or incorrect interpretations to describe technical GS terms. These modifications have implications for how LEP families understand WES results, as literal interpretations and omissions could change the meaning of the medical information. While we did not identify any modifications that changed the meaning of the information in a way that imposed serious clinical consequences or put the family’s health at risk, using non-medical language may make it difficult for parents to understand GS test results when told that their child is a ‘doorman’ or ‘porter’ of a gene or that s/he ‘holds’ or ‘takes’ a gene mutation. Additionally, omission of key GS terms like “carrier” may result in families receiving misleading information, such as being told that their child ‘has’ a condition instead of being a “carrier” for one. Previous research has found that omissions may be more likely to happen in instances where providers deliver a large amount of information, especially technical information, between interpretations [14]. In these situations, interpretations are more likely to be summaries instead of direct interpretations and thus include more omissions of words or phrases. As our analysis focused specifically on utilization of GS terms and did not explore the instances in which modifications occurred, we recommend that future studies examine in which situations misinterpretations and omissions are more likely to happen.
Of note, we found similar misinterpretations of GS terms and concepts in the six sessions during which a bilingual oncologist directly returned the results to the family in Spanish. While our bilingual oncologist data set is limited for a robust comparative analysis, we observed that bilingual oncologists were also unfamiliar with GS terminology in Spanish, especially the term “carrier.” Unlike the hospital-based interpreters who did not receive any study-specific genetics training, oncologists participated in two training sessions for the study and also had the opportunity to ask questions of a medical geneticist and pediatric oncologist expert in genomics prior to each disclosure. However, this training did not include any discussion of Spanish GS terminology. We observed that sessions with bilingual oncologists were on average shorter than interpreter-mediated sessions (16 vs. 26 minutes) and sessions with English-speaking families (16 vs. 21 minutes). This may be due to differences in the number of speakers in each session as there were two speakers in English sessions (oncologist and genetic counselor) and three speakers in interpreter-mediated sessions (oncologist, genetic counselor, and interpreter), compared to bilingual oncologists who were often the only speaker when returning results directly to Spanish-speaking families with few to no interruptions from the non-Spanish-speaking genetic counselor. Future studies should examine these different situations further to determine if there are differences in the depth and detail of the information delivered. Future research should also explore the differences in interpretation when using a bilingual clinician versus a third-party professional interpreter, and also between bilingual genetics and nongenetics clinicians.
Recommendations for Spanish Interpretation of Technical Genomic Terminology
Interpreters who provided interpretation in BASIC3 WES disclosure sessions were not given any specialized training in genetics and may have been unfamiliar with the GS terminology and concepts discussed. Given the pace at which interpretation occurs, it is not surprising that interpreters may interpret literally by using words that first come to mind or skip terms they are unfamiliar with rather than spending time searching for precise medical terms. However as we have shown, these literal interpretations and omissions may result in changes to clinical meaning that can be exacerbated if interpreters are also unfamiliar with the precise medical terminology. We recommend collaboration between interpreters and clinicians that leads to the creation and adoption of resources such as standardized genomic lexicons in languages in addition to English. We recommend using these materials to familiarize interpreters with genomic terminology, so they can immediately recall precise medical GS terms during sessions in which they must interpret genetic information. Based on our observations of frequently misinterpreted and omitted GS terms, we have compiled a list of commonly mentioned GS terms in this study and our recommendations for equivalent Spanish interpretations of those terms (Table 4). This is a valuable resource to improve interpreter and clinician interpretation accuracy, and lead the field of precision medicine toward more effective inclusion of diverse populations. Our recommendations incorporate Spanish translations from existing multilingual genomics glossaries created by two independent groups [26, 27]. These glossaries are available to the public. While there may be more than one adequate Spanish interpretation for each of the following GS terms, we offer our recommendation for the single best interpretation for each term in an effort to work toward more standardization in the Spanish GS terms used.
Table 4.
Recommended Spanish interpretations of genomic sequencing terminology.
| English Term | Recommended Spanish Interpretation | Reference |
|---|---|---|
| Actionable | Accionable | [28, 29] |
| Carrier | Portador/a | [26, 27] |
| Carrier status | Estado de portador/a | [27] |
| Genetic counselor | Asesor/a genético | [26, 27] |
| Germline report | Reporte germinal | [30] |
| Pathway | Via metabólica | [27] |
| Pathogenic variant | Variante patogénica | [27] |
| Deleterious mutation | Mutatión deletérea | [26] |
| Risk(s) | Riesgo(s) | [26, 27] |
| High risk | Alto riesgo | [26, 27] |
| Increased / Higher risk | Aumento del riesgo | [27] |
| At risk | A riesgo | [31] |
| Sequencing | Secuenciación | [27] |
| Genome sequencing | Secuenciación del genoma | [27] |
| Whole exome sequencing | Secuenciación del exoma completo | [27] |
| Variants of uncertain significance | Variantes de significado incierto | [26, 27] |
Clinics in which GS test results are frequently returned to patients with interpreters could train the interpreters by utilizing a tailored curriculum or providing bilingual genetics glossaries. As previous studies of interpreter accuracy have suggested, the frequency of interpreter modifications to clinicians’ language could be due to the lack of consistent standards for training, licensing, and certification [12, 16, 17, 32], which could result in the variability we observed among interpreters employed by the same hospital. While this is part of the larger policy issue of lack of national regulations for interpreters employed in the United States, efforts can still be made to familiarize or train medical interpreters on the GS information for which their services will be utilized. For example, one group has recently developed a curriculum for training interpreters in cancer genetics which is available online for public use [26]. We encourage use of such curricula by hospitals, clinics, and researchers to aid in creation of a standard of interpretation in genetic settings. Standardized medical genetic terms in Spanish are especially important given the variety of national and geographic differences within the Spanish language.
In addition, our findings suggest the importance of translating GS results and counseling letters into families’ preferred language. Our study returned reports written in English to families due to the additional expense of creating an individual medically-certified translated letter for each family. This practice may inadvertently contribute to a larger-scale inequity of access to personalized genomic medicine resources amongst non-English speaking populations. Returning GS results to LEP families in a language other than their preferred language potentially disempowers and disengages them in comparison to their English-speaking counterparts. At this formative stage in the field of precision medicine, an assumption of homogeneity within diverse patient groups obscures the unique communication barriers faced by minority populations. Providing results in patients’ preferred language – potentially using predesigned template language – would give LEP families opportunities outside the walls of a disclosure session to read and internalize their GS results, thus relying less on the quality of interpretation.
There are a few limitations to our study. First, as this analysis captured only instances in which a clinician uttered exact GS terms, it does not capture the accuracy of GS concepts described more broadly by the clinician. This affects the frequency of VUS the most, as many clinicians described the concept rather than use the exact term. Second, country of origin was not collected in the participants’ demographic data, and participants’ regionally-specific form of Spanish language could not be inferred, though recent population demographics report that most Latinos in Texas originate from Mexico and Central America [33]. In addition, as interpreters were not enrolled in BASIC3 as study participants, we were not able to collect any demographic information from them. Third, although a subset of parents were interviewed to explore their perspectives of the study, their understanding of the information from the disclosure session was not specifically assessed. Therefore, we only assessed whether interpretations changed the meaning of phrases on a linguistic and semantic level and were unable to determine the impact of interpreters’ modifications on families’ understanding. Future studies of interpreter accuracy should explore to what extent misinterpretations and omissions of technical genomic terms affect LEP families’ understanding of WES results. Additionally, we did not assess interpreters’ understanding of the recommended Spanish GS terms provided in Table 4. We recommend future studies assess Spanish-speakers’ understanding of these terms. Fourth, our results regarding interpretations by bilingual oncologists may not be representative as the sample size was too small for a more robust analysis. Fifth, the interpreters in this study were employed by one hospital at a single academic center, so results may not be generalizable to other interpreter populations in other clinical settings. Lastly, the results from analysis are partly subjective as investigators came to a group consensus for the categorizations of each term modification.
CONCLUSIONS
As clinical genomic sequencing becomes more prevalent and enrollment of LEP participants in genomic research increases, assessing the efficacy of communication of genomic sequencing results will help prevent disparities in the quality of information delivered to LEP populations. In this study, we examined the accuracy of interpretation of technical terminology during disclosure of whole exome sequencing results for Spanish-speaking families on a linguistic and semantic level. To our knowledge this study is the first to expand upon the accuracy of interpretation of technical medical terminology and apply it to the context of disclosure of WES results information. We observed that interpreters misinterpreted and omitted technical genomic sequencing terms and concepts when disclosing results from clinical genomic sequencing reports. In instances of misinterpretation, we identified a trend in interpreters’ use of literal terms instead of medical ones to describe genomic concepts. Omission of certain terms resulted in incomplete statements that were missing crucial clinical information. These misinterpretations and omissions of genomic terms potentially changed the clinical meaning of the genomic sequencing information. None of the modifications we observed were significant enough to put the family’s health at risk. Development of recommended terminology, better training of and certification standards for interpreters, and availability of translations of genomic sequencing terms and test results in families’ preferred language may improve the use of genomic testing in limited English proficient families.
SUMMARY POINTS.
Through discourse analysis, we analyzed 42 disclosure sessions in which Spanish-speaking parents received their child’s genomic sequencing results either from a clinician using a hospital-based interpreter, or directly from a bilingual physician.
In the 36 interpreter-mediated sessions, 76% of technical genomic terms were interpreted accordantly using equivalent terms or descriptions, 11% were misinterpreted by using nonequivalent or literal/non-medical terms or descriptions, and 13% were omitted completely.
At least one omission or misinterpretation was present in 29 (81%) of the 36 transcripts, and all 16 interpreters made at least one misinterpretation or omission.
Misinterpretations in which interpreters used literal terms instead of medical ones to describe genomic concepts like being a “carrier” potentially made it difficult for parents to understand genomic test results, for example being told that the child is a ‘doorman’ or ‘porter’ of a gene.
Omission of technical genomic terms resulted in incomplete statements that were missing crucial clinical information and in families receiving misleading information, for example being told that the child ‘has’ a condition instead of being a “carrier” for one.
Bilingual physicians used the same variations of literal non-medical terms for “carrier” as did interpreters, however they more accurately interpreted language about “risk” compared to interpreters.
The hospital-based interpreters who provided interpretation during disclosure sessions were not given any specialized training in genetics and may have been unfamiliar with the genomic terminology and concepts presented.
Based on our observations of frequently misinterpreted and omitted genomic terms, we created a list of recommendations for equivalent Spanish interpretations of the most commonly mentioned technical genomic terms in this study.
In efforts to increase interpretation accuracy, we recommend collaboration between interpreters and clinicians that leads to the creation and adoption of resources such as standardized genomic lexicons in languages in addition to English and using these materials to familiarize interpreters with precise genomic terminology.
Better training of and certification standards for interpreters as well as availability of translations of genomic sequencing terms and test results in families’ preferred language may improve the use of genomic testing in limited English proficient families.
ACKNOWLEDGEMENTS
We would like to thank Robin Raesz-Martinez, Stephanie A. Gutierrez, Uma Ramamurthy and her team at the Baylor College of Medicine Institute for Clinical and Translational Research, Priscila D. Hodges, Gabriel Lázaro-Muñoz, and Jacob D. Hofstetter. We would like to especially thank the oncologists and parents who participated in this study and all hospital interpreters for playing a crucial clinical role for limited English proficient patients and their families.
FINANCIAL & COMPETING INTERESTS DISCLOSURE
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of the Baylor Genetics Laboratories which performs exome sequencing. Dr. Plon serves on the Scientific Advisory Board of Baylor Genetics Laboratory. All other co-authors declare no conflict of interest. The BASIC3 study is a Clinical Sequencing Exploratory Research program project supported by the National Human Genome Research Institute and the National Cancer Institute (U01HG006485).
Footnotes
ETHICAL CONDUCT OF RESEARCH
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved. The Baylor College of Medicine Institutional Review Board, which also serves as the IRB for Texas Children’s Hospital, approved the BASIC3 study protocol.
REFERENCES
Papers of special note have been highlighted as:
* of interest; ** of considerable interest
- [1].Wilson E, Hm Chen A, Grumbach K, Wang F, Fernandez A. Effects of limited English proficiency and physician language on health care comprehension. J Gen Intern Med. 20(9), 800–806 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Seijo R, Gomez J, Freidenberg J. Language as a communication barrier in medical care for Hispanic patients. Hispanic J Behav Sci. 13, 363–75 (1991). [Google Scholar]
- [3].Flores G, Abreu M, Schwartz I, Hill M. The importance of language and culture in pediatric care: Case studies from the Latino community. J Pediatr. 137(6), 842–848 (2000). [DOI] [PubMed] [Google Scholar]
- [4].Cohen AL. Are language barriers associated with serious medical events in hospitalized pediatric patients? Pediatrics. 116(3), 575–579 (2005). [DOI] [PubMed] [Google Scholar]
- [5].Hampers LC, McNulty JE. Professional interpreters and bilingual physicians in a pediatric emergency department: effect on resource utilization. Arch Pediatr Adolesc Med. 156, 1108–13 (2002). [DOI] [PubMed] [Google Scholar]
- [6].Flores G The impact of medical interpreter services on the quality of health care: A systematic review. Med Care Res Rev. 62(3), 255–299 (2005). [DOI] [PubMed] [Google Scholar]
- [7].Karliner LS, Jacobs EA, Chen AH, Mutha S. Do professional interpreters improve clinical care for patients with limited English proficiency? A systematic review of the literature. Health Serv Res. 42(2), 727–754 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Haffner L Translation is not enough. Interpreting in a medical setting. West J Med. 157(3), 255–259 (1992). [PMC free article] [PubMed] [Google Scholar]
- [9].Elderkin-Thompson V, Silver RC, Waitzkin H. When nurses double as interpreters: A study of Spanish-speaking patients in a US primary care setting. Soc Sci Med. 52(9), 1343–1358 (2001). [DOI] [PubMed] [Google Scholar]
- [10].Flores G, Laws MB, Mayo SJ, et al. Errors in medical interpretation and their potential clinical consequences in pediatric encounters. Pediatrics. 111, 6–14 (2003).*Our study shows that findings by Flores et al. (2003) of interpretation errors and their clinical consequences observed in a pediatric primary care setting extend to a genetic setting, as we show errors made by interpreters can change the meaning of clinical genomic information.
- [11].Flores G, Abreu M, Barone CP, Bachur R, Lin H. Errors of medical interpretation and their potential clinical consequences: A comparison of professional versus ad hoc versus no interpreters. Ann Emerg Med. 60(5), 545–553 (2012). [DOI] [PubMed] [Google Scholar]
- [12].Pope C, Escobar-Gomez M, Davis B, Roberts J, O’Brien E, Hinton E, Darden P. The challenge of tetradic relationships in medically interpreted pediatric primary care visits: A descriptive study of communication practices. Patient Educ Couns. 99(4), 542–548 (2016).**Our study focused on the semantics of interpretation in a pediatric genetic setting builds up findings from a discourse analysis by Pope et al. (2016), which described general misinterpretations that occurred in a pediatric primary care setting.
- [13].Aranguri C, Davidson B, Ramirez R. Patterns of communication through interpreters. J Gen Intern Med. 21(6), 623–629 (2006).*Aranguri et al. (2006) reported how interpreters significantly edited primary care clinicians’ language on a linguistic level and what effects this had on the language conveyed to patients, supporting our findings of the occurrence of interpretation modifications in a genetic setting and our recommendations for why and how to lessen them when possible.
- [14].Simon CM, Zyzanski SJ, Durand E, Jimenez X, Kodish ED. Interpreter Accuracy and Informed Consent Among Spanish-speaking Families with Cancer. J of Health Commun. 11:5, 509–522 (2006).**We expand upon results reported by Simon et al. (2006) that misinterpretations increased when technical information was interpreted in a pediatric cancer research setting by showing that this extends to technical genomic terminology and by further characterizing misinterpretations.
- [15].Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 369(16), 1502–1511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].National Council on Interpreting in Health Care Standards, Training, and Certification Committee. National standards for healthcare interpreter training programs (2011). http://www.ncihc.org/assets/documents/publications/National_Standards_5-09-11.pdf.
- [17].Roat CE. Certification of health care interpreters in the United States: A primer, a status report and considerations for national certification. The California Endowment; (2006). https://ncihc.memberclicks.net/assets/certification_of_health_care_interpretors%20posted%20version.pdf. [Google Scholar]
- [18].Joseph G, Guerra C. To worry or not to worry: breast cancer genetic counseling communication with low-income Latina immigrants. J Community Genet. 6(1), 63–76 (2015).**Joseph & Guerra (2015) reported variation in the quality of interpretation and unfamiliarity of interpreters with genomic terminology during breast cancer genetic counseling, which catalyzed our undertaking of this in-depth analysis of accuracy of interpretation of genomic terms.
- [19].Scollon S, Bergstrom K, Kerstein RA, et al. Obtaining informed consent for clinical tumor and germline exome sequencing of newly diagnosed childhood cancer patients. Genome Med. 6(9), 69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Parsons DW, Roy A, Yang Y, et al. Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol. 2(5), 616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Johnstone B Discourse analysis. 2nd ed. Blackwell, Malden, MA: (2008). [Google Scholar]
- [22].Hodges BD, Kuper A, Reeves S. Discourse analysis. BMJ. 337, a879 (2008). [DOI] [PubMed] [Google Scholar]
- [23].Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 19(2), 249–55 (2017). [DOI] [PubMed] [Google Scholar]
- [24].National Cancer Institute. BRCA1 and BRCA2: Cancer Risk and Genetic Testing (2015). https://www.cancer.gov/about-cancer/causes-prevention/genetics/brca-fact-sheet.
- [25].Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17(5), 405–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Roat CE, Joseph G. Interpreting for cancer genetics. California Healthcare Interpreting Association; (2016). http://www.chiaonline.org/Interpreting-for-Cancer-Genetics.**Roat & Joseph (2016) developed a curriculum available free online to train interpreters how to work in cancer genetics settings, including bilingual glossaries for technical genomic terms. We recommend this curriculum be used to train interpreters for genetic studies and clinical settings.
- [27].Canadian Association of Genetic Counsellors. Lexigene. https://www.lexigene.com/en/.**Lexigene is an online genetics lexicon that recently published (May 2017) Spanish terms for genomic terminology, which we highly recommend be used by clinicians and interpreters.
- [28].Meléndez-Zajgla J, Lagunas VM. Genomas del cáncer: ¿Hacia dónde ir? Gac Med Mex. 150(6), 563–569 (2014). [PubMed] [Google Scholar]
- [29].European Society for Medical Oncology. Conjunto de guías ESMO para los pacientes (2013). https://www.esmo.org/content/download/46499/855050/file/ESMO-Medicina-Personalizada-Guia-para-Pacientes.pdf.
- [30].Fitze G Management of patients with hereditary medullary thyroid carcinoma. European J Pediatr Surg. 14(06), 375–383 (2004). [DOI] [PubMed] [Google Scholar]
- [31].Rodriguez G, Dorjo I, Scherer A, Desposito F. Genetic Counseling Glossary: Spanish Translation of English Terms. http://www.geneticcounselingtoolkit.com/pdf_files/GC%20Glossary.pdf
- [32].U.S. Bureau of Labor Statistics. How to Become an Interpreter or Translator (2015). http://www.bls.gov/ooh/media-and-communication/interpreters-and-translators.htm#tab-4.
- [33].Migration Policy Institute. State Demographics Data – Texas (2016). http://www.migrationpolicy.org/data/state-profiles/state/demographics/TX.