Abstract
Multiple autoimmune pathologies are associated with single nucleotide polymorphisms of the human gene TAGAP, which encodes TAGAP, a guanosine triphosphatase (GTPase)-activating protein (GAP). Here, we showed in mice that Tagap-mediated signaling by the sema3E–plexin-D1 ligand-receptor complex attenuates thymocyte adhesion to the cortex through their β1-containing integrins. By promoting thymocyte detachment within the cortex of the thymus, Tagap-mediated signaling enabled their translocation to the medulla, which is required for continued thymic selection. Tagap physically interacted with the cytoplasmic domain of plexin-D1 and directly stimulated the activity and signaling of the GTPase RhoA. In addition, Tagap indirectly mediated the activation of Cdc42 in response to the binding of sema3E to plexin-D1. Both RhoA and Cdc42 are key mediators of cytoskeletal and integrin dynamics in thymocytes. Knockdown of Tagap in mice suppressed the sema3E- and plexin-D1–mediated release of thymocytes that adhered within the cortex through β1-containing integrins. This suppression led to the impaired translocation of thymocytes from the cortex to the medulla and resulted in the formation of ectopic medullary structures within the thymic cortex. Our results suggest that TAGAP variation modulates the risk of autoimmunity by altering thymocyte migration during thymic selection.
One-sentence summary:
The GTPase-activating protein Tagap enables thymocytes to properly migrate within the thymus to undergo selection.
Editor’s summary:
Letting thymocytes go
During the process of T cell development, thymocytes must travel from the cortex of the thymus to the medulla, where any potentially autoreactive cells are removed by negative selection. This translocation is mediated by interactions between sema3E, which is secreted from the medulla, and its receptor plexin-D1, which is present on thymocytes in the cortex. Duke-Cohan et al. found that mouse thymocytes lacking the GTPase-activating protein Tagap had defective sema3E–plexin-D1 signaling and thus failed to detach from the cortex. Given that single nucleotide polymorphisms in the gene encoding TAGAP are associated with autoimmune disorders, these data suggest that Tagap facilitates the trafficking required for the efficient negative selection of autoreactive cells.
Introduction
The establishment of central tolerance in the thymus depends on tightly orchestrated chemokine-driven migratory patterns, shuttling positively selected T cells from the thymic cortex to the medulla for further selection processes (1). Initiation of thymocyte translocation from the cortex to the medulla requires sema3E, a medulla-derived, secreted semaphorin that binds to plexin-D1 on the thymocyte surface, enabling cell migration. Sema3E/plexin-D1 signaling disrupts constitutively active α4β1 and α6β1 integrin–mediated adhesion (2). The active conformation of β1-containing integrins involves stabilizing interactions with the cytoskeletal components actin, talin, and kindlin (3, 4). Adhesion occurs through the cortex-localized integrin ligands, VCAM-1 and laminin, which bind to the β1-containing integrins on thymocytes (VCAM-1 to α4β1 and laminin to α6β1) (2). In the absence of plexin-D1, sema3E cannot release the activated β1 integrin conformation and CD4+CD8+ (double positive, DP) thymocytes remain adherent to their integrin ligands and persist in the cortex, developing into CD4+ or CD8+ single-positive (CD4SP, CD8SP) thymocytes therein rather than in the medulla (2, 5, 6). Inhibiting the migration of positively selected DP thymocytes from the cortex to the medulla affects medulla-localized negative selection and results in the development of autoreactive humoral responses in Plxnd1 conditional knockout (CKO) mice (2). At the gross phenotypic level, as assessed by fluorescence-activated cell sorting (FACS) analysis, no abnormality in thymocyte subsets or T cell development is observed. Although the detailed relationship between sema3E/plexin-D1 signaling and β1 integrin conformation is not yet fully understood, cytoskeletal reorganization that releases the stabilizing interaction between the actin/talin/kindlin complex and β1-containing integrins is a likely mechanism for conversion from high- to low-affinity integrin conformational states (4).
Semaphorin signaling through plexins, a large family of transmembrane proteins, mediates guidance cues influencing directional migration in the developing nervous, vascular, and immune systems (7). The cytoplasmic tail of each plexin contains a segmented guanosine triphosphatase (GTPase)-activating protein (GAP) domain. Whereas the GAP domain of plexin-B1 regulates R-Ras activity (8, 9), the results of studies of plexin-D1 GAP domain activity are controversial. GAP activity for plexin-D1 has been reported, but only under nonphysiological conditions or when using complex, whole-cell lysates (10–12). Other studies showed no intrinsic GAP activity of the plexin-D1 cytoplasmic domain for putative downstream GTPases (13, 14). It has been suggested that plexin-D1 may function as a GTPase docking domain for other GAP proteins, thereby indirectly stimulating GTP hydrolysis and inhibition of GTPase activity (13, 14). We thus investigated whether developing thymocytes had GAPs that functioned downstream of sema3E–plexin-D1. Having previously found no alteration in Rap GTPase activity in thymocytes in response to sema3E signaling through plexin-D1 (2), we focused on the Rho GAPs that regulate members of the Cdc42, Rho, and Rac GTPase subfamilies. These subfamilies of the Rho GTPase family control the cytoskeletal and adhesion processes that are essential for initiating and maintaining cell migration (15).
Results
Evidence that thymocyte plexin-D1 GAP activity results from Rho GAP sequestration
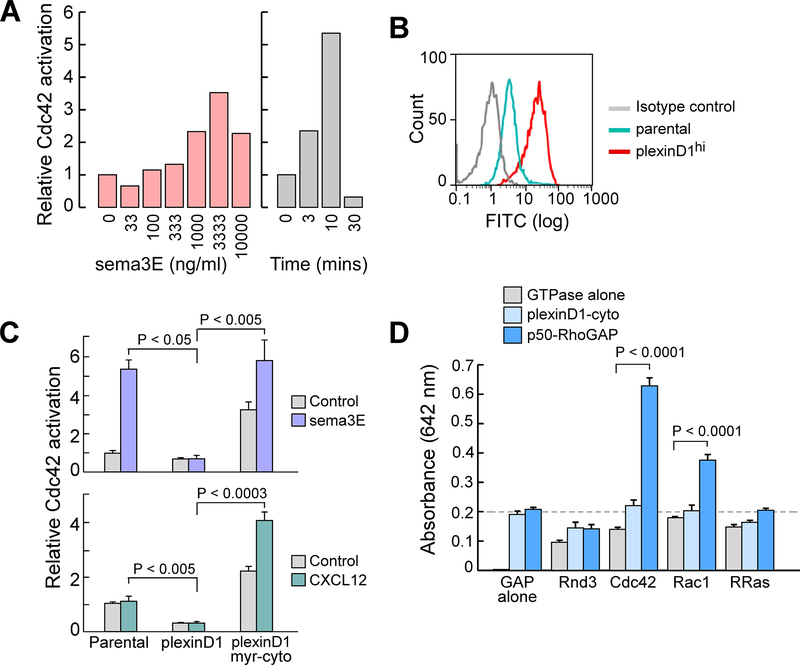
Focusing on Cdc42 as the Rho GTPase that establishes the leading edge in cells preparing to undergo directed migration, we investigated initially whether sema3E signaling through plexin-D1 altered the ratio of active (GTP-bound) to inactive (GDP-bound) Cdc42. Using the DP thymocyte-like cell line DP257–20-109 (fig. S1) (16), a time and dose response analysis detecting active Cdc42 by GST-PAK1-CRIB binding and coprecipitation indicated the maximal activation of Cdc42 at 10 min after exposure to sema3E (~3 μg/ml) (Fig. 1A). Stable overexpression of full-length plexin-D1 in DP257–20-109 cells, which increased its cell surface expression ~8-fold (Fig. 1B), impaired Cdc42 activation after sema3E binding to plexin-D1, and slightly, but consistently, reduced the basal activity of Cdc42 in the absence of sema3E (Fig. 1C, top). The increase in active Cdc42 in the parental cells stimulated with sema3E is not compatible with plexin-D1 functioning as a direct GAP for Cdc42, which would increase the hydrolysis of GTP and result in less GTP-bound active Cdc42 and more GDP-bound inactive Cdc42. However, an impaired sema3E-mediated increase in active Cdc42 could occur if the overexpressed plexin-D1 interfered with the formation of appropriately organized receptor oligomers competent to signal (17, 18). The reduction in basal Cdc42 activity in plexin-D1–overexpressing cells in the absence of sema3E suggested a ligand-independent effect of overexpressing this receptor. To differentiate between sema3E-dependent effects and the intrinsic functionality of plexin-D1 independent of sema3E-binding, we monitored the effect of overexpression of plexinD1 on the increase in active Cdc42 induced by the chemokine CXCL12 (also known as SDF-1) (19). Here, CXCL12 was unable to induce an increase in active Cdc42 in either parental or plexin-D1–overexpressing cells; however, in the latter, the basal amount of active Cdc42 was reduced compared to that in the parental cell line (Fig. 1C, bottom).
Fig. 1. GAP activity is not intrinsic to thymocyte plexinD1.
(A) Sema3E induces activated Cdc42 Rho GTPase in the DP-257–20-109 model DP thymocyte cell line. Left: The cells were treated with the indicated concentrations of Sema3E for 10 min. Right: The cells were treated with sema3E (3 μg/ml) for the indicated times. These parameters were selected as optimal for all subsequent assays. Relative activation was determined by measuring band intensities from GST-PAK1-CRIB coprecipitation assay. Data are representative of three experiments. (B) Plexin-D1 surface expression on parental DP257–20-109 cells and transfected DP257–20-109 cells stably expressing full-length plexinD1 was assessed by flow cytometry. Data are representative of four experiments. (C) Top: Measurement of active Cdc42 in DP257–20-109 cells (parental), DP257–20-109 cells over-expressing full-length plexin-D1 (plexinD1), and DP257–20-109 cells expressing myristoylated plexin-D1 cytoplasmic tail (plexinD1 myr-cyto) after treatment with sema3E (3 μg/ml) for 10 min. Bottom: Measurement of active Cdc42 in the indicated cell lines after treatment with CXCL12 (100 ng/ml) for 3 min. The responses of cells over-expressing full-length plexin-D1 represent the mean of three independent transfected clones, whereas those of cells expressing the myr-plexinD1-cyto construct are the mean of six independent clones. Data are means ± SEM and are representative of two independent experiments. (D) Assay of monomeric plexin-D1 cytoplasmic tail GAP activity for Cdc42, R-Ras, Rac1, and Rnd3 (control). We used p50-RhoGAP as a positive control. Data are means ± SEM of four replicates and are representative of three experiments.
A previous study found that the myristoylated cytoplasmic domain (myr-cyto) of plexin-D1 in transfected cells displays GAP activity for R-Ras (10). Unexpectedly, we found that DP257–20-109 cells overexpressing plexin-D1 myr-cyto exhibited both an increase in basal active Cdc42 and increases in active Cdc42 in response to both sema3E and CXCL12 (Fig. 1C). The differences between the effects of overexpression of full-length plexin-D1 and plexin-D1 myr-cyto upon active Cdc42 abundance led us to consider the proposition that the GAP activity for Cdc42 may not be intrinsic to plexin-D1, but rather may result from the signaling-dependent assembly on the plexin-D1 cytoplasmic tail of other signaling moieties. Specifically, full-length plexin-D1 may sequester Cdc42 GAPs close to the cytoplasmic face of the plasma membrane, reducing the baseline activity of Cdc42 with consequences for extracellular signal transduction. In contrast, the plexinD1 myr-cyto form may sequester the same GAPs to other membrane compartments of the cell, effectively reducing their localization to the plasma membrane, thereby enabling both enhanced basal Cdc42 activity and greater responsiveness to exogenous stimuli.
To directly test whether plexinD1 exerted direct GAP activity for Rho GTPases or R-Ras, we generated a non-myristoylated recombinant plexin-D1 cytoplasmic tail with a myc-His tag fused to the C terminus (plexinD1-cyto). This recombinant protein maintains the last helix of the C-terminal GAP domain and the putative GIPC PDZ domain containing family member 1 (GIPC1)-interacting domain of plexin-D1 (20, 21). We could not detect any GAP activity of this recombinant protein in a biochemical assay in which we measured the release of inorganic phosphate by GTP-loaded recombinant Cdc42, Rac1, or R-Ras (Fig. 1D). In contrast, the positive control p50-Rho-GAP showed GAP activity on Cdc42. Assuming that the C-terminal addition to the disordered terminal GIPC1-interacting residues (-Glu-Ala-COOH) did not affect the activity of the GAP domain (21), this reconstituted assay with recombinant proteins suggests that the direct or indirect GAP activity of plexin-D1 in cells occurs only in the context of other proteins. Furthermore, we found no direct binding of activated Rho or Rap GTPase to the plexin-D1 cytoplasmic domain in a mammalian two-hybrid assay (fig. S2).
Identification of candidate plexin-D1-interacting GAPs from thymocytes
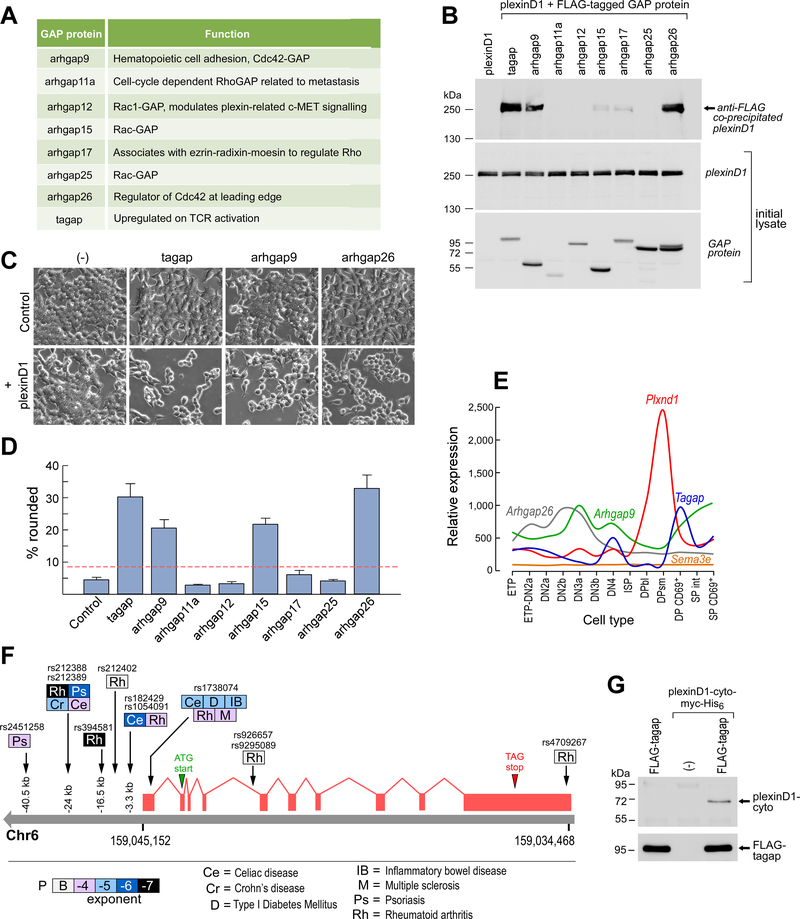
To determine the potential GAP proteins likely to interact with plexin-D1 during thymocyte development, we performed bioinformatic analysis that identified transcripts encoding 44 RhoGAPs expressed in mouse; of these, 30 were likely to be expressed in thymocytes (table S1). We screened mouse thymocytes for these 30 transcripts and identified 8 as full-length transcripts from which we could produce recombinant proteins of the expected sizes (Fig. 2A, table S1). The selected mouse GAP proteins exhibit high sequence conservation with their human counterparts (mean 82.1% identity and 87.3% similarity) with conservation of the GAP domain (mean 92.0% identity and 96.0% similarity) lending support to their similar functionality. Co-expression of each of the 8 selected RhoGAPs with plexin-D1 led to the identification of 3 proteins, Tagap, Arhgap9, and Arhgap26, that reproducibly coimmunoprecipitated with plexin-D1 (Fig. 2B). Cells expressing a GAP that coimmunoprecipitated with plexin-D1 exhibited cytoskeletal collapse, became round, and detached from the culture dish (Fig. 2, C and D). With the exception of cells expressing Arhgap15, those cells expressing GAPs that did not consistently coprecipitate with plexin-D1 showed no morphological changes (fig. S3).
Fig. 2. Thymocyte GAP proteins interact with the cytoplasmic tail of plexin-D1.
(A) List of the mouse thymocyte GAP proteins that were selected for analysis. (B) Western blotting analysis of FLAG-GAP proteins immunoprecipitated from 5 × 105 seeded cells to detect coimmunoprecipitated plexin-D1 (top). Whole-cell lysates were also analyzed by Western blotting to detect full-length plexin-D1 (middle) and the indicated GAPs (bottom). Data are representative of six experiments. (C) Analysis of cytoskeletal collapse and detachment in 5 × 105 seeded cells expressing the indicated GAPs alone or together with plexin-D1. (D) Summary analysis of the percentage of cells expressing the indicated GAPs that exhibited cell rounding or detachment from the experiments shown in (C). Data are means ± SEM of five experiments. The dotted red line marks two standard deviations above the control. For all conditions, 3109 ± 209 cells were counted. (E) Transcriptional profiles of thymocytes developing from the early thymocyte progenitor (ETP) stage to the SP stages for genes encoding the indicated GAPs, Plxnd1, and Sema3e (22). SP profiles represent the mean of pooled CD4SP and CD8SP profiles for simplicity. (F) TAGAP locus SNPs associated with human autoimmune disease. Shading intensity for each pathology represents exponent of probability of association from B (borderline) to −7 (P < 10−7). See table S2 for details. (G) Direct coimmunoprecipitation of recombinant plexinD1-cyto by FLAG-Tagap. Plexin-D1-cyto-myc-His6 (1.25 μg) was incubated with beads coated with the anti-FLAG antibody M2 (lane 2) or with similar beads saturated with recombinant FLAG-Tagap (lane 3). Lane 1 shows recombinant FLAG-Tagap beads alone. After three washes in lysis buffer, co-immunoprecipitated plexin-D1 cytoplasmic domain was eluted with SDS-PAGE buffer and detected by Western blotting with anti–plexin-D1 antibody. Bottom: Detection of recombinant Tagap in the samples of the same eluates by Western blotting with an anti-FLAG antibody. Data are representative of three experiments.
At the CD69+ stage of DP thymocyte development, where plexinD1 function is critical (5), both Tagap and Arhgap9 transcripts were abundant (Fig. 2E (22). Because Tagap mRNA abundance correlates with T cell receptor (TCR) activation (23, 24), a process that precedes the CD69+ DP stage of thymocyte development, and because multiple polymorphisms at the human TAGAP locus are associated with autoimmune diseases (Fig. 2F table S2), we investigated Tagap as a downstream effector of plexin-D1. Initially, we investigated whether there was a direct interaction between the cytoplasmic tail of plexin-D1 and Tagap. We found that FLAG-tagged Tagap interacted with the Myc-tagged cytoplasmic domain of plexin-D1 (Fig. 2G, consistent with a direct interaction. Given the caveats associated with comparing between different assay systems, the interaction between the purified recombinant proteins did not appear to be as robust as that between full-length plexin-D1 and FLAG-tagged Tagap coprecipitated from cells (Fig. 2B). This disparity raises the possibility that either other cellular proteins may stabilize the interaction, that transmembrane domain mediated-dimerization of plexin-D1 may offer a preferred binding interface for Tagap (25), or both.
Effects of inducible Tagap knockdown on thymocyte development
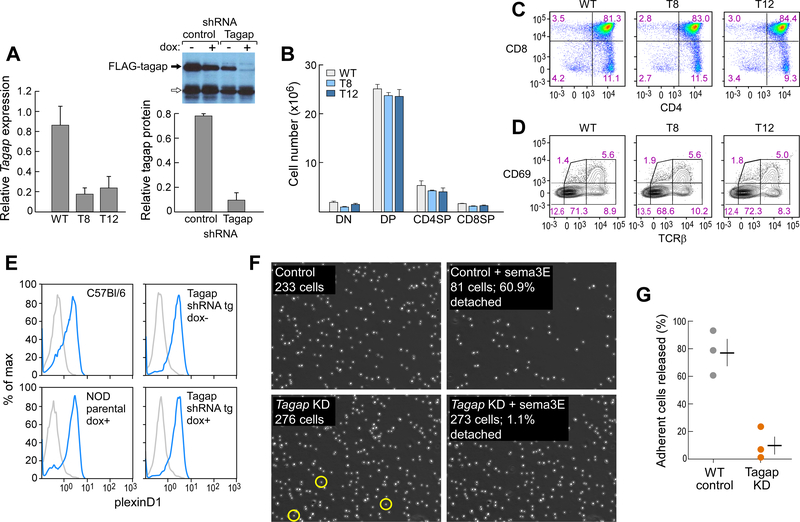
If Tagap activity were obligatory for plexinD1 signaling and other GAP proteins did not compensate for the loss of Tagap activity, some functional or developmental phenotypes consequent to the loss of plexin-D1 might be recapitulated in the absence of Tagap. We examined the phenotypes of transgenic mice in which Tagap was repressed by doxycycline-inducible RNAi (Fig. 3A). We found no obvious alteration in the total cellularity or phenotypic distribution of thymocytes in the thymus or in their surface expression of plexin-D1 (Fig. 3, B to E). Next, we examined whether sema3E regulation of thymocyte β1-integrin-mediated adhesion was altered as a result of Tagap knockdown (KD), as observed in both Plxnd1 germline and conditional knockout thymocytes (2, 5). Both WT and Tagap KD thymocytes, which were ~85% CD4+CD8+ DP cells (Fig. 3C), adhered to a VCAM-1-coated surface, but exogenously added sema3E stimulated the release of only the WT cells (Fig. 3, F and G, movies S1 and S2). These data indicate that Tagap acts as a key signal transducer downstream of sema3E-activated plexin-D1.
Fig. 3. Inducible suppression of Tagap expression by lentiviral shRNA transgenes.
(A) Left: RNAi-induced repression of Tagap mRNA in the thymus of WT (n = 3), T8 (n = 8), and T12 (n = 3) inducible KD transgenic strains treated with doxycycline for 5 to 8 weeks. Right: Loss of FLAG-Tagap expression after doxycycline-induced expression of Tagap-specific shRNA was assessed by Western blotting analysis. Bar graph shows pooled densitometry data from three experiments. The arrow indicates a cluster of nonspecific bands that served as a loading control. (B and C) Thymocyte subset representation (CD4−CD8− DN, CD4+CD8+ DP, CD4+ CD4SP, and CD8+ CD8SP) as total cell numbers (B) and FACS-determined population distribution (C) in WT mice and the T8 and T12 Tagap KD strains. Data are means ± SEM of four mice per group and are representative of five experiments. (D) Assessment of thymocyte maturation as determined by measurement of increasing TCRβ abundance and post-selection signaling as determined by cell surface expression of CD69 in WT mice and the T8 and T12 Tagap KD mice strains. Data are means ± SEM of four mice per group and are representative of five experiments. (E) Plexin-D1 abundance, as assessed by measurement of sema3E-Fcγ2c binding, was determined by flow cytometric analysis of thymocytes from the indicated mice. (blue; isotype control IgG2c in gray; thymocytes from 3 mice/sample). (F and G) Thymocytes isolated from control (top) or Tagap KD (bottom) mice were allowed to adhere to VCAM-1–coated migration chambers. Left: Adherence before the introduction of sema3E (5 μg/ml) under flow (2.5 μl/min. Right: Adherence after 40 min of exposure to sema3E and an increase in flow to 15 μl/min. Yellow circles outline Tagap KD adherent cells that detached in the presence of sema3E. Data are representative of the three experiments shown in (G). (G) Summary of the percentage of thymocytes released by sema3E. Data are means ± SEM of three experiments. P < 0.005.
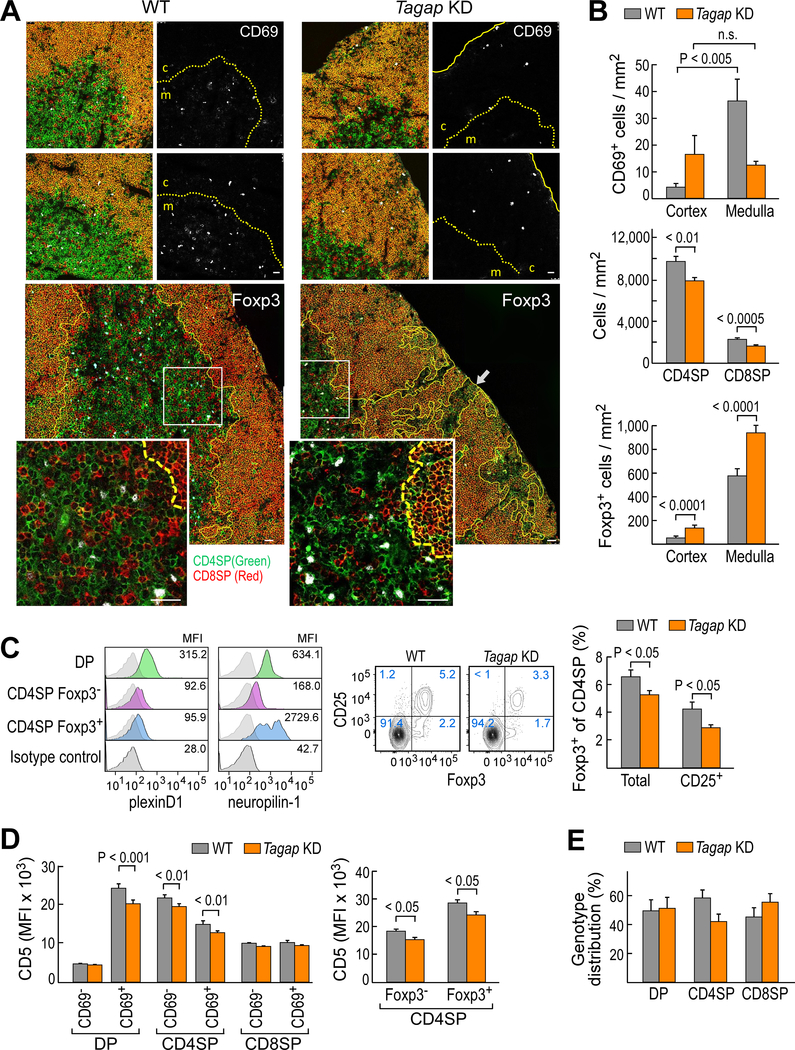
We assessed whether Tagap was also important in regulating thymocyte adhesion in vivo. We found that the migration of post-selection CD69+ DP thymocytes from the cortex to the medulla was impaired in Tagap KD mice. We observed a medullary:cortical ratio of ~9:1 for CD69+ cells in the thymus of WT mice, which was reduced to <1:1 in the thymi of Tagap KD mice (Fig. 4, A and B top panel). In contrast to the relatively homogenous appearance of the cortex in the thymus from WT mice, the cortical areas of the thymus from Tagap KD mice contained many CD4SP thymocytes (Fig. 4A). Furthermore, many of the cortical-located CD4SP and CD8SP cells were organized into discrete and relatively small medullary-like structures in addition to including CD4+Foxp3+ regulatory T (Treg) cells. The reduced medullary representation of both CD4SP and CD8SP thymocytes in the Tagap KD mice (Fig. 4B, middle panel) is consistent with impaired translocation from the cortex into the medulla.
Fig. 4. Tagap KD thymocytes show impaired cortical-medullary translocation and ectopic medullary formations.
(A) Top: Post-selection CD69+ cells (white) in WT and Tagap KD thymus. CD4 cells are in green, CD8 cells are in red, and CD4+CD8+ DP thymocytes are in yellow-orange. CD69+ signal alone in shown in the monochrome panels. The dashed yellow line shows the junction between the cortex (c) and the medulla (m). The solid yellow line shows the capsule. For both WT and Tagap KD, 22 confocal images (0.25 × 0.25 mm2) were analyzed. For the confocal images, the scale bar represents 25 μm. Ectopic medullary formations formed in the Tagap KD thymus (lower enlarged panels and insets). The intracellular Foxp3 signal is shown in white. The corticomedullary junction is shown by yellow lines (dashed in insets). White boxes outline the areas enlarged in the insets. Eighty confocal images (0.1 × 0.1 mm2) for WT mice and 61 confocal images (0.1 × 0.1 mm2) for Tagap KD were analyzed. (B) Top: The numbers of CD69+ cells in WT (n = 80 confocal sections) and Tagap KD mice (n = 61 confocal sections). Data are means ± SEM. Middle: Medullary density for CD4SP and CD8SP thymocytes in WT and Tagap KD mice. Data are means ± SEM. Bottom: Numbers of Foxp3+ CD4SP cells in WT and Tagap KD mice. Data are means ± SEM. (C) Flow cytometric analysis of plexin-D1 (left) and neuropilin1 (right) expression on WT B6 mouse thymic CD4+ subsets. The median fluorescence intensity (MFI) indicated. Data are representative of three experiments. Middle and right: Total (Foxp3+ CD4SP) and mature (CD25+Foxp3+ CD4SP) thymic Treg cells in WT and Tagap KD thymus. Data are means ± SEM of three or four mice/group and are representative of 11 experiments. (D) Flow cytometric analysis of CD5 cell surface expression on DP, CD4SP, and CD8SP cells from the indicated mice. Data are means ± SEM of four mice/group and are representative of five experiments. (E) Genotype representation of WT (GFP−) and T12 Tagap KD (GFP+) thymic subsets in mixed bone marrow chimeras. Data are means ± SEM of nine mice/group and are representative of three experiments.
The increased abundance of CD4SP cells in the cortex of the thymus of Tagap KD mice was similar to that observed in the thymus of Plxnd1 conditional knockout (CKO) mice (fig. S4, table S1), consistent with these proteins functioning in the same regulatory pathway. Both the medullary and cortical densities of CD4+Foxp3+ Treg cells were increased in the Tagap KD mice; however, because the density of medullary Treg cells was 7- to 40-fold greater than that in the cortex, the effective density in the medulla was more profoundly increased than that in the cortex (Fig. 4B, lower panel; table S1). Flow cytometry analysis of dispersed thymocytes from Tagap KD mice revealed a slight reduction in total Treg cell numbers (Fig. 4C, middle and right panels, fig. S5A). Disparities between tissue-embedded T-lineage cells as assessed by in situ staining as opposed to flow cytometry are well known (26) and are compounded further by the organ disruption used to prepare a single-cell suspension that masks the tissue geographical distribution preserved in confocal microscopic analysis. The dichotomy between the cortical retention of Foxp3− CD4SP cells and the increased medullary representation of Foxp3+ CD4SP cells led us to examine the abundance of plexin-D1 on CD4SP cell subsets. In contrast to CD8SP cells (2, 5), CD4SP cells retained some plexin-D1 after positive selection in the cortex with no difference observed in expression between the Foxp3+ and Foxp3− CD4SP subsets (Fig. 4C, left panel). Because high neuropilin-1 expression is not only considered to be a reliable marker to distinguish Foxp3+ Treg cells from other CD4SP cells (27), but is also a known modifier of sema3 family functionality (28–30), we examined its expression on the Foxp3− CD4SP and Foxp3+ CD4SP Treg cells. Relative to the precursor DP thymocytes, we observed a reduction (3.8-fold) of neuropilin-1 abundance on Foxp3− CD4SP thymocytes and an increase (4.3-fold) in neuropilin-1 on Foxp3+ CD4SP thymocytes (Fig. 4C, left panel, fig. S6).
We next determined whether Tagap KD affected thymic selection. The representation of T cell receptor (TCR) β chain clonotype diversity (Vβ) may be used as a low-resolution measure of repertoire alteration due to clonal expansion in response to infection or developmental abnormalities (31, 32). TCR Vβ chain usage in SP thymocytes was similar in WT mice and two strains of Tagap KD mice (fig. S5B), suggesting that the selected TCR repertoire was not grossly altered by the loss of Tagap. However, Tagap silencing diminished the increase in CD5 on positively selected CD69+ DP thymocytes (Fig. 4D, left panel, fig. S5C). The abundance of CD5 correlates with the strength of TCR signaling in thymocytes undergoing selection (33). We found that the impaired induction of CD5 expression specifically affected the development of CD4SP cells, including Foxp3+ Treg cells, but not that of CD8SP cells (Fig. 4D left and middle panels, fig. S5C). By generating mixed bone marrow chimeras in which Tagap KD thymocytes were in direct competition with WT cells, we observed both genotypes were equally represented at the DP pre-selection stage (Fig. 4E, fig. S5C), but the development into mature CD4SP cells was impaired in the Tagap KD mice (Fig. 4E and fig. S7). From these results, we concluded that Tagap is required for the selection and maturation of thymocytes at the developmental stage during which plexin-D1 is also most critical, consistent with a role for Tagap in plexin-D1 signaling.
Identification of Tagap targets downstream of sema3E-stimulated plexin-D1 signaling
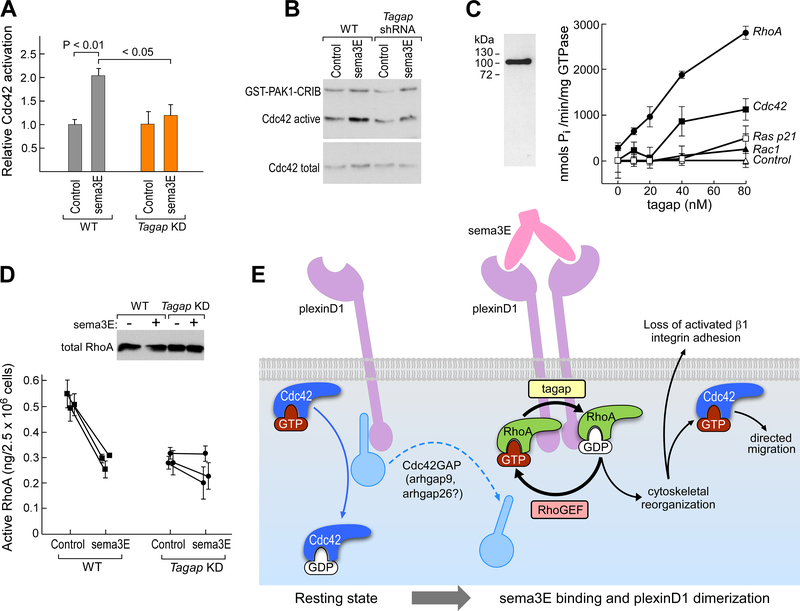
Given the observation that Tagap deficiency decreased thymocyte migration from the cortex to the medullary region of the thymus, and, in the process, impaired the maturation of CD4SP cells, we examined in WT and Tagap KD thymocytes the effect of sema3E signaling on the activity of Cdc42, Rac1, and RhoA GTPases, which regulate cell migration and adhesion. In WT thymocytes, sema3E binding to plexin-D1 induced a transient increase in the amount of the active, GTP-bound form of Cdc42 (Fig. 5, A and B). In thymocytes from Tagap KD mice, sema3E failed to increase the amount of active Cdc42. If Tagap functioned as a GAP for Cdc42, its absence would be expected to increase activated Cdc42. Instead, the observed suppression suggests that Tagap is essential for the sema3E-induced increase in active Cdc42 but that this GTPase is not a direct substrate of the GAP activity of Tagap, suggesting that sema3E/plexin-D1 signaling releases Cdc42 from GAP constraints other than that of Tagap per se.
Fig. 5. Tagap functions downstream of plexin-D1 as a RhoGAP.
(A) Assessment of the relative Cdc42 activation after sema3E binding to plexin-D1 on thymocytes from WT and Tagap KD mice. Results normalized to control for each condition and are means ± SEM of four experiments. (B) Cdc42 activation by sema3E in WT and Tagap KD thymocytes was detected by specific binding to GST-PAK1 protein-binding domain Cdc42/Rac interactive binding motif (CRIB) fusion protein. Data are representative of four experiments. The active Cdc42 signal represents total recovered active Cdc42, whereas the total Cdc42 signal (inactive and active) represents 5% of the sample. (C) Full-length mouse Tagap was expressed as an N-terminal FLAG fusion protein (left panel; silver stain) and then incubated at concentrations from 0 to 80 nM with the indicated Rho GTPases or Ras p21 loaded with GTP (right panel). GTP hydrolysis was assayed by measuring the release of inorganic phosphate. Data are means ± SEM of four samples and are representative of three experiments. (D) Regulation of active RhoA abundance by sema3E in WT and Tagap KD thymocytes. Data are means ± SEM of four samples and are representative of three experiments; For WT cells, P < 0.0001 between control and sema3E-treated; for Tagap KD cells, there was no significant difference. Top: Western blotting analysis of the relative amounts of total RhoA in each sample. (E) Proposed model for how Tagap regulates sema3e/plexinD1 function in thymocytes. Strong expression of full-length plexin-D1 in cortex-localized DP thymocytes in the absence of sema3E leads to very low basal Cdc42 activation, with no increase in response to CXCL12 (Fig. 1C) and minimal CXCL12-directed migration (2) due to sequestration of a Cdc42 GAP, possibly Arhgap9 or Arhgap26 (Fig. 2A). Upon binding to sema3E, plexin-D1 forms homodimers, generating an altered cytoplasmic domain binding interface, releasing Cdc42GAP and recruiting tagap together with other GTPase regulatory proteins. Tagap facilitates the cycling of RhoA between active and inactive states, which leads to cytoskeletal reorganization and the release of activated β1 integrins and loss of adhesion. The separation of plexin-D1 from Cdc42 GAP further enables increases in active Cdc42 abundance, which accelerates chemokine-directed migration (2).
To identify direct and specific GTPase targets for Tagap, we used a reconstituted GAP assay independent of any potential cellular intermediates (fig. S8). We tested for GAP activity for Cdc42, Rac1, and RhoA, which are all main regulators of cytoskeletal reorganization and integrin abundance (15). In this biochemical assay, Tagap acted as a GAP for RhoA at concentrations <40 nM, but it had less GAP activity for Cdc42 and negligible activity for Rac1 or RAS p21 GTPase (Fig. 5C). Despite Tagap having low, but measurable, Cdc42 GAP activity in this assay, the reduced activation of Cdc42 by sema3E in Tagap KD thymocytes suggests that in the physiological milieu of these cells, RhoA is likely the main target of Tagap. To confirm regulation of RhoA by Tagap within cells, we compared the effects upon RhoA activation of sema3E stimulation of WT and Tagap KD thymocytes (Fig. 5D). Sema3E reduced the amount of activated RhoA in WT thymocytes. Knockdown of Tagap reduced the basal amount of active RhoA, although total RhoA abundance was similar to that in WT cells (Fig. 5D). The addition of sema3E did not statistically significantly reduce the amount of active RhoA in the Tagap KD cells.
Discussion
Here, we demonstrated that Tagap, through RhoA, regulates the cytoskeletal organization of actomyosin fibrils pivotal for both cell contractility and the cycles of integrin adhesion and detachment necessary for directed thymocyte migration. The consequential downstream effect of Tagap is to release β1 integrin adhesion, which enables the migration of the thymocytes from the cortex to the medulla. Having shown that in the absence of sema3E, plexin-D1 reduces the amount of active Cdc42 (Fig. 1C) and that this inhibition is not directly mediated by Tagap (Fig. 5, A and B), we propose that monomeric plexin-D1, its natural state in the absence of sema3E (17, 18), also plays an important role in maintaining the β1 integrin–mediated adhesion status through its potential to sequester Cdc42 GAPs other than Tagap (Fig. 2B and Fig. 5E). Our screen identified two potential plexin-D1–interacting candidates, Arhgap9 and Arhgap26, both of which have Cdc42 GAP activity (34, 35). Arhgap9 additionally exhibits Rac1 GAP activity, whereas Arhgap26 also functions as a GAP for RhoA. In contrast to Arhgap26, Arhgap9 is well-expressed at the CD69+ DP thymocyte stage (Fig. 2F), and thus is a contender for blocking thymic stromal chemokine-induced induction of filopodia and lamellipodia by Cdc42 and Rac1, respectively, on DP thymocytes, thus inhibiting leading edge formation and initiation of the migratory process. We predict that sema3E-induced dimerization of plexin-D1 leads to release of the Cdc42 GAP, collectively enabling chemokine-directed formation of a leading edge and the generation of a new binding interface that favors Tagap association, cytoskeletal remodeling, release of β1 integrin–mediated adhesion, and directed thymocyte movement (Fig. 5E).
Tagap deficiency led to a block of sema3E-induced release of β1 integrin–mediated thymocyte adhesion and, consequently, cortical retention with a decrease in thymocyte migration to the medulla after positive selection. Nevertheless, we observed one population of cells, Foxp3+ CD4+ Treg cells, that exhibited increased migration to the medulla not only in Tagap KD mice, but also in Plxnd1 CKO mice. The CD4SP Treg cells had high surface amounts of neuropilin-1, an established accessory signaling protein for sema3 family directional cues (30). Neuropilin-1 in neurons negates and may even override the directional cues initiated by sema3E binding to plexin-D1 (29). Thus, the high expression of neuropilin-1 on post-selection Foxp3+ CD4SP Treg cells likely facilitates their translocation to the medulla in response to chemotactic cues independently of sema3E/plexinD1 signaling. In contrast, in the context of Tagap KD or Plxnd1 CKO, Nrp1lo Foxp3− CD4SP thymocytes would be insensitive to sema3E, thus potentiating cortical retention.
Our proposed model (Fig. 5E) is consistent with the results presented here and underpins the proposal that plexin-D1 may form a hub for the assembly of multiple GTPase regulatory proteins in thymocytes. This would further be consistent with the plexin-D1 GAP activity observed for Ras GTPases in other model cell lines in which assay of active GTPase levels in cell lysates would not differentiate between that exerted directly by plexin-D1 and that by an associated GAP. The association of binding complexes composed of protein modules specific to a cell type might form the basis for sema3E/plexin-D1 to orchestrate developmental processes in various tissues. For example, Tagap is dominantly expressed in hematopoietic cells, whereas in vascular endothelial cells, where Tagap expression is much reduced, the cytoplasmic tail of plexin-D1 associates with SH3BP1 (36), a Cdc42GAP whose expression in thymocytes is weak (table S1). While this manuscript was in review, it was reported that sema3E signaling through plexin-D1 suppresses the induction of filopodium-like lateral protrusions by activated Rac1 in neurons from newborn mice (37). However, we found no evidence to suggest that plexin-D1 is a direct GAP for Rac1 in thymocytes, which is consistent with our proposal that other tissue-specific GAPs mediate Rac1 signaling downstream of plexin-D1. Detailed characterization of plexin-D1 signaling complexes in thymocytes will lead to additional insights into the molecular controls of positive and negative selection and Treg development where polymorphisms in the TAGAP locus may impinge upon the quality of central tolerance established in the thymus. Furthermore, the reliance of plexin-D1 on divergent downstream signaling intermediates for tissue-specific effects has broad implications for plexin function in cell guidance and developmental processes more generally.
Materials and methods
Mice
C57Bl/6 mice, C57Bl/6 Plxnd1flox/flox mice, and C57Bl/6 pLck-Cre Plxnd1flox/flox mice were bred and maintained at the Dana-Farber Cancer Institute under Institutional Animal Care and Use Committee (IACUC)-approved protocol 04–034. NOD mice purchased from Jackson Laboratories and Swiss Webster mice (used for transgenesis) purchased from Taconic were bred and maintained at the Joslin Diabetes Center under IACUC-approved protocol 2014–01. Cre genotyping was used to distinguish Plxnd1 CKO (pLck-Cre+/−) mice from their WT littermates (pLck-Cre−/−); no blinding or randomization was used. Doxycycline administration in the water required, by necessity, the separation of Tagap KD mice from untreated littermates. Mice of all genetic backgrounds bore progeny with no gender distortion ratio related to genotype, and female and male mice were used without distinction in all experiments.
Recombinant protein expression
Recombinant sema3E protein was produced as a mouse γ2c Fc fusion protein as described previously (5). For screening potential interactions with plexin-D1, native, full-length plexin-D1 was expressed from pcDNA3.1 (pcDNA3.1-plexinD1), and the Arhgap proteins expressed as N-terminal fusion proteins from pFLAG-CMV-4. Each recombinant GAP protein consisted of an N-terminal FLAG sequence upstream of amino acid residues 55 to 479 of NCBI sequence NP_001139374.1 for Arhgap1, residues 23 to 332 of NP_076032.2 for Arhgap3, residues 5 to 473 of NP_666123.2 for Arhgap9, residues 3 to 987 of NP_852081.2 for Arhgap11a, residues 3 to 838 of NP_001034781.1 for Arhgap12, residues 5 to 1085 of NP_001074506.1 for Arhgap13, residues 3 to 481 of NP_722542.2 for Arhgap15, residues 3 to 846 of NP_653112.2 for Arhgap15, residues 4 to 648 of NP_001032816.1 for Arhgap25, residues 5 to 814 of NP_780373.3 for Arhgap26, residues 5 to 670 of NP_598476.2 for Arhgap27, and residues 5 to 714 of NCBI reference sequence NP_666080.1 for Tagap. For purification of recombinant Tagap, HEK 293T cells were transfected with the plasmid pFLAG-CMV-4-tagap with Lipofectamine 2000. Forty-eight hours later, cell lysates supplemented with cØmplete protease inhibitors (Roche) were prepared for affinity purification on anti-FLAG-M2 agarose beads (Sigma-Aldrich) and eluted with the 3×FLAG peptide (100 μg/ml). Purity and integrity were assessed by silver staining, Western blotting, and MS analysis of a tryptic digests. GST-PAK1-CRIB(70–117) was expressed from pGEX2-TK-PAK1-CD (Dr Jonathan Chernoff, Fox Chase Cancer Center, Philadelphia PA) and purified from a denaturing lysate of IPTG-induced (0.5 mM, 4 h) transformed E.coli culture by glutathione-agarose affinity chromatography. For the reconstituted GAP assays, the complete cytoplasmic domain of mouse plexin-D1 was subcloned into pcDNA3.1 in line with the 3’ vector-encoded myc-His(6) tag. To produce purified protein, HEK 293T cells were transfected with the construct with Lipofectamine 2000. Twenty-four hours later, cell lysates supplemented with cØmplete protease inhibitors (Roche) were prepared for affinity purification on His-select beads (Sigma-Aldrich) and eluted with 250 mM imidazole followed by buffer-exchange into Tris-buffered saline (TBS). For the co-precipitation assays examining the direct interaction of plexin-D1-cyto with recombinant Tagap, all incubations were performed in low protein-binding tubes (Eppendorf).
Cell line development
The DP257–20-109 thymocyte-like cell line (16) was used as the basis for the stable expression of large amounts of full-length, transmembrane plexin-D1 and myristoylated cytoplasmic domain plexin-D1 (parental cell line provided by Dr. Takushi Tadakuma). The surface antigen phenotypic profile of DP257–20-109 is characteristic of a pre-selection DP thymocyte (fig. S1). Full-length plexin-D1 was expressed from pcDNA3.1 (pcDNA3.1-neo-plexinD1) and clones with high cell surface expression were isolated by limiting dilution in G418 (500 μg/ml) and screened for surface plexin-D1 by FACS analysis of sema3E binding using mouse purified IgG2c as an isotype control. Cell-bound sema3E was detected with anti-mouse IgG2c-PE (Jackson Immunoresearch). For the myristoylated plexinD1 cytoplasmic domain, a myristoylation motif (MGASSS) was introduced upstream of residues 1294 to 1925 of plexinD1 (NCBI reference sequence NP_080652) and subcloned into pcDNA3.1-neoR. G418-resistant clones were isolated by limiting dilution, and lysates were prepared and screened for reactivity with a polyclonal rabbit anti-mouse plexin-D1 intracellular domain-specific antibody (anti-plexinD1-IC) by Western blotting (5).
Identification of plexin-D1–interacting GAPs by immunoprecipitation
After being seeded into 6-well plates, HEK 293T cells were grown to 50 to 75% confluence and then were transfected with a pcDNA3.1-full-length transmembrane mouse plexinD1 (wild-type, untagged) construct (1.25 μg) alone or together with pFLAG-CMV-4-GAP (1.25 μg) using Lipofectamine 2000. Incubations were continued for a further 24 to 36 hours after which the cells were imaged, which was followed by lysis with ×2 lysis buffer (2% Triton X-100, 500 mM NaCl, 50 mM Tris-HCl, (pH 7.2), 10 mM MgCl2). Lysates were clarified by centrifugation and then were incubated with anti-M2-FLAG-agarose beads (Sigma-Aldrich) for 16 hours at 4°C. After 4 washes in lysis buffer followed by 2 washes in TBS, the beads were treated with 2× Laemmli SDS-PAGE loading buffer and heated at 70°C for 10 min followed by SDS-PAGE separation under reducing conditions, transfer to nitrocellulose membranes, and blocking in TBS-BSA/Tween 20 (5%/0.5%) overnight. Blots were developed with rabbit polyclonal anti-plexinD1-intracellular domain, which was followed by anti-rabbit-HRP and chemiluminescence analysis to determine FLAG-GAP coprecipitated plexin-D1.
Lentiviral transgenesis
Lentiviral transgenic mice were generated as described previously (38, 39). Briefly, NOD zygotes were microinjected into the perivitelline space with lentiviral particles and then implanted into pseudo-pregnant Swiss-Webster female mice. Founder mice born after microinjections were screened for GFP expression macroscopically and by flow cytometry using peripheral blood samples. We generated two transgenic lines, T8 and T12, using distinct shRNA constructs to target Tagap by RNAi. Transgene expression was ubiquitous and uniform in all immune cells analyzed. Transgenic mice were bred with wild-type NOD mice and maintained as hemizygous for the transgene. Non-transgenic littermates were used as controls for all experiments, and mice were not separated by genotype after weaning. Control and transgenic mice were kept co-housed for all experiments. Comparisons between mice treated or not with doxycycline (200 μg/ml water ad libitum) were performed with mice housed in different cages.
Tagap targeting by RNAi
For lentiviral RNAi, we cloned two distinct shRNA sequences (T8: GGGAGTATGAGGTCAAGAA and T12: CACCAGAGTTTCACAACAA) into the pUTG vector (40) that contains a Tet-inducible H1 promoter driving shRNA expression and a Ubiquitin (UBC) promoter (PUb) driving the expression of a Tet-repressor-T2A-GFP transgene. For in vitro validation, shRNA expression was induced with 1 μg/ml doxycycline (from Sigma-Aldrich). For in vivo Tagap knockdown, doxycycline was provided ad libitum in the drinking water at a concentration of 200 μg/ml. To assess the efficacy of RNAi-mediated suppression of Tagap expression, quantitative RT-PCR was performed using the following primers: Tagap forward: 5’-GCTGTTTGACCAACCCTTGT-3’; and Tagap reverse: 5’-GCAGAGGATGGTGAGGATGT-3’. The extent of suppression of Tagap protein production was determined by co-transfecting HEK 293T cells with pFLAG-CMV-4-tagap and pUTG-PH1-tetO-tagap-shRNA-PUb-TetR-T2A-eGFP and determining the abundance of FLAG-tagap by Western blotting analysis of cell lysates 24 to 48 hours after transfection.
Flow cytometry
Tissues analyzed by flow cytometry were disaggregated into single-cell suspensions in phosphate-buffered saline (PBS) supplemented with 2% fetal calf serum (FCS) and 2 mM EDTA, passed through a 70-μm nylon mesh strainer, and washed several times before antibody staining. Cells (1 to 2 × 106) were stained with a viability dye for 20 min at room temperature and then for surface markers at 4°C for 20 min. For analysis of Foxp3+ CD4SP subsets, 10 to 20 × 106 cells were processed. For intracellular staining (Foxp3), cells were then washed, fixed, and permeabilized with the Foxp3/Transcriptional Factor Staining Buffer set (eBioscience) according to the manufacturer’s protocol. Flow cytometry analysis of stained cells was performed with BD LSR II and BD Fortessa flow cytometers (BD Biosciences). Data were analyzed with FlowJo software (FlowJo LLC). Antibodies used for flow cytometry were as follows with clone identifiers in parentheses: anti-CD4-BV605 (RM4–5), anti-CD4-BV421 (GK1.5), anti-CD8a-APC/Cy7 (53–6.7), anti-CD69-PE/Cy5 (H1.2F3), anti-CD5-BV510 (53–7.3), anti-TCRβ-APC (H57–597), anti-CD8a-PE/Cy7 (53–6.7), anti-CD69-Pacific Blue (H1.2F3), and Zombie Aqua, which were purchased from BioLegend. Anti-Foxp3-PE (FJK-165), anti-Foxp3-AF488 (150D/E4), anti-CD8a-APC/eFluor780 (53–6.7), anti-neuropilin-1-APC (3DS304M), anti-CD25-eFluor660, anti-CXCR4-APC (CD184; 2B11), anti-CCR7-APC (CD197; 4B12), and anti-CCR9-PE (CD199; CW-1.2) were purchased from eBioscience. PE-conjugated Vβ2 (B20.6), Vβ5.1/5.2 (MR9–4), Vβ6 (RR4–7), Vβ7 (TR310), Vβ8.1/8.2 (MR5–2), Vβ8.3 (8C1), Vβ9 (MR10–2), Vβ11 (RR3–15), Vβ12 (MR11–1), and Vβ13 (MR12–4) purchased from BioLegend, whereas Vβ4 (KT4) was from BD Biosciences. Appropriate isotype-label controls were used in all instances. To detect cell surface plexin-D1, we took advantage of the observation that the only significant receptor for sema3E on thymocytes is plexin-D1 (2), which enabled us to use the binding of sema3E as a measure of plexin-D1 expression. Sema3E-Fcγ2c was prepared as previously described (5) and directly labelled with APC using the Lynx rapid APC antibody conjugation reagent (AbD Serotec) according to manufacturer’s instructions. Mouse IgG2c-APC (Jackson Immunoresearch) was used as an isotype control.
Confocal imaging
Thymic frozen sections were analyzed as described previously (2, 5). Briefly, after embedding in OCT compound (Sakura Finetek), sectioning, and fixation in acetone for 10 min, thymic frozen sections (4 μm) were stained with the appropriate antibodies after blocking in 1% BSA-TBST overnight at 4°C. After staining and a final wash for 30 min, sections were mounted with ProLong Gold antifade (Thermo Fisher Scientific). Images were acquired with a Leica SP5X laser scanning confocal microscope equipped with an acousto-optical beam splitter (AOBS) system (Leica Camera AG) with a 40× oil objective (PL APO, NA1.25) using LAS AF software. Image processing and analyses were performed with Fiji/ImageJ software (http://rsbweb.nih.gov/ij/). Note that in all instances, images were initially captured in monochrome for maximum resolution and subsequently digitally colored for optimum differentiation. For CD69 analysis, sections were stained with anti-CD8a-eFluor660 (53–6.7) and anti-CD69-eFluor450 (H1.2F3) from eBioscience and anti-CD4-AF594 (GK1.5) from Biolegend. For Foxp3 analysis, sections were stained with anti-CD4-AF594 (GK1.5) and anti-CD8a-BV421 (53–6.7) from Biolegend, and anti-Foxp3-eFluor660 (FJK-16s) from eBioscience. For the experiments shown in fig. S4, analyzing a 200 × 150 μm2 rectangle centered on the asterisk, the brightness and contrast of the monochromatic, single-layer CD4 and CD8 signals were adjusted to the same median signal intensity and then falsely colored in green and red, respectively.
Generation of bone-marrow chimeric mice
Bone marrow cells were harvested from 7- to 9-week-old WT and Tagap KD NOD mice, and lineage-positive cells were removed using a Lineage Cell Depletion Kit (Miltenyi Biotec Inc.) according to the manufacturer’s instructions. Seven weeks-old WT NOD mice were irradiated with 1000 rad in two doses and then were injected with lineage-depleted WT and Tagap KD bone marrow cells (0.5 × 106 each) through the tail vein. Chimeric mice were analyzed after 6 weeks.
Imaging of cell adhesion
Single-cell suspensions of primary NOD WT or Tagap KD thymocytes were suspended at a density of 1 × 106 cells/ml in RPMI-1640 medium containing 1% fetal calf serum (FCS) and 15 μl (1.5 × 104 cells) was loaded onto a μ-slide V10.1 channel slide (ibidi GmbH) previously coated with recombinant mouse VCAM-1 (2.5 or 10 μg/ml) for 16 hours at room temperature followed by blocking with RPMI-1640/1% FCS. The chambers were incubated at 37°C for 30 min to permit cell settling and adhesion and then the nonadherent cells were washed away by establishing a flow of 2.5 μl/min of RPMI-1640/1% FCS (prewarmed to 37°C) across the chamber for 10 min with constant removal of unbound cells exiting the chamber. In the absence of VCAM-1, not a single cell bound to the chamber surface. Sema3E (5 μg/ml) was then introduced to establish a flow rate of 2.5 μl/min for 45 min followed by 5 min at 15 μl/min. Cell images were taken every minute and assembled in sequence to generate a single time-lapse video file. Individual cell motility was determined using ImageJ software incorporating the MTrackJ particle tracking plug-in (41).
Mammalian two-hybrid interaction assays
To determine direct interactions between the plexin-D1 cytoplasmic tail and GTPases, the Matchmaker Mammalian System 2 (Clontech) was used according to manufacturer’s instructions. Briefly, a fusion protein of the VP16 Herpes virus transcriptional activator activation domain upstream of the plexin-D1 cytoplasmic domain (AD-plexinD1) was generated. Point mutations (G12V) were introduced into the GTPases to generate constitutively active conformations except for Ralb (G23V), Ran (Q69L), and RhoH, which is constitutively active in its natural form (42). The GTPases were cloned into the pM vector to generate for each GTPase a fusion protein downstream of the GAL4 DNA-binding domain (DNA-BD-GTPase). The interaction of SV40 large T antigen (AD-SV40-T) with p53 protein (DNA-BD-p53) was used as a positive control. The interaction of the plexin-D1-cytoplasmic domain with a constitutively active GTPase brings the AD and DNA-BD into proximity, which leads to transcriptional activation of secreted alkaline phosphatase (SEAP) from the pG5-SEAP reporter vector. HEK293 cells were transfected with each DNA-BD-GTPase together with pG5-SEAP alone (GTPase-specific negative control) or with plasmid encoding AD-plexinD1 (test condition). HEK293 cells cotransfected with DNA-BD-plexinD1, pG5-SEAP, and AD-plexinD1 were used as a further control for possible plexin-D1 dimerization. Forty-eight hours after transfection, the cell culture medium was then tested for SEAP activity by hydrolysis of p-nitrophenyl phosphate and the spectrophotometric determination at 405 nm of released p-nitrophenol.
GTPase activation
For initial determination of Cdc42 activation by sema3E and optimization of conditions, the G-LISA Cdc42 assay kit (Cytoskeleton, Inc.) was used according to the manufacturer’s protocol to determine the effects of sema3E (0 to 10 μg/ml) at various times (0 to 30 min) on Cdc42 activation in the DP257–20-109 DP thymocyte cell line (Fig. 1A). Cell lysates were snap-frozen and stored at −80°C to permit protein determination. Samples were adjusted to 1 mg/ml before 50 μl was assayed through binding of activated Cdc42 to its downstream signal transducer, the Cdc42- and Rac-interactive binding (CRIB) domain (amino acid residues 70 to 117) of p21-activated kinase 1 (PAK1) immobilized on ELISA plates. Once optimal conditions of sema3E concentration (3 μg/ml) and assay time (10 min) had been established for Cdc42, a reproducible pulldown assay was used to assay Cdc42 activation. Briefly, DP257–20-109 cells or freshly isolated thymocytes were suspended at 2.5 × 107 cells in 225 μl of OptiMEM I (Life Technologies) and incubated for 3 hours at 37oC to determine basal Cdc42 activity in the absence of serum. At time zero, control medium or sema3E was added (25 μl) to each sample (to a final concentration of 3 μg/ml) for 10 min at 37oC, which was followed by the addition of 250 μl of 2× lysis buffer (as described above for GAP co-i.p. of plexinD1) on ice for 5 min. After rapid removal of cellular debris by centrifugation at 21,000g for 3 min, 25 μl of sample was removed from each clarified supernatant as a control for total Cdc42. Before the assay, glutathione beads were pre-incubated with GST-PAK1-CRIB(70–117), and 100 μl of a 50% suspension bearing 30 μg GST-PAK1-CRIB(70–117) was prepared to which thawed lysate was directly added and incubated with rotation at 4°C for 30 min followed by 4 washes in ice-cold lysis buffer. Samples were separated on a 12.5% Tris-HCl reducing SDS-PAGE gel, transferred to nitrocellulose, and blocked in TBS containing 0.05% Tween 20 and 5% BSA. Active and total Cdc42 were detected with rabbit polyclonal anti-Cdc42(P1) (Santa Cruz Biotechnology) followed by anti-rabbit-HRP (Cell Signaling Technology) and chemiluminescent development. Band densities were estimated using ImageJ software (41). To assay active RhoA in WT and Tagap KD thymocytes, the G-LISA RhoA assay kit (Cytoskeleton, Inc.) was used according to the manufacturer’s protocol, using the same sema3E incubation conditions as those determined for Cdc42. Lysis was performed by adding an equal volume of the 2× lysis buffer directly to the incubated thymocytes. All lysates were adjusted to a final protein concentration of 1 mg/ml before the assay. Comparison of total RhoA between the lysates was performed by Western blotting analysis with rabbit monoclonal anti-RhoA (clone 67B9; Cell Signaling Technology).
GAP activity assay
Direct GAP activity of recombinant plexin-D1 and recombinant Tagap was assayed using the RhoGAP assay kit (Cytoskeleton Inc.) according to the manufacturer’s instructions. GAP activity of plexin-D1 for Cdc42, Rac1, R-Ras, and Rnd3 (ProSpec-Tany TechnoGene Ltd.) and of Tagap for Cdc42, RhoA, Rac1, and Ras p21 (Cytoskeleton, Inc.) was determined by loading each GTPase with GTP and measuring the kinetics of release of inorganic phosphate (Pi) in the presence or absence of plexin-D1 or Tagap, respectively. The assay for the GAP activity for plexin-D1 used the non-myristoylated cytoplasmic domain (125 nM final concentration; 0.5 μg/well) together with GTPase (~20 μM final) in 40 μl of reaction buffer/well in 384-well microplates. GAP activity for tagap (0 to 80 nM final concentration; 0 to 0.32 μg/well) was assayed using GTPases at a final concentration of ~30 μM. p50-RhoGAP was used as a positive control GAP for RhoA, Cdc42, and Rac1; it has minimal activity for Ras p21 and R-Ras. After incubation at 37°C, CytoPhos reagent (Cytoskeleton, Inc.) was used to detect Pi in the end-reaction supernatants (10 min) and the absorbance at 650 nm measured. A Pi standard curve was generated from dilutions of KH2PO4 (100 mM) to determine Tagap specific GAP activity for each GTPase tested.
Bioinformatics and statistical analysis
Initial GAP candidates were identified in the NCBI GenBank Genome Research Consortium Build GRCm38.p4 (mouse; C57BL/6J; 3/23/2015). Candidates were refined using the Immunological Genome Project Consortium gene expression data for mouse αβTCR T cell differentiation (22) as the primary selection mechanism and refined using BioGPS mouse datasets (43). Arhgap sequence references for recombinant protein production are based on GRCm38.p4. The Chr.6 representation of the TAGAP locus and associated SNPs is based on NCBI GenBank Genome Research Consortium Build GRCh38.p7 (human; 3/21/2016). Data are presented as means ± SEM, and sample distributions were compared using the Student’s t test with two-tailed probability values derived from the t value and n-2 degrees of freedom. Regarding sample size, no statistical methods were used to predetermine requirements to achieve an adequate power or confidence level and experimental samples were not randomized in any way. The experiments were not blinded to sample identity or mouse genotype during the experiments or their outcome assessment.
Supplementary Material
Movie S1. Sema3E-mediated release of β1 integrin release from VCAM-1 for WT thymocytes.
Movie S2. Sema3E-mediated release of β1 integrin from VCAM-1 for Tagap KD thymocytes.
Fig. S1. Characterization of the DP257-20-109 DP thymocyte-like cell line.
Fig. S2. Binding of GTPases to the plexin-D1 cytoplasmic tail in a mammalian two-hybrid assay.
Fig. S3. Morphologies of cells coexpressing plexin-D1 and selected GAPs.
Fig. S4. Thymic morphology in WT and Plxnd1 CKO mice.
Fig. S5. Treg cell representation and TCR repertoire development in Tagap KD mice.
Fig. S6. Gating strategy for distinct thymocyte subsets.
Fig. S7. Thymocyte development in WT and Tagap KD hematopoietic chimeras.
Fig. S8. Calibration of GAP assay with p50 RhoGAP.
Table S1. Bioinformatically predicted mammalian RhoGAPs and protocol for target selection.
Table S2. Association of human TAGAP SNPs with autoimmunity.
Table 1. Cortical and medullary distributions of Foxp3+ Treg cells in WT, Tagap KD, and Plxnd1 CKO mice.
Plxnd1 WT and CKO thymi (three mice/genotype; B6 background) and WT and Tagap KD thymi (from two T8 mice and one T12 mouse for each condition; NOD background, all Dox-treated) were analyzed by confocal staining. Data are means ± SEM. Medulla refers to the normal medullary formations and not to the irregular, subcapsullary ectopic structures. Data are based on the analysis of 90 confocal images (0.2 × 0.15 mm2) for B6 WT and Plxnd1 CKO thymi, 80 confocal images (0.2 × 0.15 mm2) for NOD WT thymi, and 61 confocal images (0.2 × 0.15 mm2) for Tagap KD thymi. Cells were only counted as being Foxp3+ if a distinctive ring of membrane CD4 staining could be distinguished around the intracellular white Foxp3 signal.
| Genotype: | B6 WT | B6 Plxnd1 CKO | NOD WT | NOD Tagap KD |
|---|---|---|---|---|
| Foxp3+ Treg/mm2: | ||||
| Medulla | 108.5 ± 12.3 | 320.6 ± 37.3 | 558.9 ± 46.1 | 931.3 ± 68.3 |
| Cortex | 2.6 ± 0.7 | 16.9 ± 3.1 | 48.6 ± 11.5 | 131.0 ± 20.9 |
| Medulla/Cortical ratio | 41.7 | 19.0 | 11.5 | 7.1 |
| Change relative to WT medulla* | × 2.95 | × 1.67 | ||
| Change relative to WT cortex* | × 6.50 | × 2.70 |
P < 0.0001.
Acknowledgments:
The authors thank E. A. Witten for technical assistance and J. Stockton (Joslin Diabetes Center) for embryo microinjections. We thank the Joslin Flow Cytometry core facility for help with cell sorting and analyses. We thank J. Chernoff (Fox Chase Cancer Center, Philadelphia, PA) for providing pGEX2-TK-PAK1-CD and T. Tadakuma (Department of Parasitology and Immunology, National Defense Medical College, Tokorozawa, Saitama, Japan) for providing the DP-257-20-109 cell line.
Funding: This report was supported by NIH/NIAID 5R01AI115698 to E.L.R. This research was supported in part by a Pilot & Feasibility Award from the Joslin Diabetes Center DRC (P30DK036836) to S.K. Y.I. was supported by a Mary K. Iacocca Fellowship from the Iacocca Foundation. O.A. was supported by Wellcome Trust Grant WT094296MA. The Joslin Flow Cytometry core facility is supported by grants P30DK036836 and S10OD021740.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Kurd N, Robey EA, T-cell selection in the thymus: a spatial and temporal perspective. Immunol Rev 271, 114–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi YI, Duke-Cohan JS, Chen W, Liu B, Rossy J, Tabarin T, Ju L, Gui J, Gaus K, Zhu C, Reinherz EL, Dynamic control of beta1 integrin adhesion by the plexinD1-sema3E axis. Proc Natl Acad Sci U S A 111, 379–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burn P, Kupfer A, Singer SJ, Dynamic membrane-cytoskeletal interactions: specific association of integrin and talin arises in vivo after phorbol ester treatment of peripheral blood lymphocytes. Proc Natl Acad Sci U S A 85, 497–501 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larjava H, Plow EF, Wu C, Kindlins: essential regulators of integrin signalling and cell-matrix adhesion. EMBO Rep 9, 1203–1208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi YI, Duke-Cohan JS, Ahmed WB, Handley MA, Mann F, Epstein JA, Clayton LK, Reinherz EL, PlexinD1 glycoprotein controls migration of positively selected thymocytes into the medulla. Immunity 29, 888–898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi YI, Duke-Cohan JS, Tan J, Gui J, Singh MK, Epstein JA, Reinherz EL, Plxnd1 expression in thymocytes regulates their intrathymic migration while that in thymic endothelium impacts medullary topology. Front Immunol 4, 392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perala N, Sariola H, Immonen T, More than nervous: the emerging roles of plexins. Differentiation 83, 77–91 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Castellani V, Rougon G, Control of semaphorin signaling. Curr Opin Neurobiol 12, 532–541 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Oinuma I, Katoh H, Harada A, Negishi M, Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J Biol Chem 278, 25671–25677 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Uesugi K, Oinuma I, Katoh H, Negishi M, Different requirement for Rnd GTPases of R-Ras GAP activity of Plexin-C1 and Plexin-D1. J Biol Chem 284, 6743–6751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, He H, Srivastava N, Vikarunnessa S, Chen YB, Jiang J, Cowan CW, Zhang X, Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci Signal 5, ra6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negishi M, Buck M, Ras and Rap GAP function and GTPase sequestration in plexin-mediated cell signaling. Sci Signal 5, ra6 (2012).22253263 [Google Scholar]
- 13.Sakurai A, Gavard J, Annas-Linhares Y, Basile JR, Amornphimoltham P, Palmby TR, Yagi H, Zhang F, Randazzo PA, Li X, Weigert R, Gutkind JS, Semaphorin 3E initiates antiangiogenic signaling through plexin D1 by regulating Arf6 and R-Ras. Mol Cell Biol 30, 3086–3098 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worzfeld T, Swiercz JM, Senturk A, Genz B, Korostylev A, Deng S, Xia J, Hoshino M, Epstein JA, Chan AM, Vollmar B, Acker-Palmer A, Kuner R, Offermanns S, Genetic dissection of plexin signaling in vivo. Proc Natl Acad Sci U S A 111, 2194–2199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Pozo MA, Vicente-Manzanares M, Tejedor R, Serrador JM, Sanchez-Madrid F, Rho GTPases control migration and polarization of adhesion molecules and cytoskeletal ERM components in T lymphocytes. Eur J Immunol 29, 3609–3620 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Nishida T, Matsuki Y, Ono T, Oguma T, Tsujimoto K, Sato M, Tadakuma T, The novel murine CD4+CD8+ thymocyte cell line exhibits lineage commitment into both CD4+ and CD8+ T cells by altering the intensity and the duration of anti-CD3 stimulation in vitro. J Immunol 172, 6634–6641 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Janssen BJ, Robinson RA, Perez-Branguli F, Bell CH, Mitchell KJ, Siebold C, Jones EY, Structural basis of semaphorin-plexin signalling. Nature 467, 1118–1122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nogi T, Yasui N, Mihara E, Matsunaga Y, Noda M, Yamashita N, Toyofuku T, Uchiyama S, Goshima Y, Kumanogoh A, Takagi J, Structural basis for semaphorin signalling through the plexin receptor. Nature 467, 1123–1127 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Haddad E, Zugaza JL, Louache F, Debili N, Crouin C, Schwarz K, Fischer A, Vainchenker W, Bertoglio J, The interaction between Cdc42 and WASP is required for SDF-1-induced T-lymphocyte chemotaxis. Blood 97, 33–38 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Burk K, Mire E, Bellon A, Hocine M, Guillot J, Moraes F, Yoshida Y, Simons M, Chauvet S, Mann F, Post-endocytic sorting of Plexin-D1 controls signal transduction and development of axonal and vascular circuits. Nat Commun 8, 14508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang G, Brautigam CA, Chen R, Lu D, Torres-Vazquez J, Zhang X, Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, von Boehmer H, Mathis D, Benoist C, C. Immunological Genome, Best AJ, Knell J, Goldrath A, Joic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Nageswara Rao T, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S, The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 14, 619–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao M, Biery MC, Kobayashi SV, Ward T, Schimmack G, Burchard J, Schelter JM, Dai H, He YD, Linsley PS, T lymphocyte activation gene identification by coregulated expression on DNA microarrays. Genomics 83, 989–999 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Howden AJ, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Dieterich DC, Davis BG, Acuto O, QuaNCAT: quantitating proteome dynamics in primary cells. Nat Methods 10, 343–346 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Polyansky A, Buck M, Modeling transmembrane domain dimers/trimers of plexin receptors: implications for mechanisms of signal transmission across the membrane. PLoS One 10, e0121513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyarto BZ, Southern PJ, Masopust D, Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruder D, Probst-Kepper M, Westendorf AM, Geffers R, Beissert S, Loser K, von Boehmer H, Buer J, Hansen W, Neuropilin-1: a surface marker of regulatory T cells. Eur J Immunol 34, 623–630 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Zachary IC, How neuropilin-1 regulates receptor tyrosine kinase signalling: the knowns and known unknowns. Biochem Soc Trans 39, 1583–1591 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Chauvet S, Cohen S, Yoshida Y, Fekrane L, Livet J, Gayet O, Segu L, Buhot MC, Jessell TM, Henderson CE, Mann F, Gating of Sema3E/PlexinD1 signaling by neuropilin-1 switches axonal repulsion to attraction during brain development. Neuron 56, 807–822 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssen BJ, Malinauskas T, Weir GA, Cader MZ, Siebold C, Jones EY, Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat Struct Mol Biol 19, 1293–1299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miles JJ, Douek DC, Price DA, Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol 89, 375–387 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Matsutani T, Ohmori T, Ogata M, Soga H, Yoshioka T, Suzuki R, Itoh T, Alteration of T-cell receptor repertoires during thymic T-cell development. Scand J Immunol 64, 53–60 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Kearse KP, Takahama Y, Punt JA, Sharrow SO, Singer A, Early molecular events induced by T cell receptor (TCR) signaling in immature CD4+ CD8+ thymocytes: increased synthesis of TCR-alpha protein is an early response to TCR signaling that compensates for TCR-alpha instability, improves TCR assembly, and parallels other indicators of positive selection. J Exp Med 181, 193–202 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furukawa Y, Kawasoe T, Daigo Y, Nishiwaki T, Ishiguro H, Takahashi M, Kitayama J, Nakamura Y, Isolation of a novel human gene, ARHGAP9, encoding a rho-GTPase activating protein. Biochem Biophys Res Commun 284, 643–649 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Hildebrand JD, Taylor JM, Parsons JT, An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol 16, 3169–3178 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tata A, Stoppel DC, Hong S, Ben-Zvi A, Xie T, Gu C, An image-based RNAi screen identifies SH3BP1 as a key effector of Semaphorin 3E-PlexinD1 signaling. J Cell Biol 205, 573–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawada M, Ohno N, Kawaguchi M, Huang SH, Hikita T, Sakurai Y, Bang Nguyen H, Quynh Thai T, Ishido Y, Yoshida Y, Nakagawa H, Uemura A, Sawamoto K, PlexinD1 signaling controls morphological changes and migration termination in newborn neurons. EMBO J 37: e97404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kissler S, Studying autoimmunity by in vivo RNA interference. Methods Mol Biol 555, 109–118 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS, In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet 38, 479–483 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Herold MJ, van den Brandt J, Seibler J, Reichardt HM, Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci U S A 105, 18507–18512 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasband WS. (U. S. National Institutes of Health, Bethesda MD, 1997–2005). [Google Scholar]
- 42.Li X, Bu X, Lu B, Avraham H, Flavell RA, Lim B, The hematopoiesis-specific GTP-binding protein RhoH is GTPase deficient and modulates activities of other Rho GTPases by an inhibitory function. Mol Cell Biol 22, 1158–1171 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW 3rd, Su AI, BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10, R130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatzikyriakidou A, Voulgari PV, Lambropoulos A, Georgiou I, Drosos AA, Validation of the TAGAP rs212389 polymorphism in rheumatoid arthritis susceptibility. Joint Bone Spine 80, 543–544 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Chen R, Stahl EA, Kurreeman FA, Gregersen PK, Siminovitch KA, Worthington J, Padyukov L, Raychaudhuri S, Plenge RM, Fine mapping the TAGAP risk locus in rheumatoid arthritis. Genes Immun 12, 314–318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GA, Adany R, Aromaa A, Bardella MT, van den Berg LH, Bockett NA, de la Concha EG, Dema B, Fehrmann RS, Fernandez-Arquero M, Fiatal S, Grandone E, Green PM, Groen HJ, Gwilliam R, Houwen RH, Hunt SE, Kaukinen K, Kelleher D, Korponay-Szabo I, Kurppa K, MacMathuna P, Maki M, Mazzilli MC, McCann OT, Mearin ML, Mein CA, Mirza MM, Mistry V, Mora B, Morley KI, Mulder CJ, Murray JA, Nunez C, Oosterom E, Ophoff RA, Polanco I, Peltonen L, Platteel M, Rybak A, Salomaa V, Schweizer JJ, Sperandeo MP, Tack GJ, Turner G, Veldink JH, Verbeek WH, Weersma RK, Wolters VM, Urcelay E, Cukrowska B, Greco L, Neuhausen SL, McManus R, Barisani D, Deloukas P, Barrett JC, Saavalainen P, Wijmenga C, van Heel DA, Multiple common variants for celiac disease influencing immune gene expression. Nat Genet 42, 295–302 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eyre S, Hinks A, Bowes J, Flynn E, Martin P, Wilson AG, Morgan AW, Emery P, Steer S, Hocking LJ, Reid DM, Harrison P, Wordsworth P, C. Yorkshire Early Arthritis, R. A. C. C. Biologics in, Thomson W, Worthington J, Barton A, Overlapping genetic susceptibility variants between three autoimmune disorders: rheumatoid arthritis, type 1 diabetes and coeliac disease. Arthritis Res Ther 12, R175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagace C, Stokkers PC, Hommes DW, Barisani D, Palmieri O, Annese V, van Heel DA, Weersma RK, Daly MJ, Wijmenga C, Rioux JD, A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet 7, e1001283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D’Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D’Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M, Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 42, 1118–1125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, Gwilliam R, Takeuchi F, McLaren WM, Holmes GK, Howdle PD, Walters JR, Sanders DS, Playford RJ, Trynka G, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, O’Morain C, Kennedy NP, Kelleher D, Pennington DJ, Strachan DP, McArdle WL, Mein CA, Wapenaar MC, Deloukas P, McGinnis R, McManus R, Wijmenga C, van Heel DA, Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 40, 395–402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parmar AS, Lappalainen M, Paavola-Sakki P, Halme L, Farkkila M, Turunen U, Kontula K, Aromaa A, Salomaa V, Peltonen L, Halfvarson J, Torkvist L, D’Amato M, Saavalainen P, Einarsdottir E, Association of celiac disease genes with inflammatory bowel disease in Finnish and Swedish patients. Genes Immun 13, 474–480 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Patsopoulos NA,M. S. G. W. G. Bayer Pharma, I.-b. Steering Committees of Studies Evaluating, C. C. R. A. a, A. N. Consortium, GeneMsa C International Multiple Sclerosis Genetics, Esposito F, Reischl J, Lehr S, Bauer D, Heubach J, Sandbrink R, Pohl C, Edan G, Kappos L, Miller D, Montalban J, Polman CH, Freedman MS, Hartung HP, Arnason BG, Comi G, Cook S, Filippi M, Goodin DS, Jeffery D, O’Connor P, Ebers GC, Langdon D, Reder AT, Traboulsee A, Zipp F, Schimrigk S, Hillert J, Bahlo M, Booth DR, Broadley S, Brown MA, Browning BL, Browning SR, Butzkueven H, Carroll WM, Chapman C, Foote SJ, Griffiths L, Kermode AG, Kilpatrick TJ, Lechner-Scott J, Marriott M, Mason D, Moscato P, Heard RN, Pender MP, Perreau VM, Perera D, Rubio JP, Scott RJ, Slee M, Stankovich J, Stewart GJ, Taylor BV, Tubridy N, Willoughby E, Wiley J, Matthews P, Boneschi FM, Compston A, Haines J, Hauser SL, McCauley, Ivinson s, Oksenberg JR, Pericak-Vance M, Sawcer SJ, De Jager PL, Hafler DA, de Bakker PI, Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol 70, 897–912 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perkins EA, Landis D, Causey ZL, Edberg Y, Reynolds RJ, Hughes LB, Gregersen PK, Kimberly RP, Edberg JC, Bridges SL Jr., I. Consortium for the Longitudinal Evaluation of African Americans with Early Rheumatoid Arthritis, Association of single-nucleotide polymorphisms in CCR6, TAGAP, and TNFAIP3 with rheumatoid arthritis in African Americans. Arthritis Rheum 64, 1355–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasad P, Kumar A, Gupta R, Juyal RC, Thelma BK, Caucasian and Asian specific rheumatoid arthritis risk loci reveal limited replication and apparent allelic heterogeneity in north Indians. PLoS One 7, e31584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R, Alfredsson L, Amos CI, Ardlie KG, Consortium B, Barton A, Bowes J, Burtt NP, Chang M, Coblyn J, Costenbader KH, Criswell LA, Crusius JB, Cui J, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Kurreeman FA, Lee AT, Liu X, Li Y, Martin P, Morgan AW, Padyukov L, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth P, Consortium Y, Altshuler D, Karlson EW, Toes RE, de Vries N, Begovich AB, Siminovitch KA, Worthington J, Klareskog L, Gregersen PK, Daly MJ, Plenge RM, Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet 41, 1313–1318 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma A, Liu X, Hadley D, Hagopian W, Liu E, Chen WM, Onengut-Gumuscu S, Simell V, Rewers M, Ziegler AG, Lernmark A, Simell O, Toppari J, Krischer JP, Akolkar B, Rich SS, Agardh D, She JX, Group TS, Identification of Non-HLA Genes Associated with Celiac Disease and Country-Specific Differences in a Large, International Pediatric Cohort. PLoS One 11, e0152476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, Heap GA, Dubois PC, Clayton DG, Hunt KA, van Heel DA, Todd JA, Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 359, 2767–2777 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, Kang HM, Allen MH, McManus R, Novelli G, Samuelsson L, Schalkwijk J, Stahle M, Burden AD, Smith CH, Cork MJ, Estivill X, Bowcock AM, Krueger GG, Weger W, Worthington J, Tazi-Ahnini R, Nestle FO, Hayday A, Hoffmann P, Winkelmann J, Wijmenga C, Langford C, Edkins S, Andrews R, Blackburn H, Strange A, Band G, Pearson RD, Vukcevic D, Spencer CC, Deloukas P, Mrowietz U, Schreiber S, Weidinger S, Koks S, Kingo K, Esko T, Metspalu A, Lim HW, Voorhees JJ, Weichenthal M, Wichmann HE, Chandran V, Rosen CF, Rahman P, Gladman DD, Griffiths CE, Reis A, Kere J, P. Collaborative Association Study of, C. Genetic Analysis of Psoriasis, E. Psoriasis Association Genetics, C. Wellcome Trust Case Control, Nair RP, Franke A, Barker JN, Abecasis GR, Elder JT, Trembath RC, Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet 44, 1341–1348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Sema3E-mediated release of β1 integrin release from VCAM-1 for WT thymocytes.
Movie S2. Sema3E-mediated release of β1 integrin from VCAM-1 for Tagap KD thymocytes.
Fig. S1. Characterization of the DP257-20-109 DP thymocyte-like cell line.
Fig. S2. Binding of GTPases to the plexin-D1 cytoplasmic tail in a mammalian two-hybrid assay.
Fig. S3. Morphologies of cells coexpressing plexin-D1 and selected GAPs.
Fig. S4. Thymic morphology in WT and Plxnd1 CKO mice.
Fig. S5. Treg cell representation and TCR repertoire development in Tagap KD mice.
Fig. S6. Gating strategy for distinct thymocyte subsets.
Fig. S7. Thymocyte development in WT and Tagap KD hematopoietic chimeras.
Fig. S8. Calibration of GAP assay with p50 RhoGAP.
Table S1. Bioinformatically predicted mammalian RhoGAPs and protocol for target selection.
Table S2. Association of human TAGAP SNPs with autoimmunity.