Abstract
Background
l-2-aminobutyric acid (l-ABA) is an unnatural amino acid that is a key intermediate for the synthesis of several important pharmaceuticals. To make the biosynthesis of l-ABA environmental friendly and more suitable for the industrial-scale production. We expand the nature metabolic network of Escherichia coli using metabolic engineering approach for the production of l-ABA.
Results
In this study, Escherichia coli THR strain with a modified pathway for threonine-hyperproduction was engineered via deletion of the rhtA gene from the chromosome. To redirect carbon flux from 2-ketobutyrate (2-KB) to l-ABA, the ilvIH gene was deleted to block the l-isoleucine pathway. Furthermore, the ilvA gene from Escherichia coli W3110 and the leuDH gene from Thermoactinomyces intermedius were amplified and co-overexpressed. The promoter was altered to regulate the expression strength of ilvA* and leuDH. The final engineered strain E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH was able to produce 9.33 g/L of l-ABA with a yield of 0.19 g/L/h by fed-batch fermentation in a 5 L bioreactor.
Conclusions
This novel metabolically tailored strain offers a promising approach to fulfill industrial requirements for production of l-ABA.
Electronic supplementary material
The online version of this article (10.1186/s12934-019-1095-z) contains supplementary material, which is available to authorized users.
Keywords: l-2-Aminobutyric acid, l-Threonine deaminase, l-Leucine dehydrogenase, Metabolic engineering, Fed-batch fermentation
Background
l-2-aminobutyric acid (l-ABA), a nonnatural amino acid, has been used as a precursor for synthesis of many chiral drugs, such as anti-epileptic Levetiracetam, anti-tuberculotic Ethambutol and Brivaracetam [1–3]. (S)-2-amino butanol, a key intermediate of Ethambutol, can be synthesized by esterification and hydrogenation reduction of l-ABA [4]. With the increasing market demand for l-ABA in both pharmaceutical and chemical industries in recent years, the preparation of optically pure l-ABA with high efficacy has attracted much attention.
Currently, the preparation of l-ABA is mainly achieved by chemical synthesis or enzymatic conversion. In chemical methods, synthesis of l-ABA has been extensively reported including ammonolysis of α-halogen acid [5], reduction reaction [6], ammoniation hydrolysis reaction and butanone acid reduction [7]. However, the obvious disadvantages of chemical synthesis, such as poor selectivity, harsh reaction conditions, various byproducts, and the difficulty in separation and purification [8–10], limited its development. Enzymatic synthesis of l-ABA is emerged since biotransformation and bio-refining for the green production of chemicals have been attracting the increasing attention due to serious concerns about climate change and environmental problems [11–13]. It is reported that l-ABA was synthesized in a transamination reaction from α-ketobutyric acid and l-aspartic acid as substrates using aromatic aminotransferase [14] or produced from α-ketobutyric acid and benzylamine using ω-aminotransferase [1]. l-ABA could also be produced from the reduction of α-keto acids with l-leucine dehydrogenase [15] or glutamate dehydrogenase [16]. But in the enzymatic routes, the presence of by-product α-keto acid decreases the overall yield and purity of l-ABA [2]. It is well known that, most of the natural l-amino acids can now be produced from glucose by microbial fermentation [17]. Notably, l-glutamate, l-lysine, and l-threonine are produced more than 2 million tons annually [18]. Therefore, microbial production of l-ABA from cheap and clean resources has gained much attention with the potential to overcome these problems.
Escherichia coli has been reported to produce many natural amino acids owing to its clear genetic background and facile genetic manipulation [17, 19]. At present, there are few reports about biosynthesis of the nonnatural amino acid l-ABA using microbial fermentation. We attempt to construct a novel engineered E. coli strain for production of the nonnatural amino acid, l-ABA directly from glucose by expanding its metabolic pathways.
In this study, a threonine producing strain E. coli THR was firstly constructed, then the metabolic pathway was expanded via specific genes overexpression for converting l-threonine to l-ABA. Further modification for improvement of l-ABA production included l-threonine reduction, l-isoleucine synthetic pathway blockage and promoter replacement. This work provides a novel approach for the industrial production of l-ABA by fermentation using the genetically engineered E. coli strain.
Results
Construction of engineered E. coli strain for l-threonine production
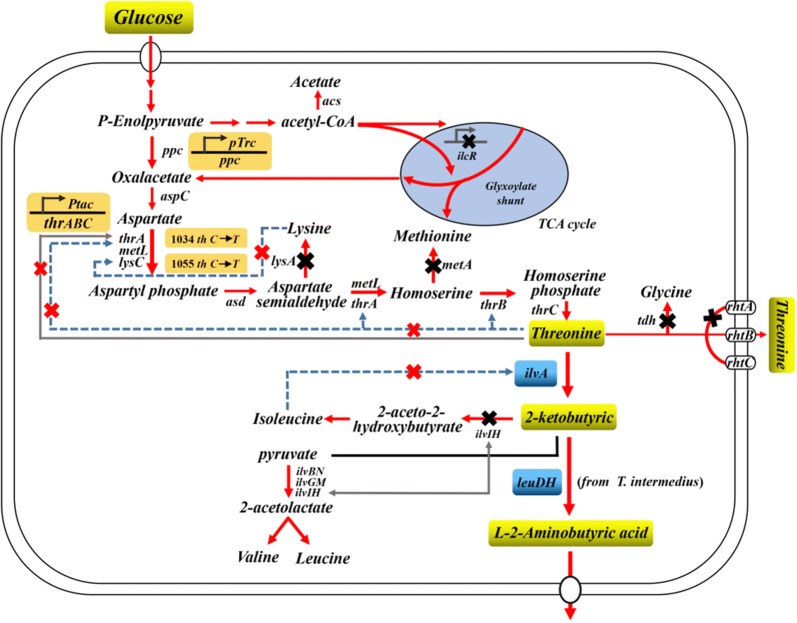
To expand metabolism for producing l-ABA, we designed a unique metabolic pathway in E. coli, where l-threonine was taken as a precursor for l-ABA synthesis (Fig. 1). In this work, E. coli THR strain (Table 1) was constructed for the overproduction of l-threonine. The feedback inhibitions of aspartokinase I and III encoded by thrA and lysC genes, respectively, were released [20]. The native promoter containing the transcriptional attenuator leader region of the thrABC operon was replaced with the tac promoter. The metA, lysA, tdh and iclR genes were deleted to make more precursors available for l-threonine formation. The native promoter of the ppc gene was replaced with the Trc promoter in the chromosome to increase the pool of oxaloacetate, a starting precursor of l-threonine biosynthesis [21]. As a result, the strain E. coli THR produced 12.45 g/L l-threonine from 50 g/L glucose in shake flask at 35 °C for 48 h in TPM medium, which was further used to construct an l-ABA producing strain.
Fig. 1.
The overall metabolic engineering strategy employed for the construction of a genetically defined l-ABA producer. Central metabolic pathways that contribute to the biosynthesis of l-ABA together with competing pathways and regulatory circuits are shown. The orange shaded boxes represent mutations introduced into the genome. The black Xs indicate the deletion of genes and the red Xs indicate the removal of inhibition or repression. Dotted lines indicate the feedback inhibition. Gray lines indicate the transcriptional attenuation regulation. The red thick arrows indicate the increased flux or activity by directly overexpressing the corresponding genes
Table 1.
Characteristics and sources of bacterial strains used in this study
| Strains | Characteristics | Sources |
|---|---|---|
| E. coli W3110 | Coli Genetic Stock Center strain (CGSC) No. 4474 | CGSCa |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697galU galK rpsL endA1 nupG | Invitrogenb |
| E. coli THR | W3110 (ΔlacI, thrAC1034T, lysCC1055T, PthrABC::Ptac, ΔlysA, ΔmetA, Δtdh, ΔiclR, Pppc::PTrc) | This study |
| E. coli THR ΔrhtA | E. coli THR derivative, ΔrhtA | This study |
| E. coli THR ΔrhtC | E. coli THR derivative, ΔrhtC | This study |
| E. coli THR ΔrhtAΔrhtC | E. coli THR derivative, ΔrhtA/ΔrhtC | This study |
| E. coli THR ΔrhtAΔilvIH | E. coli THR derivative, ΔrhtA/ΔilvIH | This study |
aInvitrogen, Crop., Carlsbad, CA
bColi Genetic Stock Center
Reprogramming of ilvA to redirect the carbon flux towards 2-KB
It is well known that l-threonine was catalyzed to form 2-KB by threonine dehydratase encoded by ilvA in E. coli. In this study, an ilvA overexpression strain was constructed, which could produce 4.38 g/L 2-KB and 7.35 g/L l-threonine (Fig. 2). The high accumulation of l-threonine intracellularly indicated that the catabolic enzyme ilvA is not active enough to fully convert l-threonine into 2-KB. The feedback inhibition of ilvA by l-isoleucine was then considered as the main factor to hinder the further improvement of the 2-KB titer [16].
Fig. 2.
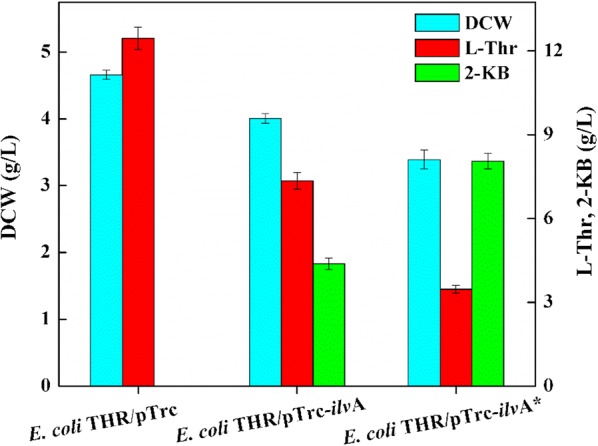
Effect of ilvA* overexpression on 2-KB titer
On the basis of previous studies, feedback inhibition of ilvA could be removed by replacing the 1054 th T with G, 1055 th T with C, 1084 th C with T, 1085 th G with T and 1086 th T with C (F352A, R362F) using site-directed mutagenesis to obtain pTrc-ilvA* [22]. The 2-KB titer of E. coli THR/pTrc-ilvA* increased by 83.8% which was up to 8.05 g/L, the value of which was nearly 2 times of that from the pTrc-ilvA strain (4.38 g/L 2-KB), and the concentration of remaining l-threonine from 7.35 g/L decrease to 3.47 g/L (Fig. 2). These results suggested that the removal of l-isoleucine which is the feedback inhibition of the ilvA gene could increase both the activity and resistance to l-isoleucine inhibition as compared to that of the wild-type strain and drive the carbon flux from l-threonine to 2-KB [22].
Enzyme selection for converting 2-KB to l-ABA
In order to gain an appropriate dehydrogenase for 2-KB production, two different sources of dehydrogenase, including leuDH from T. intermedius [23] and BleuDH from Bacillus cereus [24] were tested. Plasmids pTrc-leuDH and pTrc-BleuDH were constructed and introduced into E. coli THR. Results showed that the accumulation of l-ABA reached to 5.39 g/L and 3.16 g/L in E. coli THR/pTrc-leuDH and E. coli THR/pTrc-BleuDH, respectively, which were cultivated in TPM medium with additional feeding of 10 g/L 2-KB (Fig. 3). These results demonstrated that the dehydrogenase leuDH from T. intermedius displayed a higher specific activity in E. coli THR than that of BleuDH from Bacillus cereus.
Fig. 3.
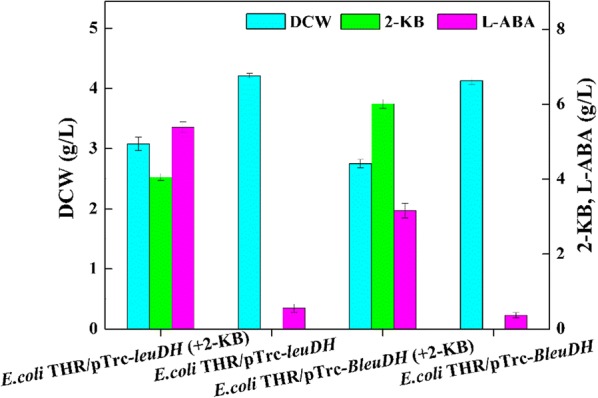
Effects of leuDH and BleuDH overexpression on l-ABA titer
Modification of bypass pathway to further improve the l-ABA titer
Based on the fermentation results from ilvA* and leuDH expression separately in E. coli THR, the co-overexpression strain E. coli THR/pTrc-ilvA*-leuDH was constructed, which could produce 3.09 g/L l-ABA from 50 g/L glucose. However, there was still 3.47 g/L l-threonine remained in the fermentation medium (Fig. 4). In E. coli, rhtA and rhtC are known to be involved in l-threonine efflux [25, 26]. When the rhtA gene was overexpressed on multicopy plasmids, the expression of the rhtA gene enhanced about tenfold [27]. The rhtC gene was induced to protect cells from toxic effects of intracellular l-threonine accumulation by exporting l-threonine out of the cell. It is reported that overexpression of the rhtC gene could increase the production of l-threonine which is 50.2% higher than that without rhtC amplification [21].
Fig. 4.
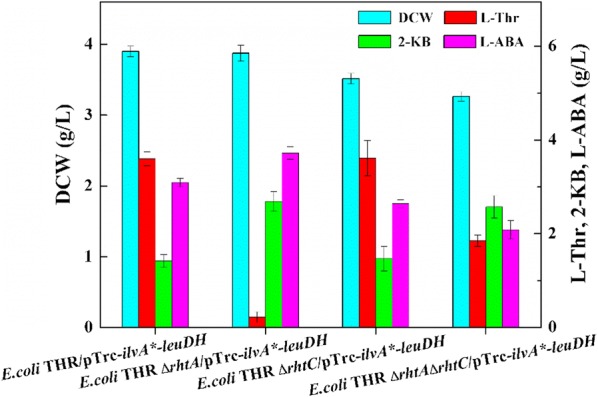
Effect of rhtA or rhtC deletion on l-ABA titer
With the purpose of decreasing the l-threonine export capacity, the gene rhtA and rhtC were deleted from the E. coli THR strain chromosome, resulting in three strains E. coli THR ΔrhtA, E. coli THR ΔrhtC and E. coli THR ΔrhtAΔrhtC. Then the recombinant plasmid pTrc-ilvA*-leuDH was respectively transformed into three strains to test the l-ABA production. The result of strain E. coli THR ΔrhtA/pTrc-ilvA*-leuDH displayed higher concentration of l-ABA than the other two strains (Fig. 4). The deletion of rhtA in the chromosome led to the increase of l-ABA concentration from 3.09 g/L to 3.72 g/L and the remaining l-threonine decrease from 3.47 g/L to 0.22 g/L, which indicated that the modification of the l-threonine transport pathway is an efficient strategy for l-ABA improvement.
In addition, three acetohydroxy acid synthase (AHAS) isoenzymes that E. coli possessed, which show different biochemical properties and regulation mechanisms, play important roles in the biosynthesis of l-isoleucine. Among them, AHAS III, encoded by ilvIH, exhibites a much higher affinity for 2-KB [28] and AHAS I, encoded by ilvBN, displays a higher affinity for pyruvate than 2-KB [28]. However, AHAS II, encoded by ilvGM, is not expressed due to the frameshift mutation of ilvG in E. coli [29]. Thus, only the gene ilvIH from the E. coli THR ΔrhtA chromosome was knocked out to reduce the metabolic flux from 2-KB to l-isoleucine, the resulting strain E. coli THR ΔrhtAΔilvIH/pTrc-ilvA*-leuDH was capable of producing 4.42 g/L l-ABA (Fig. 5).
Fig. 5.
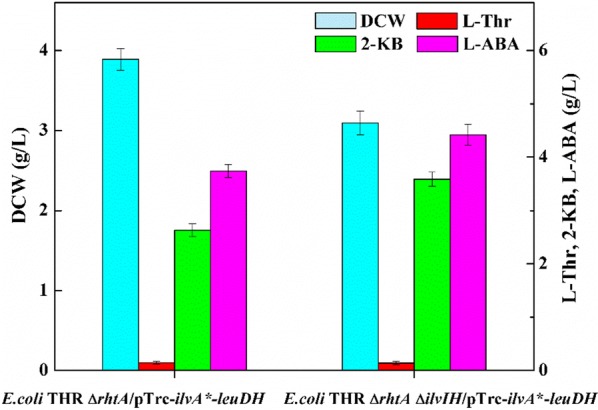
Effect of ilvIH deletion on l-ABA titer
Regulation of the expression strength of ilvA* and leuDH via promoter alteration
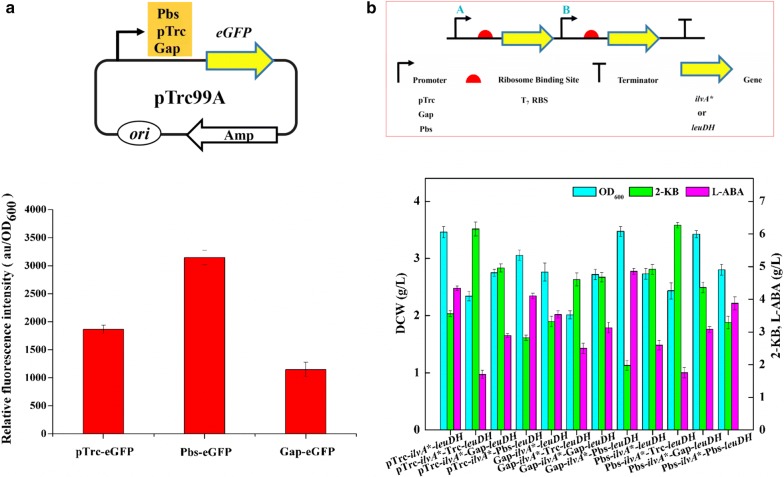
It was observed that 3.56 g/L 2-KB was remained in flask culture of E. coli THR ΔrhtAΔilvIH/pTrc-ilvA*-leuDH. We reasoned that the uncoordinated expression of ilvA* and leuDH might account for the extracellular accumulation of 2-KB. Therefore, it is necessary to regulate the expression of ilvA* and leuDH via promoter alteration. We employed different promoters with different strengths, including Pbs [30], Trc and Gap from E. coli BL21 (DE3), to regulate the expression of ilvA* and leuDH based on plasmid pTrc-ilvA*-leuDH. To investigate the strengths of different promoters, enhanced green fluorescent protein (eGFP) was introduced and used as a reporter. As a result, eGFP was successfully expressed under the control of three promoters, respectively in E. coli THR ΔrhtAΔilvIH and the relative fluorescence intensity (au/OD600) from strong to weak was Pbs-eGFP, Trc-eGFP and Gap-eGFP (Fig. 6a). Twelve recombinant plasmids were constructed when three promoters were inserted or replaced before the ilvA* or leuDH, respectively (Table 2). The results showed that 4.86 g/L of l-ABA was produced by E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH (Fig. 6b), which was the strain with the highest yield of l-ABA compared with other strains constructed in this study. Moreover, the accumulation of 2-KB was decreased to 1.98 g/L, whereas the cell growth did not change remarkably.
Fig. 6.
Regulating the expression of ilvA* and leuDH for l-ABA titer. a Relative fluorescence intensity (au/OD600) of eGFP harbored in different promoters. b Effect of promoters with different strengths on l-ABA titer
Table 2.
Descriptions and sources of plasmids used in this study
| Plasmids | Description | Sources |
|---|---|---|
| pTrc99a | AmpR, pBR322-origin, trc promoter, 4.2 kb | [35, 38] |
| pCas | KanR, Cas9 nuclease expression plasmid, temperature-sensitive origin | [39, 40] |
| pTrc-ilvA* | AmpR, ilvA* cloned in the sac I and BamH I site of pTrc99A | This study |
| Gap-ilvA* | AmpR, replacement of Trc promoter in pTrc-ilvA* with the Gap promoter | This study |
| Pbs-ilvA* | AmpR, replacement of Trc promoter in pTrc-ilvA* with the Pbs promoter | This study |
| pTrc-ilvA* -BleuDH | AmpR, BleuDH cloned in the Xba I and Hind III site of pTrc-ilvA* | This study |
| pTrc-ilvA*-leuDH | AmpR, leuDH cloned in the Xba I and Hind III site of pTrc-ilvA* | This study |
| pTrc-ilvA*-Trc-leuDH | AmpR, leuDH carrying a Trc promoter and RBS cloned in the Xba I and Hind III site of pTrc-ilvA* | This study |
| pTrc-ilvA*-Gap-leuDH | AmpR, leuDH carrying a Gap promoter and RBS cloned in the Xba I and Hind III site of pTrc-ilvA* | This study |
| pTrc-ilvA*-Pbs-leuDH | AmpR, leuDH carrying a Pbs promoter and RBS cloned in the Xba I and Hind III site of pTrc-ilvA* | This study |
| Gap-ilvA*-leuDH | AmpR, replacement of Trc promoter in pTrc-ilvA*-leuDH with the Gap promoter | This study |
| Gap-ilvA*-Trc-leuDH | AmpR, leuDH carrying a Trc promoter and RBS cloned in the Xba I and Hind III site of Gap-ilvA* | This study |
| Gap-ilvA*-Gap-leuDH | AmpR, leuDH carrying a Gap promoter and RBS cloned in the Xba I and Hind III site of Gap-ilvA* | This study |
| Gap-ilvA*-Pbs-leuDH | AmpR, leuDH carrying a Pbs promoter and RBS cloned in the Xba I and Hind III site of Gap-ilvA* | This study |
| Pbs-ilvA*-leuDH | AmpR, replacement of Trc promoter in pTrc-ilvA*-leuDH with the Pbs promoter | This study |
| Pbs-ilvA*-Trc-leuDH | AmpR, leuDH carrying a Trc promoter and RBS cloned in the Xba I and Hind III site of Pbs-ilvA* | This study |
| Pbs-ilvA*-Gap-leuDH | AmpR, leuDH carrying a Gap promoter and RBS cloned in the Xba I and Hind III site of Pbs-ilvA* | This study |
| Pbs-ilvA*-Pbs-leuDH | AmpR, leuDH carrying a Pbs promoter and RBS cloned in the Xba I and Hind III site of pbs-ilvA* | This study |
| pTarget-X | A plasmid used to transcript sgRNA targeting the particular gene X in genome. X refers to the amino acid biosynthetic pathways | This study |
Sp spectinomycin, Kan kanamycin, Amp ampicillin, R resistance
Fed-batch fermentation for l-ABA production
As the platform strains for the production of l-ABA through rational metabolic engineering was successfully constructed, fed-batch fermentation was performed to evaluate the potential of the engineered strain E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH. During the fermentation, the initially-fed glucose was exhausted at approximately 12 h. When the glucose concentration in the broth was less than 5 g/L, feed medium was injected into the broth to raise the residual glucose concentration to around 20 g/L. Production of l-ABA presented a cell growth-dependent profile in the first 36 h of the fermentation. When the growth of the cells entered the stationary phase, l-ABA was continuously accumulated, but at a lower rate than that in the exponential phase, followed by a slight decrease in the titer after 48 h of the fermentation (Fig. 7). As a result, 9.33 g/L of l-ABA was produced from glucose in a total 60 h of fed-batch fermentation, representing a 1.92-fold increase compared to the titer achieved in shake flask. Meanwhile, 2-KB was detected as the main byproduct during the fermentation in the 5 L bioreactor with the titer of 3.27 g/L. Another byproduct l-threonine was barely detected.
Fig. 7.
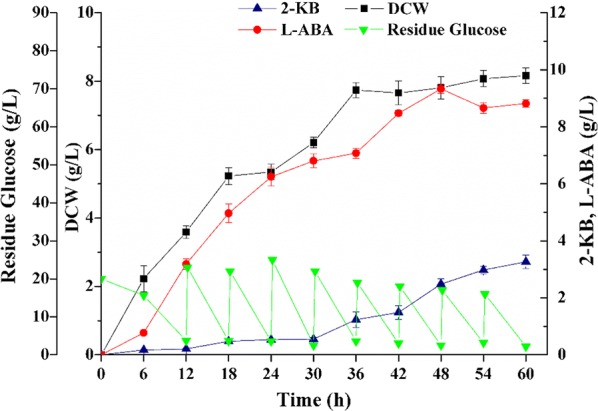
Fermentation process profiles of E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH showing DCW (filled squares), residual glucose (filled inverse triangle), 2-KB titer (filled upright triangle) and l-ABA titer (filled circles)
Discussion
In this study, we have developed a novel biosynthesis process for l-ABA production directly from glucose by fermentation using the genetically engineered E. coli strain. The results confirmed that the high expression level of leuDH and ilvA* successfully expanded the biosynthetic pathway, resulting in 3.60 g/L l-threonine, 1.42 g/L 2-KB and 3.09 g/L l-ABA in shake flask. Comparing to the traditional enzyme catalytic production method, this synthetic process possesses the advantages of simplicity in the production process and the production costs-savings, which represents an attractive approach for the production of various valuable nonnatural products from inexpensive renewable resources.
To avoid the excessive accumulation of extracellular l-threonine, a partial inactivation of the l-threonine export system was employed. In accordance with the previous reports [21, 27], rhtA and rhtC, which encoded strong l-threonine exporter, were deleted from the E. coli THR strain chromosome to inactivate the l-threonine export system. Our result indicated that it was better to knock out the rhtA gene alone rather than to knock out the rhtC gene alone or both genes at the same time regarding to the l-ABA production. The rhtA-deletion strain E. coli THR ΔrhtA/pTrc-ilvA*-leuDH showed an l-ABA production enhancement by 20.4% in comparison with that of E. coli THR/pTrc-ilvA*-leuDH. The results further indicated that the reasonable modification of l-threonine transport system was beneficial to the accumulation of l-ABA.
As competitive branches, the disruption of l-isoleucine biosynthetic pathways should be contributed to the improvement of l-ABA production [31]. Block of the l-isoleucine metabolic pathway by disrupting ilvIH made the titer of l-ABA in E. coli THR ΔrhtAΔilvIH/pTrc-ilvA*-leuDH increased by 1.19-fold compared to that in E. coli THR ΔrhtA/pTrc-ilvA*-leuDH, but the cell growth decreased. It is concluded that ilvIH disruption allowed the diversion of 2-KB flowed to l-isoleucine to l-ABA, and therefore improved the titer of l-ABA, but caused accumulation of 2-KB to the value of 3.56 g/L, which led to a reduction in cell growth [31]. Interestingly, the l-ABA titer was accumulated to 4.86 g/L and 2-KB decreased to 1.98 g/L in E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH without any obvious reduction of cell growth when promoters of different strengths including Pbs [30] and Gap from E. coli BL21 (DE3) were employed to regulate the expression of ilvA* and leuDH. These results indicated that the disruption of ilvIH drived more carbon flux to l-ABA and proper adjustments of attenuating the expression of ilvA* by a relatively weak promoter and enhancing the expression of leuDH by a strong promoter were also beneficial for the biosynthesis of l-ABA in this strain.
This work reported the development of a bacterial platform for enhanced production of a nonnatural amino acid l-ABA. The highest yield of l-ABA achieved by the engineered E. coli strain was 9.33 g/L via fed-batch fermentation, indicating a great potential for large-scale production. However, there are still some bottlenecks, including the intracellular l-ABA transfer, plasmid elimination, metabolic burden and high costs of fermentation due to the large amount requirement of glucose and high concentration of l-methionine, l-isoleucine and l-lysine, waiting to be conquered for the industrial production of l-ABA. Therefore, screening of relevant genes encoding l-ABA exporter and integrating the involved genes into the chromosome of E. coli THR will be carried out in the future. In addition, the fermentation medium and condition will be further optimized to reduce the production costs and increase the l-ABA titer.
Conclusions
In this study, a novel strain E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH for l-ABA production was constructed through metabolic engineering. The ilvA gene from E. coli W3110 and the leuDH gene from T. intermedius were firstly co-expressed in E. coli THR, and 3.09 g/L of l-ABA was achieved. Then, the rhtA gene was disrupted to decrease the extracellular secretion of l-threonine and the titer of l-ABA was improved to 3.72 g/L. To block the catabolism from 2-KB to l-isoleucine, the ilvIH gene was disrupted and the l-ABA titer was increased by 18.8% compared to the parental strain. Furthermore, in order to obtain a better expression of ilvA and leuDH, different promoters were, respectively inserted or replaced before the two enzymes to regulate their expression, resulting in l-ABA accumulation up to 4.86 g/L. Finally, l-ABA titer of the optimal strain E. coli THR ΔrhtAΔilvIH/Gap-ilvA*-Pbs-leuDH reached to 9.33 g/L in fed-batch fermentation. This study offers a possible approach for the industrial bioproduction of l-ABA and paves a way for the industrialization of other nonnatural amino acids.
Methods
Strains, media, and growth conditions
The bacterial strains used in this study were listed in Table 1. E. coli DH5α was used as host for the recombinant plasmid. E. coli W3110 and its derivatives were applied to produce l-ABA.
TPM medium was used to monitor the production of l-ABA, during the growth of cells at 35 °C. TPM medium contains per liter: glucose, 50 g; yeast extract, 6 g; MgSO4·7H2O, 2 g; KH2PO4, 4 g; (NH4)2SO4, 14 g; betaine, 1 g; l-methionine, 0.149 g; l-lysine, 0.164 g; trace metal solution, 5 mL and CaCO3, 30 g. The trace metal solution contains per liter: FeSO4·7H2O, 10 g; CaCl2, 1.35 g; ZnSO4·7H2O, 2.25 g; MnSO4·4H2O, 0.5 g; CuSO4·5H2O, 1 g; (NH4)6Mo7O24·4H2O, 0.106 g; Na2B4O7·10H2O, 0.23 g; 35% HCl, 10 mL [21]. The medium was adjusted to pH 7.0 by KOH. Ampicillin (100 mg/L; Sangon, Shanghai, China), kanamycin (50 mg/L; Solarbio, Beijing, China), and isopropyl-β-D-thio-galactopyranoside (IPTG; 100 µM; Sangon, Shanghai, China) were added when necessary.
For precultivation of E. coli W3110 and its derivatives, a single clone was grown in 5 mL Luria–Bertani (LB) medium. After incubation for 10 h, the seed culture was inoculated into 500 mL shake flask containing 30 mL cultivation media. Cells were grown at 35 °C and 100 mg/L ampicillin was supplemented when needed. For l-methionine auxotrophic and l-lysine auxotrophic mutants, l-methionine and l-lysine with final concentrations of 0.149 g/L and 0.164 g/L were added, respectively in fermentation broth to fairly compare their titer with others trains [32]. A final concentration of 0.1 mM IPTG was added to the medium for gene induction when the optical density at 600 nm (OD600) reached of 0.4–0.6. Fermentation of strains were conducted simultaneously under the same culture conditions for at least three times.
Construction of co-expression plasmids
All plasmids used in this study were listed in Table 2. The primers used for gene amplification and recombinant plasmid construction were listed in Additional file 1: Table S1. In general, genes including ilvA encoding feedback-resistant threonine dehydratase and leuDH encoding l-leucine dehydrogenase from Thermoactinomyces intermedius were overexpressed in a plasmid-based manner. The procedures of recombinant plasmid construction were described in detail shown in Additional file 1: Figure S1.
Genomic engineering: promoter replacement, site-directed mutagenesis and chromosomal gene knock-out
Donor dsDNA with 500-bp homologous arms on each side was designed based on the sequence of the gene cluster and the location of sgRNA. Two homologous arms and the Trc promoter were separately amplified and fused together by overlap-extension PCR. The PCR products were purified by gel extraction prior to electroporation. Electrocompetent cells were prepared according to a previous report [33]. A single colony was transferred into 5 mL of LB medium containing 50 mg/L kanamycin and 10 mM l-arabinose (Aladdin, Shanghai, China), and was grown at 30 °C overnight. An aliquot consisted of 100 µL resulting preculture was transferred into 50 mL of LB medium containing 50 mg/L kanamycin and 10 mM l-arabinose, and was grown at 30 °C to an OD600 value of 0.4–0.6. The cultures were chilled in ice-water slurry for 15 min, harvested by centrifugation at 4000×g for 10 min, and washed twice with ice-cold sterile ddH2O. Subsequently, 200 µL of ice-cold sterile glycerol (10%, v/v) was used to re-suspend the cells, and the glycerol suspension was separated into 100 µL aliquots for each reaction. Donor dsDNA (400 ng) and the corresponding pTarget plasmid (100 ng) were added to each electroporation reaction. A Bio-Rad MicroPulser (Bio-Rad, Hercules, CA, USA) was used for electroporation (0.1 cm cuvette, 1.8 kV). 1 mL of pre-chilled LB medium was added to the cuvette and the resulting cell suspension was transferred into a tube within 1 min. The culture was then regenerated at 30 °C for 2 h prior to plating. Positive colonies were transferred into LB medium containing 0.5 mM IPTG and cultivated at 30 °C for 8–10 h to eliminate the pTarget plasmid. The pCas plasmid was cured by cultivating at 37 °C overnight. The cultures after plasmid curing were streaked, and the colonies were tested for kanamycin (50 mg/L) and spectinomycin (50 mg/L; Sangon, Shanghai, China) sensitivity, and were confirmed by sequencing [19, 34].
Fermentation
For fed-batch fermentation, a 150 mL seed cultured in LB medium for 12 h was inoculated into a 5 L agitated bioreactor (Shanghai Baoxing biological equipment Engineering Co. Ltd, China) with 3 L TPM medium equipped with dissolved oxygen, pH and temperature probes. Cells were incubated at 35 °C with agitation at 500 rpm and 2 L/min external air flow. Ammonia or phosphate was automatically fed into the broth to keep the pH at 7.0. When the glucose concentration in the broth was less than 5 g/L, 100 mL of feed solution (500 g/L glucose, 14 g/L (NH4)2SO4, 12.5 g/L KH2PO4, 3 g/L l-methionine and 4.4 g/L l-lysine) was injected into the broth to raise the residual glucose concentration to around 20 g/L [21, 35].
Analytical methods
The cell concentration was monitored by measuring the absorbance at 600 nm which was then converted to Dry Cell Weight (DCW) by a calibration curve [36]. The fermentation supernatants were filtered through a 0.22 µm syringe filter (Nylon66; Jinteng, Tianjin, China) and used for determination of residual glucose, amino acids and organic acids. The concentration of 2-KB in the culture was determined by high pressure liquid chromatography (HPLC, Waters, Milford, MA, USA) using an Aminex HPX-87H column (7.8 × 300 mm, Bio-Rad, Hercules, CA, USA) under the following conditions: mobile phase 5 mM H2SO4; flow rate 0.5 mL/min; column temperature 30 °C; UV absorption 215 nm [31]. The residual concentration of glucose in the media was measured using a glucose analyzer (YSI model 2300, Xylem Inc., Rye Brook, NY, USA) [37] and the amino acids were determined using an amino acid analyzer (SYKAMS-433D, SYKAM, Munich, Germany). The l-Thr and l-ABA were derivatized with DNFB/Acetonitrile (DNFB = 1%, acetonitrile = 99%) and analyzed by HPLC using a LC-18DB column (5 µm, 4.6 × 250 mm, Agilent, Beijing, China). Derivatization method: 100 μL of the fermentation supernatants diluted 10 times with ddH2O were added to a 1.5 mL centrifuge tube, and then 50 μL of DNFB/Acetonitrile buffer and 100 μL of 0.5 mol/L NaHCO3 buffer were added. The reaction solution was placed in a dark water bath at 60 °C for 1 h. After the reaction was completed, 750 μL of 0.2 mol/L phosphate buffer at pH 7.0 was added. Finally, the reaction solution was filtered through a 0.22 µm syringe filter. The gradient elution profile, at 1 mL/min, was as follows: 16% A and 84% B at 0–0.18 min, 30% A and 70% B at 0.18–2.4 min, 34% A and 66% B at 2.4–4.2 min, 43% A and 57% B at 4.2–7.2 min, 55% A and 45% B at 7.2–13.3 min, 55% A and 45% B at 13.3–15 min, 98% A and 2% B at 15–20.4 min, 16% A and 84% B at 20.4–21.3 min, 16% A and 84% B at 21.3–30 min (A = 50% acetonitrile; B = 4.1 g/L sodium acetate, pH adjustment to the value of 6.4 with acetic acid); column temperature 33 °C; UV absorption at 360 nm. The retention time of the main peak of the solution should be consistent with that of the reference solution. The undiluted medium and fermentation broth of E. coli W3110/pTrc99A were used as negative controls. Amino acid standards were purchased from Sykam (Amino Acid Calibration Solution H, Sykam, Germany).
Statistical analysis
Unless otherwise specified, all experiments in this study were performed in triplicates. An analysis of variance (ANOVA) was carried out using the SAS program version 8.1 (SAS Institute Inc., Cary, NC, USA). The least significant difference (LSD) was computed at p < 0.05. All figures were drawn using Origin software version 8.5 (Origin Lab Corp., Northampton, MA, USA). Error bars denote standard deviation of the mean.
Additional file
Additional file 1: Table S1. Primers for donor DNA amplification, pTarget construction and positive colony validation. Figure S1. Structures of plasmids used in this study. Detailed description on constructions of these plasmids is shown in the section.
Authors’ contributions
JMX, JQL, ZB, ZQL and YGZ designed the study. JQL and JMX carried out the bulk of the experiments. JMX, JQL, ZB, ZQL and YGZ analyzed and interpreted the data. JQL and ZQL wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Sequences data used in this study have been deposited in the GenBank with accession number AP009048 for the gene of ilvA from Escherichia coli W3110, AE016877 for the gene of BleuDH from Bacillus cereus and X79068.1 for the gene of leuDH from Thermoactinomyces intermedius.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was financially supported by Zhejiang Provincial Public Welfare Technology Application Research Projects (2017C33157), the National Natural Science Foundation of China (No. 31500031), and the Natural Science Foundation of Zhejiang Province (No. LQ14B060004).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- l-ABA
l-2-aminobutyric acid
- 2-KB
2-ketobutyrate
- DCW
dry cell weight
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- l-Thr
l-threonine
- eGFP
enhanced green fluorescent protein
Contributor Information
Jian-Miao Xu, Email: xujianmiao@zjut.edu.cn.
Jian-Qiang Li, Email: 2111605033@zjut.edu.cn.
Bo Zhang, Email: zhangbo0305@zjut.edu.cn.
Zhi-Qiang Liu, Phone: +86-571-88320379, Email: microliu@zjut.edu.cn.
Yu-Guo Zheng, Email: zhengyg@zjut.edu.cn.
References
- 1.Shin JS, Kim BG. Transaminase-catalyzed asymmetric synthesis of l-2-aminobutyric acid from achiral reactants. Biotechnol Lett. 2009;31(10):1595–1599. doi: 10.1007/s10529-009-0057-7. [DOI] [PubMed] [Google Scholar]
- 2.Taylor PP, Pantaleone DP, Senkpeil RF, Fotheringham IG. Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends Biotechnol. 1998;16(10):412–418. doi: 10.1016/s0167-7799(98)01240-2. [DOI] [PubMed] [Google Scholar]
- 3.Zhu L, Tao R, Wang Y, Jiang Y, Lin X, Yang Y, Zheng H, Jiang W, Yang S. Removal of l-alanine from the production of l-2-aminobutyric acid by introduction of alanine racemase and d-amino acid oxidase. Appl Microbiol Biotechnol. 2011;90(3):903–910. doi: 10.1007/s00253-011-3127-4. [DOI] [PubMed] [Google Scholar]
- 4.Ragonese R, Macka M, Hughes J, Petocz P. The use of the Box-Behnken experimental design in the optimisation and robustness testing of a capillary electrophoresis method for the analysis of ethambutol hydrochloride in a pharmaceutical formulation. Biomed Anal. 2002;27(6):995–997. doi: 10.1016/s0731-7085(01)00659-8. [DOI] [PubMed] [Google Scholar]
- 5.Jeffery E, Meisters A. Electrochemical synthesis of amino acids by reductive amination of keto acids. I. Reduction at mercury electrodes. Aust J Chem. 1978;31(1):73–78. [Google Scholar]
- 6.Babievskii KK, Belikov VM, Tikhonova NA. Amino acids. Communication. 1 synthesis of dl-threonine and dl-2-aminobutyric acid based on condensations of nitroacetic ester. Bull Acad Sci USSR Div Chem Sci. 1965;14(1):76–81. [Google Scholar]
- 7.Fujii C, Yasui M, Ishimathu Y. Process for production of (+)-2-amino-1-butanol. US Patent. 3,979,457, 07 Sept 1976.
- 8.An Z, Gu X, Liu Y, Ge J, Zhu Q. Bioproduction of l-2-aminobutyric acid by a newly-isolated strain of Aspergillus tamarii ZJUT ZQ013. Catal Lett. 2017;147(4):837–844. [Google Scholar]
- 9.Liu ZQ, Lu MM, Zhang XH, Cheng F, Xu JM, Xue YP, Jin LQ, Wang YS, Zheng YG. Significant improvement of the nitrilase activity by semi-rational protein engineering and its application in the production of iminodiacetic acid. Int J Biol Macromol. 2018;116:563–571. doi: 10.1016/j.ijbiomac.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 10.Liu ZQ, Wu L, Zhang XJ, Xue YP, Zheng YG. Directed evolution of carbonyl reductase from Rhodosporidium toruloides and its application in stereoselective synthesis of tert-butyl (3R,5S)-6-Chloro-3,5-dihydroxyhexanoate. J Agric Food Chem. 2017;65(18):3721–3729. doi: 10.1021/acs.jafc.7b00866. [DOI] [PubMed] [Google Scholar]
- 11.Liu ZQ, Dong SC, Yin HH, Xue YP, Tang XL, Zhang XJ, He JY, Zheng YG. Enzymatic synthesis of an ezetimibe intermediate using carbonyl reductase coupled with glucose dehydrogenase in an aqueous-organic solvent system. Bioresour Technol. 2017;229:26–32. doi: 10.1016/j.biortech.2016.12.098. [DOI] [PubMed] [Google Scholar]
- 12.Liu ZQ, Wu L, Zheng L, Wang WZ, Zhang XJ, Jin LQ, Zheng YG. Biosynthesis of tert-butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate by carbonyl reductase from Rhodosporidium toruloides in mono and biphasic media. Bioresour Technol. 2018;249:161–167. doi: 10.1016/j.biortech.2017.09.204. [DOI] [PubMed] [Google Scholar]
- 13.Zheng YG, Yin HH, Yu DF, Chen X, Tang XL, Zhang XJ, Xue YP, Wang YJ, Liu ZQ. Recent advances in biotechnological applications of alcohol dehydrogenases. Appl Microbiol Biotechnol. 2017;101(3):987–1001. doi: 10.1007/s00253-016-8083-6. [DOI] [PubMed] [Google Scholar]
- 14.Fotheringham IG, Grinter N, Pantaleone DP, Senkpeil RF, Taylor PP. Engineering of a novel biochemical pathway for the biosynthesis of l-2-aminobutyric acid in Escherichia coli K12. Bioorg Med Chem. 1999;7(10):2209–2213. doi: 10.1016/s0968-0896(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 15.Xu JM, Cheng F, Fu FT, Hu HF, Zheng YG. Semi-rational engineering of leucine dehydrogenase for l-2-aminobutyric acid production. Appl Biochem Biotechnol. 2017;182(3):898–909. doi: 10.1007/s12010-016-2369-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhang KC, Li H, Cho KM, Liao JC. Expanding metabolism for total biosynthesis of the nonnatural amino acid l-homoalanine. Proc Natl Acad Sci. 2010;107(14):6234–6239. doi: 10.1073/pnas.0912903107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JH, Lee SY. Metabolic pathways and fermentative production of l-aspartate family amino acids. Biotechnol J. 2010;5(6):560–577. doi: 10.1002/biot.201000032. [DOI] [PubMed] [Google Scholar]
- 18.Leuchtenberger W, Huthmacher K, Drauz K. Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol. 2005;69(1):1–8. doi: 10.1007/s00253-005-0155-y. [DOI] [PubMed] [Google Scholar]
- 19.Li YF, Lin ZQ, Huang C, Zhang Y, Wang ZW, Tang YJ, Chen T, Zhao XM. Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YF, Meng QL, Ma HW, Liu YF, Cao GQ, Zhang XR, Zheng P, Sun JB, Zhang DW, Jiang WX, Ma YH. Determination of key enzymes for threonine synthesis through in vitro metabolic pathway analysis. Microb Cell Fact. 2015;14:86. doi: 10.1186/s12934-015-0275-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for l-threonine production. Mol Syst Biol. 2007;3(1):149. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Chen Z, Zheng P, Sun JB, Zeng AP. Study and reengineering of the binding sites and allosteric regulation of biosynthetic threonine deaminase by isoleucine and valine in Escherichia coli. Appl Microbiol Biotechnol. 2013;97(7):2939–2949. doi: 10.1007/s00253-012-4176-z. [DOI] [PubMed] [Google Scholar]
- 23.Galkin A, Kulakova L, Yoshimura T, Soda K, Esaki N. Synthesis of optically active amino acids from alpha-keto acids with Escherichia coli cells expressing heterologous genes. Appl Environ Microbiol. 1997;63:4651–4656. doi: 10.1128/aem.63.12.4651-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ansorge MB, Kula MR. Production of recombinant l-leucine dehydrogenase from Bacillus cereus in pilot scale using the runaway replication system E. coli [piet98] Biotechnol Bioeng. 2000;68(5):557–562. [PubMed] [Google Scholar]
- 25.Livshits VA, Zakataeva NP, Aleshin VV, Vitushkina MV. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res Microbiol. 2003;154(2):123–135. doi: 10.1016/S0923-2508(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 26.Zakataeva NP, Aleshin VV, Tokmakova IL, Troshin PV, Livshits VA. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 1999;452(3):228–232. doi: 10.1016/s0014-5793(99)00625-0. [DOI] [PubMed] [Google Scholar]
- 27.Lee JH, Sung BH, Kim MS, Blattner FR, Yoon BH, Kim JH, Kim SC. Metabolic engineering of a reduced-genome strain of Escherichia coli for l-threonine production. Microb Cell Fact. 2009;8:2. doi: 10.1186/1475-2859-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barak Z, Chipman DM, Gollop N. Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. J Bacteriol. 1987;169(8):3750–3756. doi: 10.1128/jb.169.8.3750-3756.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawther RP, Calhoun DH, Adams CW, Hauser CA, Gray J, Hatfield GW. Molecular basis of valine resistance in Escherichia coli K-12. Proc Natl Acad Sci. 1981;78(2):922–925. doi: 10.1073/pnas.78.2.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, Liu QT, Zhang YF, Du GC, Chen J, Kang Z. Construction and characterization of broad-spectrum promoters for synthetic biology. ACS Synth Biol. 2018;7(1):287–291. doi: 10.1021/acssynbio.7b00258. [DOI] [PubMed] [Google Scholar]
- 31.Zhang CL, Qi JS, Li YJ, Fan XG, Xu QY, Chen N, Xie XX. Production of α-ketobutyrate using engineered Escherichia coli via temperature shift. Biotechnol Bioeng. 2016;113(9):2054–2059. doi: 10.1002/bit.25959. [DOI] [PubMed] [Google Scholar]
- 32.Usuda Y, Kurahashi O. Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl Environ Microbiol. 2005;71:3228–3234. doi: 10.1128/AEM.71.6.3228-3234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N, Zhang B, Wang ZW, Tang YJ, Chen T, Zhao XM. Engineering Escherichia coli for fumaric acid production from glycerol. Bioresour Technol. 2014;174:81–87. doi: 10.1016/j.biortech.2014.09.147. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y, Chen B, Duan CL, Sun BB, Yang JJ, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang JF, Liu ZQ, Jin LQ, Tang XL, Shen ZY, Yin HH, Zheng YG. Metabolic engineering of Escherichia coli for microbial production of l-methionine. Biotechnol Bioeng. 2017;114(4):843–851. doi: 10.1002/bit.26198. [DOI] [PubMed] [Google Scholar]
- 36.Peng LF, Shimizu K. Effect of fadR gene knockout on the metabolism of Escherichia coli based on analyses of protein expressions, enzyme activities and intracellular metabolite concentrations. Enzyme Microb Technol. 2006;38(3-4):512–520. [Google Scholar]
- 37.Fallet C, Rohe P, Franco-Lara E. Process optimization of the integrated synthesis and secretion of ectoine and hydroxyectoine under hyper/hypo-osmotic stress. Biotechnol Bioeng. 2010;107(1):124–133. doi: 10.1002/bit.22750. [DOI] [PubMed] [Google Scholar]
- 38.Huang JF, Zhang B, Shen ZY, Liu ZQ, Zheng YG. Metabolic engineering of E. coli for the production of O-succinyl-l-homoserine with high yield. 3 Biotech. 2018;8:310. doi: 10.1007/s13205-018-1332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81(7):2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang JF, Shen ZY, Mao QL, Zhang XM, Zhang B, Wu JS, Liu ZQ, Zheng YG. Systematic analysis of bottlenecks in a multi-branched and multilevel regulated pathway: the molecular fundamentals of l-methionine biosynthesis in Escherichia coli. ACS Synth Biol. 2018;7:2577–2589. doi: 10.1021/acssynbio.8b00249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers for donor DNA amplification, pTarget construction and positive colony validation. Figure S1. Structures of plasmids used in this study. Detailed description on constructions of these plasmids is shown in the section.
Data Availability Statement
Sequences data used in this study have been deposited in the GenBank with accession number AP009048 for the gene of ilvA from Escherichia coli W3110, AE016877 for the gene of BleuDH from Bacillus cereus and X79068.1 for the gene of leuDH from Thermoactinomyces intermedius.