Abstract
Background
HOTTIP, a long non-coding RNA located in the HOXA cluster, plays a role in the patterning of tissues with mesodermal components, including the lung. Overexpression of HOXA genes, including HOTTIP, has been associated with a more aggressive phenotype in several cancers. However, the prognostic impact of HOTTIP has not yet been explored in non-small-cell lung cancer (NSCLC). We have correlated HOTTIP expression with time to relapse (TTR) and overall survival (OS) in early-stage NSCLC patients.
Methods
Ninety-nine early-stage NSCLC patients who underwent surgical resection in our center from June 2007 to November 2013 were included in the study. Mean age was 66; 77.8% were males; 73.7% had stage I disease; and 55.5% had adenocarcinoma. A validation data set comprised stage I-II patients from The Cancer Genome Atlas (TCGA) Research Network.
Results
HOTTIP was expressed in all tumor samples and was overexpressed in squamous cell carcinoma (p = 0.007) and in smokers (p = 0.018). Patients with high levels of HOTTIP had shorter TTR (78.3 vs 58 months; p = 0.048) and shorter OS (81.2 vs 61 months; p = 0.023) than those with low levels. In the multivariate analysis, HOTTIP emerged as an independent prognostic marker for TTR (OR: 2.05, 95%CI: 1–4.2; p = 0.05), and for OS (OR: 2.31, 95%CI: 1.04–5.1; p = 0.04). HOTTIP was validated as a prognostic marker for OS in the TCGA adenocarcinoma cohort (p = 0.025). Moreover, we identified a 1203-mRNA and a 61-miRNA signature that correlated with HOTTIP expression.
Conclusions
The lncRNA HOTTIP can be considered a prognostic biomarker in early-stage NSCLC.
Electronic supplementary material
The online version of this article (10.1186/s12890-019-0816-8) contains supplementary material, which is available to authorized users.
Keywords: HOTTIP, NSCLC, Early-stage, Overall survival, Lung cancer, lncRNAs
Background
According to the American Cancer Society, in 2018, lung cancer will be the second most frequent cancer in the United States of America in both males and females (14 and 13%, respectively, of all cancers) and the first leading cause of death by cancer (26 and 25%, respectively) [1]. Non-small-cell lung cancer (NSCLC), the most frequent subtype, accounts for 85% of all lung cancers. Despite years of research, the prognosis for patients with NSCLC remains dismal, with a 5-year relative survival rate of 18% for all stages combined (www.cancer.net). In the 30% of patients that debut with early-stage disease (stage I-II), the cornerstone of treatment is the surgical removal of the tumor. Moreover, in stage IB disease with a primary tumor > 4 cm and in stage II disease, adjuvant chemotherapy (usually cisplatin-vinorelbine) has proven to be beneficial, with a 4–5% absolute survival improvement at five years [2]. A large study including 1294 consecutive early-stage NSCLC patients who underwent surgery showed that after a median follow up of 35 months, 20% of patients had relapsed and 7% were diagnosed with a second primary lung cancer [3]. These data highlight the need to further investigate this disease and consolidate useful prognostic markers.
Up to 70% of our genome is transcribed into non-coding RNAs (ncRNAs) that do not serve as templates for proteins. These ncRNAs are subdivided into two major groups: small ncRNAs (< 200 nt) and long ncRNAs (lncRNAs) (> 200 nt) [4]. Although small ncRNAs, especially microRNAs (miRNAs), have been the most extensively studied [5], lncRNAs have recently emerged as worthy biomarkers, since their expression is highly cell type- and tissue-specific [6]. In NSCLC, several lncRNAs are involved in the carcinogenesis process, some of which have been associated with patient survival [7–9].
The HOX family genes are known transcription factors with a key role in embryogenesis and carcinogenesis [10, 11]. Their expression is dysregulated in several cancers, including NSCLC [11–14]. In humans, HOX genes are organized into four clusters (A, B, C, and D), which are located on different chromosomes [15]. Interestingly, several lncRNAs associated with HOX genomic regions can participate in the regulation of HOX genes and collaborate in their functions [16]. HOTTIP, also known as HOXA distal transcript antisense RNA, is an antisense lncRNA located in the HOXA cluster that coordinates the activation of several 5′ HOXA genes in vivo [17]. It is overexpressed in several cancers [18–20], including NSCLC, where its overexpression in vitro has been associated with increased proliferation and invasion of lung cancer cells through transcriptional regulation of HOXA13 [21]. Moreover, HOTTIP overexpression has been associated with worse outcome in several tumors, including hepatocellular carcinoma [22], tongue squamous cell carcinoma [18], colorectal cancer [20], osteosarcoma [23], breast cancer [24], gastric cancer [19], and even small-cell lung cancer [25]. To date, however, the prognostic impact of HOTTIP expression levels in NSCLC has not been explored [26, 27].
In the present study, we have analyzed HOTTIP expression in a cohort of 99 patients with early-stage NSCLC who underwent surgical resection in our center and have correlated HOTTIP expression levels with overall survival (OS) and time to relapse (TTR).
Methods
Patients
A total of 99 early-stage NSCLC patients who underwent complete surgical resection in Hospital Clínic (Barcelona) from May 2007 to October 2013 were included in the study. Prospectively collected tumor tissues were stored in RNALater® (Ambion) at − 80 °C until processing. Clinical data were recorded on admission: age, gender, smoking history, Eastern Cooperative Oncology Group (ECOG) performance status (PS), preoperative pulmonary function tests (PFT) and chronic obstructive pulmonary disease (COPD). COPD was defined as when forced expiratory volume in 1 s (FEV1) to forced vital capacity (FVC) ratio is below a fixed cutoff (< 70%). Type of surgical resection and pathological findings were also recorded, including tumor characteristics and the presence of emphysema (defined histopathologically in the resected non-tumoral tissue). All patient samples were studied by Sanger sequencing for mutations in TP53 (exons 5–8) and KRAS (codon 12–13), and only adenocarcinoma patients samples were studied for EGFR mutations (exons 19–21). The following primers were used: TP53 exon 5 Forward (F) 5′- GTTTCTTTGCTGCCGTCTTC-3′, TP53 exon 5 Reverse (R) 5′-GAGCAATCAGTGAGGAATCAGA-3′; TP53 exon 6 F 5′-AGAGACGACAGGGCTGGTT-3′, TP53 exon 6 R 5′-CTTAACCCCTCCTCCCAGAG-3′; TP53 exon 7 F 5′- TTGCCACAGGTCTCCCCAA-3′, TP53 exon 7 R 5′-AGGGGTCAGAGGCAAGCAGA-3′; and TP53 exon 8 F 5′-GGGACAGGTAGGACCTGATTT-3′, TP53 exon 8 R 5′-TAACTGCACCCTTGGTCTCC-3′; KRAS F 5′-TTAACCTTATGTGTGACATGTT-3′, KRAS R 5′-AGAATGGTCCTGCACCAGTAA-3′; EGFR exon 18 F 5′- GCATGGTGAGGGCTGAGGT-3′, EGFR exon 18 R 5′-TGCAAGGACTCTGGGCTCC-3′,EGFR exon 19 F 5′-TGCATCGCTGGTAACATCCA-3′, EGFR exon 19 R 5′-GAAAAGGTGGGCCTGAGGTT-3′, EGFR exon 20 F 5′-TCCTTCTGGCCACCATGC-3′, EGFR exon 20 R 5′-TGGCTCCTTATCTCCCCTCC-3′, EGFR exon 21 F 5′-ATGCAGAGCTTCTTCCCATGA-3′, EGFR exon 21 R 5′-CAGGAAAATGCTGGCTGACC-3′.
All patients signed the written consent in accordance with the Declaration of Helsinki to use their samples in the present research. The Clinical Research Ethics Committee of the Hospital Clínic de Barcelona approved the study.
NSCLC patients from The Cancer Genome Atlas (TCGA) Research Network (RNAseq data; https://cancergenome.nih.gov) were used as a validation data set. Two cohorts were used: the Lung Adenocarcinoma (TCGA-LUAD) and the Lung Squamous Cell Carcinoma (TCGA-LUSC). The patient selection criteria were: stage I-II, Caucasian, minimum OS of 35 days, and available RNAseq data. Using these selection criteria, the validation cohorts included 91 adenocarcinomas from the TCGA-LUAD and 59 squamous cell carcinomas from the TCGA-LUSC cohort. The analysis was performed separately in each cohort.
RNA extraction and lncRNA expression analysis
Total RNA was purified from frozen tissue with TriZol® Reagent (Life Technologies) as per manufacturer’s specifications. cDNA was synthetized from 500 ng of total RNA with The High Capacity cDNA Reverse Transcription Kit® (Applied Biosystems). TaqMan assays (Life Technologies) were used to quantify HOTTIP (Hs00955374_s1) in a 7500 Real Time PCR device (Applied Biosystems). CDKN1B (Hs00153277_m1) was used as endogenous control, and the mean of the HOTTIP expression in the normal tissue was used as calibrator sample to apply the2-ΔΔCt method.
Statistics
R v3.3 and IBM SPSS Statistics 22 were used for statistical analysis. TTR was calculated as the time between surgery and relapse or last follow-up and OS as the time between surgery and death from any cause or last follow-up. Kaplan-Meier survival curves and log-rank test were used for the survival analysis. All clinic-pathological factors with p-value ≤0.1 in the univariate analysis were included in the Cox multivariate regression analyses. The Optimal cutoff for the analysis of the impact on survival of the HOTTIP expression was identified using the Maxstat package (R Statistical Package) and validated by the Kaplan-Meier test. Paired (when necessary) or non-paired t-tests were used for comparisons between two groups. The independent validation of the prognostic role of HOTTIP levels were performed using TANRIC [28] (http://ibl.mdanderson.org/tanric/_design/basic/index.html) based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
Results
Patients
The main clinic-pathological characteristics of the patients are summarized in Table 1. Briefly, the mean patient age was 66 years (range, 32–84), 77 patients (77.8%) were male, 20 (20.2%) had Eastern Cooperative Oncology Group (ECOG) performance status (PS) 0 and 79 (79.8%) had PS 1. Seventy-three (60.2%) patients were diagnosed in stage I disease. Adenocarcinoma histology was found in fifty-five (55.5%) patients and 87 (87.9%) were active or former smokers. Median follow-up time was 44 months (range, 8–98). After a follow-up of 98 months, 31 (31.3%) patients experienced disease recurrence. Twenty-three patients (23.2%) received adjuvant therapy after surgery (17 stage II and six stage IB). Thirty-three percent of the patients harbored TP53 mutations and 20% harbored KRAS mutations. Adenocarcinoma patients with EGFR mutations (29.8%) showed a trend towards shorter TTR (p = 0.09) and OS (p = 0.09).
Table 1.
Main clinical characteristics of the patients
| Characteristics | Value |
N = 99 N (%) |
|---|---|---|
| Sex | Male | 77 (77.8) |
| Female | 22 (22.2) | |
| Age, yrs. | Mean (Range) | 66 (32–84) |
| <=65 | 49 (49.5) | |
| > 65 | 50 (50.5) | |
| ECOG PSa | 0 | 20 (20.2) |
| 1 | 79 (79.8) | |
| Stage | I | 73 (73.7) |
| II | 26 (26.3) | |
| T | 1 | 47 (47.5) |
| 2 | 49 (49.5) | |
| 3 | 3 (3) | |
| N | 0 | 88 (88.9) |
| 1 | 11 (11.1) | |
| Histology | Adenocarcinoma | 55 (55.5) |
| Squamous cell carcinoma | 36 (36.4) | |
| Others | 8 (8.1) | |
| Type of surgery | Lobectomy/bilobectomy | 89 (89.9) |
| Pneumonectomy | 4 (4) | |
| Atypical Resectionb | 6 (6.1) | |
| Smoking history | Current Smoker | 34 (34.4) |
| Former Smoker | 53 (53.5) | |
| Never smoker | 12 (12.1) | |
| Adjuvant chemotherapy | Yes | 23 (23.2) |
| No | 76 (76.8) | |
| Relapse | No | 68 (68.7) |
| Yes | 31 (31.3) | |
| Emphysema | Yes | 48 (48.5) |
| No | 44 (44.4) | |
| Unknown | 7 (7.1) | |
| COPDc | Yes | 64 (64.6) |
| No | 35 (35.4) | |
| DLCOd (%) | Mean (Range) | 73.4 (35–101) |
| Molecular features | TP53 mutations | 31/94 (33) |
| KRAS mutations | 20/98 (20.4) | |
| EGFR mutationse | 14/47 (29.8) |
aECOG PS, Eastern Cooperative Oncology Group performance status
bAtypical resection refers to Wedge resection, a non-anatomic limited resection of a lung portion
cCOPD, chronic obstructive pulmonary disease
dDLCO, diffusing capacity of the lung for carbon monoxide
eEGFR mutational status was assessed only in adenocarcinoma patients
HOTTIP expression and clinical characteristics
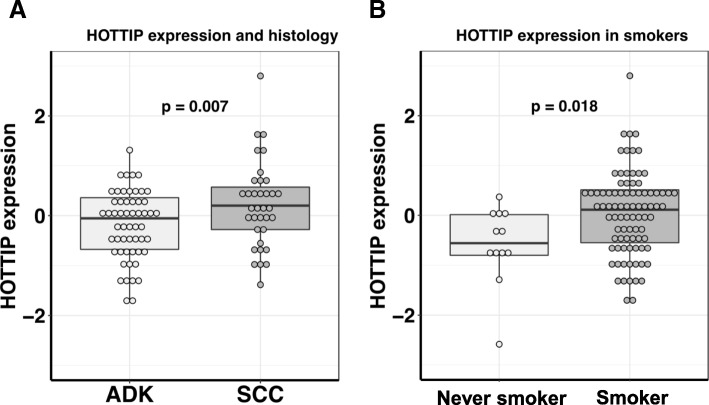
Patients with squamous cell carcinoma had significantly higher levels of HOTTIP than those with adenocarcinoma (p = 0.007; Fig. 1a). HOTTIP was also overexpressed in current and former smokers in comparison with never-smokers (p = 0.02; Fig. 1b) and in former smokers of < 15 years compared to former smokers of > 15 years although this difference was not significant (data not shown). Patients with PS 1 had higher levels of HOTTIP than those with PS 0 (p = 0.08). No association was observed between HOTTIP levels and TP53, KRAS or EGFR mutational status.
Fig. 1.
HOTTIP expression according to (a) histology and (b) smoking status. ADK, adenocarcinoma; SCC, squamous cell carcinoma
HOTTIP expression and clinical outcome
HOTTIP was expressed in all samples. Using the cutoff identified by the Maxstat package of R, patients were included depending on HOTTIP expression level into the high (n = 43) or the low (n = 56) group. The cutoff identified coincided with the Mean + SD of HOTTIP expression in the normal tissue (Additional file 1: Figure S1). Patients with high levels of HOTTIP had shorter TTR (78.3 vs 58 months; p = 0.05) and shorter OS (81.2 vs 61 months; p = 0.02) than those with low levels (Fig. 2).
Fig. 2.
a Time to relapse and (b) overall survival according to HOTTIP levels
The univariate Cox analysis for all clinical variables and for HOTTIP levels are shown in Table 2. In the multivariate analysis for TTR, including sex and HOTTIP levels, only HOTTIP levels emerged as a significant marker (Hazard ratio [HR]: 2.05; 95% confidence interval [CI]: 1–4.22; p = 0.05). The multivariate analysis for OS, including sex, age, PS, smoking status, emphysema, and HOTTIP levels, identified age > 65 (HR: 2.72; 95%CI: 1.04–5.13; p = 0.01) and HOTTIP levels (HR: 2.31, 95%CI: 1.04–5.13; p = 0.04) as independent prognostic markers (Table 3).
Table 2.
Cox univariate analyses of time to relapse and overall survival
| Hazard Ratio (95%CI) | p-value | |
|---|---|---|
| Time to Relapse | ||
| Male sex | 3.039 (0.924–9.999) | 0.067 |
| Age > 65 | 1.207 (0.596–2.443) | 0.602 |
| PS 1 | 1.738 (0.607–4.972) | 0.303 |
| Stage II | 0.888 (0.397–1.989) | 0.773 |
| T3 | 1.213 (0.159–9.275) | 0.853 |
| N1 | 1.142 (0.399–3.272) | 0.805 |
| Histology-Adenocarcinoma | 0.860 (0.402–1.840) | 0.698 |
| Type of surgery-Pneumonectomy | 4.812 (0.494–46.847) | 0.176 |
| Smoker | 1.295 (0.632–2.656) | 0.480 |
| Adjuvant chemotherapy-Yes | 0.890 (0.382–2.072) | 0.787 |
| Emphysema | 1.121 (0.540–2.329) | 0.759 |
| COPD | 1.131 (0.532–2.403) | 0.749 |
| TP53 mutated | 1.841 (0.888–3.814) | 0.101 |
| KRAS mutated | 1.217 (0.544–2.721) | 0.633 |
| EGFR mutated | 2.334 (0.845–6.450) | 0.102 |
| High HOTTIP levels | 2.047 (0.992–4.224) | 0.053 |
| Overall Survival | ||
| Male sex | 3.748 (0.889–15.8) | 0.072 |
| Age > 65 | 2.628 (1.185–5.829) | 0.017 |
| PS 1 | 6.726 (0.914–49.520) | 0.061 |
| Stage II | 0.720 (0.291–1.781) | 0.478 |
| T3 | 1.486 (0.192–11.507) | 0.705 |
| N1 | 0.585 (0.139–2.469) | 0.466 |
| Histology Adenocarcinoma | 0.763 (0.336–1.732) | 0.518 |
| Type of surgery- Pneumonectomy | 1.515 (0.213–10.797) | 0.678 |
| Smoker | 0.387 (0.156–0.959) | 0.040 |
| Adjuvant chemotherapy- Yes | 1.082 (0.459–2.549) | 0.857 |
| Emphysema | 1.948 (0.897–4.230) | 0.092 |
| COPD | 1.115 (0.504–2.467) | 0.788 |
| TP53 mutated | 1.271 (0.568–2.844) | 0.560 |
| KRAS mutated | 1.124 (0.477–2.646) | 0.789 |
| EGFR mutated | 2.551 (0.822–7.916) | 0.105 |
| High HOTTIP levels | 2.442 (1.1–5.418) | 0.028 |
Table 3.
Multivariate analyses of time to relapse and overall survival
| Hazard Ratio (95% CI) | p-value | |
|---|---|---|
| Time to Relapse | ||
| Male sex | 2.71 (0.82–8.99) | 0.10 |
| High HOTTIP expression | 2.05 (1–4.22) | 0.05 |
| Overall Survival | ||
| Male sex | 3.25 (0.76–13.84) | 0.11 |
| Age > 65 | 2.72 (1.23–6.04) | 0.01 |
| ECOG PS 1a | 4.04 (0.54–30.17) | 0.17 |
| Smoker | 0.60 (0.06–5.3) | 0.66 |
| Emphysema | 1.5 (0.68–3.34) | 0.32 |
| High HOTTIP expression | 2.31 (1.04–5.13) | 0.04 |
aECOG PS, Eastern Cooperative Oncology Group performance status
Independent validation of HOTTIP prognostic impact using TCGA data
Using TCGA data and the TANRIC web tool, we found that high levels of HOTTIP were associated with shorter OS (p = 0.025) in a cohort of 91 stage I-II patients with lung adenocarcinoma (Fig. 3a). However, the same analysis in the TCGA cohort of 59 stage I-II patients with squamous cell carcinoma of the lung showed no significant differences (p = 0.69, Fig. 3b).
Fig. 3.
Independent validation of HOTTIP as prognostic marker in (a) TCGA lung adenocarcinoma (LUAD) patients and (b) TCGA squamous cell carcinoma (LUSC) patients
Analysis of HOTTIP correlation with TCGA mRNA and miRNA data
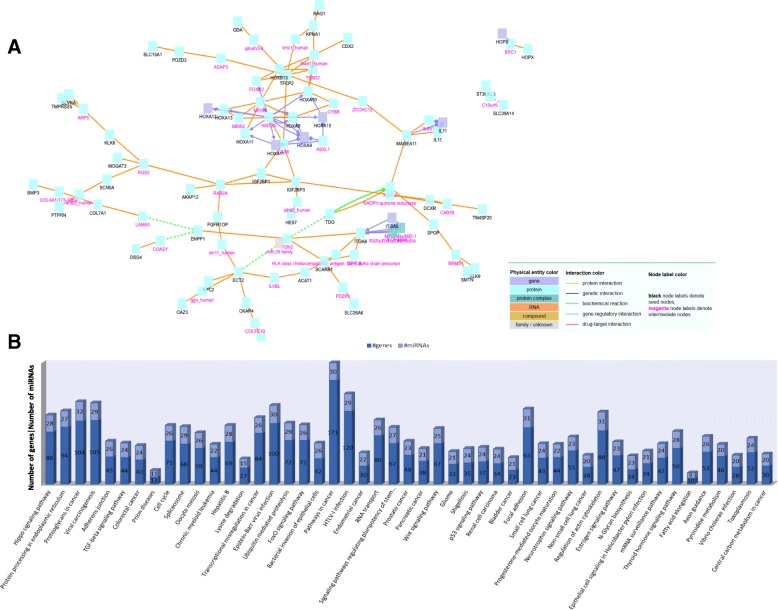
Using TANRIC and TCGA lung adenocarcinoma patients, we identified 1203 mRNAs and 61 miRNAs that significantly correlated with HOTTIP expression (p < 0.05). The most significant mRNA was HOXA13 (r = 0.7, p < 0.001). Moreover, other HOXA genes were among the top 100 mRNAs, including HOXA9 (r = 0.51, p < 0.001), HOXA11 (r = 0.48, p < 0.001), and HOXA10 (r = 0.44, p < 0.001). The Induced Network Module Analysis tool from Consensus PathDB (http://cpdb.molgen.mpg.de/CPDB) showed that most of the genes whose expression correlated with HOTTIP were closely related to the HOX gene network (Fig. 4a). One of the most significant miRNAs identified was miR-196b (r = 0.44, p < 0.001), which is the only miRNA located in the HOXA genomic region. We then used miR-Path v.3 [29] to study the putative pathways regulated by this HOTTIP-miRNA signature and identified 49 KEGG pathways (p < 0.05). The most significant pathway identified was the Hippo signaling pathway (p < 0.001), where 86 genes are Tarbase-validated targets of 28 of the miRNAs included in the signature. Other relevant KEGG pathways identified were the TGF-beta signaling pathway, Cell cycle, Signaling pathways regulating pluripotency of stem cells, Wnt signaling pathway and p53 signaling pathway (Fig. 4b). Additional file 2 includes the list of the top 100 mRNAs and all 61 miRNAs identified.
Fig. 4.
a Network analysis using the top 100 genes whose expression correlates with HOTTIP levels. The Induced Network Module Analysis tool from Consensus PathDB was used. b DIANA-miRPath analysis using the miRNAs whose expression correlates with HOTTIP levels. DIANA-mirPath is a miRNA pathway analysis web-server that examines a list of miRNAs provided by the user, identifies their potential/validated targets, and performs a pathway analysis to identify the most relevant pathways regulated by the miRNAs. In the present study, experimentally validated miRNA interactions derived from DIANA-Tarbase and KEGG analysis were used in the analysis. The KEGG signaling pathways identified are ordered from left to right in descending order of p-value. The y-axis indicates the number of genes (dark blue) and miRNAs (light blue) identified in the miRNA/target interaction analysis which are included in each KEGG pathway identified. For example, the Hippo signaling pathway is the most significant pathway identified and included 28 miRNAs from the HOTTIP-related miRNA signature, which targets 86 genes included in the Hippo signaling pathway
Discussion
HOTTIP is one of the lncRNAs located in the HOXA genomic region of chromosome 7. The HOX genes are crucial transcription factors that determine the identity of cells and tissues during embryogenesis [30]. In adult tissues, HOX genes play a role in normal hematopoiesis regulation and are overexpressed in hematological [31] and solid cancers [10], including NSCLC [14]. Specifically, the HOXA gene cluster, where HOTTIP is found, plays a critical role in the patterning of tissues with mesodermal components, such as the lung, and in the regulation of epithelial–mesenchymal interactions [32]. HOTTIP has been related to tumor metastasis through induction of epithelial-mesenchymal transition [33].
In the present study, we have examined the role of HOTTIP as a prognostic factor in early-stage NSCLC patients treated with curative surgery and found that patients with higher levels of HOTTIP had shorter TTR and shorter OS than patients with low levels. Moreover, HOTTIP emerged as an independent prognostic factor in the multivariate analysis. The prognostic role of HOTTIP levels has been described in several cancers [19, 20, 22–25] and analyzed in several meta-analyses [26, 27, 34–36], which concluded that high HOTTIP expression in cancer patients is associated with poor clinical outcome. However, to the best of our knowledge, ours is the first study to provide evidence that HOTTIP impacts prognosis in NSCLC.
Additionally, we validated our findings on the prognostic value of HOTTIP levels in another patient population using TGCA data and the TANRIC webtool [28], showing that HOTTIP impacts prognosis in NSCLC patients with adenocarcinoma but not in those with squamous cell carcinoma. In our cohort, tumor HOTTIP levels were significantly overexpressed in squamous cell carcinoma compared to adenocarcinoma. While this may suggest that the prognostic impact of HOTTIP could differ between the main histological subtypes, the sub-analysis in the different histological subtypes in our cohort (Additional file 3: Figure S2) did not produce conclusive results, probably due to the small size of the sub-groups (55 adenocarcinomas and 36 squamous cell carcinomas). However, in both cases, high HOTTIP levels were associated with shorter TTR and shorter OS. Previous studies have reported that disordered patterns of HOX gene expression – specifically, HOXA1, HOXA5 and HOXA10 – are involved not only in the development of NSCLC but also in histological diversity [13]. HOTTIP is located close to the 3′ region of HOXA10 and we observed a positive correlation in the expression of the two genes in the in silico analysis of TCGA data.
We also observed a significant upregulation of HOTTIP in current and former smokers in comparison with never smokers. An in vivo study showed that cigarette smoke increases mRNA and protein levels of HOXA in endometrial cells [37]. Interestingly, in our cohort, former smokers of > 15 years showed lower levels of HOTTIP than former smokers of < 15 years although this difference was not significant.
Finally, since regulatory interactions between lncRNAs, mRNAs and miRNAs have been described [38], we explored a possible association between the mRNAs and miRNAs whose expression correlated with HOTTIP according to TGCA data on adenocarcinoma NSCLC analyzed with TANRIC. This analysis identified a signature of 1203 mRNAs and 61 miRNAs. When we focused on the top 100 mRNAs, we observed that most of the genes whose mRNA expression correlated with HOTTIP expression were closely related to the HOX gene network, including HOXA9, HOXA10, HOXA11, HOXA13, and other HOX cluster members, such as HOXB13. Several studies have reported that interactions between HOTTIP and some of these HOX genes promote tumorigenesis. In prostate cancer, HOTTIP forms a complex with the transcription factor TWIST1 and with WDR5 and produces upregulation of HOXA9 levels through chromatin regulation which correlates with an aggressive cellular phenotype [39]. Moreover, HOTTIP modulates cancer stem cell properties by binding WDR5 and activating HOXA9, which enhances the Wnt/β-catenin pathway in prostate cancer stem cells [40]. In pancreatic cancer cells, HOTTIP regulates HOXA10, HOXB2, HOXA11, HOXA9 and HOXA1, but not HOXA13 [41]. However, HOTTIP and HOXA13 have been associated with disease progression and worse outcome in hepatocellular carcinoma [22], with progression and gemcitabine resistance in pancreatic cancer [42], and with tumorogenesis and metastasis in esophageal squamous carcinoma [43] and gastric cancer [44]. In line with our results in NSCLC patient samples, these studies have shown that HOTTIP upregulation is associated with increased levels of HOXA13. In contrast, however, an in vitro study in the A549 NSCLC cell line showed that silencing HOTTIP led to increased HOXA13 levels [21]. Although this study included only one cell line and did not analyze the correlation between HOXA13 and HOTTIP in patient samples, it showed that HOTTIP acts as an oncogene, regulating apoptosis, proliferation, and migration in NSCLC. Another study, by Zhang et al., also reported the role of HOTTIP as oncogene in A549 through regulation of the AKT signaling pathway. The authors showed that the overexpression of HOTTIP enhanced proliferation and paclitaxel resistance [45].
Further studies are needed to clarify this interaction, including analyses in other NSCLC cell lines. Interestingly, the network analysis also identified an additional node that included HOXP, a transcription factor related to alveolar differentiation, whose suppression has been linked to increased invasiveness in adenocarcinoma lung cancer [46]. There was a negative correlation between HOXP and HOTTIP levels, which could explain the more aggressive phenotype we have observed in patients with high HOTTIP levels.
When we explored the miRNAs in the 61-miRNA signature identified in the TANRIC analysis, we found a positive correlation with miR-196b, located in the distal part of the same HOXA cluster, and with miR-196a-1, located in the HOXB cluster. The study of the potential pathways regulated by the miRNA signature showed the importance of the Hippo signaling pathway, which has previously been shown to be altered in NSCLC [47].
Conclusions
Our findings provide the first indication that HOTTIP may be a prognostic biomarker in NSCLC. In line with the prognostic impact of HOTTIP levels in other cancers, high levels of HOTTIP correlated with worse TTR and worse OS in our early-stage NSCLC patients. HOTTIP may be useful for the identification of resected NSCLC patients at high risk of relapse. Further investigation in a prospective study is warranted to validate these findings and to examine potential HOTTIP-based therapeutic approaches.
Additional files
Figure S1. (A) Time to relapse and (B) overall survival according to HOTTIP levels in adenocarcinoma patients. (C) Time to relapse and (D) overall survival according to HOTTIP levels in squamous cell carcinoma patients. (PPTX 57 kb)
Excel file with the results of the analysis of HOTTIP correlation with TCGA data that includes 2 sheets: mRNAs sheet, which includes the list of the top 100 mRNAs; and miRNAs sheet with all 61 miRNAs identified. (XLSX 19 kb)
Figure S2. Bar plot showing the 99 patients ordered by HOTTIP expression level. An arrow shows the mean HOTTIP expression in the normal tissue and the cutoff used to classify the patients in high or low expression. The cutoff coincides with the Mean + SD of HOTTIP expression in the normal tissue. (PPTX 104 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by grants from AECC-Catalunya 2017 (AN), SAF2017–88606-P (AN) from the Ministry of Economy and Competition (MINECO) co-financed with the European Union FEDER funds and SDCSD from the Universitat de Barcelona (MM). SS, JC and SM are APIF fellows of the Universitat de Barcelona. None of the funding bodies had any role in the design of the study and collection, analysis, and interpretation of data, or in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbrebiations
- ECOG
Eastern Cooperative Oncology Group
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- ncRNAs
non-coding RNAs
- NSCLC
Non-small-cell lung cancer
- OR
Odds ratio
- OS
Overall survival
- PS
Performance status
- TCGA
The Cancer Genome Atlas
- TTR
Time to relapse
Authors’ contributions
AN designed the study, analyzed the data and wrote the manuscript draft; JM, SS, JJC, JC, CM have been involved in collection and assembly of data and interpretation of the results; JM, RMM, NV, JR, LM, MM collected clinical data and reviewed critically the manuscript. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
Approval for the study was obtained from the Clinical Research Ethics Committee of the Hospital Clínic de Barcelona, and written informed consent was obtained from each participant in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alfons Navarro, Phone: +34-93-402-1903, Email: anavarroponz@ub.edu.
Jorge Moises, Email: jrmoises@clinic.cat.
Sandra Santasusagna, Email: sandra.santasusagna@ub.edu.
Ramon M. Marrades, Email: marrades@clinic.cat
Nuria Viñolas, Email: nvinolas@clinic.cat.
Joan J. Castellano, Email: joan.castellano@ub.edu
Jordi Canals, Email: canals.serrat@ub.edu.
Carmen Muñoz, Email: carmen.munoz@ub.edu.
José Ramírez, Email: jramirez@clinic.cat.
Laureano Molins, Email: lmolins@clinic.cat.
Mariano Monzo, Email: mmonzo@ub.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Cortés ÁA, Urquizu LC, Cubero JH. Adjuvant chemotherapy in non-small cell lung cancer: state-of-the-art. Translational Lung Cancer Res. 2015;4(2):191. doi: 10.3978/j.issn.2218-6751.2014.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lou F, Huang J, Sima CS, Dycoco J, Rusch V, Bach PB. Patterns of recurrence and second primary lung cancer in early-stage lung cancer survivors followed with routine computed tomography surveillance. J Thorac Cardiovasc Surg. 2013;145(1):75–82. doi: 10.1016/j.jtcvs.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 5.Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan Y, Zang H, Feng J, Lu J, Chen L, Fan S. Long non-coding RNAs associated with non-small cell lung cancer. Oncotarget. 2017;8(40):69174. doi: 10.18632/oncotarget.20088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellano JJ, Navarro A, Viñolas N, Marrades RM, Moises J, Cordeiro A, Saco A, Muñoz C, Fuster D, Molins L. LincRNA-p21 impacts prognosis in resected non–small cell lung Cancer patients through angiogenesis regulation. J Thorac Oncol. 2016;11(12):2173–2182. doi: 10.1016/j.jtho.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 9.Díaz-Beyá M, Brunet S, Nomdedéu J, Pratcorona M, Cordeiro A, Gallardo D, Escoda L, Tormo M, Heras I, Ribera JM. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015;6(31):31613. doi: 10.18632/oncotarget.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med. 2014;92(8):811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 11.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 12.Calvo R, West J, Franklin W, Erickson P, Bemis L, Li E, Helfrich B, Bunn P, Roche J, Brambilla E. Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci. 2000;97(23):12776–12781. doi: 10.1073/pnas.97.23.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe M, Hamada J-i, Takahashi O, Takahashi Y, Tada M, Miyamoto M, Morikawa T, Kondo S, Moriuchi T. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. 2006;15(4):797–802. [PubMed] [Google Scholar]
- 14.Plowright L, Harrington K, Pandha H, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) Br J Cancer. 2009;100(3):470. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21(8):421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Dang Y, Liu J, Ouyang X. The function of homeobox genes and lncRNAs in cancer. Oncol Lett. 2016;12(3):1635–1641. doi: 10.3892/ol.2016.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Zhao L, Wang Y-X, Xi M, Liu S-L, Luo L-L. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumor Biol. 2015;36(11):8805–8809. doi: 10.1007/s13277-015-3645-2. [DOI] [PubMed] [Google Scholar]
- 19.Ye H, Liu K, Qian K. Overexpression of long noncoding RNA HOTTIP promotes tumor invasion and predicts poor prognosis in gastric cancer. OncoTargets Ther. 2016;9:2081. doi: 10.2147/OTT.S95414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Y-K, Xiao Y, Wan X-B, Zhao Y-Z, Li J, Li Y, Han G-S, Chen X-B, Zou Q-Y, Wang G-C. Association of long non-coding RNA HOTTIP with progression and prognosis in colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11458. [PMC free article] [PubMed] [Google Scholar]
- 21.Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao Z, Li Q, Zhang W. Up-regulation of long non-coding HOTTIP functions as an oncogene by regulating HOXA13 in non-small cell lung cancer. Am J Transl Res. 2016;8(5):2022. [PMC free article] [PubMed] [Google Scholar]
- 22.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Cao L, Hang D, Wang F, Wang Q. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8(9):11414. [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Qian J, Xiang Y, Chen Y, Qu J. The prognostic value of long noncoding RNA HOTTIP on clinical outcomes in breast cancer. Oncotarget. 2017;8(4):6833. doi: 10.18632/oncotarget.14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Zhou Y, Bai Y, Wang Q, Bao J, Luo Y, Guo Y, Guo L. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer. 2017;16(1):162. doi: 10.1186/s12943-017-0729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin N, Yang LY, Xu ZP. Long non-coding RNA HOTTIP is able to predict poor prognosis in various neoplasms: a meta-analysis. Mol Clin Oncol. 2017;7(2):263–266. doi: 10.3892/mco.2017.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Y, Yan T, Chai Y, Jiang Y, Zhu X. Long noncoding RNA HOTTIP as an independent prognostic marker in cancer. Clin Chim Acta. 2017;482:224–30. [DOI] [PubMed]
- 28.Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3. 0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iimura T, Pourquié O. Hox genes in time and space during vertebrate body formation. Develop Growth Differ. 2007;49(4):265–275. doi: 10.1111/j.1440-169X.2007.00928.x. [DOI] [PubMed] [Google Scholar]
- 31.Eklund EA. The role of HOX genes in malignant myeloid disease. Curr Opin Hematol. 2007;14(2):85–89. doi: 10.1097/MOH.0b013e32801684b6. [DOI] [PubMed] [Google Scholar]
- 32.Di-Poï N, Koch U, Radtke F, Duboule D. Additive and global functions of HoxA cluster genes in mesoderm derivatives. Dev Biol. 2010;341(2):488–498. doi: 10.1016/j.ydbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Han H, Li Y, Zhang Q, Mo K, Chen S. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget. 2016;7(51):84480. doi: 10.18632/oncotarget.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Li N, Kang X, Shi K, Chen Q. Prognostic value of the long noncoding RNA HOTTIP in human cancers. Oncotarget. 2017;8(35):59563. doi: 10.18632/oncotarget.19166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L, Li M, Ding Y, Pu L, Liu J, Xiong S. Long non-coding RNA HOTTIP, a novel potential prognostic marker in cancers. 2017. [DOI] [PubMed] [Google Scholar]
- 36.Liu F-T, Xue Q-Z, Zhang Y, Hao T-F, Luo H-L, Zhu P-Q. Long non-coding RNA HOXA transcript at the distal tip as a putative biomarker of metastasis and prognosis: a meta-analysis. Clin Lab. 2016;62(11):2091–2098. doi: 10.7754/Clin.Lab.2016.160224. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Jorgensen EM, Gan Y, Taylor HS. Cigarette smoke increases progesterone receptor and homeobox A10 expression in human endometrium and endometrial cells: a potential role in the decreased prevalence of endometrial pathology in smokers. Biol Reprod. 2011;84(6):1242–1247. doi: 10.1095/biolreprod.110.087494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA–lncRNA interactions. Methods Mol Biol. 2016;1402:271–86. 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed]
- 39.Malek R, Gajula RP, Williams RD, Nghiem B, Simons BW, Nugent K, Wang H, Taparra K, Lemtiri-Chlieh G, Yoon AR. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 2017;77(12):3181–3193. doi: 10.1158/0008-5472.CAN-16-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, Li W, Zheng S, Ye H, Wang L. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 41.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6(13):10840. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13(1):84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin C, Wang Y, Zhang S, Yu L, Guo C, Xu H. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene. 2017;36(38):5392. doi: 10.1038/onc.2017.133. [DOI] [PubMed] [Google Scholar]
- 44.Chang S, Liu J, Guo S, He S, Qiu G, Lu J, Wang J, Fan L, Zhao W, Che X. HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol Rep. 2016;35(6):3577–3585. doi: 10.3892/or.2016.4743. [DOI] [PubMed] [Google Scholar]
- 45.Zhang G, Song W, Song Y. Overexpression of HOTTIP promotes proliferation and drug resistance of lung adenocarcinoma by regulating AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:5683–5690. doi: 10.26355/eurrev_201712_14013. [DOI] [PubMed] [Google Scholar]
- 46.Cheung WK, Zhao M, Liu Z, Stevens LE, Cao PD, Fang JE, Westbrook TF, Nguyen DX. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23(6):725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhanasekaran SM, Balbin OA, Chen G, Nadal E, Kalyana-Sundaram S, Pan J, Veeneman B, Cao X, Malik R, Vats P. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and hippo pathway genes. Nat Commun. 2014;5:5893. doi: 10.1038/ncomms6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Time to relapse and (B) overall survival according to HOTTIP levels in adenocarcinoma patients. (C) Time to relapse and (D) overall survival according to HOTTIP levels in squamous cell carcinoma patients. (PPTX 57 kb)
Excel file with the results of the analysis of HOTTIP correlation with TCGA data that includes 2 sheets: mRNAs sheet, which includes the list of the top 100 mRNAs; and miRNAs sheet with all 61 miRNAs identified. (XLSX 19 kb)
Figure S2. Bar plot showing the 99 patients ordered by HOTTIP expression level. An arrow shows the mean HOTTIP expression in the normal tissue and the cutoff used to classify the patients in high or low expression. The cutoff coincides with the Mean + SD of HOTTIP expression in the normal tissue. (PPTX 104 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.