Abstract
Background
Colorectal cancer (CRC) is one of the most common cancers worldwide, and genetic variations exert distinct roles in its pathogenesis. Single nucleotide polymorphisms (SNPs) in interleukin 1 alpha (IL1A) were reported to be correlated to the susceptibility of diverse cancers. The aim of this study was to assess the association of IL1A SNPs with the risk of colorectal cancer in a Chinese Han population.
Methods
To evaluate the correlation between IL1A polymorphisms and CRC risk, Agena MassARRAY platform was used for genotype determination among 248 CRC patients and 463 controls. The relationships between IL1A variants and CRC susceptibility were examined by logistic regression analysis. Stratified analysis was conducted for the association detection in males and females. Haplotype construction and analysis were applied to evaluate the potential relationship between the genetic block and the risk of CRC. SNP functional exploration was performed with available bioinformatics datasets.
Results
After adjusting for age and gender, the “AA” genotype of rs2856838 exhibited a risk association with colorectal cancer in the recessive model (adjusted OR = 1.98, 95% CI: 1.05–3.72, p = 0.036). With stratified analysis, the recessive models of rs3783550 (OR = 2.17, 95% CI: 1.03–4.60, p = 0.043), rs2856838 (OR = 2.58, 95% CI: 1.13–5.87, p = 0.024), rs1609682 (OR = 2.20, 95% CI: 1.04–4.65, p = 0.040), and rs3783521 (OR = 2.13, 95% CI: 1.01–4.49, p = 0.048) revealed significant relationships between these variants and an increased CRC risk only in females. Bioinformatics analysis also revealed the putative functions of the selected SNPs.
Conclusions
This study demonstrated that rs2856838 could influence the susceptibility to CRC in Chinese Han population from northwest China. IL1A variants rs3783550, rs2856838, rs1609682, and rs3783521 were associated with CRC risk only in females.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-5395-9) contains supplementary material, which is available to authorized users.
Keywords: Single-nucleotide polymorphisms, IL1A, Stratification analysis, Colorectal cancer, Chinese Han population
Background
Colorectal cancer (CRC) is one of the most common cancers worldwide, and it is the third leading cause of cancer-related deaths in both genders. In the past two decades, an increasing incidence of CRC was found in China as lifestyles and eating habits have become progressively changed [16, 33]. The etiology of CRC are multifactorial, and epidemiological evidence suggests that smoking cigarettes, unhealthy eating habits and lack of exercises are three well-known causal factors for CRC development [5, 25]. It has been confirmed that long-term exposure to these adverse habits may eventually increase the individual CRC risk, however, genetic aberrations, such as single-nucleotide polymorphisms (SNPs), play more important roles in the pathogenesis of CRC [2, 8, 23, 27, 28].
Accumulating evidence has demonstrated that most cancers develop from long-time chronic inflammation, including CRC, which could be attributed to two common inflammatory bowel diseases, namely, Crohn’s disease and ulcerative colitis [7, 18]. Inflammatory and tumour cells could produce cytokines and chemokines that facilitate tumour promotion and progression [6]. Interleukin 1 (IL-1) is a family of pleiotropic cytokine with multiple functions in human innate inflammatory and immune responses [3]. It has also been reported that IL-1 proteins are involved in tumour angiogenesis, invasion, proliferation, and metastasis [22]. Together with IL-1β, IL-1α, encoded by IL1A (interleukin 1 alpha), is defined as an “alarm cytokine” that belongs to IL-1 cluster and plays dual roles in malignant tumour progression [22]. Membrane-associated IL-1α could elevate immunogenicity of the tumour cell and exert positive effect on anti-tumour immune surveillance and tumour regression. However, secretable form of IL-1α in the microenvironment of tumour cells has been proved to facilitate tumour invasiveness and angiogenesis [21].
Furthermore, it is widely recognized that the polymorphisms of IL-1A is associated with diverse diseases. Gao et al. has reported an insertion/deletion (ins/del) polymorphism (rs3783553, TTCA) in IL1A that may contribute to hepatocellular cancer susceptibility, and revealed the regulatory function of the variation on IL1α expression by disrupting the binding sites for miR-122 and miR-378 [12]. This functional polymorphism has also been demonstrated in gastric cancer and cervical carcinoma [13, 20, 31]. Additionally, the relationships between IL1A polymorphisms and the risk of papillary thyroid cancer, nasopharyngeal cancer and epithelial ovarian cancer have been reported as well [11, 29, 32].
A recent work has elucidated a decreased tumour expression of IL1A in colorectal adenocarcinoma, which indicated the potential role of IL1A in the etiology of CRC. However, few studies have examined the associations of IL1A polymorphisms with the risk of CRC. In our study, we investigated the effects of five IL1A variants (rs3783550, rs3783546, rs2856838, rs1609682, and rs3783521) on the susceptibility to CRC, which is supposed to provide more evidence for IL1A in CRC pathogenesis and contribute to early CRC risk estimation among the individuals of Chinese Han ancestry.
Methods
Study subjects
The current research involved a total of 248 CRC patients (143 males and 105 females) and 463 controls (265 males and 198 females) with unrelated Chinese Han ancestries. All CRC cases were diagnosed and confirmed by two independent pathological examinations. As for the eligible CRC cases selection, the individuals without other types of cancer, familial adenomatous polyposis, hereditary nonpolyposis colorectal cancer or undergone previous radiotherapy and chemotherapy were included. With regard to healthy controls, the subjects who had suffered from chronic or severe endocrine and metabolic diseases, digestive system disorders and malignant diseases were excluded from this study. The controls were polyp free individuals at recruitment. Moreover, the individuals with family colorectal disease and cancer history were excluded from control group as well.
SNP genotyping
By reading the previous publications of IL1A polymorphisms, we selected SNPs which could impact cancer risk, and searched the genetic data of them in dbSNP database (https://www.ncbi.nlm.nih.gov/snp/) and 1000 Genomes database (http://www.internationalgenome.org/). Only the SNPs whose minor allele frequency (MAF) beyond 5% in Asian populations were included in this study in order to achieve adequate statistical power. Finally, five candidate variations rs3783550, rs3783546, rs2856838, rs1609682, and rs3783521 in IL1A gene were selected for genotyping. Five millilitres venous blood was collected from the subjects when they were recruited in this study. Genomic DNA was extracted from the blood with the Whole Blood Genomic DNA Purification Kit (GoldMag, Xi’an, China). The PCR primers used in multiplexed PCR assay were designed by Agena Bioscience Assay Design Suite V2.0 software and are showed in Additional file 1: Table S1[10]. In order to improve the PCR efficiency and ensure that the amplification primers, extension primers and extension products could be differed by their molecular weight, a tag 10-base sequence (5′-ACGTTGGATG-3′) was added to the 5′ end of each amplification primer. SNP genotyping was carried out with the usage of the Agena Nanodispenser (Agena Bioscience, San Diego, USA) and MassARRAY iPLEX platform (Agena Bioscience, San Diego, USA) [10]. The procedure for SNP genotyping was described as follows. First, a first round PCR was performed to increase and concentrate the target genomic fragments containing the polymorphisms. Second, using the products obtained from the last step as the templates, only one ‘mass-modified’ nucleotide was added to the polymorphic locus at the end of the extension primer fragment in extension reaction. Third, the analyte mixture was dropped to a SpectroCHIP Array by Agena Bioscience Nanodispenser and further processed by the matrix-assisted laser desorption/ionization—time of flight (MALDI-TOF) mass spectrometry. With this method, the analytes could be differed according to their flight time in the system and the nucleotide at the each SNP locus was identified. Finally, the genotyping results were managed and presented in the Agena Bioscience TYPER version 4.0 software [10, 24].
Statistical analyses
SPSS 18.0 software (SPSS, Chicago, IL, USA) was employed for primary statistical analysis. The heterozygosity for the five SNPs in control cohort was assessed by the Fisher’s exact test to detect their compliance with Hardy-Weinberg equilibrium (HWE). Allele and genotype frequencies of each SNP in CRC cases and healthy controls were calculated and evaluated for the difference with Pearson’s χ2 test. Logistic regression analysis with adjustment for age and gender was applied to the odds ratio (OR) and 95% confidence interval (CI) assessment [4]. Adjustment process is usually implemented in the statistical analysis to eliminate the influence of confounding factors, such as age and gender. In all statistical analysis, p ≤ 0.05 was regarded as statistical difference. The associations of the five candidate variants with the CRC risk were examined in codominant, dominant and recessive genetic models using SNPstats online software (https://www.snpstats.net/start.htm). Additionally, we used the Haploview software package, version 4.2 (http://analysis.bio-x.cn/myAnalysis.php) for linkage disequilibrium (LD) construction and haplotype evaluation [30]. Using the genotyping results, we first constructed the LD pattern among genetic variants. The status of LD was analyzed using two parameters: r2 and D’. And D’ = 1 is defined as complete linkage disequilibrium. The allele combination of the variants in one LD block was considered as haplotype. We selected the haplotypes with frequency greater than 5% to evaluate whether the haplotypes were associated with CRC risk. These associations were detected using SHEsis software with available OR (95% CI) and p value.
Bioinformatics prediction and expression quantitative trait loci (eQTL) analysis
HaploReg v4.1 database (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was applied for SNP functional annotation using bioinformatics methods. Expression quantitative trait loci (eQTL) analysis were carried out with the data available on the Genotype-Tissue Expression (GTEx) project (https://gtexportal.org/home/). The impact of each candidate SNP on IL1A expression was determined in several human tissues and p-values were provided for statistical significance assessment.
Results
Population characteristics
The demographic information of the enrolled CRC patients and healthy controls are listed in Additional file 1: Table S2. After applicable statistical evaluation, there was no significant difference in gender observed between patients and controls. The mean age [± standard deviation (SD)] of the patient group was 58.69 ± 12.77 years at diagnosis and of the controls was 50.65 ± 11.79 years at recruitment.
SNPs and the risk of CRC
Basic information and allele frequencies of the IL1A polymorphisms are presented in Table 1. None of the five SNPs was excluded because of the deviation from HWE at the 5% p level in the control population. The minor allele of each SNP was assumed a risk factor compared to the wild-type allele. Results of the genetic model analysis are showed in Table 2. The frequency of the homozygous “A/A” genotype of the IL1A SNP rs2856838 differed significantly between patients and controls (8.9% vs. 5.2%, p < 0.05). An association between allele “A/A” of rs2856838 and an increased risk of CRC was observed under the recessive model (adjusted OR = 1.98, 95% CI: 1.05–3.72, p = 0.036) after adjustments for age and gender.
Table 1.
Basic information and allele frequencies of the five selected SNPs
| SNP_ID | Gene | Chromosome | Position | Allele | Minor allele frequency | HWE p value |
OR (95% CI) | p a | |
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||
| rs3783550 | IL1A | 2q13 | 113,532,885 | T/G | 0.358 | 0.326 | 0.399 | 1.15 (0.91–1.45) | 0.222 |
| rs3783546 | IL1A | 2q13 | 113,534,830 | C/G | 0.353 | 0.325 | 0.458 | 1.13 (0.90–1.42) | 0.296 |
| rs2856838 | IL1A | 2q13 | 113,539,972 | A/G | 0.256 | 0.247 | 0.319 | 1.04 (0.81–1.35) | 0.716 |
| rs1609682 | IL1A | 2q13 | 113,540,205 | T/G | 0.354 | 0.327 | 0.398 | 1.12 (0.90–1.42) | 0.298 |
| rs3783521 | IL1A | 2q13 | 113,543,577 | G/A | 0.356 | 0.326 | 0.399 | 1.14 (0.91–1.44) | 0.252 |
SNP Single nucleotide polymorphism, HWE Hardy-Weinberg equilibrium, OR Odds ratio, 95% CI, 95% confidence interval
HWE p-value obtained from Fisher’s exact test (p > 0.05)
pa values were calculated from Pearson’s χ2 test regarding to the allele distribution frequencies among CRC patients and healthy controls
p < 0.05 indicates SNP with statistical significance
Table 2.
Genetic model analyses of IL1A SNPs and the risk of CRC
| SNP_ID | Genotypes | Controls, n (%) | Patients, n (%) | Without adjustment | With adjustment | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p a | OR (95% CI) | p b | ||||
| rs3783550 | |||||||
| G/G | 206 (44.5%) | 103 (41.7%) | 1.00 | 1.00 | |||
| Codominant | G/T | 212 (45.8%) | 111 (44.9%) | 1.05 (0.75–1.46) | 0.330 | 1.01 (0.72–1.44) | 0.310 |
| T/T | 45 (9.7%) | 33 (13.4%) | 1.47 (0.88–2.44) | 1.49 (0.88–2.52) | |||
| Dominant (ref: G/G) | G/T-T/T | 257 (55.5%) | 144 (58.3%) | 1.12 (0.82–1.53) | 0.470 | 1.10 (0.79–1.52) | 0.580 |
| Recessive (ref: G/G + G/T) | T/T | 45 (9.7%) | 33 (13.4%) | 1.43 (0.89–2.31) | 0.140 | 1.48 (0.90–2.43) | 0.130 |
| rs3783546 | |||||||
| G/G | 206 (44.7%) | 104 (41.9%) | 1.00 | 1.00 | |||
| Codominant | G/C | 210 (45.5%) | 113 (45.6%) | 1.07 (0.77–1.48) | 0.500 | 1.02 (0.73–1.45) | 0.500 |
| C/C | 45 (9.8%) | 31 (12.5%) | 1.36 (0.82–2.28) | 1.37 (0.80–2.34) | |||
| Dominant (ref: G/G) | G/C-C/C | 255 (55.3%) | 144 (58.1%) | 1.12 (0.82–1.53) | 0.480 | 1.09 (0.78–1.51) | 0.620 |
| Recessive (ref: G/G + G/C) | C/C | 45 (9.8%) | 31 (12.5%) | 1.32 (0.81–2.15) | 0.270 | 1.35 (0.82–2.24) | 0.240 |
| rs2856838 | |||||||
| G/G | 257 (55.8%) | 143 (57.7%) | 1.00 | 1.00 | |||
| Codominant | G/A | 180 (39%) | 83 (33.5%) | 0.83 (0.60–1.15) | 0.096 | 0.83 (0.58–1.17) | 0.063 |
| A/A | 24 (5.2%) | 22 (8.9%) | 1.65 (0.89–3.04) | 1.84 (0.96–3.50) | |||
| Dominant (ref:G/G) | G/A-A/A | 204 (44.2%) | 105 (42.3%) | 0.93 (0.68–1.26) | 0.620 | 0.94 (0.68–1.31) | 0.720 |
| Recessive (ref: G/G + G/A) | A/A | 24 (5.2%) | 22 (8.9%) | 1.77 (0.97–3.23) | 0.064 | 1.98 (1.05–3.72) | 0.036 |
| rs1609682 | |||||||
| G/G | 205 (44.4%) | 105 (42.5%) | 1.00 | 1.00 | |||
| Codominant | G/T | 212 (45.9%) | 109 (44.1%) | 1.00 (0.72–1.40) | 0.350 | 0.97 (0.68–1.37) | 0.310 |
| T/T | 45 (9.7%) | 33 (13.4%) | 1.43 (0.86–2.38) | 1.45 (0.85–2.45) | |||
| Dominant (ref: G/G) | G/T-T/T | 257 (55.6%) | 142 (57.5%) | 1.08 (0.79–1.47) | 0.630 | 1.05 (0.76–1.46) | 0.770 |
| Recessive (ref: G/G + G/T) | T/T | 45 (9.7%) | 33 (13.4%) | 1.43 (0.89–2.31) | 0.150 | 1.47 (0.89–2.42) | 0.130 |
| rs3783521 | |||||||
| A/A | 206 (44.5%) | 103 (41.7%) | 1.00 | 1.00 | |||
| Codominant | G/A | 212 (45.8%) | 112 (45.3%) | 1.06 (0.76–1.47) | 0.400 | 1.02 (0.72–1.44) | 0.410 |
| G/G | 45 (9.7%) | 32 (13.0%) | 1.42 (0.85–2.37) | 1.42 (0.84–2.42) | |||
| Dominant (ref: A/A) | G/A-G/G | 257 (55.5%) | 144 (58.3%) | 1.12 (0.82–1.53) | 0.470 | 1.09 (0.79–1.51) | 0.610 |
| Recessive (ref: A/A + G/A) | G/G | 45 (9.7%) | 32 (13.0%) | 1.38 (0.85–2.24) | 0.190 | 1.41 (0.86–2.33) | 0.180 |
SNP Single nucleotide polymorphism, OR Odds ratio; 95% CI: 95% confidence interval
pa-values were calculated by logistic regression analysis with the comparison between CRC patients and healthy controls
pb-values were calculated by logistic regression analysis with adjustments for age and gender
Bold italics indicates the SNP with statistical significance (p < 0.05)
In accordance with the stratified analysis by gender, we found that the IL1A variants rs3783550, rs2856838, rs1609682 and rs3783521 did not show any significant association with CRC risk in males, whereas the statistical significance was detected in females (Table 3). The frequency of homozygous “T/T” genotype of IL1A SNPs rs3783550 and rs1609682 differed significantly between patients and controls in females (rs3783550: 15.2% vs. 8.1%; rs1609682: 15.4% vs. 8.1%, p < 0.05), respectively. Moreover, rs3783550 and rs1609682 were associated with an increased risk of CRC based on the results of the recessive model (rs3783550: OR = 2.17, 95% CI: 1.03–4.60, p = 0.043; rs1609682: OR = 2.20, 95% CI: 1.04–4.65, p = 0.040). The frequency of homozygous “A/A” genotype of rs2856838 differed significantly between patients and controls in females (13.3% vs. 6.1%, p < 0.05) as well. The recessive genetic model also provided the evidence for the role of rs2856838 in CRC susceptibility among Chinese Han women (OR = 2.58, 95% CI: 1.13–5.87, p = 0.024). Additionally, rs3783521 homozygous genotype “G/G” distributed differently between the case and control group (15.2% vs. 8.1%, p < 0.05). Further genetic model analysis revealed that SNP rs3783521 was linked to an increased risk of CRC in females according to the recessive model (OR = 2.13, 95% CI: 1.01–4.49, p = 0.048). However, no statistically significant association was detected between SNP rs3783546 and CRC risk in Chinese Han women.
Table 3.
Association of the selected SNPs in IL1A with CRC risk after stratified analysis by gender
| SNP_ID | Genotypes | Male | OR (95% CI) | p a | Female | OR (95% CI) | p b | ||
|---|---|---|---|---|---|---|---|---|---|
| Controls, n(%) | Patients, n(%) | Controls, n(%) | Patients, n(%) | ||||||
| rs3783550 | |||||||||
| G/G | 113 (42.6%) | 62 (43.7%) | 1.00 | 93 (47.0%) | 41 (39.0%) | 1.00 | |||
| Codominant | G/T | 123 (46.4%) | 63 (44.4%) | 0.85 (0.53–1.38) | 0.800 | 89 (45.0%) | 48 (45.7%) | 1.22 (0.73–2.04) | 0.096 |
| T/T | 29 (10.9%) | 17 (12.0%) | 0.96 (0.47–1.98) | 16 (8.1%) | 16 (15.2%) | 2.41 (1.09–5.34) | |||
| Dominant (ref: G/G) | G/T-T/T | 152 (57.4%) | 80 (56.3%) | 0.88 (0.56–1.38) | 0.560 | 105 (53.0%) | 64 (61.0%) | 1.40 (0.86–2.27) | 0.180 |
| Recessive (ref: G/G + G/T) | T/T | 29 (10.9%) | 17 (12.0%) | 1.05 (0.53–2.06) | 0.900 | 16 (8.1%) | 16 (15.2%) | 2.17 (1.03–4.60) | 0.043 |
| rs3783546 | |||||||||
| G/G | 113 (42.8%) | 62 (43.4%) | 1.00 | 93 (47.2%) | 42 (40.0%) | 1.00 | |||
| Codominant | G/C | 122 (46.2%) | 65 (45.5%) | 0.87 (0.54–1.40) | 0.850 | 88 (44.7%) | 48 (45.7%) | 1.21 (0.72–2.01) | 0.170 |
| C/C | 29 (11.0%) | 16 (11.2%) | 0.92 (0.44–1.90) | 16 (8.1%) | 15 (14.3%) | 2.18 (0.97–4.85) | |||
| Dominant (ref: G/G) | G/C-C/C | 151 (57.2%) | 81 (56.6%) | 0.88 (0.56–1.38) | 0.580 | 104 (52.8%) | 63 (60.0%) | 1.35 (0.83–2.20) | 0.220 |
| Recessive (ref: G/G + G/C) | C/C | 29 (11.0%) | 16 (11.2%) | 0.98 (0.49–1.95) | 0.960 | 16 (8.1%) | 15 (14.3%) | 1.98 (0.93–4.21) | 0.080 |
| rs2856838 | |||||||||
| G/G | 142 (54.0%) | 88 (61.5%) | 1.00 | 115 (58.1%) | 55 (52.4%) | 1.00 | |||
| Codominant | G/A | 109 (41.4%) | 47 (32.9%) | 0.68 (0.42–1.10) | 0.260 | 71 (35.9%) | 36 (34.3%) | 1.06 (0.63–1.77) | 0.078 |
| A/A | 12 (4.6%) | 8 (5.6%) | 1.07 (0.39–2.94) | 12 (6.1%) | 14 (13.3%) | 2.63 (1.13–6.15) | |||
| Dominant (ref: G/G) | G/A-A/A | 121 (46.0%) | 55 (38.5%) | 0.72 (0.46–1.14) | 0.160 | 83 (41.9%) | 50 (47.6%) | 1.27 (0.79–2.06) | 0.330 |
| Recessive (ref: G/G + G/A) | A/A | 12 (4.6%) | 8 (5.6%) | 1.24 (0.46–3.36) | 0.670 | 12 (6.1%) | 14 (13.3%) | 2.58 (1.13–5.87) | 0.024 |
| rs1609682 | |||||||||
| G/G | 113 (42.6%) | 64 (44.8%) | 1.00 | 92 (46.7%) | 41 (39.4%) | 1.00 | |||
| Codominant | G/T | 123 (46.4%) | 62 (43.4%) | 0.81 (0.50–1.30) | 0.670 | 89 (45.2%) | 47 (45.2%) | 1.18 (0.70–1.98) | 0.100 |
| T/T | 29 (10.9%) | 17 (11.9%) | 0.93 (0.45–1.90) | 16 (8.1%) | 16 (15.4%) | 2.39 (1.08–5.31) | |||
| Dominant (ref: G/G) | G/T-T/T | 152 (57.4%) | 79 (55.2%) | 0.83 (0.53–1.31) | 0.420 | 105 (53.3%) | 63 (60.6%) | 1.36 (0.83–2.21) | 0.220 |
| Recessive (ref: G/G + G/T) | T/T | 29 (10.9%) | 17 (11.9%) | 1.03 (0.53–2.04) | 0.920 | 16 (8.1%) | 16 (15.4%) | 2.20 (1.04–4.65) | 0.040 |
| rs3783521 | |||||||||
| A/A | 113 (42.6%) | 62 (43.7%) | 1.00 | 93 (47.0%) | 41 (39.0%) | 1.00 | |||
| Codominant | G/A | 123 (46.4%) | 64 (45.1%) | 0.86 (0.53–1.38) | 0.820 | 89 (45.0%) | 48 (45.7%) | 1.22 (0.73–2.04) | 0.110 |
| G/G | 29 (10.9%) | 16 (11.3%) | 0.92 (0.44–1.90) | 16 (8.1%) | 16 (15.2%) | 2.36 (1.07–5.22) | |||
| Dominant (ref: A/A) | G/A-G/G | 152 (57.4%) | 80 (56.3%) | 0.87 (0.55–1.37) | 0.540 | 105 (53.0%) | 64 (61.0%) | 1.39 (0.86–2.26) | 0.180 |
| Recessive (ref: A/A + G/A) | G/G | 29 (10.9%) | 16 (11.3%) | 0.99 (0.50–1.97) | 0.980 | 16 (8.1%) | 16 (15.2%) | 2.13 (1.01–4.49) | 0.048 |
SNP Single nucleotide polymorphism, OR Odds ratio; 95% CI: 95% confidence interval
pa-values were calculated by logistic regression analysis with adjustment for age in males
pb-values were calculated by logistic regression analysis with adjustment for age in females
Bold italics indicates the SNP with statistical significance (p < 0.05)
CRC and haplotypes on chromosome 2q13
Finally, five IL1A polymorphisms (rs3783550, rs3783546, rs2856838, rs1609682, rs3783521) were mapped to a 10-kb LD block and displayed three haplotypes with frequencies of more than 5% in our subjects (Table 4). In Fig. 1, the red squares of the IL1A LD block represented significant linkage between the five SNPs. The LD degree is displayed by standard color schemes with bright red for very strong LD (D’ = 1). According to the haplotype analysis, there were no statistically significant differences appearing between patients and controls among any of the IL1A haplotype frequencies.
Table 4.
IL1A haplotype frequencies and the association with CRC risk
| Block | Haplotype | Freq (case) | Freq (control) | OR (95% CI) | p a |
|---|---|---|---|---|---|
| rs3783550/rs3783546/rs2856838 /rs1609682/rs3783521 |
GGGGA | 0.640 | 0.674 | 1.00 | |
| TCATG | 0.250 | 0.249 | 1.07 (0.83–1.39) | 0.600 | |
| TCGTG | 0.100 | 0.077 | 1.33 (0.90–1.97) | 0.150 |
OR Odds ratio; 95% CI: 95% confidence interval
pa-values were obtained after adjustments for gender and age with the comparison between CRC patients and healthy controls
Fig. 1.
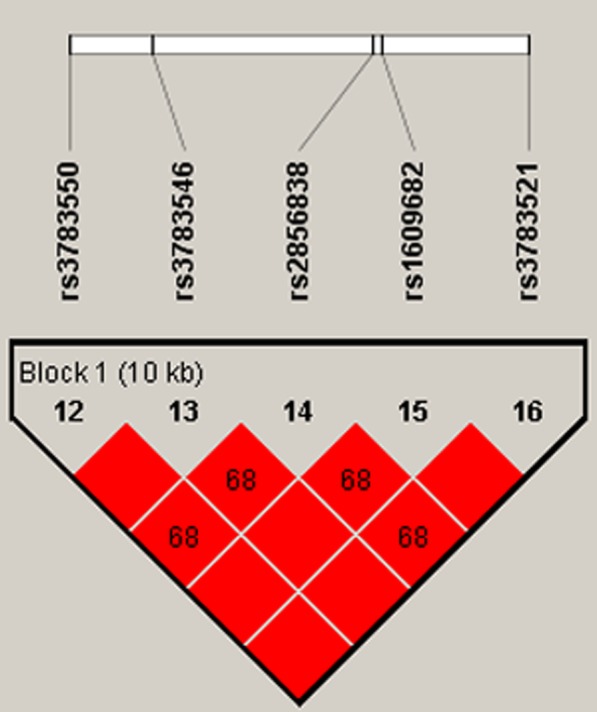
Haplotype block map formed by the SNPs in IL1A gene. Haplotype block are formed by rs3783550, rs3783546, rs2856838, rs1609682 and rs3783521 at 10 kb totally
SNP functional evaluation
In order to evaluate the possible function harbored by the five selected variants, we performed a bioinformatics analysis using HaploReg v4.1 database. All the variations were successfully predicted as regulatory SNPs with different biological functions (Additional file 1: Table S3). Furthermore, eQTL analysis uncovered remarkable relationships between these SNPs and IL1A expression in skin, testis or pituitary, which indicated the effects of different genotypes on gene expression (Additional file 1: Table S4).
Discussion
In this study, we genotyped five polymorphisms of IL1A and evaluated their correlations with the risk of CRC in a Chinese Han population. Our results first showed that four SNPs (rs3783546, rs2856838, rs1609682, rs3783521) were associated with CRC susceptibility in Chinese Han females.
Interleukin-1 alpha (IL-1α) is a major member of the interleukin-1 (IL-1) family, which has been well established as pro-inflammatory cytokine cluster involved in tumorigenesis. Several studies have showed that IL-1α contributes to tumour invasion, tumour proliferation, tumour metastases and interaction between the host immune system and malignant tumour cells [9, 19]. Kasza has reported that IL-1α serves as a pleiotropic cytokine in human cancer progression as well as various immune and inflammatory responses, and the secreted IL-1α is an active form which induces the tissue damage and tumour growth together with other regulatory factors [15]. Accordingly, it is biologically reasonable that functional IL1A polymorphisms may play causal roles in the development of cancer.
Increasing evidence has showed that IL1A polymorphisms are associated with several cancers [14]. Fei Wang et al. has found that four IL1A SNPs, rs3783550, rs3783546, rs1609682, and rs3783521, were associated with a significantly increased risk of renal cell carcinoma [26]. Xiaofeng Zeng et al. has demonstrated the relationship between the IL1A rs3783553 polymorphism and a significantly decreased risk of gastric cancer (OR = 0.48, 95% CI:0.26–0.90, p = 0.02) [31]. Abazis-Stamboulieh D et al. has proved that the IL1A rs1800587 polymorphism was related to a higher risk of multiple myeloma development [1]. Our data showed that rs2856838 was significantly associated with CRC susceptibility (p = 0.036). After stratification by gender, we found that the recessive model of rs3783550, rs2856838, rs1609682, and rs3783521 were associated with increased CRC risk in females. Moreover, it should be noted that the incidence rate of CRC differs significantly according to gender [17]. Considering the predicted functions of the selected SNPs in our study and their influences on gene expression provided by database, we speculated that SNPs might affect oncogenic processes by altering gene expression and this process could be influenced by individual gender background, thus leading to different outcomes on CRC risk between males and females. Significant IL1A variants rs3783550, rs2856838, rs1609682 were located in the intron, and rs3783521 were in promoter. As both the intron and promoter play important roles in gene expression, the alterations of the SNPs might also influence gene via modulating the functions of these regions. In addition, gender may reflect the different levels of exposure to risk factors related to diet, occupational exposure, or lifestyle. However, due to the limitations of the collected data, it was only possible to study the effect of gender rather than the complete impacts of other factors contributing to the gene environment. Therefore, it is necessary to carry out a complete analysis on the interaction of these factors with genetic polymorphisms in CRC susceptibility to verify the results of this study.
Several limitations should be considered in this research. Firstly, the influence of the selected SNPs on IL1A gene expression did not verify in our population and the underlying mechanism upon this regulatory process still need to be further elucidated. Further research will be design to investigate the influence of the promising SNPs on intron and promoter functions in gene regulation, and the gender will be considered as an important factor in future research. Secondly, although variations of age and gender were considered during our analysis, some living habits were not evaluated in this study, such as alcohol consumption and cigarette smoking, which might facilitate the progression of CRC.
Conclusion
Our study first provided objective evidence that the IL1A genetic polymorphism rs2856838 might contribute to the risk of CRC in Chinese Han population. In addition, rs3783550, rs1609682, rs3783521, and rs2856838 were susceptible pathogenic variants for CRC among females. These results could suggest new targets for CRC risk assessment, prevention and prognosis.
Additional file
Table S1. Primers used for identification of the IL1A polymorphisms. Table S2. Distributions of age and gender in CRC patients and controls. Table S3. Functional annotation of the selected variants provided by HaploReg 4.1. Table S4. eQTL analysis for the five IL1A SNPs provided by GTEx database. (DOCX 24 kb)
Acknowledgments
We are grateful to the patients and control individuals for providing blood samples. We also appreciate all editors and reviewers for their patience and suggestions to this work.
Funding
Not applicable.
Availability of data and materials
The data analyzed during the current research can be available from the corresponding author on reasonable request.
Abbreviations
- CI
Confidence interval
- CRC
Colorectal cancer
- eQTL
Expression quantitative trait loci
- GTEx
Genotype-Tissue expression; SD: Standard deviation
- HWE
Hardy-Weinberg equilibrium
- Ins/del
Insertion/deletion
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- MALDI-TOF mass spectrometry
Matrix-assisted laser desorption/ionization—time of flight mass spectrometry
- OR
Odds ratio
- SNPs
Single nucleotide polymorphisms
Authors’ contributions
HL, HJ and LL designed the study. HJ, LL, JH, YL and BZ collected the samples and performed the experiments. HT, DS and YZ managed the data and interpreted the statistical analyses. HJ, LL wrote the manuscript and HS, YL, HL revised the manuscript. All the authors reviewed the manuscript and approved the final version.
Ethics approval and consent to participate
Our research was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University. All procedures performed in this study were in accordance with the ethical standards of the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University and with the 1964 Helsinki declaration and its later amendments. Informed consents were obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hong Ji, Email: jehoung@163.com.
Le Lu, Email: luler2008@163.com.
Jingjing Huang, Email: h412279516@qq.com.
Yang Liu, Email: liuyangzuihao@126.com.
Binchao Zhang, Email: m18082274557@163.com.
Hui Tang, Email: 546471289@qq.com.
Dangze Sun, Email: sundangze@163.com.
Yafei Zhang, Email: 740228223@qq.com.
Hao Shang, Email: shanghao83@163.com.
Yiming Li, Email: liyiming2003@sina.com.
Hongwei Lu, Phone: 18700920064, Email: lhwdoc@163.com.
References
- 1.Abazis-Stamboulieh D, Oikonomou P, Papadoulis N, Panayiotidis P, Vrakidou E, Tsezou A. Association of interleukin-1A, interleukin-1B and interleukin-1 receptor antagonist gene polymorphisms with multiple myeloma. Leukemia & Lymphoma. 2007;48:2196–2203. doi: 10.1080/10428190701615892. [DOI] [PubMed] [Google Scholar]
- 2.An J, Zhao J, Zhang X, Ding R, Geng T, Feng T, Jin T. Impact of multiple alcohol dehydrogenase gene polymorphisms on risk of laryngeal, esophageal, gastric and colorectal cancers in Chinese Han population. Am J Cancer Res. 2015;5:2508–2515. [PMC free article] [PubMed] [Google Scholar]
- 3.Apte RN, Voronov E. Is interleukin-1 a good or bad ‘guy’ in tumor immunobiology and immunotherapy? Immunol Rev. 2008;222:222. doi: 10.1111/j.1600-065X.2008.00615.x. [DOI] [PubMed] [Google Scholar]
- 4.Bland JM, Altman DG. The odds ratio Bmj. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botteri E, Iodice S, Bagnardi V, Raimondi S, Lowenfels AB, Maisonneuve P. Smoking and colorectal Cancer: a meta-analysis Jama. J Am Med Assoc. 2008;102:996–997. doi: 10.1001/jama.2008.839. [DOI] [PubMed] [Google Scholar]
- 6.Burada F, Dumitrescu T, Nicoli R, Ciurea ME, Angelescu C, Mixich F, Ioana M. IL-1RN +2018T>C polymorphism is correlated with colorectal cancer. Mol Biol Rep. 2013;40:2851–2857. doi: 10.1007/s11033-012-2300-x. [DOI] [PubMed] [Google Scholar]
- 7.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan X, et al. Genetic association of PLCE1, C11orf92-C11orf93, and NOC3L with colorectal cancer risk in the Han population. Tumor Biol. 2014;35:1813–1817. doi: 10.1007/s13277-013-1242-9. [DOI] [PubMed] [Google Scholar]
- 9.Elaraj DM, et al. The role of interleukin 1 in growth and metastasis of human cancer xenografts clinical Cancer research An official journal of the American association for. Cancer Res. 2006;12:1088. doi: 10.1158/1078-0432.CCR-05-1603. [DOI] [PubMed] [Google Scholar]
- 10.Gabriel S, Ziaugra L, Tabbaa D (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform Current Protocols in Human Genetics Chapter 2:Unit 2.12. [DOI] [PubMed]
- 11.Gao L, et al. Association between a functional insertion/deletion polymorphism in IL1A gene and risk of papillary thyroid carcinoma. Tumor Biol. 2014;35:3861. doi: 10.1007/s13277-013-1512-6. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, et al. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis. 2009;30:2064–2069. doi: 10.1093/carcin/bgp283. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Ni S, Li D, He Y. An insertion/deletion polymorphism at miRNA-122 binding site in the IL1A is associated with a reduced risk of cervical squamous cell carcinoma. Genetic testing and molecular biomarkers. 2015;19:331–334. doi: 10.1089/gtmb.2015.0015. [DOI] [PubMed] [Google Scholar]
- 14.Karjalainen J, et al. The IL1A genotype is associated with nasal polyposis in asthmatic adults. Allergy. 2003;58:393–396. doi: 10.1034/j.1398-9995.2003.00118.x. [DOI] [PubMed] [Google Scholar]
- 15.Kasza A. IL-1 and EGF regulate expression of genes important in inflammation and cancer. Cytokine. 2013;62:22–33. doi: 10.1016/j.cyto.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Gu J. Changing patterns of colorectal cancer in China over a period of 20 years. World J Gastroenterol. 2005;11:4685–4688. doi: 10.3748/wjg.v11.i30.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinelli M, Scapoli L, Cura F, Rodia MT, Ugolini G, Montroni I, Solmi R. Colorectal cancer susceptibility: apparent gender-related modulation by ABCB1 gene polymorphisms. J Biomed Sci. 2014;21:89. doi: 10.1186/s12929-014-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moss SF, Blaser MJ. Mechanisms of disease: inflammation and the origins of cancer. Nat Clin Pract Oncol. 2005;2:90. doi: 10.1038/ncponc0081. [DOI] [PubMed] [Google Scholar]
- 19.Pantschenko AG, et al. The interleukin-1 family of cytokines and receptors in human breast cancer: implications for tumor progression. Int J Oncol. 2003;23:269. [PubMed] [Google Scholar]
- 20.Pu Y, et al. Association of an insertion/deletion polymorphism in IL1A 3′-UTR with risk for cervical carcinoma in Chinese Han women. Hum Immunol. 2014;75:740–744. doi: 10.1016/j.humimm.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Rider P, Carmi Y, Voronov E, Apte RN. Interleukin-1alpha. Semin Immunol. 2013;25:430–438. doi: 10.1016/j.smim.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto K, et al. Inhibitor of kappaB kinase beta regulates gastric carcinogenesis via interleukin-1alpha expression. Gastroenterology. 2010;139:226. doi: 10.1053/j.gastro.2010.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Q, Wang Y, Zhao J, Ma C, Wu T, Jin T, Xu J. Polymorphisms of PRLHR and HSPA12A and risk of gastric and colorectal cancer in the Chinese Han population. BMC Gastroenterol. 2015;15. 10.1186/s12876-015-0336-9. [DOI] [PMC free article] [PubMed]
- 24.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 25.Tsoi KK, Pau CY, Wu WK, Chan FK, Griffiths S, Sung JJ. Cigarette smoking and the risk of colorectal cancer: a meta-analysis of prospective cohort studies. Clinical Gastroenterology & Hepatology the Official Clinical Practice Journal of the American Gastroenterological Association. 2009;7:682. doi: 10.1016/j.cgh.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, et al. IL1 genes polymorphism and the risk of renal cell carcinoma in Chinese Han population. Oncotarget. 2017;8:56021–56029. doi: 10.18632/oncotarget.18715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N, et al. Multiple genetic variants are associated with colorectal cancer risk in the Han Chinese population. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2015;24:1–5. doi: 10.1097/cej.0000000000000012.. [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Gao Y, Feng T, Jin TB, Kang LL, Chen C. Meta-analysis of the rs4779584 polymorphism and colorectal cancer risk. PLoS One. 2014;9:e89736. doi: 10.1371/journal.pone.0089736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang ZH, Dai Q, Zhong L, Zhang X, Guo QX, Li SN. Association of IL-1 polymorphisms and IL-1 serum levels with susceptibility to nasopharyngeal carcinoma. Mol Carcinog. 2011;50:208–214. doi: 10.1002/mc.20706. [DOI] [PubMed] [Google Scholar]
- 30.Yong Y, Lin H. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 31.Zeng XF, Li J, Li SB. A functional polymorphism in IL-1A gene is associated with a reduced risk of gastric cancer. Tumour biology the journal of the international society for Oncodevelopmental biology & Medicine. 2014;35:265–268. doi: 10.1007/s13277-013-1034-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, et al. A polymorphism at miRNA-122-binding site in the IL-1α 3'UTR is associated with risk of epithelial ovarian cancer. Familial Cancer. 2014;13:595. doi: 10.1007/s10689-014-9739-y. [DOI] [PubMed] [Google Scholar]
- 33.Zhong R, et al. Genetic variations in the TGFβ signaling pathway, smoking and risk of colorectal cancer in a Chinese population. Carcinogenesis. 2013;34:936–942. doi: 10.1093/carcin/bgs395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for identification of the IL1A polymorphisms. Table S2. Distributions of age and gender in CRC patients and controls. Table S3. Functional annotation of the selected variants provided by HaploReg 4.1. Table S4. eQTL analysis for the five IL1A SNPs provided by GTEx database. (DOCX 24 kb)
Data Availability Statement
The data analyzed during the current research can be available from the corresponding author on reasonable request.


