A published assessment of Google Flu Trends’ (GFT) accuracy in most Latin American countries is lacking. We note inaccuracies with GFT-predicted influenza activity compared with FluNet throughout Latin America. Our findings offer lessons for future Internet-based biosurveillance tools.
Keywords: Google Flu Trends, Latin America, digital epidemiology
Abstract
Background
Latin America has a substantial burden of influenza and rising Internet access and could benefit from real-time influenza epidemic prediction web tools such as Google Flu Trends (GFT) to assist in risk communication and resource allocation during epidemics. However, there has never been a published assessment of GFT's accuracy in most Latin American countries or in any low- to middle-income country. Our aim was to evaluate GFT in Argentina, Bolivia, Brazil, Chile, Mexico, Paraguay, Peru, and Uruguay.
Methods
Weekly influenza-test positive proportions for the eight countries were obtained from FluNet for the period January 2011–December 2014. Concurrent weekly Google-predicted influenza activity in the same countries was abstracted from GFT. Pearson correlation coefficients between observed and Google-predicted influenza activity trends were determined for each country. Permutation tests were used to examine background seasonal correlation between FluNet and GFT by country.
Results
There were frequent GFT prediction errors, with correlation ranging from r = −0.53 to 0.91. GFT-predicted influenza activity best correlated with FluNet data in Mexico follow by Uruguay, Argentina, Chile, Brazil, Peru, Bolivia and Paraguay. Correlation was generally highest in the more temperate countries with more regular influenza seasonality and lowest in tropical regions. A substantial amount of autocorrelation was noted, suggestive that GFT is not fully specific for influenza virus activity.
Conclusions
We note substantial inaccuracies with GFT-predicted influenza activity compared with FluNet throughout Latin America, particularly among tropical countries with irregular influenza seasonality. Our findings offer valuable lessons for future Internet-based biosurveillance tools.
Several Internet biosurveillance tools that use population-level trends in Google and other Internet search-engine queries about infectious diseases such as dengue, pertussis, influenza, and norovirus to detect and predict epidemics have been developed in recent years [1–5]. Google Flu Trends (GFT) is one such biosurveillance tool that estimates weekly trends of influenza activity with an explanatory variable comprising the normalized number of Google searches about a set of influenza-related terms in that region and time [4]. GFT was launched in the United States in 2008 and demonstrated very low error in real-time detection (“now-casting”) of weekly US influenza-like illness activity (https://www.google.org/flutrends/about/) [4]. GFT offered a rapid, complementary surveillance signal of influenza activity days before that of traditional surveillance systems, although its optimal role in public health practice has remained an unanswered question since its launch (http://precedings.nature.com/documents/3493/version/1). GFT was then expanded to a wide range of countries including Argentina, Bolivia, Brazil, Chile, Peru, Mexico, Paraguay, and Uruguay (https://www.google.org/flutrends/about/).
GFT could provide critical information to countries in Latin America to strengthen their ability to detect and control influenza epidemics. Latin America has a substantial burden of influenza [6] and has recently dealt with an influenza pandemic [7]. Timely surveillance is crucial for the control of influenza in this region. For instance, rapid detection of changes in influenza activity can assist in risk communication, promotion of vaccinations, and healthcare resource allocation during influenza epidemics. However, existing surveillance systems in Latin American are not real time and have delays in reporting, laboratory testing, and dissemination of results to key response personnel including clinicians, epidemiologists, and public health officials. Given rising Internet access throughout Latin America [8], web-based surveillance tools like GFT could provide real-time information about influenza activity, particularly in less affluent countries with limited traditional healthcare and laboratory-based surveillance. Soon after and in the years after its launch, however, marked prediction errors were noted in the US GFT web tool despite several updates of its model [9]. Moreover, it did not perform well in detecting the initial wave of pandemic pH1N1 influenza [9]. By mid-2015, Google ceased public access to GFT, although ongoing GFT access has been granted to select US academic institutions and the Centers for Disease Control and Prevention for refinement of this web tool [10]. Despite issues with its early performance, GFT continues to be used or explored by such organizations for the real-time detection of influenza. More recently, GFT has been combined with non-Google data streams for highly accurate now-casting and forecasting of influenza epidemics [11]. In addition, several revisions to the GFT model, including those by external academic groups, have improved its performance [12–14].
With the exception of a limited study in Argentina [15], there are no published studies evaluating the accuracy of GFT in any of the Latin American countries in which the service was offered. Given that GFT was developed in the United States using North American influenza epidemiology and Internet search activity, its performance in non-US regions may be limited, particularly in tropical regions that can have irregular seasonality [16] and particularly in regions in which English is not the predominant language spoken. A careful examination of GFT's performance in Latin American countries may offer valuable lessons for the development and improvement of Internet-based biosurveillance tools. This type of evaluation may be particularly useful at a time when academic and government researchers are redesigning and rethinking GFT algorithms for the United States and beyond.
We determined the annual correlation between weekly Google-predicted and reported influenza virus activity in the 8 Latin American countries in which GFT was offered (Argentina, Bolivia, Brazil, Chile, Mexico, Peru, Paraguay, and Uruguay) and how specific GFT was in detecting influenza activity rather than just shared seasonality. We also examined possible determinants of GFT accuracy, such as international differences in population Internet access, the synchronicity of annual peaks in Google-predicted and reported influenza activity in each of these countries, and the synchronicity of epidemic onsets between Google-predicted and reported influenza activity in each of these countries. Finally, we sought to examine how well GFT performed on a finer spatial scale, using active community-based surveillance data from unique influenza cohorts in two locales in Peru.
METHODS
Data Abstraction and Consolidation
The GFT model was originally fit, validated, and refit in the United States using observed influenza-like illness activity data from ILINet [4]. ILINet measures a weekly proportion of health consultations because of influenza-like illness (ILI) among all health consultations seen that week by a network of US sentinel healthcare providers [17]. Because a system like ILINet does not exist in Latin America, we used weekly influenza test-positive proportions reported by the National Ministries of Health to the World Health Organization's FluNet as a metric of influenza activity. This metric has been used as a valid measurement of influenza activity in other Latin American influenza studies in this region [18] and has been used in the validation of GFT in the United States [19].
Weekly influenza-test positive proportions (respiratory specimens testing positive for influenza divided by the number of respiratory specimens tested for that week) for Argentina, Bolivia, Brazil, Chile, Mexico, Paraguay, Peru, and Uruguay were obtained from the public FluNet web tool [20] for the period 1 January 2011–31 December 2014. Where FluNet indicated “zero” weekly specimens were received by any of these countries for influenza testing, we coded that week as having missing data. Weekly Google-predicted influenza activity in the same countries and time period were then downloaded from the public GFT website that offers archived GFT predictions to the public up to June 2015 (https://www.google.org/flutrends/about/).
Determination of Correlation Between Google-Predicted and Observed Weekly Influenza Activity
The correlation between trends in weekly Google-predicted influenza activity and weekly FluNet reports for each country and study year was determined using Pearson correlation, which is the most frequent metric of GFT accuracy used in other studies [4, 9, 19]. We also repeated the analysis using Spearman rank correlation for robust estimates of correlation. We arbitrarily classified correlation coefficient values of <0.6 as having low accuracy, 0.6–0.8 as having moderate accuracy, 0.8–0.9 as having high accuracy, and ≥0.9 as having very high accuracy. As a sensitivity analysis, we also repeated this correlation analysis with weeks for which FluNet indicated zero weekly specimens were received coded as having zero reported influenza activity for that week.
At least part of the correlations between observed and Google-predicted weekly influenza activity data may have been driven by seasonal autocorrelation (ie, coincidentally correlating seasonal trends of two time series) and not specific for influenza activity. To explore this, we also sought to determine the background seasonal correlation between GFT and FluNet trends. This was determined by comparing the distribution of the correlation between 1000 yearly permuted versions of the weekly GFT time series and the weekly FluNet time series (for the entire period 2011–2014), yielding estimates of the mean and 95% confidence intervals of the background seasonal correlation [21].
Exploring Association Between Country Latitude and Country Internet Access With Magnitude of GFT–FluNet Correlation
We hypothesized that temperate countries with regular influenza seasonality had better GFT accuracy compared with tropical countries with more complex seasonality [16, 22]. We tested this hypothesis through autocorrelation analysis, which determined in which countries GFT relied on a regular temperate influenza seasonal pattern for a substantial part of its accuracy. We also tested the hypothesis that GFT performed better in countries with greater Internet access by comparing GFT–FluNet correlation magnitude with estimated population Internet access by Spearman rank correlation analysis. The mean published World Bank estimates of Internet access for each country for the period 2011–2014 (Supplementary Figure S1) were used as a measurement of population-level Internet access in each of the studied countries (http://data.worldbank.org/indicator/IT.NET.USER.P2).
Comparison of the Performance of GFT Versus a Simple Autoregressive Predictive Model
We further evaluated the performance of GFT in each country by comparing it with the performance of a simple autoregressive (AR1) model in which the previous week's observed influenza activity was used as a naive predictor of the following week's influenza activity, which is an example of an AR1 model. Pearson correlation between the AR1-predicted and observed influenza activity for a given week was determined for each country across the study period 2011–2014.
Assessing Synchronicity Between Google-Predicted and Observed Influenza Annual Epidemic Peak Timing and Annual Epidemic Onset Timing
We determined the synchronicity of Google-predicted peak influenza activity with reported FluNet peak influenza activity in each country by counting the number of weeks between the time of Google-predicted and observed epidemic peaks. We also determined the synchronicity of Google-predicted influenza epidemic onset timing with observed FluNet influenza epidemic onset timing by counting the number of weeks between the time of Google-predicted and observed epidemic onset. An epidemic period was defined as a period when median influenza activity was greater than the annual weekly median for at least 8 weeks, and the onset week was defined as the first week in that period [16, 22]. For countries within the Southern Hemisphere (Bolivia, Brazil, Peru, Argentina, Chile, Paraguay, and Uruguay), these assessments were conducted for each January–December yearly period. For Mexico, which experiences influenza epidemics during the Northern Hemisphere winter, these assessments were repeated for each July–June yearly period [16, 22]. Assessments of synchronicity between Google-predicted and observed peaks and epidemic onset times were only attempted in years when correlation was statistically significant and of at least a moderate magnitude (r > 0.6).
Validation Against an Alternative Measurement of Influenza Activity on a Fine Spatial Scale
Given the possible limitations of FluNet data (including those inherent to sentinel surveillance and marked differences in sampling density by country [Supplementary Figure S2]) and because FluNet does not report subnational data, we tested the correlation between subnational weekly GFT trends in Cusco and Lima, Peru (https://www.google.org/flutrends/about/) and prospectively collected weekly adjusted influenza incidence data from 2 community-based cohorts in Lima and Cusco, Peru, available from 1 January 2011 to 26 July 2014. The details of these community-based surveillance studies are described elsewhere [23, 24].
All analyses were performed with Stata version 13.1 (Stata Corporation, College Station, Texas) and R version 3.1.2 [25].
Ethical Considerations
The Naval Medical Research Unit 6 and the University of California, San Francisco, institutional review boards approved this study as non-human subjects research.
RESULTS
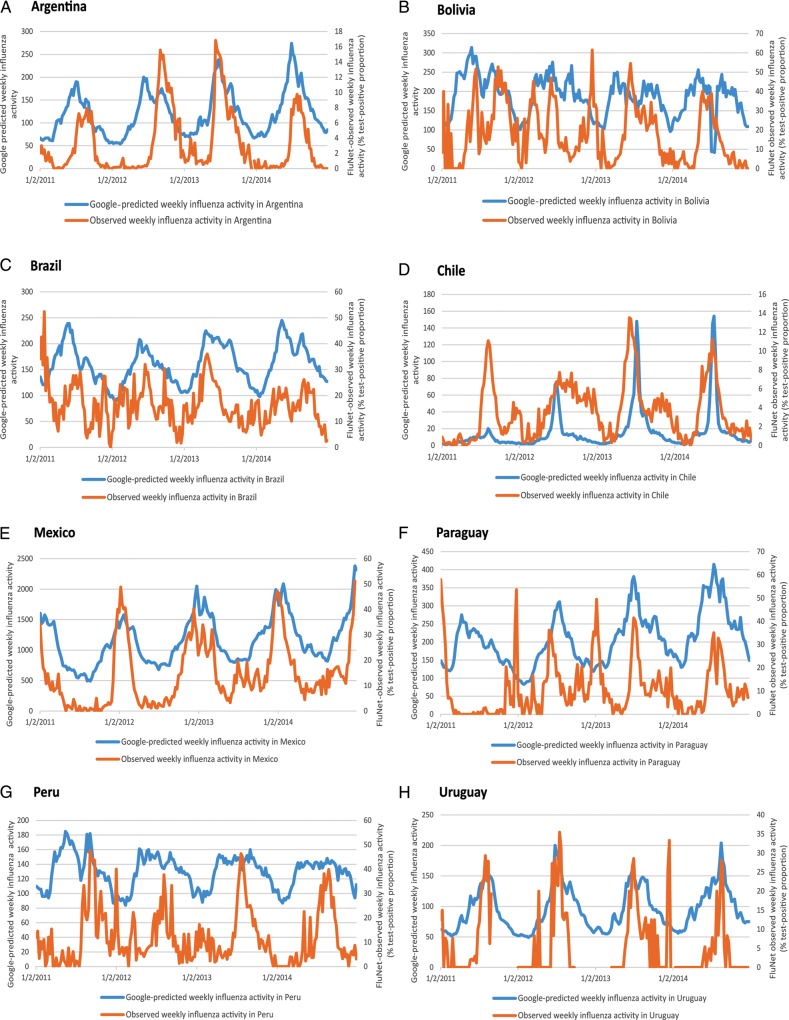
Across all countries and years, the correlation between weekly Google-predicted and observed influenza activity was highly variable, ranging from r = −0.53 to 0.91 (Table 1, Figure 1). Most correlations (24/32) were statistically significant (Table 1). Correlation was poor (r < 0.6) in 18/32, high (r = 0.8–0.9) in 3/32, and very high (r ≥ 0.9) in only 1/32 of the time periods studied (Table 1). In only four of the eight countries (Argentina, Mexico, Chile, and Uruguay), correlations were statistically significant across all years (Table 1). Supplementary Table S3 presents the same correlations determined by Spearman ρ, with broadly similar conclusions.
Table 1.
Pearson Correlation Coefficients Between Weekly Google-Predicted and Observed Influenza Activity by Year and Location in Latin America
| Location | Year |
|||
|---|---|---|---|---|
| 2011 | 2012 | 2013 | 2014 | |
| Argentina | 0.51 | 0.39 | 0.91 | 0.78 |
| Brazil | −0.07* | 0.48 | 0.63 | 0.61 |
| Bolivia | 0.16* | 0.11* | 0.16* | 0.09* |
| Chile | 0.71 | 0.49 | 0.57 | 0.78 |
| Mexico | 0.75 | 0.88 | 0.81 | 0.87 |
| Paraguay | −0.53 | 0.34 | 0.21* | 0.71 |
| Peru | ||||
| All Peru | 0.03* | 0.16* | 0.31 | 0.57 |
| Limaa | 0.21* | 0.50 | 0.17* | 0.51 |
| Cuzcoa | 0.23* | 0.40 | 0.35 | 0.36 |
| Uruguay | 0.73 | 0.73 | 0.57 | 0.75 |
Observed influenza activity determined by weekly influenza-test positivity proportion from FluNet sentinel surveillance laboratories unless otherwise indicated. Weeks with zero sentinel surveillance specimens received for influenza testing coded as missing.
a Observed influenza activity determined by prospective community-based surveillance cohort, data available from 1 January 2011 to 26 July 2014.
*All correlations were statistically significant (P < .05) except where indicated by an asterisk.
Figure 1.
Time series for Google-predicted weekly influenza activity and FluNet-observed weekly influenza activity for Argentina, Bolivia, Brazil, Chile, Mexico, Paraguay, Peru, and Uruguay. Google Flu Trends does not indicate a unit of measurement for its predicted influenza activity.
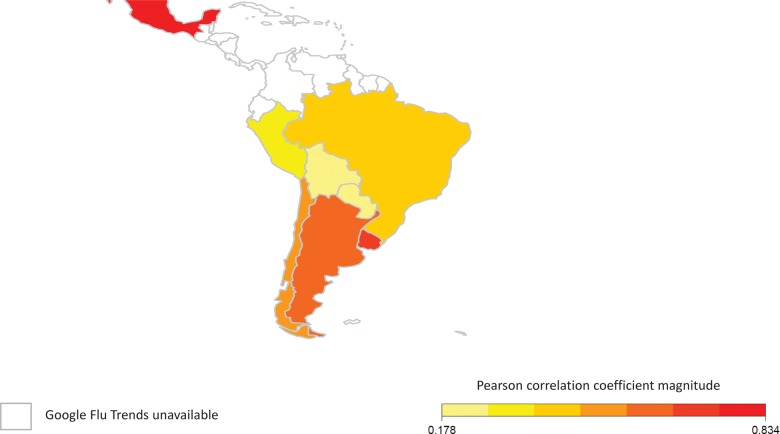
Table 2 and Figure 2 present the correlations between the weekly observed and Google-predicted influenza activity across the entire 2011–2014 study period for each country. Table 2 also presents the correlation between the permuted GFT time series data with weekly FluNet data from each country to account for possible autocorrelation. Taken across the entire study period, GFT best correlated with influenza activity in Mexico followed by Uruguay, Argentina, Chile, Brazil, Peru, Bolivia, and Paraguay (Table 2). Correlation was highest in temperate countries (Figure 2). The association between GFT–FluNet correlation and estimated population-level Internet access had borderline statistical significance (Spearman ρ = 0.69, P = .06). A sensitivity analysis with FluNet-reported influenza activity coded as zero for those weeks that reported zero specimens received for influenza testing (rather than missing) did not substantively change the FluNet-GFT correlation.
Table 2.
Pearson Correlation Between Weekly Google-Predicted and Observed Influenza Activity by Location in Latin America, 2011–2014, Using Original and Permuted Google Flu Trends Time Series Data
| Location | Original Time Series Data |
Permuted Data |
AR(1) Modela |
||||
|---|---|---|---|---|---|---|---|
| r | 95% CI | P Value | r | 95% CI | r | 95% CI | |
| Argentina | 0.61 | .52–.69 | <.001 | 0.5 | .41–.62 | 0.95 | .93–.96 |
| Bolivia | 0.19 | .05–.31 | .007 | 0.18 | .12–.22 | 0.79 | .73–.84 |
| Brazil | 0.34 | .22–.46 | <.001 | 0.28 | .21–.34 | 0.71 | .63–.77 |
| Chile | 0.59 | .49–.67 | <.001 | 0.46 | .36–.60 | 0.92 | .90–.94 |
| Mexico | 0.83 | .79–.87 | <.001 | 0.7 | .59–.83 | 0.94 | .92–.95 |
| Paraguay | 0.18 | .04–.31 | .01 | 0.05 | −.11–.23 | 0.8 | .75–.85 |
| Peru | |||||||
| All Peru | 0.24 | .11–.36 | <.001 | 0.25 | .22–.29 | 0.76 | .69–.81 |
| Limab | 0.23 | .09–.36 | .002 | 0.23 | .04–.43 | 0.68 | .59–.75 |
| Cuzcob | 0.22 | .08–.36 | .002 | 0.32 | .15–.44 | 0.79 | .74–.85 |
| Uruguay | 0.65 | .56–.73 | <.001 | 0.56 | .45–.65 | 0.66 | .57–.74 |
Pearson correlation between weekly influenza activity predicted by an AR1 model, and observed influenza activity are shown for comparison. Observed influenza activity determined by weekly influenza-test positivity proportion from FluNet sentinel surveillance laboratories unless otherwise indicated. Weeks with zero sentinel surveillance specimens received for influenza testing coded as missing.
Abbreviation: CI, confidence interval.
a Autoregressive model using the previous week's observed influenza activity as a predictor of the current week's.
b Observed influenza activity determined by prospective community-based surveillance cohort, data available from 1 January 2011 to 26 July 2014.
Figure 2.
GFT–FluNet Pearson correlation coefficient magnitude by country studied in Latin America. White colored countries do not have available Google Flu Trends data. Mexico is represented by its southern regions only.
Repeat analysis using the permuted GFT data over the entire study period (Table 2) demonstrated statistically significant autocorrelation in all countries except for Paraguay. The magnitude of seasonal autocorrelation was highest in Argentina, Mexico, Chile, and Uruguay, suggesting that GFT performance in such temperate regions is, in part, correlated with observed FluNet influenza trends because of shared winter seasonality, rather than specifically detecting weekly fluctuations in influenza activity. The AR1 model outperformed GFT in each studied country, although there was considerable overlap in the confidence intervals around the GFT–FluNet and AR1-FluNet correlations in Uruguay.
Table 3 shows the time difference between Google-predicted and observed influenza epidemic onset. Excluding years when there was poor GFT–FluNet correlation (r < 0.6), there were 4 instances when the GFT-predicted epidemic onset preceded the observed onset of an epidemic period and 1 when the GFT-predicted onset of an epidemic period was synchronous with an observed epidemic onset.
Table 3.
Time Difference Between Google-Predicted and Observed Influenza Epidemic Onset
| Location | Yeara,b |
|||
|---|---|---|---|---|
| 2011 | 2012 | 2013 | 2014 | |
| Argentina | NA | NA | 2 | 3 |
| Brazil | NA | NA | −3 | 1 |
| Bolivia | NA | NA | NA | NA |
| Chile | 12 | NA | NA | −2 |
| Mexico | … | 6 | 0 | 6 |
| Paraguay | NA | NA | NA | 5 |
| Peru | NA | NA | NA | NA |
| Uruguay | 7 | 4 | NA | 8 |
Observed influenza activity determined by weekly influenza-test positivity proportion from FluNet sentinel surveillance laboratories. Time difference in weeks. Positive difference indicates when week of Google-predicted epidemic onset precedes the week of observed epidemic onset. NA indicates no observed influenza activity data available or correlation between Google-predicted and observed influenza activity too poor (r < 0.6 or not statistically significant) to determine difference in epidemic onset timing.
a For countries within or mostly within the Southern Hemisphere, synchronicity of Google-predicted and observed epidemic onset timing assessed in a January through December calendar period.
b For countries in the Northern Hemisphere, synchronicity of Google-predicted and observed epidemic onset timing assessed in a previous July through June of stated year time period.
Table 4 shows the time difference between peak Google-predicted and observed influenza activity. Of the 15 study periods where there was enough GFT–FluNet correlation to make a meaningful inference from the calculated difference in Google-predicted and observed epidemic peaks, there were 4 instances when there was a precise synchronicity between peaks (zero weeks difference). In 4 instances, GFT preceded the peak influenza epidemic activity by 1 to 4 weeks.
Table 4.
Time Difference Between Peak Google-Predicted and Observed Influenza Activity
| Location | Yeara,b |
|||
|---|---|---|---|---|
| 2011 | 2012 | 2013 | 2014 | |
| Argentina | NA | NA | −2 | 4 |
| Brazil | NA | NA | 1 | 16 |
| Bolivia | NA | NA | NA | NA |
| Chile | 0 | 2 | −5 | −1 |
| Mexico | … | −2 | −2 | −4 |
| Paraguay | NA | NA | NA | 0 |
| Peru | NA | NA | NA | NA |
| Uruguay | 0 | 3 | NA | 0 |
Observed influenza activity determined by weekly influenza-test positivity proportion from FluNet sentinel surveillance laboratories. Time difference in weeks. Positive difference indicates when Google-predicted peak precedes the observed peak.
a The 2011 time difference not reported because Mexican influenza season runs over 2010–2011, unlike Southern Hemisphere countries studied. NA indicates correlation between Google-predicted and observed influenza activity too poor (r < 0.6) to determine differences between observed and predicted peaks.
b For countries in the Northern Hemisphere, synchronicity of Google-predicted and observed peak influenza peak activity assessed in a previous July through June of stated year time period.
When examined on a finer spatial scale in Lima and Cusco, correlation between Google-predicted and observed influenza activity was poor (r = 0.23 and 0.22, respectively; Table 2). Given such poor correlation, we were unable to assess the synchronicity between peak Google-predicted and peak observed influenza activity in these two regions of Peru, and we were unable to meaningfully compare how GFT predicted the onset of the epidemic season in these locales.
DISCUSSION
We note major, frequent inaccuracies with GFT-predicted influenza activity compared with observed FluNet influenza virus surveillance in eight Latin American countries, particularly in tropical regions and countries with limited Internet access. While FluNet also has limitations, including differences in weekly FluNet sampling density and reporting between countries, we note that GFT also performed poorly when compared with active community-based influenza surveillance. Use of national aggregate FluNet data may be limited for Brazil in particular due to its marked geographic size and broad ecological and demographic diversity. An AR1 model markedly outperformed GFT in each country. However, typical delays of 1–2 weeks for preliminary FluNet data in Latin America underscore that AR1 models in real-world practice are limited and emphasize the value of real-time estimates from Internet biosurveillance systems such as GFT.
GFT performed best in more temperate regions with more regular seasonality (ie, Argentina, Mexico, Chile, and Uruguay), with a considerable amount of its predictive performance accounted for by seasonal autocorrelation. This suggests that GFT is nonspecific for influenza and is a “part-influenza, part-winter detector” in many parts of Latin America [26].
Beyond seasonal autocorrelation, GFT's better performance in the more temperate countries of the Americas could also be explained by greater Internet access. Those nations with the poorest GFT–FluNet correlations (Bolivia, Paraguay, and Peru) also had the least population Internet access compared with the other countries studied (Supplementary Table S1). Further, all Latin American countries studied have less Internet access compared with the United States [8]; this may, in part, account for the generally superior performance of GFT in the United States compared with Latin America [4, 9]. A quantitative analysis of the apparent association between country Internet access and GFT–FluNet correlation seems to support this notion, albeit with weak statistical support (Spearman ρ = 0.69, P = .06).
There may be other reasons why GFT–FluNet correlation is higher in certain countries, and there may be many unmeasured confounders that account for GFT–FluNet correlations being higher in certain regions. For instance, the variable accuracy of GFT throughout the Americas may perhaps be the result of differences in local Google search behavior, including the language used for Google searching. This may be a particular source of error if Google merely extrapolated its US-developed model coefficients and predictor terms to non-US countries rather than fitting and validating a local Google model based on local influenza epidemiology, Internet search behavior/ontology, and languages. Future development of revamped country-specific GFT tools would be an interesting avenue for future research.
In those cases where correlation was sufficient to perform further analysis, GFT did occasionally provide a synchronous now-cast of peak influenza activity. GFT performed better at now-casting the start of an epidemic period of influenza activity, although this, too, could only be assessed in the minority of cases because of the overall poor correlation between GFT and influenza activity documented through traditional surveillance.
This validation study comes with a caveat that influenza test-positive proportions were used as the metric of observed influenza activity. Influenza test-positives are a different marker of influenza activity compared with that used for fitting and validating the GFT models in the United States, that is, the proportion of ILI among all outpatient visits to the ILINet sentinel network [4]. However, a study in the United States found that the correlation of influenza activity measured by a weekly influenza test-positive proportion metric (CDC Virus Surveillance) and ILI syndromic surveillance (ILINet) was high (0.85, 95% confidence interval, .81, .89) [19]. A further limitation of influenza test-positive proportions is that they are most useful for examining influenza trends within countries rather between countries, and we were unable to determine if relative influenza burden was a predictor of GFT performance across countries.
Our findings emphasize that caution should be used when interpreting the findings of Google-based digital influenza surveillance in Latin America. Our results also provide important lessons for the improvement of Internet-based biosurveillance methods globally, including future versions of GFT, for these and other low- to middle-income regions. Such lessons include careful assessment for seasonal autocorrelation, caution in extrapolating predictive models developed in one country to other regions of the world, and transparency of model predictor terms and coefficients as part of the validation process. Our findings also serve as a useful baseline validation of the performance of the current GFT model in Latin America. This information should be considered before potential revisions to the model are adopted and combined with non-Google data to improve its performance as exemplified in recent US studies [11–14].
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Supplementary Material
Notes
Acknowledgments. We are grateful to Rakhee Palekar (Pan-American Health Organization) for advice on the timeliness of FluNet surveillance data from Latin America.
Disclaimer. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, the US Agency for International Development, the Centers for Disease Control or Prevention (CDC), or any other US government agency. Several authors of this work are employees of the US government. This work was prepared as part of their official duties. Title 17 U.S.C. § 105 provides that “copyright protection under this title is not available for any work of the US Government.” Title 17 U.S.C. § 101 defines a US government work as a work prepared by a military service member or employee of the US government as part of that person's official duties.
Financial support. The Peruvian community-based surveillance data were originally collected in a study funded by the CDC and the US Department of Defense Global Emerging Infections Surveillance (grant I0082_09_LI).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Pollett S, Wood N, Boscardin WJ et al. Validating the use of Google trends to enhance pertussis surveillance in California. PLoS Curr 2015:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan EH, Sahai V, Conrad C, Brownstein JS. Using web search query data to monitor dengue epidemics: a new model for neglected tropical disease surveillance. PLoS Negl Trop Dis 2011; 5:e1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desai R, Hall AJ, Lopman BA et al. Norovirus disease surveillance using Google Internet query share data. Clin Infect Dis 2012; 55:e75–8. [DOI] [PubMed] [Google Scholar]
- 4. Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature 2009; 457:1012–4. [DOI] [PubMed] [Google Scholar]
- 5. Polgreen PM, Chen YL, Pennock DM, Nelson FD. Using Internet searches for influenza surveillance. Clin Infect Dis 2008; 47:1443–8. [DOI] [PubMed] [Google Scholar]
- 6. Savy V, Ciapponi A, Bardach A et al. Burden of influenza in Latin America and the Caribbean: a systematic review and meta-analysis. Influenza Other Respir Viruses 2013; 7:1017–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March-April 2009. MMWR Morb Mortal Wkly Rep 2009; 58:467–70. [PubMed] [Google Scholar]
- 8. http://data.worldbank.org/indicator/IT.NET.USER.P2. Accessed June 2015.
- 9. Olson DR, Konty KJ, Paladini M, Viboud C, Simonsen L. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: a comparative epidemiological study at three geographic scales. PLoS Comput Biol 2013; 9:e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Available at: http://googleresearch.blogspot.com/2015/08/the-next-chapter-for-flu-trends.html Accessed 29 November 2015.
- 11. Santillana M, Nguyen AT, Dredze M, Paul MJ, Nsoesie EO, Brownstein JS. Combining search, social media, and traditional data sources to improve influenza surveillance. PLoS Comput Biol 2015; 11:e1004513:12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santillana M, Zhang DW, Althouse BM, Ayers JW. What can digital disease detection learn from (an external revision to) Google Flu Trends? Am J Prev Med 2014; 47:341–7. [DOI] [PubMed] [Google Scholar]
- 13. Lampos V, Miller AC, Crossan S, Stefansen C. Advances in nowcasting influenza-like illness rates using search query logs. Sci Rep 2015; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang SH, Santillana M, Kou SC. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc Natl Acad Sci U S A 2015; 112:14473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orellano PW, Reynoso JI, Antman J, Argibay O. [Using Google Trends to estimate the incidence of influenza-like illness in Argentina]. Cad Saude Publica 2015; 31:691–700. [DOI] [PubMed] [Google Scholar]
- 16. Azziz Baumgartner E, Dao CN, Nasreen S et al. Seasonality, timing, and climate drivers of influenza activity worldwide. J Infect Dis 2012; 206:838–46. [DOI] [PubMed] [Google Scholar]
- 17. Available at: http://www.cdc.gov/flu/weekly/overview.htm Accessed 29 November 2015.
- 18. Soebiyanto RP, Clara W, Jara J et al. The role of temperature and humidity on seasonal influenza in tropical areas: Guatemala, El Salvador and Panama, 2008–2013. PLoS One 2014; 9:e100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ortiz JR, Zhou H, Shay DK, Neuzil KM, Fowlkes AL, Goss CH. Monitoring influenza activity in the United States: a comparison of traditional surveillance systems with Google Flu Trends. PLoS One 2011; 6:e18687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Available at: http://www.who.int/influenza/gisrs_laboratory/flunet/en/ Accessed June 2015.
- 21. B. F. J. Manly: Randomization, bootstrap and Monte Carlo methods in biology. 3rd ed Boca Raton, FL: Chapman & Hall/CRC, 2007. [Google Scholar]
- 22. Durand LO, Cheng PY, Palekar R et al. Timing of influenza activity and vaccine selection in the American tropics. Influenza Other Respir Viruses 2016; 10:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Razuri H, Romero C, Tinoco Y et al. Population-based active surveillance cohort studies for influenza: lessons from Peru. Bull World Health Organ 2012; 90:318–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tinoco YO, Azziz-Baumgartner E, Razuri H et al. A population-based estimate of the economic burden of influenza in Peru, 2009–2010. Influenza Other Respir Viruses 2016; 10:301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2015. ISBN 3-900051-07-0. Available at: http://www.R-project.org/. Accessed March 2015. [Google Scholar]
- 26. Lazer D, Kennedy R, King G, Vespignani A. Big data. The parable of Google Flu: traps in big data analysis. Science 2014; 343:1203–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.