Abstract
Zika virus and dengue virus serotype 2 were isolated from a patient with travel to Haiti who developed fever, rash, arthralgias, and conjunctivitis. The infecting Zika virus was related to Venezuelan and Brazilian strains but evolved along a lineage originating from strains isolated in 2014 in the same region of Haiti.
Keywords: Zika virus, dengue virus, Haiti
Zika virus (ZIKV) and the 4 dengue virus (DENV) serotypes (DENV-1 though DENV-4), are mosquito-borne RNA viruses belonging to the family Flaviviridae. DENVs are endemic to Haiti, having been first reported in 1976 [1]. In contrast, ZIKV is an emerging pathogen in Haiti and the Americas, with apparent introduction from French Polynesia and Easter Island in 2013–2014 [2]. As recently reported by our group [3], ZIKV was isolated from 3 children at a school clinic in the Gressier region of Haiti in December 2014. There was no further documentation of Zika cases until January 2016, when the Haitian Ministry of Public Health and Population reported 5 laboratory-confirmed cases to the Pan American Health Organization, as part of a large outbreak occurring in multiple parts of the country. We report a case of dual DENV/ZIKV infection in a traveler to Haiti in 2016, describe the clinical presentation, and provide a phylogenetic analysis that highlights the potential role of the Haitian epidemic in the ongoing diversification and spread of ZIKV worldwide.
CASE REPORT
A 26-year-old previously healthy female student presented to the medical outpatient clinic at the University of Florida with rash and arthralgias after visiting Haiti where she was working on a research project. She arrived in Port-au-Prince on 18 January 2016 and immediately traveled to Gressier, where she lived in a compound adjacent to a school clinic where 3 ZIKV cases were identified in 2014 [3]. During the day she worked in Carrefour, a neighboring town. Despite using a 40% DEET (diethyltoluamide) insect spray twice daily, she was bitten by mosquitos in the midafternoons while working in Carrefour, and in the mornings in Gressier prior to leaving for Carrefour, and again in the evenings in Gressier upon returning to the compound. She returned to the United States after 11 days on 29 January 2016.
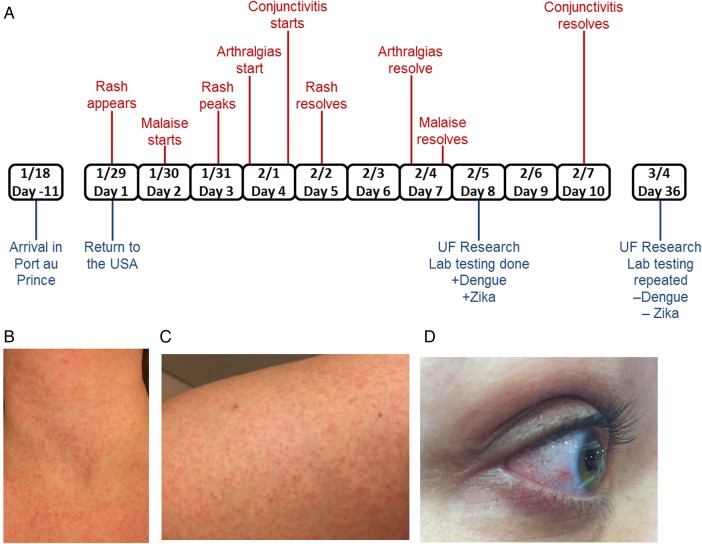
She maintained a diary of her symptoms (Figure 1A). The evening of her return, she developed her first symptom: a diffuse, nonpruritic, blanching, maculopapular rash over her abdominal area (29 January, day 1). On day 2, she developed fatigue, subjective fever, facial flushing, and malaise. On this day, she took photographs of the rash on her neck and thigh (Figure 1B and 1C). Her rash progressed to involve her entire body including palms and soles by day 3. Her subjective fever and facial flushing continued on day 3. On day 4, the patient presented to the University of Florida outpatient clinic. She denied fevers, cough, sore throat, nasal congestion, retro-orbital pain, abdominal pain, or diarrhea, and was afebrile with normal vital signs. Her physical exam was notable for a resolving rash, new bilateral conjunctival injection, and pain and swelling of her finger joints, ankles, and knees, which made it difficult to stand. The remainder of her examination was normal. The patient's rash resolved by day 5. Her joint pain resolved and she felt healthy again on day 7, although her eyes did not return to normal until day 10.
Figure 1.
Timeline, physical manifestations, and test results of patient with dual dengue virus serotype 2/Zika virus infection. A, Timeline of travel, physical manifestations, and test results. B, Fine, erythematous rash on neck and upper chest on day 2 after initial symptom onset. C, Rash on thigh, also day 2. D, Resolving conjunctivitis on day 8 after symptom onset.
Samples of serum and urine sent to the Florida State Laboratory on day 7 were negative for ZIKV viral genomic RNA (vRNA) by reverse-transcription polymerase chain reaction (RT-PCR). As per protocol, the serum was also tested for chikungunya and DENV vRNA by RT-PCR and was positive for DENV-2 only. On day 8, samples of urine and saliva were obtained for testing at our University of Florida research laboratory, and her eye showing conjunctivitis was photographed (Figure 1D). Viral genomic RNA was extracted using a QIAamp Viral RNA Mini Kit (Qiagen Inc, Valencia, California) following the manufacturer's instructions and analyzed by RT-PCR using primers as described [4] for DENV types 1–4, ZIKV [3], and chikungunya vRNAs (see Supplementary Data for more detailed viral methods). In control experiments, ZIKV primers did not amplify sequences of laboratory strains of DENV-1 through 4, and primers specific for each DENV serotype did not amplify sequences of a laboratory strain of ZIKV (African genotype). Both urine and saliva were positive for DENV-2 and ZIKV vRNAs by RT-PCR. Aliquots of the urine and saliva specimens were inoculated onto cultured cells to assess viral infectivity. Early cytopathic effects were observed 3 days postinoculation and were widespread by day 9 in mosquito and mammalian cells including LLC-MK2 (Supplementary Figure 1). Cell culture supernatants were positive for ZIKV and DENV-2 vRNAs by RT-PCR. The complete ZIKV genome was sequenced via genome walking [3]. The consensus sequence, based on vRNA purified from LLC-MK2 cells, was designated ZIKV Haiti/1/2016 (GenBank accession number KX051563.1), and was distinct from the African lineage control strain (MR 766). The dengue strain is undergoing further sequence analysis but was distinct from the New Guinea C control strain (ATCC-NR-84).
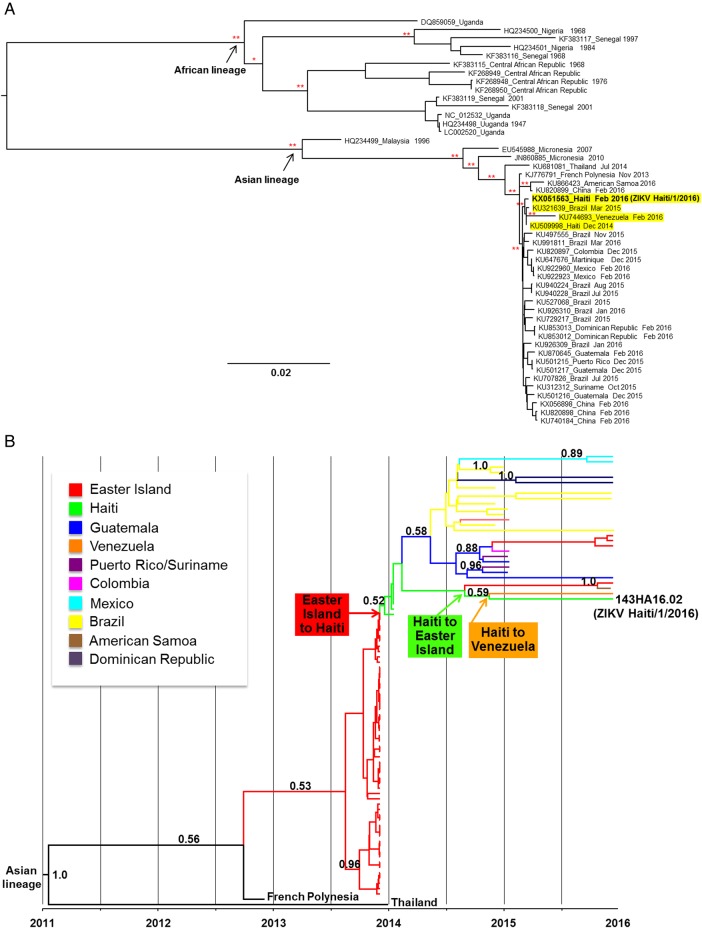
For phylogenetic analyses, all available ZIKV nucleotide sequences (http://www.ncbi.nlm.nih.gov/) were aligned [3] to generate a full genome alignment and an alignment of the NS5 gene. Using the available full genome sequences, IQ-TREE was used to infer a maximum-likelihood phylogeny [5], assess the reliability of the internal branches of the tree by bootstrapping 1000 replicates, and compute fast likelihood-based SH-like probabilities (see Supplementary Data for detailed methods). The patient's strain (ZIKV Haiti/1/2016) clustered within a well-supported clade including strains from Brazil, Venezuela, and the only other full genome Haitian strain available, isolated in 2014 in the same region of Haiti (yellow highlighted sequences, Figure 2A).
Figure 2.
Phylogenetic and phylogeography analyses of Zika virus (ZIKV) strains. A, Maximum-likelihood tree of ZIKV complete genome sequences. Branches are drawn to scale in nucleotide substitutions per site according to the bar at the bottom of the tree. *Indicates bootstrap support (>90%) or significant Shimodaira-Hasegawa-like probability (P < .01). **Indicates significant support in both tests. The 2016 Haiti sequence (ZIKV Haiti/1/2016) is in bold. Highlighted in yellow is the clade containing ZIKV Haiti/1/2016, the only other fully sequenced Haitian strain from 2014 (KU509998), a Brazilian strain from 2015 (KU321639), and a Venezuelan strain from 2016 (KU744693). B, Maximum clade credibility tree with Bayesian phylogeography reconstruction of the ZIKV NS5 gene region. Branches are scaled in time and colored according to the legend to the left where each color represents the geographic location of the sampled sequence (tip branches), as well as of the ancestral lineage (internal branches) inferred by Bayesian phylogeography. The molecular clock was calibrated by using ZIKV strains’ known sampling times and enforcing a relaxed molecular clock with a Bayesian skyline plot demographic prior (see Supplementary Methods). Posterior probabilities >0.5 are indicated along the branches. The 2016 Haitian sequence is in bold. Proposed migration events involving the Haitian strains are indicated by arrows: The red arrow indicates viral gene flow from Easter Island to Haiti; the green arrow indicates viral gene flow from Haiti back to Easter Island, and the orange arrow indicates viral gene flow from Haiti to Venezuela.
A detailed phylogeographic analysis was carried out on the NS5 alignment, which currently represents the largest dataset of ZIKV strains, with the Bayesian framework implemented in the BEAST version 1.7.4 package [6]. The analysis allowed for the reconstruction of ancestral locations (country of origin) of ZIKV lineages currently circulating in South America. Consistent with our previous findings [3], this analysis showed that South American strains emerged from Asian lineages circulating in Thailand and French Polynesia, initially spread to Easter Island, and were subsequently introduced in Haiti (Figure 2B, red arrow). Interestingly, an ancestral Haitian lineage was also at the origin of 2 strains recently sampled in Easter Island and American Samoa (Figure 2B, green and orange arrows, respectively).
On 4 March 2016, 36 days after her first symptom, urine and saliva samples were again tested and were negative for ZIKV and DENV vRNAs by RT-PCR and virus isolation.
DISCUSSION
Coinfection of ZIKV and DENV has been described, as might be expected given that the Aedes mosquito can serve as a vector for both [7]. It has been reported that approximately 80% of ZIKV infections are asymptomatic [2], as are a comparable percentage of DENV infections [8]. When symptomatic infection with either ZIKV or DENV does occur, the clinical manifestations are similar, although persons who do become ill with Zika virus usually have mild symptoms [2]. Of note, the lower extremity joint swelling our patient had has been described as unique to infection caused by ZIKV compared with that caused by DENV [9]. However, our patient with dual DENV/ZIKV infection had manifestations on the more severe end of the clinical spectrum than has been described for ZIKV infection alone, including severe arthralgias and joint swelling. While this may simply represent individual variation, it is recognized that DENV antibodies can enhance ZIKV infection, at least in an in vitro setting [10]; the impact of coinfection of ZIKV and DENV on severity of illness in patients remains to be determined.
RT-PCR–based testing for DENV and ZIKV allows for detection of dual infection if discriminatory primers are used. While detection of these viruses in blood samples is limited to approximately 1 week after symptom onset, virus may be found for longer periods of time in urine and saliva [11]. Nonetheless, low levels of viremia may affect RT-PCR results, perhaps accounting for the negative ZIKV RT-PCR State Laboratory report [12]. Our ability to detect both viruses from culture supernatants of infected cells using discriminatory primers followed by sequencing to verify the uniqueness of the viruses enabled us to establish dual infection in this patient.
The phylogenetic findings are consistent with diversification of the virus in Haiti. The maximum-likelihood tree inferred from full genome sequences shows that ZIKV Haiti/1/2016 clusters within the Asian/South American lineage, but does not share an immediate common ancestor with ZIKV Haiti/1/2016 (Figure 2A). The in-depth analysis using NS5 gene sequences based on Bayesian phylogeography suggests at least 3 separate viral gene flow (migration) events involving Haitian strains: the earliest one from Easter Island to Haiti, and 2 independent ones from Haiti back to Easter Island and Venezuela, respectively (Figure 2B). Although such events are supported by relatively low posterior probabilities in the phylogeography tree, a similar trend is strongly supported in the maximum-likelihood tree, albeit limited by the lack of available full genome sequences from several countries (ie, Easter Island). Overall, these phylogenetic analyses highlight the ongoing diversification and spread of ZIKV in the Americas, and Haiti in particular. In conclusion, the observed molecular epidemiologic patterns raise the possibility that continuous evolution and subsequent dissemination of different viral lineages in South America could eventually lead to the emergence of novel variants with distinct genomic features compared to the parent strains, which could include changes in virulence, transmissibility, and pathogenesis.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Supplementary Material
Notes
Financial support. This work was supported by institutional funding made available by the Emerging Pathogens Institute at the University of Florida.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ventura AK, Ehrenkranz NJ. Endemic dengue virus infection in Hispaniola. I. Haiti. J Infect Dis 1976; 134:436–41. [DOI] [PubMed] [Google Scholar]
- 2. Zika virus outbreaks in the Americas. Wkly Epidemiol Rec 2015; 90:609–10. [PubMed] [Google Scholar]
- 3. Lednicky J, Beau De Rochars VM, El Badry M et al. Zika virus outbreak in Haiti in 2014: molecular and clinical data. PLoS Negl Trop Dis 2016; 10:e0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santiago GA, Vergne E, Quiles Y et al. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 2013; 7:e2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 2005; 22:1185–92. [DOI] [PubMed] [Google Scholar]
- 7. Dupont-Rouzeyrol M, O'Connor O, Calvez E et al. Co-infection with Zika and dengue viruses in 2 patients, New Caledonia, 2014. Emerg Infect Dis 2015; 21:381–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Duong V, Lambrechts L, Paul RE et al. Asymptomatic humans transmit dengue virus to mosquitoes. Proc Natl Acad Sci U S A 2015; 112:14688–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goeijenbier M, Slobbe L, van der Eijk A, de Mendonca Melo M, Koopmans MP, Reusken CB. Zika virus and the current outbreak: an overview. Neth J Med 2016; 74:104–9. [PubMed] [Google Scholar]
- 10. Paul L, Carlin E, Jenkins M et al. Dengue virus antibodies enhance Zika virus infection. Florida Gulf Coast University, ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. Available at: http://biorxiv.org/content/early/2016/04/25/050112. Accessed 25 April 2016. [DOI] [PMC free article] [PubMed]
- 11. Mizuno Y, Kotaki A, Harada F, Tajima S, Kurane I, Takasaki T. Confirmation of dengue virus infection by detection of dengue virus type 1 genome in urine and saliva but not in plasma. Trans R Soc Trop Med Hyg 2007; 101:738–9. [DOI] [PubMed] [Google Scholar]
- 12. Araúz D, De Urriola L, Jones J et al. Febrile or exanthematous illness associated with Zika, dengue, and chikungunya viruses, Panama. Emerg Infect Dis 2016; 22:1515–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.