 We present a methodology for building biologically inspired, soft microelectromechanical systems (MEMS) devices.
We present a methodology for building biologically inspired, soft microelectromechanical systems (MEMS) devices.
Abstract
We present a methodology for building biologically inspired, soft microelectromechanical systems (MEMS) devices. Our strategy combines several advanced techniques including programmable colloidal self-assembly, light-harvesting with plasmonic nanotransducers, and in situ polymerization of compliant hydrogel mechanisms. We synthesize optomechanical microactuators using a template-assisted microfluidic approach in which gold nanorods coated with thermoresponsive poly(N-isopropylmethacrylamide) (pNIPMAM) polymer function as nanoscale building blocks. The resulting microactuators exhibit mechanical properties (4.8 ± 2.1 kPa stiffness) and performance metrics (relative stroke up to 0.3 and stress up to 10 kPa) that are comparable to that of bioengineered muscular constructs. Near-infrared (NIR) laser illumination provides effective spatiotemporal control over actuation (sub-micron spatial resolution at millisecond temporal resolution). Spatially modulated hydrogel photolithography guided by an experimentally validated finite element-based design methodology allows construction of compliant poly(ethylene glycol) diacrylate (PEGDA) mechanisms around the microactuators. We demonstrate the versatility of our approach by manufacturing a diverse array of microdevices including lever arms, continuum microrobots, and dexterous microgrippers. We present a microscale compression device that is developed for mechanical testing of three-dimensional biological samples such as spheroids under physiological conditions.
Introduction
Microscopic soft devices that can actively interrogate and precisely perturb the biological microenvironment through physical and chemical interactions can revolutionize therapy and biomedical research.1–3 Hydrogels have been serving as a platform to provide spatial and temporal control over the release of various therapeutic agents.4 Through appropriately designed composition and structure, they can respond to various biochemical and physical stimuli by showing conformational or optical changes.5,6 High water content and tunable mechanical properties provide physical similarity to biological matter, giving the hydrogels excellent biocompatibility and stability.7 A broad range of printing, folding, molding, and assembly methods are available for forming complex 3D hydrogel mesostructures.8 Although hydrogel microsystems have shown the unique capability of operating autonomously as sensors and actuators inside microfluidic devices,9,10 existing prototypes cannot perform complex mechanical functions with high dexterity and performance. Magnetically and acoustically actuated hydrogel structures have shown great promise for utilizing mechanical power in regenerative medicine11–15 and building miniaturized untethered machinery.16–21 However, these scaffolds cannot achieve coordination among multiple actuators, and the force and torque acting on the structures depend on their volume which does not scale down favorably. Optical manipulation methods offer high resolution22 and enable biomimetic motions of photosensitive materials.23 Yet, none of the existing formulations allow modular assembly and addressable actuation of untethered hydrogel MEMS devices within microfluidic platforms, a technological development that may become instrumental for mechanobiology.24,25
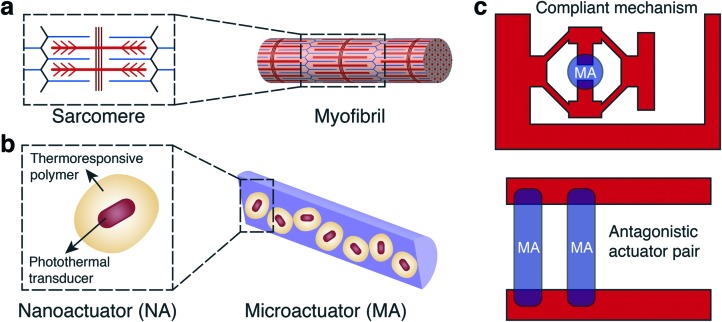
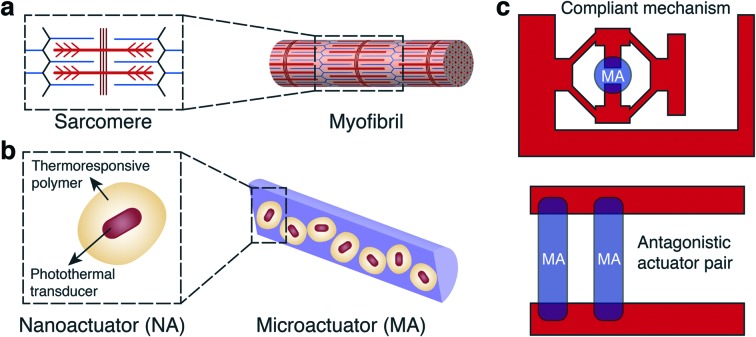
Skeletal muscle, the epitome of soft biological actuators, delivers tightly controlled actuation with unmatched power-to-weight ratio across vast size scales. The hierarchical architecture of muscle, from actomyosin motors to muscle bundles, provides scalability without sacrificing functionality. Fascicles consist of bundles of elongated muscle fibers surrounded by fibrous connective tissue, which together form a linear actuator optimized for force transmission. Artificial microactuators made out of nanoscale transducers that exhibit similar hierarchical architecture may fill this technological gap in terms of functionality and performance.26 Although assembly methods are available to organize molecular motors based on nucleic acids,27,28 proteins,29,30 or synthetic molecules31 into larger arrays, none of these constructs match physiologically relevant spatiotemporal force and deformation regimes. Another important feature underlying the spectacular performance of biological systems is the coordination among many specialized functional modules that are spatially arranged with the presence of a skeleton. The skeleton not only supports the structure of the organism, but also stores and releases energy in a controllable fashion. Inspired by these principles, we aim to physically couple hierarchical assemblies of nanoactuators with rationally-designed compliant mechanisms and develop modular soft robotic microdevices that can perform dexterous micromanipulation (Fig. 1).
Fig. 1. Inspiration, concept, and realization of bioinspired soft robotic actuators and microdevices. (a) The hierarchical sarcomere architecture within myofibrils is a great example from nature motivating the bottom up assembly of microactuators for powering larger scale machines. (b) We utilized the superior efficiency of gold plasmonic nanotransducers by either directly linking nanoactuators together using colloidal self-assembly or embedding them into soft biopolymers. (c) A variety of soft robotic microdevices are constructed by physically attaching microactuators to hydrogel mechanisms that are directly photopolymerized around the actuators without additional assembly procedures.
Here we present an experimental framework for the fabrication of soft robotic microdevices that seamlessly compliments conventional microfluidics technology. The key idea is to combine the rapid and efficient transduction of energy via the plasmonic photothermal effect at nanoscale32–35 with colloidal self-assembly36 and spatially modulated optofluidic37 additive manufacturing processes at microscale. Individual polymer coating around each gold nanorod ensures homogenous distribution of nanoactuators (NAs), thus preserving the performance of the microactuators (MAs) at multiple scales. We built a maskless microscope projection photolithography system for in situ sequential polymerization of hydrogel structures constituting monolithic compliant mechanisms. Analytical models and finite element (FE) analysis provide effective rules for designing complex machine components. Actively modulated NIR illumination enables repeatable and precise operation for the machinery.
Results and discussion
Colloidal assembly of microactuators
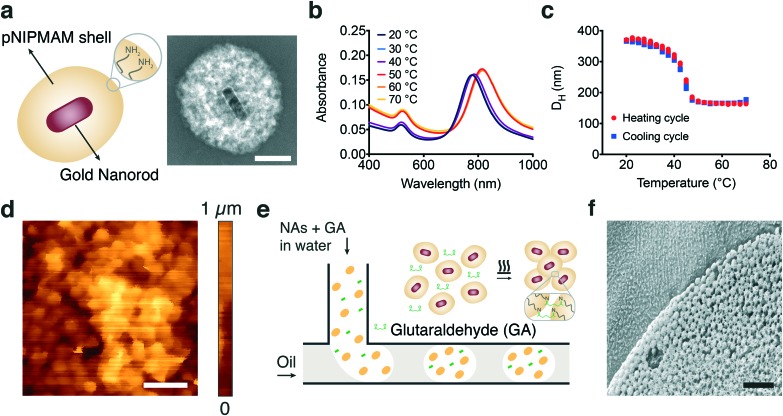
Plasmonic photothermal effect of gold nanoparticles can efficiently transduce NIR light into localized heat that drives rapid (on the order of milliseconds) and powerful (on the order of tens of piconewtons) polymer collapse at the nanoscale.38 We hypothesized that assembling MAs hierarchically from nanoscale building blocks may preserve this performance because the NAs would be uniformly distributed throughout the assembly and the pores formed in between would facilitate diffusion of water. NAs consisting of a gold nanorod core and a thermoresponsive poly(N-isopropylmethacrylamide) (pNIPMAM) shell were fabricated, as shown in Fig. 2a. Briefly, gold nanorods were synthesized using hexadecyltrimethylammonium bromide (CTAB) capped seeds in the presence of silver nitrate (AgNO3) and subsequently coated with pNIPMAM polymer using in situ free radical polymerization.38 Allylamine was added to the mixture during polymerization in order to introduce functional amine groups on the NAs for further assembly steps. The nanorods without coating were on average 95 ± 6 nm in length and 30 ± 2 nm in width and the final NA was approximately 400 nm in diameter (Fig. S1†). The NAs showed a longitudinal surface plasmon resonance peak at 785 nm as expected, while the peak shifted towards higher wavelengths with increasing temperature (Fig. 2b). This behavior is due to the increase in the refractive index during polymer collapse, which is typical of optomechanical nanomaterials.39–42 The pNIPMAM shell transitions from the hydrophilic state to the hydrophobic state above the lower critical solution temperature (LCST). This transition leads to deswelling of the hydrogel. The NAs exhibited a drastic decrease in hydrodynamic size above its LCST of 42 °C, reaching up to 50% (Fig. 2c).
Fig. 2. Fabrication and characterization of actuators. (a) A schematic representation of the nanoactuator along with the TEM image showing the thermoresponsive polymer encapsulating the gold nanorod. Scale bar, 100 nm. (b) Temperature dependent UV-vis absorbance measurements as a function of wavelength. (c) Temperature dependent DLS measurements of NA hydrodynamic size. (d) Topography scan of a representative self-assembled sheet using in-liquid atomic force microscopy. Scale bar, 1 μm. (e) Schematic illustration of the microfluidic template-assisted self-assembly process of spherical microactuators. (f) A cross-sectional cryo-SEM image of a representative microactuator. Scale bar, 2 μm.
The coupling between adjacent NAs must be strong enough to effectively transmit the force and sustain the connectivity during multiple deformation cycles. An amine–aldehyde condensation reaction was used to covalently link the NAs and assemble microactuators (MAs) in the desired form. The molecule glutaraldehyde acts as a linker between amine groups by utilizing two available reaction sites.43,44 Mixing amine functionalized NAs with glutaraldehyde and heating the solution overnight at 65 °C led to the assembly of thin-film MAs with random shapes. The surface of these films was investigated using in-liquid atomic force microscopy and the topography images revealed close-packed layers of colloidal nanoactuators forming a porous network (Fig. 2d).
A systematic characterization of actuator performance and streamlined prototyping of micromachinery requires high-throughput and repeatable synthesis of MAs with well-defined properties. Spherical MAs were fabricated using a microfluidic approach based on the break-off process of droplets in two-phase flows at a T-junction.45 An aqueous solution of NAs and glutaraldehyde forms the discontinuous phase while the continuous phase comprises the surfactant and oil mixture (Fig. 2e). The emulsion was collected and heated overnight to facilitate the crosslinking process. Consistent with the structure of the films, we observe a close-packed arrangement of NAs, while the interstices between the particles form a scaffold with uniform pores (Fig. 2f). In-liquid AFM measurements on fully swollen samples showed that the elastic modulus of actuators was 4.8 ± 2.1 kPa (n = 15) at room temperature. The microfluidic process resulted in a monodisperse suspension with less than 10% variation in size. MAs with diameters ranging from 5 ± 0.4 μm to 50 ± 4.7 μm were fabricated by tuning the channel size and flow rates.
Characterization of contraction and relaxation kinetics
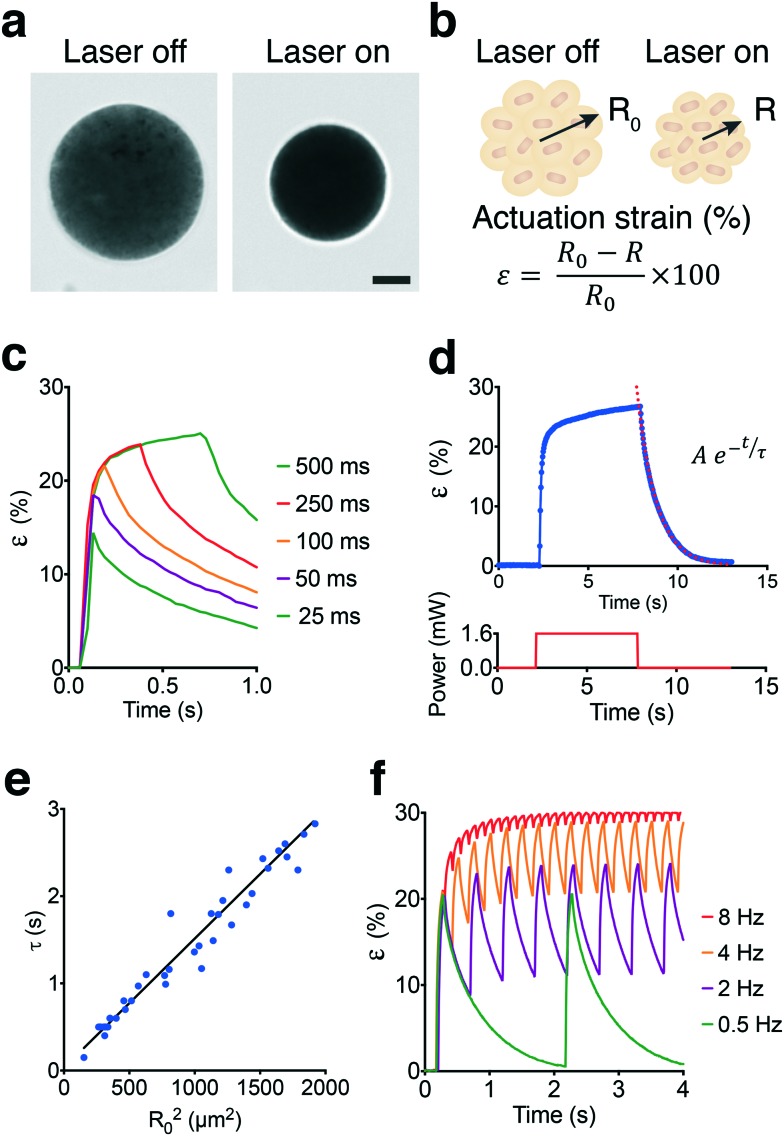
The spherical MAs were highly responsive to NIR excitation and exhibited drastic deformation upon exposure, as shown in Fig. 3a and Video S1.† Deformation was monitored by measuring the actuation strain, which is defined as percentage change in radius normalized with respect to the initial radius (Fig. 3b). The contraction phase can be divided into two periods, one representing the fast decrease in actuator size generating up to 20% strain in tens of milliseconds followed by a significantly slower period that would gradually stabilize the length within minutes. Pulse width modulation revealed that contraction was very fast as expected and predominantly completed within the first 60–80 ms of actuation at 1.4 mW of laser power (Fig. 3c). Approximately 50% of the steady-state level was reached within the first 25 ms of actuation while exceeding 70% required another 25 ms of exposure.
Fig. 3. Characterization of contraction and relaxation kinetics. (a) A representative example showing the fully contracted state of a microactuator upon NIR illumination. The laser power was set to 1.4 mW. Scale bar, 10 μm. (b) Schematic description of actuation process and measurement of strain (ε). R0 refers to the initial radius of the microactuator at the fully swollen state and R represents time-varying radius upon laser illumination. (c) Actuation strain over time for varying pulse durations. All measurements were done at 1 Hz pulse frequency with 1.6 mW laser power. (d) Actuation strain versus time plot for a single contraction–relaxation cycle. An exponential fit to the relaxation curve characterizes the relaxation constant. The lower graph shows the laser input over time. (e) Relaxation time constant (τ) with respect to the square of radius in the fully swollen state. (f) Microactuator performance is plotted over time for different actuation frequencies. All measurements were done at 100 ms pulse width with 1.6 mW laser power.
In conventional bulk hydrogel actuators, the time to swell is determined by the polymer relaxation time, τ, which characterizes the increase of size in time.46 The size of MAs followed a similar temporal evolution and increased exponentially with time during the relaxation phase until the polymer went back to its original state (Fig. 3d). From the data, we obtained τ values ranging from 0.4 to 2.6 seconds as a function of the original MA size (Fig. 3e). The linear relationship between the cross-sectional area of the actuator and the corresponding relaxation time suggests that relaxation was controlled by diffusion. The diffusion coefficient for our colloidal scaffold was estimated as 675 μm2 s–1 which is approximately two orders of magnitude higher than bulk pNIPMAM hydrogels.47 Faster actuation kinetics can be achieved by sacrificing actuation strain. We modulated the frequency of 50 ms laser pulses and recorded the time-varying response (Fig. 3f). At low frequencies, the actuator could reach 65–70% of the maximum actuation strain and had time to relax back to the fully swollen state. The amplitude of the strain decreased with increasing frequency, which corresponds to 12% strain at 2 Hz and 8% strain at 4 Hz (Video S2†). At frequencies above 8 Hz, the strain shows less than 2% deviation with respect to the baseline over time. Compared to continuous exposure, driving the actuator at high frequencies would generate a comparable strain with higher power efficiency. The actuation strain can be directed to follow an arbitrarily shaped waveform provided that the duration of signal is slow enough not to interfere with the natural contraction and relaxation kinetics.
Design and production of compliant hydrogel mechanisms
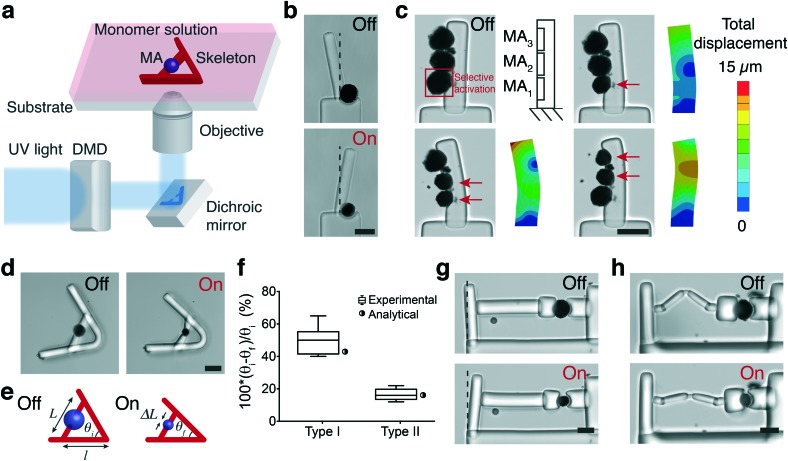
Fig. 4a shows a schematic representation of the fabrication of structures using in situ polymerization of a hydrogel monomer. We chose poly(ethylene glycol) diacrylate (PEGDA) as the hydrogel for manufacturing the skeleton due to its tunable stiffness, ease of polymerization, and temperature-independent swelling properties. Rapid and controllable polymerization kinetics at the microscale permitted formation of structures with feature sizes ranging from 10 μm to 500 μm in less than 100 ms. We first utilized actuators as active hinges to control the rotation of a cantilever beam with respect to a fixed base (Fig. 4b). The rotational motion can be precisely controlled by tuning the power and duration of NIR exposure. A 25% strain applied by the 40 μm actuator rotated the arm by 20°. Next, we polymerized a cantilever beam that has several MAs on its side (Fig. 4c). Prior to polymerization, we brought MAs together using a micromanipulator. MAs were located with 3 μm position error at their target locations within 20 s (depending on the initial and final locations) within a workspace of 400 × 400 μm2 using teleoperation. NIR illumination selectively activated MAs according to the pre-defined pattern. The longitudinal tensile strain generated by the contraction of MAs at each segment is converted to reversible bending of the hydrogel beam. The beam behaves as a bilayer bending actuator and the curvature can be actively controlled by tuning the location and power of NIR stimulation. Selective activation of MAs led to varying manipulator geometry that follows a continuous curvature kinematics, and the backbone pose can be successfully captured by the FEM analysis (Fig. 4c). The PEGDA structures were modeled as an elastic material as strains were in the linear elastic range. We applied empirical MA deformation values as boundary conditions in the model and the elastic modulus was measured as 550 ± 120 kPa using in-liquid AFM indentation tests.
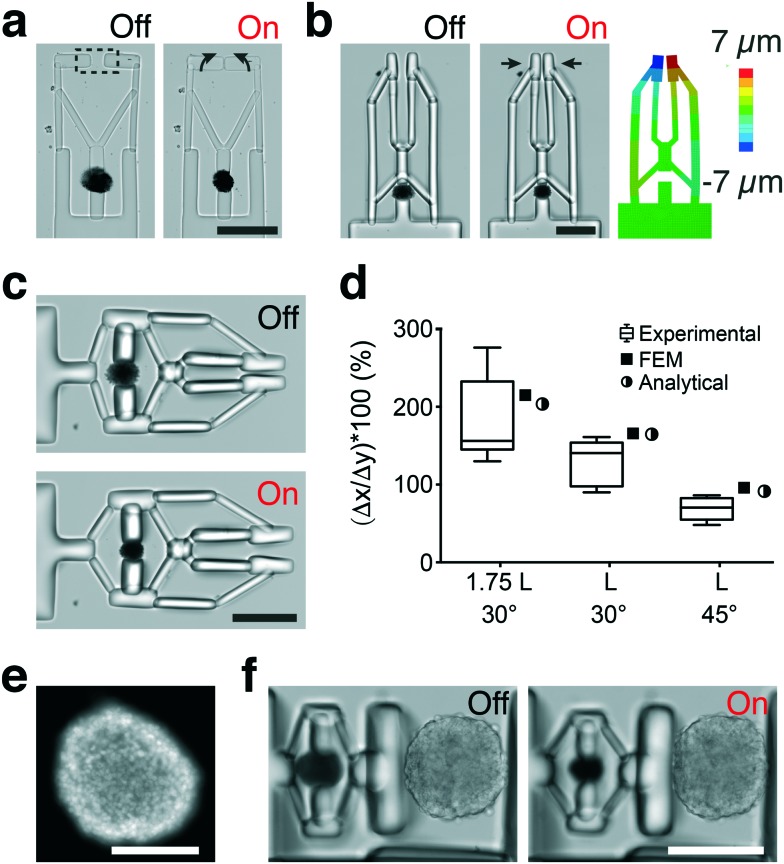
Fig. 4. In situ fabrication of hydrogel mechanisms and manipulators (a) the fabrication methodology for building multibody microsystems. Digital maskless lithography inside microchannels drives the polymerization of compliant mechanisms physically coupled to the actuators. (b) The rotation of a cantilever beam using a single MA serving as an active hinge. (c) The control of deformation of a microscale bending actuator using structured illumination. FEM analysis predicts the deformation patterns. (d) Lever system for controlling the transmission of stress and strain. (e) The schematic description of the geometrical design parameters in the lever system. (f) The change in the transmission angle θf–θi normalized with the initial value θi for two different lever arm designs. We compared two types of devices having the same transmission angle of 60° and varying arm length l (type I: 100 μm and type II: 250 μm). Experimental measurements (n = 8) are compared with the calculated values for the initial design parameters. (g) A cantilever system for measuring the force generated by a single actuator. (h) A cantilever system with a serial elastic spring connection. Spring deformation blocks force transmission to the cantilever. All scale bars represent 50 μm.
We built and tested a series of mechanisms actuated by a single MA that would demonstrate the fundamental principles for the development of advanced machinery. We constructed lever systems to gain control over the displacement by polymerizing an elastic beam with distributed compliance around the MA (Fig. 4d). The contraction of the MA moves the lever arms toward each other (flexion) while opening (extension) is achieved with the removal of the actuator force due to the stored energy, as shown in Video S3.† The angle of actuator insertion and the distance from the curved region to the point of force application determine the angular displacement according to the following trigonometric formula:
| θf = 2 sin–1(sin(θi/2) – ΔL/2l) | 1 |
where ΔL is the deformation of the actuator, l is the distance from the hinge to the point of force application, θi is the initial angle and θf is the final angle between the arms (see Fig. 4e). The final geometry of machines slightly deviates from the CAD drawings due to the swelling of MAs and the consequent extension of the associated flexible parts upon submerging in water. In the case of lever systems, the angle between the arms of the beam increases by 10° ± 2° depending on the initial design and the size of the actuator.
The experimental measurements were in accordance with the analytical calculations as shown in Fig. 4f, and these results demonstrate that the transformation of actuator deformation into arm closure can be controlled by the geometric design of the lever system (see Video S3†). As an alternative approach, we fabricated levers with slender regions serving as single-axis flexure hinges. Complex deformation patterns can be generated by increasing the number of hinges and tuning the flexibility of connectors (Fig. S2†). We next constructed a cantilever system where the deformation was applied from one side of the beam to characterize the force output of a single MA (Fig. 4g). We used linear bending theory and experimental measurements to link the measured cantilever deflection to the amount of force generated by the actuators. The bending modulus of the beams was measured using a commercially available MEMS capacitive force sensor (Fig. S3†). The modulus was found as 800 ± 85 kPa (n = 8) from the slope of the force-displacement curves. The force applied by a 40 μm actuator at 20% strain was 10.3 μN, which corresponds to a uniaxial compressive stress of 8.1 kPa. The mass of a single MA is approximately 38.2 nanograms and the actuation speed is 500 μm s–1 during the initial contraction phase, which together provides a power-to-mass ratio of 0.14 kW kg–1. The maximum stress and relative stroke of engineered skeletal muscle actuators were previously reported as 0.1–2 kPa and 0.01, respectively.48–51 Biohybrid actuators based on cardiomyocytes perform better with 10–30 kPa stress and 0.2–0.4 relative stroke.51 Our results showed that the performance of optomechanical MAs is comparable to bioengineered actuators within a similar size range. Transmission of force and subsequent deformation can be controlled by installing passive elastic elements in series or parallel with the MAs (Fig. S2†). As a representative example, the actuation is completely decoupled from the cantilever displacement due to the extensibility of the engineered spring (Fig. 4h).
Development of soft robotic microgrippers
We applied our material system to demonstrate the development of dexterous biomanipulation tools for microscopic samples. With a slight change in the blueprint, levers can be converted into an angular jaw microgripper that operates like a slider-rocker. In this configuration, the gripper jaws are synchronously opened and closed around a central pivot point, moving in an arching motion (Fig. 5a). Next, we integrated an amplification mechanism by incorporating a second pair of cantilever beams connected to both the fingers and the actuator unit (Fig. 5b). This arrangement constraints the fingers to follow a straight line, a feature that is known to improve accuracy of manipulation. FEM analysis of the second generation microgrippers showed very good quantitative agreement for strain, as shown in Fig. 5b. A 10% strain applied by the 40 μm actuator leads to full closure of the fingers with the aid of the amplification mechanism. The amplification ratio and stress distribution on the mechanism can be further improved by optimizing the design (Fig. S4†). The grippers can be operated for minutes under continuous NIR exposure without showing any sign of fatigue (Fig. S4†).
Fig. 5. Development of soft robotic microdevices actuated by spherical MAs. (a) An angular jaw gripper with rotating arms. (b) The parallel jaw gripper and the corresponding simulation results. (c) Flextensional mechanism integrated into the gripping mechanism to convert contraction of the actuator into extension of the arms. (d) The free displacement of the arm is plotted for different geometrical mechanism designs along with the simulation results and solutions of the analytical equations. (e) Fluorescence image of a representative spheroid stained for the nuclei. (f) Compression of a spheroid. The forward movement of the piston is provided by the inversion and amplification of actuator strain using a flextensional mechanism. Scale bars, 100 μm.
The mechanism shown in Fig. 5b was combined with a bridge-type flextensional mechanism to develop the microgripper shown in Fig. 5c that opened upon NIR exposure (Video S4†). Naturally closed configuration would allow holding samples and application of a constant compressive stress for extended periods in the absence of external actuation. Flextensional mechanisms provide control over the direction and amplitude of displacement. The output displacement Δx as a function of input displacement Δy is given by:
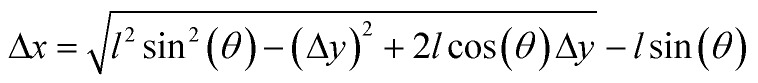 |
2 |
An increase in the nominal linkage angle θ will decrease Δx. Another important parameter is the length of the linkage arms l, the longer the arm is the larger the output displacement. We fabricated three different prototypes by varying θ and l (Fig. S5†). The measured displacement of the mechanism was found to reasonably match that predicted by the analytical equation and FE analysis (Fig. 5d).
We next focused on the design of remotely controlled machines for the application of mechanical stimuli to 3D biological samples. Previous work has shown that compressive stress generated by confined growth drives cancer cells toward invasive phenotypes.52 We developed a soft robotic compression device that allows long-term dynamically modulated mechanical stimulation of 3D culture systems. The compression device utilized the same flextensional mechanism shown in Fig. 5c to convert the contraction of the MA into extension of the arm, which as a result moves the piston forward. We monitored the displacement of the piston during repeated actuation cycles. The performance of the compressor does not change after 1000 actuation cycles, as shown in Fig. S6a.† Previous work has shown that PEGDA hydrogel structures49 and gold nanorod-pNIPMAM composites38 do not have any cytotoxic effects on cells. We verified this biocompatibility by culturing cells on the fabricated microdevices and performing a live/dead assay. We also calculated the heat distribution inside the actuators and their surface temperature. Live/dead assays confirmed cell viability after two day culture of the system (Fig. S7†). 3D simulations mapping MA temperature showed that there is minimal thermal effects on cells because the actuator core collapses without significantly altering its surface temperature (Fig. S8†). This is especially true during the operation of the compression device as the sample is located away from the actuator.
Spheroids were self-assembled from epithelial cells using a multi-well culture plate (Fig. 5e). They were placed inside the confinement using a robotically controlled microaspiration system. The device pushed the sample against an engineered PEGDA wall, which led to the compression of the sample with micrometer resolution. A single 50 μm diameter MA can deliver up to 20 μm displacement with the aid of the amplification provided by the mechanism, which corresponds to 10% deformation for a 200 μm diameter spheroid. The stress acting on the sample during compression can be modulated by changing the design of the piston (Fig. S6b†). The level of compression can be increased by changing the design of the mechanism or increasing the size of the MA. As diffusion will become problematic for actuators larger than a certain size (150 μm), once again, a programmable colloidal assembly approach must be followed. We used the same amine–aldehyde condensation reaction to covalently link MAs and form dimers, trimers, and tetramers via guided self-assembly inside microfabricated compartments (Fig. S9†).
Soft robotic devices driven by 3D printed microactuators
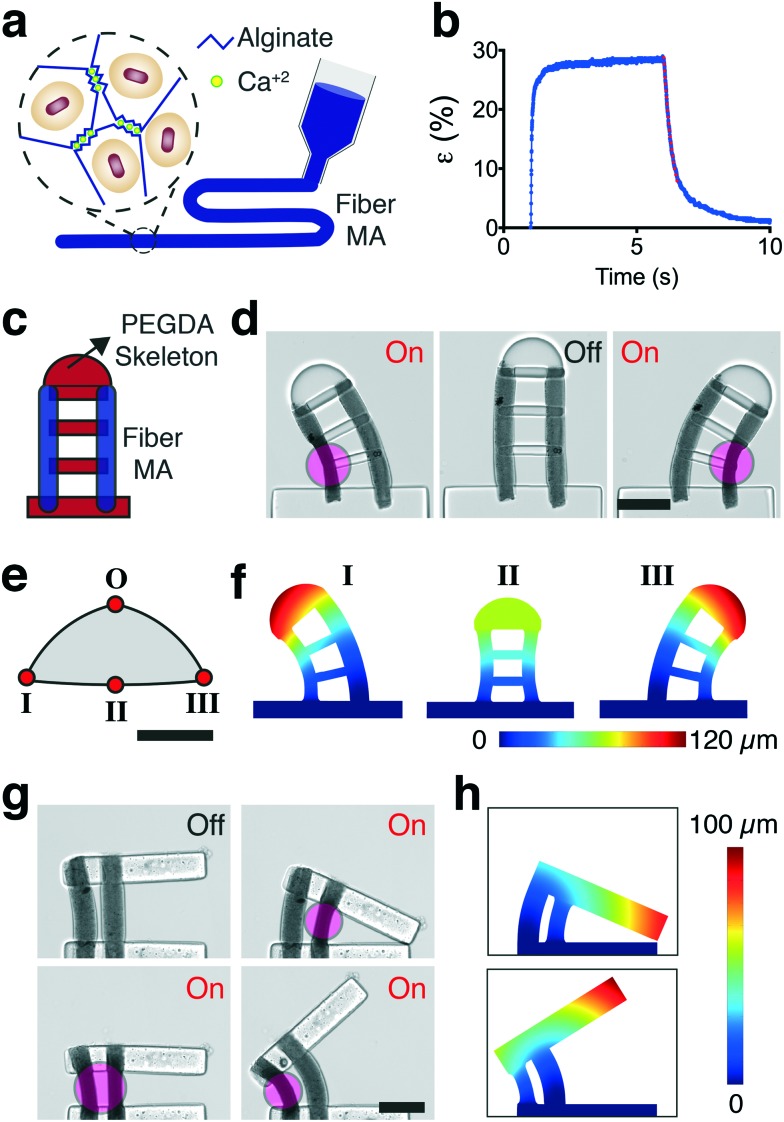
The patterning of MAs can be performed in a more systematic way with the aid of additive manufacturing techniques. We developed an alternative strategy for the fabrication of MAs in the form of nanocomposite elastic microfibers. The fiber MAs consist of NAs physically encapsulated inside ionically-crosslinked porous alginate gels (Fig. 6a). The nanocomposite was aspirated into a glass micropipette using capillary action and subsequently gelled upon exposure to calcium ions prior to extrusion. The extruded fiber MAs were highly responsive to NIR excitation and exhibited large deformation (Fig. 6b and Video S5†), comparable to the levels reached by spherical MAs even though the NA volume was lower (∼20%). The contraction kinetics also resembled the performance of the spherical MAs, 80% of the steady-state level was reached within the first 25 ms of actuation, as shown in Fig. 6b. The actuation strain quantifies percent change in the thickness normalized with respect to the initial thickness of the fiber. By tuning the laser power and alginate concentration, we could reach up to 45% strain with fiber MAs. The relaxation, on the other hand, displayed a staged process where a fast relaxation phase due to release of elastic energy was followed by a slower diffusion process. The relaxation rate of the first stage during which the fiber regains 70% of its original size was 0.5 seconds for 50 μm thick fibers. With these specifications, the fiber MAs can deliver 10% strain at 2.5 Hz and 3% strain at 10 Hz (Video S6†).
Fig. 6. Development of soft robotic microdevices actuated by fiber MAs. (a) Schematic illustration of the extrusion process and content of the nanocomposite fiber MAs. (b) Actuation strain versus time plot for a single contraction–relaxation cycle. An exponential fit to the first stage of the relaxation curve characterizes the relaxation constant. (c) The PEGDA skeleton is polymerized around the printed fiber MAs while forming physical encapsulation of fibers within the structure. (d) Selective NIR exposure leads to continuous control over the curvature of the manipulator. (e) The 2D configuration space of the device plotted by generating a polynomial fit passing through the extreme positions provided by the FEM simulations shown in (f). The non-actuated original location of the tip position is denoted by O. (g) Soft robotic microgripper controlled by an antagonistic fiber MA pair. (h) Displacement contour plot showing the displacement of the arm corresponding to the two extreme cases, fully open and fully closed. All scale bars represent 100 μm.
The pneumatic extrusion method allows us to print fibers at specified locations inside the monomer solution prior to polymerization of the PEGDA structures. We built two proof-of-concept devices actuated by multiple fiber MAs. The first device is in the class of continuum robots; continuously bending, infinite-degree-of-freedom robots with elastic structures. The device consists of two fiber MAs connected through a discrete backbone, as shown in Fig. 6c. The tip position of the robot was freely and precisely controlled within the 2D configuration space by modulating the optical power and spatial distribution of NIR exposure (Fig. 6d). We mapped the configuration space for the tip of the robot by simulating the deformation at the geometric extremes using FEM analysis (Fig. 6e). We used thermal expansion to model the strain response of the actuators. MAs were assigned an experimentally derived modulus and a negative thermal expansion coefficient according to experimentally derived strains. In-liquid AFM measurements on fully swollen samples showed that the elastic modulus of the fiber MAs was 3.5 ± 1.9 kPa (n = 15) at room temperature. The PEGDA structures were again modeled as an elastic material and assigned a thermal expansion coefficient of zero. FEM analysis successfully captured the morphological changes upon selective NIR exposure, as shown in Fig. 6f.
Second system is an advanced microgripper actuated by an antagonistic pair of fiber MAs, as shown in Fig. 6g. The robot achieves large reversible deformation in opening and closing modes by selective activation of the actuators (Video S7†). While the agonist contracts, the antagonist extends thanks to the flexibility provided by the biopolymer. FE simulations of displacement matched the experimental results, as shown in Fig. 6h. This arrangement paves the way to adjust the stiffness of the actuation machinery to achieve sophisticated motion trajectories and precise control of interaction forces when handling soft and sensitive biological samples.53
Conclusions
We present a scalable and modular framework that combines spatially resolved manufacturing and control strategies for the development of MEMS hydrogel devices. The method of fabrication is simple and flexible. By varying the number and distribution of actuators or playing with the design of the compliant mechanisms, we can generate a repertoire of complex micromachines. The MAs show outstanding performance with respect to the achievable maximum actuation strain (30%) and fast contraction–relaxation kinetics (order of milliseconds). The colloidal assembly of NAs into spherical MAs speeds the diffusion of fluid by forming a porous network with a diffusion coefficient of 675 μm2 s–1. Furthermore, the enhanced kinetics are achieved without significant loss in mechanical strength as the Young's modulus of MAs (4.8 ± 2.1 kPa) is comparable to the stiffness of bulk hydrogels. The mechanical and geometric properties of the actuators become programmable with the addition of alginate. The performance can be further improved with the reinforcement of the architecture using fibrillar proteins such as collagen or cellulose. With proper physical or chemical coupling to NAs, any soft biopolymer can serve as the matrix. This flexibility in biopolymer choice may facilitate mechanochemical interfacing of actuated matter with biological samples.
The class of programmable, soft actuated hydrogel microdevices presented in this work has potential for a wide range of applications including generation of physiologically or pathologically relevant mechanical stimuli for in vitro mechanobiology studies. The mechanical properties of the spheroids can be measured by incorporating a flexible cantilever according to the characteristics of the sample. The bending of the cantilever can be optically tracked to find the force exerted on the beam and to generate the corresponding stress–strain curves. Combination of longitudinal, transverse, and/or oblique actuator arrangements can lead to the development of highly adaptive continuum microrobots providing remotely controlled 3D motion. The hydrogel materials employed in the development of our microdevices have moduli on the order of tens of kPa which is comparable to that of biological tissues, providing an inherently safer alternative for biomedical interfacing compared to other robotic approaches. Furthermore, the in situ maskless polymerization technique allows construction of machines directly inside lab-on-a-chip devices. Combined with precise microfluidic manipulation,54 presented technology can provide simultaneous control over mass transport and biomechanics with single cell resolution. The penetration depth of NIR illumination and the biocompatibility of the ingredients also suggest potential for in vivo applications.
Experimental
A detailed description of the fabrication and characterization of the nanoactuators can be found in the supplementary information. All reagents were purchased from Sigma Aldrich and used as received, unless otherwise stated.
Fabrication and characterization of the microactuators
Self-assembled sheets and microactuators were fabricated by an amine–aldehyde condensation reaction based on a reported protocol.43 The stock NA solution was concentrated to 12 mg ml–1 by centrifuging and re-dispersing in 20 vol% glutaraldehyde in milli-Q water. The NA solution concentration was calculated based on the dry mass of a batch of stock NA solution. This solution was heated overnight in an Eppendorf tube at 65 °C to synthesize self-assembled sheets. The spherical microactuators were fabricated using a T-junction microfluidic device and the NEMESYS low pressure microfluidic pump. The discontinuous phase was the reaction solution described above while the continuous phase was a solution of 2% surfactant (Picosurf) in fluorinated oil (Novec 7500). The resulting emulsion of spherical MAs was broken with 20% 1H,1H,2H,2H-perfluoro-1-octanol (PFO, 97%, Sigma) in fluorinated oil and washed three times with ethanol to remove unreacted GA. The internal structure of the MAs was investigated using cryo-SEM (Nvision 40 Zeiss). Prior to imaging, an aqueous suspension of MAs was frozen in liquid N2, fractured, etched, and sputter coated (Baltec MED 020) and transferred to the SEM with a cold chamber (Leica EM VCT100). The nanocomposite fiber actuators were fabricated by dispersing 40 mg ml–1 NAs in 1 vol% sodium alginate in milli-Q water. The nanocomposite was transferred into a glass microcapillary by capillary forces and subsequently immersed in a 10 mM CaCl2 solution for 12 h to allow Ca2+ to diffuse in. The crosslinked fiber was extruded from the capillary using a pressure dispenser.
Experimental platform and video processing
All experiments were performed using a Nikon Ti Eclipse motorized inverted microscope and phase-contrast images were captured with an ORCA-Flash4.0 CMOS camera (Hamamatsu). A 785 nm laser (Thorlabs) beam coupled through the FRAP unit provided NIR illumination. The laser beam was guided through 10× or 20× objectives (Nikon) and an NIR filter cube (Chroma). The videos were captured at full pixel resolution (2048 × 2048) with frame rates ranging from 33 fps to 200 fps depending on the requirement of the experiment. A program based on an edge-detection algorithm in Matlab (Mathworks, MA) was used to measure the size of MAs from time-lapse videos.
Digital maskless lithography
A programmable digital micromirror device module (Andor Technology) connected to the motorized inverted microscope serves as a prototyping platform for maskless projection photolithography. The platform projects light provided by an ultraviolet LED source (365 nm) using a computer-aided design (CAD)-based digital blueprint, which initiates subsequent free-radical polymerization of photosensitive hydrogel materials. 2D drawings defined the planar shape of the structures, whereas the height was simply controlled by the channel size. Overlapping regions between the hydrogels and the MA were created in the polymerization process to ensure a mechanically stable connection between the two components. The modulated beam was projected through a 20× microscope objective onto the substrate where we had MAs suspended in poly(ethylene glycol) diacrylate (PEGDA, 700 kDa) monomer solution supplemented with 2-hydroxy-2-methylpropiophenone (DAROCUR) photoinitiator. The microfluidic devices were fabricated from poly-(dimethylsiloxane) (PDMS) using soft lithography.55
Atomic force microscopy measurements
Imaging and force spectroscopy were all performed in liquid with NanoWizard 4a Bioscience AFM (JPK Instruments). Imaging was done using qp-BioAC-Cl-10 cantilevers (Nanosensors) and CB2 tip with a measured stiffness of 0.099 N m–1. Force spectroscopy measurements were done with custom colloidal tip cantilevers at room temperature and 50 °C with 15 minutes of settling time. Polystyrene microbeads (6 μm, Polysciences) were dispersed onto a glass slide, dried and bound to cantilevers (HQ:CSC37, MikroMasch) using a glue (M-Bond 610 adhesive Kit, SPI). The colloidal tip cantilever had a measured stiffness of 0.374 N m–1. Force curves were obtained on 16 microactuators and each indentation was performed three times. Prior to both imaging and force spectroscopy, samples were transferred to a sugar solution and pipetted on to poly-lysine pretreated plastic Petri dishes to facilitate immobilization onto the surface. Each indentation measurement was analyzed by JPK data processing software using the standard Young's modulus calculation approach offered.
Mechanical characterization of hydrogel structures
A commercial MEMS force sensor (FT-S1000-LAT, FemtoTools) with a resolution of 0.05 μN was mounted on a motorized XYZ micromanipulator (SLC-2040, SmarAct GmbH) using a 3D printed adapter. The force sensor was positioned manually using the corresponding controller while being visually monitored using the microscope. The base of the PEGDA cantilevers were polymerized around PDMS pillars microfabricated on the surface of a glass chamber filled with deionized water, mounted atop the motorized microscope stage. The force sensor was powered by a programmable linear power supply (Keysight E3631A), and output was measured using a precision multimeter (Keysight 34465A). Force measurements were obtained using the factory calibration of the sensor. The Young's modulus of PEGDA structures was determined by measuring the slope of the force/deflection curves.
Finite element models
The numerical simulations were performed in COMSOL Multiphysics finite element analysis software. The models simulate the solid mechanics of the microstructures and do not take into account the hydrodynamic response of the surroundings. PEGDA structures were modeled as linear elastic substrates with a Young's modulus of 500 kPa and Poisson's ratio of 0.33. 3D CAD drawings were created in CATIA (Dassault Systemes) and imported to FEM software for further analysis. We either applied experimentally measured actuator displacements as boundary conditions or recapitulated the contraction of soft actuator with heating of materials in the simulations. See Table S1† for the list of input parameters. Accuracy of the mesh was ascertained through a mesh refinement study based on the experimental strain values. 3D heat distribution of the microactuators was calculated by finite element simulations using the heat transfer in solids physics module. We modulated the laser power and the size of the microactuator to systematically report maximum temperature values at steady state. We verified that during the transient laser heating the temperature reaches steady state conditions. We assumed that the laser intensity is uniform and heat diffuses out uniformly from the surface of the gold nanorod. The absorption cross section of gold nanorods was estimated as 20 000 nm2 based on a previously published report.41 The heat capacity and the thermal conductivity of the hydrogel was assumed to be the same as that of the water.41 Thermal conductivity of the gold was taken as 317 Wm K–1.38 A constant temperature of 37 °C was applied as the boundary condition for the surfaces of the bounding box.
Viability assay
Madin–Darby canine kidney (MDCK) cells were cultured on plastic flasks in MEM with Earle's salts supplemented with 5% FBS, 2 mM l-glutamine, and 1% penicillin–streptomycin. MDCK cells were seeded on a well-plate containing several microdevices and cultured overnight. The cells were stained using a viability/cytotoxicity assay kit from Biotium according to the manufacturer recommendations. The kit contains the cytoplasmic dyes Calcein AM for staining the living cells (green) and ethidium homodimer III for staining the dead cells (red). Briefly, 2 μM Calcein AM and 4 μM ethidium homodimer III were mixed in PBS solution. Cells were incubated inside this solution for 30 minutes at 37 °C. The solution was then replaced with complemented medium. Cells were visualized using a Nikon Ti Eclipse motorized inverted fluorescence microscope.
Spheroid culture
All reagents for cell culture were purchased from Thermo Fisher Scientific. HEK293T cells were cultured in Dulbecco's modified Eagle medium (DMEM/F-12) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Cells were passaged upon achieving confluency at a 1 : 4 ratio using 0.25% trypsin-EDTA and not kept longer than 20 passages. All experiments were done using cells tested for mycoplasma negative. A 96 well plate coated with PDMS (Sylgard) forming a concave surface was used to promote spheroid formation. The devices were sterilized in 70% ethanol followed by UV sterilization for 15 min and subsequently treated with 2% pluronic acid solution for 10 min at room temperature. Around 2000 cells were transferred to each well to obtain spheroids with a diameter between 200–250 μm. The nuclei were labeled using the cell-permeant Hoechst 33342 stain.
Author contributions
M. S. S. designed the study; B. O., R. P., A. B., L. P., and C. A. performed experiments; E. O. conducted computational analysis; B. O. and R. P. analyzed data; M. K. fabricated microfluidic devices, B. O., R. P. and M. S. S. wrote the manuscript with contributions from all authors.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the European Research Council under the ERC grant agreement ROBOCHIP (714609). B. O. gratefully acknowledges the Early Postdoc Mobility Fellowship (P2EZP2-168838) provided by the Swiss National Science Foundation (SNSF). A. B. and F. S. are grateful to the SNSF for financial support.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8lc01200h
References
- Hu C., Pané S., Nelson B. J. Annu. Rev. Control Robot Auton. Syst. 2018;1:53–75. [Google Scholar]
- Nocentini S., Parmeggiani C., Martella D., Wiersma D. S. Adv. Opt. Mater. 2018:1800207. [Google Scholar]
- Palagi S., Fischer P. Nat. Rev. Mater. 2018:1. [Google Scholar]
- Li J., Mooney D. J. Nat. Rev. Mater. 2016;1:16071. doi: 10.1038/natrevmats.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart M. A. C., Huck W. T., Genzer J., Müller M., Ober C., Stamm M., Sukhorukov G. B., Szleifer I., Tsukruk V. V., Urban M. Nat. Mater. 2010;9:101. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- Ionov L. Adv. Funct. Mater. 2013;23:4555–4570. [Google Scholar]
- Sun J.-Y., Zhao X., Illeperuma W. R., Chaudhuri O., Oh K. H., Mooney D. J., Vlassak J. J., Suo Z. Nature. 2012;489:133. doi: 10.1038/nature11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang F., Yan Z., Ma Q., Li X., Huang Y., Rogers J. A. Nat. Rev. Mater. 2017;2:17019. [Google Scholar]
- Beebe D. J., Moore J. S., Bauer J. M., Yu Q., Liu R. H., Devadoss C., Jo B.-H. Nature. 2000;404:588. doi: 10.1038/35007047. [DOI] [PubMed] [Google Scholar]
- He X., Aizenberg M., Kuksenok O., Zarzar L. D., Shastri A., Balazs A. C., Aizenberg J. Nature. 2012;487:214. doi: 10.1038/nature11223. [DOI] [PubMed] [Google Scholar]
- Zhao X., Kim J., Cezar C. A., Huebsch N., Lee K., Bouhadir K., Mooney D. J. Proc. Natl. Acad. Sci. U. S. A. 2011;108:67–72. doi: 10.1073/pnas.1007862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N., Kearney C. J., Zhao X., Kim J., Cezar C. A., Suo Z., Mooney D. J. Proc. Natl. Acad. Sci. U. S. A. 2014:201405469. doi: 10.1073/pnas.1405469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezar C. A., Roche E. T., Vandenburgh H. H., Duda G. N., Walsh C. J., Mooney D. J. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1534–1539. doi: 10.1073/pnas.1517517113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin S. Y., Poh Y. C., Kohler A.-C., Compton J. T., Hsu L. L., Lau K. M., Kim S., Lee B. W., Lee F. Y., Sia S. K. Sci. Robot. 2017;2:eaah6451. doi: 10.1126/scirobotics.aah6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P., Hu B., Leow W. R., Wang X., Loh X. J., Wu Y.-L., Chen X. Adv. Mater. 2018:1800572. doi: 10.1002/adma.201800572. [DOI] [PubMed] [Google Scholar]
- Lee H., Kim J., Kim J., Chung S. E., Choi S.-E., Kwon S. Nat. Mater. 2011;10:747. doi: 10.1038/nmat3090. [DOI] [PubMed] [Google Scholar]
- Ahmed D., Lu M., Nourhani A., Lammert P. E., Stratton Z., Muddana H. S., Crespi V. H., Huang T. J. Sci. Rep. 2015;5:9744. doi: 10.1038/srep09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-W., Sakar M. S., Petruska A. J., Pané S., Nelson B. J. Nat. Commun. 2016;7:12263. doi: 10.1038/ncomms12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed D., Dillinger C., Hong A., Nelson B. J. Adv. Mater. Technol. 2017;2:1700050. [Google Scholar]
- Kaynak M., Ozcelik A., Nourhani A., Lammert P. E., Crespi V. H., Huang T. J. Lab Chip. 2017;17:395–400. doi: 10.1039/c6lc01272h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozuyuk U., Yasa O., Yasa I. C., Ceylan H., Kizilel S., Sitti M. ACS Nano. 2018;12:9617–9625. doi: 10.1021/acsnano.8b05997. [DOI] [PubMed] [Google Scholar]
- Hu W., Ishii K. S., Fan Q., Ohta A. T. Lab Chip. 2012;12:3821–3826. doi: 10.1039/c2lc40483d. [DOI] [PubMed] [Google Scholar]
- Palagi S., Mark A. G., Reigh S. Y., Melde K., Qiu T., Zeng H., Parmeggiani C., Martella D., Sanchez-Castillo A., Kapernaum N. Nat. Mater. 2016;15:647. doi: 10.1038/nmat4569. [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Wong P. K., Park J., Levchenko A., Sun Y. Annu. Rev. Biomed. Eng. 2009;11:203–233. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- Polacheck W. J., Li R., Uzel S. G., Kamm R. D. Lab Chip. 2013;13:2252–2267. doi: 10.1039/c3lc41393d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck W. T. Mater. Today. 2008;11:24–32. [Google Scholar]
- Marras A. E., Zhou L., Su H.-J., Castro C. E. Proc. Natl. Acad. Sci. U. S. A. 2015;112:713–718. doi: 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopperger E., List J., Madhira S., Rothfischer F., Lamb D. C., Simmel F. C. Science. 2018;359:296–301. doi: 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- Hess H. Annu. Rev. Biomed. Eng. 2011;13:429–450. doi: 10.1146/annurev-bioeng-071910-124644. [DOI] [PubMed] [Google Scholar]
- Sato Y., Hiratsuka Y., Kawamata I., Murata S., Shin-ichiro M. N. Sci. Robot. 2017;2:eaal3735. doi: 10.1126/scirobotics.aal3735. [DOI] [PubMed] [Google Scholar]
- Chen J., Leung F. K.-C., Stuart M. C., Kajitani T., Fukushima T., van der Giessen E., Feringa B. L. Nat. Chem. 2018;10:132. doi: 10.1038/nchem.2887. [DOI] [PubMed] [Google Scholar]
- Ding T., Valev V. K., Salmon A. R., Forman C. J., Smoukov S. K., Scherman O. A., Frenkel D., Baumberg J. J. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5503–5507. doi: 10.1073/pnas.1524209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourran A., Zhang H., Vinokur R., Möller M. Adv. Mater. 2017;29:1604825. doi: 10.1002/adma.201604825. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Santos I., Kinnear C., Pérez-Juste J., Mulvaney P., Liz-Marzán L. M. Nat. Rev. Mater. 2018;3:375–391. [Google Scholar]
- Xin H., Namgung B., Lee L. P. Nat. Rev. Mater. 2018;3:228–243. [Google Scholar]
- Link D., Anna S. L., Weitz D., Stone H. Phys. Rev. Lett. 2004;92:054503. doi: 10.1103/PhysRevLett.92.054503. [DOI] [PubMed] [Google Scholar]
- Chung S. E., Park W., Park H., Yu K., Park N., Kwon S. Appl. Phys. Lett. 2007;91:041106. [Google Scholar]
- Liu Z., Liu Y., Chang Y., Seyf H. R., Henry A., Mattheyses A. L., Yehl K., Zhang Y., Huang Z., Salaita K. Nat. Methods. 2015;13:143. doi: 10.1038/nmeth.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg M., Pastoriza-Santos I., Pérez-Juste J., Hellweg T., Liz-Marzán L. M. Small. 2007;3:1222–1229. doi: 10.1002/smll.200700078. [DOI] [PubMed] [Google Scholar]
- Contreras-Cáceres R., Pacifico J., Pastoriza-Santos I., Pérez-Juste J., Fernández-Barbero A., Liz-Marzán L. M. Adv. Funct. Mater. 2009;19:3070–3076. [Google Scholar]
- Rodríguez-Fernández J., Fedoruk M., Hrelescu C., Lutich A. A., Feldmann J. Nanotechnology. 2011;22:245708. doi: 10.1088/0957-4484/22/24/245708. [DOI] [PubMed] [Google Scholar]
- Contreras-Cáceres R., Sánchez-Iglesias A., Karg M., Pastoriza-Santos I., Pérez-Juste J., Pacifico J., Hellweg T., Fernández-Barbero A., Liz-Marzán L. M. Adv. Mater. 2008;20:1666–1670. [Google Scholar]
- Shah R. K., Kim J.-W., Weitz D. A. Langmuir. 2009;26:1561–1565. doi: 10.1021/la9041327. [DOI] [PubMed] [Google Scholar]
- Cho E. C., Kim J.-W., Fernández-Nieves A., Weitz D. A. Nano Lett. 2008;8:168–172. doi: 10.1021/nl072346e. [DOI] [PubMed] [Google Scholar]
- Garstecki P., Fuerstman M. J., Stone H. A., Whitesides G. M. Lab Chip. 2006;6:437–446. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Sato E., Hirokawa Y., Hirotsu S., Peetermans J. Phys. Rev. Lett. 1985;55:2455. doi: 10.1103/PhysRevLett.55.2455. [DOI] [PubMed] [Google Scholar]
- Kubota K., Hamano K., Kuwahara N., Fujishige S., Ando I. Polym. J. 1990;22:1051. [Google Scholar]
- Sakar M. S., Neal D., Boudou T., Borochin M. A., Li Y., Weiss R., Kamm R. D., Chen C. S., Asada H. H. Lab Chip. 2012;12:4976–4985. doi: 10.1039/c2lc40338b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic C., Raman R., Chan V., Williams B. J., Tolish M., Bajaj P., Sakar M. S., Asada H. H., Saif M. T. A., Bashir R. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10125–10130. doi: 10.1073/pnas.1401577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. W. Annu. Rev. Biomed. Eng. 2015;17:243–265. doi: 10.1146/annurev-bioeng-071114-040632. [DOI] [PubMed] [Google Scholar]
- Ricotti L., Trimmer B., Feinberg A. W., Raman R., Parker K. K., Bashir R., Sitti M., Martel S., Dario P., Menciassi A. Sci. Robot. 2017;2:eaaq0495. doi: 10.1126/scirobotics.aaq0495. [DOI] [PubMed] [Google Scholar]
- Janet M. T., Cheng G., Tyrrell J. A., Wilcox-Adelman S. A., Boucher Y., Jain R. K., Munn L. L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:911–916. doi: 10.1073/pnas.1118910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoefer K. Nat. Rev. Mater. 2018;3:76. [Google Scholar]
- Kellogg R. A., Gómez-Sjöberg R., Leyrat A. A., Tay S. Nat. Protoc. 2014;9:1713. doi: 10.1038/nprot.2014.120. [DOI] [PubMed] [Google Scholar]
- Xia Y., Whitesides G. M. Angew. Chem., Int. Ed. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.